Abstract
Pathogen-inducible plant promoters contain multiple cis-acting elements, only some of which may contribute to pathogen inducibility. Therefore, we made defined synthetic promoters containing tetramers of only a single type of element and present evidence that a range of cis-acting elements (boxes W1, W2, GCC, JERE, S, Gst1, and D) can mediate local gene expression in planta after pathogen attack. The expression patterns of the promoters were monitored during interactions with a number of pathogens, including compatible, incompatible, and nonhost interactions. Interestingly, there were major differences in the inducibilities of the various promoters with the pathogens tested as well as differences in the speed of induction and in the basal expression levels. We also show that defense signaling is largely conserved across species boundaries at the cis-acting element level. Many of these promoters also direct local wound-induced expression, and this provides evidence for the convergence of resistance gene, nonhost, and wound responses at the level of the promoter elements. We have used these cis-acting elements to construct improved synthetic promoters and show the effects of varying the number, order, and spacing of such elements. These promoters are valuable additions to the study of signaling and transcriptional activation during plant–pathogen interactions.
INTRODUCTION
The availability of a range of defined synthetic plant promoters that direct controlled local gene expression in response to pathogens would be a major advance. These promoters could be used to help define signaling pathways, to isolate novel mutants using “targeted genetics” (Hooley, 1998), and to engineer plants with increased disease resistance. The control regions of plant genes are modular and contain a number of cis-acting elements, each of which may contribute to one or more aspects of a complex expression profile. One strategy to overcome this complexity is to produce synthetic promoters containing only defined individual elements, thereby reducing expression profile complexity (Salinas et al., 1992). However, although there are numerous reports of synthetic promoters being inducible by elicitors in transient expression systems (Rushton and Somssich, 1998), in most cases it is not known to what extent individual cis-acting elements retain their functionality in planta when removed from their native promoter context and whether we can use these individual “modules” to make synthetic promoters that direct a desired expression pattern.
Pathogen-inducible promoters represent an attractive system for the production of synthetic promoters. There are a large number of known pathogen-inducible genes (Rushton and Somssich, 1998), and their promoters are among the best studied in plants. Two groups of pathogen-inducible cis-acting elements, the GCC-like elements (Ohme-Tagaki et al., 2000) and the W boxes (Rushton et al., 1996; Eulgem et al., 2000), have been well studied. The GCC box (AGCCGCC) often is found in the promoter regions of defense genes (Ohme-Takagi and Shinshi, 1995). A similar element has been reported to direct jasmonate and elicitor-responsive expression (JERE; AGACCGCC) (Menke et al., 1999), and another (DRE; TACCGAC) directs cold-, salt stress–, and dehydration-responsive expression (Yamaguchi-Shinozaki and Shinozaki, 1994). Recently, another similar GCC-like element called box S (AGCCACC) has been identified that directs expression by fungal elicitors (Kirsch et al., 2000). It appears that minor variations in the core sequences impart responsiveness to different stimuli.
The W box [(T)TGAC(C/T)] is the binding site for members of the WRKY family of transcription factors (Rushton et al., 1996). There is increasing evidence that W boxes are a major class of cis-acting elements responsible for the pathogen inducibility of many plant genes (Raventós et al., 1995; Rushton et al., 1996; Wang et al., 1998). The importance of W boxes was illustrated recently by studies of the Arabidopsis transcriptome during systemic acquired resistance (Maleck et al., 2000; Petersen et al., 2000). In some cases, clustering of W boxes may be associated with inducibility by pathogens.
Given that GCC-like boxes and W boxes have been so well studied, it is surprising that there is almost no direct in planta evidence that they can mediate pathogen-inducible expression. Although W boxes have been shown to impart elicitor-inducible expression on a minimal promoter in transient expression systems (Raventós et al., 1995; Rushton et al., 1996; Eulgem et al., 1999), there is only one report suggesting that isolated W boxes can function alone in planta and direct pathogen-inducible expression (Kirsch et al., 2001). It was shown that the W box–containing promoter element E17 mediates gene expression at pathogen infection sites in transgenic Arabidopsis plants. Data concerning GCC-like boxes are equally scarce. A synthetic promoter containing four copies of a GCC box directs ethylene-inducible expression in tobacco (Ohme-Takagi and Shinshi, 1995), but pathogen inducibility has yet to be shown.
Both GCC-like elements (Suzuki et al., 1998) and WRKY transcription factors (Hara et al., 2000) have been implicated in gene expression in response to wounding. It was shown recently that wound- and pathogen-induced signaling consists of networks with some shared components (Romeis et al., 1999). It remains an open question, however, whether specific cis-acting elements can direct both pathogen- and wound-induced expression in planta or whether these two activities are characteristics of separate elements.
Here, we present a comprehensive study of pathogen-inducible synthetic plant promoters constructed from a range of both well-studied and novel cis-acting elements. At least seven different elements can alone direct local pathogen-inducible gene expression in transgenic Arabidopsis plants. Major differences are seen between many of the elements with regard to their background expression, their induction by different pathogens, and their speed of induction. Additionally, we demonstrate that several pathogen-inducible elements also direct local wound-inducible expression and therefore that components of pathogen- and wound-induced signaling are shared.
RESULTS
Elicitor-Inducible Synthetic Promoters
Our approach to making synthetic promoters that are induced locally by pathogens was first to test candidate elements in a parsley transient expression system for inducibility by a pathogen-derived peptide elicitor, pep25. Promising candidates then were introduced into Arabidopsis plants to evaluate their in planta expression patterns and inducibility by pathogens. Figure 1A shows how the synthetic promoters were constructed. Each element was inserted between the SpeI and XbaI restriction sites upstream of the −46 35S minimal promoter of Cauliflower mosaic virus.
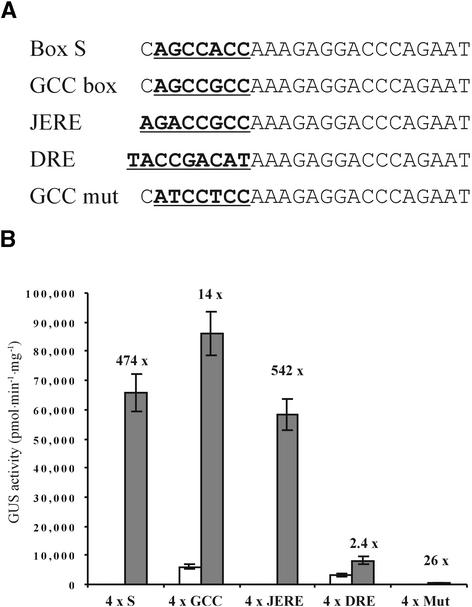
Figure 1.
Elicitor-Inducible Synthetic Plant Promoters.
(A) Scheme of the synthetic promoters. Elements were inserted between the SpeI and XbaI sites in pBT10 upstream of the −46 35S promoter of Cauliflower mosaic virus (CaMV 35S). pAnos, nos terminator.
(B) Sequence of the elicitor-inducible elements. Core sequences are shown underlined and in boldface.
(C) Elicitor inducibility of the synthetic promoters in a parsley transient expression system. Gray bars represent GUS activity 8 hr after pep25 addition. White bars show the level of GUS activity in the absence of pep25. The fold inducibility is shown, and error bars indicate ±sem. Qualitatively similar results were obtained upon normalization with a constitutively expressed Petroselinum crispum UBI4/2 ::luciferase construct (Sprenger-Haussels and Weisshaar, 2000).
Initially, synthetic promoters containing tetramers of candidate elements were constructed. The reason for the use of tetramers was the observation that promoters with multimers of elements are stronger than those with just one or two copies (see below). Because it was unknown whether any of the elements were functional in planta, we used tetramers in initial experiments to ensure that any expression would be strong enough to be detectable by β-glucuronidase (GUS) staining. The first elements tested were boxes W1 and W2 from the parsley PR1 genes (Rushton et al., 1996), box S from the parsley ELI7 genes (Kirsch et al., 2000), a novel element called box D from the parsley PR2 gene (P.J. Rushton and K. Hahlbrock, unpublished results), and an element we termed the Gst1 box from the potato gst1 gene (Strittmatter et al., 1996). The Gst1 box contains an S box and a W box separated by just 4 bp in a region of the promoter that mediates transcriptional activation in response to pathogens, during senescence, and in root apices (Figure 1B). All of the promoters showed inducibility by pep25 in transient expression studies (Figure 1C), although the strength and inducibility of the elements varied greatly. Tetramers of most elements showed inducibilities of five- to 30-fold, whereas four copies of box S had a remarkably high inducibility (>400-fold), which was attributable to an almost complete lack of expression in the absence of pep25.
Although box S is very similar in sequence to the GCC, JERE, and DRE boxes, they appear to direct different patterns of gene expression (Yamaguchi-Shinozaki and Shinozaki, 1994; Ohme-Takagi and Shinshi, 1995; Menke et al., 1999). To investigate these GCC-related elements, we constructed a series of synthetic promoters that are based on box S (Figure 2A). To determine the effect of changes within the core sequence, the flanking sequences remained identical and we altered only the bases necessary to change one variant into another. A construct containing the GCC core sequence was slightly stronger than box S but showed greatly reduced inducibility by pep25 (Figure 2B). This reduction to ∼3% of the box S value is remarkable considering that this is the result of a single base pair change and is attributable primarily to an increase in the background level of the GCC box. Results with JERE were almost identical to those with box S, both in strength and fold induction. In contrast, DRE directed a lower level of expression and showed little elicitor inducibility. A nonfunctional version of the GCC box (GCC mut) (Figure 2A) (Ohme-Takagi and Shinshi, 1995) had very low activity but was slightly inducible by pep25. These results show that minor variations in the core sequences of GCC-like elements can have profound effects on both the strength and the elicitor inducibility of these elements.
Figure 2.
Elicitor-Inducible Promoters Containing GCC-like Elements.
(A) Sequence of the elicitor-inducible GCC-like elements. Core sequences are shown underlined and in boldface.
(B) Elicitor inducibility of synthetic promoters containing GCC-like elements in a parsley transient expression system. Gray bars represent GUS activity 8 hr after pep25 addition. White bars show the level of GUS activity in the absence of pep25. The fold inducibility is shown, and error bars indicate ±sem. Qualitatively similar results were obtained upon normalization with a constitutively expressed P. crispum UBI4/2:: luciferase construct (Sprenger-Haussels and Weisshaar, 2000).
Expression Patterns of the Synthetic Promoters in Planta and in Response to Wounding
We next addressed the question of whether the synthetic promoters with defined cis-acting elements are functional in planta. Promoters containing tetramers of the elements were introduced into Arabidopsis plants. We first determined the levels of background expression for each construct. The majority of the synthetic promoters had little background expression in leaves (Figure 3A and data not shown). By comparison, the background level of expression was sometimes higher in roots (data not shown), and in the case of 4 × W2, this level was very high. Only 4 × D had no appreciable background expression in any parts of the plant.
Figure 3.
Expression Patterns of Synthetic Promoters in Planta and in Response to Wounding.
(A) Expression patterns (GUS activity) in untreated excised leaves.
(B) Expression patterns 1 hr after wounding by cutting.
(C) Local wound-induced expression from 4 × GCC 1 hr after wounding.
(D) Local wound-induced expression from 4 × S 1 hr after wounding.
Many of the leaves shown in Figure 3A manifest local induction by wounding where the leaves have been excised. This response is very rapid, being induced during the time taken to harvest the plants. Therefore, we wounded leaves by cutting through one-half of a leaf and monitored changes in promoter activity. Except for box D, expression from all promoters was induced locally by cutting (Figure 3B and data not shown). Figure 3B also illustrates that this massive damage to the leaf sometimes led to a lower level increase in GUS expression over the entire leaf (cf. Figures 3A and 3B). However, when plants grown in greenhouse conditions exhibited local wound-induced expression, presumably because of insect damage, we never observed expression in other parts of the leaf (data not shown). This finding suggests that local wounding of Arabidopsis plants leads only to highly restricted expression of these promoters. Only when more extensive damage is done (e.g., severing of vascular tissue) is there a response throughout the leaf and subsequent nonlocal induction.
We observed two different patterns of local wound-induced expression after cutting (Figures 3C and 3D). Most common was expression in a layer of cells at the actual cut site (Figure 3C). However, with the same promoters, we sometimes also observed local expression in a ring of cells some distance away from the cut site (Figure 3D). The exact conditions necessary for the production of each pattern were not investigated further.
Synthetic Promoters That Direct Local Gene Expression in Response to Peronospora parasitica
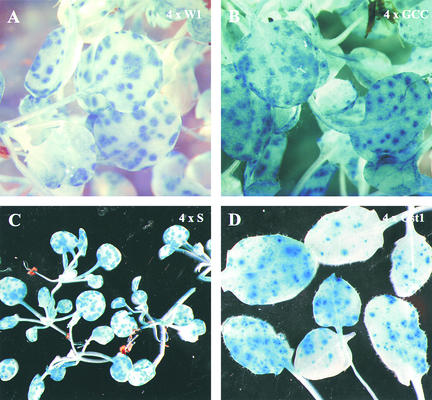
Having determined the level of background expression with the synthetic promoters, we looked to answer the following question: Do they direct pathogen-inducible expression? To do so, we investigated their expression during interactions with a range of different pathogens. We inoculated with the biotrophic oomycete P. parasitica pv Cala2, which produces an incompatible interaction with Columbia (Col-0) plants (Holub et al., 1994). Local gene expression, limited to small areas around infection sites, was seen with 4 × W1, 4 × GCC, 4 × S, 4 × Gst1 (Figure 4), 4 × W2, and JERE (data not shown). By contrast, no expression was detectable with 4 × D. Figure 4 demonstrates that although 4 × GCC is pathogen inducible, it has a much higher level of background expression in uninfected parts than does 4 × S, and these data accurately reflect the transient expression data described above (Figure 2). For all of the tested elements except box D, these results demonstrate that these elements alone can direct pathogen-inducible expression in planta when removed from their native promoter context.
Figure 4.
Local Expression in Response to P. parasitica pv Cala2.
(A) Local GUS expression 2 days after P. parasitica infection in Arabidopsis plants containing the synthetic promoter 4 × W1. Blue spots represent infection sites.
(B) Local expression in 4 × GCC plants.
(C) Local expression in 4 × S plants.
(D) Local expression in 4 × Gst1 plants.
Local Expression during Powdery Mildew Challenge
We next investigated the inducibility of the promoters during the nonhost interaction with the barley powdery mildew Blumeria graminis f. sp. hordei. Only four synthetic promoters (4 × S [Figure 5A], 2 × W2/2 × S/2 × D [Figure 5B], 4 × GST [data not shown], and 4 × W2/4 × S [data not shown]) directed significant local gene expression under the conditions tested. These promoters all contain box S, suggesting that box S may play a role during nonhost signaling. The germinating spores (sp) form appressorial germ tubes (agt), and although most do not penetrate the plant, local gene expression is found predominantly in the underlying mesophyll cells (Figure 5A) just beneath the attempted penetration site, suggesting that recognition events on the leaf surface trigger defense responses in the underlying cells. Importantly, the level of expression from these promoters and the number of expressing cells correlated directly with the extent of differentiation of the fungus. This is illustrated in Figure 5B, where at one infection site, haustorium initial formation has triggered cell death. Here, a larger number of cells show activation of the promoter, and levels of expression are higher than in the two other infection attempts, in which infection progressed only as far as papilla formation.
Figure 5.
Local Expression during Nonhost and Compatible Powdery Mildew Interactions.
(A) Light micrograph of one leaf infection site on a 4 × S plant viewed at two different planes of focus. The epidermal plane (right) shows the inducing penetration attempt by a germinated B. graminis spore (sp). The differentiation of an appressorial germ tube (agt) coincides with local cell wall thickening (pap). Focusing deeper into the tissue (mesophyll plane; left) reveals GUS expression in mesophyll cells just underneath the penetration attempt. The photograph was taken 2 days after inoculation with the nonhost pathogen B. graminis. Staining of the fungus was with Coomassie blue.
(B) Local expression in a 2 × W2/2 × S/2 × D plant upon B. graminis challenge. The rare successful penetration event at left triggered a cell death response (cd), whereas early aborted penetration attempts correlated with papilla formation (pap).
(C) Local expression 7 days after infection with the compatible powdery mildew E. cichoracearum in a 4 × S plant. Blue spots represent infection sites.
(D) Closeup of the border region of an infection site from (C). Reporter gene expression coincides with superficial mycelium (sm).
During the compatible interaction with the powdery mildew Erysiphe cichoracearum, all of the promoters except DRE directed local expression. After 7 to 10 days, areas of powdery mildew growth could be seen, and expression of the synthetic promoters was very high in these infected areas (Figure 5C). When viewed closely, intense GUS staining was apparent in these areas, and a wave of gene expression was apparent in advance of the growing hyphae (Figure 5D).
Local Gene Expression during Interactions with Pseudomonas syringae
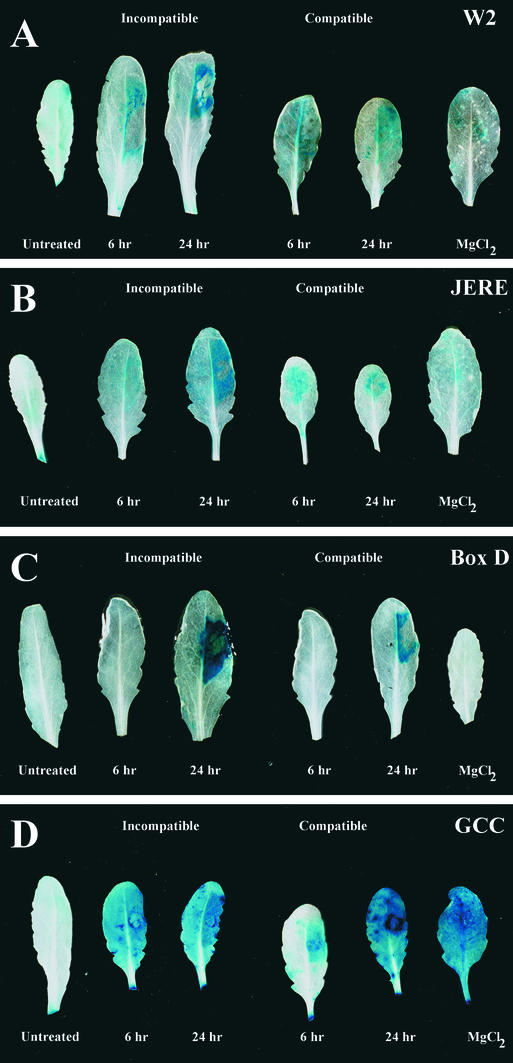
We also performed infections with the necrotrophic bacterial pathogen P. syringae pv tomato using two different isolates: a Col-0–compatible isolate (DC3000) and a Col-0–incompatible isolate (DC3000 carrying the avrRpm1 gene) (Debener et al., 1991). Figure 6 shows the expression patterns obtained with some of the synthetic promoters. Expression of 4 × W2 was induced locally within 6 hr of infection with the incompatible isolate (Figure 6A). Expression also was apparent with the compatible isolate, although the level of expression was lower. Similar results were obtained with 4 × JERE (Figure 6B), 4 × W1, 4 × S, and 4 × Gst1 (data not shown).
Figure 6.
Expression during Interactions with P. syringae.
Using a syringe, leaves were inoculated with either isolate DC3000 (compatible interaction) or isolate DC3000 carrying the avrRpm1 gene (incompatible interaction) and were harvested after 6 or 24 hr. As controls, untreated leaves and leaves 24 hr after treatment with MgCl2 buffer were harvested and stained for GUS activity.
(A) 4 × W2.
(B) 4 × JERE.
(C) 4 × D.
(D) 4 × GCC.
Although untreated leaves from plants containing 4 × GCC showed some background expression, often this was increased greatly by mock infection with MgCl2 (Figure 6D); it was difficult, therefore, to determine whether induction by P. syringae was occurring. By contrast, 4 × GCC showed clear induction by P. parasitica (Figure 4B), even though background expression was apparent. This finding suggests that the process of injecting liquid into the leaf is itself a sufficient abiotic stress to induce high-level expression of 4 × GCC. Closer inspection of plants containing other synthetic promoters (most notably, 4 × W2, 4 × Gst1, and 4 × W1) showed that background expression levels also are increased sometimes after infiltration of MgCl2 (data not shown). This is similar to the results obtained when a cut was made through one-half of a leaf (Figure 3) and suggests that wounding/stressing a large portion of a leaf can lead to a response in the entire leaf.
Infection of 4 × D–containing plants led to qualitatively different results. No expression was detected in control or mock-infected leaves or with either isolate 6 hr after infection. By contrast, strong local gene expression was seen 24 hr after infection with both isolates (Figure 6C). Again, the level of expression was higher during the incompatible interaction.
Effect of Element Number on Strength and Inducibility
Although the synthetic promoters containing tetramers of elements direct local gene expression upon pathogen attack, many are not yet ideal for our purposes because they have more background expression than desired or they respond to a number of biotic and abiotic stresses. Therefore, we have started to construct improved “second-generation” synthetic promoters using these cis-acting elements as building blocks.
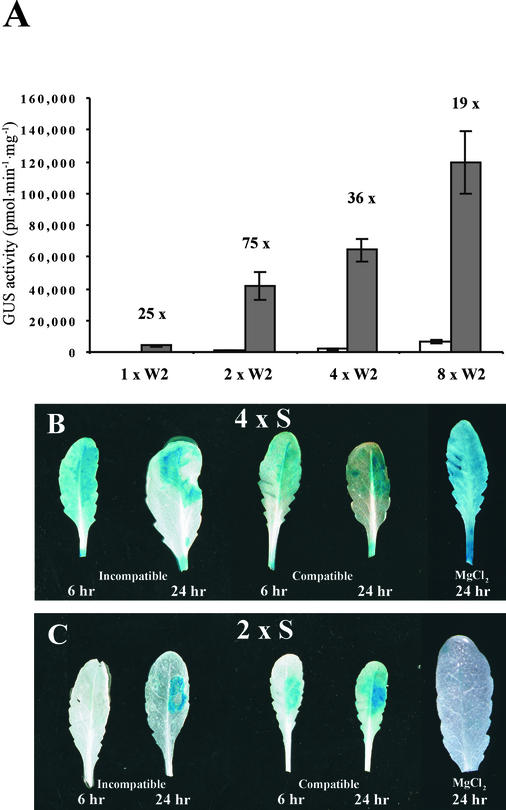
The first parameter investigated was the effect of the number of copies of an individual cis-acting element in a synthetic promoter. We constructed a series of promoters containing one, two, four, and eight copies of an element and tested these by transient assay. Figure 7A shows the results obtained with 1 × W2, 2 × W2, 4 × W2, and 8 × W2. Increasing the number of copies of W2 increased the strength of the promoter progressively. However, 2 × W2 had the best inducibility, because additional copies caused a proportionally greater increase in background expression. Similar results were obtained with boxes S and D (data not shown).
Figure 7.
Effect of Element Number on Strength and Inducibility.
(A) Elicitor inducibility of synthetic promoters containing increasing numbers of the elicitor-inducible cis-acting element box W2. Gray bars represent GUS activity 8 hr after pep25 addition. White bars show the level of GUS activity in the absence of pep25. The fold inducibility is shown, and error bars indicate ±sem.
(B) and (C) Expression patterns with plants containing the synthetic promoters 4 × S (B) and 2 × S (C) during interactions with P. syringae. Leaves were inoculated with either isolate DC3000 (compatible interaction) or isolate DC3000 carrying the avrRpm1 gene (incompatible interaction) and were harvested after 6 or 24 hr.
These findings suggest that promoters containing fewer copies of an element may be better suited to mediate pathogen-specific inducibility in planta. Accordingly, we introduced promoters with two copies of elements into Arabidopsis plants. Figures 7B and 7C show a comparison of 2 × S and 4 × S plants treated with P. syringae and demonstrate that the differences between the two promoters that were observed in the transient assay also are apparent in planta. Both promoters were inducible by pathogens, but 4 × S showed a higher background level and clear wound induction where the leaf was excised. By contrast, 2 × S, although clearly pathogen inducible, showed on average less background and in many cases no apparent wound induction.
Spacing Effects and Promoter Strength
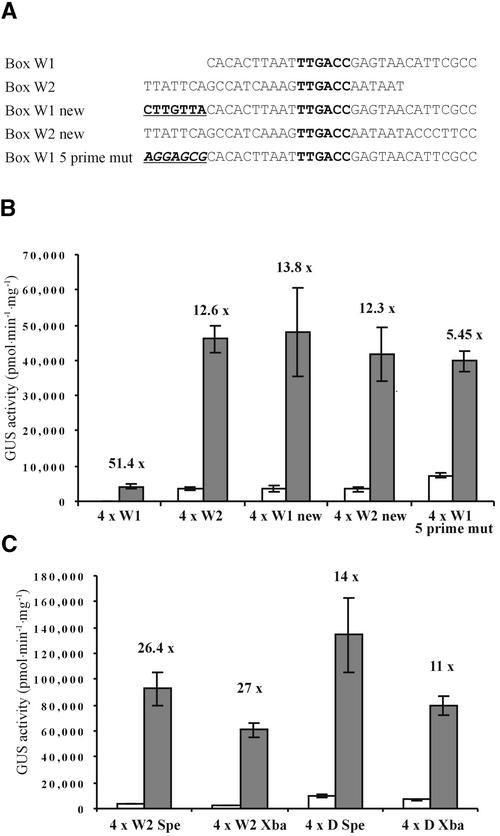
Our data show that not all W box–containing synthetic promoters behave similarly. For example, both transient expression experiments (Figure 1) and results from transgenic plants show that box W2 is much stronger than box W1, even though both contain the same TTGACC core element. Therefore, we asked the question: What makes a W box strong or weak? To eliminate possible spacing effects, we made new versions of both box W1 (box W1 new) and box W2 (box W2 new) that are identical in length and have TTGACC core sequences in identical positions (Figure 8A). The only difference between the new version of box W1 and the original is the addition of the preceding 7 bp from the PR1-1 promoter. To our surprise, both of the new elements were strong, with no detectable difference between 4 × W2, 4 × W2 new, and 4 × W1 new (Figure 8B). The seven additional bases in box W1 new have increased the strength either by adding a positive element or by altering the spacing between TTGACC core elements. Therefore, we made a version of 4 × W1 new in which an unrelated sequence was substituted for the original seven bases (Figure 8A). This construct (4 × W1 5 prime mut) directed a similar level of expression to 4 × W1 new (Figure 8B). Thus, the difference in strength between the two versions of box W1 was not sequence dependent, excluding the possibility that a new cis-acting element was generated. The pronounced difference in strength between 4 × W1 and 4 × W2, therefore, seems to be a spacing effect caused by different distances between core TTGACC elements.
Figure 8.
Spacing Effects and Promoter Strength.
(A) Sequences of the different versions of box W1 and box W2. W box core sequences are indicated in boldface. The seven additional bases in box W1 new are shown in boldface and underlined. The seven unrelated bases in box W1 5 prime mut are shown in boldface italics and underlined.
(B) Elicitor inducibility of the synthetic promoters shown in (A). Gray bars represent GUS activity 8 hr after pep25 addition. White bars show the level of GUS activity in the absence of pep25. The fold inducibility is shown, and error bars indicate ±sem.
(C) Elicitor inducibility of synthetic promoters with elements inserted at different positions. Elements were inserted into either the SpeI site (−74) or the XbaI site (−56).
To establish whether spacing from the TATA box had any effect on promoter strength or inducibility, we inserted elements 18 bp farther upstream into the SpeI site instead of between the SpeI and XbaI sites (Figure 1A). Figure 8C shows that this resulted in a small increase in strength but had little effect on inducibility. Placing the elements at other positions farther upstream had little effect (data not shown). Together, our results suggest that positioning an element closer to the TATA box can result in some change in promoter strength but that there is little effect on inducibility.
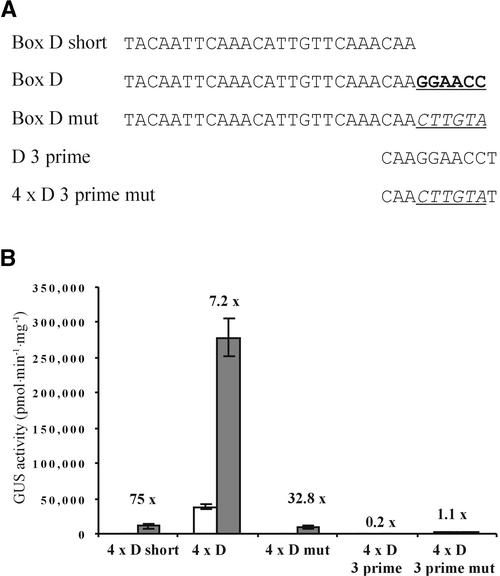
Box D and the Role of Flanking Sequences
Box D is of considerable interest because box D–containing synthetic promoters have no appreciable background expression or wound inducibility in planta, and they also show different kinetics of induction by pathogens. Box D was discovered as a DNase1 footprint from approximately −76 to −52 in the parsley PR2 promoter (P.J. Rushton and K. Hahlbrock, unpublished results). This region (box D short) (Figure 9A) had a high elicitor inducibility but was weak (Figure 9B). Because the exact extent of the element was unclear, a longer version (box D) containing the next six bases from the PR2 promoter at the 3′ end was constructed (Figure 9A). Box D was almost 30 times stronger than 4 × D short (Figure 9B), although inducibility was reduced as a result of increased background levels. We then constructed a version of box D in which the six additional bases were exchanged (Figure 9B, box D mut), and this was almost identical to box D short (Figure 9A), demonstrating that the considerable increase in strength is a sequence-specific effect that is dependent on the six bases GGAACC. Therefore, we believe that box D consists of at least two elements: an elicitor-responsive element and a positive-acting element. This positive element cannot function alone (Figure 9, 4 × D 3 prime and 4 × D 3 prime mut) but appears to be a coupling element that forms a functional unit with the elicitor-inducible element(s). Alternatively, the shortened version of box D simply may lack crucial 3′ nucleotides of a single element required for selective high-affinity binding of a specific transcription factor, thus allowing binding to other related factors, leading to the observed differences in inducibility.
Figure 9.
Box D.
(A) Sequences of the different versions of box D. The six additional bases in box D are shown in boldface and underlined. The six unrelated bases in box D mut and 4 × D 3 prime mut are shown in boldface italics and underlined.
(B) Elicitor inducibility of the synthetic promoters shown in (A). Gray bars represent GUS activity 8 hr after pep25 addition. White bars show the level of GUS activity in the absence of pep25. The fold inducibility is shown, and error bars indicate ±sem.
Promoters with Combinations of Elements
Having taken a reductionist approach, reducing synthetic promoters to a single type of cis-acting element to demonstrate functionality, we next took the first steps toward making improved synthetic promoters that contain more than one type of defined cis-acting element. The question that we asked was the following: What effect does the insertion of a second element have? To answer this question, we inserted four copies of box S either upstream (4 × S/4 × W2) or downstream (4 × W2/4 × S) of 4 × W2. Figure 10A shows that this insertion had little effect on the strength of the promoters and a relatively minor effect on inducibility. Mixing up the elements to make 2 × S/2 × W2/2 × S/2 × W2 led to only a slight increase in promoter strength.
Figure 10.
Synthetic Promoters Containing Combinations of Elements.
(A) Elicitor inducibility of synthetic promoters containing combinations of box W2 and box S. Gray bars represent GUS activity 8 hr after pep25 addition. White bars show the level of GUS activity in the absence of pep25. The fold inducibility is shown, and error bars indicate ±sem.
(B) GUS expression pattern of the synthetic promoter 2 × W2/2 × S/2 × D 3 days after treatment with B. graminis (nonhost interaction).
(C) GUS expression pattern of the synthetic promoter 4 × W2/4 × S 6 and 12 hr after treatment with P. syringae carrying the avrRpm1 gene (incompatible interaction).
In contrast to the transient expression results, in planta studies produced encouraging results. We introduced two synthetic promoters (2 × W2/2 × S/2 × D and 4 × W2/4 × S) that contain combinations of different elements into Arabidopsis plants. Both showed good inducibility by a range of pathogens and were among the best promoters tested, combining high inducibility with low background (Figures 10B and 10C). It seems likely that all of the cis-acting elements contribute to the overall expression of the synthetic promoter and that promoters containing carefully selected combinations of elements may be among the best pathogen-inducible promoters.
DISCUSSION
Pathogen-Inducible Synthetic Promoters
In this report, we provide direct evidence that a range of pathogen-inducible cis-acting elements can alone mediate pathogen-inducible expression in planta. When taken out of their native promoter contexts, they retain pathogen inducibility as components of synthetic promoters, directing expression that is local and that correlates with the extent of growth of the pathogen. The cis-acting elements tested fall into three groups: W boxes, GCC-like boxes, and box D. Our observations suggest that binding sites for WRKY (W box) or AP2/ERF (GCC-like box) transcription factors can be sufficient to confer pathogen inducibility on a promoter. This is an important observation because these represent two of the three largest families of plant-specific transcription factors (Riechmann and Ratcliffe, 2000). It will be interesting to extend these studies to include other pathogen-inducible elements such as Myb Recognition Elements (MREs) and as-1–like elements, although evidence suggests that these elements might not be able to function alone (Rushton and Somssich, 1998).
Interestingly, although more than one type of cis-acting element is not required for pathogen inducibility, some pathogen-inducible promoters contain elements of more than one type. An example is the Gst1 box, which contains both a W box and an S box. This places the gst1 gene under the control of both WRKY and AP2/ERF transcription factors. The potato gst1 promoter has been well studied (Strittmatter et al., 1996), and our work provides the first clear evidence of how this gene is activated transcriptionally in response to pathogens. It may be common that signaling pathways operating via different transcription factors can target the same gene; another example is the parsley WRKY1 gene (a W box and a GCC box) (Eulgem et al., 1999).
Our results suggest that defense signaling is largely conserved across species boundaries, because an element from a potato gene (the Gst1 box) is active in a parsley transient expression system and in Arabidopsis plants. In fact, none of the elements tested originate from Arabidopsis promoters, yet all retain their functionality. These results also validate our use of a parsley transient expression system for the initial characterization of elements, because we have successfully identified and characterized numerous pathogen-inducible elements and synthetic promoters in a way that would not have been possible in planta. In almost all cases, elicitor inducibility in the transient expression system is accompanied by pathogen inducibility in planta.
Signaling in Different Types of Plant–Pathogen Interactions
Table 1 summarizes the responses of the synthetic promoters to pathogens under the conditions tested. Importantly, not all of the elements respond in the same way to pathogens and wounding. Even elements that are bound by the same family of transcription factors (e.g., GCC, JERE, S, and DRE) show different expression patterns. This is consistent with reports that these elements may respond to different hormones, for example, salicylic acid (W boxes) (Yang et al., 1999), jasmonate (JERE) (Menke et al., 1999), and ethylene (GCC) (Ohme-Takagi and Shinshi, 1995). Therefore, we have generated a spectrum of synthetic promoters useful for studying plant–pathogen interactions at the molecular level.
Table 1.
Expression Patterns of Synthetic Promoters
| Synthetic Promoter |
P. parasitica Incompatible |
P. syringae Incompatible |
P. syringae Compatible |
E. cichoracearum Compatible |
B. graminis Nonhost |
Wounding Abiotic |
|---|---|---|---|---|---|---|
| 4 × W2 | + | + | + | + | − | + |
| 4 × W1 | + | + | + | + | − | + |
| 4 × D | − | + | + | + | − | − |
| 4 × GCC | + | ± | ± | + | − | + |
| 4 × S | + | + | + | + | + | + |
| 4 × JERE | + | + | + | + | − | + |
| 4 × GST | + | + | + | + | + | + |
| 4 × DRE | ND | − | − | − | − | + |
| 4 × W2/4 × S | + | + | + | + | + | + |
| 4 × W2/2 × S/2 × D | + | + | + | + | + | + |
(+), high-level induction; (±), lower level of induction attributable to lower induced expression or high background; (−), no expression; ND, not determined.
During compatible interactions, the synthetic promoters accurately report the in vivo situation in which gene activation is slower and perhaps weaker (Lo et al., 1999). In this context, the observed high-level expression of the synthetic promoters during the compatible interaction with a powdery mildew (E. cichoracearum) illustrates their potential usefulness during plant–pathogen interactions of commercial importance.
Under the conditions tested, only promoters containing box S consistently showed local expression during the nonhost interaction with barley powdery mildew. Box S, therefore, could be a useful tool for studying nonhost responses and for engineering plants with broad-spectrum resistance. These results are remarkable considering that there is only a single basepair difference between box S and the GCC box (Figure 2). This single alteration results in box S having much lower background levels both in the transient expression system and in planta. Given that both elements probably are bound by AP2/ERF transcription factors, experiments designed to identify family members with high affinities for box S could yield useful data concerning signaling during plant defense.
Box D is an extremely interesting novel element, unlike any of the others tested, because it combines an apparent lack of background expression or induction by wounding with strong induction during some, but not all, plant–pathogen interactions (Table 1). Box D has been found to be responsive to P. syringae, E. cichoracearum, the bacterially derived peptide elicitor flg22 (data not shown), and the oomycete-derived peptide elicitor pep25 in protoplasts. Box D also responds with different kinetics than other elements, being induced later than the other elements (Figure 6). The molecular characterization of box D and the identification of its cognate transcription factors would provide new insights into defense gene activation during plant–pathogen interactions.
Pathogen and Wound Signaling through the Same cis-Acting Elements
Recently, work on plant defense signaling has demonstrated the convergence of resistance gene, elicitor, wound, and salicylate responses at the level of mitogen-activated protein kinase (MAPK) activation (Romeis et al., 1999). It is not clear at present whether these signaling pathways merge at the MAPKs or upstream thereof or whether the same MAPK can mediate disparate responses by interacting with other proteins (Bent, 2001). Our results showing that tetramers of a range of pathogen-inducible cis-acting elements also direct local wound-induced gene expression demonstrate the convergence of resistance gene, nonhost, and wound responses at the level of promoter elements. This observation is in agreement with recent data demonstrating an extensive overlap in the transcriptional response of plants to race-specific elicitors and mechanical stress (Durrant et al., 2000). Studies of 290 Avr9/Cf-9 rapidly elicited genes demonstrated that many of these also are induced by wounding (e.g., by cutting or infusion of liquids).
The results presented here extend these observations from the level of the entire promoter to that of individual cis-acting elements. In addition, the identities of the elements suggest that pathways involving salicylic acid, jasmonate, and ethylene may converge at this level. One key question that remains is whether the same or different members of transcription factor families are responsible for both pathogen and wound responses. In other words, whether convergence occurs at the level of transcription factors as well as at the level of cis-acting elements and promoter architecture. Most likely, these signaling networks have shared components, with proteins at many nodes being capable of receiving inputs from multiple pathways (Bent, 2001). Many pathogen- and wound-inducible cis-acting elements could represent such nodes.
Improving Synthetic Promoters
We used synthetic promoters containing tetramers of elements to ensure that the promoters were strong enough to detect activity by GUS staining. These promoters, however, are not optimal for all purposes; therefore, we set about making improved second-generation promoters by varying several parameters. Most important was the number of copies of an individual element in a promoter. Both the strength and the inducibility of a promoter can be modulated by varying the number of copies of an element. Importantly, this also can have the effect of reducing/eliminating some background expression because pathogen inducibility appears stronger than basal or wound-induced expression (Figure 7B). Spacing between individual cis-acting elements and/or between these elements and the preinitiation complex also can have a profound effect (Figure 8), but spacing is difficult to predict (Wray, 1998), and the optimal spacing, like the optimal number of elements, needs to be determined experimentally.
Promoters with combinations of different elements may be among the best pathogen-inducible promoters (Figures 10B and 10C), because they often combine good inducibility with low background. Together, our results suggest that the optimal pathogen-inducible synthetic promoters may consist of combinations of one or two copies of defined cis-acting elements. Perhaps the best synthetic promoters, although made up of carefully chosen components, will not be that dissimilar to natural promoters after all.
Biological Importance and Applications
The pathogen-inducible synthetic promoters could have major applications, first, as molecular markers, and second, in engineering crops with increased disease resistance. As molecular markers, the promoters are attractive because they consist of one cis-acting element and therefore one defined end point of signaling pathways. This could have major advantages over the full-length promoters that are used commonly. Through introduction into mutant backgrounds, the synthetic promoters can be used to better characterize these mutants, and the use of defined synthetic promoters for this purpose could become a standard practice. Differences in responses during different plant–pathogen interactions also can be investigated at the molecular level. Synthetic promoters can be used as reporters for mutant screens using targeted genetics (Hooley, 1998), and this may prove a powerful approach.
The spread of plant pathogens and insect pests is increasing worldwide (Moffat, 2001). Researchers have identified numerous plant and pathogen genes that can be used to increase crop resistance toward invading pathogens. These strategies involve interfering with the replication of viruses in the plant, expression of gene products toxic to certain pathogens, and enhancement of the plant's own natural resistance mechanisms. Such introduced genes usually are placed under the control of strong promoters, yielding constitutive expression of the gene product in all tissues of the plant. This can have detrimental effects on plant growth, development, and crop yield. Use of the synthetic promoters presented here and future improvements thereof may prove valuable in engineering plants with increased resistance, because the use of such defined regulatory sequences may allow highly restricted expression of the desired gene product exclusively at the sites of attempted pathogen invasion. Thus, expression of the gene product is limited to cells surrounding an infection site and is not found in healthy parts of the plant. Moreover, expression via such promoters can be triggered by a range of different pathogens, including during compatible interactions with pathogens of commercial importance, such as powdery mildew. This expression may be sufficient to abort the progression of the invader even in such compatible interactions.
METHODS
Construction of Synthetic Promoters
Promoter constructs were produced by annealing phosphorylated upper and lower strand oligonucleotides to create elements containing a SpeI restriction site at the 5′ end and an XbaI restriction site at the 3′ end. These were introduced into pBT10–β-glucuronidase (GUS) (Sprenger-Haussels and Weisshaar, 2000) between the SpeI and XbaI sites (Figure 1). Promoters containing multiple copies of elements or combinations of elements in any desired order were obtained by digesting the constructs with either SpeI or XbaI together with SacI, which cuts the plasmid at a site outside of the synthetic promoter. Ligation of two such fragments recreates the plasmid with an increased number of elements. This can be repeated as the 5′ SpeI and the 3′ XbaI sites are recreated, but internal SpeI-XbaI ligations result in the loss of these restriction sites. For analysis in Arabidopsis thaliana, the entire synthetic promoter was excised as a HindIII-SacI fragment and ligated into the binary vector pGPTV-GUS-KAN (Becker et al., 1992).
Transient Expression
Transient expression analysis was performed as described previously (van de Löcht et al., 1990) using 20 μg of ScaI-linearized DNA per assay. Protoplasts were harvested 8 hr after transfection. All results represent a minimum of seven independent experiments. Normalization control experiments were performed after cotransfection with a constitutively expressed Petroselinum crispum UBI4/2::luciferase construct (Sprenger-Haussels and Weisshaar, 2000).
Transgenic Arabidopsis Lines
Agrobacterium tumefaciens–mediated plant transformation was performed as described (Bechtold et al., 1993). Approximately 10 independent lines were isolated for each synthetic promoter-GUS reporter transgene, and between two and four representative lines (typically between T2 and T5) were subjected to detailed analysis. Tested lines varied between T2 and T5, depending on the promoter tested. Histochemical staining for GUS activity was performed as described (Jefferson, 1987).
Wounding of Plants
Leaves were cut with scissors and harvested 1 hr thereafter. Control leaves were excised and immediately stained for GUS activity.
Infection of Plants with Pathogens
Plants were infected with Pseudomonas syringae pv tomato strain DC3000 without an avr gene (compatible) or with the avrRpm1 gene (incompatible) or with Peronospora parasitica pv Cala2 as described (Kirsch et al., 2001) and stained for GUS activity. Four- to 6-week-old Arabidopsis plants were infected with Erysiphe cichoracearum UCSC1 according to the method of Vogel and Somerville (2000) and with Blumeria graminis f. sp. hordei K1 according to Peterhänsel et al. (1997).
Microscopic Analyses
Clearing of leaves and staining of fungal structures were performed according to Peterhänsel et al. (1997).
Acknowledgments
We thank all of those who supported and encouraged this work, especially Günther Strittmatter, Dietmar Stahl, Jonathan Phillips, Ursula Cordier, Silke Fontein, and Luca Santi. We also thank Maret Kalda for photography and Paul Schulze-Lefert and Klaus Hahlbrock for comments on the manuscript.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010412.
References
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. Mol. Biol. Genet. 316, 1194–1199. [DOI] [PubMed] [Google Scholar]
- Becker, D., Kemper, E., Schell, J., and Masterson, R. (1992). New plant binary vector with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol. 20, 1195–1197. [DOI] [PubMed] [Google Scholar]
- Bent, A.F. (2001). Plant mitogen–activated protein kinase cascades: Negative regulatory roles turn out positive. Proc. Natl. Acad. Sci. USA 98, 784–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debener, T., Lehnackers, H., Arnold, H., and Dangl, J.L. (1991). Identification and molecular mapping of a single Arabidopsis thaliana locus determining resistance to a phytopathogenic Pseudomonas syringae isolate. Plant J. 1, 289–302. [DOI] [PubMed] [Google Scholar]
- Durrant, W.E., Rowland, O., Piedras, P., Hammond-Kosack, K.E., and Jones, J.D.G. (2000). cDNA-AFLP reveals a striking overlap in race-specific resistance and wound response gene expression profiles. Plant Cell 12, 963–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem, T., Rushton, P.J., Schmelzer, E., Hahlbrock, K., and Somssich, I.E. (1999). Early nuclear events in plant defense: Rapid gene activation by WRKY transcription factors. EMBO J. 18, 4689–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem, T., Rushton, P.J., Robatzek, S., and Somssich, I.E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. [DOI] [PubMed] [Google Scholar]
- Hara, K., Yagi, M., Kusano, T., and Sano, H. (2000). Rapid systemic accumulation of transcripts encoding a tobacco WRKY transcription factor on wounding. Mol. Gen. Genet. 263, 30–37. [DOI] [PubMed] [Google Scholar]
- Holub, E.B., Beynon, J.L., and Crute, I.R. (1994). Phenotypic and genotypic characterization of interactions between isolates of Peronospora parasitica and accessions of Arabidopsis thaliana. Mol. Plant-Microbe Interact. 7, 223–239. [Google Scholar]
- Hooley, R. (1998). Auxin signalling: Homing in with targeted genetics. Plant Cell 10, 1581–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R.J. (1987). Assaying chimaeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 5, 387–405. [Google Scholar]
- Kirsch, C., Takamiya-Wik, M., Schmelzer, E., Hahlbrock, K., and Somssich, I.E. (2000). A novel regulatory element involved in rapid activation of parsley ELI7 gene family members by fungal elicitor or pathogen infection. Mol. Plant Pathol. 1, 243–251. [DOI] [PubMed] [Google Scholar]
- Kirsch, C., Logemann, E., Lippok, B., Schmelzer, E., and Hahlbrock, K. (2001). A highly specific pathogen-responsive promoter element from the immediate-early activated CMPG1 gene in Petroselinum crispum. Plant J. 26, 1–12. [DOI] [PubMed] [Google Scholar]
- Lo, S.-C.C., Hipskind, J.D., and Nicholson, R.L. (1999). cDNA cloning of a sorghum pathogenesis-related protein (PR-10) and differential expression of defense-related genes following inoculation with Cochliobulus heterostrophus or Colletotrichum sublineolum. Mol. Plant-Microbe Interact. 12, 479–489. [DOI] [PubMed] [Google Scholar]
- Maleck, K., Levine, A., Eulgem, T., Morgen, A., Schmid, J., Lawton, K., Dangl, J.L., and Dietrich, R.A. (2000). The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26, 403–410. [DOI] [PubMed] [Google Scholar]
- Menke, F.L.H., Champion, A., Kijne, J.W., and Memelink, J. (1999). A novel jasmonate- and elicitor-responsive element in the periwinkle secondary metabolite biosynthetic gene Str interacts with a jasmonate- and elicitor-inducible AP2-domain transcription factor, ORCA2. EMBO J. 18, 4455–4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat, A.S. (2001). Finding new ways to fight plant disease. Science 292, 2270–2273. [DOI] [PubMed] [Google Scholar]
- Ohme-Takagi, M., and Shinshi, H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Tagaki, M., Suzuki, K., and Shinshi, H. (2000). Regulation of ethylene-induced transcription of defense genes. Plant Cell Physiol. 41, 1187–1192. [DOI] [PubMed] [Google Scholar]
- Peterhänsel, C., Freialdenhoven, A., Kurth, J., Kolsch, R., and Schulze-Lefert, P. (1997). Interaction analyses of genes required for resistance responses to powdery mildew in barley reveal distinct pathways leading to leaf cell death. Plant Cell 9, 1397–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, M., et al. (2000). Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 103, 1111–1120. [DOI] [PubMed] [Google Scholar]
- Raventós, D., Jensen, A.B., Rask, M.-B., Casacuberta, J.M., Mundy, J., and San Segundo, B. (1995). A 20-bp cis-acting element is both necessary and sufficient to mediate elicitor response of a maize PRms gene. Plant J. 7, 147–156. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L., and Ratcliffe, O.J. (2000). A genomic perspective on plant transcription factors. Curr. Opin. Plant Biol. 3, 423–434. [DOI] [PubMed] [Google Scholar]
- Romeis, T., Piedras, R., Zhang, S., Klessig, D.F., Hirt, H., and Jones, J.D.G. (1999). Rapid Avr9- and Cf9-dependent activation of MAP kinases in tobacco cell cultures and leaves: Convergence of resistance gene, elicitor, wound, and salicylate responses. Plant Cell 11, 273–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton, P.J., and Somssich, I.E. (1998). Transcriptional control of plant genes responsive to pathogens. Curr. Opin. Plant Biol. 1, 311–315. [DOI] [PubMed] [Google Scholar]
- Rushton, P.J., Torres, J.T., Parniske, M., Wernert, P., Hahlbrock, K., and Somssich, I.E. (1996). Interaction of elicitor-induced DNA binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 15, 5690–5700. [PMC free article] [PubMed] [Google Scholar]
- Salinas, J., Oeda, K., and Chua, N.-H. (1992). Two G-box-related sequences confer different expression patterns in transgenic tobacco. Plant Cell 4, 1485–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger-Haussels, M., and Weisshaar, B. (2000). Transactivation properties of parsley proline-rich bZIP transcription factors. Plant J. 22, 1–8. [DOI] [PubMed] [Google Scholar]
- Strittmatter, G., Gheysen, G., Gianinazzi-Pearson, V., Hahn, K., Niebel, A., Rohde, W., and Tacke, E. (1996). Infections with various types of organisms stimulate transcription from a short promoter fragment of the potato gst1 gene. Mol. Plant-Microbe Interact. 9, 68–73. [DOI] [PubMed] [Google Scholar]
- Suzuki, K., Suzuki, N., Ohme-Takagi, M., and Shinshi, H. (1998). Immediate early induction of mRNAs for ethylene-responsive transcription factors in tobacco leaf strips after cutting. Plant J. 15, 657–665. [DOI] [PubMed] [Google Scholar]
- van de Löcht, U., Meier, I., Hahlbrock, K., and Somssich, I.E. (1990). A 125-bp promoter fragment is sufficient for strong elicitor-mediated gene activation in parsley. EMBO J. 9, 2945–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, J., and Somerville, S. (2000). Isolation and characterization of powdery mildew-resistant Arabidopsis mutants. Proc. Natl. Acad. Sci. USA 97, 1897–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Yang, P., Fan, B., and Chen, Z. (1998). An oligo selection procedure for identification of sequence-specific DNA-binding activities associated with plant defense. Plant J. 16, 515–522. [DOI] [PubMed] [Google Scholar]
- Wray, G.A. (1998). Promoter logic. Science 279, 1871–1872. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A novel cis-acting element in an Arabidopsis gene involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6, 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, P., Wang, Z., Fan, B., Chen, C., and Chen, Z. (1999). A pathogen- and salicylic acid–induced WRKY DNA-binding activity recognizes the elicitor response element of tobacco class I chitinase gene promoter. Plant J. 18, 141–149. [Google Scholar]