Abstract
TATA binding protein (TBP) and transcription factor IIB (TFIIB) are key factors for the assembly of eukaryotic transcription initiation complexes. We used a rice whole-cell extract in vitro transcription system to characterize the functional interactions of recombinant plant TBP and TFIIB. Bacterially expressed rice TBP (OsTBP2) bound to the TATA box of the rice pal gene encoding phenylalanine ammonia-lyase, caused DNA bending, and enhanced basal transcription from the pal promoter in a TATA box–dependent manner. Recombinant rice TFIIB (OsTFIIB) stimulated the DNA binding and bending activities of OsTBP2 and synergistically enhanced OsTBP2-mediated transcription from the pal promoter and the promoter of Rice tungro bacilliform virus but not from the barley pr1 promoter. We also demonstrate a physical interaction between OsTBP2 and RF2a, a rice bZIP transcription factor that bound to the box II cis element of the promoter of Rice tungro bacilliform virus, resulting in enhanced transcription from the viral promoter. Enhancement of rice whole-cell extracts with recombinant transcription factors thus provides a powerful tool for the in vitro determination of plant gene regulation mechanisms. We conclude that OsTBP2 undergoes promoter-specific functional interactions with both the basal transcription factor OsTFIIB and the accessory transcription factor RF2a.
INTRODUCTION
Selective expression of sets of functionally related genes governs plant development, cellular differentiation, and responses to environmental stimuli (Brunelle and Chua, 1993). In other eukaryotes, the in vitro assembly of the transcription complex is initiated by the interaction between the TATA binding protein (TBP), which is the central component of transcription factor IIB (TFIIB), and the TATA box. This is followed by TFIIB binding to the TBP promoter complex to give a more stable ternary complex, which acts as the scaffold for RNA polymerase II and accessory factors (Nikolov et al., 1995; Orphanides et al., 1996; Tansey and Herr, 1997). However, although the plant RNA polymerase II transcription machinery appears to be very similar to that in other eukaryotes, little is known about the functional interactions underlying the selective transcription of plant genes and the mechanisms by which sequence-specific trans factors regulate the basal transcription machinery.
Analysis of RNA polymerase II–mediated transcription initiation in vitro is a powerful approach for the functional analysis of the transcription machinery and its regulation by specific cis element–trans factor interactions (Zhu, 1996; Sugiura, 1997). Efficient in vitro transcription by rice whole-cell or tobacco nuclear extracts giving authentic initiation from the in vivo start site was reported recently (Fan and Sugiura, 1995; Zhu et al., 1995a). The rice whole-cell extract supports approximately four cycles of transcription (Zhu et al., 1995a), which is comparable to the performance of systems derived from yeast, Drosophila, and HeLa cells (Kadonaga, 1990). Transcription from the promoter of a rice phenylalanine ammonia-lyase (pal) gene by the rice whole-cell extract was TATA box dependent (Zhu et al., 1995b). Using this in vitro transcription system, it was demonstrated that the pal initiator sequence was required for the selection of the transcription start site and that the spacing between the TATA box and the initiator was critical for their functional interaction (Zhu et al., 1995b). Moreover, the rice bZIP protein RF2a, which binds to the box II cis element of the promoter of Rice tungro bacilliform virus (RTBV), stimulates RTBV transcription by the rice whole-cell extract in a box II–dependent fashion (Yin et al., 1997), demonstrating the potential utility of this system in the study of functional interactions between specific cis elements and their cognate trans factors.
In aggregate, these studies indicate that the rice whole-cell extract provides the basis for a convenient and reliable homologous in vitro transcription system for the determination of the molecular mechanisms underlying selective gene expression in plants. To reconstitute a more defined system, we cloned cDNAs encoding rice TBP and TFIIB and used the recombinant proteins to stimulate transcription by the rice whole-cell extract. Using this enhanced plant in vitro transcription system, we demonstrate that rice TBP undergoes promoter-specific functional interactions with both the basal transcription factor TFIIB and the accessory transcription factor RF2a.
RESULTS
Rice cDNAs Encoding TBP and TFIIB
Two partially degenerate oligonucleotides, TBP-F and TBP-R, corresponding to conserved regions of plant TBPs, were used for polymerase chain reaction (PCR) amplification of rice TBP sequences. The PCR products were obtained with nucleotide sequences homologous with other plant TBP genes, and one of these amplicons, OsTBP, was used to probe a λZAPII rice cDNA library. Nine positive clones correspond-ing to two different TBP genes, designated OsTBP1 and OsTBP2, were identified. The OsTBP2 cDNA contains 5′ and 3′ untranslated sequences of 170 and 301 bp, respectively, flanking an open reading frame of 609 bp encoding a 203–amino acid polypeptide of 22 kD, with >90% identity to the TBPs from other plant species (Gasch et al., 1990; Haas and Feix, 1992; Hawata et al., 1992; Holdsworth et al., 1992; Apsit et al., 1993; Vogel et al., 1993). The deduced amino acid sequences of OsTBP1 and OsTBP2 are ∼95% identical, and OsTBP2 was selected for further study.
Similarly, two partially degenerate oligonucleotides, TFIIB-F and TFIIB-R, were used for PCR amplification of rice TFIIB sequences. One PCR product, which showed high homology with TFIIB genes from other plants, was used to screen the λZAPII cDNA library, and three positive plaques were purified that corresponded to a single rice TFIIB gene, designated OsTFIIB. The coding region of OsTFIIB was 936 bp and encoded a 312–amino acid polypeptide of 24 kD. The deduced amino acid sequence of OsTFIIB showed ∼78% identity to TFIIB from soybean and Arabidopsis (Baldwin and Gurley, 1996) and ∼46% identity to yeast and human TFIIB (Ha et al., 1991; Pinto et al., 1992). The OsTFIIB cDNA contains 5′ and 3′ untranslated sequences of 82 and 182 bp, respectively.
Expression of OsTBP2 and OsTFIIB in Escherichia coli
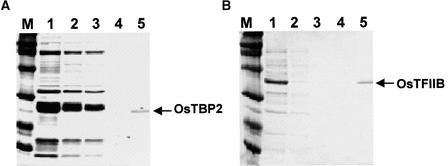
Constructs pET-OsTBP2 and pET-OsTFIIB were made to generate recombinant proteins with N-terminal S tags and C-terminal His6 tags. The recombinant tagged proteins were expressed in E. coli BL21 (DE3) cells and purified by Ni and S protein affinity chromatography before treatment with biotinylated thrombin to excise the S tag. The purified OsTBP2 and OsTFIIB proteins contained six His residues at their C termini and two additional amino acid residues (Gly-Ser) at their N termini. Gel electrophoresis indicated molecular masses of 23 kD for OsTBP2 and 35 kD for OsTFIIB (Figure 1), in agreement with their respective deduced molecular masses. Moreover, no other proteins were detected by Coomassie blue staining after gel electrophoresis of the affinity-purified fractions.
Figure 1.
Expression of OsTBP2 and OsTFIIB in E. coli.
(A) OsTBP2.
(B) OsTFIIB.
Lane 1, Ni affinity-purified fraction; lanes 2 to 4, sequential washes from the S-protein affinity matrix; lane 5, recombinant protein after excision of the S tag. Lane M, Molecular mass markers.
Physical Interaction of OsTBP2 and OsTFIIB with the pal TATA Box
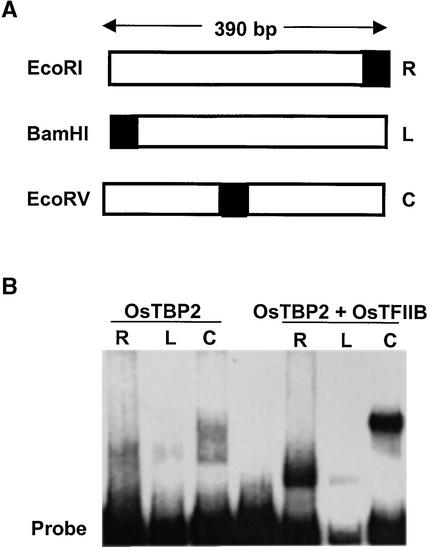
The DNA binding and bending activities of OsTBP2 and OsTFIIB were analyzed by gel retardation assays of complexes formed with 390-bp EcoRI, BamHI, and EcoRV fragments of pCY4-TA containing the rice pal TATA region. The TATA box is positioned in the center of the EcoRV fragment such that any DNA bending activity resulting from factor binding to the TATA region would decrease the migration of the DNA-protein complex compared with equivalent binding complexes with the same-size EcoRI and BamHI fragments in which the TATA box is located peripherally. Affinity-purified recombinant OsTBP2 exhibited weak binding to the TATA box region of the pal promoter, in agreement with the findings of Gasch et al. (1990). The binding complexes with the EcoRV fragment migrated more slowly than the complexes with the EcoRI or BamHI fragment (Figure 2), indicating that this weak binding nonetheless caused some DNA bending. In parallel gel retardation assays, affinity-purified recombinant OsTFIIB exhibited no binding to the pal TATA box (data not shown). However, when added together with OsTBP2, OsTFIIB markedly stimulated not only TATA binding but also DNA bending, as judged by the marked gel retardation of the ternary binding complex formed with the EcoRV fragment compared with the migration of the equivalent complexes formed with the EcoRI or BamHI fragments (Figure 2).
Figure 2.
DNA Binding and Bending Activities of OsTBP2 and OsTFIIB.
(A) Structure of DNA fragments generated from pCY4-TATA. The black region denotes the location of the pal TATA box.
(B) Gel retardation assay of the effect of OsTFIIB on the binding complexes formed between OsTBP2 and the pCY4-TATA fragments containing the pal promoter TATA box in the center (EcoRV) or at either terminus (BamHI and EcoRI). Recombinant protein (50 ng) was used as indicated. The reaction products were separated by electrophoresis on a 5% polyacrylamide gel in Tris-Gly buffer.
C, center; L, left, R, right.
OsTBP2 Stimulation of pal Transcription
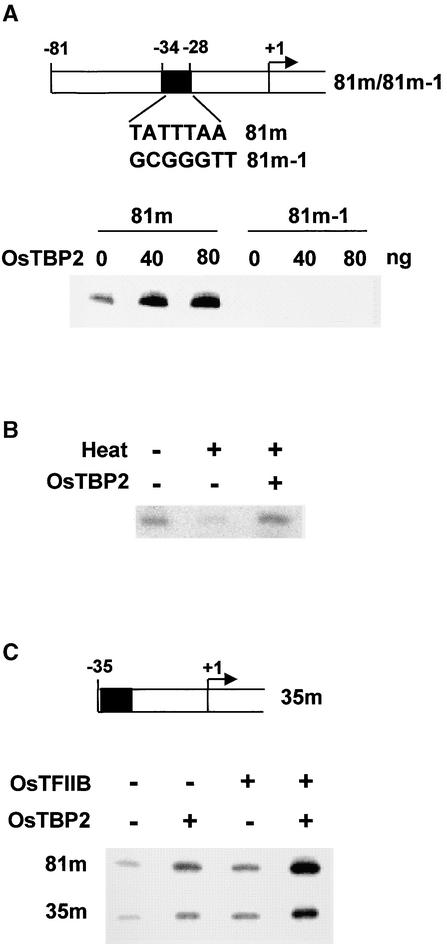
To study the functional activity of OsTBP2, we used a homologous in vitro system that accurately initiates transcription from a template containing pal promoter sequences from −81 to +45 fused with the uidA reporter gene encoding β-glucuronidase (Zhu et al., 1995a). Affinity-purified recombinant OsTBP2 markedly enhanced the accurate transcription of the pal promoter template by rice whole-cell extracts in a dose-dependent manner (Figure 3A; see also Figure 5A). In contrast, OsTBP2 failed to stimulate transcription from the pal 81m-1 promoter template, in which the 7-bp TATA element TATTTAA, extending from −34 to −28, was substituted by the sequence GCGGGTT (Figure 3A), indicating that OsTBP2 stimulation of pal transcription was TATA box dependent. As reported previously for tobacco nuclear extracts (Iwataki et al., 1997), the transcriptional activity of rice whole-cell extracts is heat labile (Figure 3B). The addition of recombinant OsTBP2 to rice whole-cell extracts inactivated by incubation at 46°C for 15 min completely restored accurate transcription initiation; hence, TBP appears to be a labile, rate-limiting factor for transcription by rice whole-cell extracts.
Figure 3.
OsTBP2 Stimulation of pal Transcription by Rice Whole-Cell Extracts.
(A) OsTBP2 stimulation of pal transcription depends on the TATA element.
(B) The transcriptional activity of a heat-inactivated rice whole-cell extract was restored by the addition of OsTBP2.
(C) OsTBP2 stimulation of transcription from the minimal pal promoter 5′ truncated to −35.
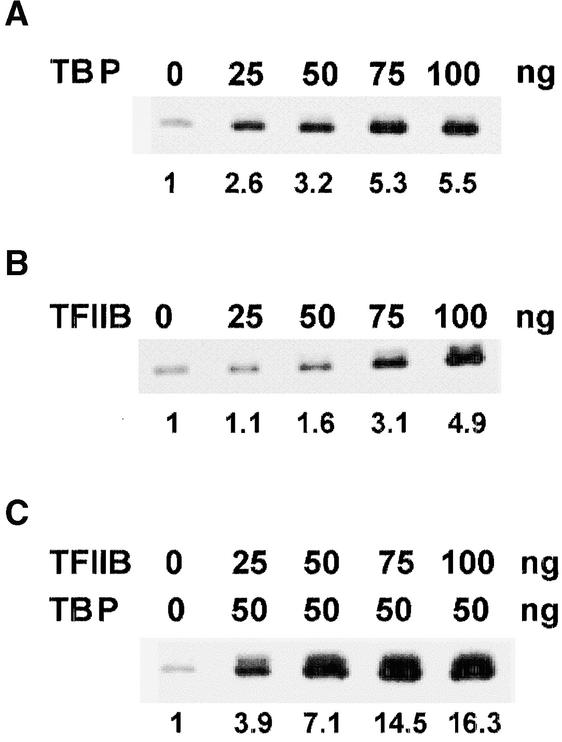
Figure 5.
Dose Responses of OsTBP2 and OsTFIIB Functional Interactions.
(A) Dose responses of OsTBP2.
(B) Dose responses of OsTFIIB.
(C) Dose responses of OsTBP2 and OsTFIIB functional interactions.
Synergistic Activity of OsTBP2 and OsTFIIB
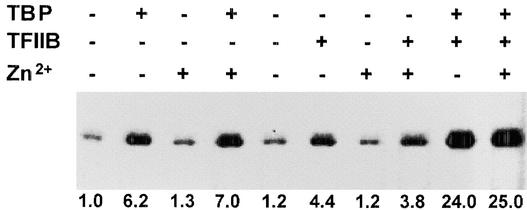
Because OsTFIIB promoted OsTBP2 binding to the pal TATA box and DNA bending activity, we next examined whether OsTFIIB modulated the functional activity of TBP. The addition of affinity-purified recombinant OsTFIIB to the rice whole-cell extract stimulated pal transcription up to fourfold (Figure 4), and the enhancement occurred in a dose-dependent manner (Figure 5B). TFIIBs are Zn-finger transcription factors (Baldwin and Gurley, 1996). However, the addition of Zn2+ had no further effect on OsTFIIB-mediated transcription by the rice whole-cell extract, indicating that the trace amounts of Zn2+ in the reaction system were sufficient for the function of OsTFIIB. In contrast, the simultaneous addition of recombinant OsTFIIB with recombinant OsTBP2 resulted in a marked further enhancement of pal transcription. Thus, the addition of nearly saturating amounts (100 ng) of both OsTBP2 and OsTFIIB resulted in a 24-fold stimulation of pal transcription compared with the basal rate observed with the unenhanced rice whole-cell extract (Figure 5).
Figure 4.
Functional Interaction between OsTBP2 and OsTFIIB in the Initiation of pal Transcription.
81m was used as the template for in vitro transcription reactions supplemented with recombinant OsTBP2, OsTFIIB, and Zn2+, as indicated.
This response was substantially greater than the sum of the effects of the two factors tested separately. Similar synergistic interactions between OsTBP2 and OsTFIIB also were observed with nonsaturating levels of the recombinant factors (Figure 5C). Moreover, strong functional interactions between OsTBP2 and OsTFIIB were observed with the pal promoter 5′ truncated to −35 as a template. With this minimal promoter, which contains the TATA box but no upstream pal promoter sequences, the addition of either OsTBP2 or OsTFIIB alone stimulated transcription by the rice whole-cell extract ∼3-fold, whereas the simultaneous addition of both factors resulted in a >10-fold enhancement (Figure 3C). Stronger transcription from the −81 promoter compared with the minimal pal promoter presumably reflects the activities of cis elements upstream of −35 and associated trans factors in the whole-cell extract.
Selective Promoter Activation by OsTFIIB
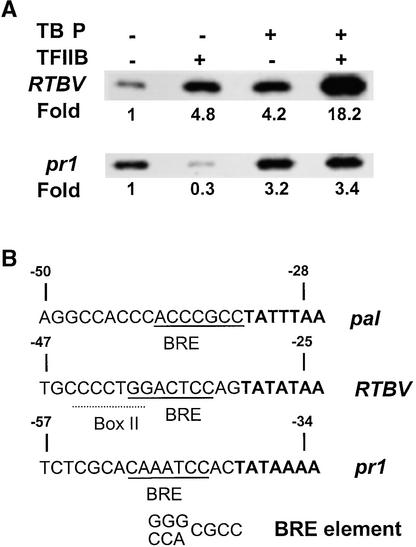
To study the functional activities of OsTBP2 and OsTFIIB in different settings, we examined the effects of the recombinant factors on the in vitro transcription of the RTBV (Yin and Beachy, 1995) and barley pr1 (Mouradov et al., 1994) promoters. Previous work established that the rice whole-cell extract accurately initiates transcription from the RTBV promoter (Yin et al., 1997). Separate addition of OsTBP2 and OsTFIIB stimulated transcription from this promoter 4.8- and 4.2-fold, respectively, whereas simultaneous addition of the two factors stimulated transcription 18-fold (Figure 6). Thus, as with the pal promoter, OsTBP2 and OsTFIIB interact functionally to give a synergistic enhancement of transcription from the viral promoter. In contrast, there was no functional interaction between OsTBP2 and OsTFIIB when the barley pr1 promoter was used as a template. Rice whole-cell extracts initiate transcription more efficiently from the pr1 promoter than from the pal or RTBV promoters. Although the addition of OsTBP2 stimulated transcription by more than threefold, recombinant OsTFIIB had no enhancement effect on pr1 transcription by the rice whole-cell extracts either when added alone or when added together with OsTBP2 (Figure 6).
Figure 6.
Promoter-Specific and Selective Interaction between OsTBP2 and OsTFIIB.
(A) Selective interaction between OsTBP2 and OsTFIIB in the transcription of RTBV::uidA and pr1::uidA templates by rice whole-cell extracts.
(B) DNA sequences upstream of the TATA element in the pal, RTBV, and pr1 promoters. BRE is the TFIIB-recognized element.
OsTBP2–RF2a Interactions in RTBV Transcription
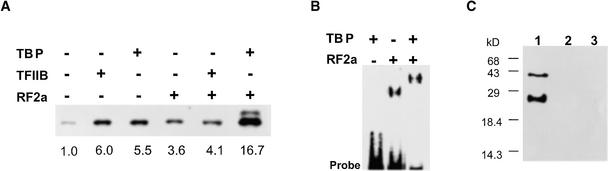
The observation that functional interactions between OsTBP2 and OsTFIIB were promoter specific prompted us to examine whether OsTBP2 also interacted functionally with a promoter-selective accessory transcription factor. To address this question, we monitored the effects of RF2a on OsTBP2 stimulation of transcription from the RTBV promoter. We reported previously that recombinant RF2a stimulates RTBV in vitro transcription in a box II–dependent manner (Yin et al., 1997). However, when the rice whole-cell extract was supplemented with both recombinant OsTBP2 and RF2a, there was a synergistic stimulation of RTBV transcription compared with the effects of either factor alone (Figure 7A). In contrast, there was no synergistic or even additive interaction between RF2a and OsTFIIB, with RTBV transcription remaining at approximately the level observed with either factor alone. When the rice whole-cell extract was supplemented with recombinant OsTBP2, OsTFIIB, and RF2a together, the stimulation was approximately the same as that observed with just OsTBP and RF2a together, and there was no further effect of OsTFIIB on the transcription of the RTBV promoter (data not shown).
Figure 7.
Physical and Functional Interactions between OsTBP2 and the RF2a bZIP Transcription Factor.
(A) Functional interactions between RF2a and OsTBP2 in the transcription of the RTBV::uidA template by rice whole-cell extracts. Recombinant proteins (100 ng) were added as indicated 10 min before the initiation of the transcription reaction by addition of the whole-cell extract.
(B) Effect of OsTBP2 on binding complexes formed between RF2a and box II of the RTBV promoter.
(C) Protein gel blot analysis. Recombinant OsTBP2 (lane 1), coat protein of Tobacco mosaic virus (lane 2), and BSA (lane 3) were subjected to SDS–12% PAGE, blotted onto a nitrocellulose membrane, and incubated with recombinant RF2a. RF2a binding was visualized by alkaline phosphatase activity after sequential incubation of the membrane with rabbit anti-RF2a antibody and alkaline phosphatase–conjugated goat anti-rabbit IgG.
The synergistic functional interaction between OsTBP2 and RF2a in the initiation of transcription from the RTBV promoter apparently was accompanied by a direct physical interaction. Thus, in gel retardation assays, OsTBP2 did not bind to a small fragment of the RTBV promoter containing the box II cis element, but OsTBP2 further retarded electrophoretic migration of the RF2a–box II binding complex (Figure 7B). Protein gel blot analysis indicated that the supershifted DNA protein complex reflected, at least in part, a direct, DNA-independent interaction between RF2a and OsTBP2. In this experiment, recombinant OsTBP2 was subjected to SDS-PAGE and then transferred to a nitrocellulose membrane and incubated with recombinant RF2a. Bound RF2a was visualized by anti-RF2a antibody in conjunction with alkaline phosphatase–conjugated goat anti-rabbit IgG. Figure 7C shows that RF2a bound strongly to the 23-kD OsTBP2 and more weakly to a second, more slowly migrating species, which may be a partially denatured OsTBP2 dimer. No RF2a binding was observed with either BSA or the coat protein of Tobacco mosaic virus as a negative control.
DISCUSSION
TBP and TFIIB are two key components of the RNA polymerase II transcription initiation complex (Buratowski, 1994; Orphanides et al., 1996). Like other plants (Gasch et al., 1990; Haas and Feix, 1992; Hawata et al., 1992; Holdsworth et al., 1992; Apsit et al., 1993), rice has two distinctive isoforms of TBP, the C-terminal sequences of which are highly homologous with those in yeast and human TBP (Horikoshi et al., 1990; Peterson et al., 1990). Studies in nonplant systems indicate that the binding of TBP to the TATA element facilitates promoter recognition by the machinery for transcription initiation and that the TBP-TATA complex locally disrupts chromatin packaging (Burley and Roeder, 1996; Orphanides et al., 1996). The in vitro TATA binding and DNA bending activity of bacterially expressed OsTBP2 (Figure 2) is entirely consistent with this model. Moreover, recombinant OsTBP2 strongly stimulated the initiation of transcription by the rice whole-cell extract. Likewise, bacterially expressed recombinant OsTFIIB appears to be functionally active. Although OsTFIIB alone was not able to bind to the TATA element of the pal promoter or cause DNA bending, OsTFIIB stimulated the DNA binding and bending activities of OsTBP2. These observations are consistent with the current model, in which TFIIB interacts with the TBP-DNA complex to form the TBP-TFIIB-DNA complex, which docks the RNA polymerase II machinery (Buratowski, 1994; Orphanides et al., 1996).
The stimulation by recombinant OsTBP2 and OsTFIIB reflects initiation from the in vivo start site and is dose dependent, indicating that transcription in the enhanced system remains authentic. The stimulation of transcription by OsTBP2 is TATA box dependent and was observed with all three promoters tested, each of which contains a clearly discernible TATA element (Figure 6B). In contrast, OsTFIIB stimulated only the pal and RTBV promoters but not the pr1 promoter. The crystallographic structures of the ternary complex of the TFIIB core domain, the TBP core domain, and a 16-bp DNA fragment containing the TATA element show that TFIIB interacts with TBP, the DNA major groove immediately upstream of the TATA element, and the DNA minor groove immediately downstream of the TATA element (Nikolov et al., 1995).
TFIIB was shown recently to bind to a specific G/C-G/C-G/A-C-G-C-C element (BRE) that often is located immediately upstream of the TATA element (Lagrange et al., 1998). Sequence comparisons (Figure 6B) show that there is a BRE-like sequence located immediately upstream of the pal TATA element. There also is a BRE-like element in the RTBV promoter 2 bp upstream from the TATA element. The BRE-like element overlaps with the box II element recognized by RF2a (Yin and Beachy, 1995). The barley pr1 promoter (Mouradov et al., 1994) also has a BRE-like element sequence (CAAATCC) located 2 bp upstream of the TATA element. However, OsTFIIB does not stimulate transcription from the pr1 promoter, and this may reflect in part the fact that the pr1 element matches only 4 of 7 bp of the canonical BRE element. Moreover, the rice whole-cell extracts support much higher rates of transcription from the pr1 promoter than from the pal or RTBV promoters (Figure 6A), and the pr1 promoter may have a strong TATA element, so that transcription from this promoter may be less dependent on TFIIB.
OsTFIIB also stimulates the minimal pal promoter, which lacks the BRE-like element. TFIIB is involved in the recruitment of RNA polymerase II–TFIIF and selection of the correct transcription start site (Roeder, 1996), and the selective action of TFIIB also may depend in part on the structure of the minimal promoter. TFIIB also interacts with the DNA minor groove immediately downstream of the TATA element (Nikolov et al., 1995), and TATA-flanking sequences can influence the rate and stability of TBP and TFIIB binding (Wolner and Gralla, 2001). Our data indicate that the synergistic functional interactions between OsTBP2 and OsTFIIB likewise depend in part on the sequences flanking the TATA element.
The effect of RF2a on RTBV promoter transcription also reveals selective functional interactions. Thus, recombinant RF2a stimulated transcription from the RTBV promoter and synergistically enhanced OsTBP2 activity but not OsTFIIB activity with this promoter. Because the box II element and the BRE-like element overlap in the RTBV promoter, TFIIB and RF2a may compete with each other for the binding site, thereby precluding additive or synergistic effects on transcription from the RTBV promoter. Nevertheless, these data confirm the function of RF2a as a transcription activator (Yin et al., 1997) and suggest that the basal transcription factor OsTBP2 mediates the action of the cis element–specific RF2a transcription factor.
The selective use of TBP and TFIIB to activate transcription is a common mechanism in all eukaryotic systems. In HeLa cells, activators such as CTF, with a Pro-rich domain, and VP16, with an acidic amino acid–rich domain, use the TBP–TFIIB interaction to stimulate transcription. The activator Sp1, which has a Gln-rich domain, can stimulate transcription independently of TFIIB (Tansey and Herr, 1997). RF2a has Pro-rich and acidic amino acid–rich domains at its N terminus and a Gln-rich domain at its C terminus (Yin et al., 1997) as structural components potentially contributing to its transcription activation function. Because RF2a apparently functions independently of OsTFIIB, it will be of interest to compare the mechanism of RF2a action with that of Sp1.
In yeast, activation domains of trans factors can stimulate the recruitment of TBP to promoters, and several activators, such as Epstein-Barr virus Zta, adenovirus E1A, Herpes simplex virus VP16, and Human immunodeficiency virus 1 Tat, have direct functional interactions with TBP (Horikoshi et al., 1991; Ingles et al., 1991; Lee et al., 1991; Lieberman and Berk, 1991; Kashanchi et al., 1994; Xiao et al., 1997). Likewise, our data indicate that the synergistic functional interactions between OsTBP2 and RF2a in the initiation of RTBV transcription reflect a direct physical interaction. Moreover, protein gel blot analysis indicates that the physical interaction between the two transcription factors does not require the corresponding cis elements. The selective use of TBP and TFIIB provides a mechanism by which activators can selectively stimulate transcription from different promoters on which the basal transcription machinery might be arranged differently.
Use of the rice whole-cell extract in vitro transcription has allowed us to demonstrate that OsTBP2 undergoes promoter-specific functional interactions with both the basal transcription factor OsTFIIB and the accessory transcription factor RF2a. The introduction of bacterially expressed recombinant factors markedly stimulates transcription initiation by the whole-cell extract, indicating that although this extract is transcriptionally competent, specific factors are limiting. The enhanced system provides markedly more robust transcription of all promoters examined while retaining key functional attributes, including transcription start site, TATA box dependence, and promoter selectivity. Thus, the rice whole-cell extract supplemented with recombinant OsTBP2 and OsTFIIB represents a powerful tool for ex-ploring the functional interactions underlying the selective activation of plant genes, including the rapid functional elucidation of specific cis element–trans factor interactions and analysis of the functional attributes of orphan transcription factors disclosed by plant genome sequencing.
METHODS
Nucleic Acid Manipulations
General molecular biological techniques were performed according to standard protocols (Sambrook et al., 1989). Site-directed mutagenesis was performed as described previously (Zhu et al., 1995c).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
PCR and cDNA Library Screening
Partially degenerate oligonucleotides corresponding to the conserved peptide sequences CDVKFPI and IYRMKVP of TATA binding protein (TBP) and TSEWRTF and KKEIGRA of transcription factor IIB (TFIIB) in other plants were used for polymerase chain reaction amplification of rice (Oryza sativa) TBP and TFIIB sequences. The polymerase chain reaction fragments were cloned into pGEM7 (Promega) and used to screen a rice λZAPII cDNA library (S.R. McCouch, Cornell University, Ithaca, NY). pBluescript KS− plasmids (Stratagene) containing rice TBP and TFIIB cDNAs were recovered from the purified λZAPII clones, and nucleotide sequences were determined by the dideoxy chain-termination method (Sambrook et al., 1989).
Expression of Recombinant Proteins in Escherichia coli
NcoI and XhoI sites were introduced into OsTBP2 and OsTFIIB cDNAs around their translation initiation and stop codons, respectively. The NcoI and XhoI OsTBP2 and OsTFIIB coding region fragments were ligated into pET-29a (Novagen, Madison, WI) to generate pET-OsTBP2 and pET-OsTFIIB, which were transformed into E. coli BL21 (DE3). The transformed cells were grown at 30°C to an OD600 of 1.0 and then induced with 0.2 mM isopropyl-β-d-thiogalactopyranoside for 3 hr. Recombinant proteins were purified from bacterial cell extracts using His-Trap (Pharmacia Biotech) and S tag agarose beads (Novagen) according to the manufacturers' instructions. Purified recombinant OsTBP2 and OsTFIIB were released from the S tag–agarose bead complex by biotinylated thrombin, which was removed by streptavidin. To increase the activities of purified recombinant proteins, they were denatured with 2 M urea and then dialyzed in 20 mM Hepes-KOH, pH 7.9, 1 mM MgCl2, 50 mM KCl, 1 mM DTT, 20% glycerol, and 0.02% Nonidet P-40 overnight. The recombinant proteins were concentrated with Centriprep10 (Amicon, Beverly, MA), snap frozen in liquid N2, and stored at −70°C. Recombinant RF2a was prepared as described previously (Yin et al., 1997).
DNA Binding and Bending Assays
A 23-bp fragment of the rice pal promoter (−42 to −19) containing the TATA element (Zhu et al., 1995b) was cloned into the SmaI site of pCY4 (Prentki et al., 1987) to generate pCY4-TA. The BamHI, EcoRI, and EcoRV fragments of pCY4-TA, each of 390 bp, were labeled with 32P-γ-ATP and used as probes in gel retardation assays for the interaction with OsTBP and OsTFIIB. The −53 to −39 element (5′-CCAGTGTGGCGCTGG-3′) of the promoter of Rice tungro bacilliform virus was used as a probe in gel retardation assays of RF2a binding. Binding reactions were performed in 20 μL of 20 mM Hepes-KOH, pH 8.0, 50 mM KCl, 1 mM DTT, 10% glycerol, 0.3 μg of poly(dI-dC): poly(dI-dC), and recombinant proteins. The mixtures were incubated at room temperature for 15 min and then separated by electrophoresis on 5% native polyacrylamide gels. The gels were dried and exposed to x-ray film overnight.
Protein Gel Blot Analysis
Protein gel blot analysis was performed as described by Fischer et al. (1997) with modifications. Briefly, 1 μg each of partially purified recombinant OsTBP2, coat protein of Tobacco mosaic virus, and BSA was electrophoresed separately on a 12% polyacrylamide gel and transferred to a nitrocellulose membrane, which was denatured in 20 mM Hepes-KOH, pH 7.9, 10% glycerol, 60 mM KCl, 6 mM MgCl2 0.6 mM EDTA, and 2 mM DTT with 6 M guanidine chloride for 30 min. The proteins then were renatured twice by incubation in the same buffer containing 100 mM guanidine chloride for 2 hr. After further incubation for 1 hr in this renaturation buffer containing 4% skim milk, 10 μg of purified recombinant RF2a was added to the buffer and incubated overnight. The membrane was washed for 1 hr in 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 0.1% Tween-20 (TBST buffer) plus 4% skim milk and then incubated for 2 hr with rabbit anti-RF2a antibody (Petruccelli et al., 2001) at a dilution of 1:1000 in TBST buffer plus 1% skim milk. The membrane was washed three times with TBST buffer, and alkaline phosphatase–conjugated goat anti-rabbit IgG (1:5000 dilution) was added and incubated for 1 hr. The membrane was washed three times with TBST buffer. Binding was visualized by alkaline phosphatase activity (Harlow and Lane, 1988).
In Vitro Transcription
Whole-cell extracts of rice cv IR72 suspension cultures were used for in vitro transcription reactions as described previously (Zhu et al., 1995b). Within each experiment, the amounts of template and primer were the same in each individual reaction. The indicated amounts of recombinant proteins were added to the reaction mixture 10 min before the addition of the whole-cell extract. Transcription products were analyzed by primer extension (Zhu et al., 1995a).
Accession Numbers
The GenBank accession numbers for the sequences described in this article are AF464907 (OsTBP2) and AF464908 (OsTFIIB).
Acknowledgments
M.I.O. was a fellow of the Fundacion para la Investigacion Cientifica y Tecnologica Asturias, Spain. This research was supported by grants to R.N.B. and C.L. from the Rockefeller Foundation Rice Biotechnology Program.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010364.
References
- Apsit, V., Freeberg, J.A., Chase, M.R., Davis, E.A., and Ackerman, S. (1993). Wheat TFIID TATA-binding protein. Nucleic Acids Res. 21, 1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin, D., and Gurley, W.B. (1996). Isolation and characterization of cDNA encoding transcription factor IIB from Arabidopsis and soybean. Plant J. 10, 561–568. [DOI] [PubMed] [Google Scholar]
- Brunelle, A.N., and Chua, N.-H. (1993). Transcription regulatory proteins in higher plants. Curr. Opin. Genet. Dev. 3, 254–258. [DOI] [PubMed] [Google Scholar]
- Buratowski, S. (1994). The basics of basal transcription by RNA polymerase II. Cell 77, 1–3. [DOI] [PubMed] [Google Scholar]
- Burley, S.K., and Roeder, R.G. (1996). Biochemistry and structural biology of transcription factor IID (TFIID). Annu. Rev. Biochem. 65, 769–799. [DOI] [PubMed] [Google Scholar]
- Fan, H., and Sugiura, M. (1995). A plant basal in vitro transcription system supporting accurate transcription of both RNA polymerase II- and III-dependent genes: Supplement of green leaf components drives accurate transcription of a light-responsive rbcS gene. EMBO J. 14, 1024–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, N., Kremmer, E., Lautscham, G., Mueller-Lantzsch, N., and Grasser, F. (1997). Epstein-Barr virus nuclear antigen 1 forms a complex with the nuclear transporter karyopherin α2. J. Biol. Chem. 272, 3999–4005. [DOI] [PubMed] [Google Scholar]
- Gasch, A., Hoffmann, A., Horikoshi, M., Roeder, R.G., and Chua, N.-H. (1990). Arabidopsis thaliana contains two genes for TFIID. Nature 346, 390–394. [DOI] [PubMed] [Google Scholar]
- Ha, I., Lane, W.S., and Reinberg, D. (1991). Cloning of a human gene encoding the general transcription initiation factor IIB. Nature 352, 689–695. [DOI] [PubMed] [Google Scholar]
- Haas, M.M., and Feix, G. (1992). Two different cDNAs encoding TFIID proteins of maize. FEBS Lett. 301, 294–298. [DOI] [PubMed] [Google Scholar]
- Harlow, E., and Lane, D. (1988). Antibodies: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Hawata, T., Minami, M., Tamura, T., Sumita, K., and Iwabuchi, M. (1992). Isolation and characterization of a cDNA clone encoding the TATA box-binding protein (TFIID) from wheat. Plant Mol. Biol. 19, 867–872. [DOI] [PubMed] [Google Scholar]
- Holdsworth, M.J., Grierson, C., Schuch, W., and Bevan, M. (1992). DNA binding properties of cloned TATA-binding protein from potato tubers. Plant Mol. Biol. 19, 455–464. [DOI] [PubMed] [Google Scholar]
- Horikoshi, M., Yamamoto, T., Okhuma, Y., Wei, P.A., and Roeder, R.G. (1990). Analysis of structure-function relationships of yeast TATA-binding factor TFIID. Cell 61, 1171–1178. [DOI] [PubMed] [Google Scholar]
- Horikoshi, N., Maguire, K., Kralli, A., Maldonado, E., Reinberg, D., and Weinmann, R. (1991). Direct interaction between adenovirus E1A protein and the TATA box binding transcription factor IID. Proc. Natl. Acad. Sci. USA 88, 5124–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingles, C.J., Shales, M., Cress, W.D., Triezenberg, S.J., and Greenblatt, J. (1991). Reduced binding of TFIID to transcriptionally compromised mutants of VP16. Nature 351, 588–590. [DOI] [PubMed] [Google Scholar]
- Iwataki, N., Hoya, A., and Yamazaki, K.-I. (1997). Restoration of TATA-dependent transcription in a heat-inactivated extract of tobacco nuclei by recombinant TATA-binding protein (TBP) from tobacco. Plant Mol. Biol. 34, 69–79. [DOI] [PubMed] [Google Scholar]
- Kadonaga, J.T. (1990). Assembly and disassembly of the Drosophila RNA polymerase II complex during transcription. J. Biol. Chem. 265, 2624–2631. [PubMed] [Google Scholar]
- Kashanchi, F., Piras, G., Radonovich, M.F., Durall, J.F., Fattaey, A., Chiang, C.-M., Roeder, R.G., and Brady, J.N. (1994). Direct interaction of human TFIID with the HIV-1 transactivator Tat. Nature 367, 295–299. [DOI] [PubMed] [Google Scholar]
- Lagrange, T., Kapanidis, A.N., Tang, H., and Reinberg, D. (1998). New core promoter element in RNA polymerase II-dependent transcription: Sequence-specific DNA binding by transcription factor IIB. Genes Dev. 12, 34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, W.S., Kao, C.C., Bryant, G.O., Liu, X., and Berk, A.J. (1991). Adenovirus E1A activation domain binds the basic repeat in the TATA box transcription factor. Cell 67, 365–376. [DOI] [PubMed] [Google Scholar]
- Lieberman, P.M., and Berk, A.J. (1991). The Zta trans-activator protein stabilizes TFIID association with promoter DNA by direct protein-protein interaction. Genes Dev. 5, 2441–2454. [DOI] [PubMed] [Google Scholar]
- Mouradov, A., Mouradova, E., and Scott, K.J. (1994). Gene family encoding basic pathogenesis-related 1 proteins in barley. Plant Mol. Biol. 26, 503–507. [DOI] [PubMed] [Google Scholar]
- Nikolov, D.B., Chen, H., Halay, E.D., Usheva, A.A., Hisatake, K., Lee, D.K., Roeder, R.G., and Burley, S.K. (1995). Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature 377, 119–128. [DOI] [PubMed] [Google Scholar]
- Orphanides, G., Lagrange, T., and Reinberg, D. (1996). The general transcription factors of RNA polymerase II. Genes Dev. 10, 2657–2683. [DOI] [PubMed] [Google Scholar]
- Peterson, M.G., Tanese, N., Pugh, B.F., and Tjian, R. (1990). Functional domains and upstream activation properties of cloned human TATA binding protein. Science 248, 1625–1630. [DOI] [PubMed] [Google Scholar]
- Petruccelli, S., Dai, S., Carcamo, R., Yin, Y., Chen, S., and Beachy, R.N. (2001). Transcription factor RF2a alters expression of the rice tungro bacilliform virus promoter in transgenic tobacco plants. Proc. Natl. Acad. Sci. USA 98, 7635–7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto, I., Ware, D.E., and Hampsey, M. (1992). The yeast SUA7 gene encodes a homolog of human transcription factor TFIIB and is required for normal start site selection. Cell 68, 977–988. [DOI] [PubMed] [Google Scholar]
- Prentki, P., Pham, M.-H., and Galas, J. (1987). Plasmid perturbation vectors to monitor DNA bending. Nucleic Acids Res. 15, 10060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder, R.G. (1996). The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21, 327–335. [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sugiura, M. (1997). Plant in vitro transcription systems. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 383–398. [DOI] [PubMed] [Google Scholar]
- Tansey, W.P., and Herr, W. (1997). Selective use of TBP and TFIIB revealed by a TATA-TBP-TFIIB array with altered specificity. Science 275, 829–831. [DOI] [PubMed] [Google Scholar]
- Vogel, J.M., Roth, B., Cigan, M., and Freeling, M. (1993). Expression of the two maize TATA binding protein genes and functions of the encoded TBP proteins by complementation in yeast. Plant Cell 5, 1627–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolner, B.S., and Gralla, J.D. (2001). TATA-flanking sequences influence the rate and stability of the TATA-binding protein and TFIIB binding. J. Biol. Chem. 276, 6260–6266. [DOI] [PubMed] [Google Scholar]
- Xiao, H., Lis, J.T., and Jeang, K.T. (1997). Promoter activity of Tat at steps subsequent to TATA-binding protein recruitment. Mol. Cell. Biol. 17, 6898–6905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, Y., and Beachy, R.N. (1995). The regulatory regions of the rice tungro bacilliform virus promoter and interacting nuclear factors in rice (Oryza sativa L.). Plant J. 7, 969–980. [DOI] [PubMed] [Google Scholar]
- Yin, Y., Zhu, Q., Da, S., Lamb, C., and Beachy, R.N. (1997). RF2a, a bZIP transcriptional activator of the phloem-specific rice tungro bacilliform virus promoter, functions in vascular development. EMBO J. 16, 5247–5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Q. (1996). RNA polymerase II-dependent plant in vitro transcription systems. Plant J. 10, 185–188. [Google Scholar]
- Zhu, Q., Chappell, J., Hedrick, S.A., and Lamb, C. (1995. a). Accurate in vitro transcription from circularised plasmid templates by plant whole cell extracts. Plant J. 7, 1021–1030. [DOI] [PubMed] [Google Scholar]
- Zhu, Q., Dabi, T., and Lamb, C. (1995. b). TATA box and initiator functions in the accurate transcription of a plant minimal promoter in vitro. Plant Cell 7, 1681–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Q., Dabi, T., Beeche, A., Yamamoto, R., Lawton, M.A., and Lamb, C. (1995. c). Cloning and properties of a rice gene encoding phenylalanine ammonia-lyase. Plant Mol. Biol. 29, 535–550. [DOI] [PubMed] [Google Scholar]