Abstract
The Pti4, Pti5, and Pti6 proteins from tomato were identified based on their interaction with the product of the Pto disease resistance gene, a Ser-Thr protein kinase. They belong to the ethylene-response factor (ERF) family of plant-unique transcription factors and bind specifically to the GCC-box cis element present in the promoters of many pathogenesis-related (PR) genes. Here, we show that these tomato ERFs are localized to the nucleus and function in vivo as transcription activators that regulate the expression of GCC box–containing PR genes. Expression of Pti4, Pti5, or Pti6 in Arabidopsis activated the expression of the salicylic acid–regulated genes PR1 and PR2. Expression of jasmonic acid– and ethylene-regulated genes, such as PR3, PR4, PDF1.2, and Thi2.1, was affected differently by each of the three tomato ERFs, with Arabidopsis-Pti4 plants having very high levels of PDF1.2 transcripts. Exogenous application of salicylic acid to Arabidopsis-Pti4 plants suppressed the increased expression of PDF1.2 but further stimulated PR1 expression. Arabidopsis plants expressing Pti4 displayed increased resistance to the fungal pathogen Erysiphe orontii and increased tolerance to the bacterial pathogen Pseudomonas syringae pv tomato. These results indicate that Pti4, Pti5, and Pti6 activate the expression of a wide array of PR genes and play important and distinct roles in plant defense.
INTRODUCTION
Plants respond to pathogen attack by activating multiple defense mechanisms to protect themselves from infection. These rapid cellular responses often are triggered by the recognition of specific pathogens and the activation of highly regulated signal transduction pathways. A major target of these pathways is the cell nucleus, where signals lead to the transcriptional activation of a large array of defense genes (Maleck et al., 2000; Schenk et al., 2000). The products of these genes include pathogenesis-related (PR) proteins as well as enzymes involved in the biosynthesis of protective secondary metabolites. Although the functions of many PR proteins remain unknown, some PR proteins, such as β-1,3-glucanase (PR2) and chitinase (PR3), are hydrolytic enzymes that have been shown to degrade fungal cell walls and to inhibit fungal growth both in vivo and in vitro (Broglie et al., 1991; Sela-Buurlage et al., 1993; Zhu et al., 1994). It was shown recently that osmotin (PR5) induces apoptosis in yeast, and it may act similarly toward plant fungal pathogens (Narasimhan et al., 2001).
Several signaling molecules, such as salicylic acid (SA), ethylene (ET), and jasmonic acid (JA), have been shown to be important components of defense response pathways (Dong, 1998; Reymond and Farmer, 1998; Dempsey et al., 1999; Pieterse and van Loon, 1999). Infection by microbial pathogens results in an increase in the levels of these molecules in plants, and many PR genes that are induced upon pathogen infection also are upregulated by one or more of these signaling molecules (Malamy et al., 1990; Thomma et al., 1998; Dempsey et al., 1999). The SA-dependent defense signaling pathway regulates the expression of acidic PR proteins such as PR1, PR2, and PR5. The ET/JA-dependent signaling pathway(s) regulates the expression of vacuole-localized basic PR proteins such as PR3, PR4, and PDF1.2. Genetic and biochemical evidence exists for communication between the different pathways (Feys and Parker, 2000), which could be either coregulatory or antagonistic responses (Maleck and Dietrich, 1999). In addition, the nature of this communication appears to be pathogen dependent, which is consistent with the finding that activation of the SA- and/or ET/JA-dependent defense pathways also is pathogen specific (Thomma et al., 1998).
The promoters of several PR genes have been studied to identify the specific cis-acting elements that confer responsiveness to SA, ET, or JA. In some cases, the identification of these cis-acting elements has led to the isolation of the cognate transcription factors. Many ET-inducible PR genes contain an 11-bp GCC box (TAAGAGCCGCC) in their promoter regions (Eyal et al., 1993). Transcription factors that bind the GCC box specifically, the ET-responsive element binding proteins (EREBPs), were first isolated from tobacco and shown to be ET induced and involved in the expression of GCC box–containing PR genes. These proteins were later renamed ET response factors (ERFs) (Ohme-Takagi and Shinshi, 1995; Suzuki et al., 1998). The ERF domain was believed previously to be closely related to the AP2 domain, which is found in AP2 domain transcription factors involved in plant development (Okamuro et al., 1997; Riechmann and Meyerowitz, 1998). It has now been shown that ERFs possess a highly conserved DNA binding domain, and the solution structure of this domain shows that it is novel, with a unique form of DNA recognition (Ohme-Takagi et al., 2000). ERF-encoding genes are present only in higher plants.
ERFs are present in plant species from phylogenetically distinct taxa. These genes have been characterized in Arabidopsis (ERF1, AtEBP, and AtERFs; Büttner and Singh, 1997; Solano et al., 1998; Fujimoto et al., 2000), tomato (Pti4/5/6; Zhou et al., 1997), and soybean (GmEREBP1; M. Mazarei, D.P. Puthoff, J.K. Hart, S.R. Rodermel, and T.J. Baum, unpublished data) in addition to tobacco (EREBPs; see above). They all share common features, such as being induced by biotic and abiotic stresses and mediating the expression of GCC box–containing genes such as PDF1.2. ERFs in Arabidopsis have been shown to be both activators and repressors of GCC box–mediated gene expression (Fujimoto et al., 2000). Interestingly, a novel JA- and elicitor-responsive element consisting of a GCC box–like element also is involved in the regulation of secondary metabolite biosynthetic genes in Catharanthus roseus (Menke et al., 1999; Memelink et al., 2001). The transcription factors that bind this element, the ORCAs, belong to the AP2 family (Menke et al., 1999; Memelink et al., 2001), suggesting a global role for the AP2/ERFs and related transcription factors in both ET and JA signaling pathways. Therefore, the elucidation of the role of ERFs in the defense response in plants is an emerging, important field of study.
In tomato, resistance to bacterial speck disease is governed by the Pto resistance gene, which encodes a Ser/Thr protein kinase (Martin et al., 1993). Previously, we identified three tomato ERF transcription factors, Pti4, Pti5, and Pti6, by virtue of their specific interaction with Pto kinase in a yeast two-hybrid screen (Zhou et al., 1997). Because Pti4/5/6 interact with Pto and also specifically bind the GCC-box cis element, a role for these genes in the activation of PR genes during the plant defense response was proposed (Zhou et al., 1997; Gu and Martin, 1998; Gu et al., 2000). The expression of Pti4/5/6 is enhanced in response to infection by Pseudomonas syringae pv tomato bacterial pathogens and by treatment with different signaling molecules (Thara et al., 1999; Gu et al., 2000). Furthermore, the Pto kinase phosphorylates Pti4 in vitro, and this phosphorylation enhances the GCC box binding activity of Pti4 (Gu et al., 2000). These observations suggested that Pti4, activated by the gene-for-gene avrPto–Pto interaction, may regulate the induction of defense-related genes that result in plant disease resistance.
In this study, we first characterized the localization and transactivation activity of Pti4/5/6. Then, we expressed the tomato ERFs in Arabidopsis to take advantage of the many mutants of defense signaling pathways in this species and its well-characterized plant–microbe interactions. Our results indicate that Pti4/5/6 play a direct role in mediating PR gene expression in vivo and that Pti4 activity, in particular, activates PR gene expression, resulting in enhanced defense against certain bacterial and fungal pathogens.
RESULTS
Pti4, Pti5, and Pti6 Are Localized to the Nucleus
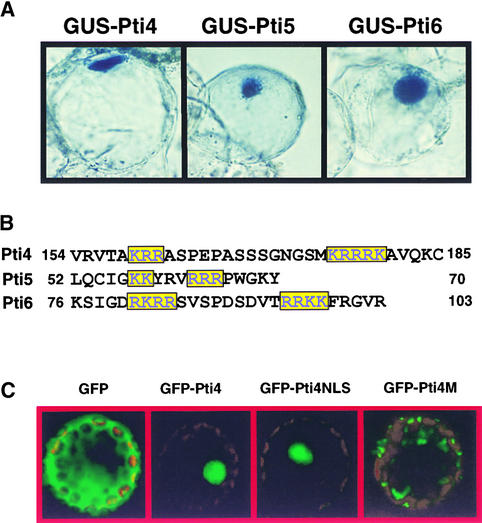
Inspection of the amino acid sequences of Pti4/5/6 revealed that each one contains typical nuclear localization sequences (NLSs) of the bipartite class, with two clusters of basic residues separated by 4 to 20 amino acids (Figure 1B) (Zhou et al., 1997; Gu et al., 2000). To examine the subcellular localization of Pti4/5/6, their coding regions were fused to the 3′ end of a β-glucuronidase (GUS) reporter gene and expressed under the control of the 35S promoter of Cauliflower mosaic virus (CaMV). Each GUS fusion construct was introduced by particle bombardment into W-38 tobacco suspension cells. Transient expression of Pti4/5/6::GUS fusions showed that GUS activity was localized to the nucleus of transformed tobacco cells (Figure 1A).
Figure 1.
Nuclear Localization of Pti4/5/6.
(A) Transient expression of GUS-Pti4/5/6 fusions in tobacco suspension cells. 35S::GUS-Pti4/5/6 constructs were introduced into W-38 tobacco suspension cells by particle bombardment. Localization of the fusion proteins was visualized after the addition of 5-bromo- 4-chloro-3-indolyl-β-d-glucuronide substrate to the cells.
(B) Putative bipartite nuclear localization signals of Pti4/5/6. The two basic clusters are colored and boxed. Residue numbers corresponding to the Pti4/5/6 amino acid sequences in GenBank are given for each peptide.
(C) Arabidopsis protoplasts were transfected with the indicated constructs, as described in the text. GFP fluorescence was visualized using a confocal microscope, as described in Methods.
Pti4 Nuclear Localization Does Not Require Pto
To determine if the nuclear localization of Pti4 is Pto dependent, a green fluorescent protein (GFP)–Pti4 fusion was developed, and the construct was transfected into protoplasts isolated from Arabidopsis ecotype Columbia (Col-0). We used Arabidopsis Col-0 because, unlike W-38 tobacco, Pto-like activity has not been identified in this ecotype (Thilmony et al., 1995; Y. Gu and G. Martin, unpublished data). Protoplasts transfected with a GFP control construct showed green fluorescence throughout the entire cytoplasm and nucleus. In contrast, the GFP-Pti4 fusion proteins were localized exclusively to the nucleus, indicating that Pti4 contains an NLS that is sufficient to target the fusion proteins into the nucleus (Figure 1C).
To further characterize the bipartite NLS domain of Pti4 (Figure 1B), a DNA segment encoding the 32 amino acids spanning the putative Pti4 NLS was fused to the GFP gene, and the construct was expressed transiently in Arabidopsis protoplasts. As shown in Figure 1C, the GFP-Pti4-NLS fusion was localized to the nucleus, indicating that this region spanning 32 amino acids functions as an NLS. To examine this functional domain further, the two clusters of the basic charged residues of the NLS from the full-length Pti4 sequence were deleted, and the resulting mutant (Pti4M) was fused to GFP. This deletion caused the loss of nuclear localization of Pti4, and the green fluorescence was visualized as patches in the protoplasts (Figure 1C). Therefore, this bipartite domain region is the sole NLS necessary to target Pti4 protein into the nucleus.
Pti4/5/6 Are Transcription Activators
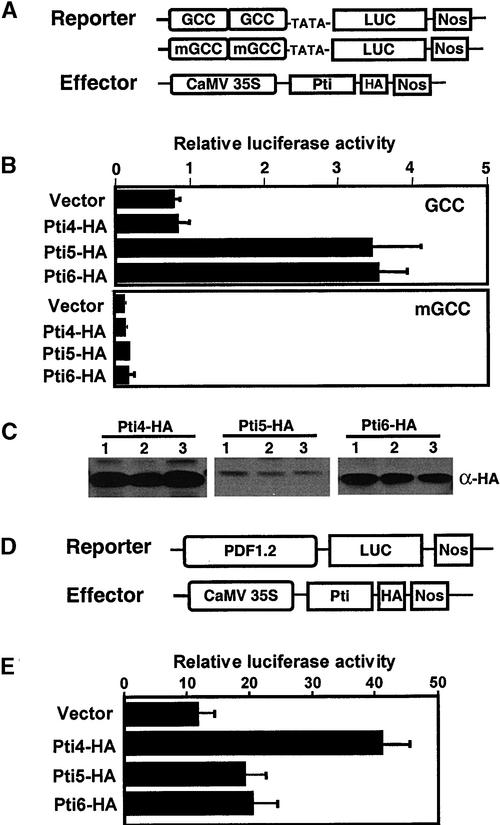
To determine if Pti4/5/6 can function as activators of GCC box–mediated transcription, Arabidopsis protoplasts were cotransfected with a GCC box–luciferase (LUC) construct and a vector expressing Pti4, Pti5, or Pti6 under the CaMV 35S promoter (Figure 2A). Compared with expression of the control, the expression of Pti5 and Pti6 increased the GCC box–mediated transcription by threefold and fourfold, respectively. To determine whether Pti5 or Pti6 activates the reporter gene via interaction with the GCC-box cis element, cotransfection of Pti5/6 with a mutated mGCC-LUC construct was performed; transactivation activity of Pti5/6 of this construct was not observed (Figure 2B). Surprisingly, cotransfection with Pti4 did not result in the activation of GCC box–mediated transcription. We also observed that the Pti4 protein has lower affinity in binding to a synthetic GCC box in vitro than Pti5 and Pti6 (Y. Gu, unpublished data).
Figure 2.
Transactivation of GCC Box–Mediated Transcription by Pti4/5/6.
(A) Schemes of the effector and reporter constructs used in the cotransfection experiments. The reporter constructs contain two copies of the GCC box or the mutated mGCC box in tandem that were fused upstream to the CaMV 35S minimal TATA promoter, the coding region from the LUC gene, and the nopaline synthase (NOS) terminator. The effector plasmids contain a CaMV 35S promoter fused to HA-tagged Pti4, Pti5, or Pti6 cDNA.
(B) Transactivation of the GCC-LUC reporter gene by Pti4/5/6. Arabidopsis protoplasts were cotransfected with a mixture of plasmids containing reporter, effector (empty vector with no insert was used as a control), and internal control constructs (35S-LUC). Dual luciferase activity was measured 20 hr after transfection of the protoplasts, as described in Methods. The data shown are derived from triplicate samples and three independent experiments.
(C) Expression of effector proteins in transfected Arabidopsis protoplasts. Twenty micrograms of total protein from transfected Arabidopsis protoplasts was separated by SDS-PAGE. The expression of effector proteins was detected by protein gel blot analysis using an anti-HA antibody (α-HA). Lanes 1, 2, and 3 contain protein samples from three independent transfections.
(D) Scheme of the PDF1.2 promoter–LUC construct. A 1.2-kb segment of the PDF1.2 promoter (Manners et al., 1998) was amplified by PCR from Arabidopsis genomic DNA and fused to the coding region of the LUC gene. The effector construct containing the Pti4 coding region is the same as in (A).
(E) Transactivation of the PDF1.2-LUC reporter gene by Pti4. Arabidopsis protoplasts were cotransfected with PDF1.2-LUC, 35S::Pti4-HA, and the internal control construct. Empty vector was used as a control for the effector plasmid. Dual luciferase activity in protoplasts was measured 20 hr after transfection. The data are derived from triplicate samples and three independent experiments.
We verified the expression of Pti4/5/6 proteins in Arabidopsis protoplasts by immunoblot analysis using an anti-hemagglutinin (HA) antibody (Figure 2C). Although the expression level varied for each effector protein, Pti4 was detected consistently in the protoplasts from each transfection. To further study the possible transactivation function of Pti4, the promoter of a known ET-regulated gene, PDF1.2 from Arabidopsis, was isolated. The PDF1.2 promoter, which contains a GCC box, was fused to a LUC reporter gene (Figure 2D). Cotransfection of this reporter construct with the Pti4 effector plasmids resulted in threefold enhanced transcription of the reporter gene (Figure 2E). However, neither Pti5 nor Pti6 significantly increased transcription mediated by the PDF1.2 promoter. Based on these results, we postulate that Pti4/5/6 bind GCC boxes differently depending on the flanking nucleotide sequences.
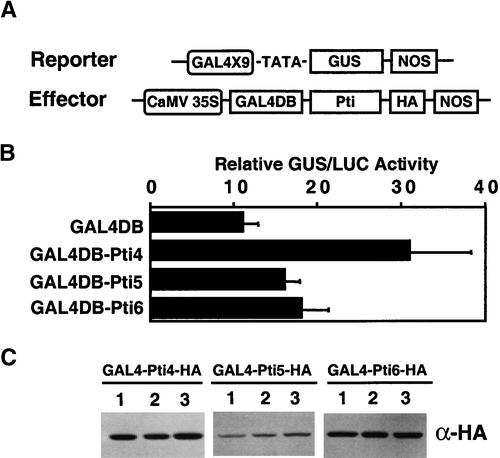
To analyze the transactivation function of Pti4/5/6 without the complications caused by the differential binding of the GCC box, the DNA binding domain of the yeast GAL4 protein (GAL4DB) was fused to the full-length Pti4/5/6 (Figure 3A). The reporter construct (UASGAL4-GUS) contained nine copies of the GAL4 upstream activation sequence fused to the GUS reporter gene. As shown in Figure 3B, GAL4DB-Pti4 gave a 3.4-fold increase in activation of the reporter gene over the control construct, and GAL4DB-Pti5 and GAL4DB-Pti6 increased the expression of the reporter gene by 1.7- and 1.8-fold, respectively.
Figure 3.
Transactivation of the GAL4X9-GUS Reporter Gene by GAL4DB-Pti4/5/6 Fusion Proteins.
(A) Schemes of reporter and effector constructs. The reporter construct contains nine copies of the GAL4 DNA binding site linked to a minimal CaMV 35S promoter, the GUS gene, and the NOS terminator. The effector constructs contain the CaMV 35S promoter fused to the GAL4 DNA binding domain (GAL4DB), Pti4/5/6-HA, and the NOS terminator.
(B) Transactivation of the GAL4X9-GUS reporter gene by GAL4DB-Pti4/5/6. Arabidopsis protoplasts were cotransfected with a mixture of plasmids containing GAL4X9-GUS, GAL4DB-Pti4/5/6-HA, and internal control constructs. The internal control plasmid used to normalize for transfection efficiency contains the CaMV 35S promoter fused to the LUC gene. Protoplasts were incubated for 20 hr after transfection. GUS and LUC activity in the protoplasts were determined according the method described by Sprenger-Haussels and Weisshaar (2000).
(C) Expression of GAL4DB-Pti4/5/6-HA fusion proteins. Twenty micrograms of total protein from transfected Arabidopsis protoplasts was separated by SDS-PAGE. The expression of the HA-tagged fusion proteins was detected by protein gel blot analysis using an anti-HA antibody (α-HA).
Expression of Pti4 in Arabidopsis Causes Phenotypic Changes Associated with the Response to ET Treatment
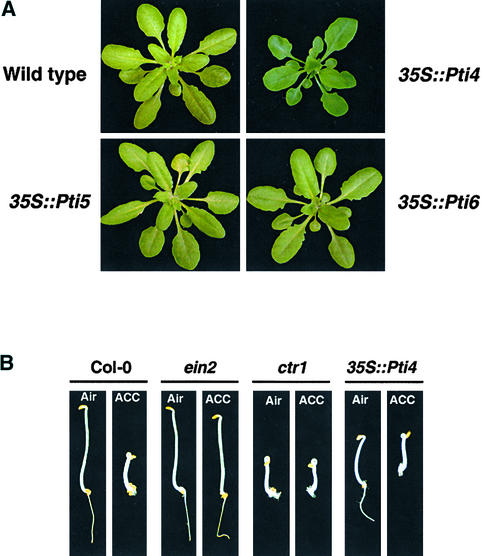
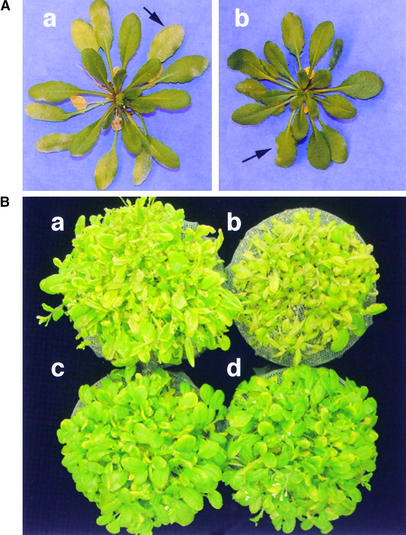
To analyze the role of Pti4/5/6 in the regulation of GCC-box PR gene expression, Arabidopsis transgenic plants were developed that constitutively express Pti4, Pti5, or Pti6 mRNAs from the 35S promoter. Approximately 30 individual transgenic plants were generated for each construct. RNA gel blot analysis was performed on primary transformants to determine the expression of the transgenes (data not shown). Homozygous lines (from the T3 generation) with a single insertion of the transgene were selected for each construct and used for further analysis. Of six independent homozygous lines expressing Pti4, plants from five lines displayed phenotypic changes. These plants were slightly smaller and darker green compared with wild-type plants (Figure 4A). Transgenic plants expressing Pti5 or Pti6 appeared no different than wild-type plants.
Figure 4.
Ectopic Expression of Pti4/5/6 in Arabidopsis and Triple-Response Assay of the Transgenic Plants.
(A) Phenotypes of Arabidopsis transgenic plants carrying the 35S::Pti4, 35S::Pti5, or 35S::Pti6 transgene. An untransformed wild-type Col-0 plant is shown for comparison.
(B) Overexpression of Pti4 caused constitutive activation of the ET response phenotype. Three-day-old seedlings overexpressing Pti4/5/6 were germinated on agar plates in the dark with or without 20 μM 1-aminocyclopropane-d-carboxylic acid (ACC). Untransformed wild type, ein2, and ctr1 mutants are shown as controls.
Because Pti4 is known to be involved in the ET signaling pathway (Gu et al., 2000), we used the triple-response assay (Solano et al., 1998) to determine whether the expression of Pti4/5/6 activated ET responses. Compared with control wild-type seedlings, etiolated Pti4-expressing seedlings in the absence of ET showed inhibition of hypocotyl elongation, a phenotype caused by ET treatment (Figure 4B). However, unlike the constitutive ET response mutant ctr, the Pti4-expressing seedlings did not display severe inhibition of root growth or exaggerated apical hook curvature. Therefore, the expression of Pti4 appears to activate a subset of ET responses. This partial seedling triple-response phenotype was not observed in transgenic plants expressing Pti5 or Pti6 (data not shown).
Expression of Pti4/5/6 in Arabidopsis Upregulates Different Sets of PR Genes
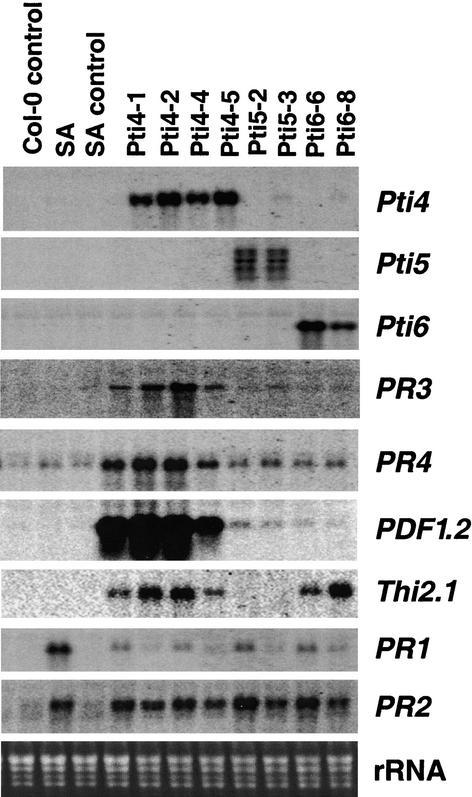
Our previous results suggested that Pti4/5/6 play a direct role in the regulation of tomato GCC-box PR genes (Thara et al., 1999; Gu et al., 2000). To determine whether Pti4/5/6 play a role in activating PR genes in Arabidopsis, the expression of different classes of PR genes was examined in transgenic plants expressing Pti4, Pti5, or Pti6. As shown in Figure 5, expression of Pti4 in Arabidopsis caused an increase in the steady state abundance of PR3, PR4, and PDF1.2 transcripts, which are known to be ET inducible and to contain GCC boxes in their promoters. The expression of PDF1.2 transcripts was induced 20- to 40-fold in the Pti4-expressing plants examined. Transgenic plants expressing Pti5 or Pti6 showed weak or no increase in the expression of these ET-regulated PR genes. This finding is consistent with the results described above that Pti4 (and not Pti5 or Pti6) significantly enhanced the transcription of the Arabidopsis PDF1.2 promoter (Figure 2E).
Figure 5.
Overexpression of Pti4/5/6 in Arabidopsis Causes Constitutive Upregulation of PR Genes.
Two to four individual transgenic lines (homozygous, from the T3 generation) were chosen for analysis for each construct expressing Pti4/5/6. Total RNA was isolated from leaves of 4-week-old Arabidopsis plants. Duplicated RNA gel blots were hybridized with the probes indicated. Equal loading was verified by visualizing rRNA on a gel stained with ethidium bromide.
JA regulates the expression of a subset of PR genes, some of which also are ET inducible, such as PDF1.2 (Thomma et al., 1998; Schenk et al., 2000). The effect of the expression of Pti4/5/6 on the expression of Thi2.1, which encodes the potent fungal defense protein thionin, was examined (Epple et al., 1997; Bohlmann et al., 1998). Thi2.1 is induced by JA but not by ET (Epple et al., 1995) and requires a functional JA signal transduction pathway (Bohlmann et al., 1998; Xie et al., 1998). The transcript abundance of Thi2.1 was increased in Pti4- and Pti6-expressing plants but not in Pti5-expressing plants (Figure 5).
The transcript abundance of two known SA-regulated PR genes, PR1 and PR2, in transgenic plants also was assessed. Expression of Pti4/5/6 in Arabidopsis increased the expression of the PR2 gene substantially, with the transcript abundance being similar to that in plants treated with SA (Figure 5). The abundance of PR1 transcripts was induced only minimally in most of the transgenic plants compared with wild-type plants.
Pti4/5/6 Enhance the Expression of SA-Induced PR Genes, and Pti4 May Mediate SA Antagonism of ET-Regulated PR Genes
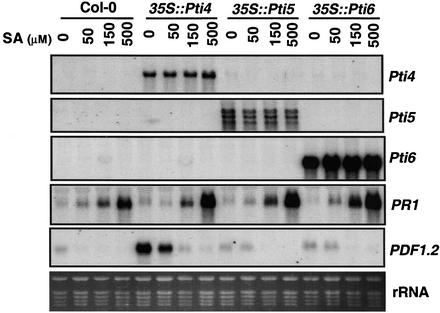
To further study the effect of Pti4/5/6 on the expression of SA-regulated PR genes, RNA isolated from plants treated with SA was hybridized with the PR1 gene probe. An increase in the level of expression of PR1 was observed in Pti4/5/6 transgenic plants compared with SA treated wild-type plants, and this increase became clear at higher (150 and 500 μM) concentrations of SA (Figure 6). PR1 transcripts were approximately threefold more abundant in Pti4/5/6 plants compared with wild-type plants when treated with 500 μM SA.
Figure 6.
Overexpression of Pti4/5/6 Sensitized the SA Signaling Pathway, and SA Suppressed the Expression of PDF1.2 Transcripts in the Pti4-Overexpressing Line.
Three-week-old seedlings of wild-type plants or Pti4/5/6-overexpressing lines were treated with different concentrations of SA as indicated for 16 hr. Total RNA was extracted from treated leaf tissues, and duplicated blots were hybridized with the probes indicated. Equal loading was verified by visualizing rRNA on a gel stained with ethidium bromide.
We reported previously that SA suppresses the ET induction of GCC box–containing PR genes in tomato (Gu et al., 2000). To further elucidate this suppression mechanism, Arabidopsis plants expressing Pti4, Pti5, or Pti6 were treated with different concentrations of SA. As shown in Figure 6, the abundance of PDF1.2 transcripts decreased in Pti4-expressing plants upon SA treatment. This suppression was observed even at a low concentration of SA (50 μM), whereas at a high SA concentration (500 μM), the accumulation of PDF1.2 transcripts was abolished completely. In Pti5, Pti6, and wild-type plants, the low levels of PDF1.2 transcripts present also were suppressed by treatment with higher concentrations of SA.
Transcriptional Regulation of PDF1.2 by Overexpression of Pti4 Is Not Affected by the jar1 or ein2 Mutation
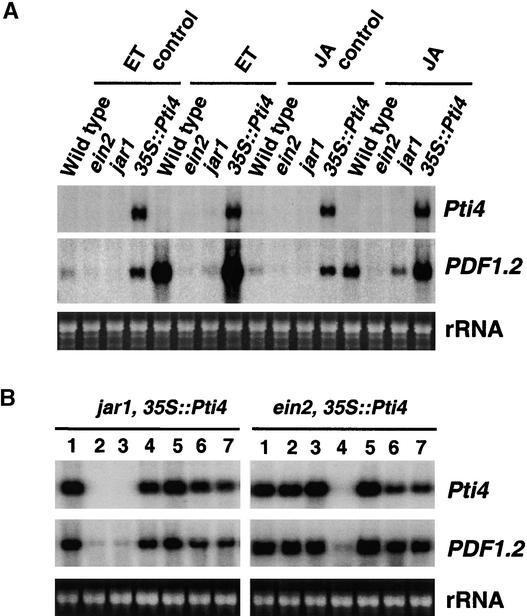
In Arabidopsis, the expression of the PDF1.2 gene is dependent on functional ET and JA signaling pathways (Penninckx et al., 1998). As shown in Figure 5, the expression of Pti4 caused an increase of PDF1.2 transcripts. To study the role of Pti4 in the ET/JA induction of defense gene expression, we treated wild-type plants, the ET-insensitive mutant ein2, the JA-insensitive mutant jar1, and Pti4-expressing plants with ET and JA. As shown in Figure 7A, both ET and JA induced the expression of PDF1.2 in wild-type plants, and ET failed to induce the expression of PDF1.2 in ein2 or jar1 mutant plants. JA did not induce the expression of the PDF1.2 gene in the ein2 mutant but weakly induced its expression in the jar1 mutant. Plants expressing Pti4 showed an additional marked increase of PDF1.2 transcripts when treated with either ET or JA compared with wild-type control plants.
Figure 7.
Activation of PDF1.2 by the Overexpression of Pti4 Is Independent of the ET and JA Signaling Pathways.
(A) Induction of PDF1.2 by JA and ET in Arabidopsis wild-type plants, jar1 and ein2 mutants, and Pti4-expressing plants. Treatment of plants with ET and JA was performed as described in Methods. Total RNA was extracted from treated leaf tissues, and the RNA gel blot was hybridized with a PDF1.2 probe. Equal loading was verified by visualizing the rRNA on a gel stained with ethidium bromide.
(B) Activation of PDF1.2 by overexpression of Pti4 in the jar1 and ein2 backgrounds. Total RNAs were extracted from seven putative T1 transgenic lines for each mutant background. Duplicated blots were hybridized with the Pti4 and PDF1.2 probes. Equal loading was verified by visualizing the rRNA on a gel stained with ethidium bromide.
To further elucidate the transcriptional activation mechanism of PDF1.2 by Pti4, Arabidopsis plants that express Pti4 in ein2 and jar1 mutant backgrounds were generated. As shown in Figure 7B, of seven primary transformants in each mutant background, five plants of ein2 and six plants of jar1 expressed the Pti4 transgene, and these transformants also accumulated PDF1.2 transcripts constitutively. Homozygous T3 progeny overexpressing Pti4 were derived from two primary transformants in each mutant background (i.e., lines jar1-1/35S::Pti4 and jar1-4/35S::Pti4 and lines ein2-2/35S:: Pti4 and ein2-5/35S::Pti4). The T3 homozygous lines also showed constitutive expression of the PDF1.2 gene (data not shown).
Arabidopsis Plants Expressing Pti4 Are Resistant to Erysiphe orontii and Show Increased Tolerance to Pseudomonas
As shown in Figure 5, the expression of Pti4/5/6 in Arabidopsis led to the constitutive expression of several PR genes. This raised the possibility that defense responses are activated in these plants and that resistance to certain pathogens might be increased. The Pti4/5/6 plants were first tested with Erysiphe orontii, a biotrophic fungal pathogen (recently renamed Golovinomyces orontii; Braun, 1999) that infects wild-type Arabidopsis Col-0 and typically does not induce the ET/JA-dependent PR genes PDF1.2 and Thi2.1 (Plotnikova et al., 1998). As shown in Figure 8A and Table 1, when two independent lines of Arabidopsis expressing Pti4 were infected with Erysiphe, greatly enhanced resistance was observed in the inoculated leaves at 14 days after inoculation. In contrast, in wild-type plants, Erysiphe growth was observed as powdery mildew covering 50% or more of the infected leaf area. The stronger resistance observed in line Pti4-2 compared with Pti4-5 is consistent with the greater abundance of PR3, PR4, PDF1.2, and Thi2.1 transcripts in the former line. Transgenic plants expressing Pti5 or Pti6 did not show enhanced resistance against the Erysiphe pathogen compared with wild-type plants (Table 1).
Figure 8.
Expression of Pti4 in Arabidopsis Confers Increased Resistance to Erysiphe and Tolerance to Pseudomonas.
(A) Increased resistance to the biotrophic fungal pathogen Erysiphe. Representative wild-type Arabidopsis Col-0 (a) and Pti4-2 (b) transgenic plants from the same inoculation box are shown at 14 days after inoculation with Erysiphe. Arrows highlight areas of powdery mildew infection. Three boxes each containing nine transgenic plants and three wild-type plants were assessed for each transgenic line. Experiments on each line were repeated once with similar results. Average Erysiphe disease scores for wild-type and Pti4-2 transgenic plants were +3 and +1, respectively, based on the scoring system of Reuber et al. (1998) as follows: 0, no growth; +1, isolated spots of infection; +2, ∼20% coverage of leaves; +3, ∼50% coverage of leaves; and +4, nearly 100% coverage of leaves.
(B) Increased tolerance to infection by Pseudomonas strain DC3000. Four-week-old plants were inoculated by dipping them into a suspension of virulent Pseudomonas strain DC3000 (106 colony-forming units/mL). Four days after inoculation, differences in the development of disease symptoms on the plants were observed. Arabidopsis wild-type Col-0 (a) and NahG plants (b) showed extensive and complete chlorosis, respectively, whereas Pti4-2 (c) and Pti4-5 (d) plants showed mild chlorosis.
Table 1.
Quantitative Analysis of Plant Responses to Erysiphe
| No. of Plants with Each Disease Score
|
|||||
|---|---|---|---|---|---|
| Arabidopsis Line | 0.0 | 1.0 | 2.0 | 3.0 | 4.0 |
| Pti4-2 | 27 | ||||
| Wild type | 9 | ||||
| Pti4-5 | 9 | 14 | |||
| Wild type | 9 | ||||
| Pti5-2 | 27 | ||||
| Wild type | 9 | ||||
| Pti6-6 | 27 | ||||
| Wild type | 9 | ||||
Wild-type Arabidopsis and Arabidopsis-Pt4/5/6 plants grown for 4.5 weeks were infected with an inoculum of Erysiphe using a settling tower. Three inoculation boxes were used for each line to account for variability in the inoculum within and between boxes. Each box contained three wild-type and nine transgenic plants. Plants were scored at 14 days after inoculation using the scale described in the legend to Figure 8A. Experiments with each line were repeated once with similar results.
Pti4/5/6 plants also were inoculated with a bacterial pathogen, Pseudomonas strain DC3000, that is known to induce both the SA-dependent genes PR1 and PR2 and the ET/JA-dependent PR genes PDF1.2 and Thi2.1. Pseudomonas strain DC3000 is a virulent pathogen that causes lesions and chlorosis on many Arabidopsis ecotypes, including Col-0. In three independent experiments, bacterial growth in leaves of Pti4-2 and Pti4-5 plants was not significantly different from that in wild-type plants 4 days after infection (data not shown). However, leaves of both independent transgenic lines showed markedly less chlorosis compared with wild-type plants (Figure 8B). NahG plants also were infected to serve as another susceptible control to evaluate response to the pathogen. These plants showed the most serious disease symptoms among all of the plants studied (Figure 8B). No enhanced tolerance to Pseudomonas-induced chlorosis was observed for the Pti5 and Pti6 overexpressers (data not shown).
DISCUSSION
In an effort to identify signaling components of the Pto disease resistance pathway, we previously discovered three transcription factors, Pti4/5/6, which interact physically with Pto kinase and bind the GCC-box cis element present in the promoters of many PR genes. The discovery of Pti4/5/6 established a direct molecular link between pathogen recognition and the activation of PR gene expression involved in host defense responses (Zhou et al., 1997; Gu and Martin, 1998). In this study, we have demonstrated an in vivo function for Pti4/5/6 in defense by expressing them in Arabidopsis plants. We found that Pti4/5/6 mediate the expression of both SA- and ET/JA-regulated PR genes and that Pti4 may play a role in the communication between these pathways. Pti4 expression also leads to enhanced resistance to a fungal pathogen and to increased tolerance to a bacterial pathogen. This is an example of the expression of ERF genes in a heterologous background, and it suggests that these genes might be useful generally in engineering diverse plant species for increased disease resistance.
Nuclear Localization and Transcription Activation by Pti4/5/6
As expected for transcription factors, Pti4/5/6 were localized to the nucleus in both tobacco and Arabidopsis cells. Pti4/5/6 all contain typical bipartite NLSs, and this NLS is sufficient to target Pti4 to the nucleus. This finding raises two interesting questions. (1) Is the phosphorylation of Pti4 (by Pto in tomato or by a functional Pto homolog in Arabidopsis) required for the nuclear localization of Pti4? (2) Where does the physical interaction between Pto (or a homolog) and Pti4 occur? Although experiments in both tobacco and Arabidopsis protoplasts indicate that Pto itself is not required for Pti4 nuclear localization, we cannot exclude a role for phosphorylation in Pti4 localization.
Tobacco is known to express a Pto-like activity that is effective in recognizing AvrPto (Thilmony et al., 1995), and this activity may play a role in Pti4 localization in this species. This possibility is consistent with the observation that PR genes often are expressed more rapidly and to a higher degree during incompatible plant–pathogen interactions involving specific resistance genes and their cognate avirulence proteins (Voisey and Slusarenko, 1989; Jia and Martin, 1999). As for the physical interaction between Pti4 and Pto (or a related kinase in Arabidopsis), this could occur either in the cytoplasm or in the nucleus. The ethylene-responsive MAP (ERM) kinase in parsley provides one precedent for the latter possibility. This kinase is activated upon recognition of an elicitor, leading to its translocation into the nucleus, where it interacts with transcription factors that induce the expression of defense genes (Ligterink et al., 1997). At present, we are using GFP fusions and cellular fractionation studies to investigate the subcellular localization of Pto kinase.
By using Arabidopsis protoplasts and reporter constructs carrying a GCC box or a GAL4 DNA binding sequence, transactivation activity for Pti4/5/6 was demonstrated. As with nuclear localization, it is possible that the phosphorylation of Pti4 by Pto (in tomato) or by a functionally analogous kinase (in Arabidopsis and tobacco) facilitates its transactivation activity. A similar mechanism is known for other transcription factors (Hunter and Karin, 1992). In fact, we have shown previously that specific phosphorylation of Pti4 by Pto kinase enhances its DNA binding activity in vitro (Gu et al., 2000). We are mapping the phosphorylation sites of Pti4 to further investigate the role of phosphorylation activity in Pti4 activity.
Activation of ET/JA-Regulated Genes by Pti4/5/6
The expression of Pti4 in Arabidopsis caused the activation of ET-regulated PR genes, such as PDF1.2, and phenotypic changes associated with the plant response to ET, suggesting that Pti4 can play a role in regulating the expression of genes in the ET signaling pathway. It was observed that Pti4 causes an additive increase of PDF1.2 expression in transgenic plants exposed to ET and JA compared with wild-type plants. We also observed that the expression of Pti4 in ein2 and jar1 mutant plants still led to the activation of PDF1.2. These results suggest that Pti4 acts either independent of or downstream of the EIN2 and JAR1 genes.
Pti4 is known to play a role in tomato in regulating the expression of GCC-box PR genes in defense responses against Pseudomonas (Thara et al., 1999; Gu et al., 2000). Such regulatory functions for Pti4 appear not to require the ET signaling pathway (Thara et al., 1999). Taking into consideration the observations in the current study, we propose that the disease resistance and ET signaling pathways converge at transcription factors such as Pti4. Available experimental evidence on the function of different Arabidopsis ERFs (Solano et al., 1998; Fujimoto et al., 2000) does not indicate clearly which gene might be the functional homolog of Pti4. Moreover, the possible role of Arabidopsis ERFs in defense response pathways has yet to be reported.
The expression of Pti5 or Pti6 did not cause strong constitutive expression of ET/JA-regulated PR genes in Arabidopsis. However, both Pti5 and Pti6 transactivated GCC box–mediated transcription in transient assays conducted with Arabidopsis protoplasts. It is possible that the synthetic GCC-box cis element used in our transient assay is present in a different nucleotide context than those in the promoters of most ET-regulated PR genes in Arabidopsis. Upon examination of the promoter sequence of the PDF1.2 gene, it was found that the nucleotides flanking the GCCGCC sequence in this promoter are not the same as those in the synthetic GCC box (Y. Gu and G. Martin, unpublished data). It is well known that flanking nucleotides can contribute strongly to the binding affinity of transcription factors to their respective target sequences and can serve to discriminate among closely related factors. For example, several Arabidopsis ERFs (AtERFs) have been shown to have distinct DNA binding preferences (Fujimoto et al., 2000). Therefore, Pti5 and Pti6 may upregulate other, as yet unidentified, GCC box–containing defense-related genes.
Activation of SA-Regulated Genes by Pti4/5/6
The expression of Pti4/5/6 in Arabidopsis enhanced the expression level of the SA-regulated genes PR1 and PR2. In addition, upon SA treatment, the transcript levels of PR1 were induced to higher levels in Pti4/5/6 plants than in wild-type SA-treated plants. Although Pti4/5/6 are not known to bind cis elements of SA-regulated PR genes directly, they may act indirectly by interacting with protein factors that are involved in SA-regulated PR gene expression. In animals, cross-coupling of transcription factors is known to play an important role in mediating responses to various signaling events (Schule and Evans, 1991). Interestingly, AtEBP, an AtERF transcription factor, was identified because of its interaction with OBF4, the ocs element binding factor that belongs to the class of bZIP proteins that includes Arabidopsis TGA transcription factors (Büttner and Singh, 1997). Several TGA transcription factors have been shown to bind specifically to the SA-responsive elements in the promoters of PR1 genes (Zhang et al., 1999; Després et al., 2000). Thus, it is possible that TGA transcription factors interact with Pti4/5/6 directly or indirectly and thereby enhance SA-regulated PR gene expression.
We found that with increasing concentrations of SA, the increase in PR1 expression was accompanied by a decrease in PDF1.2 transcripts. This effect was most noticeable in Pti4 plants, in which PDF1.2 transcripts were most abundant. There are two possible explanations for this result. First, SA may act independently of Pti4 to suppress PDF1.2 expression. There are previous reports of SA-mediated suppression of PDF1.2 expression. For example, in Arabidopsis, the expression of the PDF1.2 gene is higher in NahG plants in which a bacterial SA-degrading enzyme is overexpressed (Penninckx et al., 1998). We have demonstrated previously that SA also suppresses the ET induction of GCC-box PR gene expression in tomato (Gu et al., 2000). A second possibility, as discussed below (see model), is that SA might play a role in inhibiting Pti4 activity toward the PDF1.2 promoter.
Pti4 Enhances Host Responses to Pathogens
Plants expressing Pti4 supported bacterial growth comparable to wild-type plants but showed significantly less chlorosis. Such decreased symptom development after bacterial infection is referred to as tolerance and has been observed previously in Arabidopsis disease signaling mutants and certain ecotypes (Bent et al., 1992; Buell and Somerville, 1995). Chlorosis caused by Pseudomonas is attributable primarily to the bacterial toxin coronatine, because strains unable to produce this toxin cause much decreased disease symptoms (Mittal and Davis, 1995). There is evidence that both the Arabidopsis EIN2 protein and the coronatine-insensitive COI1 protein are involved in the development of bacterial disease symptoms (Bent et al., 1992; Feys et al., 1994). The increased tolerance in Pti4 plants, therefore, may reflect interference in the activity of EIN2 or COI1 or another component of the coronatine perception response. Increases in bacterial resistance have been found in plants that overexpress other ERF genes, including Pti5 and a close tobacco homolog of Pti6 (He et al., 2001; Park et al., 2001). No studies have been reported in which multiple ERF-like genes are overexpressed together in a single plant, although this would appear to be a reasonable strategy to increase levels of resistance even more.
Plants expressing Pti4 showed significant resistance to Erysiphe, a fungal biotroph that is virulent on Arabidopsis Col-0. Infection of Arabidopsis by Erysiphe typically results in the induction of the SA-dependent PR genes PR1, PR2, PR5, and GST1 (Reuber et al., 1998). Interestingly, a few studies also show that genes belonging to the ET/JA pathway may contribute to resistance against Erysiphe (Dewdney et al., 2000; Ellis and Turner, 2001). For example, the Arabidopsis cev1 mutant, which appears to have constitutively active ET and JA signaling pathways and which constitutively expresses PDF1.2 and Thi2.1, exhibits increased resistance to Erysiphe and other powdery mildews (Ellis and Turner, 2001). The observation that the Pti4-expressing Arabidopsis lines displayed enhanced resistance to Erysiphe suggests that there is an additive effect of defense genes belonging to both the SA and ET/JA signaling pathways.
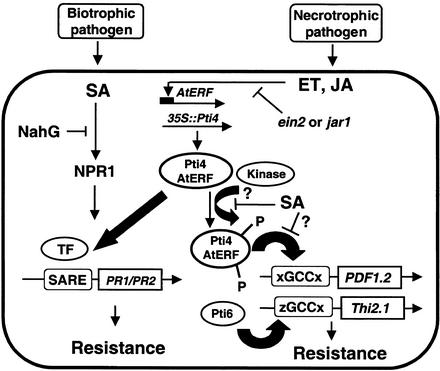
A Model for the Role of Pti4 in PR Gene Expression
Our current and previous results suggest the following model for the role of Pti4/5/6 (and possibly the functionally analogous AtERFs) in defense response (Figure 9). First, pathogen attack and/or the associated increase in ET activates the expression of the Pti4 gene (or the AtERF genes) (Fujimoto et al., 2000; Gu et al., 2000) (Figures 3 and 6). In the Arabidopsis Pti4 line, this expression is constitutive, so the level of Pti4 protein already is increased and induction by pathogen or ET is not needed. When Pti4 (or AtERF) becomes available, Pto kinase (or an analogous Arabidopsis kinase) phosphorylates the protein, which might facilitate its localization, DNA binding, and/or interaction with other transcription factors. The putative kinase phosphorylating Pti4 likely is not dependent on ET or JA, because the Pti4-mediated activation of PDF1.2 is not affected in the ein2 and jar1 mutants. The specificity of Pti4/5/6 promoter binding may be conferred by nucleotides that flank the GCC box. This context-specific DNA binding would account for the different subsets of PR genes that are regulated by Pti4/5/6 and might provide the plant with additional control of the defense responses it deploys.
Figure 9.
Model for the Proposed Role of Pti4 and AtERFs in Mediating Cross-Talk between the SA and ET/JA Pathways.
NahG refers to the salicylate hydroxylase protein that degrades SA. NPR1 refers to the “non-expresser of PR” protein. ein2 and jar1 refer to mutations in Arabidopsis that affect ET perception (ein2) and JA biosynthesis ( jar1). Also shown is a hypothetical transcription factor (TF) that might interact with the Pti4/5/6 or AtERF proteins and play a role in binding an SA-responsive element (SARE). The SA-inhibited protein kinase refers to either Pto (in tomato) or an analogous kinase in Arabidopsis. The possibility that Pti4/AtERF and Pti6 differentially recognize the GCC box when it is flanked by different nucleotides is shown as xGCCx and zGCCx. The possible roles of other signaling components shown are discussed in the text.
The stimulation of PR1 gene expression by SA is accompanied by a concomitant inhibition of expression of PDF1.2. As mentioned above, these changes might be independent of Pti4. However, based on the observed involvement of Pti4 in the expression of both of these genes, it is possible that Pti4 plays a role in the communication between the SA and ET signaling pathways. If that is the case, it seems unlikely that SA affects Pti4 activity directly; rather, it might play an indirect role (e.g., by attenuating Pti4 phosphorylation). What are the possible mechanisms of this attenuation? One possibility is that unphosphorylated Pti4 plays a role in SA-regulated PR gene expression, whereas phosphorylated Pti4 is more effective in the activation of JA/ET-regulated PR gene expression. This notion is supported by our observation that the phosphorylation of Pti4 enhances its binding to the GCC box in vitro. SA then might inhibit a protein kinase that phosphorylates Pti4, which could act as a “switch” to divert Pti4 toward the activation of SA-regulated PR gene expression (e.g., by interaction with TGA factors, as discussed above).
In this model, lower levels of SA would not inhibit this kinase; instead, they would allow Pti4 to exist in both the phosphorylated and unphosphorylated forms, leading to the activation of both SA- and JA/ET-regulated gene expression, as seen in the Pti4-expressing line. A possible candidate for a protein kinase fulfilling this role is the recently described MPK4. The loss of MPK4 activity in an mpk4 mutant leads to the constitutive activation of SA-regulated PR genes and the inability to induce PDF1.2 and Thi2.1 (Petersen et al., 2000). We are testing this model at present by determining the phosphorylation state of Pti4 in plant cells with and without the application of SA and by examining whether the activity of these transcription factors is affected in the mpk4 mutant.
In conclusion, the tomato ERFs Pti4, Pti5, and Pti6 each plays a distinct role in the activation of defense responses in tomato and in Arabidopsis (Zhou et al., 1997; Thara et al., 1999; Gu et al., 2000). This study demonstrates the expression of ERFs in a heterologous background and indicates that ERFs can play a role in the expression of SA-regulated genes. Based on these results and our previous report (Gu et al., 2000), we propose that the phosphorylation of Pti4/5/6 proteins may facilitate their nuclear localization and/or transactivation properties for GCC-box promoters. We also hypothesize that Pti4 may play a role in mediating the communication between the SA and ET/JA signaling pathways and that the phosphorylation of Pti4 might act as a switch for this communication. Finally, the demonstration that Pti4 expression in Arabidopsis confers enhanced resistance to Erysiphe and tolerance to Pseudomonas suggests that ERFs from diverse species might be useful for engineering increased disease resistance in plants.
METHODS
Plant Materials, Growth Conditions, and Treatments with Salicylic Acid, Jasmonic Acid, and Ethylene
Arabidopsis thaliana (ecotype Columbia) plants were grown at 22°C with a daylength of 16 hr. For salicylic acid treatments, 4-week-old plants were sprayed with different concentrations of salicylic acid in water, as indicated in Results. Water was used as a control. Jasmonic acid treatment was performed by spraying the plants with 50 μM jasmonic acid dissolved in 0.01% ethanol (control was 0.01% ethanol alone). The control and treated plants were placed in a sealed plexiglass chamber for 24 hr before leaf tissue was harvested. Treatment of plants with ethylene was performed in a gas-tight plexiglass chamber by injecting a volume of ethylene gas to give a final concentration of 20 μL/L. Control plants were handled in an identical manner but without the injection of ethylene. Leaf tissues were harvested 24 hr after treatment.
Plasmid Constructions
All of the plasmid constructs generated in this study were made using standard recombinant DNA techniques and verified by DNA sequencing.
Constructs Used in the Nuclear Localization Assay
The coding regions of Pti4, Pti5, and Pti6 were amplified by polymerase chain reaction (PCR) to introduce BglII at the 5′ end and BamHI at the 3′ end. The resulting fragments were digested with BglII and BamHI and subcloned into the expression vector pRTL2-GUS-NIa (Restrepo et al., 1990) by replacing the NIa sequence to yield the in-frame fusion plasmids pRTL2-GUS-Pti4, pRTL2-GUS-Pti5, and pRTL2-GUS-Pti6. To generate green fluorescent protein (GFP) and Pti4 fusions, the coding region of GFP was amplified by PCR to introduce a NcoI site at the 5′ end and a BglII site at the 3′ end. The PCR products were digested with NcoI and BglII and subcloned into pRTL2-GUS-Pti4 plasmid to replace the β-glucuronidase (GUS) coding region. The resulting plasmid, pRTL2-GFP-Pti4, was engineered further to delete the two clusters of basic residues in the nuclear localization sequence domain of Pti4 (Figure 1) using the Quick Exchange Kit (Stratagene) to generate the pRTL2-GFP-Pti4M construct.
Constructs Used in the Protoplast Transient Assay
The reporter construct (GCC-LUC) contains two GCC-box repeats that were placed upstream of the minimal −42 to +8 TATA box from the 35S promoter of Cauliflower mosaic virus (CaMV) and then joined as a transcriptional fusion to the coding region of the firefly luciferase gene (LUC). The construct (mGCC-LUC) with the replacement of the GCC box by a mutated GCC box (mGCC box) was used as a control. To generate the effector constructs, the coding regions of Pti4, Pti5, and Pti6 were amplified by PCR, tagged with the double hemagglutinin (HA) epitope, and inserted into a plant expression vector containing the CaMV 35S promoter and the nopaline synthase terminator (Kovtun et al., 2000). For the reporter construct with GAL4 cis elements (GAL4-GUS), the sequence containing nine tandem repeats of the 17-mer yeast GAL4 DNA binding site (Ma and Ptashne, 1988) was placed upstream of the CaMV minimal 35S promoter and then fused to the coding region of the GUS gene. The corresponding effector constructs contain the GAL4 DNA binding domain (amino acids 1 to 94) fused in frame to Pti4-HA, Pti5-HA, or Pti6-HA, and expression of these fusion genes was driven by the 35S promoter.
Constructs Used in the Generation of Transgenic Arabidopsis
The coding regions of Pti4, Pti5, and Pti6 were amplified by PCR to introduce a BamHI site at both the 5′ and 3′ ends. The fragments were digested with BamHI and ligated into BamHI-digested pBTEX binary vector (Frederick et al., 1998) to yield plasmid constructs pBTEX35S-Pti4, pBTEX35S-Pti5, and pBTEX35S-Pti6.
Subcellular Localization of Pti4/5/6
Particle bombardment was performed using a Bio-Rad Biolistic PDS1000/He system to transiently express the GUS constructs in tobacco W-38 suspension cells. Plasmid DNA (0.66 μg) was coated onto tungsten particles as described by Varagona et al. (1992). DNA-coated particles were bombarded at 1100 p.s.i. into 200 mg of W-38 suspension cells laid on filter paper at a target distance of 9 cm. After bombardment, the cells were incubated in Murashige and Skoog (1962) medium containing 0.5 mg/L 2,4-D, 0.5 mg/L kinetin, and 0.3 mg/L indoleacetic acid for 24 hr at 25°C in the light. GUS activity was determined by histochemical staining. Cells were viewed with a light microscope, and micrographs taken 2 to 4 hr after the addition of substrate.
Subcellular localization of GFP fusions was performed by transiently expressing the GFP constructs in Arabidopsis protoplasts as described below and monitoring the localization of GFP with a confocal laser scanning microscope (Bio-Rad MRC-600). Excitation light at 488 and 514 nm was attenuated to 10% transmittance. Detectors were set at 610 nm for chlorophyll and 530 nm for GFP fluorescence. Serial confocal sections (2 μm thick) were collected. Images were exported as TIFF files and processed for printing using Adobe Photoshop (Mountain View, CA).
Arabidopsis Protoplast Transient Expression and Reporter Gene Activity Assay
Isolation and transfection of Arabidopsis protoplasts were performed according to a modified polyethylene glycol method as described by Abel and Theologis (1994). Typically, in a cotransfection assay, 5 × 105 protoplasts in 200 μL were transfected with 16 μg of effector plasmids, 8 μg of reporter plasmids, and 2 μg of internal control plasmids. The transfected protoplasts were incubated at 22°C for 16 to 20 hr, harvested by centrifugation at 80g for 3 min, and then quickly frozen and stored at −80°C. For reporter gene activity assays, either the protoplasts were lysed in passive lysis buffer (Promega) and luciferase activity was measured using a dual-luciferase assay kit according to the manufacturer's instructions (Promega), or proteins were extracted in extraction buffer (100 mM potassium phosphate and 1 mM DTT, pH 7.5) and GUS and LUC activity were determined as described by Sprenger-Haussels and Weisshaar (2000).
Arabidopsis Transformation
The plasmids pBTEX35S::Pti4, pBTEX35S::Pti5, and pBTEX35S:: Pti6 were introduced into Agrobacterium tumefaciens strain GV3101 and used to transform Arabidopsis (ecotype Columbia) using an in planta transformation method (Bechtold et al., 1993). Putative transformants (T1 plants) were selected by plating seed on Murashige and Skoog (1962) medium containing 50 mg/L kanamycin. After selection for 2 weeks, kanamycin-resistant seedlings were transferred to soil. Homozygous lines for the transgenes were identified in the T3 generation by segregation for kanamycin resistance and confirmed by DNA gel blot analysis.
SDS-PAGE and Immunoblotting Assay
Transfected protoplasts were harvested as described above, and total proteins were extracted by adding 200 μL of 1 × SDS sample buffer. Protein electrophoresis and transfer to polyvinylidene difluoride membranes were described previously (Frederick et al., 1998). For the immunoblotting assay, the blots were incubated overnight with anti-HA antibody at a concentration of 0.1 μg/mL, and proteins were visualized using an enhanced chemiluminescence kit (Amersham).
RNA Extraction and RNA Gel Blot Analysis
Total RNA was isolated according to the method described previously (Gu et al., 2000), separated by electrophoresis on formaldehyde-agarose gels, and transferred onto nylon membranes (Hybond N+; Amersham). 32P labeling of cDNA probes was performed using a random hexamer labeling kit (Ambion, Austin, TX). The procedure for RNA gel blot hybridization has been described previously (Gu et al., 2000). Radioactivity was detected by either autoradiography or phosphorimaging (for the qualitative assays).
Bacterial and Fungal Infection
For the infection of plants with the fungal pathogen Erysiphe orontii (recently renamed Golovinomyces orontii; Braun, 1999), Arabidopsis plants were grown in Metro-Mix 200 (Scotts-Sierra Horticultural Products, Marysville, OH) under a 12-hr light/dark cycle in a greenhouse with supplemental fluorescent lighting (19 ± 2°C). Four- to 4.5-week-old plants were infected with a moderately heavy inoculum (conidia from two infected leaves) of Erysiphe using a settling tower and scored for disease symptoms at 14 days after infection as described by Reuber et al. (1998). Transgenic plants were compared with wild-type Columbia plants in the same box. Three boxes, each containing nine transgenic plants and three wild-type plants, were assessed per transgenic line. The experiment was repeated with similar results.
For bacterial inoculation, plants were grown in a light room under a 16-hr photoperiod at 22°C. Four-week-old plants were dipped for 30 sec in a suspension of virulent Pseudomonas syringae pv tomato strain DC3000 (106 colony-forming units/mL). Leaves were photographed 4 days after inoculation.
Acknowledgments
We are grateful to Dr. Jen Sheen for providing us with the expression vectors for the protoplast transient assay and to Dr. Ray Bressan for help with Arabidopsis transformation. This research was supported by National Science Foundation Grants 98-96308 and 00-90402 and by a David and Lucile Packard Foundation fellowship to G.B.M. M.C.W. was supported by U.S. Department of Agriculture National Research Initiative Competitive Grants Program postdoctoral fellowship 2001-35319-09850 and by National Institutes of Health Grant GM48707 (awarded to F.M. Ausubel).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.000794.
References
- Abel, S., and Theologis, A. (1994). Transient transformation of Arabidopsis leaf protoplasts: A versatile experimental system to study gene expression. Plant J. 5, 421–427. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris 316, 1194–1199. [Google Scholar]
- Bent, A.F., Innes, R.W., Ecker, J.R., and Staskawicz, B.J. (1992). Disease development in ethylene-insensitive Arabidopsis thaliana infected with virulent and avirulent Pseudomonas and Xanthomonas pathogens. Mol. Plant-Microbe Interact. 5, 372–378. [DOI] [PubMed] [Google Scholar]
- Bohlmann, H., Vignutelli, A., Hilpert, B., Miersch, O., Wasternack, C., and Apel, K. (1998). Wounding and chemicals induce expression of the Arabidopsis thaliana gene Thi2.1, encoding a fungal defense thionin, via the octadecanoid pathway. FEBS Lett. 437, 281–286. [DOI] [PubMed] [Google Scholar]
- Braun, U. (1999). Some critical notes on the classification and generic concept of the Erysiphaceae. Schlechtendalia 3, 48–54. [Google Scholar]
- Broglie, K.E., Chet, I., Holliday, M., Cressman, R., Biddle, P., Knowlton, S., Mauvais, C.J., and Broglie, R. (1991). Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani. Science 254, 1194–1197. [DOI] [PubMed] [Google Scholar]
- Buell, C.R., and Somerville, S.C. (1995). Expression of defense-related and putative signaling genes during tolerant and susceptible interactions of Arabidopsis with Xanthomonas campestris pv. campestris. Mol. Plant-Microbe Interact. 8, 435–443. [Google Scholar]
- Büttner, M., and Singh, K. (1997). Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc. Natl. Acad. Sci. USA 94, 5961–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey, D.A., Shah, J., and Klessig, D.F. (1999). Salicylic acid and disease resistance in plants. Crit. Rev. Plant Sci. 18, 547–575. [Google Scholar]
- Després, C., DeLong, C., Glaze, S., Liu, E., and Fobert, P.R. (2000). The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12, 279–290. [PMC free article] [PubMed] [Google Scholar]
- Dewdney, J., Reuber, T.L., Wildermuth, M.C., Devoto, A., Cui, J., Stutius, L.M., Drummond, E.P., and Ausubel, F.M. (2000). Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. Plant J. 24, 205–218. [DOI] [PubMed] [Google Scholar]
- Dong, X. (1998). SA, JA, ethylene, and disease resistance in plants. Curr. Opin. Plant Biol. 1, 316–323. [DOI] [PubMed] [Google Scholar]
- Ellis, C., and Turner, J.G. (2001). The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13, 1025–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple, P., Apel, K., and Bohlmann, H. (1995). An Arabidopsis thaliana thionin gene is inducible via a signal transduction pathway different from that of pathogenesis-related proteins. Plant Physiol. 109, 813–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple, P., Apel, K., and Bohlmann, H. (1997). Overexpression of an endogenous thionin enhances resistance of Arabidopsis against Fusarium oxysporum. Plant Cell 9, 509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyal, Y., Meller, Y., Lev-Yadum, S., and Fluhr, R. (1993). A basic-type PR-1 promoter directs ethylene responsiveness, vascular and abscission zone-specific expression. Plant J. 4, 225–234. [DOI] [PubMed] [Google Scholar]
- Feys, B.J., and Parker, J.E. (2000). Interplay of signaling pathways in plant disease resistance. Trends Genet. 16, 449–455. [DOI] [PubMed] [Google Scholar]
- Feys, B.J.F., Benedetti, C.E., Penfold, C.N., and Turner, J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6, 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick, R.D., Thilmony, R.L., Sessa, G., and Martin, G.B. (1998). Recognition specificity for the bacterial avirulence protein AvrPto is determined by Thr-204 in the activation loop of the tomato Pto kinase. Mol. Cell 2, 241–245. [DOI] [PubMed] [Google Scholar]
- Fujimoto, S.Y., Ohta, M., Usui, A., Shinshi, H., and Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box–mediated gene expression. Plant Cell 12, 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Y.-Q., and Martin, G.B. (1998). Molecular mechanisms involved in bacterial speck disease resistance of tomato. Philos. Trans. R. Soc. Lond. B 353, 1455–1461. [Google Scholar]
- Gu, Y.-Q., Yang, C., Thara, V.K., Zhou, J., and Martin, G.B. (2000). Pti4 is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. Plant Cell 12, 771–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, P., Warren, R.F., Zhao, T., Shan, L., Zhu, L., Tang, X., and Zhou, J.M. (2001). Overexpression of Pti5 in tomato potentiates pathogen-induced defense gene expression and enhances disease resistance to Pseudomonas syringae pv. tomato. Mol. Plant Microbe Interact. 14, 1453–1457. [DOI] [PubMed] [Google Scholar]
- Hunter, T., and Karin, M. (1992). The regulation of transcription by phosphorylation. Cell 70, 375–387. [DOI] [PubMed] [Google Scholar]
- Jia, Y., and Martin, G.B. (1999). Rapid transcript accumulation of pathogenesis-related genes during an incompatible interaction in bacterial speck disease-resistant tomato plants. Plant Mol. Biol. 40, 455–465. [DOI] [PubMed] [Google Scholar]
- Kovtun, Y., Chiu, W.-L., Tena, G., and Sheen, J. (2000). Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. USA 97, 2940–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligterink, W., Kroj, T., Zur Nieden, U., Hirt, H., and Scheel, D. (1997). Receptor-mediated activation of a MAP kinase in pathogen defense of plant. Science 276, 2054–2057. [DOI] [PubMed] [Google Scholar]
- Ma, J., and Ptashne, M. (1988). Deletion analysis of GAL4 defines two transcription activating segments. Cell 48, 847–853. [DOI] [PubMed] [Google Scholar]
- Malamy, J., Carr, L.P., Klessig, D.F., and Raskin, I. (1990). Salicylic acid: A likely endogenous signal in the resistance response of tobacco to viral infection. Science 250, 1002–1004. [DOI] [PubMed] [Google Scholar]
- Maleck, K., and Dietrich, R.A. (1999). Defense on multiple fronts: How do plants cope with diverse enemies? Trends Plant Sci. 4, 215–219. [DOI] [PubMed] [Google Scholar]
- Maleck, K., Levine, A., Eulgem, T., Morgan, A., Schmid, J., Lawton, K.A., Dangl, J.L., and Dietrich, R.A. (2000). The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26, 403–410. [DOI] [PubMed] [Google Scholar]
- Manners, J.M., Penninckx, I.A., Vermaere, K., Kazan, K., Brown, R.L., Morgan, A., Maclean, D.J., Curtis, M.D., Cammue, B.P., and Broekaert, W.F. (1998). The promoter of the plant defensin gene PDF1.2 from Arabidopsis is systemically activated by fungal pathogens and responds to methyl jasmonate but not to salicylic acid. Plant Mol. Biol. 38, 1071–1080. [DOI] [PubMed] [Google Scholar]
- Martin, G.B., Brommonschenkel, D., Chunwongse, J., Frary, A., Ganal, M.W., Spivey, R., Wu, T., Earle, E.D., and Tanksley, S.D. (1993). Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 265, 966–970. [DOI] [PubMed] [Google Scholar]
- Memelink, J., Verpoorte, R., and Kijne, J.W. (2001). ORCAnization of jasmonate-responsive gene expression in alkaloid metabolism. Trends Plant Sci. 6, 212–219. [DOI] [PubMed] [Google Scholar]
- Menke, F.L., Champion, A., Kijne, J.W., and Memelink, J. (1999). A novel jasmonate- and elicitor-responsive element in the periwinkle secondary metabolite biosynthetic gene Str interacts with a jasmonate- and elicitor-inducible AP2-domain transcription factor, ORCA2. EMBO J. 18, 4455–4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal, S., and Davis, K.R. (1995). Role of the phytotoxin coronatine in the infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Mol. Plant-Microbe Interact. 8, 165–171. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Narasimhan, M.L., Damsz, B., Coca, M.A., Ibeas, J.I., Yun, D.J., Pardo, J.M., Hasegawa, P.M., and Bressan, R.A. (2001). A plant defense response effector induces microbial apoptosis. Mol. Cell 8, 921–930. [DOI] [PubMed] [Google Scholar]
- Ohme-Takagi, M., and Shinshi, H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi, M., Suzuki, K., and Shinshi, H. (2000). Regulation of ethylene-induced transcription of defense genes. Plant Cell Physiol. 41, 1187–1192. [DOI] [PubMed] [Google Scholar]
- Okamuro, J.K., Caster, B., Villarroel, R., Van Montagu, M., and Jofuku, K.D. (1997). The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 94, 7076–7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J.M., Park, C.J., Lee, S.-B., Ham, B.K., Shin, R., and Paek, K.-H. (2001). Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2–type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell 13, 1035–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx, I.A.M.A., Thomma, B.P.H.J., Buchala, A., Metraux, J.P., and Broekaert, W.F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10, 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, M., Brodersen, P., et al. (2000). Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103, 1111–1120. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M.J., and van Loon, L.C. (1999). Salicylic acid-independent plant defence pathways. Curr. Rev. Plant Biol. 4, 52–58. [DOI] [PubMed] [Google Scholar]
- Plotnikova, J.M., Reuber, T.L., Ausubel, F.M., and Pfister, D.H. (1998). Powdery mildew pathogenesis of Arabidopsis thaliana. Mycologia 90, 1009–1016. [Google Scholar]
- Restrepo, M.A., Freed, D.D., and Carrington, J.C. (1990). Nuclear transport of plant potyviral proteins. Plant Cell 2, 987–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuber, T.L., Plotnikova, J.M., Dewdney, J., Rogers, E.E., Wood, W., and Ausubel, F.M. (1998). Correlation of defense gene induction defects with powdery mildew susceptibility in Arabidopsis enhanced disease susceptibility mutants. Plant J. 16, 473–485. [DOI] [PubMed] [Google Scholar]
- Reymond, P., and Farmer, E.D. (1998). Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 1, 404–411. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L., and Meyerowitz, E.M. (1998). The AP2/EREBP family of plant transcription factors. Biol. Chem. 379, 633–646. [DOI] [PubMed] [Google Scholar]
- Schenk, P.M., Kazan, K., Wilson, I., Anderson, J.P., Richmond, T., Somerville, S.C., and Manners, J.M. (2000). Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 97, 11655–11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schule, R., and Evans, R.M. (1991). Cross-coupling of signal transduction pathways: Zinc finger meets leucine zipper. Trends Genet. 7, 377–381. [DOI] [PubMed] [Google Scholar]
- Sela-Buurlage, M., Ponstein, A.S., Bres-Vloemans, S.A., Melchers, L.S., van den Elzen, P.J.M., and Cornelissen, B.J.C. (1993). Only specific tobacco (Nicotiana tabacum) chitinase and β-1,3-glucanases exhibit antifungal activity. Plant Physiol. 101, 857–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano, R., Stepanova, A., Chao, Q., and Ecker, J.R. (1998). Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSIVE-FACTOR1. Genes Dev. 12, 3703–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger-Haussels, M., and Weisshaar, B. (2000). Transactivation properties of parsley proline-rich bZIP transcription factors. Plant J. 22, 1–8. [DOI] [PubMed] [Google Scholar]
- Suzuki, K., Suzuki, N., Ohme-Takagi, M., and Shinshi, H. (1998). Immediate early induction of mRNAs for ethylene-responsive transcription factors in tobacco leaf strips after cutting. Plant J. 15, 657–665. [DOI] [PubMed] [Google Scholar]
- Thara, K.V., Tang, X., Gu, Y.-Q., Martin, G.B., and Zhou, J.-M. (1999). Pseudomonas syringae pv. tomato induces the expression of tomato EREBP-like genes Pti4 and Pti5 independent of ethylene, salicylate and jasmonate. Plant J. 20, 475–484. [DOI] [PubMed] [Google Scholar]
- Thilmony, R.L., Chen, Z., Bressan, R.A., and Martin, G.B. (1995). Expression of tomato Pto gene in tobacco enhances resistance to Pseudomonas syringae pv. tabacci expressing avrPto. Plant Cell 7, 1529–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B.P.H.J., Eggermont, K., Penninckx, I.A.M.A., Mauch-Mani, B., Vogelsang, R., Cammue, B.P.A., and Broekaert, W. (1998). Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA 95, 15107–15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varagona, M.J., Schmidt, R.J., and Raikhel, N.V. (1992). Nuclear localization signal(s) required for nuclear targeting of the maize regulatory protein Opaque-2. Plant Cell 4, 1213–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisey, C.R., and Slusarenko, A.J. (1989). Chitinase mRNA and enzyme activity in Phaseolus vulgaris (L.) increase more rapidly in response to avirulent than to virulent cells of Pseudomonas syringae pv. phaseolicola. Physiol. Mol. Plant Pathol. 35, 403–412. [Google Scholar]
- Xie, D.X., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Fan, W., Kinkema, M., Li, X., and Dong, X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. USA 96, 6523–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J., Tang, X., and Martin, G.B. (1997). The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind a cis-element of pathogenesis-related genes. EMBO J. 16, 3207–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Q., Maher, E.A., Masoud, S., Dixon, R.A., and Lamb, C.J. (1994). Enhanced protection against fungal attack by constitutive co-expression of chitinase and glucanase in transgenic tobacco. Bio/Technology 12, 807–812. [Google Scholar]