Abstract
A wide spectrum of soil heterocyclic nitrogen compounds are potential nutrients for plants. Here, it is shown that Arabidopsis plants are able to use allantoin as sole nitrogen source. By functional complementation of a yeast mutant defective in allantoin uptake, an Arabidopsis transporter, AtUPS1 (Arabidopsis thaliana ureide permease 1), was identified. AtUPS1 belongs to a novel superfamily of plant membrane proteins with five open reading frames in Arabidopsis (identity, 64 to 82%). UPS proteins have 10 putative transmembrane domains with a large cytosolic central domain containing a “Walker A” motif. Transport of 14C-labeled allantoin by AtUPS1 in yeast exhibited saturation kinetics (Km ∼ 52 μM), was dependent on Glc and a proton gradient, and was stimulated by acidic pH. AtUPS1 transports uric acid and xanthine, besides allantoin, but not adenine. Protons are cosubstrates in allantoin transport by AtUPS1, as demonstrated by expression in Xenopus laevis oocytes. In plants, AtUPS1 gene expression was dependent on the nitrogen source. Therefore, AtUPS1 presumably is involved in the uptake of allantoin and other purine degradation products when primary sources are limiting.
INTRODUCTION
In most soils, nitrogen is an important limiting factor for plant growth. Under agricultural conditions, nitrate and ammonium are the predominant nitrogen sources available for plant nutrition (Von Wirén et al., 2000). Many natural soils contain a broad spectrum of organic compounds, including amino acids, amino sugars, and nitrogen heterocycles, particularly under conditions in which bacterial activity is inhibited. It has been estimated that heterocyclic nitrogen compounds represent ∼30% of the reduced nitrogen in soils (Schulten and Schnitzer, 1998). Therefore, it is important to study the relevance of this organic nitrogen for plant nutrition. The molecular identities of distinct heterocyclic nitrogen compounds and their relative contributions to the total soil nitrogen content remain controversial (Schulten and Schnitzer, 1998). One group of heterocyclic nitrogen compounds, the nucleobases and their degradation products, has been identified in several soils as representing on average 3% of total nitrogen (Cortez and Schnitzer, 1979). These compounds are produced during the natural degradation of organic matter by soil microorganisms or are derived from animal excretion and may represent an alternative low-cost or backup source of nitrogen for plants. Adenine, guanine, hypoxanthine, xanthine, and urate are good nitrogen sources for the growth of green algae such as Chlamydomonas (Pineda and Cardenas, 1996). It appears plausible that, to use these compounds as nitrogen source, specialized transport mechanisms are present in plants.
The transport of purine oxidation products has been demonstrated in tropical legumes (Schubert, 1986). In these plants, N2 is fixed by symbiotic bacteria and then transported as ammonium to the cytosol of infected host cells. There, a substantial fraction of the fixed nitrogen is used for the synthesis of purines and converted to uric acid. After transport to neighboring uninfected cells, uric acid is used for allantoin synthesis in peroxisomes. In addition, allantoin can be converted to allantoic acid in the cytosol. Both allantoin and allantoic acid are nitrogen-rich compounds with a C:N ratio of 1:1 and serve as principal long-distance transport molecules in tropical legumes, representing up to 90% of the transported nitrogen. In nonlegume species of the genera Acer, Platanus, and Aesculus, the sieve tube sap contains ∼15% of the total nitrogen in the form of allantoin and allantoic acid (Ziegler, 1975). To our knowledge, neither long-distance nor intracellular transport mechanisms of these purine catabolites have been studied.
Except for the well-characterized family of plant amino acid transporters, information on transport systems for other nitrogen compounds is scarce (D. Wipf, U. Ludewig, M. Tegeder, D. Rentsch, W. Koch, and W.B. Frommer, unpublished data). Two classes of plant nucleobase transporters designated purine permeases and nucleobase ascorbate transporters were described recently (De Konig and Diallinas, 2000; Gillissen et al., 2000). Maize Lpe1, the only functionally characterized plant transporter of the nucleobase ascorbate transporter family, preferentially transports xanthine and uric acid (Argyrou et al., 2001) and is necessary for the proper development of chloroplasts (Schultes et al., 1996). In contrast, transporters for allantoin have been identified and characterized only in yeast and bacteria.
Saccharomyces cerevisiae accumulates and uses allantoin as a nitrogen source (Chisholm et al., 1987). This may seem surprising, because to date little is known about the occurrence of allantoin in grapes, the natural host of yeast. However, if nitrogen limits yeast growth in its natural habitat, the uptake and degradation of allantoin or related compounds may provide an additional source of nitrogen. The carrier responsible for allantoin uptake has been identified as the product of the dal4 gene (Yoo et al., 1992). DAL4 localizes to the plasma membrane, spans the membrane 10 times, and functions as a proton cotransporter. DAL4 belongs to the purine-related transporter family that is known only in microorganisms.
Assuming that nitrogen heterocyclic compounds such as allantoin may serve as nitrogen sources or nitrogen transport compounds in plants that are not able to fix nitrogen, allantoin transport was studied in Arabidopsis. In this article, we show that Arabidopsis can use allantoin as an alternative nitrogen source. Suppression cloning in an allantoin transport-deficient yeast mutant and expression in Xenopus laevis oocytes were used to identify and characterize a new superfamily of polytopic membrane proteins mediating H+-coupled transport of a wide spectrum of oxo derivatives of heterocyclic nitrogen compounds, including allantoin, uric acid, and xanthine.
RESULTS
Arabidopsis Plants Are Able to Use Allantoin as Sole Nitrogen Source
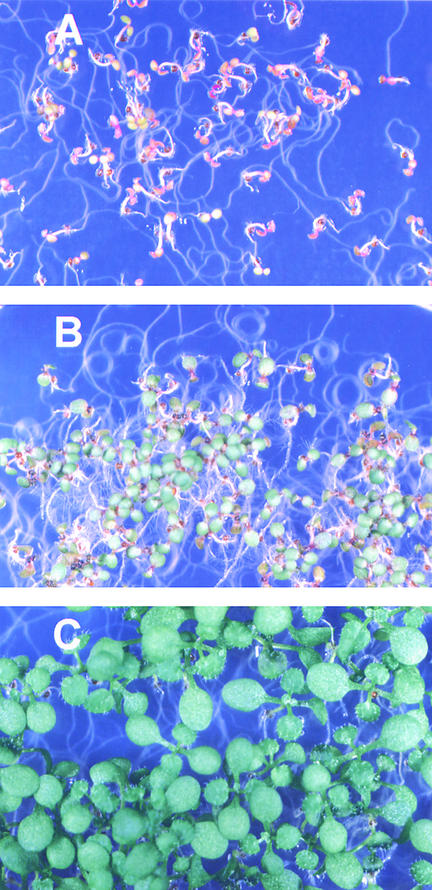
To determine whether naturally occurring oxidation products of purines can serve as a nitrogen source for plants, Arabidopsis seed were germinated on medium supplied with different nitrogen compounds as sole nitrogen sources. In the presence of 5 mM allantoin, seedlings were able to grow and exhibited a phenotype characterized by small, green leaves and strong root elongation (Figure 1B). Because a purity of 99.9% was certified by the manufacturer for the chemical used (Sigma), a maximum nitrogen concentration of 20 μM in other chemical form than allantoin was assumed. Therefore, 10 μM ammonium nitrate was included on control plates. On these plates, seed germinated, but seedlings did not develop beyond the cotyledon stage (Figure 1A). The growth rate, however, was reduced significantly compared with that of plants grown on 10 mM ammonium nitrate (Figure 1C), indicating nitrogen limitation for plants grown on allantoin as sole nitrogen source. Nevertheless, plants grown on allantoin completed a life cycle and produced normal seed (data not shown). Other related compounds (e.g., 10 mM hydantoin or 5 mM xanthine) supported growth less efficiently (data not shown) Although we cannot completely exclude degradation in the medium, these observations might indicate that oxo derivatives of heterocyclic nitrogen compounds such as allantoin can support the growth of Arabidopsis as sole nitrogen source.
Figure 1.
Phenotypes of 11-Day-Old Arabidopsis Plants Grown with 10 μM Ammonium Nitrate (A), 5 mM Allantoin (B), or 10 mM Ammonium Nitrate (C) as Sole Nitrogen Source.
Cloning of Putative Allantoin Transporters
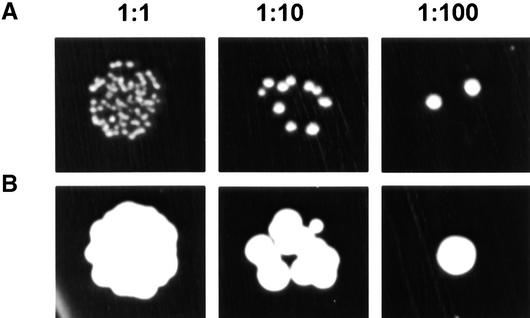
The apparent ability of Arabidopsis to take up allantoin prompted us to investigate transporters potentially mediating root uptake of nitrogen heterocycles. For this purpose, functional complementation of a yeast mutant deficient in allantoin uptake was used. First, the dal4 gene was deleted in an ura3 strain of S. cerevisiae. Because the dal5 gene product, an allantoic acid transporter, mediates the uptake of small but significant amounts of allantoin (Chisholm et al., 1987), this gene was disrupted in a subsequent step. The resulting dal4 dal5 double mutant was deficient in allantoin uptake and grew slowly with allantoin as sole nitrogen source (Figure 2). Heterologous expression of an Arabidopsis cDNA library (Minet et al., 1992) in this strain and subsequent selection of suppressors on medium containing allantoin as sole nitrogen source led to the identification of several cDNAs able to complement the functional deficiency (Figure 2). Retransformation of the dal4 dal5 mutant with the isolated plasmids demonstrated that no second-site mutation or reversion was responsible for the suppression. Sequence analysis confirmed that all clones were derived from the same gene, which was named AtUPS1 (Arabidopsis thaliana ureide permease 1).
Figure 2.
Functional Complementation of a Yeast Mutant Deficient in Allantoin Uptake.
The dal4 dal5 mutant was transformed with the empty vector pFL61 (A) or with pFL61 carrying AtUPS1 (B). Three cell dilutions (1:1, 1:10, and 1:100) were plated on minimal medium containing 0.1% allantoin as sole nitrogen source and incubated for 5 days at 28°C.
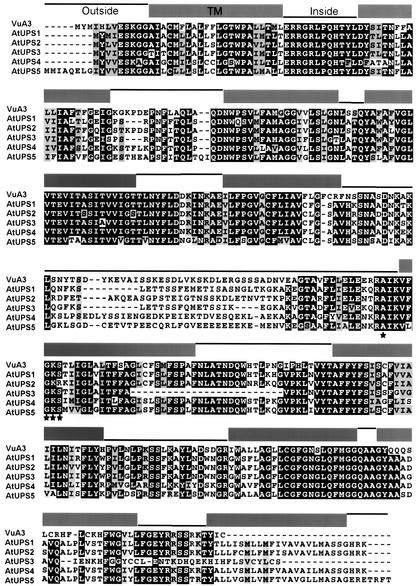
AtUPS1 contains an open reading frame of 1173 bp encoding a 390–amino acid protein with a calculated molecular mass of 42.7 kD and is localized on chromosome II (At2 g03590). Four putative Arabidopsis open reading frames with 64 to 82% identity to AtUPS1 (AtUPS2, At2 g03530; AtUPS3, At2 g03600; AtUPS4, At2 g03520; AtUPS5, At1 g26440), one cDNA sequence from Vigna unguiculata, and expressed sequence tags from several plant species were identified by database searches. Hydrophobicity analysis of these sequences predicts 10 putative transmembrane domains conserved in all members (Sonnhammer et al., 1998). The C and N termini are predicted to protrude into the extracellular space defining a large cytosolic domain between transmembrane helices 5 and 6. This central domain contains a consensus sequence for a P loop, also designated a “Walker A” motif (Saraste et al., 1990), which is highly conserved in all UPS proteins.
Figure 3 shows the alignment of the five amino acid sequences from Arabidopsis and the only additional full-length sequence from Vigna and a graphic representation of the hydrophobicity analysis. No transit peptides for chloroplasts or mitochondria were predicted. A PSI-BLAST analysis of the nonredundant National Center for Biotechnology Information database (December 21, 2001) yielded only the Arabidopsis and Vigna homologs with likelihood values of E < 1 (Altschul and Gish, 1996). In addition, bacterial sequences from Lactococcus lactis and Streptococcus pyrogenes with E values > 1 were obtained. The Lactococcus protein is structurally related to the UPS family, because the hydrophobicity analysis predicts 10 transmembrane-spanning domains separated by a small cytosolic loop, and the N and C termini are predicted to be directed to the extracellular space. These features imply that AtUPS1 is a member of a new class of plant membrane proteins potentially localized on the plasma membrane.
Figure 3.
Amino Acid Sequence Alignment of Members of the UPS Family.
Amino acids are given with standard single-letter designation, and dashes indicate gaps. Residues are shown in white letters on black if five or more sequences have identical residues at the aligned position. Gray background indicates similar amino acids, and asterisks indicate conserved amino acids sharing the consensus sequence A-x(4)-G-K-S (“Walker A” motif). Gray boxes above the sequences indicate the positions of 10 potential transmembrane domains (TM), and the connecting lines indicate potential outside and inside domains as predicted by TMHMM version 2.0 (http://www.cbs.dtu.dk/services/TMHMM/). VuA3, protein A3 from Vigna unguiculata.
Biochemical Properties of AtUPS1
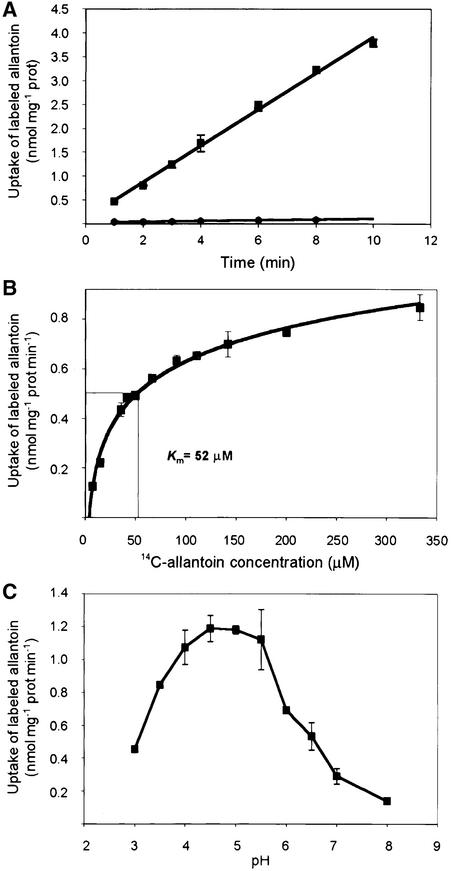
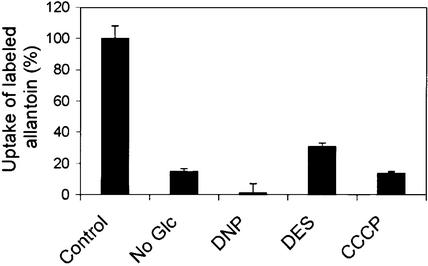
To determine the transport properties of AtUPS1, radiotracer uptake studies were performed using 14C-labeled allantoin. Yeast dal4 dal5 cells expressing AtUPS1 showed >50-fold increased uptake of 14C-allantoin compared with cells transformed with vector pFL-61 alone (Figure 4A). Under standard assay conditions, the 14C-allantoin uptake rate remained constant during the first 10 min, was concentration dependent, and displayed saturation kinetics with an apparent Km of 52 μM (Figure 4B). AtUPS1 activity was strictly pH dependent, with an optimum at pH ∼4.75 (Figure 4C). 14C-allantoin uptake depended on the presence of Glc and was sensitive to protonophores and a plasma membrane H+-ATPase inhibitor, indicating that energization is required for transport (Figure 5). Competition with a 10-fold molar excess of unlabeled allantoin (99.9% pure) led to a reduction of the uptake rate by 90%, strongly supporting the notion that AtUPS1 transports allantoin and that the measured uptake of radiolabel is not attributable to impurities in the labeled product.
Figure 4.
Uptake of 14C-Allantoin by dal4 dal5 Mutants.
(A) Time dependence. Mutants were transformed with the empty vector pFL61 (circles) or with pFL61 carrying AtUPS1 (squares). Incubations were performed in 100 mM potassium phosphate buffer, pH 4.5, 100 mM Glc, and 200 mM 14C-allantoin at 30°C.
(B) Michaelis-Menten kinetics. The uptake rate of cells expressing AtUPS1 was determined with the indicated 14C-allantoin concentrations.
(C) pH dependence. Incubations were performed in 100 mM potassium phosphate buffer adjusted to the indicated pH values, 100 mM Glc, and 200 mM 14C-allantoin at 30°C. Results represent means of three independent experiments ±sd.
Figure 5.
Influence of Plasma Membrane Energization on the Uptake Rate of 14C-Allantoin in dal4 dal5 Mutants Expressing AtUPS1.
Yeast cells were preincubated for 5 min in the presence of 100 mM Glc (control), without Glc, or with Glc and 0.1 mM 2,4-dinitrophenol (DNP), 0.1 mM diethylstilbestrol (DES), or 0.1 mM carbonyl cyanide m-chlorphenyl-hydrazone (CCCP). Data are expressed as percentage of control values (0.85 μmol 14C-allantoin·mg−1 protein·min−1) and represent means of three independent experiments ±sd.
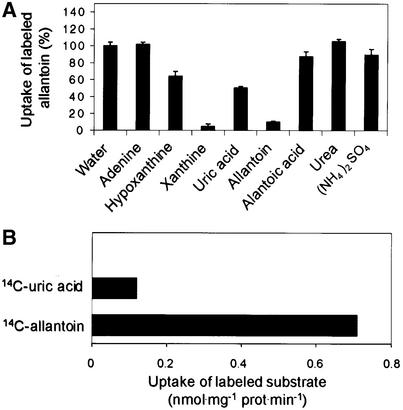
To study the substrate specificity of AtUPS1, 14C-labeled allantoin uptake was determined in the presence of a 10-fold molar excess of purines or purine derivatives (Figure 6A). Xanthine and uric acid were efficient competitors for 14C-allantoin uptake, whereas hypoxanthine was less efficient. Adenine, allantoic acid, urea, and ammonium showed no or very weak uptake inhibition. These data suggest that besides allantoin, AtUPS1 also transports other oxo derivatives of heterocyclic nitrogen compounds (e.g., xanthine and uric acid). The transport of uric acid was confirmed using 14C-uric acid (Figure 6B).
Figure 6.
Substrate Specificity of AtUPS1.
(A) Inhibition of 200 μM 14C-allantoin uptake by a 10-fold molar excess of purines or purine degradation products. Data are expressed as percentage of the control values (0.79 μmol 14C-allantoin·mg−1 protein·min−1).
(B) Uptake rate of 200 μM 14C-labeled uric acid or allantoin. Results represent means of three independent experiments ±sd.
Transport Mechanism of AtUPS1
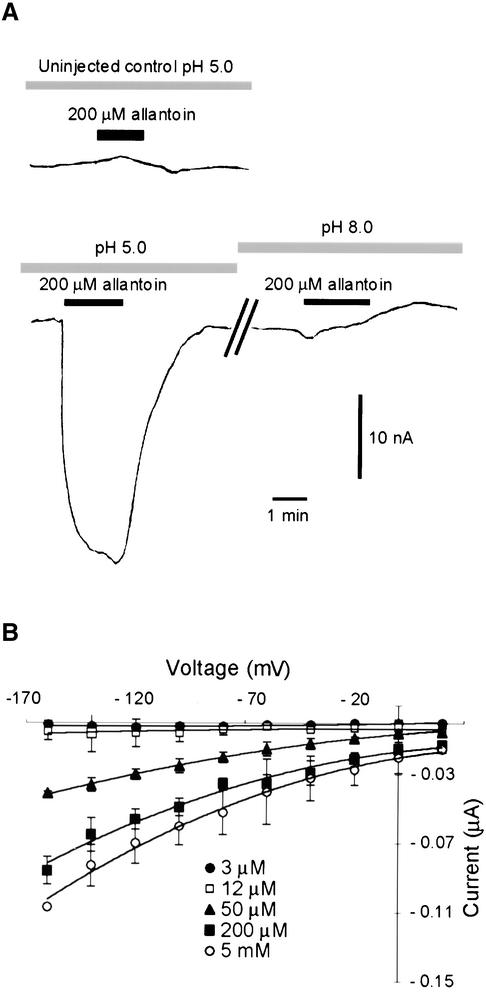
To study the transport mechanism of the carrier, AtUPS1 was expressed functionally in Xenopus oocytes. Superfusion of AtUPS1-injected oocytes with 200 μM allantoin specifically induced net inward currents at pH 5.0 (Figure 7A), corresponding to a net influx of positive charge. Allantoin-mediated currents were absent in uninjected control oocytes and were abolished at alkaline pH (Figure 7A). As at pH 5.0, the neutral form of allantoin was dominant (pK = 8.9), which strongly supports the notion that protons are cotransported with the substrate.
Figure 7.
Transport of Allantoin by AtUPS1 Expressed in Xenopus Oocytes.
(A) Inward current induced by 200 mM allantoin in uninjected oocytes (top) or oocytes injected with AtUPS1 mRNA (bottom) at pH 5.0 or pH 8.5. Voltage was clamped at −70 mV throughout the experiment.
(B) Voltage and concentration dependence of AtUPS1 currents.
Allantoin-induced currents were time independent (data not shown) and voltage and concentration dependent (Figure 7B). Allantoin transport was saturable at submillimolar concentrations, and the Km was ∼70 μM at all voltages tested, which is in accord with the substrate affinity calculated in the yeast system.
Expression Analysis of AtUPS1
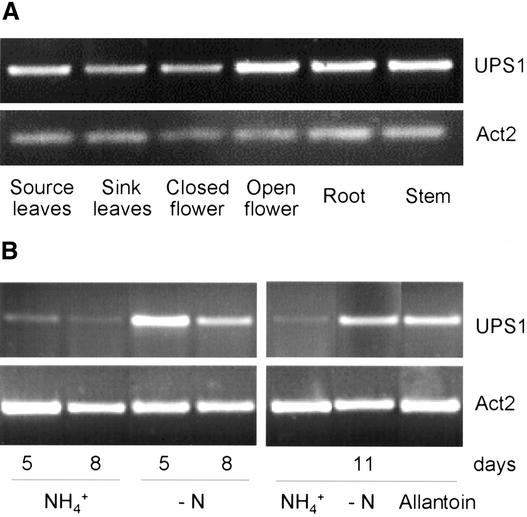
AtUPS1 mRNA from Arabidopsis plants was not detectable by RNA gel blot hybridization experiments. Therefore, reverse transcriptase–mediated polymerase chain reaction (RT-PCR) was performed with total RNA from different plant organs. Using primers specific for AtUPS1, the transcript was detected in RNA from all organs tested without noticeable differences (Figure 8A). In addition, RT-PCR was performed on total RNA from seedlings grown on medium containing ammonium nitrate, allantoin, or without any nitrogen source. Compared with seedlings grown on medium con-taining ammonium, seedlings grown on allantoin showed significantly higher levels of AtUPS1 transcript (Figure 8B). However, AtUPS1 expression also was clearly enhanced in the absence of nitrogen. Because seedlings grown on allantoin showed symptoms of nitrogen deficiency (e.g., increased root:shoot mass ratios), AtUPS1 expression presumably is induced by nitrogen deficiency rather than by allantoin itself.
Figure 8.
Expression of AtUPS1 mRNA in Different Plant Organs (A) and in Whole Plants Supplied with Different Nitrogen Sources (B).
RNAs were extracted and converted to cDNA by reverse transcriptase, and a 288-bp AtUPS1 fragment was amplified by 25 PCR cycles. To ensure that equal amounts of cDNA were added to each PCR reaction, an Actin2 fragment was amplified simultaneously by 20 PCR cycles (Act2).
DISCUSSION
A Novel Superfamily of Plant Transporters
In this study, AtUPS1, the first plant transporter for allantoin and other oxo derivatives of nitrogen heterocyclic compounds, was identified and characterized. AtUPS1 is a member of a protein superfamily with 10 predicted membrane-spanning domains. The function of AtUPS1 was addressed initially by complementation studies in yeast, which indicated that AtUPS1 mediates the transport of allantoin across the plasma membrane that is necessary for the yeast dal4 dal5 mutant to grow on allantoin as sole nitrogen source. In addition, the uptake of radiotracers in yeast and functional expression in Xenopus oocytes demonstrated the ability to transport allantoin and provide crucial information about kinetics, substrate specificity, and mechanism of transport.
The Arabidopsis UPS family consists of five highly conserved members, and related genes are present in various other plant species (e.g., cowpea, tomato, and rice). The UPS family does not share significant homology with any known allantoin transport proteins from either yeast or bacteria and is not related to the recently identified families of plant nucleobase transporters (De Konig and Diallinas, 2000; Gillissen et al., 2000). Furthermore, no significant homology with DNA or protein sequences outside of the plant kingdom were found. It remains to be determined whether the weak sequence similarities to the structurally related bacterial membrane proteins are based on an evolutionary relationship.
Transport Mechanism of AtUPS1
UPS proteins contain a conserved Walker A motif for ATP binding, which makes them candidates for primary active allantoin pumps. On the other hand, the strong dependence on the presence of Glc and a proton gradient indicates that AtUPS1-mediated transport needs to be energized and preliminarily suggests a secondary active transport mechanism. The functional expression of plant transporters in Xenopus oocytes allows a more direct analysis of their mode of action, but it has been achieved only in a limited number of cases (Miller and Zhou, 2000).
Two-electrode voltage-clamp analysis of oocytes expressing AtUPS1 showed that allantoin influx was simply stimulated at acidic external pH and inhibited at alkaline pH. Superfusion with different pH solutions is unlikely to immediately change the metabolic state and ATP concentration of the oocyte cytosol, suggesting a secondary active transport mechanism for AtUPS1. Inward currents induced by allantoin (at pH 5.0, the neutral form of allantoin is dominating and <1% is negatively charged) directly show that protons are cotransported with the substrate. The affinity to allantoin is largely independent of the applied voltage, which may indicate that the neutral form of allantoin binds to AtUPS1 or that the binding site is located outside the electric field that decreases over the membrane.
Together, the data from yeast and oocytes argue in favor of a simple proton/substrate cotransport as the transport mechanism in AtUPS1. The exact stoichiometry of transport, however, has yet to be determined. Further studies will address the significance of the conserved Walker A motif for the activity of UPS proteins.
Putative Function of AtUPS1
Uptake and competition studies using 14C-labeled allantoin and uric acid allowed the identification of substrates of AtUPS1. Among the substances tested, xanthine and uric acid, allantoin precursors in the purine degradation pathway, are able to inhibit allantoin uptake competitively. Uptake experiments with 14C-labeled uric acid showed that it is in fact a substrate for AtUPS1. Interestingly, a common structural feature of compounds transported by AtUPS1 is the oxo derivation of the purine or imidazol rings that might be involved in substrate recognition. In support of this notion, adenine and imidazol, which do not contain oxo groups, are not transported by AtUPS1. Future investigations should expand the number of compounds tested to clarify the mechanism of substrate recognition and, most importantly, to identify other potential substrates.
Because the low expression level of AtUPS1 prevented the use of conventional RNA hybridization experiments, gene-specific RT-PCR was used to detect AtUPS1 transcript in comparable amounts in RNA from different plant organs. RT-PCR of RNA from seedlings grown on different media revealed that the level of AtUPS1 expression depends strongly on the nitrogen supply. Compared with its expression in seedlings grown on ammonium nitrate, AtUPS1 expression was significantly higher in nitrogen-deprived seedlings as well as in seedlings grown on allantoin as a nitrogen source. These results are similar to nitrogen catabolic gene expression in S. cerevisiae, in which the expression of allantoin pathway genes is induced in the presence of allantoin or its degradative metabolites and is repressed when a good nitrogen source (e.g., Asn or Gln) is provided (Cooper and Sumrada, 1983). It will be interesting to determine whether, as in the yeast system, AtUPS1 activity is subject to feedback regulation by allantoin (Chisholm et al., 1987).
In conjunction with our finding that allantoin can support the growth of Arabidopsis seedlings, the nitrogen-dependent expression suggests strongly that AtUPS1 plays a role in the uptake of allantoin in planta in situations in which primary nitrogen sources are limiting. Although heterocyclic nitrogen compounds have been identified in several natural soils, their relative contributions to total soil nitrogen are controversial. Because allantoin is a highly polar molecule, it is not detected easily by conventional methods, so it remains to be determined if allantoin, together with the other AtUPS1 substrates, contributes significantly to soil nitrogen levels. In this context, it is noteworthy that allantoin is the final product of purine degradation excreted by all mammals except primates, whereas uric acid is the end product of purine degradation excreted by primates and the final product of nitrogen metabolism in terrestrial reptiles and birds. More detailed studies addressing soil and in planta concentrations of allantoin and other related heterocyclic nitrogen compounds may help to determine whether the main physiological substrates of AtUPS1 (Km = 52 μM) have been identified.
In certain tropical legumes, allantoin and allantoic acid serve as the principal long-distance transport molecules, representing up to 90% of transported nitrogen. It has been postulated that relatively few additional functions are necessary to engineer nonlegumes into hosts for nitrogen-fixing bacteria (Parniske, 2000). The results presented here suggest that the model plant Arabidopsis can be of use in the identification of proteins involved in symbiosis. Studying members of the UPS family in legumes such as soybean could lead to the identification of transporters required for the long-distance transport of allantoin. In this way, crucial information can be obtained to improve nitrogen translocation in legumes (Streeter, 1993).
In conclusion, AtUPS1, the first allantoin transporter identified from a higher plant, is likely to be involved in the uptake of allantoin and other purine degradation products, which might serve as backup nitrogen sources in situations in which primary sources become limiting. The identification of the novel UPS transporter superfamily might facilitate the identification of the major nitrogen transporters from important crop species such as soybean.
METHODS
Yeast Strains, Mutant Generation, and Complementation with an Arabidopsis thaliana cDNA Library
The yeast (Saccharomyces cerevisiae) strain Σ 23346c (Mat a, ura3) (Grenson, 1969) was used to generate a mutant defective in allantoin uptake. The DAL4 gene was deleted by homologous integration of a disruption cassette containing loxP sites flanking the marker gene kan (Güldener et al., 1996). Selection was performed on solid plates containing 100 mg/L G418, and gene deletion was confirmed by polymerase chain reaction (PCR). The dal4::kan strain was used for deletion of the DAL5 gene. A disruption cassette with the selection marker nat was used for this purpose. Transformed cells were selected using 200 mg/L Clonat (Hans-Knöll-Institut e.V., Jena, Germany), and gene deletion was confirmed by PCR. The deficiency of the mutant in allantoin uptake was determined on solid plates containing different concentrations of allantoin as sole nitrogen source, and optimal selective conditions were found at 1 g/L.
The dal4 dal5 mutant was transformed with an expression library derived from Arabidopsis seedlings in the episomal plasmid pFL61 containing the URA3 gene (Minet et al., 1992). Transformants were selected on solid plates containing yeast nitrogen base, 2% Glc, and 1 g/L allantoin (Sigma) as sole nitrogen source. Colonies able to grow were reselected under the same conditions. Plasmids were prepared, and the dal4 dal5 mutant was retransformed and reselected. As a negative control, the mutant was transformed with the empty plasmid.
Transport Measurements
Because labeled allantoin was not available commercially, 14C-labeled allantoin was produced from 8-14C-uric acid (ARC 513; Biotrends, Köln, Germany; specific activity, 50 mCi/mmol) as described by Bergmeyer (1970) with unlabeled uric acid. Briefly, 10 μmol of 8-14C-uric acid was incubated with 1 unit of uricase (Sigma; catalog No. U1878) in 1 mL of reaction mixture containing 20 mM Tris-HCl, pH 8.5, at room temperature for 2 hr. The reaction was monitored spectrophotometrically after the decrease in absorption of uric acid (maximum at 293 nm). Samples of the product were analyzed by HPLC using a NAK 50,927 Spherisorb 5 NH2 column (Kontron, Neufahrn, Germany). Solvent system A (60% acetonitrile and 0.1% acetic acid) and solvent system B (92% acetonitrile and 0.1% acetic acid) were used for isocratic separation of uric acid and allantoin, respectively. The retention times of 14C-labeled compounds were monitored with an LB 507B radiodetector (Berthold, Wildbad, Germany). Using solvent system A, 14C-uric acid could not be detected. Using solvent system B, a major peak corresponding to allantoin and two additional minor peaks with lower retention times were observed. Integration of these radioactive peaks indicated that of the total labeled compounds, 89.9% was 14C-labeled allantoin. Allantoin (Sigma; catalog No. A7878) and uric acid (Sigma; catalog No. U0881) were used as standards.
For uptake studies, yeast cells were harvested at an OD600 of 0.8 by centrifugation for 5 min at 4500g. Cells were washed and resuspended in double-distilled water to a final OD600 of 4. One hundred microliters of cells was mixed with 20 μL of 1 M potassium phosphate, pH 4.5, 20 μL of 1 M Glc, and 60 μL of double-distilled water and preincubated for 5 min at 30°C. To start the reaction, 20 μL of 2 mM 14C-labeled allantoin (specific activity, 1.85 to 2.22 MBq/μmol) was added. Samples of 50 μL were removed after 1, 2, 3, and 4 min, transferred to 4 mL of ice-cold water, filtered on fiberglass filters, and washed with 8 mL of water. Radioactivity incorporated in cells was determined by liquid scintillation spectrometry (Beckman). Transport measurements were repeated independently and represent the means of at least three experiments.
Functional Expression of Xenopus laevis Oocytes
Oocyte handling and recording was performed essentially as described by Stühmer (1998). Briefly, AtUPS1 cDNA was subcloned into pOO2, an oocyte expression plasmid derived from pSP64T (Promega) that contains 5′ and 3′ untranslated β-globin sequences from Xenopus and a 92-bp poly(A) tail. Capped copy RNA was transcribed in vitro using the mMessage mMachine kit (Ambion, Austin, TX). Oocytes were removed from adult female frogs by surgery and manual defolliculation. Oocytes were injected with 50 nL of copy RNA (∼50 ng/oocyte). Currents were measured by two-electrode voltage-clamp analysis 3 days after injection using a Dagan (Minneapolis, MN) CA1 amplifier and pClamp6.0 software (Axon Instruments, Union City, CA). Standard bath solutions contained 100 mM choline chloride, 2 mM CaCl2, 2 mM MgCl2, 4 mM Tris/Mes, pH 5.0 or pH 8.5, and appropriate amounts of allantoin. For current-voltage analysis, background currents (recorded before and after the addition of substrate) were subtracted.
Plant Material and mRNA Expression
Arabidopsis ecotype Columbia was used for plant studies. With the exception of roots, which were obtained from plants cultured on agar plates, all plant organs were harvested from plants grown in soil in the greenhouse. For plant growth and AtUPS1 mRNA expression studies under different nitrogen regimens, Arabidopsis seed were placed on Murashige and Skoog (1962) medium with Glc and allantoin or ammonium nitrate as a nitrogen source. All procedures were performed under sterile conditions. After 5, 8, and 11 days, plants were harvested and total RNA was extracted. For reverse transcriptase–mediated PCR analysis, RNAs were converted to cDNAs by reverse transcriptase using the manufacturer's protocol (Ambion, Austin, TX), and a 288-bp cDNA fragment was amplified by 25 PCR cycles using specific primers for AtUPS1 (forward primer, 5′-GCACAATAATCGGATTGGTG-3′; reverse primer, 5′-ATGTTAAGTATC-AGAGCAACTACAAATGC-3′). To ensure that equal amounts of cDNA were added to each PCR reaction, a cDNA fragment of the constitutively expressed actin 2 gene (ACT2) was amplified simultaneously by 20 PCR cycles (An et al., 1996). The identities of the amplicons were confirmed by sequencing.
Accession Numbers
The GenBank accession numbers for the sequences mentioned in this article are as follows: Vigna unguiculata A3 (X90487), Lactococcus lactis transporter (NC002662), and Streptococcus pyrogenes putative transporter (NC002737).
Acknowledgments
We are grateful to N. von Wirén for critical reading of the manuscript. We also thank Bettina Stadelhofer and Harald Stransky for HPLC analysis and Diana Dembinsky, Anja Schmidt, and Michael Fitz for technical assistance. This work was supported by the German Bundesministerium für Bildung und Forschung in the framework of (Genomanalyse im Biologischen System Pflanze). We also gratefully acknowledge support by Kleinwanzlebener Saatzucht and Südzucker.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010458.
References
- Altschul, S.F., and Gish, W. (1996). Local alignment statistics. Methods Enzymol. 266, 460–480. [DOI] [PubMed] [Google Scholar]
- An, Y.-Q., McDowell, J.M., Huang, S., McKinney, E.C., Chambliss, S., and Meagher, R.B. (1996). Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 10, 107–121. [DOI] [PubMed] [Google Scholar]
- Argyrou, E., Sophianopoulou, V., Schultes, N., and Diallinas, G. (2001). Functional characterization of a maize purine transporter by expression in Aspergillus nidulans. Plant Cell 13, 953–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmeyer, H.U. (1970). Methoden der Enzymatischen Analyse, Vol. 1. (Weinheim, Germany: Verlag Chemie), pp. 478–479.
- Chisholm, V.T., Lea, H.Z., Rai, R., and Cooper, T.G. (1987). Regulation of allantoate transport in wild-type and mutant strains of Saccharomyces cerevisiae. J. Bacteriol. 169, 1684–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, T.G., and Sumrada, R.A. (1983). What is the function of nitrogen catabolite repression in Saccharomyces cerevisiae? J. Bacteriol. 155, 623–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez, J., and Schnitzer, M. (1979). Purines and pyrimidines in soils and humic substances. Soil Sci. Soc. Am. J. 43, 958–961. [Google Scholar]
- De Konig, H., and Diallinas, G. (2000). Nucleobase transporters. Mol. Membr. Biol. 75, 75–94. [DOI] [PubMed] [Google Scholar]
- Gillissen, B., Bürkle, L., André, B., Kühn, C., Rentsch, D., Brandl, B., and Frommer, W.B. (2000). A new family of high-affinity transporters for adenine, cytosine, and purine derivatives in Arabidopsis. Plant Cell 12, 291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenson, M. (1969). The utilization of exogenous pyrimidines and recycling of the uridine-5′-phosphate derivatives in Saccharomyces cerevisiae, as studied by means of mutants affected in pyrimidine uptake and metabolism. Eur. J. Biochem. 11, 249–260. [DOI] [PubMed] [Google Scholar]
- Güldener, U., Heck, S., Fiedler, T., Beinhauer, J., and Hegemann, J.H. (1996). A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24, 2519–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, A.J., and Zhou, J.J. (2000). Xenopus oocytes as an expression system for plant transporters. Biochim. Biophys. Acta 1465, 343–358. [DOI] [PubMed] [Google Scholar]
- Minet, M., Millington, W.R., and Wurtman, R.J. (1992). Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNA. Plant J. 2, 417–422. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Parniske, M. (2000). Intracellular accommodation of microbes by plants: A common developmental program for symbiosis and disease? Curr. Opin. Plant Biol. 3, 320–328. [DOI] [PubMed] [Google Scholar]
- Pineda, M., and Cardenas, J. (1996). Transport and assimilation of purines in Chlamydomonas reinhardtii. Sci. Mar. 60, 195–201. [Google Scholar]
- Saraste, M., Sibbald, P.R., and Wittinghofer, A. (1990). The P-loop: A common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 15, 430–434. [DOI] [PubMed] [Google Scholar]
- Schubert, K. (1986). Products of biological nitrogen fixation in higher plants: Synthesis, transport and metabolism. Annu. Rev. Plant Physiol. 37, 539–574. [Google Scholar]
- Schulten, H.R., and Schnitzer, M. (1998). The chemistry of soil organic nitrogen: A review. Biol. Fertil. Soils 26, 1–15. [Google Scholar]
- Schultes, N.P., Brutnell, T.P., Allen, A., Dellaporta, S.L., Nelson, T., and Chen, C. (1996). Leaf permease 1 gene of maize is required for chloroplast development. Plant Cell 8, 463–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer, E.L.L., von Heijne, G., and Krogh, A. (1998). A hidden Markov model for predicting transmembrane helices in protein sequences. In Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology, J. Glasgow, T. Littlejohn, F. Major, R. Lathrop, D. Sankoff, and C. Sensen eds (Menlo Park, CA: American Association for Artificial Intelligence Press), pp. 175–182. [PubMed]
- Streeter, J.G. (1993). Translocation: A key factor limiting the efficiency of nitrogen fixation in legume nodules. Physiol. Plant. 87, 616–623. [Google Scholar]
- Stühmer, W. (1998). Electrophysiologic recordings from Xenopus oocytes. Methods Enzymol. 293, 280–300. [DOI] [PubMed] [Google Scholar]
- Von Wirén, N., Gazzarrini, S., Gojon, A., and Frommer, W.B. (2000). The molecular physiology of ammonium uptake and retrieval. Curr. Opin. Plant Biol. 3, 254–261. [PubMed] [Google Scholar]
- Yoo, H.S., Cunningham, T.S., and Cooper, T.G. (1992). The allantoin and uracil permease gene sequences of Saccharomyces cerevisiae are nearly identical. Yeast 8, 997–1006. [DOI] [PubMed] [Google Scholar]
- Ziegler, H. (1975). Nature of transported substances. In Encyclopedia of Plant Physiology. Transport in Plants. I. Phloem Transport, M.H. Zimmermann and J.A. Milburn eds (Berlin, Germany: Springer-Verlag), pp. 59–100.