Abstract
RNA silencing is a sequence-specific RNA degradation process that follows the recognition of double-stranded RNA. Here, we show that virus vectors carrying parts of a green fluorescent protein (GFP) transgene targeted RNA silencing in Nicotiana benthamiana and Arabidopsis against the entire GFP RNA. These results indicate that there was spreading of RNA targeting from the initiator region into the adjacent 5′ and 3′ regions of the target gene. Spreading was accompanied by methylation of the corresponding GFP DNA. It also was dependent on transcription of the transgene and on the putative RNA-dependent RNA polymerase, SDE1/SGS2. These findings indicate that SDE1/SGS2 produces double-stranded RNA using the target RNA as a template. RNA silencing of ribulose-1,5-bisphosphate carboxylase/oxygenase and phytoene desaturase was not associated with the spreading of RNA targeting or DNA methylation, indicating that these endogenous RNAs are not templates for SDE1/SGS2.
INTRODUCTION
RNA silencing is a nucleotide sequence–specific process of RNA degradation in higher plants (post-transcriptional gene silencing), animals (RNA interference [RNAi]), and fungi (quelling) as well as in unicellular eukaryotic algae. In higher plants, a natural role of RNA silencing is to protect against viruses (Covey et al., 1997; Ratcliff et al., 1997; Al-Kaff et al., 1998; Hamilton and Baulcombe, 1999). A role in genome protection also is likely because there is enhanced transposon mobility in RNA silencing–defective mutants of Chlamydomonas reinhardtii and Caenorhabditis elegans and because DNA transposition is suppressed by RNA silencing in Drosophila melanogaster (Jensen et al., 1999; Ketting and Plasterk, 2000; Wu-Scharf et al., 2000).
Double-stranded RNA (dsRNA) is a potent activator of RNA silencing in C. elegans, D. melanogaster, and mammals (Fire et al., 1998; Zamore et al., 2000; Elbashir et al., 2001). Biochemical analyses of RNA silencing in D. melanogaster have shown that an RNase III (DICER) cleaves the dsRNAs into 21- to 25-nucleotide RNAs (siRNAs) that then associate with a second RNase in an RNA-induced silencing complex (RISC) (Hammond et al., 2000; Bernstein et al., 2001). RISC cleaves target single-stranded RNAs (ssRNAs) at a site that is complementary to the (antisense) siRNA. Thus, siRNAs provide sequence specificity to the RNA degradation process (Elbashir et al., 2001). Plant DICER and RISC have not yet been identified. However, siRNAs are present in plants (Hamilton and Baulcombe, 1999), suggesting that mechanisms are conserved across kingdoms.
In plants, RNA silencing is activated by viral RNAs that replicate via double-stranded intermediates and by transgenes with inverted repeat (IR) structures that produce dsRNA (Chuang and Meyerowitz, 2000; Smith et al., 2000; Waterhouse et al., 2001). Single-copy transgenes without IR structures also can activate RNA silencing (Elmayan and Vaucheret, 1996; Jorgensen et al., 1996). In these cases, it is possible that promoters in the transgene and the flanking plant DNA result in the transcription of both strands of the transgene DNA. However, it is possible as well that ssRNA is converted to dsRNA by an RNA-dependent RNA polymerase (RdRP) (Lindbo et al., 1993). In support of this hypothesis, the activity of a putative RdRP encoded by the SDE1/SGS2 locus is required for RNA silencing in Arabidopsis (Dalmay et al., 2000b; Mourrain et al., 2000).
Virus-induced gene silencing (VIGS) is a type of RNA silencing that is initiated by virus vectors carrying portions of host genes (Lindbo et al., 1993; Ruiz et al., 1998). When plant transgenes are targeted by VIGS, sequence-specific methylation of the transgene DNA occurs (Jones et al., 1998, 1999). It is likely that this DNA methylation is directed by RNA–DNA interactions, because it is induced by RNA viruses that do not have DNA intermediates in their replication cycles. The interacting RNA species in RNA-directed DNA methylation (RdDM) could be either dsRNA or siRNA derived from the initiator of silencing (Wassenegger, 2000).
In some cases of VIGS, RNA silencing leads to the elimination of the viral RNA (Lindbo et al., 1993; Ruiz et al., 1998). However, despite the absence of the viral initiator, RNA silencing of the target gene persists (Lindbo et al., 1993; Ruiz et al., 1998; Jones et al., 1999). To explain this initiator-independent silencing, we proposed that there is a maintenance phase of RNA silencing that is distinct from initiation (Ruiz et al., 1998). Initiation requires the viral RNA initiator of silencing, whereas maintenance does not.
In Arabidopsis, these two phases are differentiated by mutation analysis. In wild-type plants, RNA silencing of a green fluorescent protein (GFP) transgene can be initiated by viruses and maintained in the absence of the viral initiator. However, in silencing-deficient (sde) mutants, RNA silencing can be initiated but the transition to maintenance does not occur (Dalmay et al., 2001). Initiation and maintenance also are observed as distinct processes when systemic RNA silencing is initiated by the localized delivery of ectopic DNA into Nicotiana benthamiana and tobacco (Voinnet and Baulcombe, 1997; Voinnet et al., 1998; Palauqui and Balzergue, 1999). The systemic effect is caused by a signal that moves through the plant, and the maintenance phase is inferred from the persistence of silencing after removal of the tissue that received the ectopic DNA (Voinnet et al., 1998). Thus, as with VIGS, the persistence of silencing in the maintenance phase did not require the continued presence of the initiator molecule. Moreover, also as in VIGS of transgenes, the maintenance of systemic silencing was associated with the methylation of the targeted gene (Jones et al., 1999).
These and other examples involving the grafting of a nonsilenced scion onto a silenced stock (Palauqui et al., 1997; Palauqui and Vaucheret, 1998) indicate that initiator-independent maintenance is a feature of many RNA-silencing systems. However, systems also exist in which maintenance does not occur. For example, the VIGS of the phytoene desaturase (PDS) and ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit (Rubisco) endogenous genes was not maintained in the absence of the virus and did not lead to RdDM of the corresponding DNA (Jones et al., 1999; Dalmay et al., 2001; Thomas et al., 2001).
In principle, the initiator-independent maintenance of RNA silencing could be achieved by direct amplification of the initiator dsRNA/siRNA molecules. Alternatively, maintenance could reflect the recruitment of the target gene or its RNA as a source of dsRNA/siRNA. The latter hypothesis seems more likely than direct amplification because an initiator from the 5′ or 3′ part of a target GFP sequence caused systemic RNA silencing to be targeted along the entire length of the transgene transcript (Voinnet et al., 1998). Correspondingly, the entire transcribed region of the GFP transgene was methylated (Jones et al., 1999). Thus, associated with the RNA silencing of a GFP transgene, there is a process that allows the influence of silencing to spread beyond the initiator sequence. This effect is referred to as spreading of RNA targeting because it is similar to the previously described spreading of gene silencing and DNA methylation between adjacent regions of chromosomal DNA (Jahner and Jaenisch, 1985; Jones and Takai, 2001).
Here, we describe experiments that were designed to investigate the relationship of the spreading of RNA targeting and the maintenance of RNA silencing. Our results confirm that the maintenance of RNA silencing involves the spreading of RNA targeting and of DNA methylation in Arabidopsis and N. benthamiana. Target site spreading and maintenance both are features of GFP RNA silencing, but both of them are absent in the RNA silencing of two endogenous genes. We further show that target site spreading is dependent on the SDE1/SGS2 putative RdRP and on the transcription of the target RNA. From these data, we conclude that the maintenance of silencing involves the synthesis of dsRNA by SDE1/SGS2 using the full-length target RNA as a template.
RESULTS
Spreading of siRNA Production
Spreading of RNA targeting has been described previously in systemic RNA silencing that was triggered by DNA bombardment (Voinnet et al., 1998). However, in VIGS, although it is known that DNA methylation spreads beyond the initiator sequence (Jones et al., 1999), the distribution of RNA-silencing targets had not been investigated previously. Therefore, to investigate target site spreading, we performed VIGS of a 35S:GFP:nopaline synthase (NOS) transgene in N. benthamiana (Ruiz et al., 1998) using a viral vector carrying part of a GFP RNA. When the silencing of GFP was established, we challenge-inoculated these plants with an unrelated viral vector that also carried a region of GFP RNA. We anticipated that the second virus would not accumulate if it carried an insert that is a target for the silencing triggered by the first virus.
The initiators of RNA silencing in this experiment were vectors of Tobacco rattle virus (TRV) carrying the 5′ or 3′ half of the GFP coding region (TRV:GF or TRV:P, respectively) or the 3′ untranslated region (TRV:NOS). As controls, plants were infected with TRV without transgene sequence (TRV:00) or with a portion of the promoter (TRV:35S). TRV:00 would not cause silencing of the transgene, whereas TRV:35S would trigger transcriptional silencing of the 35S:GFP transgene (Jones et al., 1999, 2001). By 21 days after inoculation (DAI), there was loss of green fluorescence, indicative of silencing in plants infected with TRV:35S, TRV:GF, TRV:P, and TRV:NOS, whereas plants infected with TRV:00 remained fully green fluorescent (Figure 1A).
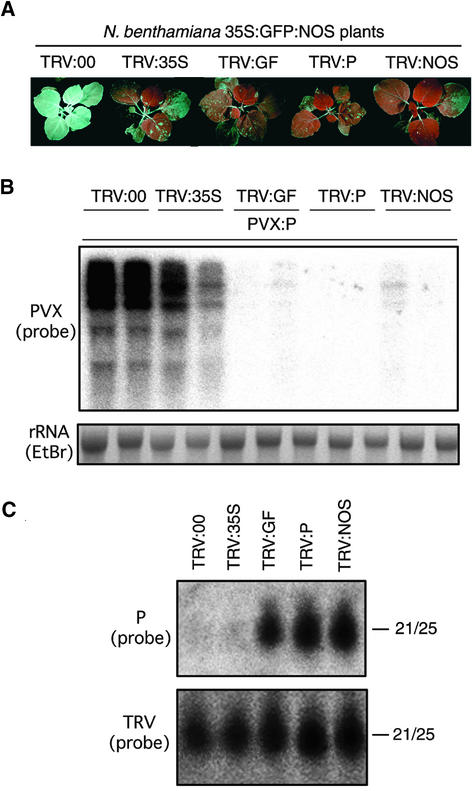
Figure 1.
Spreading of siRNA Production.
(A) N. benthamiana 35S:GFP:NOS transgenic plants (line 16c) infected with TRV:00, TRV:35S, TRV:GF, TRV:P, or TRV:NOS and photographed 21 days later under UV light. Silencing of GFP is evident as red fluorescence, which is caused by the chlorophyll. Infection with TRV:35S induced transcriptional gene silencing of the transgene, and infection with TRV:GF, TRV:P, or TRV:NOS induced post-transcriptional gene silencing. Infection with the TRV vector carrying no sequences of the transgene (TRV:00) did not trigger any GFP silencing.
(B) PVX RNA levels in TRV-infected plants challenge-inoculated with PVX:P. RNA samples were extracted from upper leaves of PVX:P-challenged plants at 10 DAI, and a probe specific for PVX was used in RNA gel blot analysis. At this time, TRV:00-infected plants were not yet showing GFP silencing by PVX:P. Ethidium bromide (EtBr)–stained rRNAs are shown at bottom.
(C) RNA gel blot analysis of siRNAs (21 to 25 nucleotides in length [21/25]) from TRV-inoculated plants at 21 DAI. Sense RNA probes were specific for antisense RNAs corresponding to the P region of GFP (top) or TRV (bottom).
Correspondingly, there was less GFP mRNA in silenced plants than in nonsilenced plants (data not shown). The TRV-infected plants (21 DAI) then were challenge-inoculated with a vector of Potato virus X (PVX) carrying the 3′ half of the GFP sequence (PVX:P), and levels of PVX:P RNA were assessed 10 days later. Figure 1B shows that PVX:P accumulated to high levels in TRV:00- and TRV:35S-infected plants and to low levels in TRV:GF-, TRV:P-, and TRV:NOS-infected plants. Thus, the P region of GFP was a target irrespective of whether the initiator of RNA silencing was GF, P, or NOS. Because there was no overlap of P with either GF or NOS, this result shows that the spreading of RNA targeting is associated with VIGS.
To further investigate the spreading of RNA targeting, we characterized the siRNA population. RNA was isolated from TRV-infected tissue (at 21 DAI), and RNA gel blot analysis was performed using a probe specific for the antisense 3′ part of GFP (the P region). As shown in Figure 1C (top), antisense P-specific siRNAs (P-siRNAs) were present in samples from TRV:GF-, TRV:P-, and TRV:NOS-infected plants but not in TRV:00- or TRV:35S-infected plants. Similarly, GF-siRNAs and NOS-siRNAs were present in samples from TRV:GF-, TRV:P-, and TRV:NOS-infected plants (data not shown). TRV-siRNAs were detected in all of the TRV-infected plants (Figure 1C, bottom), as expected from the finding that RNA silencing is a natural mechanism for virus resistance in plants (Ratcliff et al., 1997, 1999; Hamilton and Baulcombe, 1999).
These combined results demonstrate in a VIGS system that the target sites and the production of siRNA can spread within the transcribed region of the GFP transgene from the initiator region in both 3′ (from GF to P) and 5′ (from NOS to P) directions. Moreover, because NOS-siRNAs were produced after initiation with GF and vice versa, we have shown that spreading of RNA targeting can extend at least through the 332 nucleotides corresponding to the P region of the GFP RNA.
Spreading of RNA Targeting and the PDS and Rubisco Endogenous Genes
VIGS of GFP undergoes the transition from initiation to virus-independent maintenance (Ruiz et al., 1998). In contrast, during VIGS of PDS and Rubisco endogenous genes, there is no maintenance phase, because the silencing phenotype persists only as long as the virus infection (Ruiz et al., 1998; Jones et al., 1999; Thomas et al., 2001). If spreading of RNA silencing is an integral process in maintenance, the silencing of these endogenous genes would not exhibit spreading of RNA targeting and siRNA would correspond only to the initiator region. To test this prediction, we used the same approach we used with the 35S:GFP:NOS transgene. First, we initiated VIGS in N. benthamiana plants with a TRV vector carrying part of the transcribed region of an endogenous gene. Then, when silencing was established, plants were challenge-inoculated with a PVX vector also carrying part of the transcribed region of the same gene. Spreading of RNA targeting would cause plants to be resistant to the challenge inoculum even if there were nonoverlapping inserts in the initiator and challenge inocula.
The TRV vectors used in these experiments were TRV:PD and TRV:S, which carry contiguous but nonoverlapping regions of the N. benthamiana PDS transcribed region. Plants infected with TRV:00 were used as a control for nonspecific effects of the virus inoculations. By 21 DAI, PDS silencing was observed as photobleached tissue in plants infected with TRV:PD or TRV:S, whereas the TRV:00-infected plants remained nonsilenced (Figure 2A). The TRV-infected plants then were challenge-inoculated with the PVX:PD or PVX:S vector (carrying the PSD fragments of TRV:PD and TRV:S, respectively), and the levels of PVX RNA were assessed 4 days later by RNA gel blot analysis.
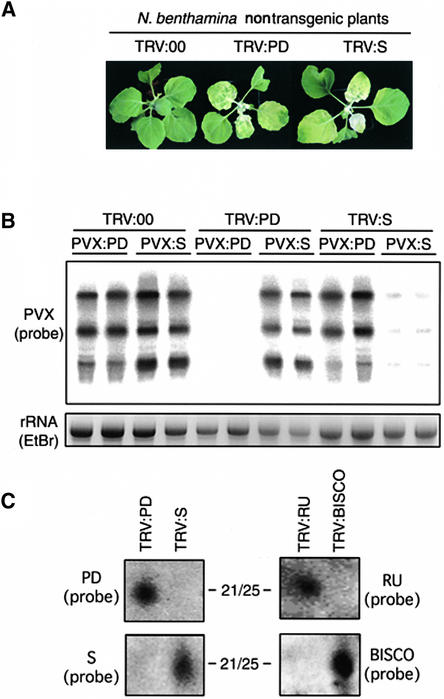
Figure 2.
Spreading and Endogenous Genes.
(A) N. benthamiana nontransgenic plants inoculated with TRV:00, TRV:PD, or TRV:S and photographed at 21 DAI. Only infection with TRV vectors carrying fragments of the PDS cDNA (TRV:PD or TRV:S) induces PDS silencing, which was manifested as photobleaching of the infected tissue.
(B) PVX RNA levels in TRV:00-, TRV:PD-, or TRV:S-infected plants challenge-inoculated with PVX:PD or PVX:S. RNA samples were extracted from PVX-inoculated leaves at 4 DAI, and a probe specific for PVX was used in RNA gel blot analysis. At this time, TRV:00-infected plants were not yet showing PDS silencing by PVX:PD or PVX:S. Ethidium bromide (EtBr)–stained rRNAs are shown at bottom.
(C) RNA gel blot analysis of siRNAs (21 to 25 nucleotides in length [21/25]) from TRV:PD-, TRV:S-, TRV:RU-, or TRV:BISCO-inoculated plants at 21 DAI. Sense RNA probes were specific for antisense RNAs corresponding to the PD, S, RU, and BISCO regions of the PDS and Rubisco genes.
Figure 2B shows that, in TRV:00-infected plants, both PVX:PD and PVX:S accumulated to high levels. In TRV:PD-infected plants, PVX:S accumulated to high levels. In contrast, PVX:PD accumulated to low levels as a consequence of RNA silencing (Ratcliff et al., 1997, 1999). Likewise, in TRV:S-infected plants, PVX:PD accumulated to high levels and PVX:S accumulated to low levels. Similar results were obtained when the endogenous target gene was the highly expressed Rubisco endogenous gene (data not shown). From these results, as predicted, it was inferred that VIGS of PDS or Rubisco does not lead to target site spreading.
To confirm that spreading of RNA targeting had not occurred, we characterized the siRNA population in plants infected with the TRV vectors carrying the Rubisco and PDS inserts. At 21 DAI, PD-siRNAs were present in samples from TRV:PD-infected plants and absent in TRV:S-silenced plants. Likewise, S-siRNAs were detected in TRV:S-infected plants but not in TRV:PD-infected plants (Figure 2C, left). Analogous results were obtained from plants infected with TRV constructs carrying contiguous nonoverlapping fragments from the 5′ (RU) or 3′ (BISCO) regions of the Rubisco transcript. Plants infected with a TRV:RU vector produced only RU-siRNAs, and plants infected with a TRV:BISCO vector produced only BISCO-siRNAs (Figure 2C, right). Therefore, the finding that the silencing target corresponds only to the initiator sequence shows that spreading does not occur if the target genes do not support mainitenance.
Spreading of RNA Targeting and DNA Methylation Requires Transgene Transcription
To determine whether the spreading of RNA targeting and DNA methylation depends on transcription, we silenced the promoter of the 35S:GFP:NOS transgene. This transcriptional gene silencing was induced by infection with TRV:35S, as described previously (Jones et al., 1999, 2001). After 21 days, these silenced plants were inoculated with PVX vectors carrying the 5′ or 3′ half of the GFP sequence (PVX:GF and PVX:P, respectively). siRNA and DNA methylation were assessed 28 days later.
As shown in Figure 3A, both GF- and P-siRNAs were produced in TRV:00-infected plants after PVX:GF or PVX:P inoculation. Thus, the spreading of RNA targeting occurred with PVX vectors, as was observed for TRV vectors, and was not affected by the presence of TRV:00. In contrast, in TRV:35S-infected plants, only GF-siRNAs were detected in PVX:GF-infected tissue, and likewise, only P-siRNAs were detected in PVX:P-infected tissue (Figure 3A). From these results, we conclude that the spreading of RNA targeting requires transcription of the target GFP transgene.
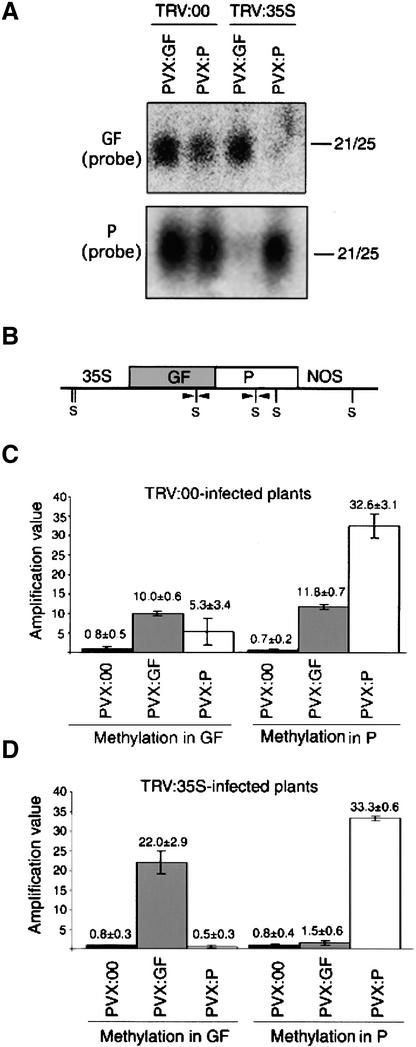
Figure 3.
Spreading Requires Transgene Transcription.
(A) RNA gel blot analysis of siRNAs (21 to 25 nucleotides in length [21/25]) from nontranscriptionally silenced (TRV:00-infected) or transcriptionally silenced (TRV:35S-infected) 35S:GFP:NOS N. benthamiana plants (line 16c) infected with PVX:GF or PVX:P. RNA samples were extracted from upper leaves at 28 days after PVX infection. At this time, TRV:00-infected control plants were showing full GFP silencing by PVX:GF or PVX:P. Sense RNA probes were specific for antisense RNAs corresponding to the GF or P region of GFP (top and bottom, respectively).
(B) Structure of the GFP transgene, including the 35S promoter (35S), the GF and P regions, the NOS terminator, and Sau96I sites (S). Sau96I sites analyzed by TaqMan quantitative PCR are indicated with arrowheads.
(C) and (D) Analysis of DNA methylation within the GF and P regions by Sau96I digestion from TaqMan quantitative PCR. DNA samples were prepared from TRV:00 (C) or TRV:35S (D) plants infected with PVX:00, PVX:GF, or PVX:P at 28 DAI. There is a linear relationship between the amplification values and the amount of amplifiable starting material. Because cytosine methylation inhibits Sau96I digestion, the greater the degree of DNA methylation, the higher the amplification value. Amplification values are the average of three PCRs of three independent DNA samples. Error bars indicate ±sd.
Spreading of DNA methylation was assessed by real-time quantitative polymerase chain reaction (PCR) (TaqMan; PE–Applied Biosystems) of DNA that had been digested with the methylation-sensitive restriction enzyme Sau96I. A TaqMan probe was designed to span a Sau96I site in either GF or P (Figure 3B), and the progression of the PCR reaction was monitored in real time using the TaqMan system. The threshold cycle number, representing the cycle in which fluorescence increases above background, is inversely related to the amount of amplifiable DNA in the initial sample. An amplification value is calculated based on the inverse log of the threshold cycle number and is directly proportional to the amount of amplifiable DNA in the initial sample (see Methods). If the template DNA is unmethylated, it would be digested with Sau96I, and the amplification value would be lower than the value for methylated DNA, which would be undigested.
Figure 3C shows that, after PVX:GF or PVX:P inoculation of TRV:00-infected plants, the methylation of both GF and P DNA was significantly higher than in the negative control (PVX:00-infected) plants. Thus, when the 35S:GFP:NOS transgene is transcribed, DNA methylation spreads from the targeted region to adjacent sequences. It should be emphasized that the TaqMan procedure indicates only relative differences between samples rather than the absolute level of DNA methylation. However, equivalent results were previously obtained by Southern blot analysis (Jones et al., 1999). Thus, the results shown in Figure 3C confirm the previous findings and validate the TaqMan approach for the analysis of relative DNA methylation levels.
When the 35S:GFP:NOS transgene was silenced by TRV:35S, DNA methylation was detected only in the GFP region being targeted by the recombinant PVX vector (Figure 3D). Thus, DNA methylation was restricted to GF after PVX:GF infection and to P after PVX:P infection. Therefore, the spreading of DNA methylation, like the spreading of RNA targeting, is dependent on the transcription of the 35S:GFP:NOS transgene.
Spreading of RNA Targeting Requires SDE1/SGS2
To determine whether the SDE1/SGS2 putative RdRP is required for the spreading of RNA silencing, we analyzed VIGS in wild-type or sde1/sgs2 Arabidopsis plants carrying a 35S:GFP:NOS transgene (Dalmay et al., 2001). RNA silencing was initiated with TRV:GF, and nucleic acid samples were taken for siRNA analysis at 10 to 15 DAI when GFP silencing was evident in both wild-type and mutant plants. In wild-type plants, GFP silencing was maintained throughout the life of the plant. However, as reported previously, GFP silencing was transient in the sde1/sgs2 background, and the leaves were fully green fluorescent at 25 DAI and later (Dalmay et al., 2001).
Figure 4A shows that, at 7 to 10 DAI, GFP mRNA levels were lower in silenced plants than in nonsilenced, mock-inoculated plants. The GF-siRNAs were present in both samples (Figure 4B, bottom), but, corresponding to the level of TRV:GF RNA (Figure 4A), they were more abundant in sde1/sgs2 plants. These results confirm our earlier finding that SDE1/SGS2 is not necessary for siRNA production (Dalmay et al., 2000b). The P-siRNAs were present in TRV:GF-infected wild-type plants (Figure 4B, top), indicating that spreading of siRNA production takes place in Arabidopsis as in N. benthamiana. However, P-siRNAs were not detected in the TRV:GF-infected sde1/sgs2 plants (Figure 4B, top).
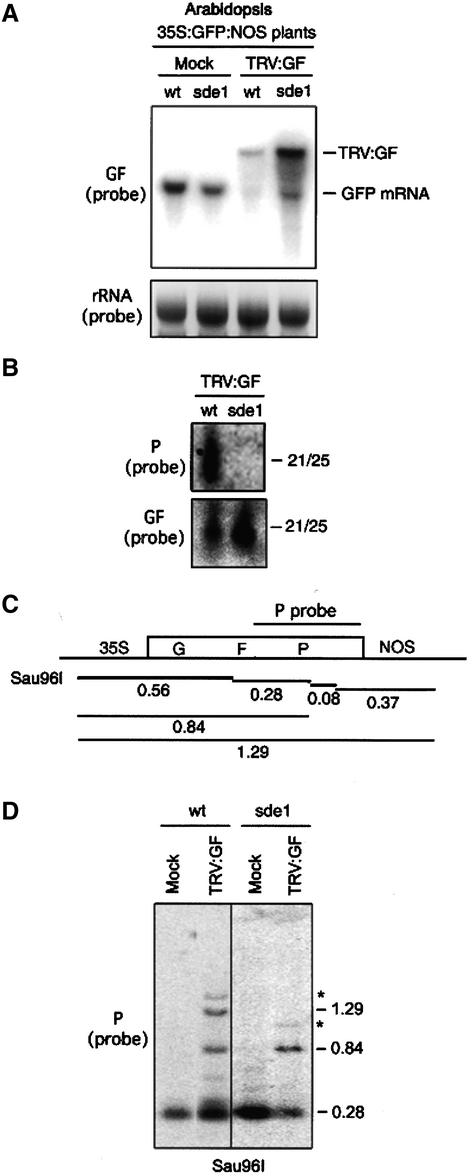
Figure 4.
Spreading Requires SDE1/SGS2.
(A) Detection of GFP and TRV:GF RNAs in mock-inoculated or TRV:GF-inoculated wild-type (wt) or sde1/sgs2 mutant (sde1) 35S:GFP:NOS Arabidopsis plants (top). RNA preparations were made from pools of 10 plants at 7 to 10 DAI, and the probe used was specific to the GF region of the GFP RNA. Ethidium bromide (EtBr)–stained rRNAs are shown at bottom.
(B) RNA gel blot analysis of siRNAs (21 to 25 nucleotides in length [21/25]) in wild-type (wt) and sde1/sgs2 (sde1) TRV:GF-inoculated plants at 7 to 10 DAI. Sense RNA probes were specific for antisense RNAs corresponding to the GF or P region of GFP (bottom and top, respectively).
(C) Scheme of the 35S:GFP:NOS transgene in Arabidopsis. Expected sizes (kb) for total and relevant partial Sau96I restriction enzyme digestions and the P probe used for DNA gel blot analysis are indicated.
(D) DNA gel blot analysis of Sau96I-digested DNA extracted from pooled plants as described in (A). Sizes (kb) of relevant DNA fragments are indicated. Fragments marked with asterisks are attributable to a low level of methylation at the Sau96I site within the 35S promoter.
To analyze the pattern of GFP DNA methylation in the sde1/sgs2 plants, the DNA was digested with Sau96I and hybridized with a P-specific probe. Figure 4C shows the organization of the 35S:GFP:NOS transgene, the location of Sau96I restriction enzyme sites, and the sizes of total and relevant partial digestion products of the GFP transgene. Figure 4D shows that, in mock-inoculated plants, there was a single 0.28-kb P-specific fragment corresponding to the unmethylated GFP DNA. In TRV:GF-infected wild-type plants, the same probe detected 0.28-, 0.84-, and 1.29-kb fragments. The 0.84-kb fragment indicates methylation in the GF region, and the 1.29-kb fragment reflects methylation in both GF and P regions. However, in the TRV:GF-infected sde1/sgs2 plants, the only fragment diagnostic of transgene methylation (0.84 kb) was indicative of methylation in the GF DNA. Thus, the spreading of DNA methylation, like the spreading of targeting, is dependent on SDE1/SGS2. Results leading to the same conclusion were generated with HaeIII-digested DNA as well (data not shown).
DISCUSSION
Spreading of RNA Targeting and Maintenance Mechanisms
From previous analyses, it was concluded that the initiator-independent maintenance of RNA silencing in plants requires the putative RdRP encoded by SDE1/SGS2 (Dalmay et al., 2000b). Previous work also had described a spreading phenomenon in which RNA-silencing target sites and DNA methylation corresponded to all parts of a target transgene transcript irrespective of whether the initiator was only a part of the transgene (Voinnet et al., 1998; Jones et al., 1999). However, there was no conclusive evidence that spreading and the maintenance phase of RNA silencing were related directly. From the data presented here, we show that maintenance is a feature of systems that show spreading of RNA targeting, whereas RNA silencing that does not progress to maintenance is specific for the initiator region. We also show that spreading is dependent on the transcription of the target RNA and the putative RdRP SDE1/SGS2.
These data can be explained if single-stranded GFP transcripts are converted to dsRNA by the putative SDE1/SGS2 RdRP. This double-stranded transgene RNA would be processed by a DICER homolog, and the resulting siRNAs would confer sequence specificity on RISC. The effects of RNA silencing would spread beyond the initiator because the dsRNA synthesized by SDE1/SGS2 would be produced on the transgene RNA template. The siRNAs generated by processing of the dsRNA would target RISC to sequences adjacent to the initiator region. In addition, either siRNAs or dsRNAs would mediate RdDM throughout the transcribed zone of the transgene. According to this mechanism, the spreading of RNA targeting is an inevitable consequence when amplification and maintenance of RNA silencing are attributable to the SDE1/SGS2-mediated synthesis of dsRNA.
Why does SDE1/SGS2 use the transgene RNA as a template only in the presence of the initiator of silencing? One possibility is that siRNAs produced from the initiator anneal to the GFP RNA and serve as primers of dsRNA synthesis. This primer-dependent process was invoked recently to explain the spreading of RNA-silencing target sites in C. elegans and D. melanogaster (Lipardi et al., 2001; Sijen et al., 2001). However, primer-mediated production of dsRNA on a sense RNA template would account for spreading only in the 3′→5′ direction. To explain the observed primer-dependent spreading in the 5′→3′ direction (Figures 1, 3, and 4) (Voinnet et al., 1998), a full-length antisense RNA would be needed as a template for SDE1/SGS2. Formally, we cannot exclude the possibility that these antisense RNAs are present. However, they have not been detected (A. Hamilton, unpublished data), and it is notable that tomato RdRP does not require primers to synthesize RNA in vitro (Schiebel et al., 1993a, 1993b). Therefore, it is possible that a primer-independent mechanism of spreading operates in addition to, or in some systems, instead of, a primer-dependent mechanism.
How could SDE1/SGS2 mediate spreading in a primer-independent manner? One possibility is that sense transcripts interact with the antisense siRNAs produced from the initiator dsRNA molecule. This interaction, irrespective of whether the initiator was from the 5′ or the 3′ region, might change the structure of the RNA or of a ribonucleoprotein complex and thereby allow SDE1/SGS2 to access the 3′ end of the target RNA. Synthesis of dsRNA from the 3′ end would result in siRNA production corresponding to the entire transcript sequence. A second possibility requires that the dsRNA/siRNA from the initiator of silencing interact directly with the DNA transgene and induce changes in the structure of chromatin. Perturbation of transcription caused by the chromatin change could lead to the production of aberrant RNAs that could be templates for SDE1/SGS2.
RNA-Silencing Systems without Spreading of RNA Targeting
To explain the absence of maintenance in the RNA silencing of PDS and Rubisco endogenous genes, we propose that their RNA or DNA is protected from the mechanism that leads to spreading. For example, the endogenous RNAs or their associated proteins could have characteristics that inhibit SDE1/SGS2 or that prevent interactions with the initiator of silencing. Alternatively, if spreading follows from RNA–DNA interactions, it is possible that these endogenous genes are resistant to interactions with RNA or do not exhibit the RNA-mediated changes in chromatin that lead to the perturbation of transcription.
In addition to silencing in these PDS and Rubisco genes, there are examples of transgene RNA silencing that do not exhibit spreading of RNA targeting and DNA methylation. For example, in tobacco plants carrying a β-glucuronidase (GUS):viral RNA chimeric transgene, replication of the viral RNA triggers RNA silencing. However, the siRNAs and DNA methylation correspond only to the viral part of the transgene sequence. They do not correspond to the GUS region, as would be expected if spreading occurred (Wang et al., 2001). In this system, the lack of spreading could be caused by the factors that prevent spreading in PDS and Rubisco RNA silencing. It also could be explained if the replicating viral RNA causes strong silencing of the target RNA. Low levels of the target RNA would mean that the template RNA for the SDE1/SGS2 RdRP is scarce, which would reduce the efficiency of spreading.
A second transgene system without spreading involves GUS-silenced tobacco and Arabidopsis lines in which DNA methylation and the target of silencing are restricted to the 3′ region of the transgene (English et al., 1996; Elmayan et al., 1998). Perhaps there are primary/secondary structure features of the GUS transcript that would prevent SDE1/SGS2 from using the 5′ region as a template. Alternatively, SDE1/SGS2 may be able to synthesize RNA only for a distance of a few hundred nucleotides from the transcript 3′ end. This distance would be a localized region of the ∼1800-nucleotide GUS RNA but would extend over most of the ∼800-nucleotide GFP RNA.
Spreading in Nonplant Systems
The initial analysis of RNAi in animals indicated that, unlike RNA silencing in plants (Voinnet et al., 1998), there was no spreading of RNA targeting (Fire et al., 1998). However, more recently, it has become apparent that there is short-range spreading of RNA targeting associated with RNAi in both C. elegans and D. melanogaster (Lipardi et al., 2001; Sijen et al., 2001). It seems likely that the plant and animal phenomena are mechanistically similar because both are dependent on putative RdRP. However, in animals, the spreading operates over shorter distances than in our plant systems, and only in the 3′→5′ direction. We discussed above how these differences may reflect the existence of primer-dependent and primer-independent mechanisms. However, we cannot say whether this difference reflects fundamental differences in plants and animals. At present, there are only a few examples of spreading, and it remains possible that there are animal systems with primer-mediated spreading in the 3′→5′ direction and primer-independent plant systems operating in both directions.
A second example of target site spreading in an animal system is nonhomologous cosuppression in D. melanogaster (Pal-Bhadra et al., 1999). The initiator sequence and target sequences were transgenes with nonoverlapping but contiguous Adh inserts, and the spreading was dependent on the presence of the endogenous Adh gene. However, this nonhomologous cosuppression appears to be RNA independent and is unlikely to involve an RdRP.
The spreading of silencing target sites and GFP DNA methylation is similar to certain epigenetic phenomena in animals. For example, in heterochromatinization and X chromosome inactivation, there are gene-silencing effects that involve the spreading of DNA methylation from an initiator region (reviewed by Jones and Takai, 2001). Similarly, genome-integrated retroelements nucleate DNA methylation that spreads to the flanking host DNA and mediates silencing of the adjacent genes (Jahner and Jaenisch, 1985). The current models for the spreading of methylation are based on de novo methylation by DNA cytosine methyltransferases (Lindsay and Adams, 1996). However, in the light of our recent demonstration that RdDM can impose a heritable methylation imprint in transgene DNA (Jones et al., 2001), other mechanisms cannot be excluded. It is possible, for example, that spreading of DNA methylation is mediated in mammalian systems by transient production of dsRNA or siRNA. Once established, the imprint could be maintained by DNA cytosine methyltransferases so that continued production of the dsRNA or siRNA would not be necessary.
Conclusion
At present, the evidence that SDE1/SGS2 is an RdRP is based on the analysis of a related protein in tomato (Schiebel et al., 1993a, 1993b). Also, there is no direct evidence that SDE1/SGS2 and homologs have RdRP activity (Baulcombe, 1999). It is imperative, therefore, that the models of silencing mechanisms be reinforced by direct analysis of the enzymatic activity of SDE1/SGS2 and related proteins.
An intriguing question remains regarding the identity of natural targets of SDE1/SGS2 and why a mechanism for the maintenance of RNA silencing exists. Our observation that the spreading of RNA targets occurred on transgenes may reflect a mechanism that allows foreign sequences to be targeted and silenced permanently. However, it has been reported recently that RNA silencing in animals is involved in the regulation of endogenous genes (Aravin et al., 2001; Hutvagner et al., 2001). It will be interesting to determine whether these examples of natural RNA silencing are RdRP dependent and associated with the spreading of RNA targeting.
METHODS
Biomaterials
The Nicotiana benthamiana 16c line and the Arabidopsis thaliana green fluorescent protein (GFP) wild-type and sde1/sgs2 lines were described previously (Ruiz et al., 1998; Dalmay et al., 2000a, 2001). GFP fluorescence was observed under UV light as described previously (Voinnet et al., 1998).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described here that would limit their use for noncommercial research purposes. Requests should follow the procedures available on our World Wide Web site (http://www.sainsbury-laboratory.ac.uk).
Viral Vectors and Virus Inoculations
The primers used in polymerase chain reaction (PCR) of the different regions of the GFP, nopaline synthase (NOS), phytoene desaturase (PDS), and ribulose-1,5-bisphosphate carboxylase/oxygenase genes (and the sizes of the PCR products) are as follows: GFP1 and GFP4 for GF (400 bp), GFP5 and GFP8 for P (332 bp), TNOS1 and TNOS3 for NOS (154 bp), PDS-5-AS and PDS-MID3 for PD (213 bp), PDS-MID5 and PDS-3-AS for S (216 bp), Rub-5-AS and Rub-MID3 for RU (272 bp), and Rub-MID5 and Rub-3-AS for BISCO (250 bp) (see Table 1 for sequences). These fragments were cloned into pGEM-T Easy (Promega), excised with SalI-ApaI, and inserted into SalI-ApaI–digested pTRV.00 (Ratcliff et al., 2001) to produce pTRV:GF, pTRV:P, pTRV:NOS, pTRV:PD, pTRV:S, pTRV:RU, and pTRV:BISCO, respectively. The TRV:35S vector was described previously (Jones et al., 2001). Tobacco rattle virus (TRV) inoculations of N. benthamiana have been described previously (Ratcliff et al., 2001). For Arabidopsis TRV:GF inoculations, 7-day-old seedlings were vacuum-infiltrated with a pTRV:GF/pBINTRA6 Agrobacterium tumefaciens suspension mixture (Ratcliff et al., 2001). pPVX:PD and pPVX:S vectors were obtained by cloning the PD and S fragments into the SmaI site of pGR107 (Jones et al., 1999). PVX:GF and PVX:P vectors were described previously (Ruiz et al., 1998; Voinnet et al., 1998). Potato virus X (PVX) inoculations of N. benthamiana were as described by Ruiz et al. (1998), Voinnet et al. (1998), and Jones et al. (1999).
Table 1.
Oligonucleotide Sequences
| Name | Sequence |
|---|---|
| GFP1 | AGTAAAGGAGAAGAACTTTTCACT |
| GFP4 | TTCCGTCCTCCTTGAAATCGA |
| GFP5 | AACATCCTCGGCCACAAGTT |
| GFP8 | GAGCTCTTAGAGTTCGTCATG |
| TNOS1 | CGTTCAAACATTTGGCAATAA |
| TNOS3 | CTCTAATCATAAAAACCCATC |
| PDS-5-AS | CAGGGATCCGGCACTCAACTTTATAAACC |
| PDS-3-AS | CAGTCTAGATCCCTTCAGTTTTCTGTCAAA |
| PDS-MID5 | AATGAGGATGGAAGTGTCAAAT |
| PDS-MID3 | CAGCTCGATCTTTTTTATTCGT |
| Rub-5-AS | CAGGGATCCTGGCTTCCTCAGTTCTTTCC |
| Rub-3-AS | CAGTCTNGACACTTGACGCACGGGTC |
| Rub-MID5 | GAAAAATGGATGGGTTCCTTG |
| Rub-MID3 | AAAAGGTACTCAACTTCACTA |
| GF-5 | ATACGTGCAGGAGAGGACCATTCT |
| GF-3 | ACGAGGGTGTCTCCCTCAAACT |
| GF-probe | ACGACGGGAACTACAAGACACGTGCTGA |
Nucleic Acid Analysis
RNA was extracted using Tri-reagent (Sigma) according to the manufacturer's instructions. Total RNA was used for both high-molecular-weight RNA and 21- to 25-nucleotide RNA (siRNA) analysis. RNA gel blot analyses were performed as described previously (Jones et al., 1998); siRNA analyses were performed as described (Hamilton and Baulcombe, 1999). For siRNA detection, probes were made by in vitro transcription from pKS (Stratagene) carrying the corresponding fragments cloned into the SmaI site. The TRV probe corresponds to the movement protein (encoded by the TRV RNA1) cloned into pKS. Genomic DNA was extracted using the DNeasy plant DNA extraction kit (Qiagen, Valencia, CA) according to the manufacturer's instructions.
DNA methylation analysis by Sau96I digestion and TaqMan quantitative PCR was performed as described previously (Jones et al., 2001) using DNA prepared from upper leaves. PCR amplification was performed using an ABI Prism 7700 sequence detection system (PE-Applied Biosystems, Foster City, CA), and reactions were performed in triplicate for each sample. Real-time plots were used to determine the threshold cycle number (CT), The CT value then was used to calculate an amplification value, which is related directly to the amount of amplifiable template DNA in the sample (TaqMan PCR protocol; PE–Applied Biosystems, Foster City, CA). For all DNA samples, it was confirmed that the levels of Sau96I digestion were approximately equal by using primer/probe combinations corresponding to DNA with an unmethylated Sau96I site (Jones et al., 2001). Minor variations in the level of digestion were corrected by normalization of CT to CT of the nonmethylated DNA (Jones et al., 2001). Minor corrections allowing for nonspecific effects of Sau96I were based on CT for DNA fragments that do not contain a Sau96I site (Jones et al., 2001). The two primers and the probe used for the analysis of the GF region were GF-5, GF-3, and GF probe, respectively (see Table 1 for sequences). The primers and probes used for analysis of the P region have been described (Jones et al., 2001).
Acknowledgments
We thank all members of the laboratory for providing valuable comments on aspects of the manuscript and especially Olivier Voinnet for his input in this work. Bart Feys provided help and advice about the TaqMan system. We also thank Mike Hill and his team for excellent plant care. Funding for this work was provided by the Gatsby Charitable Foundation. Use of genetically modified plant viruses was licensed by Department of Environment, Food and Rural Affairs license PHL 24B/3654 (3/2001).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010480.
References
- Al-Kaff, N.S., Covey, S.N., Kreike, M.M., Page, A.M., Pinder, R., and Dale, P.J. (1998). Transcriptional and posttranscriptional plant gene silencing in response to a pathogen. Science 279, 2113–2115. [DOI] [PubMed] [Google Scholar]
- Aravin, A.A., Naumova, N.M., Tulin, A.V., Vagin, V.V., Rozovsky, Y.M., and Gvozdev, V.A. (2001). Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11, 1017–1027. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D.C. (1999). Gene silencing: RNA makes RNA makes no protein. Curr. Biol. 9, R599–R601. [DOI] [PubMed] [Google Scholar]
- Bernstein, E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366. [DOI] [PubMed] [Google Scholar]
- Chuang, C.-H., and Meyerowitz, E.M. (2000). Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97, 4985–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covey, S.N., Al-Kaff, N.S., Langara, A., and Turner, D.S. (1997). Plants combat infection by gene silencing. Nature 385, 781–782. [Google Scholar]
- Dalmay, T., Hamilton, A.J., Mueller, E., and Baulcombe, D.C. (2000. a). Potato virus X amplicons in Arabidopsis mediate genetic and epigenetic gene silencing. Plant Cell 12, 369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmay, T., Hamilton, A.J., Rudd, S., Angell, S., and Baulcombe, D.C. (2000. b). An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101, 543–553. [DOI] [PubMed] [Google Scholar]
- Dalmay, T.D., Horsefield, R., Braunstein, T.H., and Baulcombe, D.C. (2001). SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J. 20, 2069–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., Lendeckel, W., and Tuschl, T. (2001). RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15, 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmayan, T., and Vaucheret, H. (1996). Expression of single copies of a strongly expressed 35S transgene can be silenced post-transcriptionally. Plant J. 9, 787–797. [Google Scholar]
- Elmayan, T., Balzergue, S., Beon, F., Bourdon, V., Daubremet, J., Guenet, Y., Mourrain, P., Palauqui, J.C., Vernhettes, S., Vialle, T., Wostrikoff, K., and Vaucheret, H. (1998). Arabidopsis mutants impaired in cosuppression. Plant Cell 10, 1747–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English, J.J., Mueller, E., and Baulcombe, D.C. (1996). Suppression of virus accumulation in transgenic plants exhibiting silencing of nuclear genes. Plant Cell 8, 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Hamilton, A.J., and Baulcombe, D.C. (1999). A novel species of small antisense RNA in post-transcriptional gene silencing. Science 286, 950–952. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M., Bernstein, E., Beach, D., and Hannon, G. (2000). An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cell extracts. Nature 404, 293–296. [DOI] [PubMed] [Google Scholar]
- Hutvagner, G., McLachlan, J., Pasquinelli, A.E., Balint, E., Tuschl, T., and Zamore, P.D. (2001). A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293, 834–838. [DOI] [PubMed] [Google Scholar]
- Jahner, D., and Jaenisch, R. (1985). Retrovirus-induced de novo methylation of flanking host sequences correlates with gene inactivity. Nature 315, 594–597. [DOI] [PubMed] [Google Scholar]
- Jensen, S., Gassama, M.P., and Heidmann, T. (1999). Taming of transposable elements by homology-dependent gene silencing. Nat. Genet. 21, 209–212. [DOI] [PubMed] [Google Scholar]
- Jones, A.L., Thomas, C.L., and Maule, A.J. (1998). De novo methylation and co-suppression induced by a cytoplasmically replicating plant RNA virus. EMBO J. 17, 6385–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, L., Hamilton, A.J., Voinnet, O., Thomas, C.L., Maule, A.J., and Baulcombe, D.C. (1999). RNA–DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell 11, 2291–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, L., Ratcliff, F., and Baulcombe, D.C. (2001). RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr. Biol. 11, 747–757. [DOI] [PubMed] [Google Scholar]
- Jones, P.A., and Takai, D. (2001). The role of DNA methylation in mammalian epigenetics. Science 293, 1068–1070. [DOI] [PubMed] [Google Scholar]
- Jorgensen, R.A., Cluster, P.D., English, J.J., Que, Q., and Napoli, C.A. (1996). Chalcone synthase cosuppression phenotypes in petunia flowers: Comparison of sense vs. antisense constructs and single-copy vs. complex T-DNA sequences. Plant Mol. Biol. 31, 957–973. [DOI] [PubMed] [Google Scholar]
- Ketting, R.F., and Plasterk, R.H.A. (2000). A genetic link between co-suppression and RNA interference in C. elegans. Nature 404, 296–298. [DOI] [PubMed] [Google Scholar]
- Lindbo, J.A., Silva-Rosales, L., Proebsting, W.M., and Dougherty, W.G. (1993). Induction of a highly specific antiviral state in transgenic plants: Implications for regulation of gene expression and virus resistance. Plant Cell 5, 1749–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay, H., and Adams, R.L.P. (1996). Spreading of methylation along DNA. Biochem. J. 320, 473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipardi, C., Wei, Q., and Paterson, B.M. (2001). RNAi as random degradative PCR: siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell 107, 297–307. [DOI] [PubMed] [Google Scholar]
- Mourrain, P., et al. (2000). Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101, 533–542. [DOI] [PubMed] [Google Scholar]
- Palauqui, J.-C., and Balzergue, S. (1999). Activation of systematic acquired silencing by localised introduction of DNA. Curr. Biol. 9, 59–66. [DOI] [PubMed] [Google Scholar]
- Palauqui, J.-C., and Vaucheret, H. (1998). Transgenes are dispensable for the RNA degradation step of cosuppression. Proc. Natl. Acad. Sci. USA 95, 9675–9680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palauqui, J.-C., Elmayan, T., Pollien, J.-M., and Vaucheret, H. (1997). Systemic acquired silencing: Transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 16, 4738–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhadra, M., Bhadra, U., and Birchler, J.A. (1999). Cosuppression of nonhomologous transgenes in Drosophila involves mutually related endogenous sequences. Cell 99, 35–46. [DOI] [PubMed] [Google Scholar]
- Ratcliff, F., Harrison, B.D., and Baulcombe, D.C. (1997). A similarity between viral defense and gene silencing in plants. Science 276, 1558–1560. [DOI] [PubMed] [Google Scholar]
- Ratcliff, F., MacFarlane, S., and Baulcombe, D.C. (1999). Gene silencing without DNA: RNA-mediated cross protection between viruses. Plant Cell 11, 1207–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff, F., Martin-Hernandez, A.M., and Baulcombe, D.C. (2001). Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 25, 237–245. [DOI] [PubMed] [Google Scholar]
- Ruiz, M.T., Voinnet, O., and Baulcombe, D.C. (1998). Initiation and maintenance of virus-induced gene silencing. Plant Cell 10, 937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiebel, W., Haas, B., Marinkovic, S., Klanner, A., and Sanger, H.L. (1993. a). RNA-directed RNA polymerase from tomato leaves. I. Purification and physical properties. J. Biol. Chem. 268, 11851–11857. [PubMed] [Google Scholar]
- Schiebel, W., Haas, B., Marinkovic, S., Klanner, A., and Sanger, H.L. (1993. b). RNA-directed RNA polymerase from tomato leaves. II. Catalytic in vitro properties. J. Biol. Chem. 268, 11858–11867. [PubMed] [Google Scholar]
- Sijen, T., Fleenor, J., Simmer, F., Thijssen, K.L., Parrish, S., Timmons, L., Plasterk, R.H.A., and Fire, A. (2001). On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107, 465–476. [DOI] [PubMed] [Google Scholar]
- Smith, N.A., Singh, S.P., Wang, M.B., Stoutjesdijk, P.A., Green, A.G., and Waterhouse, P.M. (2000). Total silencing by intron-spliced hairpin RNAs. Nature 407, 319–320. [DOI] [PubMed] [Google Scholar]
- Thomas, C.L., Jones, L., Baulcombe, D.C., and Maule, A.J. (2001). Size constraints for targeting post-transcriptional gene silencing and for RNA-directed methylation in Nicotiana benthamiana using a potato virus X vector. Plant J. 25, 417–425. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., and Baulcombe, D.C. (1997). Systemic signalling in gene silencing. Nature 389, 553. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., Vain, P., Angell, S., and Baulcombe, D.C. (1998). Systemic spread of sequence-specific transgene RNA degradation is initiated by localised introduction of ectopic promoterless DNA. Cell 95, 177–187. [DOI] [PubMed] [Google Scholar]
- Wang, M.B., Wesley, S.V., Finnegan, E.J., Smith, N.A., and Waterhouse, P.M. (2001). Replicating satellite RNA induces sequence-specific DNA methylation and truncated transcripts in plants. RNA 7, 16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger, M. (2000). RNA-directed DNA methylation. Plant Mol. Biol. 43, 203–220. [DOI] [PubMed] [Google Scholar]
- Waterhouse, P.M., Wang, M.B., and Lough, T. (2001). Gene silencing as an adaptive defence against viruses. Nature 411, 834–842. [DOI] [PubMed] [Google Scholar]
- Wu-Scharf, D., Jeong, B.-r., Zhang, C., and Cerutti, H. (2000). Transgene and transposon silencing in Chlamydomonas reinhardtii by a DEAH-box RNA helicase. Science 290, 1159–1162. [DOI] [PubMed] [Google Scholar]
- Zamore, P.D., Tuschl, T., Sharp, P.A., and Bartel, D.P. (2000). RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101, 25–33. [DOI] [PubMed] [Google Scholar]