Abstract
Cyclin-dependent kinases and cyclins regulate with the help of different interacting proteins the progression through the eukaryotic cell cycle. A high-quality, homology-based annotation protocol was applied to determine the core cell cycle genes in the recently completed Arabidopsis genome sequence. In total, 61 genes were identified belonging to seven selected families of cell cycle regulators, for which 30 are new or corrections of the existing annotation. A new class of putative cell cycle regulators was found that probably are competitors of E2F/DP transcription factors, which mediate the G1-to-S progression. In addition, the existing nomenclature for cell cycle genes of Arabidopsis was updated, and the physical positions of all genes were compared with segmentally duplicated blocks in the genome, showing that 22 core cell cycle genes emerged through block duplications. This genome-wide analysis illustrates the complexity of the plant cell cycle machinery and provides a tool for elucidating the function of new family members in the future.
INTRODUCTION
Cell proliferation is controlled by a universally conserved molecular machinery in which the core key players are Ser/Thr kinases, known as cyclin-dependent kinases (CDKs). CDK activity is regulated in a complex manner, including phosphorylation/dephosphorylation by specific kinases/phosphatases and association with regulatory proteins. Although many cell cycle genes of plants have been identified in the last decade (for review, see Stals and Inzé, 2001), the correct number of CDKs, cyclins, and interacting proteins with a role in cell cycle control is unknown. Now that the complete sequence of the nuclear genome of Arabidopsis is available (Arabidopsis Genome Initiative, 2000), it is possible to scan an entire plant genome for all of these core cell cycle genes and determine their numbers, positions on the chromosomes, and phylogenetic relationships. From an evolutionary point of view, this core cell cycle gene catalog would be extremely interesting because it allows us to determine which processes are specific to plants and which are conserved among all eukaryotes. Furthermore, there is a unique opportunity to unravel in future experiments the functions and interactions of newly found family members of primary cell cycle regulators, thus expanding our knowledge of how the cell cycle is regulated in plants.
Nevertheless, a genome-wide inventory of all core cell cycle genes is possible only when the available raw sequence data are annotated correctly. Although genome-wide annotations of organisms sequenced by large consortia have produced huge amounts of information that benefits the scientific community, this automated high-throughput annotation is far from optimal (Devos and Valencia, 2001). For this reason, it is not easy to extract clear biological information from these public databases. When high-quality annotation is needed, a supervised semiautomatic annotation may be a good compromise between quality and speed.
Generally, annotation is performed in two steps: first, structural annotation, which aims to find and characterize biologically relevant elements within the raw sequence (such as exons and translation starts); and second, functional annotation, in which biological information is attributed to the gene or its elements. Unfortunately, there are some problems inherent to both.
When structural annotation is performed, the first problem occurs when no cDNA or expressed sequence tag (EST) information is available, which is the case for 60% of all Arabidopsis genes (Arabidopsis Genome Initiative, 2000). Then, one has to resort to intrinsic gene prediction software, which remains limited, although much improvement has been made in the last few years. Errors range from wrongly determined splice sites or start codons, to so-called spliced (one gene predicted as two) or fused (two genes predicted as one) genes, to completely missed or nonexistent predicted genes (Rouzé et al., 1999). In addition, no general and well-defined prediction protocol is used by the different annotation centers, which results in the generation of redundant, nonuniform structural annotations. Furthermore, clear information is lacking on the methods and programs used as well as the motivation for applying special protocols, making it impossible to trace the annotation process.
The problem with functional annotation is related to the difficulty of linking biological knowledge to a gene. Such a link is made generally on the basis of sequence similarity that is derived either from full-length sequence comparisons or by means of multiple alignments, patterns, and domain searches. Of major concern is the origin of the assigned function, because the transfer of low-quality or faulty functional annotation information propagates incorrect annotations in the public databases. Even correct annotations can be disseminated erroneously: one can easily imagine the transfer of a good functional assignment from a multidomain protein to a protein that has only one of the domains. This problem can be avoided using only experimentally derived information to predict unambiguously a gene's structure and function.
Here, we applied a homology-based annotation using experimental references to build a full catalog with 61 core cell cycle genes of Arabidopsis. In total, 30 genes are either new or genes for which the previous annotation was incorrect. Based on phylogenetic analysis, we updated and rationalized their nomenclature. Furthermore, relations between gene family members were correlated with large segmental duplications.
RESULTS
Strategy
To correctly annotate all core cell cycle genes, a strategy was defined that uses as much reliable information as possible, combining experimentally derived data with the best prediction tools available for Arabidopsis (see Methods). First, experimental representatives for each family were used as bait to locate regions of interest on the different chromosomes. For these selected regions, genes were predicted and candidate genes were validated; the presence of mandatory domains in their gene products was determined by aligning them with the experimental representatives; if necessary, the predicted gene structure was modified using the family-related characteristics or ESTs. In some cases, however, this approach did not allow us to conclude whether a region of interest really coded for a potential gene or whether a candidate gene was a core cell cycle gene.
To clarify such situations, a more integrated analysis was performed. First, the members of every family were used to build a profile for that specific family. By taking the new predicted genes into account when creating the profile, a more “flexible” (i.e., all diversity within a class/subclass being represented) and plant-specific profile could be established. With this new profile, novel family members were sought within a collection of genome-wide predicted Arabidopsis proteins. Subsequently, the predicted gene products were again validated or modified by comparing them with those of other family members in a multiple alignment. With this additional approach, we could determine clearly whether the predicted genes were similar to a certain class of cell cycle genes.
To characterize subclasses within the gene families, phylogenetic trees were generated that included reference cell cycle genes from other plants and known genes from Arabidopsis. By different methods and statistical analysis of nodes, the significance of the derived classification was tested. Based on the position on the tree and the presence of class-specific signatures, genes were named according to the proposed nomenclature rules for cell cycle genes (Renaudin et al., 1996; Joubès et al., 2000). A complete list of core cell cycle genes in Arabidopsis is presented in Table 1. Additional data regarding nomenclature and gene models can be found at http://www.plantgenetics.rug.ac.be/bioinformatics/coreCC/.
Table 1.
Characteristics of All 61 Core Cell Cycle Genes in Arabidopsis
| Gene | Chromosome | Starta | Stopb | Strand | Statusc | Featuresd | Open Reading Frame Name |
|---|---|---|---|---|---|---|---|
| Arath;CDKA;1 | 3 | 18,368,303 | 18,370,279 | + | EXP | PSTAIRE | AT3g48750 |
| Arath;CDKB1;1 | 3 | 20,355,861 | 20,357,226 | + | EXP | PPTALRE | AT3g54180 |
| Arath;CDKB1;2 | 2 | 16,301,446 | 16,302,758 | + | EXP | PPTALRE | AT2g38620 |
| Arath;CDKB2;1 | 1 | 28,430,923 | 28,429,129 | − | EXP | PSTTLRE | AT1g76540 |
| Arath;CDKB2;2 | 1 | 7,294,679 | 7,292,770 | − | EXP | PPTTLRE | AT1g20930 |
| Arath;CDKC;1 | 5 | 3,224,679 | 3,221,723 | − | AI993037 | PITAIRE | AT5g10270 |
| Arath;CDKC;2 | 5 | 25,955,460 | 25,958,387 | + | AV439592 | PITAIRE | AT5g64960 |
| Arath;CDKD;1 | 1 | 27,423,792 | 27,425,694 | + | PRED | NVTALRE | AT1g73690 |
| Arath;CDKD;2 | 1 | 24,603,461 | 24,605,698 | + | AV554642 | NFTALRE | AT1g66750 |
| Arath;CDKD;3 | 1 | 6,206,888 | 6,209,316 | − | AF344314 | NITALRE | AT1g18040 |
| Arath;CDKE;1 | 5 | 25,465,021 | 25,463,612 | − | BG459367 | SPTAIRE | AT5g63610 |
| Arath;CDKF;1 | 4 | 13,494,330 | 13,495,958 | + | EXP | None | AT4g28980 |
| Arath;CYCA1;1 | 1 | 16,354,762 | 16,352,618 | − | AV556475 | LVEVxEEY | AT1g44110 |
| Arath;CYCA1;2 | 1 | 28,792,710 | 28,790,480 | − | PRED | LVEVxEEY | AT1g77390 |
| Arath;CYCA2;1 | 5 | 8,885,657 | 8,887,990 | + | EXP | LVEVxEEY | AT5g25380 |
| Arath;CYCA2;2 | 5 | 3,604,472 | 3,601,820 | − | EXP | LVEVxDDY | AT5g11300 |
| Arath;CYCA2;3 | 1 | 5,363,054 | 5,365,235 | + | EXPe | LVEVxEEY | AT1g15570 |
| Arath;CYCA2;4 | 1 | 29,923,266 | 29,925,430 | + | AV558333 | LVEVxEEY | AT1g80370 |
| Arath;CYCA3;1 | 5 | 17,293,193 | 17,294,681 | + | PRED | LVEVxEEY | AT5g43080 |
| Arath;CYCA3;2 | 1 | 17,022,212 | 17,023,757 | + | AT50514 | LVEVxEEY | AT1g47210 |
| Arath;CYCA3;3 | 1 | 17,024,852 | 17,026,370 | + | PRED | LVEVxEEY | AT1g47220 |
| Arath;CYCA3;4 | 1 | 17,027,927 | 17,029,762 | + | PRED | LVEVxEEY | AT1g47230 |
| Arath;CYCB1;1 | 4 | 16,830,051 | 16,827,976 | − | EXP | HxRF | AT4g37490 |
| Arath;CYCB1;2 | 5 | 1,861,577 | 1,859,551 | − | EXP | HxKF | AT5g06150 |
| Arath;CYCB1;3 | 3 | 3,627,150 | 3,625,489 | − | EXPf | HxKF | AT3g11520 |
| Arath;CYCB1;4 | 2 | 11,548,850 | 11,552,088 | + | PRED | HxKF | AT2g26760 |
| Arath;CYCB2;1 | 2 | 7,813,050 | 7,815,144 | + | EXP | HxKF | AT2g17620 |
| Arath;CYCB2;2 | 4 | 16,107,598 | 16,109,617 | + | EXP | HxKF | AT4g35620 |
| Arath;CYCB2;3 | 1 | 7,137,288 | 7,135,091 | − | PRED | HxKF | AT1g20610 |
| Arath;CYCB2;4 | 1 | 28,338,772 | 28,336,622 | − | PRED | HxKF | AT1g76310 |
| Arath;CYCB3;1 | 1 | 5,584,476 | 5,582,409 | − | PRED | HxKF | AT1g16330 |
| Arath;CYCD1;1 | 1 | 26,148,702 | 26,150,664 | + | EXP | LxCxE | AT1g70210 |
| Arath;CYCD2;1 | 2 | 9,704,757 | 9,703,043 | − | EXP | LxCxE | AT2g22490 |
| Arath;CYCD3;1 | 4 | 15,563,758 | 15,565,156 | + | EXP | LxCxE | AT4g34160 |
| Arath;CYCD3;2 | 5 | 26,836,277 | 26,837,626 | + | AI995751 | LxCxE | AT5g67260 |
| Arath;CYCD3;3 | 3 | 18,862,632 | 18,861,289 | − | AV527915 | LxCxE | AT3g50070 |
| Arath;CYCD4;1 | 5 | 26,143,713 | 26,141,558 | − | EXP | LxCxE | AT5g65420 |
| Arath;CYCD4;2 | 5 | 3,282,347 | 3,280,801 | + | PRED | no LxCxE | AT5g10440 |
| Arath;CYCD5;1 | 4 | 16,885,341 | 16,886,338 | + | AI998509 | LFLCxE | AT4g37630 |
| Arath;CYCD6;1 | 4 | 1,432,497 | 1,431,184 | − | PRED | no LxCxE | AT4g03270 |
| Arath;CYCD7;1 | 5 | 417,084 | 418,547 | + | PRED | LxCxE | AT5g02110 |
| Arath;CYCH;1 | 5 | 9,813,161 | 9,816,075 | + | AV560893 | None | AT5g27620 |
| Arath;CKS1 | 2 | 12,060,430 | 12,059,793 | − | EXP | None | AT2g27960 |
| Arath;CKS2 | 2 | 12,061,999 | 12,061,350 | − | AV553882 | None | AT2g27970 |
| Arath;DEL1 | 3 | 18,079,607 | 18,081,809 | + | EXP | None | AT3g48160 |
| Arath;DEL2 | 5 | 4,858,640 | 4,861,044 | + | PRED | None | AT5g14960 |
| Arath;DEL3 | 3 | 126,812 | 124,606 | − | EXP | None | AT3g01330 |
| Arath;DPa | 5 | 544,155 | 844,977 | − | EXP | None | AT5g02470 |
| Arath;DPb | 5 | 842,841 | 845,196 | + | EXP | None | AT5g03410 |
| Arath;E2Fa | 2 | 15,268,582 | 15,271,784 | + | EXP | None | AT2g36010 |
| Arath;E2Fb | 5 | 7,431,826 | 7,434,541 | + | EXP | None | AT5g22220 |
| Arath;E2Fc | 1 | 17,356,113 | 17,358,730 | + | EXP | None | AT1g47870 |
| Arath;KRP1 | 2 | 10,126,806 | 10,125,908 | − | EXP | None | AT2g23430 |
| Arath;KRP2 | 3 | 19,096,470 | 19,097,325 | + | EXP | None | AT3g50630 |
| Arath;KRP3 | 5 | 19,794,310 | 19,792,575 | − | EXP | None | AT5g48820 |
| Arath;KRP4 | 2 | 14,022,387 | 14,024,238 | + | EXP | None | AT2g32710 |
| Arath;KRP5 | 3 | 9,060,905 | 9,061,654 | + | EXP | None | AT3g24810 |
| Arath;KRP6 | 3 | 6,617,597 | 6,616,567 | − | EXP | None | AT3g19150 |
| Arath;KRP7 | 1 | 18,087,625 | 18,086,761 | − | EXP | None | AT1g49620 |
| Arath;Rb | 3 | 3,919,344 | 3,913,685 | − | AF245395 | None | AT3g12280 |
| Arath;WEE1 | 1 | 673,409 | 676,125 | + | EXPg | None | AT1g02970 |
Position of start codon on the chromosome.
Position of stop codon on the chromosome.
Expression status of the gene. EXP, experimentally characterized; PRED, prediction. Numbers are EST accession numbers.
Family-specific protein signatures.
EST BE528080 found for the first exon completes the structural annotation.
Gene structure was determined using two cDNA sequences, confirming the manual annotation.
Annotation and Nomenclature
CDK
In yeast, one CDK is sufficient to drive cells through all cell cycle phases, whereas multicellular organisms evolved to use a family of related CDKs, all with specific functions. In plants, two major classes of CDKs, known as A-type and B-type CDKs, have been studied to date. The A-type CDKs regulate both the G1-to-S and G2-to-M transitions, whereas the B-type CDKs seem to control the G2-to-M checkpoint only (Hemerly et al., 1995; Magyar et al., 1997; Porceddu et al., 2001). In addition, the presence of C-type CDKs and CDK-activating kinases (CAKs) have been reported (Magyar et al., 1997; Umeda et al., 1998; Joubès et al., 2001). Whereas the latter were shown to regulate the activity of the A-type CDKs, the function of the C-type CDKs remains unknown. To date, one A-type and four B-type CDKs have been described for Arabidopsis (Joubès et al., 2000; Boudolf et al., 2001). Furthermore, C-type CDKs and one CAK have been reported as well (Umeda et al., 1998; Lessard et al., 1999). In alfalfa, one E-type CDK has been identified, but no counterparts had been found previously in Arabidopsis (Magyar et al., 1997). By the homology-based annotation method used here, we identified eight CDKs (one A type, four B type, two C type, and one E type) and four CAKs (three D type and one F type).
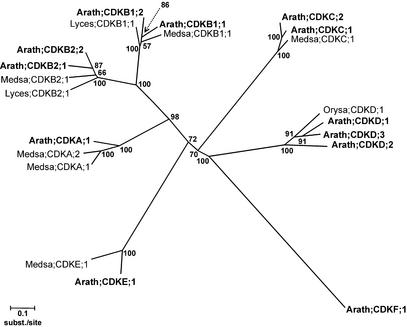
The previously described CAK homolog of Arabidopsis (cak1At) differs substantially from the known rice CAK, R2 (Umeda et al., 1998; Yamaguchi et al., 1998). R2 has been suggested to be specific for monocots (Yamaguchi et al., 1998). However, with the rice sequence as an experimental reference, three related sequences were identified in Arabidopsis, designated CDKD;1, CDKD;2, and CDKD;3, with 75, 68, and 79% sequence similarity at the protein level to R2 from rice, respectively. These genes are only distantly related to cak1At, indicating that Arabidopsis has two functional classes of CAK. To stress this functional difference and to have a more uniform nomenclature, cak1At was renamed CDKF;1. The phylogenetic relationships among CDKs of Arabidopsis are shown in Figure 1.
Figure 1.
Unrooted Neighbor-Joining Tree of the A, B, C, D, E, and F Classes of CDKs with the Poisson Correction for Evolutionary Distance Calculation.
Bootstrap values of 500 bootstrap iterations are shown. Numbers indicate evolutionary distance. Arath, Arabidopsis; Lyces, tomato (Lycopersicon esculentum); Medsa, alfalfa (Medicago sativa); Orysa, rice (Oryza sativa). Reference genes are Medsa;CDKC;1, Orysa;CDKD;1, Medsa;CDKE;1, Medsa;CDKA;1, Medsa;CDKA;2, Medsa;CDKB1;1, Lyces;CDKB1;1, Lyces;CDKB2;1, and Medsa;CDKB2;1.
Cyclins
Monomeric CDKs have no kinase activity and must associate with regulatory proteins called cyclins to be activated. Because cyclin protein levels fluctuate in the cell cycle, cyclins are the major factors that determine the timing of CDK activation. Cyclins can be grouped into mitotic cyclins (designated A- and B-type cyclins in higher eukaryotes and CLBs in budding yeast) and G1-specific cyclins (designated D-type cyclins in mammals and CLNs in budding yeast). H-type cyclins regulate the activity of the CAKs. All four types of cyclins known in plants were identified, mostly by analogy to their human counterparts. For Arabidopsis, at present, four A-type, five B-type, five D-type, but no H-type cyclins have been described (Soni et al., 1995; Renaudin et al., 1996; De Veylder et al., 1999; Swaminathan et al., 2000). Using the known plant cyclin sequences as probes, a total of 30 cyclins were detected in the Arabidopsis genome. For 19 cyclins, an EST was found (Table 1).
Three different subclasses of plant A-type cyclins (A1, A2, and A3) have been described (Renaudin et al., 1996), and all of them were found in Arabidopsis, comprising 10 cyclins. Two A1-type genes (CYCA1;1 and CYCA1;2), four A2-type genes (CYCA2;1, CYCA2;2, CYCA2;3, and CYCA2;4), and four A3-type genes (CYCA3;1, CYCA3;2, CYCA3;3, and CYCA3;4) were detected.
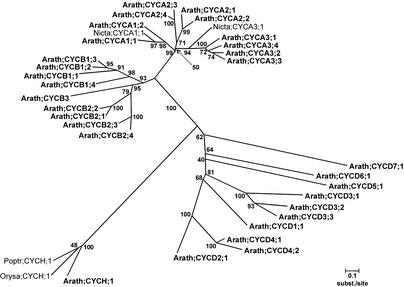
B-type cyclins are subdivided into two subclasses, B1 and B2. In total, Arabidopsis contains nine B-type cyclins, of which four belong to the B1 class (CYCB1;1, CYB1;2, CYCB1;3, and CYCB1;4) and four belong to the B2 class (CYCB2;1, CYCB2;2, CYCB2;3, and CYCB2;4). One gene could not be attributed to either the B1 or the B2 class, although it clearly contained a B-type–like cyclin box in combination with the B-type–specific HxKF signature. On the other hand, no B1- or B2-like destruction box was detected. The phylogenetic position of this gene within the B cluster depended on the number of positions used for the analysis. Because cyclin sequences are known to be saturated with substitutions (Renaudin et al., 1996), a technique was applied to construct trees on unsaturated positions only (Van de Peer et al., 2002). No support was found for assigning this gene to one of the two classes of B-type cyclins (data not shown). Thus, it seemed justified to create a new subclass of cyclins, the B3 type (Figure 2).
Figure 2.
Unrooted Neighbor-Joining Tree of the A, B, D, and H Subgroups of the Cyclin Family with Poisson Correction for Evolutionary Distance Calculation.
Bootstrap values of 500 bootstrap iterations are shown. Scales indicate evolutionary distance. Arath, Arabidopsis; Nicta, tobacco (Nicotiana tabacum); Orysa, rice; Poptr, poplar (Populus tremula × Populus tremuloides). Reference genes are Nicta;CYCA1;1, Nicta;CYCA3;1, Poptr;CYCH, and Orysa;CYCH.
In addition to the five D-type cyclins described previously (CYCD1;1, CYCD2;1, CYCD3;1, CYCD3;2, and CYCD4;1), five new D-type genes were detected. Based on their phylogenetic positions, two of these genes were assigned to the D3 class (CYCD3;3 and CYCD3;4) and one was assigned to the D4 class (CYCD4;2). The remaining new D-type cyclins were subdivided further into classes CYCD5, CYCD6, and CYCD7 according to their phylogenetic positions. It is remarkable that CYCD4;2 and CYCD6;1 do not contain the LxCxE retinoblastoma (Rb) binding motif, whereas CYCD5;1 contains a divergent Rb binding motif (FxCxE) located at the N terminus. The biological functions of cyclins lacking the conserved Rb binding motif remain unclear. One Arabidopsis gene was found with high sequence similarity to cyclin H of poplar (71%) and rice (66%).
Aligning all cyclins allowed us to identify the cyclin and destruction box consensus sequences for A-, B-, D-, and H-type cyclins (Table 2). Although A- and B-type cyclin boxes are very similar, these two types of cyclins can be discriminated by their destruction boxes. For two genes within the A- and B-type cyclins (CYCA3;1 and CYCB3;1), no destruction box could be detected. In addition, these genes have a highly diverged cyclin box compared with their subclass consensus. The low overall sequence similarity within D-type cyclins also is reflected in their cyclin boxes.
Table 2.
Consensus Sequences for Cyclin and the Destruction Box in Arabidopsis Cyclins
| Subclass | Cyclin Box Signature | Destruction Box |
|---|---|---|
| Cyclin A1 | MR-(I/V)L(I/V)DW | RAPL(G/S)(D/N)ITN |
| Cyclin A2 | MR-(I/V)L(I/V)DW | RAVL(K/G)(D/E)(I/V)(T/S)N |
| Cyclin A3a | MR-(I/V)L(I/V)DW | RVVLGEL(P/L)N |
| Cyclin B1 | MR-IL(I/V/F)DW | R-(A/V)LGDIGN |
| Cyclin B2 | MR-IL(I/V/F)DW | RR(A/V)L–IN |
| Cyclin B3 | TRGILINW | N.D.b |
| Cyclin D1 | REDSVAW | N.D. |
| Cyclin D2 | RNQALDW | N.D. |
| Cyclin D3 | R(E/K)(E/K)A(L/V)(D/G)W | N.D. |
| Cyclin D4 | R(R/I)(D/Q)AL(N/G)W | N.D. |
| Cyclin D5 | RLIAIDW | N.D. |
| Cyclin D6 | RNQAISS | N.D. |
| Cyclin D7 | RFHAFQW | N.D. |
| Cyclin Hc | MRAFYEAK | N.D. |
a In CYCA3;1, cyclin box KRGVLVDW was not included in the consensus; no destruction box was detected.
b N.D., not detected.
c Plant cyclin H consensus for cyclin box MR(A/V)(F/Y)YE-K (based on the sequence of Arath;CYCH, Orysa;CYCH, and cyclin H of poplar).
In addition to the cyclins described above, two presumed pseudogenes were predicted that were very similar to B-type cyclins. The precise number of pseudogenes for the seven selected families remains unclear, because the detection of pseudogenes depends on the degree of conservation in the gene structure and the degree of detection by prediction tools of these degenerated structures.
CDK/Cyclin Interactors and Regulatory Proteins
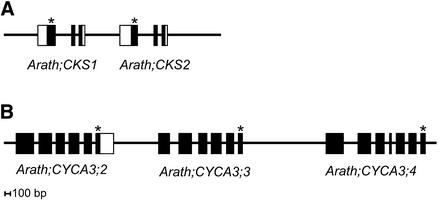
CDK subunit (CKS) proteins act as docking factors that mediate the interaction of CDKs with putative substrates and regulatory proteins. Besides the CDK subunit gene in Arabidopsis described previously (Arath;CKS1; De Veylder et al., 1997), a second CKS gene was found (Arath;CKS2) with sequence (83% identical and 90% similar amino acids) and gene structure (number and size of exons and introns) very similar to those of Arath;CKS1 (Figure 3A). The two CKS gene products do not contain both the N- and C-terminal extensions compared with the yeast Suc1p/Cks1p homologs (De Veylder et al., 1997).
Figure 3.
Gene Tandem Duplication of CKS and A3-Type Cyclin Genes.
Black rectangles represent protein-encoding exons, and white rectangles represent untranslated regions based on hits with ESTs or mRNA. Asterisks denote the exon with the stop codon.
(A) Gene structure of CKS1 and CKS2 on chromosome 2. The indicated chromosome region spans from 12,059 to 12,063 kb.
(B) Gene structure of CYCA3;2, CYCA3;3, and CYCA3;4 on chromosome 1. The indicated region spans from 17,022 to 17,030 kb. ESTs AT50714, AT50514, and AT37419 hit with CYCA3;2 (data not shown).
Upon the occurrence of stress or the perception of antiproliferation agents, the CDK/cyclin complexes are repressed by the CDK inhibitor (CKI) proteins. In mammals, two different classes of CKIs exist (the INK4 and the Kip/Cip families), each with its own CDK binding specificity and protein structure. Seven CKI genes belonging to the group of Kip/Cip CKIs have been described previously for Arabidopsis, designated KRP1 to KRP7 (De Veylder et al., 2001). No extra KRPs were detected in the complete genome, and no plant counterparts of the INK4 family were found.
CDK/cyclin activity is regulated negatively by phosphorylation of the CDK subunit by the WEE1 kinase and positively when the inhibitory phosphate groups are removed by the CDC25 phosphatase. A single WEE1 gene was identified on chromosome 1. The WEE1 kinase was annotated using two cDNA sequences that were at our disposal (L. De Veylder, unpublished results) and has its highest homology with the WEE1 kinase of maize, showing 56% similarity to the gene product of a partial mRNA (Sun et al., 1999). No CDC25 phosphatase could be identified.
Rb and E2F/DP
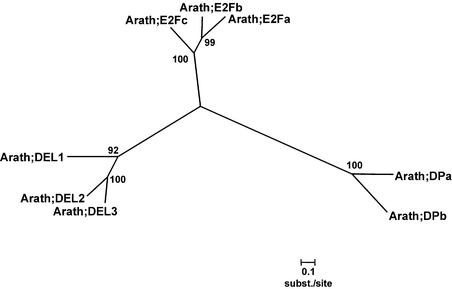
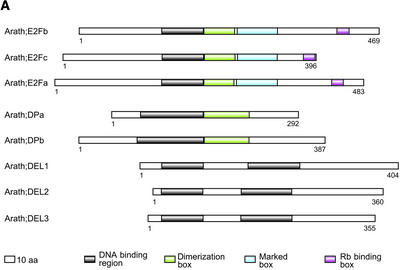
Rb and the E2F/DP proteins are key regulators that control the start of DNA replication. When the E2F/DP transcription factors are bound to Rb, they are inactive, but they become active when Rb is phosphorylated by G1-specific CDK/cyclin complexes, stimulating the transcription of genes needed for G1-to-S and S-phase progression. Only one Rb could be identified in the Arabidopsis genome; it was located on chromosome 3. E2F genes are known for tobacco, carrot, and wheat (Ramírez-Parra et al., 1999; Sekine et al., 1999; Albani et al., 2000; Magyar et al., 2000), but no Arabidopsis family members have been described until now, whereas two Arabidopsis DP genes (DPa and DPb) have been reported. The E2F and DP genes were analyzed in a combined approach, because the sequences of both types of proteins are partially similar (22% overall similarity). In total, eight genes were detected in Arabidopsis. Although the sequence similarity between these eight members of the E2F/DP family is rather low (20% overall mean similarity), three groups had emerged based on previous experimental information (Magyar et al., 2000) and phylogenetic analysis (Figure 4). The first group comprises the E2F transcription factors that are most similar to the mammalian E2F factors and were designated E2Fa, E2Fb, and E2Fc (46% overall similarity). The second group consists of the two already known DP factors.
Figure 4.
Unrooted Neighbor-Joining Tree of the E2F, DP, and DEL Families with Poisson Correction for Evolutionary Distance Calculation.
Bootstrap values of 500 bootstrap iterations are shown. Scales indicate evolutionary distance. Arath, Arabidopsis.
The third group contains three new genes with an internal similarity of 59% and a sequence similarity with both E2F (21%) and DP genes (18%), initially indicating some kind of relation with the E2F/DP genes. When the boxes present in the E2F genes (DNA binding, dimerization, Marked, and Rb binding boxes) and the DP genes (DNA binding and dimerization boxes) were compared with those in the three new genes, only a DNA binding domain was found, but in duplicate (Figure 5A). Both DNA binding domains are highly similar to the E2F DNA binding domain. Because of their phylogenetic positions, they form a distinct class, which we designated DP-E2F–like (DEL). The DNA binding domains of the E2F and DP genes have a limited across-family homology (Figure 5B), including the RRxYD DNA recognition motif (in their α3-helices), which interacts with half of the palindromic promoter binding site (CGCGCG and CGCGCG). Within all three DEL genes, the conserved DNA recognition motif RRxYD also is present in two copies. The E2F/DP heterodimer binds and recognizes the palindromic sequence of the binding site in an essentially symmetric arrangement (Zheng et al., 1999). Protein secondary structure prediction for the DEL genes showed that the winged-helix DNA binding motif, a fold found in the cell cycle transcription factors E2F/DP (three α-helices and a β-sheet), is present in duplicate in all of these DEL genes. The first and second DEL DNA binding domains have an overall similarity of 61 and 47%, respectively, with the E2F DNA binding domain. Currently, no experimental data are available regarding the putative function and role of the DEL genes in cell cycle regulation.
Figure 5.
Structural Organization of the E2F, DP, and DEL Families at the Protein Level.
(A) Scheme of the DNA binding, dimerization, Marked, and Rb binding boxes in E2F, DP, and DEL genes of Arabidopsis.
(B) Alignment of putative DNA binding domains of E2F, DP, and DEL proteins. All DEL proteins were split in two (parts a and b) to compare both DNA binding motifs with those of E2F and DP. The RRxYD DNA binding motif is indicated by asterisks.
Numbers indicate protein length in amino acids (aa).
Gene/Genome Organization
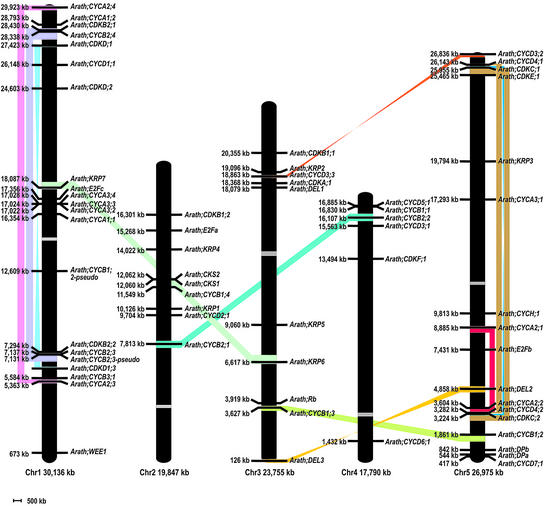
To determine whether the segmental or genomic duplications and the acquisition of new cell cycle regulation mechanisms are linked, we mapped all cell cycle genes on the five different chromosomes (Figure 6). Subsequently, all duplicated regions in the Arabidopsis genome were defined, and the position of every cell cycle gene was compared with the coordinates of each duplicated block.
Figure 6.
Physical Positions of Core Cell Cycle Genes on the Arabidopsis Genome.
Segmental duplicated regions are shown only when a cell cycle gene is present in a duplication event. Colored bands connect corresponding duplicated blocks. Duplicated blocks in reverse orientation are connected with twisted colored bands. Centromeres are represented as gray boxes. Chr1 to Chr5, chromosomes 1 to 5.
Comparison of the positions of A2 cyclin genes with the positions of duplicated blocks in the Arabidopsis genome revealed that all four members are located in duplicated blocks: one internal duplication on chromosome 1 (CYCA2;3 linked with CYCA2;4) and one on chromosome 5 (CYCA2;2 linked with CYCA2;1). The three CYCA3 genes are organized in tandem (CYCA3;2, CYCA3;3, and CYCA3;4 spanning a region of <8 kb) and have a highly similar gene structure (number and size of exons and introns) as well as highly similar protein sequences (74.3% overall similarity). Only CYCA3;2 had one significant EST hit, whereas CYCA3;4 had an additional small predicted exon (33 nucleotides) compared with the other CYCA3 genes that occur in the same tandem (Figure 3B).
Like the A2-type cyclins, all four B2-type cyclins were located within duplicated blocks: one duplicated block between chromosomes 2 and 4 (linking CYCB2;1 and CYCB2;2) and one internal duplication on chromosome 1 (linking CYCB2;3 and CYCB2;4). Although 10 D-type cyclins were detected in total, few of them were located in duplicated blocks. CYCD3;2 and CYCD3;3 are members of an inverted block between chromosomes 5 and 3, whereas CYCD4;1 and CYCD4;2 are located within an internal block of chromosome 5. The two CKS genes were located in a gene tandem duplication in which the stop codon of CKS2 was separated by only 916 bp from the start codon of CKS1 (Figure 3A).
Special attention is required for two duplication events. On chromosome 1, a large internal duplication occurred (spanning an area of ∼4890 kb, or 16% of chromosome 1) that was followed by several inversions (data not shown), leading to the formation of multiple smaller blocks, one of which contained two pairs of cell cycle genes: CDKB2;2 linked with CDKB2;1 and CYCB2;3 linked with CYCB2;4. The CYCB2;3 gene was present in tandem (interspersed by one gene), and the second copy was designated Arath; CYCB2;3–pseudo, because its gene structure was degraded and imperfect with respect to CYCB2;3. We conclude that this tandem duplication occurred after the segmental duplication event, because in the region linked to the duplicated block, no trace of another extra B2-like cyclin was found.
Another special, internally duplicated event was found on chromosome 5. Two duplicated blocks (Figure 5, brown blocks) were detected that connected both extremities of the chromosome. Although these blocks could be regarded as one, we clearly distinguished an invertedly duplicated block between them (Figure 6, blue block). CYCD4;1 and CYCD4;2 both fit nicely into the first block. CDKC;1 and CDKC;2 mapped to this region as well, located in the small invertedly duplicated block. It is remarkable that, although both pairs of linked genes were located in duplicated blocks with different orientations, their relative positions were the same (i.e., at the bottom and at the top of chromosome 5, a C-type CDK was followed by a D4-type cyclin). This configuration suggests that, initially, one large duplication event occurred (Figure 6, the region spanning the brown and blue blocks) that was reshuffled later by inversions (and perhaps some deletions), resulting in adjacent, duplicated blocks with different orientations and sizes.
DISCUSSION
The members of the Arabidopsis genome sequencing consortia use different tools to perform automated genome annotations and determine similarities to ESTs and known protein sequences to refine gene models. This procedure has generated a large quantity of information on the Arabidopsis genome. However, the extraction of clear biological information for a particular process from these public databases is not always easy (for instance, the word “cyclin” as a query in the Martiensried Institute for Protein Sequences database returned 37 hits, with 23 putative cyclin or cyclin-like hits). To solve this problem, we designed a protocol focused mainly on high-quality, homology-based annotation. We used a combination of two selected high-quality Arabidopsis prediction tools (Pavy et al., 1999; Schiex et al., 2001; C. Mathé and P. Rouzé, personal communication) together with pure experimental information as reference material. One advantage of this method is that the chance of finding new and rarely expressed genes is maximized, because they are structurally characterized by tools with higher specificity and sensitivity than those used by the different consortia to generate genome annotations (Gopal et al., 2001). Second, the focus on families with available experimental references allows comparisons with functionally well-characterized genes and diminishes the risk of the propagation of incorrect annotations. In addition, the use of hidden Markov profiles, which represent the complete diversity within a family, clearly is more powerful than the use of a single sequence for remote homolog detection (Karplus et al., 1998).
With this strategy, we have built a catalog of 61 core cell cycle genes belonging to seven selected families. From these, 30 genes had not been described before, and for 22 of them the gene prediction provided by the Arabidopsis Genome Initiative (2000) was incorrect. Corrected gene models have been submitted to The Arabidopsis Information Resource (TAIR) and also can be found on the World Wide Web at http://www.plantgenetics.rug.ac.be/bioinformatics/coreCC/. These results highlight the complexity of cell cycle regulation in Arabidopsis, indicating a larger variety of genes than was known experimentally.
Like mammals, plants evolved to use different classes of CDKs to regulate their cell cycle. In Arabidopsis, six different CDK classes can be identified, designated A through F. Although some of these CDKs have been proven to be active during specific phases of the cell cycle (Magyar et al., 1997; Porceddu et al., 2001; Sorrell et al., 2001), no functional correlation can be made with CDKs of other eukaryotes on the basis of protein sequences. For example, no clear orthologs can be identified for the mammalian G1/S-specific CDK4 and CDK6, suggesting that plants developed independently additional CDKs for more specialized functions in cell cycle control. This hypothesis is in agreement with the observation that the cyclin binding motifs found in the plant B-type CDKs cannot be found in any CDK of other eukaryotes.
Within the CDK family, we identified three new CAK members that are close homologs of the rice R2 gene (Hata, 1991). These CAKs (CDKD;1, CDKD;2, and CDKD;3) differ structurally from the previously isolated Arabidopsis cak1At, which was renamed CDKF;1. The high sequence diversity (35% overall sequence similarity between D- and F-type CDKs) suggests that plants use two distinct classes of CAKs. When the Arabidopsis CDKF;1 is compared with the rice R2, both classes are functionally different. They both can complement yeast CAK mutant strains, but they show different substrate specificity: the rice R2 phosphorylates both CDKs and the C-terminal domain of the largest subunit of RNA polymerase II, whereas CDKF;1 phosphorylates CDKs only (Umeda et al., 1998; Yamaguchi et al., 1998).
The complexity of the cyclin gene family appears to be higher in plants than in mammals. Compared with human, Arabidopsis has ∼14 more A- and B-type cyclins and seven more D-type cyclins. A major part of the A-type cyclins originated through large segmental duplications. For the 10 A-type cyclins, all 4 members of the A2-type subclass are part of duplicated blocks, and 3 of the 4 A3-type cyclins are organized in tandem. Several analyses of the Arabidopsis genome sequence had already concluded that genes had duplicated extensively in the history of the model plant. More than 50% of the genes in Arabidopsis belong to a gene family with three or more members. After analyzing regions of chromosomes 2, 4, and 5, Blanc et al. (2000) estimated that more than 60% of the genome consisted of duplicated regions and suggested the possibility that Arabidopsis was an ancient tetraploid. In a later analysis, Vision et al. (2000) concluded that several large independent duplications of chromosome segments had happened at different times during the plant's evolution. This view was blurred by extensive deletions, inversions, and translocations of genes and chromosome segments as well as smaller and tandem gene duplications (Arabidopsis Genome Initiative, 2000; Vision et al., 2000). In our analysis, we determined that 22 core cell cycle genes are part of a segmental duplication in the Arabidopsis genome. Whether there is functional redundancy within A- and B-type cyclins or different regulation (and expression) of some cyclin subclasses remains to be analyzed.
In contrast to the A- and B-type cyclins, D-type cyclins lack high sequence similarity to each other, which is reflected in the phylogenetic analysis resulting in seven D-type subclasses. Compared with A- and B-type cyclins, of which some complete subclasses (A2 and B2) are located within segmentally duplicated blocks, no large duplications can be found for the D-type cyclins. Only the D3 and D4 subclasses have different members. Redundancy of the D3-type cyclins has been proposed previously as an explanation for the failure to observe mutant phenotypes when knocking out a single D3-type cyclin (Swaminathan et al., 2000). Our analysis clearly confirms this hypothesis: the fact that two D3-type cyclins are linked via a recent segmental duplication strengthens our belief that these D3-type cyclins are functionally redundant. A similar hypothesis could hold for D4-type cyclins, because two out of three are located in a duplicated block.
The much larger divergence seen for D-type cyclins compared with A- and B-type cyclins might reflect the presumed role of D-type cyclins in integrating developmental signals and environmental cues into the cell cycle. For example, D3-type cyclins have been shown to respond to plant hormones, such as cytokinins and brassinosteroids, whereas CYCD2 and CYCD4 are activated earlier in G1 and react to sugar availability (for review, see Stals and Inzé, 2001). Because of the large number of various D-type cyclins with different responses to developmental and environmental signals, cell division and growth in sessile plants might be more flexible than what is observed in other eukaryotes.
Although plants clearly share all of the elements needed for G1/S entry with other higher eukaryotes, they lack the typical class of E-type cyclins, which are known to be essential regulators of DNA replication (Duronio et al., 1996). Presumably, some of the A- or D-type cyclins assume the role of the E-type cyclins. Also, the lack of a consensus Rb binding motif in some D-type cyclins suggests that some cyclins might have gained other novel functions during evolution. Alternatively, some of the core cell cycle genes might have undergone such dramatic changes during evolution that they can no longer be recognized as functional homologs of animal and yeast counterparts; the CDC25 gene is the most likely example of this phenomenon. Both the presence of the antagonistic WEE1 kinase and accumulating biochemical evidence suggest the existence of a CDC25 phosphatase in plants (Zhang et al., 1996; Sun et al., 1999), although it could not be identified as such in the Arabidopsis genome.
It is surprising that mammals and plants have approximately the same number of core cell cycle genes, with the exception of the difference in cyclin number described above. Complex, multicellular organisms may need many more cell cycle genes to coordinate cell cycle progression with their diverse developmental pathways. However, the pool of mammalian cell cycle genes probably is larger than expected because of the frequent occurrence of alternative splicing. For example, spliced variants of cyclin E are known, with an expression profile and substrate specificity different from those of cyclin E itself (Mumberg et al., 1997; Porter and Keyomarsi, 2000). At least five distinct DP-2 mRNAs are synthesized in a tissue-specific manner (Rogers et al., 1996). Depending on the splice variant, the DP family members lack a nuclear localization signal, and when associated with E2F, these different DP molecules have opposing effects on E2F/DP activity (de la Luna et al., 1996). Furthermore, alternative splicing in humans is known for CDKs, CDC25, and CKIs (Wegener et al., 2000; Herrmann and Mancini, 2001; Hirano et al., 2001). For cell cycle genes of plants, only one case of alternative splicing has been reported (Sun et al., 1997).
E2F/DP transcription factors are characterized by the presence of both a DNA binding domain and a transcription activation domain. Binding of these transcription factors to the E2F/DP palindromic binding site is mediated by a small DNA recognition motif (RRxYD). By scanning the genome for E2F/DP-related proteins, a putatively novel class of cell cycle–regulating genes was identified, designated DEL. The DEL proteins have two E2F-like DNA binding boxes, each including the RRxYD motif, but they have no activation domain. By competing for the same DNA binding sites, monomeric DEL proteins could act as competitors of the E2F/DP proteins, and because they lack an activation domain, they would act as repressors of E2F/DP-regulated genes. This mechanism would avoid the G1-to-S transition in cases in which conditions are not appropriate for entry into the S phase (such as DNA damage and stress). This new class of putative cell cycle regulators seems not to be plant specific, because one homolog was found in Caenorhabditis elegans (data not shown).
In conclusion, our genome-wide analysis demonstrated an unexpected complexity of the core cell cycle machinery in plants that is comparable with that seen in mammals. The major challenge for the future is to understand the specific role of these individual genes in regulating cell division during plant development.
METHODS
Annotation of Arabidopsis thaliana Cell Cycle Genes
The genome version of January 18, 2001 (version 180101), was downloaded from the ftp site (ftp://ftpmips.gsf.de/cress/) of the Martiensried Institute for Protein Sequences (Martiensried, Germany). Regions of interest on the chromosomes were localized with BLAST software (Altschul et al., 1997) with experimental representatives as query sequences. For the regions returned by BLAST, chromosome sequences were extracted with 15 kb upstream and downstream from the hit to prevent unreliable predictions caused by border effects.
Gene prediction was performed with EuGene (Schiex et al., 2001) in combination with GeneMark.hmm (Lukashin and Borodovsky, 1998), because the latter had been reported previously to give the best scores for Arabidopsis (Pavy et al., 1999). New analysis (C. Mathé, personal communication), however, showed that EuGene has become the best gene prediction tool for Arabidopsis. The EuGene program combines NetGene2 (Tolstrup et al., 1997) and SplicePredictor (Brendel and Kleffe, 1998) for splice site prediction, NetStart (Pedersen and Nielsen, 1997) for translation initiation prediction, interpolated Markov model–based content sensors, and information from protein, expressed sequence tag, and cDNA matches to predict the final gene model.
The predicted candidate gene products were aligned with the experimental representatives using CLUSTAL W (Thompson et al., 1994). On the final alignments, HMMer (Eddy, 1998) was used to generate profiles for each specific gene family with hidden Markov models. These profiles then were used to search for new family members (Eddy, 1998). The genome-wide, nonredundant collection of Arabidopsis protein-encoding genes was predicted with GeneMark.hmm. Based on these predictions, we built a database of virtual transcripts (and a corresponding protein database), which we designated genome-predicted transcripts. Manual annotation was performed with Artemis (Rutherford et al., 2000).
Phylogeny and Nomenclature
Phylogenetic analysis was performed on more conserved positions of the alignment. Editing of the alignment and reformatting were performed with BioEdit (Hall, 1999) and ForCon (Raes and Van de Peer, 1999). Similarity between proteins was based on a BLOSUM62 matrix (Henikoff and Henikoff, 1993). Trees were constructed using various distance and parsimony methods. Distance matrices were calculated based on Poisson, Kimura, or point accepted mutation (PAM) correction, and trees were constructed with the neighbor-joining algorithm using the software packages TREECON (Van de Peer and De Wachter, 1994) and PHYLIP (Felsenstein, 1993). The latter also was used for the parsimony analysis. Bootstrap analysis with 500 replicates was performed to test the significance of nodes.
Protein Structure Analysis
Protein secondary structure prediction was performed with PSIpred version 2.0 (Jones, 1999).
Segmental Duplications in the Arabidopsis Genome
For the detection of large segmental duplications, duplicated blocks were identified by a method similar to that described by Vision et al. (2000). Initially, protein-coded genes predicted by GeneMark.hmm (26,352 present in our genome-predicted transcript database) were ordered according to their locations on the corresponding chromosome. BLASTP was used to identify genes with high sequence similarity, and all BLASTP scores were stored in a matrix to be analyzed. Initially, filtering was performed to reduce low-similarity hits (E value < 1e−50; Friedman and Hughes, 2001), followed by a procedure to define duplicated blocks in the scoring matrix. Finally, by postprocessing, only blocks of appropriate size (i.e., blocks containing more than seven genes) were selected.
Accession Numbers
Accession number for the genes referred to in the figures are CAA65979.1 (Medsa;CDKC;1), CAA41172.1 (Orysa;CDKD;1), CAA65981.1 (Medsa;CDKE;1), AAB41817.1 (Medsa;CDKA;1), CAA50038.1 (Medsa;CDKA;2), CAA65980.1 (Medsa;CDKB1;1), CAC15503.1 (Lyces;CDKB1;1), CAC15504.1 (Lyces;CDKB2;1), and CAA65982.1 (Medsa;CDKB2;1) (all in Figure 1); BAA09366.1 (Nicta;CYCA1;1), CAA63540.1 (Nicta;CYCA3;1), AAD02871.1 (Poptr;CYCH), and BAB11694.1 (Orysa;CYCH) (all in Figure 2); and AF242582 (Arath;E2Fa), AD242580 (Arath;E2Fb), AF242581 (Arath;E2Fc), AJ294531 (Arath;DPa), and AJ294532 (Arath;DPb) (all in Figure 4).
NOTE ADDED IN PROOF
The postulated function of the DEL proteins has recently been confirmed (Mariconti, L., Pellegrini, B., Cantoni, R., Stevens, R., Bergounioux, C., Cella, R., and Albani, D. [January 10, 2002] J. Biol. Chem. 10.1074/jbc.M110616200), but the gene prediction for one DEL family member (E2Ff~DEL3) differs from the one we present here. The gene structure we propose has been validated experimentally in our laboratory.
Acknowledgments
We especially thank Yvan Saeys for providing us with the necessary programs to define duplicated blocks in the Arabidopsis genome, Dr. Yves Van de Peer for help with the analysis of saturated positions in the cyclin alignments, Dr. Catherine Mathé for additional information regarding EuGene, Patrice Déhais for the programs developed to run EuGene, Sébastien Aubourg for helpful discussions, and Martine De Cock and Rebecca Verbanck for help in preparing the manuscript and artwork, respectively. This work was supported by grants from the Interuniversity Poles of Attraction Programme (Belgian State, Prime Minister's Office—Federal Office for Scientific, Technical and Cultural Affairs; P4/15), the European Union (ECCO QLG2-CT1999-00454), Génoplante (Project BI1999087), and CropDesign N.V. (0235). K.V. is indebted to the Vlaams Instituut voor de Bevordering van het Wetenschappelijk–Technologisch Onderzoek in de Industrie for a predoctoral fellowship, and L.D.V. is a postdoctoral fellow of the Fund for Scientific Research (Flanders).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010445.
References
- Albani, D., Mariconti, L., Ricagno, S., Pitto, L., Moroni, C., Helin, K., and Cella, R. (2000). DcE2F, a functional plant E2F-like transcriptional activator from Daucus carota. J. Biol. Chem. 275, 19258–19267. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Blanc, G., Barakat, A., Guyot, R., Cooke, R., and Delseny, M. (2000). Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell 12, 1093–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf, V., Rombauts, S., Naudts, M., Inzé, D., and De Veylder, L. (2001). Identification of novel cyclin-dependent kinases interacting with the CKS1 protein of Arabidopsis. J. Exp. Bot. 52, 1381–1382. [PubMed] [Google Scholar]
- Brendel, V., and Kleffe, J. (1998). Prediction of locally optimal splice sites in plant pre-mRNA with applications to gene identification in Arabidopsis thaliana genomic DNA. Nucleic Acids Res. 26, 4748–4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Luna, S., Burden, M.J., Lee, C.-W., and La Thangue, N.B. (1996). Nuclear accumulation of the E2F heterodimer regulated by subunit composition and alternative splicing of a nuclear localization signal. J. Cell Sci. 108, 2443–2452. [DOI] [PubMed] [Google Scholar]
- De Veylder, L., Segers, G., Glab, N., Casteels, P., Van Montagu, M., and Inzé, D. (1997). The Arabidopsis Cks1At protein binds to the cyclin-dependent kinases Cdc2aAt and Cdc2bAt. FEBS Lett. 412, 446–452. [DOI] [PubMed] [Google Scholar]
- De Veylder, L., De Almeida Engler, J., Burssens, S., Manevski, A., Lescure, B., Van Montagu, M., Engler, G., and Inzé, D. (1999). A new D-type cyclin of Arabidopsis thaliana expressed during lateral root primordia formation. Planta 208, 453–462. [DOI] [PubMed] [Google Scholar]
- De Veylder, L., Beeckman, T., Beemster, G.T.S., Krols, L., Terras, F., Landrieu, I., Van Der Schueren, E., Maes, S., Naudts, M., and Inzé, D. (2001). Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell 13, 1653–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos, D., and Valencia, A. (2001). Intrinsic errors in genome annotation. Trends Genet. 17, 429–431. [DOI] [PubMed] [Google Scholar]
- Duronio, R.J., Brook, A., Dyson, N., and O'Farrell, P.H. (1996). E2F-induced S phase requires cyclin E. Genes Dev. 10, 2505–2513. [DOI] [PubMed] [Google Scholar]
- Eddy, S.R. (1998). Profile hidden Markov models. Bioinformatics 14, 755–763. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. (1993). PHYLIP (Phylogeny Inference Package), Version 3.5c. (Seattle, WA: Department of Genetics, University of Washington).
- Friedman, R., and Hughes, A.L. (2001). Gene duplication and the structure of eukaryotic genomes. Genome Res. 11, 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal, S., Schroeder, M., Pieper, U., Sczyrba, A., Aytekin-Kurban, G., Bekiranov, S., Fajardo, J.E., Eswar, N., Sanchez, R., Sali, A., and Gaasterland, T. (2001). Homology-based annotation yields 1,042 new candidate genes in the Drosophila melanogaster genome. Nat. Genet. 27, 337–340. [DOI] [PubMed] [Google Scholar]
- Hall, T.A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98. [Google Scholar]
- Hata, S. (1991). cDNA cloning of a novel cdc2+/CDC28-related protein kinase from rice. FEBS Lett. 279, 149–152. [DOI] [PubMed] [Google Scholar]
- Hemerly, A., de Almeida Engler, J., Bergounioux, C., Van Montagu, M., Engler, G., Inzé, D., and Ferreira, P. (1995). Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO J. 14, 3925–3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff, S., and Henikoff, J.G. (1993). Performance evaluation of amino acid substitution matrices. Proteins 17, 49–61. [DOI] [PubMed] [Google Scholar]
- Herrmann, C.H., and Mancini, M.A. (2001). The Cdk9 and cyclin T subunits of TAK/P-TEFb localize to splicing factor-rich nuclear speckle regions. J. Cell Sci. 114, 1491–1503. [DOI] [PubMed] [Google Scholar]
- Hirano, K., Hirano, M., Zeng, Y., Nishimura, J., Hara, K., Muta, K., Nawata, H., and Kanaide, H. (2001). Cloning and functional expression of a degradation-resistant novel isoform of p27Kip1. Biochem. J. 353, 51–57. [PMC free article] [PubMed] [Google Scholar]
- Jones, D.T. (1999). Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202. [DOI] [PubMed] [Google Scholar]
- Joubès, J., Chevalier, C., Dudits, D., Heberle-Bors, E., Inzé, D., Umeda, M., and Renaudin, J.-P. (2000). CDK-related protein kinases in plants. Plant Mol. Biol. 43, 607–620. [DOI] [PubMed] [Google Scholar]
- Joubès, J., Lemaire-Chamley, M., Delmas, D., Walter, J., Hernould, M., Mouras, A., Raymond, P., and Chevalier, C. (2001). A new C-type cyclin-dependent kinase from tomato expressed in dividing tissues does not interact with mitotic and G1 cyclins. Plant Physiol. 126, 1403–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus, K., Barrett, C., and Hughey, R. (1998). Hidden Markov models for detecting remote protein homologies. Bioinformatics 14, 846–856. [DOI] [PubMed] [Google Scholar]
- Lessard, P., Bouly, J.-P., Jouannic, S., Kreis, M., and Thomas, M. (1999). Identification of cdc2cAt: A new cyclin-dependent kinase expressed in Arabidopsis thaliana flowers. Biochim. Biophys. Acta 1445, 351–358. [DOI] [PubMed] [Google Scholar]
- Lukashin, A.V., and Borodovsky, M. (1998). GeneMark.hmm: New solutions for gene finding. Nucleic Acids Res. 26, 1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magyar, Z., et al. (1997). Cell cycle phase specificity of putative cyclin-dependent kinase variants in synchronized alfalfa cells. Plant Cell 9, 223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magyar, Z., Atanassova, A., De Veylder, L., Rombauts, S., and Inzé, D. (2000). Characterization of two distinct DP-related genes from Arabidopsis thaliana. FEBS Lett. 486, 79–87. [DOI] [PubMed] [Google Scholar]
- Mumberg, D., Wick, M., Bürger, C., Haas, K., Funk, M., and Müller, R. (1997). Cyclin ET, a new splice variant of human cyclin E with a unique expression pattern during cell cycle progression and differentiation. Nucleic Acids Res. 25, 2098–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavy, N., Rombauts, S., Déhais, P., Mathé, C., Ramana, D.V.V., Leroy, P., and Rouzé, P. (1999). Evaluation of gene prediction software using a genomic data set: Application of Arabidopsis thaliana sequences. Bioinformatics 15, 887–899. [DOI] [PubMed] [Google Scholar]
- Pedersen, A.G., and Nielsen, H. (1997). Neural network prediction of translation initiation sites in eukaryotes: Perspectives for EST and genome analysis. In Proceedings of the Fifth International Conference on Intelligent Systems for Molecular Biology, T. Gaasterland, P. Karp, K. Karplus, C. Ouzounis, C. Sander, and A. Valencia, eds (Menlo Park, CA: American Association for Artificial Intelligence Press), pp. 226–233. [PubMed]
- Porceddu, A., Stals, H., Reichheld, J.-P., Segers, G., De Veylder, L., De Pinho Barrôco, R., Casteels, P., Van Montagu, M., Inzé, D., and Mironov, V. (2001). A plant-specific cyclin-dependent kinase is involved in the control of G2/M progression in plants. J. Biol. Chem. 276, 36354–36360. [DOI] [PubMed] [Google Scholar]
- Porter, D.C., and Keyomarsi, K. (2000). Novel splice variants of cyclin E with altered substrate specificity. Nucleic Acids Res. 28, e101–e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes, J., and Van de Peer, Y. (1999). ForCon: A software tool for the conversion of sequence alignments. http://www.ebi.ac.uk/embnet.news/vol6_1/ForCon/body_forcon.html.
- Ramírez-Parra, E., Xie, Q., Boniotti, M.B., and Gutierrez, C. (1999). The cloning of plant E2F, a retinoblastoma-binding protein, reveals unique and conserved features with animal G1/S regulators. Nucleic Acids Res. 27, 3527–3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaudin, J.-P., et al. (1996). Plant cyclins: A unified nomenclature for plant A-, B- and D-type cyclins based on sequence organization. Plant Mol. Biol. 32, 1003–1018. [DOI] [PubMed] [Google Scholar]
- Rogers, K.T., Higgins, P.D.R., Milla, M.M., Phillips, R.S., and Horowitz, J.M. (1996). DP-2, a heterodimeric partner of E2F: Identification and characterization of DP-2 proteins expressed in vivo. Proc. Natl. Acad. Sci. USA 93, 7594–7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouzé, P., Pavy, N., and Rombauts, S. (1999). Genome annotation: Which tools do we have for it? Curr. Opin. Plant Biol. 2, 90–95. [DOI] [PubMed] [Google Scholar]
- Rutherford, K., Parkhill, J., Crook, J., Horsnell, T., Rice, P., Rajandream, M.-A., and Barrell, B. (2000). Artemis: Sequence visualization and annotation. Bioinformatics 16, 944–945. [DOI] [PubMed] [Google Scholar]
- Schiex, T., Moisan, A., and Rouzé, P. (2001). EuGène: An eukaryotic gene finder that combines several sources of evidence. In Computational Biology: Selected Papers (Lecture Notes in Computer Science, Vol. 2066), O. Gascuel and M.-F. Sagot, eds (Berlin: Springer-Verlag), pp. 111–125
- Sekine, M., Ito, M., Uemukai, K., Maeda, Y., Nakagami, H., and Shinmyo, A. (1999). Isolation and characterization of the E2F-like gene in plants. FEBS Lett. 460, 117–122. [DOI] [PubMed] [Google Scholar]
- Soni, R., Carmichael, J.P., Shah, Z.H., and Murray, J.A.H. (1995). A family of cyclin D homologs from plants differentially controlled by growth regulators and containing the conserved retinoblastoma protein interaction motif. Plant Cell 7, 85–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrell, D.A., Menges, M., Healy, J.M.S., Deveaux, Y., Amano, X., Su, Y., Nakagami, H., Shinmyo, A., Doonan, J.H., Sekine, M., and Murray, J.A.H. (2001). Cell cycle regulation of cyclin-dependent kinases in tobacco cultivar Bright Yellow-2 cells. Plant Physiol. 126, 1214–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stals, H., and Inzé, D. (2001). When plant cells decide to divide. Trends Plant Sci. 6, 359–364. [DOI] [PubMed] [Google Scholar]
- Sun, Y., Flannigan, B.A., Madison, J.T., and Setter, T.L. (1997). Alternative splicing of cyclin transcripts in maize endosperm. Gene 195, 167–175. [DOI] [PubMed] [Google Scholar]
- Sun, Y., Dilkes, B.P., Zhang, C., Dante, R.A., Carneiro, N.P., Lowe, K.S., Jung, R., Gordon-Kamm, W.J., and Larkins, B.A. (1999). Characterization of maize (Zea mays L.) Wee1 and its activity in developing endosperm. Proc. Natl. Acad. Sci. USA 96, 4180–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan, K., Yang, Y., Grotz, N., Campisi, L., and Jack, T. (2000). An enhancer trap line associated with a D-class cyclin gene in Arabidopsis. Plant Physiol. 124, 1658–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolstrup, N., Rouzé, P., and Brunak, S. (1997). A branch point consensus from Arabidopsis found by non-circular analysis allows for better prediction of acceptor sites. Nucleic Acids Res. 25, 3159–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda, M., Bhalerao, R.P., Schell, J., Uchimiya, H., and Koncz, C. (1998). A distinct cyclin-dependent kinase-activating kinase of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 95, 5021–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer, Y., and De Wachter, R. (1994). TREECON for Windows: A software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10, 569–570. [DOI] [PubMed] [Google Scholar]
- Van de Peer, Y., Frickey, T., Taylor, J.S., and Meyer, A. (2002). Dealing with saturation at the amino acid level: A case study based on anciently duplicated zebrafish genes. Gene, in press. [DOI] [PubMed]
- Vision, T.J., Brown, D.G., and Tanksley, S.D. (2000). The origins of genomic duplications in Arabidopsis. Science 290, 2114–2117. [DOI] [PubMed] [Google Scholar]
- Wegener, S., Hampe, W., Herrmann, D., and Schaller, H.C. (2000). Alternative splicing in the regulatory region of the human phosphatases CDC25A and CDC25C. Eur. J. Cell Biol. 79, 810–815. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, M., Umeda, M., and Uchimiya, H. (1998). A rice homolog of Cdk7/MO15 phosphorylates both cyclin-dependent protein kinases and the carboxy-terminal domain of RNA polymerase II. Plant J. 16, 613–619. [DOI] [PubMed] [Google Scholar]
- Zhang, K., Letham, D.S., and John, P.C.L. (1996). Cytokinin controls the cell cycle at mitosis by stimulating the tyrosine dephosphorylation and activation of p34cdc2-like H1 histone kinase. Planta 200, 2–12. [DOI] [PubMed] [Google Scholar]
- Zheng, N., Fraenkel, E., Pabo, C.O., and Pavletich, N.P. (1999). Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F-DP. Genes Dev. 13, 666–674. [DOI] [PMC free article] [PubMed] [Google Scholar]