Abstract
Often during flowering plant evolution, ribosomal protein genes have been lost from the mitochondrion and transferred to the nucleus. Here, we show that substitution by a duplicated, divergent gene originally encoding the chloroplast or cytosolic ribosomal protein counterpart accounts for two missing mitochondrial genes in diverse angiosperms. The rps13 gene is missing from the mitochondrial genome of many rosids, and a transferred copy of this gene is not evident in the nucleus of Arabidopsis, soybean, or cotton. Instead, these rosids contain a divergent nuclear copy of an rps13 gene of chloroplast origin. The product of this gene from all three rosids was shown to be imported into isolated mitochondria but not into chloroplasts. The rps8 gene is missing from the mitochondrion and nucleus of all angiosperms examined. A divergent copy of the gene encoding its cytosolic counterpart (rps15A) was identified in the nucleus of four angiosperms and one gymnosperm. The product of this gene from Arabidopsis and tomato was imported successfully into mitochondria. We infer that rps13 was lost from the mitochondrial genome and substituted by a duplicated nuclear gene of chloroplast origin early in rosid evolution, whereas rps8 loss and substitution by a gene of nuclear/cytosolic origin occurred much earlier, in a common ancestor of angiosperms and gymnosperms.
INTRODUCTION
Mitochondrial genomes are derived from the genome of a bacterial endosymbiont, with many genes having been lost or transferred to the nucleus early in mitochondrial evolution (reviewed by Gray, 1992; Gray et al., 1999). The transfer of mitochondrial genes to the nucleus and functional activation has been an ongoing and frequent process during flowering plant evolution. Several separate transfers of the same gene have been documented (Adams et al., 2000, 2001b). The process of functional gene transfer in angiosperms involves several steps, including reverse transcription of mRNA (usually; the cellular location in which reverse transcription occurs is not known), movement of the nucleic acid to the nucleus, chromosomal integration, gain of a nuclear promoter and other regulatory elements, gain of a mitochondrial targeting sequence (usually), and silencing and loss of the mitochondrial copy (reviewed by Brennicke et al., 1993; Thorsness and Weber, 1996; Martin and Herrmann, 1998; Adams et al., 1999; Palmer et al., 2000). Nonfunctional gene transfers (i.e., transfers that do not result in functional activation of the transferred gene) omit many of these steps and can occur by direct DNA transfers of varying sizes (Knoop and Brennicke, 1994; Blanchard and Schmidt, 1995), including the extraordinary transfer of most of a mitochondrial genome in an ecotype of Arabidopsis (Arabidopsis Genome Initiative, 2000).
Repeated transfer to the nucleus has led to a highly variable distribution of ribosomal protein and succinate de-hydrogenase genes among mitochondrial genomes of angiosperms. This variability is highlighted by the complete sequence determination of the mitochondrial genomes of Arabidopsis (Unseld et al., 1997) and sugar beet (Kubo et al., 2000) and by comprehensive DNA gel blot hybridization surveys of mitochondrial gene contents across angiosperms (Adams et al., 2002). For maize, there is complete correspondence between gene loss from the mitochondrion and gene transfer to the nucleus: eight ribosomal protein and succinate dehydrogenase genes that are present in at least one other angiosperm mitochondrial genome have been inferred to have been lost from the maize mitochondrial genome (Adams et al., 2002), and transferred copies of all eight genes have been discovered in the nucleus (Figueroa et al., 1999a; Adams et al., 2000, 2001a, 2001b, 2002). In contrast, the completely sequenced Arabidopsis mitochondrial genome (Unseld et al., 1997) lacks functional copies of nine genes that are present in other angiosperm mitochondria, but only seven of these genes have been discovered in the nucleus of Arabidopsis (Wischmann and Schuster, 1995; Sánchez et al., 1996; Perotta et al., 1998; Figueroa et al., 1999b; Adams et al., 2001b; rps11 expressed sequence tags [ESTs] in the National Center for Biotechnology Information [NCBI] databases), despite its nearly complete sequence. Furthermore, there are two genes (rps8 and rpl6) in the mitochondrial genome of the liverwort Marchantia polymorpha that are not present in the mitochondrion of Arabidopsis or any other angiosperm examined. rpl6 has been identified in the Arabidopsis nucleus (Arabidopsis Genome Initiative, 2000), but a transferred rps8 gene has not been found.
We have studied the fate of two genes, rps13 and rps8, that are missing from the mitochondrion of diverse angiosperms. Our results indicate that these two genes have been lost entirely from the cell and replaced by duplicated nuclear genes of cytosolic or chloroplastic origin.
RESULTS
Arabidopsis, Cotton, and Legumes Have Lost rps13 from the Mitochondrion but Contain an rps13-Like Gene in the Nucleus
As part of a DNA gel blot hybridization survey of mitochondrial gene losses in 280 angiosperm genera, the rps13 gene was inferred to have been lost from the mitochondrial genome 30 times among the surveyed angiosperm DNAs (Adams et al., 2002). Most of the losses (24) are in the rosids (one of the major groups of eudicots). These losses encompass 32 of the 69 examined genera of core rosids, including Gossypium, Glycine, and Medicago, and the previously reported loss in Arabidopsis (Unseld et al., 1997). The high concentration of rps13 gene losses among rosids contrasts with the more uniform distribution of losses across angiosperms of 13 other ribosomal protein genes and of two sdh genes (Adams et al., 2002). In Arabidopsis, the missing mitochondrial rps13 does not appear to have been transferred to the nucleus and instead was hypothesized to have been substituted functionally by a ribonucleoprotein (RNP) domain that was acquired by the transferred rps19 gene (Sánchez et al., 1996). In contrast, the transferred rps19 genes in cotton and legumes do not contain the RNP domain (Adams et al., 2002), suggesting either gene transfer to the nucleus or substitution by another protein to account for the missing rps13. Although not essential for ribosomal function in bacteria, ribosomal protein S13 nonetheless is necessary for normal translational efficiency and cell growth rate (reviewed by Faxén et al., 1994), indicating that loss of RPS13 function from the cell is unlikely to account for the missing rps13 in the mitochondria of cotton and legumes.
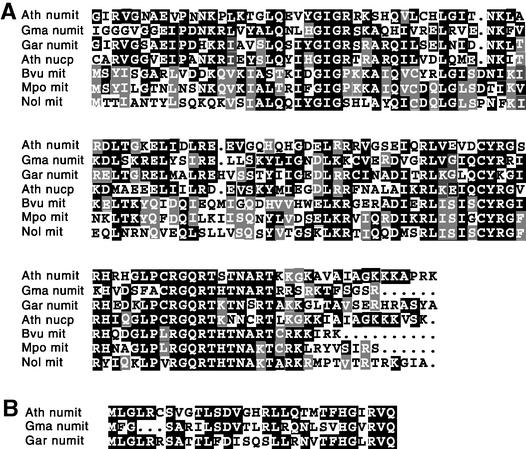
To identify transferred rps13 genes in the nucleus of cotton and legumes, tBLASTn searches of the NCBI EST databases were performed. An rps13 gene of eubacterial origin was identified in cotton, Medicago trunculata, and Lotus japonicus (see Methods for accession numbers) that was highly divergent from both the mitochondrially located rps13 in other angiosperms and the nuclear gene for chloroplast RPS13 from Arabidopsis (Kumar et al., 1995), cotton, and soybean. Considering that the last common ancestor of cotton and legumes also was an ancestor of Arabidopsis, we predicted that the Arabidopsis nucleus also might contain a homologous rps13 gene. Searches of the Arabidopsis genome sequence revealed an annotated 30S rps13 on chromosome 1 (Theologis et al., 2000). The products of these newly identified rps13 genes (Figure 1A) were predicted by three mitochondrial protein prediction programs, Mitoprot (Claros and Vincens, 1996), TargetP (Emanuelsson et al., 2000), and Predotar version 0.5 (www.inra.fr/Internet/Produits/Predotar), to be mitochondrial proteins. Mitoprot and TargetP predict that all three gene products contain a cleavable mitochondrial targeting presequence (Predotar does not predict cleavage sites). Furthermore, the predicted presequences are readily alignable (Figure 1B) and hence homologous. These nuclear rps13 genes for putative mitochondrial proteins are referred to herein as “numit rps13” to distinguish them from other rps13 genes. There are 69 conserved amino acid positions in the numit and mitochondrial RPS13 sequences (Figure 1A), with 17 amino acids being conserved completely among all six numit and mitochondrial genes.
Figure 1.
RPS13 Sequence Alignments.
(A) Alignment of RPS13 sequences. Identical amino acids are shown on black and gray backgrounds.
(B) Predicted targeting presequences of numit RPS13 sequences. Identical amino acids are shown in white on a black background. Dots indicate gaps inserted to improve alignment.
Ath, Arabidopsis; Gma, Glycine max; Gar, Gossypium arboreum; Bvu, Beta vulgaris; Mpo, liverwort Marchantia polymorpha; Nol, green alga Nephroselmis olivacea.
Numit rps13 from Arabidopsis contains two introns. The genomic sequence of numit rps13 from soybean was determined by polymerase chain reaction amplification and sequencing, revealing two introns in the same positions as those in Arabidopsis. Because the rps13 genes from legumes and Arabidopsis have homologous putative mitochondrial targeting sequences (Figure 1B) and also share two intron positions, they are clearly homologous genes. Surprisingly, though, the numit rps13 genes are rather dissimilar, with only 36 to 45% amino acid identity to each other. However, mitochondrial RPS13 amino acid sequences from the liverwort Marchantia (Takemura et al., 1992) and the green alga Prototheca wickerhamii (Wolff et al., 1994) are only 42% identical, indicating the sequence flexibility of this protein. Thus, it is not at all surprising that numit RPS13A from Arabidopsis, a gene of chloroplast ancestry (see below), is 31% identical to RPS13 synthesized in the mitochondria of P. wickerhamii.
To determine if homologs of numit rps13 are present in other angiosperms, the NCBI EST databases were searched for homologous sequences in tomato and several grasses (maize, rice, barley, wheat, and sorghum). The searches revealed the nucleus-encoded chloroplast rps13 gene, referred to herein as nucp rps13, and the cytosolic counterpart of rps13 (rps18), but no numit rps13 genes. Considering the large number of ESTs that are available publicly for tomato (>116,000) and the five grasses (>321,000), numit rps13 may be restricted to rosids.
Numit rps13 Genes Are Derived from the Nucleus-Encoded Chloroplast rps13, and Their Products Are Imported into Mitochondria but Not Chloroplasts
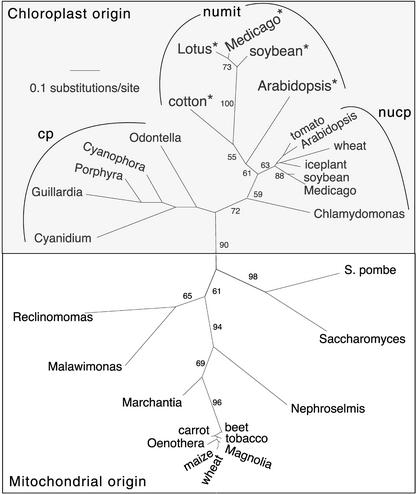
The BLAST similarity scores of the numit rps13 genes generally were higher in relation to nucp rps13 genes than to mitochondrial rps13 genes of other angiosperms. Also, the percent amino acid identities were higher between the numit rps13 genes and nucp rps13 from Arabidopsis (43 to 51%) than between the numit rps13 genes and the angiosperm mitochondrial rps13 sequences (35 to 37%). To determine if the numit rps13 genes were derived from a mitochondrial gene transfer to the nucleus or from the nucp rps13, phylogenetic analyses were performed with a variety of genes for mitochondrial and chloroplast RPS13 proteins. Analyses were performed using maximum parsimony and maximum likelihood methods, and bootstrapping was performed to assess the support at each node (see Methods for details).
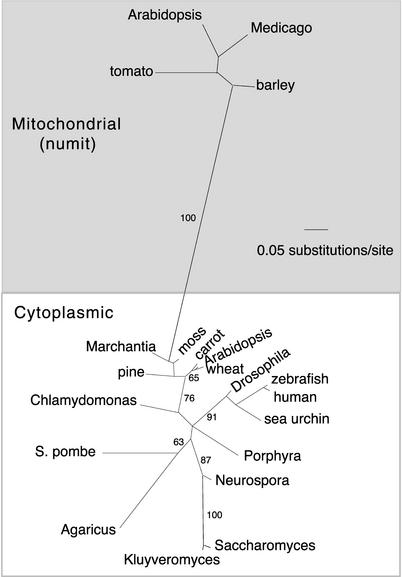
Essentially identical results were obtained with both methods; the likelihood results are shown in Figure 2. The numit rps13 genes from cotton, Arabidopsis, and legumes all branched together, as expected for orthologous genes, and their sequences have diverged rapidly, as indicated by their long branch lengths compared with the branch lengths of nucp rps13 genes from a similar set of angiosperms (Figure 2). The numit rps13 genes from angiosperms branched with the set of angiosperm nucp rps13 genes, and these two sets of angiosperm genes branched with the Chlamydomonas nucp rps13. The rps13 genes still resident in angiosperm mitochondrial genomes (bottom) branched within the larger clade of mitochondrially located rps13 genes. Together, these results clearly indicate that the numit rps13 genes of angiosperms are derived via duplication and substitution from the nucp rps13 rather than from a recent transfer of mitochondrial rps13 to the nucleus. The predicted targeting sequences of numit rps13 and nucp rps13 do not have any sequence similarity.
Figure 2.
Phylogenetic Analysis of Mitochondrial and Chloroplast rps13 Genes.
Unrooted phylogram depicting the results of maximum likelihood analyses of the first and second nucleotide positions. The genes for numit rps13 from Arabidopsis, cotton, and legumes are indicated by asterisks; note the long branch lengths among these sequences compared with those for the nucp rps13 genes (genes for chloroplast rps13, located in the nucleus) from angiosperms. Genes located in the chloroplast genome are indicated by cp. The rps13 genes from the yeasts are located in the nucleus; other genes in the mitochondrial origin box are in the mitochondrial genome. Numbers indicate bootstrap values. See Methods for details of the analysis.
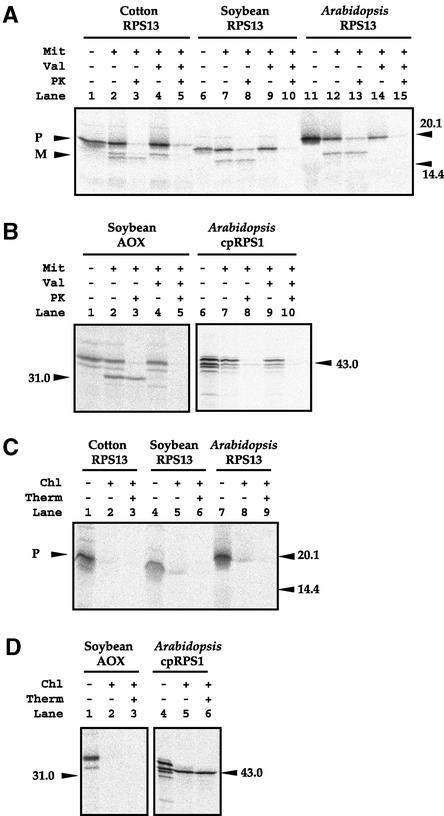
To determine if the numit rps13 genes actually encode mitochondrial proteins, and to determine if any sequence is cleaved from the precursor protein upon import, their gene products were tested for the ability to be imported in vitro into isolated mitochondria. When incubated with isolated soybean mitochondria under conditions that support mitochondrial import, each of the RPS13 proteins from cotton, soybean, and Arabidopsis was imported into soybean mitochondria (Figure 3A; note the proteinase K–insensitive bands in lanes 3, 8, and 13) and processed into a smaller form, most likely by cleavage of the mitochondrial targeting presequence. In contrast, when incubated with isolated pea chloroplasts under conditions that support chloroplast protein import, none of the three RPS13 proteins was imported (note that there were no thermolysin-insensitive proteins detected; Figure 3C, lanes 3, 6, and 9).
Figure 3.
RPS13 Protein Import Experiments.
(A) Import of RPS13 into isolated mitochondria. Lane 1, 35S-labeled precursor protein; lane 2, precursor protein incubated with soybean mitochondria; lane 3, as in lane 2 except that proteinase K was added after mitochondrial incubation to degrade unimported proteins; lanes 4 and 5, as in lanes 2 and 3, respectively, except that valinomycin was added before import to dissipate the membrane potential. Lanes 6 to 10 and 11 to 15 are equivalent to lanes 1 to 5. Markers are given in relative molecular mass (kD). Precursor (P) and mature (M) forms are indicated at left.
(B) Import of the mitochondrial AOX protein (positive control) and the chloroplast protein RPS1 (cpRPS1; negative control) into isolated soybean mitochondria. Lanes are as in (A).
(C) Import of RPS13 into isolated chloroplasts. Lane 1, 35S-labeled RPS13 precursor protein; lane 2, RPS13 precursor protein incubated with pea chloroplasts; lane 3, as in lane 2 except that thermolysin was added after chloroplast incubation to degrade unimported proteins. Lanes 4 to 6 and 7 to 9 are equivalent to lanes 1 to 3.
(D) Import of the mitochondrial protein AOX (negative control) and the chloroplast protein RPS1 (positive control) into isolated pea chloroplasts. Lanes are as in (C).
Chl, chloroplast; Mit, mitochondria; PK, proteinase K; Therm, thermolysin; Val, valinomycin; (+), presence; (−), absence.
To examine further the specificity of protein import into mitochondria and chloroplasts using the in vitro import assays, the mitochondrial protein alternative oxidase (AOX) and the chloroplast protein RPS1 were tested in the assays. As expected, there was strong import of AOX into mitochondria (Figure 3B, lane 3) but no import into chloroplasts (Figure 3D, lane 3). Conversely, there was essentially no import of RPS1 into mitochondria (Figure 3B, lane 8) but strong import into chloroplasts (Figure 3D, lane 6). (When chloroplast RPS1 was tested in the mitochondrial import assay, a very weak band was seen in this figure but not in Figure 6B; most likely, this is caused by a process known as bypass import [Pfaller et al., 1989] rather than authentic import, because the intensity of the signal is extremely weak compared with the signal normally seen in the mitochondrial import assay and compared with the amount of RPS1 imported into chloroplasts.) Overall, these results indicate that the RPS13 proteins from cotton, soybean, and Arabidopsis function in the mitochondrion but not in the chloroplast.
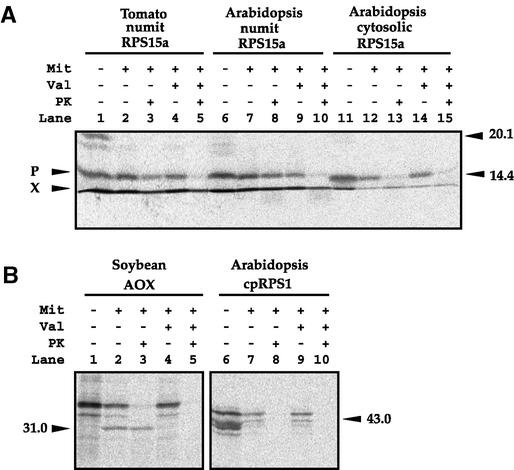
Figure 6.
Import of RPS15A into Mitochondria.
(A) Import of RPS15A into soybean mitochondria. Lanes are the same as in Figure 3A. X denotes the ion front of the gel.
(B) Import of the mitochondrial protein AOX and the chloroplast protein RPS1 (cpRPS1) into isolated soybean mitochondria. Lanes 1 to 5 and 6 to 10 are equivalent to lanes 1 to 5 of (A), except that the AOX and cpRPS1 precursor proteins were used.
Mit, mitochondria; P, precursor; PK, proteinase K; Val, valinomycin.
A Eubacteria-Like rps8 Gene Is Missing from the Mitochondrion and Nucleus of Angiosperms
rps8 is present in the mitochondrial genome of the liverwort Marchantia (Oda et al., 1992), and DNA gel blot hybridization indicated that it is present also in the mitochondria of several other bryophytes and lycopods (data not shown). The rps8 gene has not been discovered in the mitochondrion of any angiosperm; one hypothesis is that rps8 was transferred to the nucleus before the emergence of angiosperms. However, our searches of the Arabidopsis genome and the NCBI EST databases have not revealed a transferred rps8 gene of mitochondrial origin in the nucleus of any angiosperm. Considering that the Arabidopsis genome is 92% sequenced, including all of the gene-rich regions (Arabidopsis Genome Initiative, 2000), and that >65,000 EST sequences are available publicly from each of nine flowering plants (soybean, M. trunculata, Arabidopsis, tomato, maize, wheat, barley, rice, and sorghum), for a total of >895,000 ESTs, the absence of a detectable transferred rps8 gene in the NCBI databases suggests that this gene was not transferred to the nucleus in angiosperms.
Instead, RPS8 probably was replaced by another protein. Note that RPS8 is thought to be an essential ribosomal protein in bacteria, because rps8 mutants cannot assemble the large and small ribosomal subunits (Wower et al., 1992). Might a divergent copy of the nucleus-encoded chloroplast RPS8 be functioning in the mitochondrion, as appears to be the case for RPS13? The gene for chloroplast RPS8 is located in the chloroplast genome in all of the plants examined, and we found no evidence in any sequence databases for transferred copies of chloroplast rps8 in the nucleus of angiosperms. If chloroplast RPS8 itself has taken the place of mitochondrial RPS8 in the mitochondrion, then the protein would have to be exported from the chloroplast and imported into the mitochondrion, an unprecedented and highly unlikely scenario.
A Divergent Copy of Cytosolic RPS15A Is Imported into Mitochondria
We next considered the possibility that a copy of the cytosolic counterpart of rps8, rps15A (also named rps22 and rps24 in some eukaryotes), might provide a product to the mitochondrion. Cytosolic rps15A has been characterized from Arabidopsis (Bonham-Smith and Moloney, 1994), and we used this sequence as a query to the Arabidopsis genome sequence to identify all rps15A-like genes. Two genes with 53 to 54% identity to Arabidopsis cytosolic rps15A were discovered on chromosomes 2 and 4 (Figure 4) (Lin et al., 1999; Mayer et al., 1999; see Methods for accession numbers); these genes were identified concurrently as rps15A genes (RPS15aB and RPS15aE) by Barakat et al. (2001). The two genes are equal in length and are closely related to each other (90% amino acid identity), probably reflecting a recent gene duplication; the copy on chromosome 4 was used for further experiments and analyses. Although the product of this rps15A-like gene product has no N-terminal extension relative to cytosolic RPS15A, it is predicted to be a mitochondrial protein by three prediction programs (Mitoprot, TargetP, and Predotar), with part of the putative mature coding sequence as the targeting domain. Thus, the rps15A-like gene, referred to herein as numit rps15A, is a good candidate as a replacement for the missing rps8 gene product in Arabidopsis mitochondria.
Figure 4.
Alignment of RPS15A and RPS8 Sequences.
Identical amino acids are shown in white on a black or gray background. Dots indicate gaps inserted to improve alignment. The N-terminal extension of Marchantia RPS8 is not shown. Ath cp, chloroplast RPS8 from Arabidopsis; Ath cyto, cytosolic RPS15A from Arabidopsis; Ath numit, numit RPS15A from Arabidopsis; Les numit, numit RPS15A from tomato; Mpo mit, mitochondrial RPS8 from Marchantia; Nol mit, mitochondrial RPS8 from N. olivacea.
To identify homologs of the numit rps15A gene in other angiosperms, we used tBLASTn searches of the NCBI EST databases; complete sequences of homologs were identified in tomato (Figure 4), barley, and M. trunculata (see Methods for accession numbers), representing a diversity of angiosperms. Each of the numit RPS15A amino acid sequences is ∼75 to 80% identical to the others. To verify that each of the numit rps15A genes is derived from a common ancestor and to assess the amount of divergence among the numit rps15A genes compared with each other and compared with the cytosolic rps15A genes, phylogenetic analyses were performed using a variety of rps15A genes from eukaryotes. The four numit rps15A genes branch together in a well-supported clade (Figure 5), and this group branches with the plant cytosolic rps15A sequences. The exact placement of the numit rps15A genes, as sister to the Marchantia gene, probably is a phylogenetic artifact caused by the short length of the gene and the very long branch leading to the mitochondrial sequences relative to the short-branched cytosolic sequences. This long branch, together with the much longer branches within the clade of angiosperm numit rps15A sequences compared with those of the angiosperm cytosolic rps15A genes, indicates much higher rates of diversification of the mitochondrial ribosomal proteins than the cytosolic proteins after the gene duplication event that gave rise to these two lineages of genes. We also identified in the NCBI EST databases numit rps15A ESTs from Pinus taeda (see Methods for accession numbers); although incomplete at the 5′ end, this gene branches with numit rps15A genes in phylogenetic trees (data not shown).
Figure 5.
Phylogenetic Analysis of rps15A Genes.
Unrooted phylogram of maximum likelihood analyses of the first and second nucleotide positions of an rps15A nucleotide alignment. Numbers indicate bootstrap values. See Methods for details of the analyses.
The rather low sequence identity of numit rps15A to its mitochondrial counterparts in other plants (rps8) is to be expected, because it is derived from the counterpart cytosolic ribosomal protein gene (Figure 4). Mitochondrial rps8 is not highly conserved in general. For example, mitochondrial RPS8 amino acid sequences from Marchantia and the green alga Nephroselmis olivacea (i.e., from genes that still reside in the mitochondrion in both organisms) are only 27% identical. Considering that these are both mitochondrially encoded proteins from relatively closely related organisms (land plants having arisen from green algae), it is not surprising that the Marchantia mitochondrial RPS8 would show only 18% identity to a ribosomal protein of far greater evolutionary distance (i.e., numit RPS15A) that is of cytosolic/nuclear ancestry. Nonetheless, there are 34 conserved amino acids in the numit and mitochondrial genes shown in Figure 4, with 11 being conserved completely.
To determine if the numit rps15A genes encode mitochondrial proteins and if they contain a cleavable presequence, numit RPS15A from Arabidopsis and tomato were tested for importability into isolated soybean mitochondria. When incubated with mitochondria under conditions that support protein import into mitochondria, both numit RPS15A proteins clearly were imported into mitochondria, and the imported proteins appear to be the same size as the precursor proteins (Figure 6A, lanes 3 and 8). To determine if targeting was specific for the numit RPS15A but not for the cytosolic RPS15A, the import experiments were repeated with the cytosolic RPS15A from Arabidopsis. When incubated with mitochondria under conditions that support protein import into mitochondria, it was evident that that cytosolic RPS15A did not import into mitochondria (Figure 6, lane 13). Again, the specificity of the in vitro import assay was verified with the mitochondrial protein AOX and the chloroplast protein RPS1. Thus, the numit RPS15A gained mitochondrial targeting ability as a result of sequence divergence from the cytosolic RPS15A.
Expression Patterns of Numit rps13 and Numit rps15A in Arabidopsis
The number of mitochondria per cell increases in anthers in response to the high energy demands of the process of pollen development (Huang et al., 1994). Several studies have shown an increased level of steady state transcripts in floral versus vegetative organs for nuclear genes encoding subunits of respiratory complex I (Grohmann et al., 1996; Heiser et al., 1996; Zabaleta et al., 1998), complex II (Figueroa et al., 2001), complex III (Huang et al., 1994), and cytochrome c (Felitti et al., 1997), along with a few metabolic enzymes (reviewed by Mackenzie and McIntosh, 1999). Therefore, increased expression in flowers appears to be a hallmark of nuclear genes for mitochondrial respiratory and metabolic proteins. In contrast, much less is known about the tissue-specific expression patterns of genes for mitochondrial ribosomal proteins; to our knowledge, only one such study has been reported. Handa et al. (2001) found comparable levels of rpl11 transcripts in flowers, shoots, and roots of wheat, but the transcript levels in flowers and shoots decreased upon cold exposure.
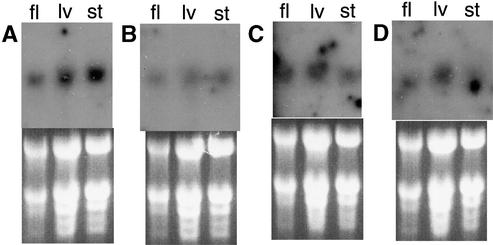
To determine if steady state transcript levels of numit rps13 and numit rps15A are increased in flowers of Arabidopsis relative to other organs, we performed RNA gel blot hybridizations using probes for these genes along with the recently transferred nuclear genes for mitochondrial rps10 (Wischmann and Schuster, 1995) and 3′ rpl2 (Adams et al., 2001a). Figure 7 shows that, for all four ribosomal protein genes tested, transcript accumulation is comparable in whole flowers, leaves, and stems. (Note that the flower lanes are underloaded, as indicated by comparison with the rRNA bands on the ethidium bromide–stained gels.) These data and those of Handa et al. (2001) suggest that steady state transcript levels of genes for mitochondrial ribosomal proteins are not increased in flowers, in contrast to genes for other mitochondrial proteins.
Figure 7.
Expression Patterns of Four Nuclear Genes for Mitochondrial Ribosomal Proteins.
(A) 3′ rpl2.
(B) rps10.
(C) Numit rps15A.
(D) Numit rps13.
Top panels show hybridizations, and bottom panels show the corresponding ethidium bromide–stained gels with prominent rRNA bands. The rps10 and 3′ rpl2 transcripts are ∼1.8 kb, the rps15A transcripts are ∼1.0 kb, and the rps13 transcripts have an approximate range of 900 to 1000 bp (with smaller transcripts present in flowers). fl, whole flowers; lv, rosette leaves; st, stems.
DISCUSSION
Substitution of Two Mitochondrial Ribosomal Proteins by Highly Derived Copies of Their Chloroplast or Cytosolic Counterparts
The results of this study indicate that a divergent copy of the cytosolic rps15A gene, termed numit rps15A, is present in the nucleus of diverse angiosperms and a gymnosperm and that the gene products from tomato and Arabidopsis are imported into mitochondria. We also have shown that a divergent copy of the chloroplast-derived rps13 gene (numit rps13) is present in the nucleus of cotton, Arabidopsis, and legumes and that its product is imported into mitochondria but not chloroplasts.
Three lines of evidence indicate that the gene products of numit rps13 and rps15A are mitochondrial ribosomal proteins. First, rps13 and rps8 (the organellar homolog of rps15A) clearly are absent from the mitochondrial genomes of the plants in question, and recently transferred copies are absent from the Arabidopsis genome and from the NCBI EST databases containing extensive cotton and legume ESTs. Second, feasible alternative ribosomal proteins are encoded by nuclear genes (numit rps13 and numit rps15A), and their products are predicted to be mitochondrial proteins by all three mitochondrial protein prediction programs used. Third, the products of numit rps13 and numit rps15A were shown to be imported by isolated plant mitochondria. Numit RPS13 was not imported by chloroplasts, and cytosolic RPS15A was not imported by mitochondria; thus, numit rps13 and rps15A have evolved specific signals for mitochondrial targeting. Most likely, therefore, numit RPS13 and RPS15A have partially or completely replaced the function of the missing ribosomal proteins RPS8 and RPS13 of mitochondrial origin. To convincingly prove that numit RPS13 and RPS15A function in mitochondrial ribosomes in the plants in question would require a substantial undertaking, one that is well beyond the scope of the current study. Immunoblot analyses of mitochondrial ribosomes would not suffice, because homologous ribosomal proteins of different compartmental origins will sometimes cross-react. Instead, it probably would be necessary to purify and microsequence the candidate proteins from mitochondrial ribosomes.
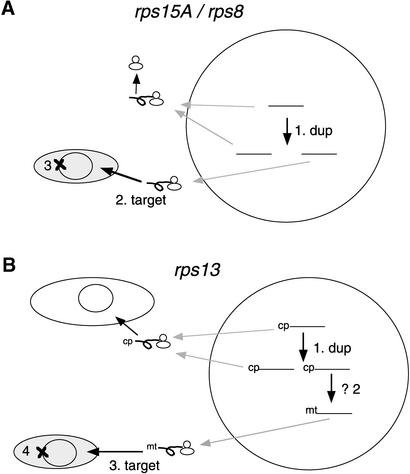
A model of the numit rps15A gene substitution process is shown in Figure 8A. The region used to target the protein to the mitochondria probably is within the mature coding region, given the apparent lack of processing of the protein upon import into mitochondria. A small number of mitochondrial proteins in plants have been shown to be targeted and imported without a mitochondrial targeting presequence (Adams et al., 2000, and references therein). The numit rps15A gene has diverged considerably from the very highly conserved cytosolic rps15A genes (Figure 5), consistent with functional divergence in the new ribosomal environment. This gene substitution event is accompanied by, and presumably led to, loss of the rps8 gene from the mitochondrion. This seldom-used pathway of mitochondrial gene content evolution is in contrast to the many examples of mitochondrial gene transfer to the nucleus that seem to account for most mitochondrial gene losses (see Introduction).
Figure 8.
Models of Gene Substitution Events in the Evolution of Mitochondrial Ribosomal Proteins.
Large circles indicate nuclei, gray ovals indicate mitochondria, and white ovals indicate chloroplasts. Black arrows indicate gene duplication (dup) or protein targeting to mitochondria (target); gray arrows indicate movement of gene transcripts to the cytosol. Bars indicate the mature coding regions of genes; targeting presequences are indicated by cp (chloroplast) and mt (mitochondrial). Proteins are indicated by loops attached to the ribosomes. Gene deletion (or pseudogene formation) is indicated by ×.
(A) rps8/rps15A substitution. Step 1, cytosolic rps15A duplication; step 2, targeting of the duplicate to mitochondria; step 3, loss of rps8 from mitochondrial DNA.
(B) rps13 substitution. Step 1, duplication of nucp rps13; step 2, possible acquisition of mitochondrial targeting presequence; step 3, targeting of the duplicate to mitochondria; step 4, loss of rps13 from mitochondrial DNA.
A model for the complex history of the organellar rps13 genes is shown in Figure 8B. Chloroplast rps13 was transferred to the nucleus (Kumar et al., 1995), and this nuclear gene was duplicated, followed by sequence and functional divergence of one copy to allow replacement of the mitochondrial rps13. Numit rps13 has a mitochondrial targeting sequence that has no similarity to the targeting sequence of nucp rps13. Thus, either there has been extensive sequence divergence in the numit rps13 presequence, even more than for the mature coding region (Figure 2), so that the sequence is unrecognizable, or the numit rps13 gene acquired a mitochondrial presequence that replaced the original chloroplast presequence. Eventually, the mitochondrial rps13 gene was lost multiple times in different descendant lineages (Figure 8B). Numit RPS13 represents an example of the substitution of a chloroplast-derived ribosomal protein for a mitochondrial ribosomal protein and illustrates another pathway by which the nuclear genome can usurp mitochondrial genes.
Our results differ from, but in a sense are consistent with, those of two previous studies on rps13. A maize mitochondrial RPS13 antibody and a nucp RPS13 antibody used by Kumar et al. (1995) and Sánchez et al. (1996), respectively, did not react with Arabidopsis mitochondrial ribosomal proteins, leading these authors to hypothesize that a eubacteria-like ribosomal protein, S13, is absent from Arabidopsis mitochondria. The extensive sequence divergence of the numit rps13 from both the nucp rps13 and the mitochondrial rps13 of other angiosperms (Figure 2) might explain why neither antibody recognized the numit rps13 gene product reported here. The mitochondrial RPS13 antibody did react with mitochondrial ribosomal proteins from bean (Kumar et al., 1995), and the nucp RPS13 antibody reacted with mitochondrial ribosomal proteins from pea (Sánchez et al., 1996), leading the authors to hypothesize that the gene was transferred to the nucleus or that the nucp RPS13 might have replaced the mitochondrial RPS13 in pea. Sánchez et al. (1996) proposed that the supposedly missing eubacteria-derived RPS13 protein in Arabidopsis mitochondria was functionally replaced by the RNP domain on the transferred RPS19. In light of the identification of numit rps13 in Arabidopsis, one must reconsider this proposal. It now seems entirely possible, perhaps even likely, that this RNP domain carries no RPS13 function. Alternatively, the function of RPS13 in Arabidopsis mitochondria might have been partitioned between the numit RPS13 and the RNP domain of RPS19 after the divergence of Arabidopsis and cotton from a common ancestor.
Evolutionary Timing of the Gene Substitutions
The losses of rps13 from angiosperm mitochondria are highly clustered in core rosids (Adams et al., 2002), and numit rps13 genes have been found from three diverse groups of core rosids (cotton, legumes, and Arabidopsis), the common ancestor of which is at the base of core rosids (Soltis et al., 1999, 2000). Conversely, there have been very few losses of rps13 from the mitochondria of nonrosid angiosperms, and numit rps13 genes have not been found in any nonrosid species (including tomato and the grasses, with many EST sequences available; see Results). From these observations, we infer that the gene substitution event occurred more or less at the base of the core rosid lineage.
The results of rps13 phylogenetic analyses (Figure 2) provide little insight into the timing of the gene substitution, probably because rps13 is a short gene and has a small number of informative characteristics. The chloroplast rps13 sequences from angiosperms do not branch according to their expected phylogenetic relationships. For example, Arabidopsis and legumes (both rosids) do not branch together but instead are interspersed with other eudicots and even monocots. Thus, it is completely unsurprising that the divergent numit rps13 genes from rosids do not branch as sister to a clade of rosid nucp rps13 sequences, as would be expected for a gene substitution at the base of core rosids.
rps13 has been sequenced from the mitochondrial genomes of several nonrosid angiosperms, including maize, wheat, Magnolia, carrot, tobacco, and beet. Interestingly, an intact, transcribed, and RNA-edited rps13 gene also is present in the mitochondrial genome of the rosid Oenothera (Wissinger et al., 1990). It is possible, therefore, that Oenothera mitochondria might contain RPS13 proteins produced by both nuclear (numit rps13) and mitochondrial rps13 genes. Alternatively, numit rps13 might have been lost or become a pseudogene in Oenothera, or the transcribed and edited mitochondrial rps13 mRNA might not be translated.
rps8 has not been found in the mitochondria of any angiosperm, but it is present in the mitochondrial genome of the liverwort Marchantia (Oda et al., 1992). DNA gel blot hybridizations of ∼30 nonseed plants using a Marchantia rps8 probe revealed good hybridization to the DNAs of many liverworts, mosses, and lycopods (data not shown), suggesting the widespread presence of mitochondrial rps8 in these basal land plant lineages. The substitution of rps8 by the numit rps15A occurred minimally in a common ancestor of angiosperms and gymnosperms, as shown by the presence of an orthologous numit rps15A gene in both groups. Together, these observations indicate that the gene substitution occurred after the divergence of lycopods (the first branch of vascular plant evolution) and before the diversification of angiosperms and gymnosperms from a common ancestor. (The rps15A phylogenetic tree [Figure 5] shows little meaningful resolution among the land plant rps15A sequences and thus is uninformative regarding the timing of gene substitution.)
Gene Substitution in the Evolution of Organellar Proteins
As with the gene losses from mitochondria, most investigated chloroplast gene losses in plants also appear to reflect gene transfer to the nucleus (Gantt et al., 1991; Martin et al., 1998; Millen et al., 2001). We are aware of only two described cases of gene substitution in the evolution of chloroplast ribosomal proteins in land plants. A long-standing controversy regarding the origin of the angiosperm nuclear gene for chloroplast RPL21 (Smooker et al., 1990; Martin et al., 1990) was settled recently by Gallois et al. (2001), who showed that this gene was derived by substitution from a nuclear rpl21 gene of mitochondrial origin. The chloroplast-derived mitochondrial RPS13 (numit rps13) and the mitochondrially derived chloroplast RPL21 essentially represent the same type of substitution but in opposite directions: a chloroplast protein for a mitochondrial protein in one case, and a mitochondrial protein for a chloroplast protein in the other.
The second case of chloroplast gene substitution parallels the cytosol-to-mitochondria substitution described in this study for rps8 and rps15A. Spinach chloroplast DNA contains only a pseudogene for rpl23, and the nuclear gene that supplies the spinach chloroplast ribosome with RPL23 evidently is derived by substitution from a duplicated copy of cytosolic rpl23 (Bubunenko et al., 1994). (Although there was biochemical evidence for this conclusion, no cytosolic rpl23 gene had been isolated from plants at the time. The sequence of cytosolic rpl23 from spinach has since been determined [X92367], and homologous sequences from Arabidopsis and other angiosperms also are available in the NCBI sequence databases. The nucleus-encoded chloroplast rpl23 from angiosperms is much more similar to the angiosperm cytosolic rpl23 [∼51%] than to the angiosperm chloroplast-encoded rpl23 [∼20%]. Thus, the inference of Bubunenko et al. [1994] about the origin of the spinach chloroplast rpl23 from the mitochondrial rpl23 seems to be correct.) An interesting difference between the rps15A and rpl23 substitutions is that rpl23 gained an N-terminal chloroplast targeting presequence but rps15A did not gain a mitochondrial presequence and relies on an internal signal that has evolved as a result of sequence divergence. These four cases of substituted organellar ribosomal protein genes in land plants provide clear and dramatic examples of the gain of new function by one gene copy after gene duplication, one of the major possible fates of duplicated genes (Lynch and Conery, 2000, and references therein).
Although gene substitution appears to be relatively rare in the evolution of organellar ribosomal proteins in plants, and gene transfer appears to be the predominant mode, in the case of metabolic and other enzymes, gene substitution is rather common (reviewed by Martin and Schnarrenberger, 1997; Small et al., 1998). To a significant extent, this probably reflects differences in the degree to which the different classes of proteins are coadapted to interact over large portions of their length with other molecules. Ribosomal proteins often interact with multiple other ribosomal proteins and the rRNA within the ribosome, and it may be relatively difficult for a ribosomal protein that is adapted to the ribosome of the chloroplast or cytosol to substitute effectively for a missing mitochondrial ribosomal protein. In contrast, many metabolic enzymes interact with the same few, relatively simple substrates and cofactors regardless of their disparate evolutionary histories, so functional substitution may be achieved more easily. Fundamentally, this is the same argument—the “complexity hypothesis” of Jain et al. (1999)—used to explain the observation that lateral gene transfer is much more common in the evolution of metabolic enzymes in bacteria than for components of the genetic apparatus.
The considerable divergence of the substituted copies of rps13 and rps15A relative to those that still function in their original compartments (Figures 2 and 5) may be related to the apparent difficulty of substitution of ribosomal proteins. That is, it may reflect selection for a highly altered form of the protein, one that functions better in its new ribosome. In contrast, in a few cases, a single nuclear gene (of either chloroplast or mitochondrial origin) encodes a tRNA synthetase (Akashi et al., 1998; Menand et al., 1998; Small et al., 1998) or a metabolic enzyme (Creissen et al., 1995) that is targeted to and functions in both the chloroplast and the mitochondrion, highlighting the ready interchangeability of these enzymes.
Further indication of the interchangeability of some metabolic enzymes and tRNA synthetases is shown by cytosolic proteins that are derived from their organellar counterparts (e.g., Flechner et al., 1996; Mireau et al., 1996; Peeters et al., 2000; Krepinsky et al., 2001; reviewed by Martin and Schnarrenberger, 1997). Perhaps the most intriguing cases of gene substitution in organellar evolution involve the single-subunit, phage-type RNA polymerase (nucleus encoded but of uncertain origin) that has completely or partly substituted for the multisubunit eubacteria-type polymerase that was encoded originally by the mitochondrial or the chloroplast genome, respectively (Hedtke et al., 1997, 2000; Gray and Lang, 1998; Cahoon and Stern, 2001). These cases are intriguing both because they involve an enzyme (RNA polymerase) that interacts with so many other players (i.e., promoters) and because they include at least one case of dual targeting of a single enzyme to both organelles.
METHODS
Gene Amplification, Cloning, and Sequencing
A soybean (Glycine max) mitochondrial rps13 cDNA clone (BG047149) was purchased from Incyte Genomics (Palo Alto, CA) and sequenced fully using M13 forward and reverse primers, along with the internal forward primer 5′-AGATGTGGGAAGGTTGGTGG-3′. The genomic sequence of soybean mitochondrial rps13 was isolated by polymerase chain reaction (PCR) using primers 5′-CAACTCCACCATGTTCGG-TTC-3′ and 5′-TCACATCATCATAGTTTTCCGGCTCC-3′.
To construct cDNA clones for protein import experiments, complete cDNAs were isolated by reverse transcriptase–mediated RT-PCR (Adams et al., 1999), and additional Met codons were added to the 3′ end of each cDNA using the following primers: Arabidopsis thaliana rps13 (F1, 5′-ATGGGTATCTCTCGTGATTCA-3′; R2, 5′-TTCGGGGAGCGTACTACTACTACACT-3′; and R3, 5′-TCACATCATCATCATCATCATCATCATG-3′), cotton (Gossypium arboreum) rps13 (F1, 5′-TCAGCGTCTAAGAATGTTGGG-3′; R1, 5′-TCACATCATCATCATAGCATAAGAAGC-3′; and R2, 5′-TCACATCATCATCATCATCATCATAGCA-3′), soybean rps13 (F1, 5′-CAACTCCACCATGTT-CGGTTC-3′; R1, 5′-TCACATCATCATAGTTTTCCGGCTCC-3′; and R2, 5′-TCACATCATCATCATCATCATCATCATAGT-3′), tomato (Lycopersicon esculentum) rps15A (F1, 5′-AATGGGGAGGAGAATATT-GAAC-3′; R1, 5′-TCACATATAAAAGTAGCCAAGAAC-3′; and R2, 5′-TCACATCATCATCATCATCATATAAAAGT-3′), Arabidopsis mitochondrial rps15A (F1, 5′-ATGGGGAGGAGGATTTTGAAC-3′; R1, 5′-TCACATCATGTAAAAGAAGCCAAGAAC-3′; and R2, 5′-TCACATCATCATCATCATGTAAAAGAAG-3′), and Arabidopsis cytosolic rps15A (F1, 5′-AAGGCAAGATGGTAAGAATCAG-3′; and R1, 5′-TCA-CATATAGAAGAAGCCGAGAACCT-3′).
All RT-PCR products were cloned into vector pCR2.1 TOPO (Invitrogen, Carlsbad, CA), and some inserts were subcloned into pBluescript KS− (Stratagene). All clones were sequenced using an ABI 3700 DNA sequencer (Foster City, CA) to verify the orientation of the insert relative to the T7 site and to detect Taq polymerase misincorporation errors.
Sequence and Phylogenetic Analyses
Sequence alignments were made using the xPileUp program from the Genetics Computer Group (Madison, WI) and refined by eye. Pairwise distance comparisons were performed using PAUP* version 4.0b8 (Swofford, 2001) and represent uncorrected pairwise distances.
Parsimony analyses were performed with PAUP* version 4.0b8 (Swofford, 2001) using heuristic searches with random taxon addition (10 replicates) and tree bisection reconnection (TBR) branch swapping. Bootstrapping was performed as described above, with stepwise addition and 100 replicates. Maximum likelihood analyses were conducted with PAUP* version 4.0b8. The HKY85 model was used, assuming a discrete gamma distribution with four categories of site-to-site rate variability. For each analysis, the transition/transversion ratio and base frequencies were estimated using Tree-PUZZLE version 4.02 (Strimmer and von Haeseler, 1996) under the HKY model of evolution with gamma-distributed rates and parameter estimation set to “approximate.” Analyses used heuristic searches with random taxon addition (10 replicates) and TBR branch swapping. Bootstrapping was performed using PAUP* version 4.0b8 as described above, with stepwise addition and 100 replicates.
RNA Isolation and RNA Gel Blot Hybridization
Total RNA was isolated from Arabidopsis ecotype Columbia using the Qiagen Plant RNAeasy kit (Valencia, CA) according to the manufacturer's instructions. The following tissues and organs were used: whole flowers at various stages, rosette leaves, and stems. Approximately 10 μg of total RNA per lane was electrophoresed, blotted, and hybridized with 32P-labeled probes as described by Adams et al. (1999).
Organelle Isolation and in Vitro Import Assays
Mitochondria were prepared from 7-day-old soybean cotyledons (cv Stevens) as described by Day et al. (1985). 35S-labeled precursor proteins were synthesized from cDNA clones as described by Whelan et al. (1995). In vitro import assays were performed as described by Whelan et al. (1996). Proteins were separated by 16% SDS-PAGE, and gels were dried and exposed to a BAS TR2040S plate for 24 hr. Detection was performed on a BAS 2500 phosphorimager according to the manufacturer's instructions (Fuji, Tokyo, Japan).
Chloroplasts were prepared from 10-day-old pea leaves (Pisum sativum cv Greenfeast) as described by Bruce et al. (1994). In vitro import assays were performed as described by Bruce et al. (1994) and Waegemann and Soll (1995). Protein separation and detection were performed as for mitochondrial in vitro import assays.
In some cases, smaller precursor proteins were denatured before in vitro import into mitochondria or chloroplasts. 35S-labeled precursor proteins were centrifuged at 128,000g for 15 min at 4°C. The supernatant was precipitated by the addition of 4 volumes of water-saturated (NH4)2SO4, incubated on ice for 30 min, and centrifuged at 14,000g for 15 min at 4°C. The protein pellet containing the 35S-labeled precursor protein was resuspended in half of its original volume with 8 M urea, incubated on ice for 30 min, and then diluted with an equal volume of 20 mM Hepes, pH 7.7.
Accession Numbers
The soybean mitochondrial rps13 genomic sequence has been assigned GenBank accession number AY044157. Accession numbers for EST and Arabidopsis genomic sequences used in this study are as follows. Numit rps13: Arabidopsis (AC012193, AV440855, AV442527, and AV552576), cotton (BF272717), soybean cDNAs (BG047149 and BG237013), M. trunculata (BG449364, AL388944, and AL388196), and L. japonicus (AV423337). Numit rps15A: Arabidopsis (AC005169 and AL161575), tomato (AW220956 and BG135881), M. trunculata (AW776943, AW559986, and BF519104), and P. taeda (BE607125, BE607249, and BE758611).
NOTE ADDED IN PROOF
Another report of numit rps13 from Arabidopsis was recently published online: Mollier, P., Hoffmann, B., Debast, C., and Small, I. (2002). The gene encoding Arabidopsis thaliana mitochondrial ribosomal protein S13 is a recent duplication of the gene encoding plastid S13. Curr. Genet. DOI 10.1007/s00294-002-0271-5 (http://link.springer.de/link/service/journals/00294/contents/02/00271). These authors also concluded that the gene is derived from nucp rps13 and that its product functions in mitochondria.
Acknowledgments
We thank Mark Stoutemyer for help with sequencing, Beth Fatland for providing Arabidopsis plants, and Jonathan Wendel for reading the manuscript. K.L.A. was supported by a William Ogg fellowship from Indiana University and is a Fellow of the Jane Coffin Childs Memorial Fund for Medical Research. Funding for this work came from National Institutes of Health Grant GM-35087 to J.D.P. and a grant from the Australian Research Council to J.W.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010483.
References
- Adams, K.L., Song, K., Roessler, P.G., Nugent, J.M., Doyle, J.L., Doyle, J.J., and Palmer, J.D. (1999). Intracellular gene transfer in action: Dual transcription and multiple silencings of nuclear and mitochondrial cox2 genes in legumes. Proc. Natl. Acad. Sci. USA 96, 13863–13868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, K.L., Daley, D.O., Qiu, Y.-L., Whelan, J., and Palmer, J.D. (2000). Repeated, recent and diverse transfers of a mitochondrial gene to the nucleus in flowering plants. Nature 408, 354–357. [DOI] [PubMed] [Google Scholar]
- Adams, K.L., Ong, H.C., and Palmer, J.D. (2001. a). Mitochondrial gene transfer in pieces: Fission of a ribosomal protein gene and partial or complete gene transfer to the nucleus. Mol. Biol. Evol. 18, 2289–2297. [DOI] [PubMed] [Google Scholar]
- Adams, K.L., Rosenblueth, M., Qiu, Y.-L., and Palmer, J.D. (2001. b). Multiple losses and transfers to the nucleus of two mitochondrial succinate dehydrogenase genes during angiosperm evolution. Genetics 158, 1289–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, K.L., Qiu, Y.-L., Stoutemyer, M., and Palmer, J.D. (2002). Punctuated evolution of mitochondrial gene content: High and variable rates of mitochondrial gene loss and transfer during angiosperm evolution. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- Akashi, K., Grandjean, O., and Small, I. (1998). Potential dual targeting of an Arabidopsis archaebacterial-like histidyl-tRNA synthetase to mitochondria and chloroplasts. FEBS Lett. 431, 39–44. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Barakat, A., Szick-Miranda, K., Chang, I.F., Guyot, R., Blanc, G., Cooke, R., Delseny, M., and Bailey-Serres, J. (2001). The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiol. 27, 398–415. [PMC free article] [PubMed] [Google Scholar]
- Blanchard, J.L., and Schmidt, G.W. (1995). Pervasive migration of organellar DNA to the nucleus in plants. J. Mol. Evol. 41, 397–406. [DOI] [PubMed] [Google Scholar]
- Bonham-Smith, P.C., and Moloney, M.M. (1994). Nucleotide and protein sequences of a cytoplasmic ribosomal protein S15a gene from Arabidopsis thaliana. Plant Physiol. 106, 401–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennicke, A., Grohmann, L., Hiesel, R., Knoop, V., and Schuster, W. (1993). The mitochondrial genome on its way to the nucleus: Different stages of gene transfer in higher plants. FEBS Lett. 325, 140–145. [DOI] [PubMed] [Google Scholar]
- Bruce, B.D., Perry, S., Froelich, J., and Keegstra, K. (1994). In vitro import of proteins into chloroplasts. In Plant Molecular Biology Manual, 2nd ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. J1–J15.
- Bubunenko, M.G., Schmidt, J., and Subramanian, A.R. (1994). Protein substitution in chloroplast ribosome evolution: A eukaryotic cytosolic protein has replaced its organelle homologue (L23) in spinach. J. Mol. Biol. 240, 28–41. [DOI] [PubMed] [Google Scholar]
- Cahoon, A.B., and Stern, D.B. (2001). Plastid transcription: A menage à trois? Trends Plant Sci. 6, 45–46. [DOI] [PubMed] [Google Scholar]
- Claros, M.G., and Vincens, P. (1996). Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241, 779–786. [DOI] [PubMed] [Google Scholar]
- Creissen, G., Reynolds, H., Xue, Y., and Mullineaux, P. (1995). Simultaneous targeting of pea glutathione reductase and of a bacterial fusion protein to chloroplasts and mitochondria in transgenic tobacco. Plant J. 8, 167–175. [DOI] [PubMed] [Google Scholar]
- Day, D.A., Neuberger, M., and Douce, R. (1985). Biochemical characterization of chlorophyll-free mitochondria from pea leaves. Aust. J. Plant Physiol. 12, 219–228. [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Faxén, M., Walles-Granberg, A., and Isaksson, L.A. (1994). Antisuppression by a mutation in rpsM (S13) giving a shortened ribosomal protein S13. Biochim. Biophys. Acta 1218, 27–34. [DOI] [PubMed] [Google Scholar]
- Felitti, S.A., Chan, R.L., Gago, G., Valle, E.M., and Gonzalez, D.H. (1997). Expression of sunflower cytochrome c mRNA is tissue-specific and controlled by nitrate and light. Physiol. Plant. 99, 342–347. [Google Scholar]
- Figueroa, P., Gómez, I., Holuigue, L., Araya, A., and Jordana, X. (1999. a). Transfer of rps14 from the mitochondrion to the nucleus in maize implied integration within a gene encoding the iron-sulphur subunit of succinate dehydrogenase and expression by alternative splicing. Plant J. 18, 601–609. [DOI] [PubMed] [Google Scholar]
- Figueroa, P., Gómez, I., Carmona, R., Holuigue, L., Araya, A., and Jordana, X. (1999. b). The gene for mitochondrial ribosomal protein S14 has been transferred to the nucleus in Arabidopsis thaliana. Mol. Gen. Genet. 262, 139–144. [DOI] [PubMed] [Google Scholar]
- Figueroa, P., Leon, G., Elorza, A., Holuigue, L., and Jordana, X. (2001). Three different genes encode the iron-sulfur subunit of succinate dehydrogenase in Arabidopsis thaliana. Plant Mol. Biol. 46, 241–250. [DOI] [PubMed] [Google Scholar]
- Flechner, A., Dreben, U., Westhoff, P., Henze, K., Schnarrenberger, C., and Martin, W. (1996). Molecular characterization of transketolase (EC 2.2.1.1) active in the Calvin cycle of spinach chloroplasts. Plant Mol. Biol. 32, 475–484. [DOI] [PubMed] [Google Scholar]
- Gallois, J.-L., Achard, P., Green, G., and Mache, R. (2001). The Arabidopsis chloroplast ribosomal protein L21 is encoded by a nuclear gene of mitochondrial origin. Gene 274, 179–185. [DOI] [PubMed] [Google Scholar]
- Gantt, J.S., Baldauf, S.L., Calie, P.J., Weeden, N.F., and Palmer, J.D. (1991). Transfer of rpl22 to the nucleus greatly preceded its loss from the chloroplast and involved the gain of an intron. EMBO J. 10, 3073–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, M.W. (1992). The endosymbiont hypothesis revisited. Int. Rev. Cytol. 141, 233–357. [DOI] [PubMed] [Google Scholar]
- Gray, M.W., and Lang, B.F. (1998). Transcription in chloroplasts and mitochondria: A tale of two polymerases. Trends Microbiol. 6, 1–3. [DOI] [PubMed] [Google Scholar]
- Gray, M.W., Burger, G., and Lang, B.F. (1999). Mitochondrial evolution. Science 283, 1476–1481. [DOI] [PubMed] [Google Scholar]
- Grohmann, L., Rasmusson, A.G., Heiser, V., Thieck, O., and Brennicke, A. (1996). The NADH-binding subunit of respiratory chain complex I is nuclear-encoded in plants and identified only in mitochondria. Plant J. 10, 793–803. [DOI] [PubMed] [Google Scholar]
- Handa, H., Kobayashi-Uehara, A., and Murayama, S. (2001). Characterization of a wheat cDNA encoding mitochondrial ribosomal protein L11: Qualitative and quantitative tissue-specific differences in its expression. Mol. Genet. Genomics 265, 569–575. [DOI] [PubMed] [Google Scholar]
- Hedtke, B., Borner, T., and Weihe, A. (1997). Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis thaliana. Science 277, 809–811. [DOI] [PubMed] [Google Scholar]
- Hedtke, B., Borner, T., and Weihe, A. (2000). One RNA polymerase serving two genomes. EMBO Rep. 1, 435–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiser, V., Brennicke, A., and Grohmann, L. (1996). The plant mitochondrial 22 kDa (PSST) subunit of respiratory chain complex I is encoded by a nuclear gene with enhanced transcript levels in flowers. Plant Mol. Biol. 31, 1195–1204. [DOI] [PubMed] [Google Scholar]
- Huang, J., Struck, F., Matzinger, D.F., and Levings, C.S. (1994). Flower-enhanced expression of a nuclear-encoded mitochondrial respiratory protein is associated with changes in mitochondrion number. Plant Cell 6, 439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain, R., Rivera, M.C., and Lake, J.A. (1999). Horizontal gene transfer among genomes: The complexity hypothesis. Proc. Natl. Acad. Sci. USA 96, 3801–3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop, V., and Brennicke, A. (1994). Promiscuous mitochondrial group II intron sequences in plant nuclear genomes. J. Mol. Evol. 39, 144–150. [DOI] [PubMed] [Google Scholar]
- Krepinsky, K., Plaumann, M., Martin, W., and Schnarrenberger, C. (2001). Purification and cloning of chloroplast 6-phosphogluconate dehydrogenase from spinach: Cyanobacterial genes for chloroplast and cytosolic isoenzymes encoded in eukaryotic chromosomes. Eur. J. Biochem. 268, 2678–2686. [DOI] [PubMed] [Google Scholar]
- Kubo, T., Nishizawa, S., Sugawara, A., Itchoda, N., Estiati, A., and Mikami, T. (2000). The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNA Cys (GCA). Nucleic Acids Res. 28, 2571–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, R., Drouaud, J., Raynal, M., and Small, I. (1995). Characterization of the nuclear gene encoding chloroplast ribosomal protein S13 from Arabidopsis thaliana. Curr. Genet. 28, 346–352. [DOI] [PubMed] [Google Scholar]
- Lin, X., et al. (1999). Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature 402, 761–768. [DOI] [PubMed] [Google Scholar]
- Lynch, M., and Conery, J.S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. [DOI] [PubMed] [Google Scholar]
- Mackenzie, S., and McIntosh, L. (1999). Higher plant mitochondria. Plant Cell 11, 571–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, W., and Herrmann, R.G. (1998). Gene transfer from organelles to the nucleus: How much, what happens, and why? Plant Physiol. 118, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, W., and Schnarrenberger, C. (1997). The evolution of the Calvin cycle from prokaryotic to eukaryotic chromosomes: A case study of functional redundancy in ancient pathways through endosymbiosis. Curr. Genet. 32, 1–18. [DOI] [PubMed] [Google Scholar]
- Martin, W., Lagrange, T., Li, Y.F., Bisanz-Seyer, C., and Mache, R. (1990). Hypothesis for the evolutionary origin of the chloroplast ribosomal protein L21 of spinach. Curr. Genet. 18, 553–556. [DOI] [PubMed] [Google Scholar]
- Martin, W., Stoebe, B., Goremykin, V., Hansmann, S., Hasegawa, M., and Kowallik, K.V. (1998). Gene transfer to the nucleus and the evolution of chloroplasts. Nature 393, 162–165. [DOI] [PubMed] [Google Scholar]
- Mayer, K., et al. (1999). Sequence and analysis of chromosome 4 of the plant Arabidopsis thaliana. Nature 402, 769–776. [DOI] [PubMed] [Google Scholar]
- Menand, B., Marechal-Drouard, L., Sakamoto, W., Dietrich, A., and Wintz, H. (1998). A single gene of chloroplast origin codes for mitochondrial and chloroplastic methionyl-tRNA synthetase in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 95, 11014–11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millen, R.S., et al. (2001). Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell 13, 645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mireau, H., Lancelin, D., and Small, I.D. (1996). The same Arabidopsis gene encodes both cytosolic and mitochondrial alanyl-tRNA synthetases. Plant Cell 8, 1027–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda, K., Yamato, K., Ohta, E., Nakamura, Y., Takemura, M., Nozato, N., Akashi, K., Kanegae, T., Ogura, Y., Kohchi, T., and Ohyama, K. (1992). Gene organization deduced from the complete sequence of liverwort Marchantia polymorpha mitochondrial DNA. J. Mol. Biol. 223, 1–7. [DOI] [PubMed] [Google Scholar]
- Palmer, J.D., Adams, K.L., Cho, Y., Parkinson, C.L., Qiu, Y.L., and Song, K. (2000). Dynamic evolution of plant mitochondrial genomes: Mobile genes and introns and highly variable mutation rates. Proc. Natl. Acad. Sci. USA 97, 6960–6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters, N.M., Chapron, A., Giritch, A., Grandjean, O., Lancelin, D., Lhomme, T., Vivrel, A., and Small, I. (2000). Duplication and quadruplication of Arabidopsis thaliana cysteinyl- and asparaginyl-tRNA synthetase genes of organellar origin. J. Mol. Evol. 50, 413–423. [DOI] [PubMed] [Google Scholar]
- Perotta, G., Grienenberger, J.M., and Gualberto, J.M. (1998). Plant mitochondrial rps2 genes code for proteins with a C-terminal extension that apparently is processed. In Plant Mitochondria: From Gene to Function, I.M. Møller, P. Gardeström, K. Glimelius, and E. Glaser, eds (Leiden, The Netherlands: Backhuys Publishers), pp. 37–41.
- Pfaller, R., Pfanner, N., and Neupert, W. (1989). Mitochondrial protein import: Bypass of proteinaceous surface receptors can occur with low specificity and efficiency. J. Biol. Sci. 264, 34–39. [PubMed] [Google Scholar]
- Sánchez, H., Fester, T., Kloska, S., Schroder, W., and Schuster, W. (1996). Transfer of rps19 to the nucleus involves the gain of an RNP-binding motif which may functionally replace RPS13 in Arabidopsis mitochondria. EMBO J. 15, 2138–2149. [PMC free article] [PubMed] [Google Scholar]
- Small, I., Wintz, H., Akashi, K., and Mireau, H. (1998). Two birds with one stone: Genes that encode products targeted to two or more compartments. Plant Mol. Biol. 38, 265–277. [PubMed] [Google Scholar]
- Smooker, P.M., Kruft, V., and Subramanian, A.R. (1990). A ribosomal protein is encoded in the chloroplast DNA in a lower plant but in the nucleus of angiosperms. J. Biol. Chem. 265, 16699–16703. [PubMed] [Google Scholar]
- Soltis, P.S., Soltis, D.E., and Chase, M. (1999). Angiosperm phylogeny inferred from multiple genes as a tool for comparative biology. Nature 402, 402–404. [DOI] [PubMed] [Google Scholar]
- Soltis, P.S., et al. (2000). Angiosperm phylogeny inferred from 18S rDNA, rbcL, and atpB sequences. Bot. J. Linn. Soc. 133, 381–461. [Google Scholar]
- Strimmer, K., and von Haeseler, A. (1996). Quartet puzzling: A quartet maximum likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13, 964–969. [Google Scholar]
- Swofford, D.L. (2001). PAUP*, Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0B8. (Sunderland, MA: Sinaur Associates).
- Takemura, M., Oda, K., Yamato, K., Ohta, E., Nakamura, Y., Nozato, N., Akashi, K., and Ohyama, K. (1992). Gene clusters for ribosomal proteins in the mitochondrial genome of a liverwort, Marchantia polymorpha. Nucleic Acids Res. 20, 3199–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theologis, A., et al. (2000). Sequence and analysis of chromosome 1 of the plant Arabidopsis thaliana. Nature 408, 816–820. [DOI] [PubMed] [Google Scholar]
- Thorsness, P.E., and Weber, E.R. (1996). Escape and migration of nucleic acids between chloroplasts, mitochondria, and the nucleus. Int. Rev. Cytol. 165, 207–233. [DOI] [PubMed] [Google Scholar]
- Unseld, M., Marienfeld, J.R., Brandt, P., and Brennicke, A. (1997). The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 15, 57–61. [DOI] [PubMed] [Google Scholar]
- Waegemann, K., and Soll, J. (1995). Characterization and Isolation of the Chloroplast Import Machinery. (San Diego, CA: Academic Press), pp. 255–267. [DOI] [PubMed]
- Whelan, J., Hugosson, M., Glaser, E., and Day, D.A. (1995). Studies on the import and processing of the alternative oxidase precursor by isolated soybean mitochondria. Plant Mol. Biol. 27, 769–778. [DOI] [PubMed] [Google Scholar]
- Whelan, J., Tanudji, M.R., Smith, M.K., and Day, D.A. (1996). Evidence for a link between translocation and processing during import into soybean mitochondria. Biochim. Biophys. Acta 1312, 48–54. [DOI] [PubMed] [Google Scholar]
- Wischmann, C., and Schuster, W. (1995). Transfer of rps10 from the mitochondrion to the nucleus in Arabidopsis thaliana: Evidence for RNA-mediated transfer and exon shuffling at the integration site. FEBS Lett. 374, 152–156. [DOI] [PubMed] [Google Scholar]
- Wissinger, B., Schuster, W., and Brennicke, A. (1990). Species-specific RNA editing patterns in the mitochondrial rps13 transcripts of Oenothera and Daucus. Mol. Gen. Genet. 224, 389–395. [DOI] [PubMed] [Google Scholar]
- Wolff, G., Plante, I., Lang, B.F., Kuck, U., and Burger, G. (1994). Complete sequence of the mitochondrial DNA of the chlorophyte alga Prototheca wickerhamii: Gene content and genome organization. J. Mol. Biol. 237, 75–86. [DOI] [PubMed] [Google Scholar]
- Wower, I., Kowaleski, M.P., Sears, L.E., and Zimmermann, R.A. (1992). Mutagenesis of ribosomal protein S8 from Escherichia coli: Defects in regulation of the spc operon. J. Bacteriol. 174, 1213–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabaleta, E., Heiser, V., Grohmann, L., and Brennicke, A. (1998). Promoters of nuclear-encoded respiratory chain complex I genes from Arabidopsis thaliana contain a region essential for anther/pollen-specific expression. Plant J. 15, 49–59. [DOI] [PubMed] [Google Scholar]