Abstract
Analysis of a collection of 120,892 single-pass ESTs, derived from 26 different tomato cDNA libraries and reduced to a set of 27,274 unique consensus sequences (unigenes), revealed that 70% of the unigenes have identifiable homologs in the Arabidopsis genome. Genes corresponding to metabolism have remained most conserved between these two genomes, whereas genes encoding transcription factors are among the fastest evolving. The majority of the 10 largest conserved multigene families share similar copy numbers in tomato and Arabidopsis, suggesting that the multiplicity of these families may have occurred before the divergence of these two species. An exception to this multigene conservation was observed for the E8-like protein family, which is associated with fruit ripening and has higher copy number in tomato than in Arabidopsis. Finally, six BAC clones from different parts of the tomato genome were isolated, genetically mapped, sequenced, and annotated. The combined analysis of the EST database and these six sequenced BACs leads to the prediction that the tomato genome encodes ∼35,000 genes, which are sequestered largely in euchromatic regions corresponding to less than one-quarter of the total DNA in the tomato nucleus.
INTRODUCTION
Currently, the only plant genome to have been sequenced fully is that of Arabidopsis—a major milestone for plant biology. The availability of this sequence provides us with a detailed view of the gene content and genome organization of one plant species. Yet, the degree to which gene content, gene number, and genome organization are conserved among plant species remains unresolved. To answer these questions and to allow us to begin to understand the forces that have shaped plant genome evolution will require the sequencing of multiple plant genomes. Because of the relatively large size of most plant genomes and the associated high cost of sequencing, it is unlikely that we will have the full genomic sequence for many plant species in the near future.
A less expensive alternative is to sequence or partially sequence cDNA clones, which can reveal a substantial portion of the expressed genes of a genome at a fraction of the cost of genomic sequencing. As a result, extensive EST efforts are under way in a wide variety of plant species (National Science Foundation Plant Genome Research Program [http://www.nsf.gov/bio/dbi/dbi_pgr.htm]; Pennisi, 1998; Adam, 2000; Paterson et al., 2000). One such species is tomato, a member of the family Solanaceae.
Solanaceae, the nightshade family, is the third most valuable crop family in the United States, exceeded only by the grasses and the legumes, and is the most valuable family in terms of vegetable crops. In addition to its economic value, the family is unique with respect to the number of species that have been domesticated and the wide variety of uses to which they have been put. Solanaceous species have been domesticated for edible fruit (tomato, eggplant, pepper, tomatillo, and tamarindo), leafy vegetables (S. macrocarpon in Africa), tubers (potato), secondary compounds (tobacco), and ornamental flowers (petunia, Nicotiana spp). Tomato is the centerpiece for genetic and molecular research for the Solanaceae, attributable in part to inherent features of the species, including diploidy, modestly sized genome (950 Mb), tolerance of inbreeding, amenability to genetic transformation, and the availability of well-characterized genetic resources.
Through a National Science Foundation–funded project, we have generated a database for tomato comprising >120,000 ESTs (http://sgn.cornell.edu/; http://www.tigr.org/tdb/lgi). In addition, BAC clones corresponding to six selected regions of the tomato genome were sequenced. In this report, we describe the analysis of both the tomato EST database and the BAC sequences. Computational comparisons are made against the Arabidopsis genomic sequence and a similar high-density EST database from another dicot species, Medicago truncatula (http://www.tigr.org/tdb/mtgi/). As a result of these analyses, we have been able to address a number of issues, including the content, number, and organization of genes in the tomato genome and the degree to which genes have diverged since tomato, Arabidopsis, and M. truncatula diverged from their last common ancestor.
RESULTS
Contig Assembly of ESTs and Establishment of a Tomato Unigene Set
EST data sets of randomly sequenced cDNA libraries are redundant for many gene transcripts. This redundancy approximately represents gene transcript levels in the tissues that were used for library construction and can be used to assemble ESTs into contiguous overlapping clusters, with each cluster potentially representing a single unique gene. A substantial number of the low-frequency transcripts occur as single ESTs (singletons) and hence are not incorporated into contig assemblies. The combined set of contigs and singletons is referred to as a unigene set. This unigene set is believed to represent the minimal gene content for a species, with the caveat that in certain instances multiple unigenes could represent a single gene transcript, for example, as a result of nonoverlapping EST sequences.
In this study, a high stringency for matching was applied in the clustering to ensure a high level of confidence that each sequence in the unigene set represents a unique gene transcript. The specifications for clustering and unigene construction were as described in Quackenbush et al. (2000). The current unigene set is available through the TIGR World Wide Web site (http://www.tigr.org/tdb/lgi/) and the Solanaceae Genome Network database (http://sgn.cornell.edu) and comprises the EST sequences from 26 different libraries totaling 120,892 single-pass sequences.
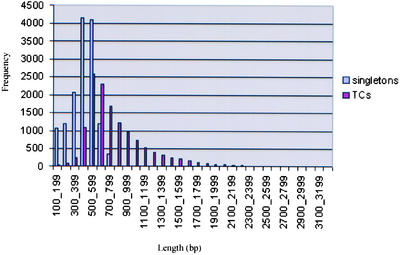
From each library, between 2000 and 10,000 directional clones were sequenced from the 5′ end; in addition, 5998 3′ end sequences from a flower tissue library were included. Details of the individual libraries are available through the Solanaceae Genome Network. The data set of 120,892 ESTs was reduced to 27,274 unigenes, comprising approximately equal numbers of contigs (also referred to as “tentative consensus” sequences) and singletons (Table 1). The contig sequence length ranged from 107 to 3285 bp, with an average of 823 bp, and the singleton length ranged from 101 to 823 bp, with an average of 447 bp (Figure 1).
Table 1.
Tomato Unigene Set Statistics
| No. of Sequences | Average Length (bp) | Role Category Assigneda | |
|---|---|---|---|
| Total number of ESTs | 120,892 | NDb | ND |
| ESTs in contigs | 106,833 | ND | ND |
| Singleton ESTs | 14,059 | 447 bp | 3687 (26%) |
| Total number of contigs | 13,215 | 823 bp | 5912 (45%) |
| Total number of unique sequences (unigenes) | 27,274 | ND | 9599 (35%) |
Role categories were derived from BLASTX matches of tomato unigenes against the annotated Arabidopsis proteome.
ND, not determined.
Figure 1.
Distribution of Sequence Length (bp) of Consensus Sequences (TCs) and Singletons That Constitute the Tomato Unigene Set.
Functional Annotation of the Unigene Set
Annotation of the EST-derived unigene set was approached in two ways. First, a surrogate annotation approach was applied in which the unigene set was annotated on the basis of the existing annotation available for the proteome of Arabidopsis. BLASTX was used to screen the entire tomato unigene set against the subset of the Arabidopsis proteome to which functional categories have been assigned (Arabidopsis Genome Initiative; 2000; http://wwww.Arabidopsis.org). Tomato unigenes with an expect value (E-value) of <1.0 E-10 were assigned to the corresponding Arabidopsis annotation. In doing so, the assumption was made that functionality is transferable based on sequence conservation, to which there are many exceptions. Annotation followed the Munich Information Center for Protein Sequences (http://mips.gsf.de) role categorization.
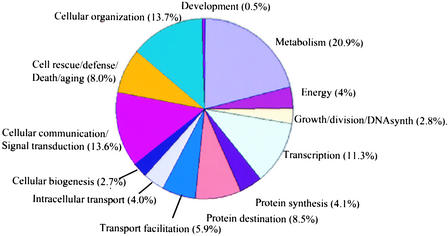
A total of 65% of the unigenes did not have a significant Arabidopsis match at this threshold and thus were considered “unclassified.” Another 5% had Arabidopsis matches, but their matching genes were listed as “classification unclear.” Thus, only 30% of the 27,274 tomato unigenes were assigned a putative function using this method (Figure 2). As a control, a subset of ∼1000 tomato unigenes, representing a set of conserved and putatively orthologous genes between Arabidopsis and tomato, was annotated manually (Fulton et al., 2002).
Figure 2.
Distribution of Tomato Unigenes Whose Putative Functions Could Be Assigned through Annotation.
Role categories are according to the Munich Information Center for Protein Sequences (http://mips.gsf.de) and are as follows: metabolism (r.c. 1); energy (r.c. 2); cell growth, cell division, and DNA synthesis (r.c. 3); transcription (r.c. 4); protein synthesis (r.c. 5); protein destination (r.c. 6); transport facilitation (r.c. 7); intracellular transport (r.c. 8); cellular biogenesis (r.c. 9); cellular communication/signal transduction (r.c. 10); cell rescue, defense, death, and aging (r.c. 11); cellular organization (r.c. 30); and development (r.c. 50).
This set of genes, referred to as conserved ortholog set markers, was annotated based on matches against the GenBank protein database (http://www.ncbi.nlm.nih.gov/Database/index.html). The assignment of functional categories was very similar for the entire unigene set and for the conserved ortholog set markers that were annotated manually, providing support for the surrogate annotation approach used for the entire unigene set (data not shown).
The largest proportion of functionally assigned unigenes fell into four role categories (r.c.): metabolism (r.c. 1), transcription (r.c. 4), cellular organization (r.c. 30), and cellular communication/signal transduction (r.c. 10). Together, these classes accounted for more than half of the assignable unigenes (Figure 2). These categories also are the largest for the Arabidopsis proteome and may represent a general tendency for all plant species (Arabidopsis Genome Initiative, 2000).
Comparing Tomato Gene Content with That of Other Plant Species
Having large-scale EST and genomic sequence databases for multiple organisms makes it feasible to ask questions about the relationship between species evolution and gene evolution. Specifically, one can identify the subset of highly conserved genes that likely serve common functional roles across plant species. Alternatively, one can identify the subset of genes that are fast evolving and hence might be key to species divergence and adaptation.
With these issues in mind, we computationally compared the tomato unigene set with the gene repertoire of Arabidopsis and M. truncatula. Tomato, Arabidopsis, and M. truncatula each belongs to a different plant family (Solanaceae, Brassicaceae, and Leguminoseae, respectively). However, M. truncatula and Arabidopsis are much more closely related to each other than to tomato, which diverged from Arabidopsis and M. truncatula as much as 150 million years ago, early in the period of dicot diversification (Yang et al., 1999).
Tomato-Arabidopsis Comparisons
To study the extent of gene conservation between the tomato and Arabidopsis genomes, all tomato unigenes were screened in all translated frames (tBLASTX) against the complete Arabidopsis genomic sequence (http://www.Arabidopsis.org). For this analysis, the E-values of BLAST similarity searches were used as an estimate of sequence conservation. However, we acknowledge that two factors may compromise this assumption to varying degrees. First, many of the unigene sequences are not full length, which generally decreases the potential E-values relative to those of full-length sequences. Second, BLAST performs local alignments, resulting in potentially high E-values only over short stretches of sequence conservation, favoring the conservation of domains rather than complete genes.
Nonetheless, because the E-values are summarized over a large number of sequence comparisons and used primarily to reveal general trends in the conservation of sequence and functionality, such drawbacks are unlikely to affect the overall conclusions. The analysis was made directly against the Arabidopsis genomic sequence (rather than the predicted proteome), so that genes previously unidentified in the Arabidopsis genome also could be detected via homology with tomato ESTs.
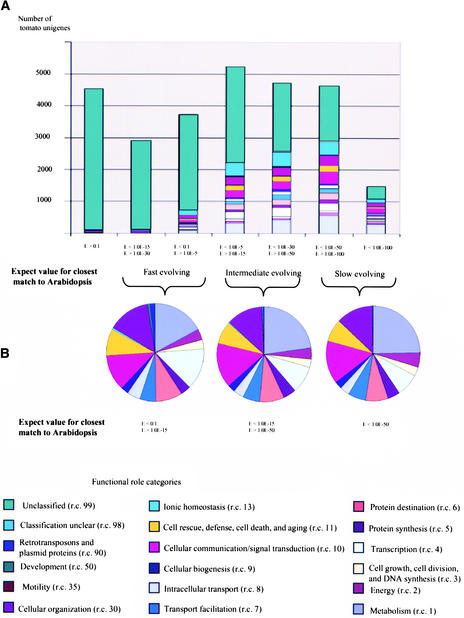
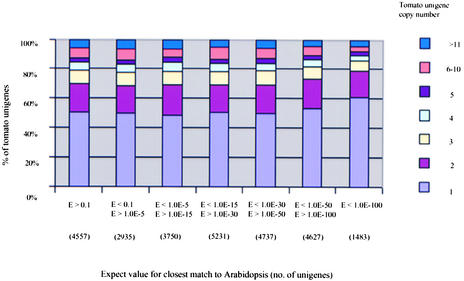
Figure 3 displays the distribution of E-value matches in conjunction with functional role categories for the tomato unigene set. Nearly 70% of the tomato unigenes have significant matches at the amino acid level (E-value < 1.0 E-5) to one or more translated portions of the Arabidopsis genomic sequence. The majority (52%) of the tomato unigenes with matches to the Arabidopsis genome hit Arabidopsis genes for which no putative functions have been assigned. The highest proportion of these fell into categories that showed the weakest homology with their Arabidopsis counterparts. For example, for those unigenes that had weak homology with their Arabidopsis counterparts, 80% matched unclassified Arabidopsis genes (Figure 3). In contrast, for unigenes that had high homology (tBLASTX E-value < 1.0 E-100) with their Arabidopsis counterparts, only 20% matched unclassified Arabidopsis genes (Figure 3).
Figure 3.
Distribution of Conservation between Tomato Unigenes and Genes in the Arabidopsis Genome Based on tBLASTX scores.
(A) All tomato genes.
(B) Only those tomato genes for which putative function could be established through annotation.
To further analyze the nature of both fast- and slow-evolving genes identified by the tomato-Arabidopsis comparisons, we simultaneously examined more closely the putative functional role and the degree of sequence similarity (to its closest Arabidopsis counterpart) of each tomato unigene (Figure 3). The goal of this exercise was to determine whether certain functional classes of genes have evolved more rapidly since tomato and Arabidopsis diverged from their last common ancestor. Such information might provide clues to which types of genes/gene functions are more constant across plant taxa (more ancestral gene functions) and which genes/gene functions tend to evolve rapidly as species evolve (more derived gene functions). A summary of this analysis is presented below.
Of the >27,000 tomato unigenes, 22% show very high conservation (E-value < 1.0 E-50) with Arabidopsis genes (Figure 3). Within this “slow-evolving” category, by far the highest proportion (24%) belonged to the metabolism category (r.c. 1; Figure 3). The proportion of genes assigned to this category decreased to 19 and 12%, respectively, as one moved to the “intermediate-evolving” (E < 1.0 E-15 to E ≥ 1.0 E-50; 37% of unigenes) and the “fast-evolving” (E > 1.0 E-15; 24% of unigenes) categories, suggesting that metabolic functions have remained more highly conserved in plant evolution (Figure 3). Genes encoding transcription factors appear to be faster evolving, changing from 15% in the fast-evolving category to 13 and 8% in the intermediate- and slow-evolving categories (Figure 3; see Discussion). Genes involved in cell rescue, defense, cell death and aging, and cellular communication/signal transduction do not appear to be fast evolving as a group, showing similar frequencies in the fast-, intermediate-, and slow-evolving categories (Figure 3).
Identification of Tomato-Specific Genes
Of the ∼27,000 tomato unigenes, 4529 (17%) had no detectable homolog (E ≥ 0.1) in the Arabidopsis genome (Figure 3). This set of unigenes was further searched against the GenBank protein database to identify putative matches. A very small proportion of these unigenes (233 [5%]; E-value < 10 E-15) showed similarity to any protein sequence in GenBank. Of those that showed homology with one or more GenBank entries, a subset of the 114 with the most significant matches (E-value < 1.0 E-30) were annotated for putative gene functions.
A large proportion of these 114 sequences (75%) revealed perfect matches with Escherichia coli DNA, bacteriophage λDNA, or other contaminating sequences, obviously representing sequences that were missed initially in cleanup procedures of the EST collection. For 11 other unigenes (9%), it was determined that the sequences probably were too short to contain domains that would give significant matches with Arabidopsis. Therefore, it was assumed that these unigenes may not represent genes unique to tomato.
Only 28 of the original 114 unigenes had no detectable counterpart in the Arabidopsis genome but matched genes from other species (as present in the GenBank protein database) (Table 2). Eleven of these unigenes (39%) corresponded to three gene families that appear to be specific to Solanaceae, having matches with other solanaceous plants but not with other plant families. These three Solanaceae-unique gene families are type II proteinase inhibitors (TC67527; six unigenes assigned), fruit-specific proteins/metallocarboxypeptidase inhibitors (TC63650; two unigenes assigned), and extensin-like proteins (TC63390; three unigenes assigned).
Table 2.
Putative Functions of Genes Not Conserved between Tomato and Arabidopsis
| Unigene | Match Description, Species (GenBank Number) | E-Value | Length Query (Amino Acids) |
No. of Isoforms |
|---|---|---|---|---|
| TC58703 | Polyphenoloxidase, tomato (CAA78300) | 0 | 585 | 6 |
| TC67527 | Proteinase inhibitor II, tomato (AAC37397) | 1.0 E-130 | 223 | 6 |
| TC63650 | Fruit-specific protein, tomato (CAA32007)/metallocarboxypeptidase inhibitor, potato (BAA21500) |
1.0 E-39 | 96 | 2 |
| TC67742 | Ornithine decarboxylase, tomato (AF029349) | 1.0 E-103 | 431 | 3 |
| TC59463 | Putative translation activator/inclusion body protein, tobacco (CAB42623) | 1.0 E-98 | 402 | 3 |
| TC63390 | Extensin-like protein potato (CAA06000) | 1.0 E-69 | 221 | 3 |
| TC59945 | pSTH2-protein, potato (AAA03019) | 1.0 E-50 | 155 | 1 |
| TGSAY39TH | TSI-1 protein, tomato (CAA75803) | 1.0 E-35 | 178 | 1 |
| TC66719 | Similar to human hypothetical protein, rice (BAA92729) | 1.0 E-46 | 377 | 1 |
| TOVCB02THB | WREBP-2, tobacco (BAA75685) | 1.0 E-59 | 371 | 1 |
| TC69096 | Specific tissue protein 2, chickpea (CAA66109) | 1.0 E-37 | 348 | 1 |
These three gene families appear to be specific to the Solanaceae; however, we cannot determine from these data whether these genes were lost in the Arabidopsis lineage or subjected to accelerated evolution (hence, their uniqueness to the Solanaceae lineage). Proteinase inhibitors (type II and metallocarboxypeptidase inhibitor) generally are known to be involved in resistance against herbivory (Johnson et al., 1989; Duan et al., 1996).
The other 17 tomato unigenes (not found in Arabidopsis but found in other species) had matches not only in solanaceous species (a member of the asterid clade) but also in species belonging to the rosid clade, to which Arabidopsis belongs (Chase et al., 1993) (data not shown). Therefore, these genes likely were present in the last common ancestor of tomato and Arabidopsis and were lost subsequently in the Arabidopsis lineage. Some examples of these unigenes that may have been lost specifically from the Arabidopsis lineage are presented below (Table 2).
Polyphenoloxidases (TC58703) generally are known to be involved in resistance against herbivory and are found in many plant species, including many rosids (such as apple and bean); therefore, they appear to have been lost specifically from the Arabidopsis genome or ancestral lineage (Cary et al., 1992; Murata et al., 1997). It is unclear why other rosids have retained these genes but the ancestral line of Arabidopsis did not. The absence of polyphenoloxidases in Arabidopsis raises interesting questions with respect to the ecology of Arabidopsis and the selective pressure of herbivory on Arabidopsis.
Ornithine decarboxylase (TC67742) catalyzes the second step in putrescine biosynthesis from the amino acid Arg. Putrescine is a precursor for the biosynthesis of the polyamines spermine and spermidine, which are essential for the growth and development of plants. Interestingly, the pathway from l-Arg to putrescine involving ornithine decarboxylase is redundant in many plants with a pathway involving Arg decarboxylase (Kumar et al., 1997; Tiburcio et al., 1997). The Arabidopsis genome appears to have lost one of the redundant pathways.
TC59945, TGSAY39TH, and TC69096 are tomato unigenes with a high degree of similarity to genes from species belonging to the rosid clade but lack matches in the Arabidopsis genome. They share high similarity with pathogenesis-related genes identified previously in potato and tomato (pSTH2 and TS-1; Matton and Brisson, 1989).
TOVCB02THB shares considerable sequence similarity with a range of transcription factors. Matches are found with tobacco and maize and several nonplant eukaryotic genes, including mammalian species and Drosophila. This may represent a case in which genes were lost in the Arabidopsis lineage.
TC59463 and TC66179 do not display matches with species in the rosid clade but have matches outside of the plant kingdom, which suggests a more complicated ancestry. TC59463 is highly similar to a pararetroviral sequence integrated into the tobacco genome. Integration of this type of pararetrovirus may be specific to members of the Solanaceae simply because of the host range for these viruses. TC66179 appears to be highly conserved in a rice gene and has some weak sequence similarity with a gene from human (E-value = 5 E-5), but no other matches with plant species were identified.
Evolutionary Comparison of Tomato Unigenes with Those in Arabidopsis and M. truncatula
By computationally comparing the unigene set of tomato with the gene repertoire of Arabidopsis, a picture emerges about how gene evolution has proceeded since these two species diverged from their last common ancestor ∼100 million years ago. To understand the evolution of plant genes and plant gene functions in the larger context of plant evolution, it is necessary to extend this question to other plant species. Specifically, we need to determine whether genes that are fast or slow evolving in some branches of plant evolution are likely to have evolved in a similar manner throughout plant evolution.
A definitive answer to this question awaits the full genomic sequencing of a wide variety of species throughout the evolutionary tree of plants. However, in an attempt to elucidate this issue, we wondered if genes that are highly conserved between tomato and Arabidopsis also are highly conserved in other plants and whether genes that have diverged rapidly since tomato and Arabidopsis diverged from their last common ancestor are likely to have evolved rapidly in other plant lineages.
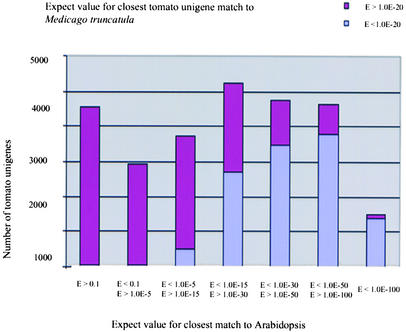
To attempt to address these questions, we computationally compared the tomato unigene set not only with that of Arabidopsis but also with the comprehensive EST data set now available for M. truncatula, which belongs to a third dicot family, Leguminoseae (http://www.tigr.org/tdb/mtgi). The entire tomato unigene set was compared with the entire M. truncatula gene index (http://www.tigr.org) at the amino acid level using tBLASTX. Figure 4 depicts these results in a manner whereby the similarity of tomato–M. truncatula matches can be compared with the similarity of tomato-Arabidopsis matches.
Figure 4.
Distribution of Tomato Unigene Sequences That Are Conserved with M. truncatula (threshold of ≤1.0 E-20; tBLASTX) Plotted against the Conservation of These Genes with Arabidopsis Genes (as in Figure 3).
Approximately 90% of the genes that were most conserved between tomato and Arabidopsis (tBLASTX E-value < 1.0 E-100) have a highly significant detectable counterpart in M. truncatula with a tBLASTX E-value threshold of <1.00 E-20 (Figure 4). As one moves down to sets of genes that are less well conserved between Arabidopsis and tomato, the proportion with significant matches to M. truncatula genes decreases dramatically (Figure 4).
These results strongly suggest that genes well conserved between a given pair of plant species also are likely to be well conserved in other plants species, presumably as a result of strong negative selection pressure to maintain essential, and hence less mutable, gene functions (e.g., basic metabolism; see above). These results also support the notion that highly conserved orthologs detected in pairwise species comparisons will have similar conserved orthologs in other plant genomes, a finding that has implications for both comparative gene mapping and molecular phylogenetic studies in plants (Fulton et al., 2002).
Finally, it is worth noting that virtually no tomato unigenes displayed a match with M. truncatula but not with Arabidopsis. The only exception was TC59945, which matched a pathogenesis-related gene isolated from potato (pSTH2). Good matches for this particular protein also are found in other members of the rosid clade, such as Prunus species, cowpea, and birch. However, as mentioned above, this gene was not detected in the Arabidopsis genome; hence, it may have been lost relatively recently in the Arabidopsis lineage. However, we cannot dismiss the possibility that this gene exists in Arabidopsis in one of the gene-poor centromeric regions that have not been sequenced fully.
Characteristics of Tomato Multigene Families
Analysis of the complete Arabidopsis genomic sequence has revealed that 65% of Arabidopsis genes belong to multicopy gene families (Arabidopsis Genome Initiative, 2000). This raises the questions of whether the genes in other plant species are organized into similar gene families and whether the size of gene families has stayed relatively constant across plant species or is highly dynamic. To describe the gene family organization in tomato computationally, and to answer these questions, gene copy number for each unigene was determined by counting the number of tBLASTX matches against the unigene data set itself with an E-value threshold of <1.00 E-20. These calculated unigene copy numbers were used to describe the sizes of multigene families. Note that gene families were not defined by functional groupings, because this would require expert annotators to define family membership.
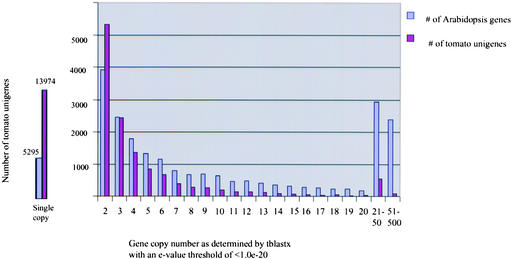
To have comparable results for comparisons, the Arabidopsis gene set (available from TAIR at http://www.Arabidopsis.org) was subjected to the same analysis using the same threshold. Figure 5 presents the distribution of unigenes into gene families of various copy numbers for both the tomato and Arabidopsis genomes. It is important to note that, because the unigene set does not represent all tomato genes (especially those with low expression levels), the copy numbers of tomato unigenes may be underestimated, which would account also for the large excess of singletons in the tomato gene copy number distribution.
Figure 5.
Comparison of Multigene Family Copy Numbers between Tomato and Arabidopsis.
Comparison of Multigene Families between Tomato and Arabidopsis
To address the question of whether gene copy number in tomato is correlated with the degree of conservation with other species, the gene copy numbers for tomato unigenes were plotted against the conservation (as determined by tBLASTX) with their Arabidopsis counterparts. In other words, are conserved genes more or less likely to be multicopy than less conserved genes?
As described above, genes were assigned a gene copy number based on matches with an E-value threshold of <1.0 E-20 using tBLASTX. Of the 27,274 tomato unigenes, ∼56% (15,387 unigenes) had significant matches at this threshold in Arabidopsis and were used for this analysis. Figure 6 displays the results, suggesting a weak positive correlation between the copy number of gene families and the degree of sequence conservation between these families in tomato and Arabidopsis. These results suggest that higher-copy gene families are not likely the result of slower rates of divergence after gene duplication.
Figure 6.
Percentage of Tomato Unigenes Belonging to Single versus Multigene Families and Categorized on the Basis of the Level of Sequence Conservation between Each Tomato Gene Family and the Corresponding Arabidopsis Family Members as Measured by tBLASTX Scores (as in Figure 3).
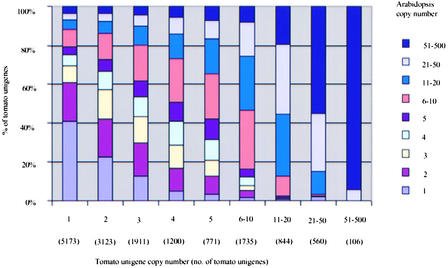
A second question that can be addressed with these data is whether the copy number of highly conserved genes is conserved between tomato and Arabidopsis. In an attempt to answer this question, the copy number of each tomato gene family (as determined by tBLASTX E-values of <1.00 E-30) was plotted against the copy number of the corresponding Arabidopsis gene family. The results demonstrate a significant correlation (r = 0.49) between the copy numbers of Arabidopsis and tomato multigene families. The results from this analysis are depicted in the form of a histogram in Figure 7.
Figure 7.
Distribution of Copy Numbers of Tomato Gene Families and Comparison of Copy Numbers for Corresponding Gene Families in Arabidopsis.
Both tomato and Arabidopsis genes were assigned to gene families based on tBLASTX scores with E-values of <1.00 E-20. Tomato and Arabidopsis gene family correspondence was based on the best tomato and Arabidopsis gene match for each family returning tBLASTX scores with E-values of <1.00 E-30.
To illustrate these observations, the genes with the 10 highest copy numbers for both the tomato unigene set and Arabidopsis are listed in Tables 3 and 4. Protein kinases and cytochrome P450s represent the largest families in tomato and represent very large families in Arabidopsis as well. Other large families in common are genes with similarity to Myb-like transcription factors and NAC domain–containing proteins (Souer et al., 1996) and glucosyltransferases. It should be noted that one difference between a gene set based on genomic sequence (Arabidopsis) as opposed to ESTs (tomato) is the high copy number of transposon-based reverse transcriptases in Arabidopsis, which are much less abundant in the tomato EST data set, presumably as a result of low levels of expression of the transposon-related genes.
Table 3.
The 10 Highest Copy Number Gene Families in Tomato
| Tomato Unigene Family Representative |
Putative Function of the Gene Family |
Tomato Copy Number |
Corresponding Arabidopsis Gene Family Representative (Best Match) |
Arabidopsis Copy Number |
|---|---|---|---|---|
| TC59932 | Protein kinases | 139 | 68156.m00140#F25P12.104 | 307 |
| TC65270 | Cytochrome P450s | 60 | 67603.m00019#F13G24_190 | 151 |
| TC66702 | Peroxidases | 53 | 67589.m00013#K18I23_14 | 75 |
| TC63395 | RAS-related GTP binding protein | 48 | 51050.m00114#T7I23.6 | 64 |
| TMEBK31TH | Myb-like transcription factor | 41 | 67258.m00015#F1P2_150 | 126 |
| TC58771 | E8 protein homolog | 39 | 51438.m00063#F12K11.9 | 20 |
| TC67841 | Ubiquitin-conjugating proteins | 37 | 67845.m00010#K19E1_10 | 28 |
| TC65109 | Glucosyltransferases | 35 | 67199.m00013#F28A23_110 | 103 |
| TC66239 | NAC domain proteins | 34 | 51786.m00083#T1N6.22 | 79 |
| TC60901 | Glutathione S-transferases | 32 | 42492.m00098#F16P2.20 | 29 |
Table 4.
The 10 Highest Copy Number Gene Families in Arabidopsis
| Arabidopsis Gene Family Representative |
Putative Function of the Gene Family |
Arabidopsis Copy Number |
Corresponding Tomato Gene Family Representative |
Tomato Copy Number |
|---|---|---|---|---|
| 60116.m00109#F5O11.26 | Protein kinases | 451 | TC71301 | 72 |
| 67584.m00014#T1E3_140 | Selenium binding protein-like | 201 | TC60645 | 6 |
| 67317.m00023#T20K12_230 | Copia-type polyprotein | 196 | TC62263 | 3 |
| 8264.m00077#F4L23.26 | Non-LTR retroelement reverse transcriptase | 191 | TC66035 | 1 |
| 67603.m00019#F13G24_190 | Cytochrome P450s | 151 | TC65270 | 60 |
| 60510.m00064#MBK21.8 | Myb-related transcription factors | 126 | TAIAA88TH | 38 |
| 67043.m00014#T7B11_19 | Putative CHP-rich zinc finger Protein/ Ta11-like non-LTR retroelement protein |
133 | No good tomato match TCAHZ13TH, 2e-09 |
8 |
| 49257.m00071#F9O13.3 | Glucosyltransferase | 111 | TC65109 | 35 |
| 67207.m00009#F8D20_90 | NAM/CUC2/NAC domain proteins | 85 | TC63626 | 32 |
| 38795.m00065#T32F12.24 | Peroxidases | 78 | TC66702 | 53 |
The only group of unigenes, represented by TC58771, that seems to have a higher copy number in tomato than in Arabidopsis (39 versus 20 copies) is a group composed of E8-like genes (23 copies with E8-like protein as best match) and smaller numbers of different oxidoreductases such as dioxygenases (five), hydroxylases (three), flavonal synthases (three), and 1-aminocyclopropane-1-carboxylate oxidases (five). Although the function of E8-like proteins is unknown, they share extensive similarity with numerous oxidoreductases and are known to be expressed specifically during fruit ripening (Deikman and Fischer, 1988). The expansion of this gene family in tomato may represent an evolutionary adaptation important for fruit ripening.
Analysis of Gene Content and Organization Based on Genomic Sequencing
Although analysis of EST data sets provides information about the content of expressed genes, it does not address the question of the chromosomal organization of those genes. The issue of genome organization is best addressed by either physical mapping or genomic sequencing. At present, the tomato genome has not been sequenced; however, genomic sequence is available for six BAC clones corresponding to 592 kb of sequenced tomato genomic DNA (Table 5). Moreover, the map position is known for each of these BACs, providing a chromosomal context for this genomic sequence (Figure 8). We have annotated these six BAC sequences for putative coding regions and used this information, in combination with the EST data set, to make inferences about the overall gene content and organization of the tomato genome.
Table 5.
Analysis of Gene Content and EST Coverage of Genomic Sequences from Six Tomato BACS
| BAC Number |
Genbank Accession Number |
Predicted Genes |
Number of Predicted Genes with Perfect EST Matches | Number of Predicted Genes with Paralogous (Imperfect) EST Matches |
No. of Matches |
Transposon Sequences |
Total Length (bp) |
Gene Density (kb/gene) |
References |
|---|---|---|---|---|---|---|---|---|---|
| 62O11a | AF411808 | 7 | 4 | 1 | 2 (0/2)b | 0 | 70347 | 10 | R. McCombie and M. Katari (unpublished data) |
| 127E11a | AF411807 | 19 | 8 | 9 | 2 (1/1) | 0 | 95845 | 5 | T. Fulton, R. Van der Hoeven, and S. Tanksley (unpublished data) |
| FW2.2a | AF411809 | 20 | 14 | 1 | 5 (3/2) | 0 | 127892 | 6 | T.C. Nesbitt, R. Van der Hoeven, and S. Tanksley (unpublished data) |
| 2O7a |
AF411805, AF411806 |
6 | 0 | 3 | 3 (2/1) | 4 | 92221 | 15 | R. Van der Hoeven and S. Tanksley (unpublished data) |
| 47I13a | AF411804 | 6 | 1 | 3 | 2 (0/2) | 6 | 100810 | 17 | R. Van der Hoeven and S. Tanksley (unpublished data) |
| BAC19 | AF27333 | 18 | 9 | 4 | 5 (3/2) | 0 | 105308 | 6 | Ku et al., 2000 |
| Total/average | 76 | 36 | 21 | 19 (9/10) | 9 | 592423 | 9.8 |
Clemson University Genomics Institute tomato BAC number (http://www.genome.clemson.edu/).
Numbers in parentheses indicate whether the predicted genes have a match with sequences in the Arabidopsis genomic sequence or are based solely on prediction by the gene prediction program FGENESH (Arabidopsis match/FGENESH prediction). BAC 47I13 contains three copia- and three gypsy-like retrotransposon reverse transcriptases, whereas 2O7 contains only ty3–gypsy-like transposon sequences (reverse transcriptases).
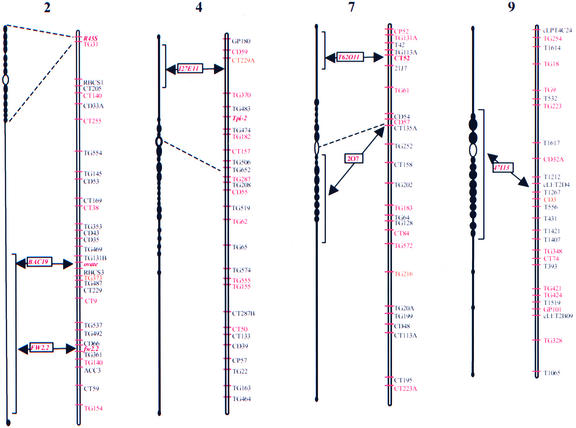
Figure 8.
Genetic Map Position of Each of the Six Sequenced BAC Clones.
Genetic linkage map based on the work of Tanksley et al. (1992). At left of each linkage map is the corresponding pachytene chromosome. Open ovals indicate centromeres. Corresponding positions of centromeres on the genetic map are indicated by dashed lines. Dark knobs adjacent to centromeres represent the heterochromatin of each pachytene chromosome. The approximate position of each BAC clone in the corresponding pachytene chromosome is indicated by brackets and is based on previous deletion mapping of genetically mapped markers (Khush and Rick, 1968).
Variation of Gene Density across BACs
The predicted gene density of the various BACs varied from 5 kb/gene to 17 kb/gene, a threefold difference (Table 5). For the two BACs with the lowest gene density (2O7 and 47I13), 12 putative genes were identified, only 1 of which had an exact match in the EST unigene set. By contrast, for the other four BACs with higher predicted gene densities, nearly half of the putative genes had corresponding unigene matches. Besides being low in gene density, 2O7 and 47I13 were the only BACs that contained transposon-type sequences, mostly reverse transcriptase sequences similar to copia- and gypsy-like retrotransposons.
Gene Number Estimate Based on BAC Sequences and the EST Database
A total of 76 putative genes were identified computationally on the six BAC sequences, and 36 (47%) had perfect matches in the EST-derived unigene set (Table 5). For another 21 genes, nonperfect matches were found, most likely representing transcripts from paralogous genes. An extrapolation of the average gene content of these six BACs to the entire tomato genome (950 Mb) (Arumuganathan and Earle, 1991) would predict a total gene content of >97,000 genes.
This is almost certainly a gross overestimate of the gene density (9.8 kb/gene; see below), being much larger than the gene content of any of the recently completely sequenced eukaryotic genomes, all of which contain <40,000 protein-coding sequences (Arabidopsis Genome Initiative, 2000; International Human Genome Sequencing Consortium, 2001). Therefore, it seems likely that the average gene content of the sequenced BACs is not representative of the entire tomato genome and reflects a bias toward gene-rich genomic regions.
A more accurate estimate of the total gene content of tomato can be made by comparing the size of the EST-derived unigene set and the percentage of predicted genes in genomic DNA (e.g., BAC sequences) that are represented by a unigene match. As described above, the current EST-derived unigene set is composed of ∼27,000 gene sequences. However, in using this estimate, one must take into account the fact that ESTs from transcripts of a single gene are not always assembled into a single contig because of insufficient or lacking sequence overlap (attributable to non-full-length clones or nonoverlapping 5′ and 3′ derived ESTs), sequencing errors, or chimeric cDNA clones.
Hence, the actual number of unique genes represented by an EST-derived unigene set usually is less than the number of unigenes. For example, in Arabidopsis, the EST-derived unigene set leads to a 35% overestimate of the actual number of genes ultimately revealed by genomic sequencing (Arabidopsis Genome Initiative, 2000). Undoubtedly, the same situation is true for tomato. If we assume that the number of unigenes is a 35% overestimate of the actual number of genes, then the unigene set would represent only 17,500 unique genes instead of 27,000. Considering that this unigene set contains matches to approximately half of the predicted genes in the six sequenced BACs, we estimate the total gene content of tomato to be ∼17,500 × 2, or 35,000 genes. Thus, the predicted gene content of tomato is 40% greater than that of Arabidopsis; however, this increase in gene content is not proportional to the sevenfold larger size of the tomato genome (950 Mb for tomato versus 125 Mb for Arabidopsis).
DISCUSSION
How well gene number, gene organization, and gene function in Arabidopsis will predict those of other plant species is unknown at present. In this respect, tomato is a useful species for comparison. It belongs to a plant family (Solanaceae) that diverged from the lineage leading to Arabidopsis as much as 150 million years ago—early in the radiation of dicots. Thus, by identifying features conserved between the tomato and Arabidopsis genomes, one would expect to identify gene/genome features that might be conserved in other plants.
However, because of the long period of divergence between these species, one might expect to reveal trends in gene and genome divergence, which also can be instructive about plant genome evolution. For these reasons, we analyzed and annotated a large EST data set and genomic sequences of BAC clones and used the results to make deductions about the composition and organization of the tomato genome. These analyses were made using Arabidopsis, and to a lesser degree M. truncatula, as points of comparison.
Characteristics of the Tomato Gene Repertoire
The analysis of the large EST database and BAC sequences described in this article leads to the estimate that the tomato genome contains ∼35,000 genes, considerably more than the 25,500 genes in the Arabidopsis genome. However, the majority of the tomato genes (70%) have significant matches to Arabidopsis genes and may reflect conserved gene functions. Of the 30% without matches in Arabidopsis, the majority have unknown functions and are without matches in other current genome databases. These may represent fast-evolving genes that have acquired new functions in tomato and related taxa. Examples of such novel genes include those encoding type II proteinase inhibitors and a class of extensin-like proteins that are confined to tomato and other solanaceous species.
Examination of the tomato gene content also provides evidence for selective gene loss in the Arabidopsis lineage. For example, polyphenoloxidases and ornithine decarboxylase are found in tomato as well as many other plant taxa but not in Arabidopsis. Thus, Arabidopsis probably has lost some gene functions still retained in other plant species and hence is not a ready model for the exploration of such functions.
As a lower limit, we estimate that at least 50% of the tomato genes belong to multigene families. This estimate is based on the observation that approximately half of the EST-derived tomato unigenes have significant matches to one or more other unigenes. However, the actual proportion of tomato genes that belong to multigene families probably is larger than this because the unigene set is estimated to represent no more than half of the tomato genes. It is worth noting that previous studies based on probing of random cDNAs on genomic DNA gel blots led to the estimate that 47% of tomato genes belong to multigene families (Bernatzky and Tanksley, 1986).
Because DNA gel blot hybridization cannot readily detect genes with >30% nucleotide divergence, such estimates will be inherently less than those derived from computational comparisons that do not have this restriction. Overall, however, the percentage of genes that belong to multigene families in tomato does not appear to be significantly higher than that in Arabidopsis (65%). This observation lends support to the hypothesis that the evolutionary lineage leading to tomato did not experience any recent whole-genome duplication events (e.g., polyploidy). Rather, any whole-genome duplications occurred in the distant past, near the time that tomato and Arabidopsis diverged from their last common ancestor (Ku et al., 2000). If this hypothesis proves correct, then the larger gene number of tomato may be attributable to a slower rate of gene loss after ancient polyploidization events rather than to the occurrence of recent duplications (Ku et al., 2000).
The copy numbers of specific multigene families are correlated significantly between tomato and Arabidopsis, which may be a result of the duplication/diversification of these families before the divergence of the tomato and Arabidopsis lineages. Alternatively, selection pressure may have been exerted independently in each lineage to maintain a relatively stable copy number, even in the face of continuing duplication and deletion. An exception was found for the E8 gene family, whose functions are not well elucidated but that often is associated with tomato fruit development/ripening. In this instance, the E8 gene family is larger in tomato than in Arabidopsis and may reflect a more complex fruit development/ripening process in tomato compared with Arabidopsis.
Characteristics of Plant Gene Evolution as Deduced from Comparisons of Tomato, Arabidopsis, and M. truncatula
A comparison of the Arabidopsis, tomato, and M. truncatula gene repertoires indicates that there is a set of highly conserved genes (∼17%) common to all three genomes. The fact that the majority of these genes have retained a high degree of similarity despite the long divergence times among these species suggests that these genes represent conserved functions that predate plant diversification. Although proteins of all functional classes appear in this conserved set, it is significantly biased for genes encoding metabolic functions. This finding is consistent with the idea that basic metabolic pathways are largely conserved among plant species.
Although genes encoding metabolism appear to evolve more slowly, genes encoding transcription factors appear to diverge more rapidly among species. The transcription factor category nearly doubles in frequency as one moves from the slow-evolving to the fast-evolving category (Figure 3). This result suggests that changes in gene regulation (through the accelerated evolution of transcription factors) have been a significant force in plant evolution. The sequencing of the Arabidopsis genome revealed that plants have developed a number of transcription factor gene families that are unique to plants and not present in other eukaryotic lineages and that plants contain a significantly greater proportion of transcription factors that exhibit rapid evolution rates in regions outside the core conserved domains (Arabidopsis Genome Initiative, 2000; Lagercrantz and Axelsson, 2000). The abundance of transcription factors in the less conserved categories confirms these notions.
The finding that transcription factors, as a group, evolve more rapidly than other classified groups of genes also is consistent with the idea that the morphological evolution of species is highly dependent on changes in the regulatory patterns of gene expression (King and Wilson, 1975; Keys et al., 1999; Stern, 2000). In this regard, recent studies have shown that natural variations affecting traits involved in crop domestication often are associated with changes in gene regulation rather than changes that affect the proteins themselves (e.g., maize [Doebley and Lukens, 1998; Wang et al., 1999] and tomato [Frary et al., 2000]).
Tomato Genes Probably Are Concentrated in Euchromatin, Which Represents the Minority of DNA in the Tomato Genome
Analysis of the six sequenced genomic BACs revealed a gene density ranging from 5 to 17 kb/gene and averaging 9.8 kb/gene, compared with ∼4.5 kb/gene for Arabidopsis (Table 5). Extrapolation of the gene densities from these BACs to the entire 950 kb of the tomato genome led to an estimated total number of 55,000 to 190,000 genes, much higher than that estimated from the combined EST and BAC data (see above). In considering these results, it is important to consider the fact that the six BACs used in this analysis were isolated with specific gene probes and hence were known a priori to contain one or more gene(s) (our unpublished data). Hence, they are not random BACs but biased for gene-containing regions.
The fact that the gene densities on all of these BACs are much higher than necessary to account for the estimated 35,000 tomato genes suggests that genes are contained on only a relatively small portion of the tomato chromosomes. Tomato chromosomes, like those of other species in the Solanaceae family, are composed of centromeric heterochromatin with more distal euchromatic regions (Khush and Rick, 1968; Rick, 1975) (Figure 8). The centromeric heterochromatic regions of tomato constitute ∼77% of the chromosomal DNA and, based on deletion studies, contain few genes (Khush et al., 1964; Peterson et al., 1996, 1998).
From the current study, the two BACs with the lowest gene densities (15 and 17 kb/gene) probably are located in or near the centromeric heterochromatin and also contain numerous retrotransposon-like sequences, which are characteristic of the centromeric regions of Arabidopsis (Figure 8) (Arabidopsis Genome Initiative, 2000). We hypothesize that these two BACs represent transition regions between euchromatin and genetically inactive centromeric heterochromatin. The remaining four BACs appear to be from euchromatin and have on average a higher gene density (7 kb/gene).
If 7 kb/gene is characteristic of euchromatin, which constitutes 23% of the tomato genome (Peterson et al., 1996), then the euchromatic portions of the tomato chromosomes should contain ∼31,000 genes [(950,000 kb/7 kb) × 0.23)], which is remarkably similar to the 35,000 genes estimated for the entire genome (see above). These results have implications not only for genome organization in tomato and other solanaceous species but also with respect to sequencing of the tomato genome. Sequencing only the euchromatic regions of the tomato genome would reveal the majority of the genes.
METHODS
cDNA Library Construction
The primary phage cDNA libraries were constructed and excised into bacterial cultures as phagemids according to the manufacturer's instructions (Stratagene; http://www.stratagene.com). The bacterial cultures were arrayed subsequently into 384-well plates and used for sequencing. Specific information pertaining to the individual libraries can be found online at http://sgn.cornell.edu. From each library, between 2500 and 10,000 high-quality sequence runs were produced, with various success rates for different libraries. The library collection was designed to maximize gene discovery, currently consisting of a considerable variety of >26 different libraries, capturing genes expressed in different tissue types and developmental stages or expressed during pathogen-elicited responses.
All sequencing of the cDNA clones was performed at the Institute for Genomic Research (http://www.tigr.org); the entire data set used in this report can be downloaded from the Solanaceae Genome Network through anonymous file transfer protocol (http://sgn.cornell. edu). The unigene set was constructed in accordance with TIGR's gene indexes, as described in Quackenbush et al. (2000). All BLAST analyses were run on nodes (four central processing units, 4 gigabytes of random-access memory) of the Dell/Intel cluster of the Cornell Theory Center running under Microsoft Windows 2000 (http:// www.tc.cornell.edu).
Data Sets Used for Analyses
The tomato (Lycopersicon esculentum) unigene set and the ESTs used for this unigene set build are available on the TIGR World Wide Web site as the tomato gene index version 7.0 (May 2001). This gene index will be replaced periodically by newer builds including more tomato ESTs, but the “heritability” function of the gene index makes it possible to trace forward from the “TC” numbers discussed in the text. The Arabidopsis thaliana genomic sequence data set consists of a minimal tiling path of BAC sequences covering the Arabidopsis genome. More information on the BAC tiling path is available on the Solanaceae Genome Network World Wide Web site (http://www.sgn.cornell.edu). The Arabidopsis gene set as predicted from the genomic sequence is the December 2000 version of Arabidopsis coding sequences (ATH1.cds), which contains 27,427 sequences, and is available through the Arabidopsis Information Resource (http://www.Arabidopsis.org).
BAC Sequences
All six BACs were derived from tomato cv Heinz 1706 (Budiman et al., 2000) and are available through the Clemson University Genomics Institute (http://www.genome.clemson.edu). The BACs were annotated by analysis with FGENESH, a gene prediction program developed by A.A Salamov and V.V. Solovyev (http://genomic.sanger.ac.uk/papers/AN_dro_paper.html). Furthermore, each BAC was screened against the tomato EST database (http://www.sgn.cornell.edu) (BLASTN) and the Arabidopsis BAC tiling path (tBLASTX) to identify matches to putative genes in each BAC as well as against the GenBank protein database maintained by the National Center for Biological Information (http://www.ncbi.nlm.nih.gov/).
Acknowledgments
The tomato EST database was derived from cDNA libraries constructed at Cornell University and sequenced at TIGR. Sequencing of BAC T62O11 was accomplished by Richard McCombie and Manpreet Katari at the Cold Spring Harbor Laboratory. Thanks to Anne Frary and Todd Vision for critical review of the manuscript. This work was supported by Grant DBI-9872617 from the Plant Genome Program of the National Science Foundation to S.T. (Cornell University), J.G. (U.S. Department of Agriculture/Agricultural Research Service), and G.M. (Boyce Thompson Institute for Plant Research) and by Grant DBI-9813392 to TIGR.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010478.
References
- Adam, D. (2000). Now for the hard ones. Nature 408, 792–793. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Arumuganathan, K., and Earle, E.D. (1991). Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9, 208–219. [Google Scholar]
- Bernatzky, R., and Tanksley, S.D. (1986). Majority of random cDNA clones correspond to single loci in the tomato genome. Mol. Gen. Genet. 203, 8–14. [Google Scholar]
- Budiman, M.A., Mao, L., Wood, T.C., and Wing, R.A. (2000). A deep-coverage tomato BAC library and prospects toward development of an STC framework for genome sequencing. Genome Res. 10, 129–136. [PMC free article] [PubMed] [Google Scholar]
- Cary, J.W., Lax, A.R., and Flurkey, W.H. (1992). Cloning and characterization of cDNAs coding for Vicia faba polyphenol oxidase. Plant Mol. Biol. 20, 245–253. [DOI] [PubMed] [Google Scholar]
- Chase, M.W., Soltis, D.E., Olmstead, R.G., Morgan, D., Les, D.H., Mishler, B.D., Duvall, M.R., Price, R.A., Hills, H.G., and Qiu, Y.-L. (1993). Phylogenetics of seed plants: An analysis of nucleotide sequences from the plastid gene rbcL. Ann. Mo. Bot. Gard. 80, 528–580. [Google Scholar]
- Deikman, J., and Fischer, R. (1988). Interaction of a DNA binding factor with the 5′-flanking region of an ethylene-responsive fruit ripening gene from tomato. EMBO J. 7, 3315–3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J., and Lukens, L. (1998). Transcriptional regulators and the evolution of plant form. Plant Cell 10, 1075–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, X., Li, X., Xue, Q., Abo-el-Saad, M., Xu, D., and Wu, R. (1996). Transgenic rice plants harboring an introduced potato proteinase inhibitor II gene are insect resistant. Nat. Biotechnol. 14, 494–498. [DOI] [PubMed] [Google Scholar]
- Frary, A., Nesbitt, T.C., Grandillo, S., Knaap, E., Cong, B., Liu, J., Meller, J., Elber, R., Alpert, K.B., and Tanksley, S.D. (2000). Fw2.2: A quantitative trait locus key to the evolution of tomato fruit size. Science 289, 85–88. [DOI] [PubMed] [Google Scholar]
- Fulton, T., van der Hoeven, R., Eannetta, N., and Tanksley, S. (2002). Identification, analysis, and utilization of conserved ortholog set markers for comparative genomics in higher plants. Plant Cell 14, 1457–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- Johnson, R., Narvaez, J., An, G., and Ryan, C. (1989). Expression of proteinase inhibitors I and II in transgenic tobacco plants: Effects on natural defense against Manduca sexta larvae. Proc. Natl. Acad. Sci. USA 86, 9871–9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keys, D.N., Lewis, D.L., Selegue, J.E., Pearson, B.J., Goodrich, L.V., Johnson, R.L., Gates, J., Scott, M.P., and Carrol, S.B. (1999). Recruitment of a hedgehog regulatory circuit in butterfly eyespot evolution. Science 283, 532–534. [DOI] [PubMed] [Google Scholar]
- Khush, G.S., and Rick, C.M. (1968). Cytogenetic analysis of the tomato genome by means of induced deficiencies. Chromosoma 23, 452–484. [Google Scholar]
- Khush, G.S., Rick, C.M., and Robinson, R.W. (1964). Genetic activity in a heterochromatic chromosome segment of the tomato. Science 145, 1432–1434. [DOI] [PubMed] [Google Scholar]
- King, M.C., and Wilson, A.C. (1975). Evolution at two levels in humans and chimpanzees. Science 188, 107–116. [DOI] [PubMed] [Google Scholar]
- Ku, H.K., Vision, T., Liu, J., and Tanksley, S.D. (2000). Comparing sequenced segments of the tomato and Arabidopsis genomes: Large-scale duplication followed by selective gene loss creates a network of synteny. Proc. Natl. Acad. Sci. USA 97, 9121–9126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, A., Altabella, T., Taylor, M.A., and Tiburcio, A.F. (1997). Recent advances in polyamine research. Trends Plant Sci. 2, 124–130. [Google Scholar]
- Lagercrantz, U., and Axelsson, T. (2000). Rapid evolution of the family of CONSTANS like genes in plants. Mol. Biol. Evol. 17, 1499–1507. [DOI] [PubMed] [Google Scholar]
- Matton, D.P., and Brisson, N. (1989). Cloning, expression, and sequence conservation of pathogenesis-related gene transcripts of potato. Mol. Plant-Microbe Interact. 2, 325–331. [DOI] [PubMed] [Google Scholar]
- Murata, M., Tsurutani, M., Hagiwara, S., and Homma, S. (1997). Subcellular location of polyphenol oxidase in apples. Biosci. Biotechnol. Biochem. 61, 1495–1499. [DOI] [PubMed] [Google Scholar]
- Paterson, A.H., Bowers, J.E., Burow, M.D., Draye, X., Elsik, C.G., Jiang, C.-X., Katsar, C.S., Lan, T.-H., Lin, Y.-R., Ming, R., and Wright, R.J. (2000). Comparative genomics of plant chromosomes. Plant Cell 12, 1523–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi, E. (1998). A bonanza for plant genomics. Science 282, 652–654. [DOI] [PubMed] [Google Scholar]
- Peterson, D.G., Pearson, W.R, and Stack, S.M. (1998). Characterization of the tomato (Lycopersicon esculentum) genome using in vitro and in situ DNA reassociation. Genome 41, 346–356. [Google Scholar]
- Peterson, D.G., Price, H.J., Johnston, J.S., and Stack, S.M. (1996). DNA content of heterochromatin and euchromatin in tomato (Lycopersicon esculentum) pachytene chromosomes. Genome 39, 77–82. [DOI] [PubMed] [Google Scholar]
- Quackenbush, J., Liang, F., Holt, I., Pertea, G., and Upton, J. (2000). The TIGR gene indices: Reconstruction and representation of expressed gene sequences. Nucleic Acids Res. 28, 141–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rick, C.M. (1975). The tomato. In Handbook of Genetics, Vol. 2, R.C. King, ed (New York: Plenum Press), pp. 247–280.
- Souer, E., van Houwelingen, A., Kloos, D., Mol, J., and Koes, R. (1996). The no apical gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85, 159–170. [DOI] [PubMed] [Google Scholar]
- Stern, D.L. (2000). Evolutionary developmental biology and the problem of variation. Evolution 54, 1079–1091. [DOI] [PubMed] [Google Scholar]
- Tanksley, S.D., et al. (1992). High density molecular linkage maps of the tomato and potato genomes. Genetics 132, 1141–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiburcio, A.F., Altabell, T., Borrell, A., and Masgrau, C. (1997). Polyamine biosynthesis and its regulation. Physiol. Plant. 100, 664–674. [Google Scholar]
- Wang, R., Stec, A., Hey, J., Lukens, L., and Doebley, J. (1999). The limits of selection during maize domestication. Nature 398, 236–239. [DOI] [PubMed] [Google Scholar]
- Yang, Y.W., Lai, K.N., Tai, P.Y., and Li, W.H. (1999). Rates of nucleotide substitution in angiosperm mitochondrial DNA sequences and dates of divergence between Brassica and other angiosperm lineages. J. Mol. Evol. 48, 597–604. [DOI] [PubMed] [Google Scholar]