Abstract
We have screened a large tomato EST database against the Arabidopsis genomic sequence and report here the identification of a set of 1025 genes (referred to as a conserved ortholog set, or COS markers) that are single or low copy in both genomes (as determined by computational screens and DNA gel blot hybridization) and that have remained relatively stable in sequence since the early radiation of dicotyledonous plants. These genes were annotated, and a large portion could be assigned to putative functional categories associated with basic metabolic processes, such as energy-generating processes and the biosynthesis and degradation of cellular building blocks. We further demonstrate, through computational screens (e.g., against a Medicago truncatula database) and direct hybridization on genomic DNA of diverse plant species, that these COS markers also are conserved in the genomes of other plant families. Finally, we show that this gene set can be used for comparative mapping studies between highly divergent genomes such as those of tomato and Arabidopsis. This set of COS markers, identified computationally and experimentally, may further studies on comparative genomes and phylogenetics and elucidate the nature of genes conserved throughout plant evolution.
INTRODUCTION
In the past 10 years, we have seen great progress in linking plant genomes through comparative genetic maps, especially for species belonging to the same family (for review, see Paterson et al., 2000). For example, most of the economically important species in the grass family (e.g., maize, wheat, barley, rice, millet, and sorghum) have detailed comparative maps such that both gene content and gene order often can be predicted across species (Bennetzen et al., 1998; Gale and Devos, 1998; Wilson et al., 1999). Similarly, genetic and genomic information can be shared among many leguminous species (e.g., soybean and mung bean) (Menancio-Hautea et al., 1993; Boutin et al., 1995) or among species in the nightshade family (e.g., tomato, pepper, and potato) (Tanksley et al., 1992; Livingstone et al., 1999).
In all of these instances, the species within families have been linked by a common set of orthologous genes detected through DNA gel blot hybridization. The ability to detect single-copy orthologous genes among plant genomes has permitted comparative plant genomics to advance as rapidly as it has.
By contrast, during this same period, relatively little progress was made in comparative genomics among more divergent plant species, that is, those belonging to different plant families. Evolutionary divergence time among plant families is greater, allowing for more genomic rearrangements. Moreover, comparisons between plant families have been impeded further by the technical difficulties in identifying conserved orthologous genes that can be used to link these plant genomes. Specifically, reduced gene similarities between plant families have made comparative mapping, via common probes and DNA gel blot hybridization, difficult at best and often impossible. As a result, at present, there is no framework for clearly interpreting genomic similarities among higher plants.
With the Arabidopsis genome having been sequenced and major genomic efforts under way on other plant species (National Science Foundation Plant Genome Research Program [http://www.nsf.gov/bio/dbi/dbi_pgr.htm]; Pennisi, 1998; Adam, 2000; Paterson et al., 2000), the challenge will be to find the manner in which map, sequence, and eventually functional genomic information from one species can be accessed, compared, and exploited across all plant species. To do so will require the identification of a subset of plant genes that have remained relatively stable in both sequence and copy number since the radiation of flowering plants from their last common ancestors. Identification of such a set of genes also would facilitate taxonomic and phylogenic studies in higher plants that are based at present on a very small set of highly conserved sequences, especially those of chloroplast and mitochondrial genes.
We have attempted to remedy this situation and provide the basis for more robust comparative genomics and phylogenetic studies across plant taxa by identifying a set of genes conserved throughout evolution in both sequence and copy number. This set of >1000 conserved genes, which we refer to as conserved ortholog set (COS) markers, was identified by computationally comparing the Arabidopsis genomic sequence with the EST database of tomato, which comprises 130,000 ESTs representing approximately half of the tomato gene content (Van der Hoeven et al., 2002; http://sgn.cornell.edu, http://www.tigr.org). The computational screening criteria required that the tomato EST have a single best match in the Arabidopsis genome, avoiding problems with matches to multigene families for which orthology and paralogy cannot be distinguished readily. To ensure that these putative orthologs also were single or low copy in the tomato genome (and hence likely to be orthologs and not paralogs), the majority of COS markers also were screened against tomato genomic DNA via DNA gel blot analysis.
Tomato and Arabidopsis are both dicots, but they belong to different families (Brassicaceae and Solanaceae) that diverged early in flowering plant evolution, ∼100 to 150 million years ago (Gandolfo et al., 1998; Yang et al., 1999). Because Arabidopsis and tomato diverged early in plant evolution, the COS markers reported here should be useful for comparative genomics and taxonomy studies in a wide array of plant species (Figure 1). We show the utility of these COS markers for comparative mapping between tomato and Arabidopsis. In addition, we demonstrate two strategies by which these COS markers can be used for comparative mapping in other plant species, the result of which eventually could provide the basis for comparing genome sequence and map information across many plant species.
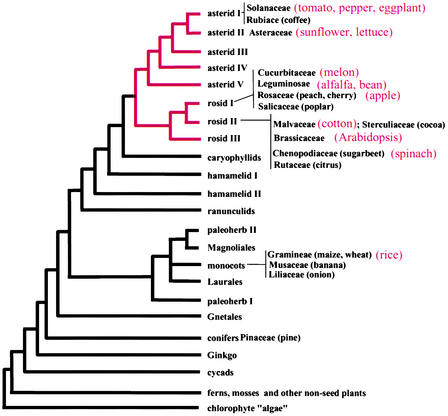
Figure 1.
Dendrogram Depicting Phylogenetic Relationships of Higher Plant Taxa.
Common names are given in parentheses. Species in red are those used on the garden blot. (Figure reprinted from Ku et al. [2000], based on a figure in Chase et al. [1993].)
RESULTS
Selection of COS Markers
The 1025 COS markers described here were identified initially by manually screening tomato EST sequences against the Arabidopsis BAC tiling path database (http://www.Arabidopsis.org) using the criteria described in Methods. The purpose of this screen was to identify single-copy tomato genes that have a single best match to one region of the Arabidopsis genome and hence would qualify as potential orthologs. This method inherently selects against multigene families for which orthologs between specific genes may not be readily distinguishable. Genes meeting these criteria are referred to herein as putative orthologs, with the disclaimer that we recognize that these data are not sufficient to prove orthology in the strictest evolutionary context, but nevertheless they can be a useful tool.
To obtain the 1025 COS markers described here, >20,000 tomato ESTs were screened as described above. The COS markers described were identified and characterized during the past 2 years, during which time the Arabidopsis genome and tomato ESTs were being sequenced. To standardize all results, the COS marker set was rescreened against both the current Arabidopsis tiling path and the tomato EST/unigene set as of April 2001. To estimate the percentage of tomato unigenes that meet COS criteria, the entire tomato unigene set was rescreened against the Arabidopsis tiling path at the same time.
Of the 27,000 tomato unigenes, 55% had a match to the Arabidopsis genome with a tBLASTX score of <E-15. Of those, 16% met the second criterion (no close second match in the Arabidopsis genome). Hence, ∼10% of all tomato unigenes could be classified as COS markers. A set of 1025 such COS markers was identified and annotated (see below), and 927 of those were screened against tomato genomic DNA via DNA gel blot hybridization (stringency of 0.5 × SSC at 65°C) (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate). The majority (85%) were classified as single or low copy (>95% of the hybridization signal assigned to three or fewer restriction fragments). Table 1 contains descriptions of the 10 COS markers most conserved with Arabidopsis (based on tBLASTX scores). The full set of 1025 COS markers, associated annotations, and map positions can be found at the Solanaceae Genome Network World Wide Web site (http://sgn.cornell.edu).
Table 1.
The 10 COS Markers with Expect Values (from tBLASTX) Given for the Three Most Significant Matches to an Arabidopsis (At) Sequence and M. truncatula (Mt) ESTs
| COS No. | Tomato EST Sequence No. |
At First | At BAC Accession No. |
Location | Start | At Second | Mt First | Mt EST Sequence No. |
Mt Second | Mt Third |
|---|---|---|---|---|---|---|---|---|---|---|
| COS1335 | TPTAN80TH | −145 | AC008075 | 1.134 | 93215 | −127 | −111 | TC11357 | −51 | −29 |
| COS94 | TSHAB35TH | −132 | AC007168 | 2.127 | 50839 | −120 | −143 | TC8674 | −123 | −21 |
| COS1358 | TPTAP09TH | −130 | AL034567 | 4.165 | 42561 | −111 | −139 | TC6272 | −114 | −99 |
| COS1923 | TRZCX40TH | −125 | AL137898 | 3.24 | 12698 | −112 | −81 | TC6527 | −28 | −27 |
| COS1039 | TRXCA53TH | −123 | AL080283 | 4.16 | 44327 | −129 | TC8858 | |||
| COS1850 | TRZCJ56TH | −120 | AB025622 | 5.209 | 15473 | −106 | −104 | TC5734 | −80 | −45 |
| COS34 | TOVAS39TH | −117 | AB019228 | 5.247 | 42855 | −59 | −140 | TC13434 | −74 | −73 |
| COS1928 | TPTAD69TH | −116 | AB006700 | 5.011 | 81181 | −97 | −17 | TC5201 | −13 | |
| COS1683 | TRZBF08TH | −116 | AB025631 | 3.056 | 13692 | −120 | TC7038 | |||
| COS1263 | TPTAE91TH | −115 | AF080121 | 5.075 | 28443 | −100 | −87 | TC14399 | −40 | −38 |
Location indicates Arabidopsis chromosomal location and consecutive numbers of BACs in the tiling path (e.g., 1.134 = chromosome 1, BAC No. 134) (for details, see http://soldb.cit.cornell.edu/). Start indicates approximate base pair position on the BAC for the start of match with tomato EST.
Annotation of COS Markers and Functional Role Categories
The significant sequence conservation between the COS markers and Arabidopsis genes, coupled with the fact that these genes are single or low copy in both species, raises the possibility of conserved functional roles in both species and potentially in all plant species. Therefore, the COS marker genes may fulfill roles that are universally important to all plant species. In addition, these genes have remained stable during the course of plant speciation, suggesting that many of them were present in a similar form before the radiation of plant species.
The COS markers were searched (BLASTX) against the GenBank protein database maintained by the National Center for Biotechnology Information, and the results were used for annotation and assignment to functional role categories (r.c.). It is important to realize that this analysis was limited in its scope with respect to the collection of ESTs used to initially identify the COS markers and will reflect, to a great extent, the types of genes in the database that have been characterized previously. In addition, the sequence information for each COS marker represents on average 553 high-quality nucleotides (range of 178 to 832 nucleotides) from the 5′ end of the gene transcript. As a result of the variation in EST length, expect value scores of sequence similarity searches against Arabidopsis cannot be used reliably to compare the degree of sequence conservation between COS markers. The complete COS marker annotation and functional categorization are available online (http://sgn.cornell.edu).
For the majority (751; 73%) of the 1025 COS markers, the most significant match was against predicted or characterized genes in Arabidopsis. Solanaceous species or other plant species represented another 151 (14%) and 109 (11%) best matches, respectively. In only 15 instances (2%), nonplant species provided the best match. However, when these markers were analyzed again by BLASTX (tBLASTX) against the Arabidopsis genome (versus the predicted gene set), a more significant Arabidopsis match could be found.
These instances most likely represent previously unidentified genes in the Arabidopsis genome. This result also is consistent with the fact that the COS markers were selected initially based on the screening of tomato ESTs against the entire Arabidopsis genomic sequence, rather than only the predicted Arabidopsis gene set. These results also demonstrate the utility of non-Arabidopsis EST databases in the further annotation of the Arabidopsis genome.
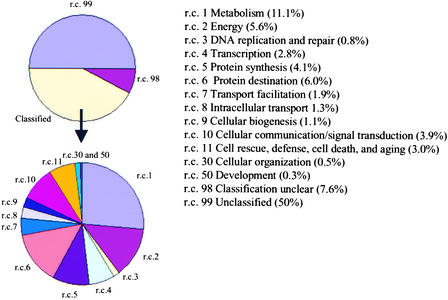
Of the 1025 COS markers, 514 (50%) had matches to genes or sequences of unknown function and hence were assigned to the unclassified (r.c. 99) role category. Another 76 (7%) were placed in the classification unclear (r.c. 98) category. The classification unclear (r.c. 98) category contains a significant number of COS markers with matches against genes that have been described previously, but uncertainty about their putative function prevented categorization. These include COS markers with matches against a number of cytochrome P450s and transferases for which the target substrates are unclear and genes that are known to be expressed only under certain environmental conditions (e.g., auxin induced) and developmental stages or in specific tissues. The remaining 435 COS markers (42%) were assigned to various functional role categories based on significant matches to proteins already assigned functional roles (Figure 2).
Figure 2.
Relative Distribution of COS Markers over Functional Role Categories.
Assignments of COS markers to specific subcategories on the Solanaceae Genome Network online database (http://sgn.cornell.edu).
A large proportion (42%) of the 435 assigned COS markers represent genes that appear to be involved in basic metabolic processes, such as energy-generating processes and the biosynthesis and degradation of cellular building blocks. Genes involved with the cellular transcriptional and translational machinery represent ∼17% of the assigned COS markers, those involved in protein processing and destination represent 14%, and those involved in signal transduction represent 9%. These types of genes, representing many aspects of plant cellular processes and metabolism of cellular structural components, are part of the set of genes that have remained highly conserved across plant species and at an approximately equal copy number since the divergence of Arabidopsis and tomato 100 to 150 million years ago (Yang et al., 1999).
Use of COS Markers in Comparative Plant Genomics
Comparative Mapping between the Tomato and Arabidopsis Genomes
One of the primary motivations for identifying the COS markers was to provide a set of putatively orthologous genes for comparative genome mapping between tomato and Arabidopsis (and eventually other plant species). At present, there are two problems with comparative mapping between highly divergent plants such as Arabidopsis and tomato. First, until now, there has not been a large set of putatively orthologous genes useful for mapping. Previous studies of comparative mapping within plant families relied largely on DNA gel blot hybridization using heterologous probes between species. Before identifying COS markers computationally, we attempted to use Arabidopsis gene probes as heterologous probes for tomato mapping with mixed results.
The second problem is that many Arabidopsis genes do not hybridize with tomato genomic DNA under standard stringency conditions, and in the cases in which hybridization was detected, the signals often were weak, making interpretation difficult. DNA gel blot hybridization works well when sequences share >70% nucleic acid similarity, but this threshold often is violated when making comparisons across plant families as distant as those of tomato and Arabidopsis. By computationally identifying putative ortholog sets (composed of a single tomato EST and its best Arabidopsis match), one can use the tomato probe/sequence for mapping on tomato, resulting in clear results with DNA gel blots.
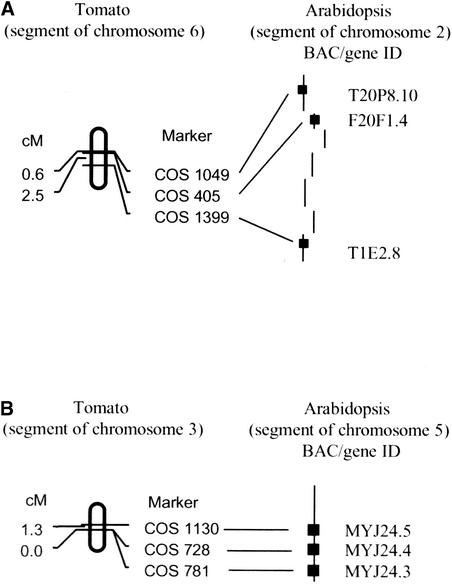
Currently, we have mapped >550 COS markers in tomato and expect to map up to 1000 to elucidate the syntenic relationships between these two genomes; the results from this study will be the topic of a future publication. The current COS marker map can be viewed at http://sgn.cornell.edu. However, what we have discovered to date is as follows: (1) the COS markers can reveal segments of conserved linkage between these two genomes; (2) the size of these conserved segments usually is restricted to <10 centimorgan (Figure 3); and (3) polyploidization events that occurred both before and near the time that plant families radiated (including Solanaceae and Brassicaceae) have resulted in networks of synteny both within and between plant genomes (Ku et al., 2000; Vision et al., 2000). Having a large set of genome-wide COS markers available provides the means and strategy to decipher the syntenic relationships among widely divergent plant genomes such as those of tomato and Arabidopsis.
Figure 3.
Microsynteny between Tomato and Arabidopsis Genomes.
(A) A region of tomato chromosome 6 showing conservation of synteny with a region of Arabidopsis chromosome 2.
(B) A region of tomato chromosome 3 showing conservation of synteny with a region of Arabidopsis chromosome 5.
Strategies for Using COS Markers for Comparative Mapping in Other Plant Species
Direct Hybridization
Several strategies can be imagined for using the COS marker sequences for comparative mapping in other plants. First, because the COS markers were selected to be both highly conserved and single/low copy, it is possible that some portion of them may be useful directly as hybridization probes for restriction fragment length polymorphism mapping in other species. Depending on whether the species in question is more closely related to tomato or Arabidopsis, one might choose either the tomato or the Arabidopsis probe.
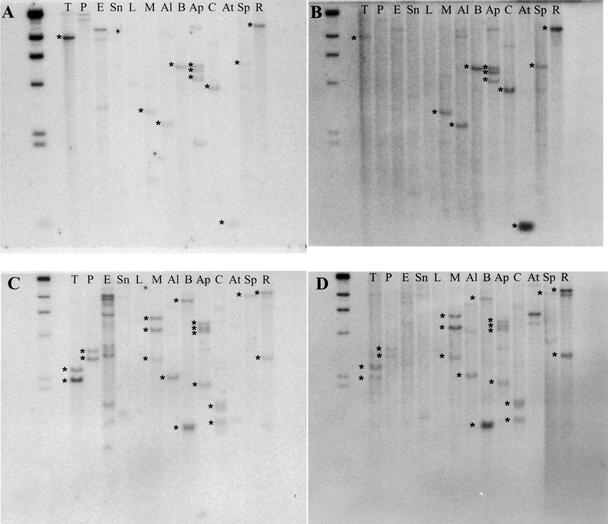
To test this possibility and to determine whether these COS markers are single/low copy in most other plant genomes, we constructed a “garden blot” composed of DNA from a wide range of plant species (Figures 1 and 4). The blots were probed with the COS markers listed in Table 2, first with a tomato EST clone corresponding to the COS marker and then with the counterpart Arabidopsis COS probe. The COS markers selected for testing were among the most conserved (at the amino acid level) based on tomato-Arabidopsis comparisons. Hybridization results for two of the nine tested COS markers are depicted in Figure 4. Although these were selected for display based on the quality of the DNA gel blots, the qualitative results are representative.
Figure 4.
Autoradiographs from the Hybridization of Garden Blots with COS Markers.
T, tomato (Lycopersicon esculentum); P, garden pepper (Capsicum annuum); E, eggplant (Solanum melongena); Sn, sunflower (Helianthus annuus); L, lettuce (Lactuca sativa); M, melon (Cucumis melon); Al, alfalfa (Medicago sativa); B, field bean (Phaseolus vulgaris); Ap, apple (Malus domestica); C, cotton (Gossypium hirsutum); At, Arabidopsis thaliana; Sp, spinach (Spinacia oleracea); and R, rice (Oryza sativa).
(A) A garden blot filter probed with COS1039 tomato probe.
(B) The same filter probed with the Arabidopsis counterpart of COS1039.
(C) A garden blot filter probed with COS1263 tomato probe.
(D) The same filter probed with the Arabidopsis counterpart of COS1263.
Table 2.
Tomato COS Markers and Primers Used to Amplify Their Putative Orthologous Counterparts from the Arabidopsis Genome
| COS No. | Tomato EST Sequence No. |
Matching Arabidopsis BAC Accession No. |
tBLASTX Expect Value |
Arabidopsis Probe Forward Primer |
Arabidopsis Probe Reverse Primer |
Arabidopsis Probe (bp) |
|---|---|---|---|---|---|---|
| COS1335 | TPTAN80TH | AC008075 | −145 | TCTTGGTGGGGTGATGAAAT | TTGTGAGTTGCGATGGTCTC | 341 |
| COS1358 | TPTAP09TH | AL034567 | −130 | AGGACAATGCCGACTGAAGA | TGGATGGATCTATGGTTCTGTG | 199 |
| COS1039 | TRXCA53TH | AL080283 | −123 | GGAGAATTCACCAAGGACGA | CATTCAAACTCTGCCCACAT | 443 |
| COS1263 | TPRAE91TH | AF080121 | −115 | AATCCCCGCTCAGAAATACC | AGCATGATAGCCAGGACCAT | 384 |
| COS276 | TOVCE01THE | AF058914 | −108 | CTCCTCGACGCTATGATTCC | GCTCCAGAGCCAAGTGGTTA | 348 |
| COS270 | TOVBP41TH | AL096860 | −101 | CAAAGCTCTCCACCAATTTGA | TCATCAAAGGCATTGCGTAG | 189 |
| COS1006 | TRXAM55TH | AC006248 | −98 | ATGTGTGTGTGGTGGGGACT | TCTGTCGGTTTCCTCGAGTT | 243 |
| COS1106 | TFBAU87THB | AB024033 | −97 | CCTTCGGTAGAAGCATGAGC | CCAGAAGTGGAAAGCTTGGT | 185 |
| COS55 | TOVBR32TH | AC005824 | −97 | CAAGAGCGAGACGAGGAAGT | TCATTGGAAGCAAAGGTGAA | 156 |
Markers are listed in order of tomato-Arabidopsis tBLASTX expect value.
Two aspects of these hybridization results are worth noting. (1) In the majority of cases, both the tomato and Arabidopsis COS probes detected single- or low-copy genes in most of the species tested (Figure 4). The only exception was COS1358, for which the Arabidopsis probe hybridized to three to seven restriction fragments in many of the genomes, reflecting a small gene family (data not shown). (2) Both the tomato and Arabidopsis probes detected many if not most of the same fragments in the genomes to which they both hybridized (Figure 4). For example, with both COS1039 and COS1263, the tomato probe and the Arabidopsis probe detected nearly identical restriction fragments in the lanes for which hybridization was detected (Figure 4). However, the tomato probe gave a much stronger signal, not only with tomato but also with other species in the Solanaceae family (e.g., pepper and eggplant). Lettuce and sunflower gave weak signals (or no signal) with all probes (both tomato and Arabidopsis), a result possibly attributable to insufficient DNA loading and/or quality of DNA for these samples (Figure 4). Rice was the only monocot included in the survey; in a number of instances, it showed clear hybridization signals with both Arabidopsis and tomato probes (Figure 4).
The combined results from these hybridization experiments suggest that at least the more conserved COS markers can be used directly as hybridization probes for restriction fragment length polymorphism mapping. The advantage of this strategy is that species that do not have sequence databases at present (either genomic or ESTs) still can be mapped with some COS markers. However, it is important to note that the COS markers chosen for hybridization experiments were those with the highest tomato-Arabidopsis tBLASTX values. Hence, it is possible that less conserved COS markers may be less useful for direct mapping through hybridization.
Computational Screens with COS Marker Consensus Sequences
A second strategy for the application of COS markers in other species would be to use the sequence of each COS marker (or a consensus sequence derived from aligning the corresponding tomato and Arabidopsis sequences) to search EST or genomic databases of other species to find their corresponding COS marker sequences. Although this strategy is restricted to species with substantial sequence databases, it has the advantage that one can use the homologous probes and/or sequence primers for mapping in the species of interest.
We tested this strategy by using the tomato sequences for 10 COS markers as queries against the Medicago truncatula unigene database, which is one of the largest EST databases for a dicot species, containing >30,000 tentative consensus sequences (http://www.tigr.org). As a control, the most similar M. truncatula unigene, identified by screening with the tomato COS sequence, was screened against the Arabidopsis BAC tiling path (tBLASTX). The goal was to determine whether the same Arabidopsis BAC would be identified by the M. truncatula EST sequence that was identified by the tomato during the original screen for COS markers (see above).
Table 1 lists the tBLASTX expect values for the top three M. truncatula EST matches to each tomato COS sequence. In all cases, M. truncatula ESTs with highly significant matches to each COS sequence were identified (Table 1). Furthermore, when a COS marker had only one significant match in Arabidopsis, it had only one significant match in M. truncatula as well. In all 10 cases, using the M. truncatula counterpart of each COS marker to screen the Arabidopsis tiling path identified the same segment of the same Arabidopsis BAC that was identified originally using the tomato EST. In the majority (6 of 10) of these, this Arabidopsis BAC was the most significant hit; in the other four cases, it was one of the top four most significant hits.
Three-way alignments were made for each set of tomato, Arabidopsis, and M. truncatula putative orthologs to determine the relative divergence among the three (Figure 5). In addition, pairwise distances (shown as mean character differences) were calculated for these three sets of COS markers using PAUP software (Swofford, 1999) and are listed in Table 3. The sequence similarities for sequences within each set ranged from 82 to 95% similarity, whereas there was no sequence similarity among sets (i.e., between the three COS markers, etc.) (Table 3).
Figure 5.
Alignment of Protein Sequences of COS1335 and Its M. truncatula and Arabidopsis Counterparts (A), COS1358 and its M. truncatula and Arabidopsis Counterparts (B), and COS94 and its M. truncatula and Arabidopsis Counterparts (C).
The consensus sequences are given at top. Triangles indicate approximate positions of Arabidopsis introns according to the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/).
Table 3.
Pairwise Divergence Values for Protein Sequence Comparisons between the Tomato, M. truncatula, and Arabidopsis Sequence Counterparts of Three COS Marker Sets (COS1335, COS1358, and COS94)
| COS No. | Tomato-Arabidopsis | Tomato-M. truncatula | M. truncatula-Arabidopsis |
|---|---|---|---|
| COS1335 | 0.162 | 0.162 | 0.185 |
| COS1358 | 0.120 | 0.134 | 0.128 |
| COS94 | 0.074 | 0.051 | 0.080 |
Values were calculated as percentage mean character differences by PAUP (Swofford, 1999).
For each COS set, the level of amino acid sequence divergence among tomato, M. truncatula, and Arabidopsis was similar, despite the fact that the tomato lineage is thought to have diverged before the M. truncatula–Arabidopsis lineage diverged (Figure 1). However, a comparison of divergence values among COS sets showed remarkable variation among these gene sets. For example, for COS1335, the divergence values for pairwise comparisons of tomato, M. truncatula, and Arabidopsis ranged from 0.162 to 0.185; the values for COS1358 ranged from 0.120 to 0.134; and the values for COS94 ranged from 0.051 to 0.074 (Table 3).
Although the computational screen was limited to only a few COS markers and against a single database (M. truncatula), these results, combined with the DNA gel blot hybridization results (see above), suggest that orthologous counterparts to many if not most COS markers exist in the genomes of other plant species. As plant EST (and genomic) databases expand, the computational approach to finding the COS marker counterparts in plant genomes will increase. Eventually, it may be possible to identify sufficiently conserved consensus primers that could be used to amplify COS markers from a wide variety of plant genomes. This would facilitate mapping in plant genomes that lack genomic/EST databases and also could be used to generate multiple sequence comparisons across plant taxa for phylogenetic reconstructions.
DISCUSSION
Once a number of species have been sequenced fully, it will be possible to computationally classify the corresponding proteins into probably clusters of orthologs and paralogs (Tatusov et al., 1997, 2000). Unfortunately, identifying putative orthologs in plants, and distinguishing them from paralogs, is difficult for several reasons. First, Arabidopsis is the only plant species for which the complete genomic sequence is available. The majority of other plants have only partial sequences available (e.g., EST databases). When comparing a sequence from one plant species (e.g., species X) against Arabidopsis, it often is possible to identify a highly significant homolog(s) in the Arabidopsis genome. However, one cannot assume that it is an ortholog, because the “true” ortholog in species X may reside in the unsequenced portion of that genome. We guarded against this possibility by probing filters of tomato nuclear DNA, via DNA gel blot analysis, each putative tomato ortholog to determine whether it is a member of a multigene family.
Second, because plant genomes have experienced extensive gene duplication events, most genes belong to multigene families. Thus, orthologs may not be distinguished easily from paralogs. This is why we required that there be only one best match in the Arabidopsis genome when screening for putative orthologs of tomato genes.
Here, a large EST database from one plant species has been screened computationally against the Arabidopsis genome and tested experimentally in a manner that could yield a large set of genes that have a high probability of being orthologs. Although there are databases/algorithms, such as TOGA (available at http://www.tigr.org/tdb/toga/orth_search.shtml), that can search for and cluster homologous sequences across multiple genome databases, the results from these analyses do not automatically distinguish between paralogs and orthologs. Although straightforward and useful for gene alignments, such an approach for establishing orthology is highly risky.
The COS markers reported here can be used for comparative mapping studies between highly divergent genomes such as those of tomato and Arabidopsis. The consensus sequences of COS markers (from tomato-Arabidopsis alignments) also can be used to search genome databases of other plants to find corresponding putative orthologous genes. Therefore, these COS markers may be useful for comparative mapping across plant families and may facilitate the development of the syntenic networks across plant taxa necessary for understanding the evolution of genes, genomes, and gene functions. This set of COS markers also may serve as the basis for extending plant phylogenetic studies that are limited at present by the availability of genes for which putative orthologs can be identified readily across plant taxa.
METHODS
Tomato EST Database
The tomato (Lycopersicon esculentum) EST collection is stored and accessible through the online Solanaceae Genome Network database (http://sgn.cornell.edu ). The EST collection is derived from a variety of >25 different cDNA libraries, capturing genes expressed in different tissue types and developmental stages or expressed during pathogen-elicited responses (Van der Hoeven et al., 2002). The collection comprises >130,000 ESTs, representing >27,000 unique gene transcripts. All information pertaining to the conserved ortholog set (COS) markers is available online, including gene annotation, contig membership, and positional coordinates against the Arabidopsis tiling path (http://sgn.cornell.educos_list.html).
Computational Screening of Conserved Ortholog Set Markers
To identify conserved orthologs between Arabidopsis thaliana and tomato but to avoid misidentifying gene families or paralogs, a very conservative computational strategy was followed. Tomato ESTs were scanned against the Arabidopsis genome (specifically, the genomic sequence ordered tiling path from TAIR [http://www.Arabidopsis.org/]) using tBLASTX. A unique tomato EST was selected as a conserved ortholog if it met the following criteria: it matched a single Arabidopsis BAC at an expect value of ≤E-15, and the next best Arabidopsis match was of lower significance (i.e., there was a difference of ≥10 between expect scores).
ESTs that met both of these criteria were classified as conserved orthologs; all others were considered potentially paralogous and eliminated. The ESTs selected as conserved orthologs then were screened computationally against the tomato unigene set currently composed of 27,000 contigs and/or singletons (Van der Hoeven et al., 2002; http://sgn.cornell.edu) to ensure that all COS markers chosen represent unique tomato genes.
The 10 COS markers with the highest expect values against the Arabidopsis genome also were used to screen the Medicago truncatula EST database (http://www.tigr.org) using tBLASTX.
Mapping of the COS Markers in Tomato
The mapping population used was an F2 population from a tomato × Lycopersicon pennellii cross consisting of 83 plants with an average map resolution <1 centimorgan. This population was chosen after preliminary results indicated that possibly syntenic blocks consisted of several map units only; thus, a good mapping resolution would be needed (T.M. Fulton, Y. Xu, N. Eannetta, R. Van der Hoeven, and S.D. Tanksley, unpublished data). The tomato EST clones corresponding to each ortholog were surveyed against tomato genomic DNA of the two parents via DNA gel blot analysis using the restriction enzymes EcoRI, EcoRV, DraI, HaeIII, and ScaI. Random hexamer labeling, hybridization, and washing methods were as described previously (Feinberg and Vogelstein, 1983). Only those clones determined to be single or very low copy at a stringency of 0.5 × SSC at 65°C (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) and polymorphic in this cross were mapped using Mapmaker software (Lander et al., 1987).
To date, >550 of the COS markers that meet these criteria have been mapped. In addition, 200 restriction fragment length polymorphisms from the tomato high-density map (Tanksley et al., 1992) also have been mapped on this population to anchor the two maps. The current tomato-Arabidopsis comparative map, based on COS markers, can be viewed on the Solanaceae Genome Network (SGN) World Wide Web site (http://www.soldb.cit.cornell.edu). Upon request, COS marker clones described in this article will be made available in a timely manner for noncommercial research purposes. Sequences of COS markers can be found on the SGN World Wide Web site (http://www.sgn.cornell.edu). No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Hybridization of COS Markers to Other Species
A subset of nine COS markers (Table 2) with highly significant tBLASTX scores to Arabidopsis BACs and identified previously as being low copy in tomato were used as hybridization probes for DNA gel blots containing genomic DNA from a wide variety of plant species (tomato, pepper, eggplant, sunflower, lettuce, melon, alfalfa, bean, apple, cotton, Arabidopsis, spinach, and rice). The first three species belong to the Solanaceae family. The other species were chosen to represent a diverse set of families throughout the plant kingdom (Figure 1).
Genomic DNA of each species was digested with EcoRI; ∼3 μg of tomato, 1.5 μg of Arabidopsis, and 10 μg of the other species was run on an electrophoresis gel and DNA gel blotted. The amounts of DNA loaded were not strictly proportional to the genome sizes of each of the species. However, larger amounts of DNA were used for species other than Arabidopsis and tomato because all probes were heterologous (from tomato or Arabidopsis COS sequences) with respect to these species; hence, weaker signals were expected on DNA gel blots.
Tomato EST clones corresponding to each of the nine selected COS markers were radiolabeled, probed onto filters of these DNA gel blots, and washed at a stringency of 1.0 × SSC at 65°C. After exposure to film, the same blots were stripped and rehybridized with probes from the corresponding region in Arabidopsis. For these probes, genomic Arabidopsis DNA was amplified with primers specific to the coding regions of Arabidopsis that correspond to each of the nine COS markers (Table 2). The blots hybridized with the Arabidopsis probes were washed at a stringency of 1.0 × SSC at 65°C.
Annotation of COS Markers
The COS markers were annotated by analyzing the results of BLASTX analysis of the COS markers against the GenBank protein database maintained at the National Center for Biological Information (http://www.ncbi.nlm.nih.gov/). The definition line of the best match was stored as a description of the putative function of the gene transcript, but in many cases in which less strong hits were more informative, this information was included in a separate field for comments.
Functional annotation was achieved by assigning functional role categories as developed for the analysis of the Arabidopsis genome and used in conjunction with the numerical index for categories and subcategories as defined by TIGR (http://www.tigr.org). Annotation followed the Munich Information Center for Protein Sequences (http://mips.gsf.de) role categorization. A list of the role categories can be found on the SGN World Wide Web site (http://www.sgn.cornell.edu). Criteria used for role assignment required an approximate expect value of <E-30 against an experimentally characterized gene. However, in cases in which a COS marker matched a number of characterized genes of similar function with an expect value of >E-30, occasionally a role category was assigned.
Acknowledgments
Special thanks to Dan Ilut, Mark Wright, and Damon Little for computational support, Yimin Xu and Eloisa Tedeschi for technical support, and Nevin Young, Anne Frary, and Todd Vision for reviewing the manuscript. DNA for the garden blots was received from Susan Brown (Geneva Experimental Station, Geneva, NY) (apple), Susan McCouch (Cornell University) (rice), Molly Jahn (Cornell University) (pepper), Shunxue Tang (Oregon State University) (sunflower), and John Yu (U.S. Department of Agriculture, College Station, TX) (cotton). This project was supported by grants from the National Science Foundation (DBI-9872617) and the U.S. Department of Agriculture Plant Genome Program (97-35300-4384).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010479.
References
- Adam, D. (2000). Now for the hard ones. Nature 408, 792–793. [DOI] [PubMed] [Google Scholar]
- Bennetzen, J., et al. (1998). Grass genomes. Proc. Natl. Acad. Sci. USA 95, 1975–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutin, S.R., Young, N.D., Olson, T., Yu, Z.-H., Shoemaker, R.C., and Vallejos, C. (1995). Genome conservation among three legume genera detected with DNA markers. Genome 38, 928–937. [DOI] [PubMed] [Google Scholar]
- Chase, M.W., Soltis, D.E., Olmstead, R.G., Morgan, D., Les, D.H., Mishler, B.D., Duvall, M.R., Price, R.A., Hills, H.G., and Qiu, Y.-L. (1993). Phylogenetics of seed plants: An analysis of nucleotide sequences from the plastid gene rbcL. Ann. Mo. Bot. Gard. 80, 528–580. [Google Scholar]
- Feinberg, A.P., and Vogelstein, B. (1983). A technique for radiolabelling DNA restriction fragments to a high specific activity. Anal. Biochem. 132, 6–13. [DOI] [PubMed] [Google Scholar]
- Gale, M., and Devos, K. (1998). Comparative genetics in the grasses. Proc. Natl. Acad. Sci. USA 95, 1971–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandolfo, M.A., Nixon, K.C., and Crepet, W.L. (1998). A phylogenetic analysis of modern and cretaceous Triuridaceae (Monocotyledoneae). Am. J. Bot. 85, 964–974. [Google Scholar]
- Ku, H.M., Vision, T., and Liu, J. (2000). Comparing sequenced segments of the tomato and Arabidopsis genomes: Large-scale duplication followed by selective gene loss creates a network of synteny. Proc. Natl. Acad. Sci. USA 97, 9121–9126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander, E.S., Green, P., Abrahamson, J., Barlow, A., Daly, M.J., Lincoln, S.E., and Newburg, L. (1987). MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1, 174–181. [DOI] [PubMed] [Google Scholar]
- Livingstone, K.D., Lackney, V.K., Blauth, J.R., van Wijk, R., and Jahn, M.K. (1999). Genome mapping in Capsicum and the evolution of genome structure in the Solanaceae. Genetics 152, 1183–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menancio-Hautea, D., Fatokun, C., Kumar, L., Danesh, D., and Young, N. (1993). Comparative genome analysis of mungbean (Vigna radiata [L.] Wilczek) and cowpea (V. unguiculata [L.] Walpers) using RFLP mapping data. Theor. Appl. Genet. 86, 797–810. [DOI] [PubMed] [Google Scholar]
- Paterson, A.H., Bowers, J.E., Burow, M.D., Draye, X., Elsik, C.G., Jiang, C.-X., Katsar, C.S., Lan, T.-H., Lin, Y.-R., Ming, R., and Wright, R.J. (2000). Comparative genomics of plant chromosomes. Plant Cell 12, 1523–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi, E. (1998). A bonanza for plant genomics. Science 282, 652–654. [DOI] [PubMed] [Google Scholar]
- Swofford, D.L. (1999). PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0. (Sunderland, MA: Sinauer Associates).
- Tanksley, S.D., et al. (1992). High density molecular linkage maps of the tomato and potato genomes. Genetics 132, 1141–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov, R.L., Galperin, M.Y., Natale, D.A., and Koonin, E.V. (2000). The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28, 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov, R.L., Koonin, E.V., and Lipman, D.L. (1997). A genomic perspective on protein families. Science 278, 631–636. [DOI] [PubMed] [Google Scholar]
- Van der Hoeven, R., Ronning, C., and Tanksley, S.D. (2002). Deductions about the number, organization, and evolution of genes in the tomato genome based on analysis of a large expressed sequence tag collection and selective genomic sequencing. Plant Cell 14, 1441–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vision, T., Brown, D., and Tanksley, S.D. (2000). The origins of genome duplications in Arabidopsis. Science 290, 2114–2117. [DOI] [PubMed] [Google Scholar]
- Wilson, A., et al. (1999). Inferences on the genome structure of progenitor maize through comparative analysis of rice, maize and the domesticated panicoids. Genetics 153, 453–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y.W., Lai, K.N., Tai, P.Y., and Li, W.H. (1999). Rates of nucleotide substitution in angiosperm mitochondrial DNA sequences and dates of divergence between Brassica and other lineages. J. Mol. Evol. 48, 597–604. [DOI] [PubMed] [Google Scholar]