Abstract
We explored genomic expression patterns in the yeast Saccharomyces cerevisiae responding to diverse environmental transitions. DNA microarrays were used to measure changes in transcript levels over time for almost every yeast gene, as cells responded to temperature shocks, hydrogen peroxide, the superoxide-generating drug menadione, the sulfhydryl-oxidizing agent diamide, the disulfide-reducing agent dithiothreitol, hyper- and hypo-osmotic shock, amino acid starvation, nitrogen source depletion, and progression into stationary phase. A large set of genes (∼ 900) showed a similar drastic response to almost all of these environmental changes. Additional features of the genomic responses were specialized for specific conditions. Promoter analysis and subsequent characterization of the responses of mutant strains implicated the transcription factors Yap1p, as well as Msn2p and Msn4p, in mediating specific features of the transcriptional response, while the identification of novel sequence elements provided clues to novel regulators. Physiological themes in the genomic responses to specific environmental stresses provided insights into the effects of those stresses on the cell.
INTRODUCTION
Cellular organisms require specific internal conditions for optimal growth and function. Myriad strategies have evolved to maintain these internal conditions in the face of variable and often harsh external environments. Whereas multicellular organisms can use specialized organs and tissues to provide a relatively stable and homogenous internal environment, unicellular organisms such as the yeast Saccharomyces cerevisiae have evolved autonomous mechanisms for adapting to drastic environmental changes. Yeasts regularly withstand fluctuations in the types and quantities of available nutrients, temperature, osmolarity and acidity of their environment, and the variable presence of noxious agents such as radiation and toxic chemicals. The genomic expression program required for maintenance of the optimal internal milieu in one environment may be far from optimal in a different environment. Thus, when environmental conditions change abruptly, the cell must rapidly adjust its genomic expression program to adapt to the new conditions.
The complexity of the yeast cell's system for detecting and responding to environmental variation is only beginning to emerge. Genes whose transcription is responsive to a variety of stresses have been implicated in a general yeast response to stress (Mager and De Kruijff, 1995; Ruis and Schuller, 1995). Other gene expression responses appear to be specific to particular environmental conditions. Several regulatory systems have been implicated in modulating these responses, but the complete network of regulators of stress responses and the details of their actions, including the signals that activate them and the downstream targets they regulate, remain to be elucidated.
We used DNA microarrays to analyze changes in transcript abundance in yeast cells responding to a panel of diverse environmental stresses. Our analysis of this large body of gene expression data allowed us to define stereotyped patterns of gene expression during the adaptation to stressful environments, and to compare and contrast the gene expression responses to different stresses. Here, we present three key results. First, we describe the global expression programs in response to a diverse set of stresses, including their specific features and a common response to all of the stressful conditions, termed the “environmental stress response” (ESR). Second, several sets of coregulated genes share promoter elements, which point to the involvement of specific transcription factors in the regulation of those genes. The roles of the transcription factor Yap1p and the related factors Msn2p and Msn4p are examined by analyzing the expression responses of strains deleted for or overproducing these factors. Third, we interpret the responses of genes with known functions to gain insights into the physiological effects of each of the stresses as well as the mechanisms that yeast cells use to cope with these stresses. The complete data set, as well as supplemental materials, is available at http://www-genome.stanford.edu/yeast_stress.
MATERIALS AND METHODS
(Additional details, including descriptions of duplicated experiments and appropriate reference citations, can be found on the web supplement, at the address given above.)
Strains and Growth Conditions
The strains used in this study are listed in Table 1. Unless otherwise noted, cells were grown in rich medium (YPD) (Sherman, 1991) at 30°C and shaken at 250–300 rpm.
Table 1.
Strains used in this study
| Name | Genotype | Source |
|---|---|---|
| DBY7286 | MATaura3-52 GAL2 | Ferea et al., 1999 |
| DBY8768 | ura3-52/ura3-52 GAL2/GAL2 | Ferea et al., 1999 |
| DBY9434 | MATa ura3-52 yap1::KANMX4 GAL2 | This study |
| DBY9435 | MATa msn2::KANMX4 msn4::URA3 GAL2 | DeRisi |
| DBY9439 | DBY7286 harboring pRS416 | Tae Bum Shin |
| DBY9440 | DBY7286 harboring pTS1 | Tae Bum Shin |
| DBY9441 | DBY7286 harboring pTS2 | Tae Bum Shin |
Sample Collection, Cell Lysis, and RNA Isolation
In most cases, cells were grown to early log phase (OD600 0.2 to 0.4), and an aliquot of cells was collected to serve as the time-zero reference. Cells were collected by centrifugation at 3000 ×g for 3 to 7 min at room temperature. Each 50-ml cell pellet was resuspended in 3 to 10 ml of lysis buffer (10 mM Tris-Cl pH 7.4, 10 mM EDTA, 0.5% SDS), and stored at −80°C until RNA preparation. Total RNA was collected by acid lysis similar to that previously described (Spellman et al., 1998; see web supplement). Where indicated, mRNA was purified using oligo-dT cellulose (Ambion, Austin, TX), precipitated and resuspended in Tris-EDTA (TE) at a final concentration of ∼ 0.5–1 μg/μL.
Probe Preparation, Microarray Hybridization, and Data Acquisition
Probe preparation and microarray construction and analysis were performed as previously described (Shalon et al., 1996; DeRisi et al., 1997; Spellman et al., 1998; see web supplement). Arrays were scanned using a commercially available scanning laser microscope (GenePix 4000) from Axon Instruments (Foster City, CA). Full details on using the GenePix 4000 can be obtained from Axon. All arrays were analyzed using the program ScanAlyze (available from http://rana.stanford.edu/), as described in the manual.
Heat Shock from 25°C to 37°C
Cells grown continuously at 25°C were collected by centrifugation, resuspended in an equal volume of 37°C medium, and returned to 37°C for growth. Samples were collected at 5, 15, 30, and 60 min. For array analysis, each Cy5-labeled sample was compared with a Cy3-labeled reference pool, consisting of an equal mass of all of the RNA samples. Following data acquisition and clustering analysis, the data were mathematically “zero transformed” for visualization by dividing the expression ratios for each gene measured on a given array by the corresponding ratios measured for the unshocked, time-zero cells. Therefore, in all figures, the ratios represent the expression level at each time point relative to the expression level in the unshocked, time-zero sample.
Heat Shock from Various Temperatures to 37°C and Steady-State Temperature Growth
Six cultures were grown continuously at 17°, 21°, 25°, 29°, 33°, or 37°C for ∼20 h. Half of each culture was collected to serve as the unstressed reference, and the remainder of each culture was collected by centrifugation and immediately resuspended in 37°C medium. After 20 min at 37°C, the cells were harvested, and total RNA was isolated.
To measure steady-state expression at each temperature, RNA collected from cells grown continuously at each temperature was also compared directly to RNA from cells grown at 33°C.
Temperature Shift from 37°C to 25°C
Cells grown at 37°C for ∼20 h were collected by centrifugation, resuspended in two volumes of 25°C medium, and returned to 25°C for growth. Samples were collected at 5, 15, 30, 45, 60, and 90 min, and total RNA was collected. Gene expression in cells growing continuously at 37°C was also compared directly to expression in cells growing at 25°C.
Mild Heat Shock at Variable Osmolarity
To compare the effects of mild heat shock at different osmolarities, three experiments were performed. In the first, a YPD culture of DBY7286 was grown at 29°C to OD600 0.3. Cells were collected by centrifugation, the culture was resuspended in 33°C medium, and samples were collected at 5, 15, 30 min after resuspention. A second time series was performed nearly identically, except that cells were grown in YPD supplemented with 1 M sorbitol throughout the experiment. In the third experiment, cells growing in YPD with 1 M sorbitol at 29°C were collected and resuspended in YPD without sorbitol at 33°C, and serial samples were collected. Total RNA was isolated for array analysis.
Response of Mutant Cells to Heat Shock
Wild-type and mutant strains were exposed to heat shock in triplicate experiments. Wild-type, yap1, and msn2 msn4 cultures, grown at 30°C, were collected and resuspended in an equal volume of medium preheated to 37°C. After 20 min at 37°C the cells were collected, and total RNA was isolated.
Hydrogen Peroxide Treatment
Cells were grown to early-log phase at which point H2O2 (Sigma, St. Louis, MO) was added for a final concentration of 0.30 mM. Samples were collected at 10, 20, 30, 40, 50, 60, 80, 100, and 120 min. The culture volume and the concentration of H2O2 were maintained throughout the experiment. The H2O2 concentration was monitored every 3 min using a horseradish-peroxidase based assay (Green and Hill, 1984), which showed that the concentration of H2O2 was maintained at 0.32 +/−0.03 mM H2O2 over the course of the experiment (data not shown).
Response of Mutant Cells to H2O2 Exposure
Wild-type, yap1, and msn2 msn4 cultures were exposed to 0.3 mM H2O2 in duplicate experiments. A single dose of H2O2 was added to 0.3 mM of each culture, and after 20 min, the cultures were collected, and mRNA was isolated.
Menadione Exposure
Menadione bisulfite (Sigma) was suspended immediately before use in water at a concentration of 1 M and was filter-sterilized. Menadione bisulfite was added to the culture at a concentration of 1 mM, samples were removed at 10, 20, 30, 40, 50, 60, 80, 105, and 120 min, and mRNA was isolated.
Diamide Treatment
1.5 mM diamide (Sigma) was added to the culture, and samples were recovered at 5, 10, 20, 30, 40, 60, 90 min. Polyadenylated RNA was isolated for array analysis.
DTT Exposure
Cells were grown at 25°C and dithiothrietol (DTT) (Boeringer Manheim, Indianapolis, IN) was added for a final concentration of 2.5 mM. Samples were removed at 15, 30, 60, 120, 240, 480 min. Total RNA recovered from each time point, as well as the unstressed sample, was labeled with Cy5-dUTP and compared with a reference pool, consisting of equal mass of total RNA from each sample that was labeled with Cy3-dUTP. The array data were “zero-transformed” subsequent to clustering analysis.
Hyper-osmotic Shock
A YPD culture was inoculated and grown to OD600 0.6. One volume of 30°C YPD supplemented with 2 M sorbitol was added to the culture for a final concentration of 1 M sorbitol. Samples were collected at 5, 15, 30, 45, 60, 90, and 120 min, and mRNA was isolated.
Hypo-osmotic Shock
Cells were grown for ∼20 h in YPD supplemented with 1 M sorbitol. The cells were grown to OD600 0.15, collected by centrifugation, and resuspended in YPD without sorbitol. Samples were collected at 5, 15, 30, 60 min, and total RNA was isolated for array analysis.
Amino Acid Starvation
Cells were grown in complete minimal medium (SCD) to early-log phase. Cells were collected by centrifugation and resuspended in an equal volume of minimal medium lacking amino acids and adenine (YNB−AA, 2% glucose, 20 mg/L uracil) and allowed to grow. Samples were then harvested after 0.5 h, 1 h, 2 h, 4 h, and 6 h, and total RNA was collected.
Nitrogen Depletion
Cells were grown in SCD medium, collected by centrifugation and resuspended in an equal volume of minimal medium without amino acids or adenine and with limiting concentrations of ammonium sulfate (YNB−AA−AS, 2% glucose, 20 mg/L uracil, 0.025% ammonium sulfate) and returned to the 30°C shaker. Samples were subsequently harvested after 0.5 h, 1 h, 2 h, 4 h, 8 h, 12 h, 1 d, 2 d, 3 d, and 5 d of culture incubation, and mRNA was isolated.
Stationary Phase
A YPD culture was grown to OD600 0.3, at which point a sample was collected to serve as the time-zero reference. Samples were recovered at 2 h, 4 h, 6 h, 8 h, 10 h, 12 h, 1 d, 2 d, 3 d, and 5 d of culture incubation. Total RNA was isolated for array analysis.
Steady-state Growth on Alternative Carbon Sources
Cells were grown continuously in YP media supplemented with 2% weight to volume of glucose, galactose, raffinose, fructose, sucrose, or ethanol as a carbon source. Total RNA harvested from each of the samples was labeled with Cy5-dUTP and compared with a reference pool, consisting of an equal mass of RNA from each sample that was labeled with Cy3-dUTP. The data were mathematically transformed subsequent to clustering analysis by dividing the expression ratios for each gene measured on a given array by the corresponding ratios measured for the cells grown in glucose.
Overexpression Studies
Overexpression constructs pRS-MSN2 and pRS-MSN4, as well as the parent vector pRS416 (Mumberg et al., 1994), were received from Tae Bum Shin (postdoctoral fellow in Brown lab). Wild-type DBY7286 cells harboring each plasmid were grown in SCD medium supplemented with 2% galactose for ∼ 6 h. Total RNA collected from cells harboring pTS1 (MSN2 vector) and pTS2 (MSN4 vector) was compared directly to RNA collected from cells containing pRS416.
Hierarchical Clustering
Hierarchical clustering of the data was performed as previously described (Eisen et al., 1998) using the program Cluster (available at http://rana.stanford.edu). Data from 142 microarray analyses of RNA samples isolated from wild-type cells under various conditions were clustered, along with previously-published data (DeRisi et al., 1997). The cluster analysis was performed without the “time-zero” mathematical transformation of the data from experiments in which a reference pool was used. The data from each array experiment were weighted by the program Cluster (available at http://rana.stanford.edu/software) according to the overall similarity of each array to others in the data set, which served to under-weight arrays that were highly similar. The resulting cluster was visualized using the program TreeView (available at http://rana.stanford.edu/software/).
Promoter Analysis
For coregulated genes, either 600 bp or 1000 bp, as indicated, upstream of each gene start site was recovered using Yeast Tools (http://copan.cifn.unam.mx/∼jvanheld/rsa-tools). Sequence motifs common to the upstream sequences were identified by the MEME algorithm (http://www.sdsc.edu/MEME/meme/website/meme.html [Bailey and Elkan, 1994]). Upstream sequences were searched for specific sequence motifs using Yeast Tools.
RESULTS
Overview
We characterized genomic expression programs in yeast responding to environmental changes in three ways. First, we characterized the temporal program of gene expression in the response of cells to heat shock, hydrogen peroxide, superoxide generated by menadione, a sulfhydryl oxidizing agent (diamide), and a disulfide reducing agent (dithiothreitol), hyper-osmotic shock, amino acid starvation, nitrogen source depletion, and progression into stationary phase. The severity of each condition was calibrated to preserve more than 80% cell viability, so that we could observe the expression programs in viable cells adapting successfully to a changing environment. For most of the environmental changes we studied, samples were collected over the course of 2–3 h; in our investigation of the responses to nitrogen depletion and stationary phase, samples were collected over a period of 5 d. Second, we examined the dose response to heat shock in a series of experiments in which cells were subjected to temperature shifts of variable magnitude. Third, we compared the genomic expression programs in cells already adapted to steady-state growth at different temperatures and on alternative carbon sources.
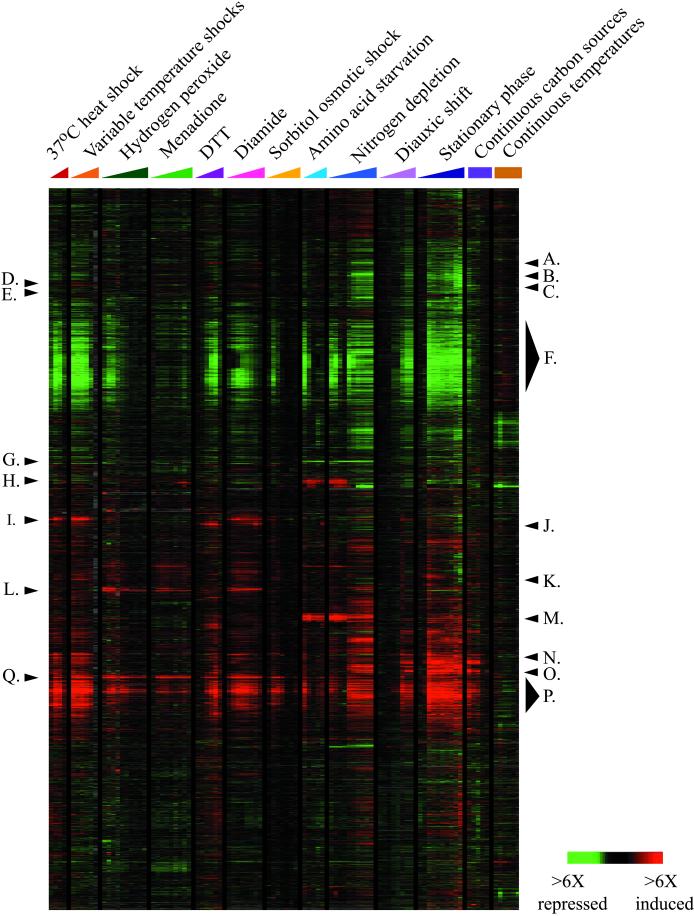
In our initial experiments, 142 different mRNA samples were analyzed by whole-genome microarray hybridization. Each microarray used in this study contained ∼ 6,200 known or predicted yeast genes that had been identified at the time of our analysis (Ball et al., 2000). The resulting table of ∼ 9 × 105 quantitative measurements of transcript levels was organized by hierarchical clustering and displayed as previously described (Eisen et al., 1998) (Figure 1). Briefly, the clustering algorithm arranges genes according to their similarity in expression profiles across all of the array experiments, such that genes with similar expression patterns are clustered together. The data are graphically displayed in tabular format in which each row of colored boxes represents the variation in transcript abundance for each gene, and each column represents the variation in transcript levels of every gene in a given mRNA sample, as detected on one array. The variations in transcript abundance for each gene are depicted by means of a color scale, in which shades of red represent increases and shades of green represent decreases in mRNA levels, relative to the unstressed culture, and the saturation of the color corresponds to the magnitude of the differences. A black color indicates an undetectable change in transcript level, and a gray color represents missing data. A dendrogram constructed during the clustering process depicts the relationships between genes: the branch lengths represent the degree of similarity between genes based on their expression profiles. Genes that display similar patterns of gene expression over multiple experiments are thus grouped together on a common branch of the dendrogram and can also be recognized by an obvious pattern of contiguous patches of color in the cluster diagram.
Figure 1.
Genomic expression programs in response to environmental changes. The entire set of yeast genes identified at the time of our analysis (∼6,200) was clustered based on their expression patterns in 142 array experiments that followed wild-type yeast responding to environmental changes. Here, data from 94 arrays are shown, omitting duplicate experiments that were considered in the clustering analysis (see web supplemental MATERIALS AND METHODS for details). Experiments are labeled according to the color key depicted: time course experiments are indicated with colored triangles, while steady-state experiments are labeled with colored squares. Individual clusters of coregulated genes that are discussed in the text and web supplements are labeled as follows: A. WSC2 cluster, B. CIS3 cluster, C. LHS1 cluster, D. Glycolysis cluster, E. SSA2 cluster, F. Repressed ESR cluster, G. Amino acid transporters, H. Amino acid and purine genes, I. Chaperone cluster, J. KAR2 cluster, K. Proteasome and Endocytosis cluster L. TRR1 and TRR2 clusters, M. DAL cluster, N. Oxidative phosphorylation cluster, O. Glyoxylate and TCA cycle cluster, P. Induced ESR cluster, Q. TRX2 cluster.
Several general features in the global expression pattern can be recognized from the results of hierarchical clustering (Eisen et al., 1998). First, genes that are coregulated under the conditions examined will correspondingly cluster together, and analysis of their promoters often identifies common sequence motifs, in some cases suggesting regulation by known transcription factors and in others identifying novel promoter elements (Eisen, Derisi, Brown - personal communication, and unpublished data). Second, because genes involved in the same cellular processes are usually similarly expressed, the functions of characterized genes in a given cluster can suggest hypothetical functions for uncharacterized genes in the same cluster. Third, the choreography of expression of the various gene clusters can be related to the series of events occurring during each experiment, suggesting links between specific sets of genes and specific features of the experimental conditions. Finally, in many cases, a physiological picture of the cellular response can be sketched by considering the expression of genes of known function and regulation, in turn suggesting specific effects of each condition on the cell.
An overview of the microarray results is presented in Figure 1. The large-scale features of the expression programs visible in this display vividly illustrate the massive and rapid genome-wide changes in gene expression in response to each environmental shift. Some sets of genes responded in a stereotypical manner to many different environmental changes, whereas the response of other sets of genes was unique to specific conditions. Although there were shared features between the responses to different conditions, no two expression programs were identical in terms of the genes affected, the magnitude of expression alteration, and the choreography of expression. The uniqueness of each program highlights the precision with which yeast respond to changes in their environment.
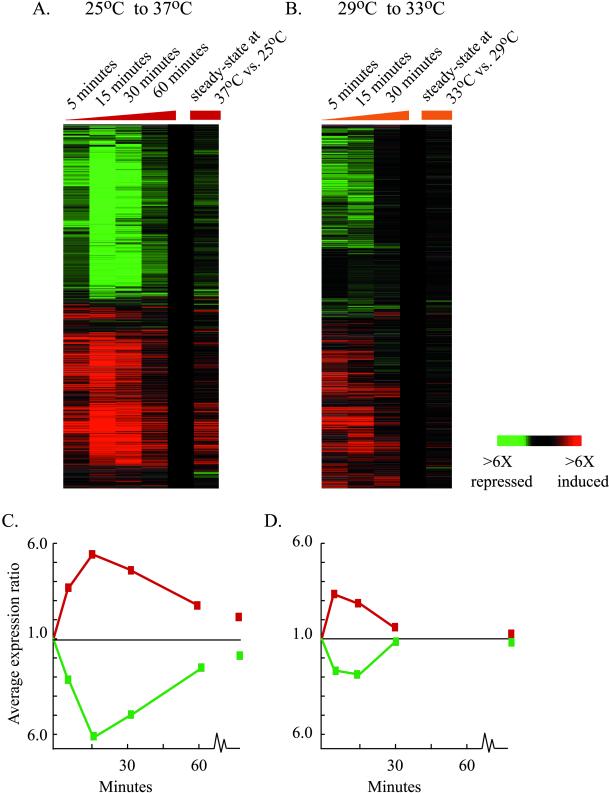
One of the remarkable features of the genomic expression programs shown in Figure 1 is that, with the exception of adaptation to starvation conditions, the global changes in transcript abundance were largely transient (Figure 2, A and C). Immediately after most of the environmental shifts, the cells responded with large changes in the transcript levels of hundreds of genes. However, genomic expression adapted over time to new steady-state transcript levels, with far smaller differences in transcript abundance between the steady-state programs at each condition. The duration and amplitude of the transient changes in transcript levels varied with the magnitude of the environmental change. Furthermore, the magnitude of differences in the corresponding steady-state gene expression programs also correlated with the magnitude of environmental shift. This trend was evident in a series of experiments in which cells subjected to temperature shifts of varying magnitude responded with correspondingly graded transcriptional changes (Figure 2). Cells subjected to a larger shift in temperature responded with larger and more prolonged alterations in gene expression before adapting to their new steady-state expression levels, relative to cells exposed to smaller temperature changes.
Figure 2.
Transient changes in genomic expression following environmental change. (A-B) The expression of ∼ 1000 genes that changed by a factor of at least twofold in response to heat shock is shown as the cells responded to (A) 25°C to 37°C heat shock and (B) 29°C to 33°C heat shock. (C-D) The average expression changes of genes displayed in (A) and (B), respectively, are shown. The average expression of genes induced in each response is depicted by a red curve, while the average expression of genes repressed in each response is depicted by a green curve.
The Environmental Stress Response
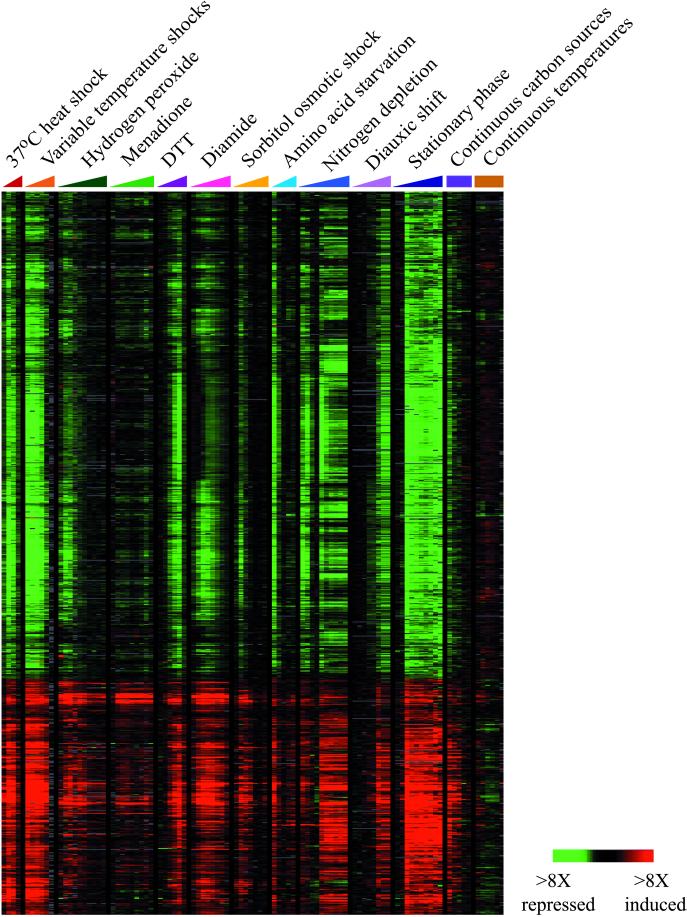
A striking feature of the expression programs displayed in Figure 1 is the large fraction of the genome that responded in a stereotypical manner to each of the stressful conditions we tested. Two large clusters of genes, one consisting of repressed genes and one consisting of induced genes, displayed reciprocal but otherwise nearly-identical temporal profiles (Figure 3). These clusters amounted to ∼ 900 genes, more than 14% of the currently-predicted genes in the yeast genome (Ball et al., 2000). This stereotypical response shared features with the previously-recognized general response to stress, which typically refers to the response of a set of ∼ 50 genes induced by a variety of stresses through the stress response element (STRE) promoter sequence, recognized by the transcription factors Msn2p and Msn4p (Kobayashi and McEntee, 1993; Marchler et al., 1993; Martinez-Pastor et al., 1996). Our results reveal that, although genes in this large program showed a similar response to the conditions tested here, the regulation of their expression is not general, but is instead dependent on many different signaling systems that act in a condition-specific and gene-specific manner (see below). Therefore, while it is important to recognize the similarities between this program and the previously-described general stress response, to avoid confusion we refer to the stereotyped response of this entire set of induced and repressed genes as the environmental stress response (ESR).
Figure 3.
Overview of the Environmental Stress Response (ESR). Genes that participate in the ESR were chosen based strictly on the genomic cluster analysis shown in Figure 1. Two extended clusters of genes, one corresponding to repressed genes and one corresponding to induced genes, displayed nearly identical but opposite patterns of gene expression in response to environmental stress. Color scale and array identification are indicated.
Genes Repressed in the ESR
Within the large cluster of ∼ 600 genes that were repressed in the ESR, two clusters with distinct expression profiles are evident (see web supplement for details). The first cluster consists of genes involved in growth-related processes, various aspects of RNA metabolism (such as RNA processing and splicing, translation initiation and elongation, tRNA synthesis and processing), nucleotide biosynthesis, secretion, and other metabolic processes. These genes appeared to be coregulated, and promoter analysis revealed the presence of two novel and conserved motifs in the upstream elements of these genes (see web supplement for details), one of which was similar to a site identified in the promoters of RNA processing genes by Hughes et al. (2000). The second cluster is distinguished from the first by a slight delay in the decline in transcript levels, and it consists almost entirely of genes encoding ribosomal proteins. The repression of ribosomal protein genes has previously been observed during multiple stress responses (Warner, 1999) and is known to be regulated by the transcription factor Rap1p (Moehle and Hinnebusch, 1991; Li et al., 1999). Our results show that the repression of the ribosomal genes, along with the large set of genes involved in RNA metabolism, protein synthesis, and aspects of cell growth, is a general feature of the ESR.
Genes Induced in the ESR
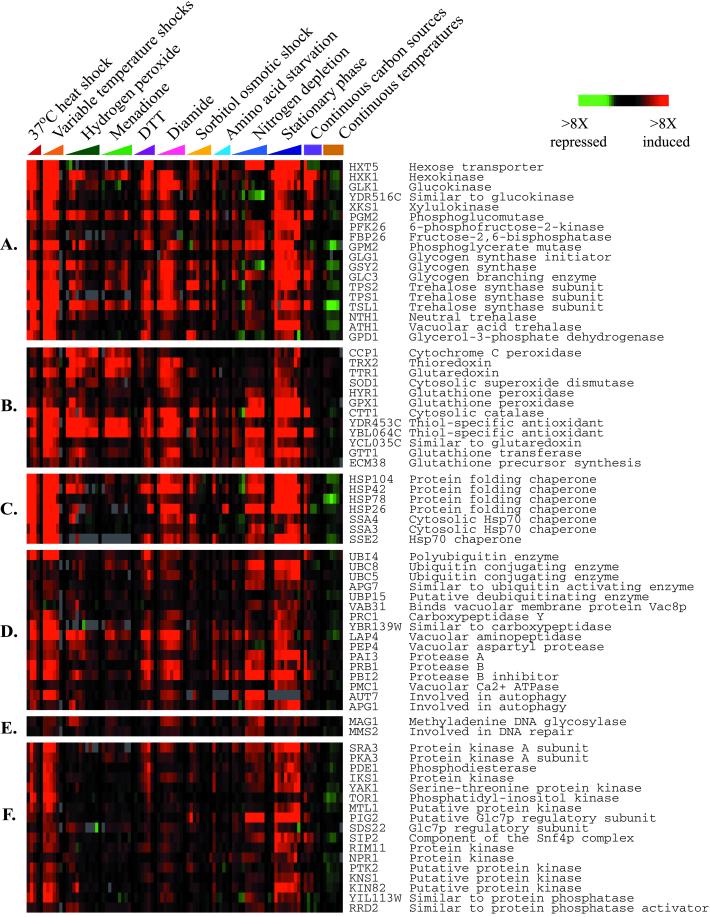
Approximately 300 genes, of which nearly 60% are completely uncharacterized, were induced in the ESR (see web supplement for details). The functional themes represented by these genes are likely to provide many clues to the ways cells fortify themselves for survival in inhospitable environments. The genes in this group with known molecular functions are involved in a wide variety of processes, including carbohydrate metabolism, detoxification of reactive oxygen species, cellular redox reactions, cell wall modification, protein folding and degradation, DNA damage repair, fatty acid metabolism, metabolite transport, vacuolar and mitochondrial functions, autophagy, and intracellular signaling (Figure 4). Many of the genes induced in the ESR have previously been proposed to offer cellular protection during stressful conditions, such as oxidative stress, heat shock, osmotic shock, and starvation (Hohmann and Mager, 1997; Mager and De Kruijff, 1995). More than half of the 50 genes that were previously reported to be STRE-regulated (see Moskvina et al., 1998 and Yeast Protein Database for references) were induced in the ESR.
Figure 4.
Characterized genes induced in the ESR. Characterized genes that are induced in the ESR are displayed according to their involvement in (A) carbohydrate metabolism, (B) cellular redox reactions and defense against reactive oxygen species, (C) protein folding, (D) protein degradation and vacuolar functions, (E) DNA damage repair, and (F) intracellular signaling. Additional functional categories can be viewed on the web supplement.
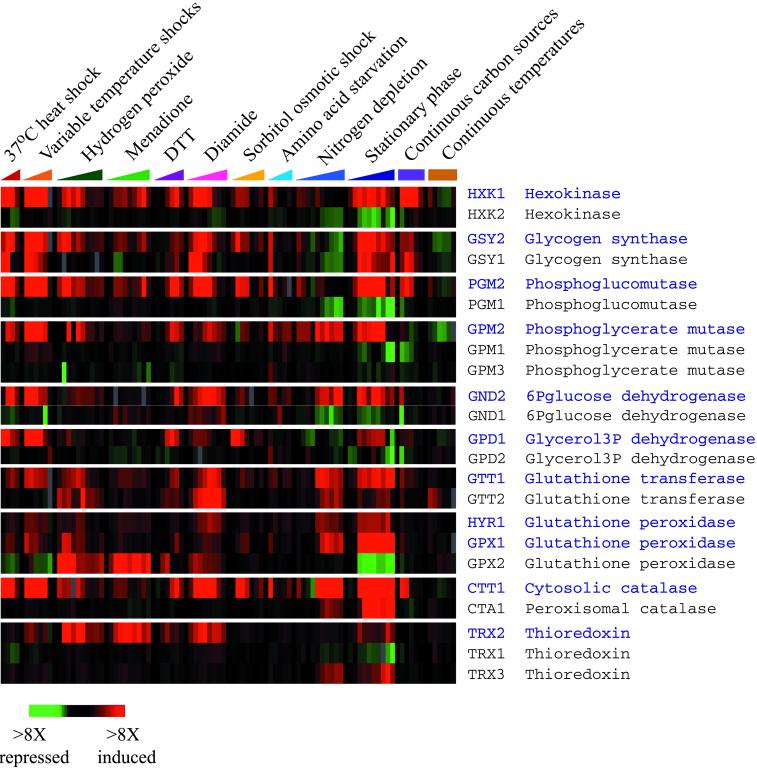
One notable feature of the ESR was the differential expression of isozymes. For example, various enzymes involved in carbon metabolism, protein folding, and defense against reactive oxygen species were specifically induced in the ESR, while their counterparts were not (Figure 5). This result confirms and expands previous observations that isozymes involved in carbohydrate metabolism are differentially expressed in response to osmotic shock (Norbeck and Blomberg, 1997; Rep et al., 2000). One possible explanation for this divergence in regulation is that the putative isozymes might possess different properties, including biochemical function, substrate specificity, and physical location, that make one isozyme optimized to the ESR and the other to more specific conditions. Alternatively, the existence of differentially-regulated isozymes possessing similar properties might allow ESR isozymes to be regulated as part of this more general program, while their related counterparts are regulated by specialized signals.
Figure 5.
Differentially regulated isozymes in the ESR. The expression of genes encoding differentially regulated isozymes is shown. Isozymes regulated in the ESR are labeled in purple. Color scale and array identification are shown.
Among the genes induced in the ESR were many whose products play reciprocal metabolic roles. An example is provided by genes involved in the metabolism of trehalose and glycogen, whose roles in stress responses have been linked to storage of energy reserves, protein stabilization, and osmolyte balance (Hounsa et al., 1998; Singer and Lindquist, 1998). Genes encoding enzymes that synthesize trehalose, glycogen, and their precursors, as well as genes that encode catabolic enzymes for degrading these carbohydrates, are jointly induced in the ESR (Figure 4A), consistent with the previously-observed induction of many of these genes in response to numerous stresses (Parrou et al., 1999; Parrou et al., 1997; Rep et al., 2000; Zahringer et al., 1997). The simultaneous induction of both synthetic and catabolic enzymes seems paradoxical. However the activity of many of these enzymes is sensitively controlled at the posttranslational level (Hwang et al., 1989; Marchase et al., 1993; Dey et al., 1994; Huang et al., 1998; Parrou et al., 1999). We propose that the coinduction of these genes renders the cell poised to rapidly and sensitively modulate the activity of the corresponding enzymes and thereby increases the cell's capacity for regulated flux of carbohydrates into and out of its glycogen and trehalose stores. Induction of these genes presumably enhances the cell's ability to rapidly buffer and manage osmotic instability and energy reserves.
The induction of genes encoding reciprocally-related functions is also evident in regulatory networks, including those that may be involved in regulation of the ESR itself. Components of the PKA pathway, including both positive effectors (SRA3, PKA3) and negative regulators (PDE2, SRA1) of PKA signaling, were coordinately induced in the ESR (Figure 4F). Induction of PKA signaling components in the ESR is particularly noteworthy, as activity of the pathway inhibits Msn2p and Msn4p activity by triggering relocalization of the factors to the cytosol (Gorner et al., 1998; Smith et al., 1998). In fact, induction of the PKA components in the ESR was dependent on Msn2p and Msn4p (see more below). The cell may concomitantly increase protein levels of positive and negative regulators of PKA signaling to allow sensitive posttranslational control of signaling through the pathway. Among the likely effects of this adaptation would be an enhancement of the cell's ability to rapidly and precisely modulate expression of genes in the ESR.
What Triggers the ESR?
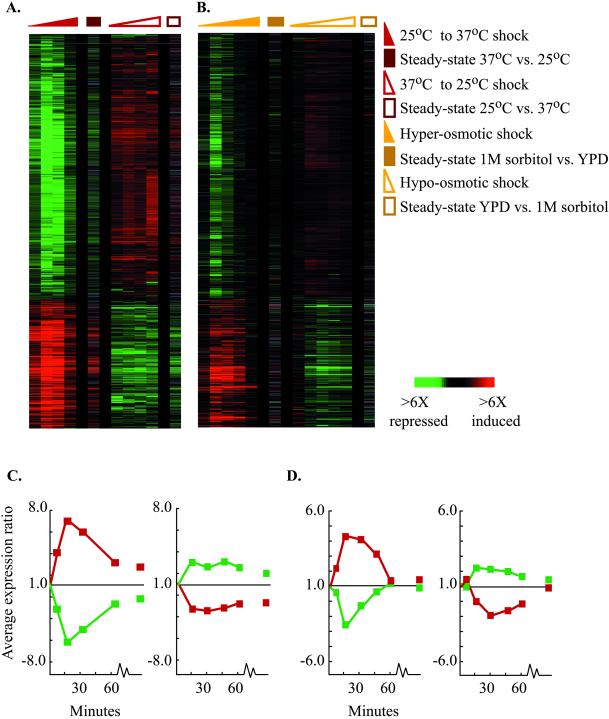
Given the universal induction of the ESR in our initial experiments, we hypothesized that the ESR might be initiated in response to any abrupt change in the cells' environment. According to this hypothesis, the response would be triggered by transferring cells in either direction between two environments. To test this, we examined the pattern of ESR expression following an abrupt shift in temperature from 37°C to 25°C (Figure 6A and 6C). The response to this shift was fundamentally different from the reverse shift from 25°C to 37°C in two ways. First, in response to the 37°C to 25°C temperature shift, the cells responded with reciprocal changes in the expression of ESR genes relative to the response to a 25°C to 37°C heat shock, reflecting the suppression, rather than initiation, of the ESR. Second, unlike the response to a 25°C to 37°C heat shock, which elicited massive and transient changes in ESR expression, the transition from 37°C to 25°C resulted in a simple, rapid transition to the gene expression program characteristic of steady-state growth at 25°C, with essentially no transient features. A similar result was observed when cells were transferred between medium of standard osmolarity and medium supplemented with 1 M sorbitol: when cells were transferred to hyperosmolar medium, they initiated the ESR with transient changes in expression, whereas when cells adapted to growth in 1 M sorbitol were transferred to medium of standard osmolarity, they suppressed the ESR with only subtle transient features (Figure 6, B and D). These results reveal that the ESR is not initiated in response to all environmental changes and that the large, transient changes in expression that are characteristic of the ESR are only seen when this response is initiated and not in the reciprocal response to diminished environmental stress.
Figure 6.
Reciprocal expression of the ESR following reciprocal environmental changes. (A-B) The expression of ESR genes is shown for (A) cells transferred from 25°C to 37°C (▴) and cells transferred from 37°C to 25°C (▵), and (B) cells exposed to hyper-osmotic shock (▴) and reverse osmotic shock (▵) over the course of 1 h. Steady-state expression at each condition is also shown and is indicated by a closed or open square. The genes represented are the same as those shown in Figure 3. (C-D) The average expression changes of genes displayed in (A) and (B), respectively, over time are graphed. Genes normally induced in the ESR are represented by a red curve, and genes normally repressed in the ESR are represented by a green curve on each plot.
We also characterized the response of cells shifted between two different environments that were equally stressful, in the sense that steady-state expression of genes in the ESR was comparable at each of the conditions. Cells adapted to growth at 29°C in the presence of 1 M sorbitol were shifted to 33°C medium lacking sorbitol, resulting in a mild temperature shift in combination with a shift to lower osmolarity. The resulting expression of ESR genes closely approximated the sum of the individual responses to heat shock and a transition to lower osmolarity, with only a few exceptions (see web for supplemental details). This result suggests that the cell responds independently to the unique features of each of the environmental transitions, i.e. the effects of temperature shift and the consequences of osmolarity change, resulting in additive effects on gene expression.
Regulation of the ESR
Because the ESR unfolds in a stereotypical manner in response to diverse environmental stresses, it might be supposed that the response is governed by one all-purpose regulatory system. However, several lines of evidence suggest that the ESR is not controlled by a single system but by different regulatory systems evoked under different environmental conditions.
Numerous subclusters of genes within the large cluster of induced ESR genes showed subtly different expression patterns, suggesting differences in the regulation of those genes. For example, genes in the TRX2 cluster were induced in the ESR but were super-induced relative to other ESR genes in response to agents that alter the cellular redox potential. Similarly, a group of protein folding chaperones induced in the ESR were super-induced in response to heat shock, relative to other stresses (Figure 4C). These results suggest that subsets of genes within the ESR are governed by condition-specific regulatory mechanisms.
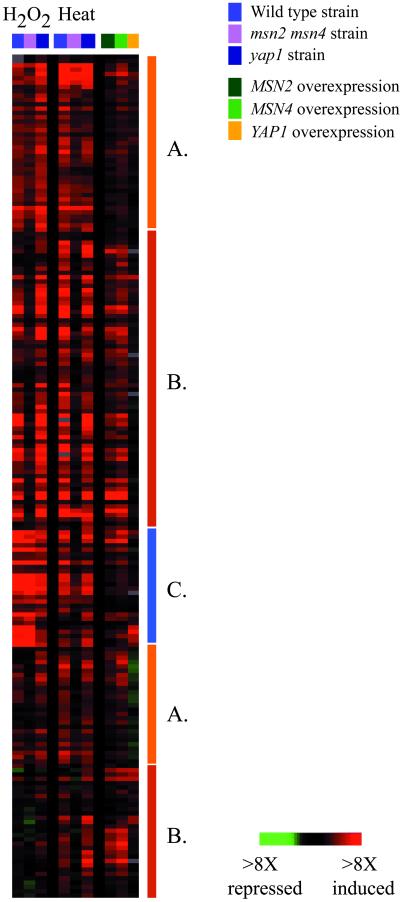
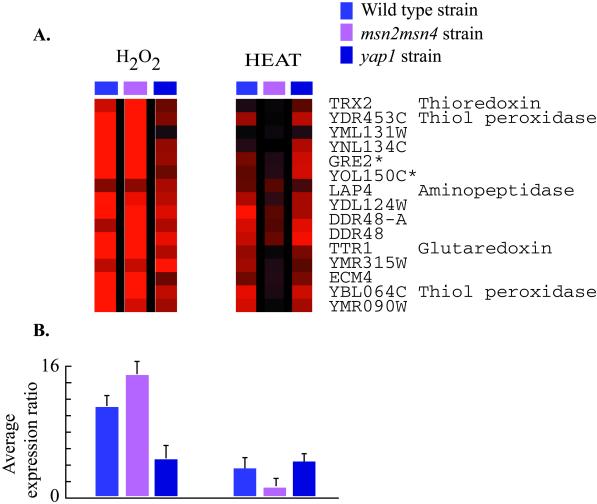
The expression of some of the genes induced in the ESR has previously been shown to be governed by Msn2p and/or Msn4p (Msn2/Msn4p) in response to stressful conditions (see Moskvina et al., 1998 and Yeast Protein Database for review and references). We characterized genomic expression in cells lacking these factors, identifying additional targets of Msn2/Msn4p and providing evidence for alternative regulators of ESR gene expression. The expression of roughly 180 genes was affected in an msn2 msn4 double deletion strain responding to heat shock or H2O2 treatment, relative to the isogenic wild-type, and the genes affected varied in their dependence on Msn2/Msn4p (Figure 7). One large class of genes, including previously known targets such as CTT1, HSP12, and carbohydrate metabolism genes (Martinez-Pastor et al., 1996; Schmitt et al., 1996; Boy-Marcotte et al., 1998; Moskvina et al., 1998), was largely dependent on Msn2/Msn4p in response to both heat shock and H2O2, while the induction of a second class of genes was partially dependent on these factors in response to both conditions. A third class of genes was dependent on Msn2/Msn4p in response to one of the two conditions, but not the other. For example, the induction of genes in the TRX2 cluster was dependent on Msn2/Msn4p in response to heat shock, but the expression of these genes in response to H2O2 treatment was unaffected by deletion of the factors (Figure 8). Because the genes in this group contained within their promoters the consensus binding site for the transcription factor Yap1p (Fernandes et al., 1997), we reasoned that Yap1p may also play a role in governing their expression. Characterization of gene expression in a yap1 deletion strain revealed that the induction of genes in the TRX2 cluster was dependent on Yap1p in response to H2O2 treatment, but their expression in response to heat shock was unaffected by deletion of YAP1. These data reveal that genes in the TRX2 cluster were induced through Msn2/Msn4p in response to heat shock but were induced through Yap1p in response to H2O2. Thus, genes in the ESR can be regulated by different transcription factors depending on the specific environmental shock. This conclusion confirms and expands that of a recent study by Rep et al. (1999), in which the induction of the ESR genes GPD1, HSP12, and CTT1 was shown to be governed by different transcription factors, namely Msn1p, Msn2p, Msn4p, or Hot1p, depending on the exact environmental conditions (Rep et al., 1999).
Figure 7.
Genes dependent on Msn2/Msn4p. Wild-type, msn2 msn4, and yap1 strains were exposed to H2O2 and heat shock, and genes whose expression was affected in the msn2 msn4 mutant strain are shown. The display represents data averaged from duplicate experiments. The effects on these genes of overexpression of MSN2, MSN4 (this study), and YAP1 (DeRisi et al., 1997) are also shown. Genes fell into three classes based on their expression patterns, marked by bars to the right of the cluster diagram: (A) genes partially dependent on Msn2/Msn4p in response to both stresses; (B) genes largely dependent on Msn2/Msn4p in response to both stresses; and (C) genes dependent on Msn2/4p only in response to heat shock.
Figure 8.
ESR genes dependent on Msn2/Msn4p and Yap1p. (A) Gene expression in the wild-type, msn2 msn4, and yap1 strains was monitored following heat shock and H2O2 treatment as described. The expression ratios for each gene in this diagram represent the average from duplicate array experiments. Transcripts of the GRE2 and YOL150C genes (*) are highly homologous and are likely to cross-hybridize on the microarrays. (B) The bar graphs depict the average change in expression for the genes shown in (A) in each of the strains tested.
More than 90% of the genes whose expression was dependent on Msn2/Msn4p in response to heat shock or H2O2 exposure were also induced by overexpression of MSN2 or MSN4 (Figure 7), but significantly more ESR genes were affected by overexpression of the factors than by their deletion. Approximately 80 additional ESR genes were induced by MSN2 or MSN4 overexpression, as were other genes that do not participate in the ESR, whose expression in response to either heat shock or H2O2 treatment was unaffected by deletion of the factors. Some of these genes may be induced through indirect effects of MSN2 or MSN4 overexpression, but many may be legitimate targets of the factors. That more putative Msn2/4p targets were affected by overexpression of the factors than by their deletion, under the conditions we examined, is consistent with the hypothesis that, in response to stressful environmental changes, the dependence of gene induction on Msn2/4p is condition-specific. Deletion of these factors therefore reveals only the subset of their gene targets whose activation is Msn2/4p-dependent under the specific conditions examined. A substantial fraction of the genes in the ESR were unaffected by overexpression or deletion of MSN2 or MSN4 and did not contain the STRE promoter element recognized by the factors, further implicating additional regulators of ESR expression.
Specific Responses to Specific Environmental Changes
In addition to the common ESR, many of the gene expression responses to different environmental changes were specific to individual conditions. Thus, the global expression response to each of the environmental transitions was unique. The physiological themes represented by the gene expression changes in each global response sketched a picture of the physiological effects of each condition and suggested directions for future investigation of the molecular adaptation to these conditions. We present a brief synopsis of each genomic response, and we encourage readers to visit the companion website to explore the complete data set and view supplemental details.
Heat
Sudden heat shock elicited massive and rapid alterations in genomic expression. The ESR was initiated within minutes of a temperature shift, and numerous specialized responses were also triggered. Most notably, the concurrent induction of protein folding chaperones localized to the cytoplasm, mitochondria, and ER supports the notion that one of the primary effects of heat shock is protein unfolding. In addition, the genomic response to heat shock was strikingly similar to that triggered by stationary phase, including the induction of genes involved in respiration and alternative carbon source utilization. Because extracellular glucose concentrations did not change during the course of the heat shock experiment (data not shown), we propose that chaperone-dependent protein folding in the immediate aftermath of heat shock causes a sudden decrease in cellular ATP concentrations. A shift in the ATP:AMP ratio might then lead to the observed expression alterations in central energy metabolism genes, similar to the response seen in mammalian cells (Hardie and Carling, 1997; Hardie, 1999).
H2O2 and Menadione
The gene expression programs following H2O2 and menadione treatment were largely identical, despite the fact that these agents are thought to generate different reactive oxygen species within the cell. The responses to both agents were characterized by the strong induction of genes known to be involved in the detoxification of both H2O2 and superoxide (such as superoxide dismutases, glutathione peroxidases, and thiol-specific antioxidants), as well as genes involved in oxidative and reductive reactions within the cell (thioredoxin, thioredoxin reductases, glutaredoxin, and glutathione reductase). Many of the genes most strongly induced in response to H2O2 and menadione were dependent on the transcription factor Yap1p for their induction (Schnell et al., 1992; Stephen et al., 1995; Jamieson, 1998) (see web for supplemental details).
DTT
The transcriptional profile of the DTT response was quite distinct from the responses to other stresses, particularly in its temporal pattern. The initial induction response, which occurred within 30 min of DTT exposure, included protein disulfide isomerases and protein folding chaperones localized to the ER and genes implicated in the response to alterations in the cellular redox potential. These observations are consistent with the hypothesis that DTT-dependent reduction inhibits protein folding in the ER, triggering the unfolded protein response (Cox et al., 1993; Jamsa et al., 1994; Travers et al., 2000). Surprisingly, initiation of the ESR did not occur until hours after DTT exposure, suggesting that secondary effects of DTT treatment eventually triggered this response. Indeed, the late induction of genes involved in cell wall synthesis, concomitant with the induction of signaling systems involved in the response to cell wall damage, suggests that the accumulation of cell wall defects ultimately initiated the ESR and that, in response to DTT treatment, ESR expression may be governed by regulatory systems specific to cell wall perturbations. Cell wall defects may result from prolonged impairment of secretion, and they may be exacerbated by direct effects of DTT on cell wall disulfide linkages (Cappellaro et al., 1998).
Diamide
The expression response elicited by the sulfhydryl-oxidant diamide resembled a composite of the responses to heat shock, H2O2 and menadione, and DTT. For example, genes involved in protein folding and respiration were induced by diamide in a manner similar to heat shock. Genes whose products are implicated in the response to altered cellular redox potential and defense against reactive oxygen species were also strongly induced, as they were during H2O2 and menadione treatment. Finally, like DTT treatment, diamide induced many putative cell wall biosynthesis genes, as well as genes involved in protein secretion and processing in the ER. These observations suggest that diamide has pleiotropic effects, including protein unfolding following oxidation of protein sulfhydryl groups, oxidative stress resulting from the sulfhydryl modification, and defects in secretion and, ultimately, cell wall damage due to improper disulfide bond formation in the ER.
Hyperosmotic Shock
The genomic expression response to sorbitol osmotic shock included only a few genes whose expression was specifically affected by this condition, but there were two unique features to this response. First, the global expression response to sorbitol was extremely transient, perhaps indicative of the relatively minor cellular changes required for adaptation to hyperosmolarity. Second, numerous genes that were generally induced in the ESR appeared to be super-induced in response to sorbitol, pointing to systems that were selectively called into play in this response. Among the earliest and strongest responses was the induction of ESR genes involved in the synthesis and regulation of critical internal osmolytes, including glycerol and trehalose. Interestingly, other ESR genes, including oxidoreductases and cytosolic catalase, were superinduced in response to sorbitol, for reasons that are not understood.
Starvation
Carbon and nitrogen starvation elicited dramatic global changes in the gene expression program. A more extensive discussion of genomic responses to starvation will be presented elsewhere (Kao et al., unpublished data). While many of the metabolic changes during starvation have been described previously, thousands of genes that are known to participate in other cellular processes or have completely unknown functions showed significant, and previously unrecognized, expression changes during the response to starvation. Many of the starvation-specific expression alterations may be rationalized by the fact that starvation involves a transition from active growth to growth arrest, in contrast to the response to other stresses in which cells resume growth after adapting to the new conditions. Furthermore, gradual nutrient starvation also involves changes in multiple environmental parameters over time, such as cell density, pH, and the successive depletion of various nutrients, which contribute to the complex temporal pattern of gene expression during starvation (Kao et al., unpublished data).
DISCUSSION
To survive in natural environments, microorganisms must be able to respond swiftly and appropriately to sudden environmental changes, adapting to the unique features of each environment. The genomic expression programs characterized in this study reveal that yeast cells respond to environmental changes by altering the expression of thousands of genes, creating a genomic expression program that is customized for each environment. These genomic programs include features that are specific to each stress, reflecting gene products specifically called into play under those conditions. In addition, a remarkable fraction of the genome responds in a stereotypical manner following environmental stress, as part of a program we refer to as the ESR.
Role of the ESR
The ESR is initiated not only by conditions known to threaten cellular viability (data not shown), but also by small environmental changes that do not detectably impair viability and growth. Nonetheless, the response appears to be specific to transitions to environments less optimal for growth and survival. It is not triggered, for example, when cells adapted to growth at elevated temperatures or hyperosmolarity are suddenly shifted to standard growth conditions. Based on these observations, we propose that the ESR is a general adaptive response to suboptimal environments. We hypothesize that, when a cell is shifted to an environment for which its physiological systems are not optimized, the specific cellular consequences resulting from the shift can lead to a series of secondary instabilities within the cell, potentially threatening many key physiological systems. Thus, the genome has evolved to initiate the ESR to protect and maintain critical features of the yeast cell's internal system in response to diverse signs of potential trouble.
The functions of the characterized genes in the ESR provide clues to cellular features that are protected under stressful conditions. The requirement to conserve energy is likely an important feature of all stress responses, and the ESR presumably aids this effort by rapidly repressing hundreds of genes involved in protein synthesis and cellular growth. The characterized genes induced in the ESR participate in a diverse range of cellular processes, including energy generation and storage, defense against reactive oxygen species, synthesis of internal osmolytes, protein folding and turnover, and DNA repair, and together these may represent physiological systems that must be protected under any circumstance. Indeed, the broad protection of these systems by the ESR probably accounts for the observed cross-resistance to various stresses, in which cells exposed to a low dose of one stress become resistant to an otherwise low dose of a second, unrelated stress (Hohmann and Mager, 1997).
The ESR is a graded response. The magnitude of the changes in gene expression, as well as the duration and amplitude of the transient expression changes seen when the response is initiated, is graded to the severity of the environmental stress (this work and data not shown). This correlation suggests that the ESR responds in proportion to the deviation of key physiological systems from a homeostatic set-point. The signals from different pathways that respond to distinct physiological perturbations appear to be integrated in transducing an overall measure of this deviation from homeostasis. Thus, initiation of the ESR may provide a useful operational definition of suboptimal environments, and expression of the program can therefore serve as a molecular gauge of the level of stress experienced by the cell.
Regulation of the Environmental Stress Response
While the ESR displays stereotypical expression changes under diverse types of environmental shifts, we have shown that its regulation is both gene-specific and condition-specific. The expression of genes in the ESR is regulated by different transcription factors depending on the conditions, and the response is governed by several different upstream signaling pathways. For example, the repression of genes encoding ribosomal proteins, and the induction of some of the genes we find induced in the ESR, have previously been shown to be regulated by the PKA pathway in response to nutritional signals and by the PKC pathway following inhibition of secretion (Klein and Struhl, 1994; Neuman-Silberberg et al., 1995; Nierras and Warner, 1999), suggesting that the PKA pathway may govern the entire ESR in response to nutritional signals, while the PKC pathway plays a key role in ESR initiation when secretion is impaired. The induction of many genes we find induced in the ESR was also shown to be dependent on the high osmolarity glycerol (HOG) pathway in response to osmotic stress (Rep et al., 2000), suggesting the involvement of the HOG pathway in ESR regulation under those conditions. In response to DNA-damaging agents, the ESR is governed by the DNA damage-specific Mec1 pathway; the Mec1 pathway appears to play no role in ESR regulation in response to heat shock, suggesting the specific involvement of this pathway following DNA damage (Gasch, Huang, Botstein, Elledge, Brown; manuscript in preparation). In addition to regulating the ESR, each of these signaling systems has also been implicated in regulating more specialized gene expression responses. Thus, these pathways simultaneously regulate the expression of both the ESR and specialized responses specific to the stimuli that activate the pathways.
Our results suggest that, in response to each environmental change, yeast cells simultaneously yet independently detect many distinct cellular signals and create a genomic expression program that integrates the individual responses to each of these signals. Evidence for composite expression programs is provided by the response of cells subjected to a shift to lower osmolarity in combination with mild temperature shock, which can be closely approximated as the sum of the individual responses. The additive response to multiple signals of physiological stress may allow the cell to customize its response to the specific features of the new environment. An example of such emergent genomic expression programs is provided by the response to diamide, which shares specific features of the responses to several other stresses, and which suggests that the pleiotropic effects of this agent trigger specific responses to misfolded proteins, redox stress, and secretion and cell wall defects.
Accounting for the Large Transient Changes in Genomic Expression following Environmental Changes
Immediately following stressful environmental changes, the cell responds with rapid and dramatic alterations in global gene expression, but as the cell adapts to growth at the new conditions, the gene expression program adjusts to a new steady-state that may be only slightly altered from the program seen before the environmental change. We consider two models for the physiological role of the large, transient changes in gene expression. One possibility is that the gene products affected play important roles mainly during the transient period of adaptation to the new conditions. In this model, transient changes in transcript levels would be accompanied by transient changes in the corresponding protein levels. We favor an alternative model, in which the large, transient changes in transcript levels serve as a loading dose, providing rapid, but relatively small, alterations in the corresponding protein levels to the new steady-state concentrations appropriate to the new environment. After the new optimal protein concentrations are achieved, only subtle differences in transcript levels are required to maintain those subtly-altered protein concentrations. The latter model is supported by the observation that, in response to heat shock, the transient changes in transcripts encoding protein folding chaperones do not lead to transient changes in the corresponding protein levels, but rather result in a steady increase in the levels of chaperones until they reach the appropriate steady-state levels (S. Lindquist, personal communication). Future experiments evaluating the changes in the levels of protein products of genes regulated by environmental stress will further test the validity of this model.
CONCLUSION
The detailed characterization of global expression programs triggered by environmental stress is a first step toward defining the role of each gene and each physiological system in cellular adaptation to environmental change. This study suggests hypotheses for the mechanisms yeast employ to survive environmental stress, and raises many questions regarding the role and regulation of the observed genomic expression responses. How initiation of the ESR contributes to cellular resistance to various stresses is an important question in understanding the role of this program in the yeast life cycle. This work has provided a partial sketch of the complex regulation of this critical physiological program. More complete identification and mapping of the regulatory circuits that govern the ESR and the more specialized genomic responses to stress will help us understand the remarkable ability of yeast and other organisms to recognize and survive stressful and unstable environments.
Online References
The following references are cited in the supplemental material available online at: http://www.genome.stanford.edu/yeast_stress.
Jung, U.S., and Levin, D.E. (1999). Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signaling pathway [In Process Citation]. Mol. Microbiol. 34, 1049–1057.
Kawahara, T., Yanagi, H., Yura, T., and Mori, K. (1997). Endoplasmic reticulum stress-induced mRNA splicing permits synthesis of transcription factor Hac1p/Ern4p that activates the unfolded protein response. Mol. Biol. Cell 8, 1845–1862.
Liu, X.D., Morano, K.A., and Thiele, D.J. (1999). The yeast Hsp110 family member, Sse1, is an Hsp90 cochaperone. J. Biol. Chem. 274, 26654–26660.
Mattison, C.P., Spencer, S.S., Kresge, K.A., Lee, J., and Ota, I.M. (1999). Differential regulation of the cell wall integrity mitogen-activated protein kinase pathway in budding yeast by the protein tyrosine phosphatases Ptp2 and Ptp3. Mol. Cell. Biol. 19, 7651–7660.
Morano, K.A., Santoro, N., Koch, K.A., and Thiele, D.J. (1999). A trans-activation domain in yeast heat shock transcription factor is essential for cell cycle progression during stress. Mol. Cell. Biol. 19, 402–411
Mori, K., Ogawa, N., Kawahara, T., Yanagi, H., and Yura, T. (1998). Palindrome with spacer of one nucleotide is characteristic of the cis- acting unfolded protein response element in Saccharomyces cerevisiae. J Biol Chem. 273, 9912–9920.
Payne, W.E., and Garrels, J.I. (1997). Yeast protein database (YPD): a database for the complete proteome of Saccharomyces cerevisiae. Nucleic. Acids. Res. 25, 57–62.
Schmitt, A.P., and McEntee, K. (1996). Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 93, 5777–5782.
Tamai, K.T., Liu, X., Silar, P., Sosinowski, T., and Thiele, D.J. (1994). Heat shock transcription factor activates yeast metallothionein gene expression in response to heat and glucose starvation via distinct signaling pathways. Mol. Cell. Biol.14, 8155–8165.
Winzeler, E.A., Shoemaker, D.D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J.D., Bussey, H., and et al. (1999). Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906.
ACKNOWLEDGMENTS
We thank Joe DeRisi for the original msn2 msn4 double-deletion strain and Tae Bum Shin for overexpression constructs. Special thanks to Christian Rees, Gavin Sherlock, and especially Ash Alizadeh for invaluable help with construction of the companion website, and Mark Schroeder, Gavin Sherlock, and the curators of Saccharomyces Genome Database (SGD) for computer support. We thank Susan Lindquist, Sean O'Rourke, Ira Herskowitz, Jonathan Warner, Anders Blomberg, Judith Frydman, Jim Garrels, Christoph Schuller, Max Diehn, Ash Alizadeh, Jennifer Boldrick, Oliver Rando, and members of the Brown and Botstein labs for helpful discussions. Much of the analysis presented here was possible due to genome databases, in particular SGD and the Yeast Protein Database (YPD). This work was supported by grants from the National Institutes of Health (HG-00450 and HG-00983) and by the Howard Hughes Medical Institute. P.O.B is an associate investigator of the Howard Hughes Medical Institute.
Abbreviations:
- dithiothrietol (DTT)
environmental stress response (ESR), hydrogen peroxide (H2O2), Msn2p and/or Msn4p (Msn2/Msn4p), stress response element (STRE)
Footnotes
Online version of this article contains data set material, and is available at www.molbiolcell.org.
REFERENCES
- Bailey TL, Elkan C. Conference on Intelligent systems for Molecular Biology. Menlo Park, CA: AAAl Press; 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers; pp. 28–36. [PubMed] [Google Scholar]
- Ball CA, Dolinski K, Dwight SS, Harris MA, Issel-Tarver L, Kasarskis A, Scafe CR, Sherlock G, Binkley G, Jin H, Kaloper M, Orr SD, Schroeder M, Weng S, Zhu Y, Botstein D, Cherry JM. Integrating functional genomic information into the Saccharomyces genome database. Nucleic Acids Res. 2000;28:77–80. doi: 10.1093/nar/28.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boy-Marcotte E, Perrot M, Bussereau F, Boucherie H, Jacquet M. Msn2p and Msn4p control a large number of genes induced at the diauxic transition which are repressed by cyclic AMP in Saccharomyces cerevisiae. J Bacteriol. 1998;180:1044–1052. doi: 10.1128/jb.180.5.1044-1052.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappellaro C, Mrsa V, Tanner W. New potential cell wall glucanases of Saccharomyces cerevisiae and their involvement in mating. J Bacteriol. 1998;180:5030–5037. doi: 10.1128/jb.180.19.5030-5037.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Dey NB, Bounelis P, Fritz TA, Bedwell DM, Marchase RB. The glycosylation of phosphoglucomutase is modulated by carbon source and heat shock in Saccharomyces cerevisiae. J Biol Chem. 1994;269:27143–27148. [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes L, Rodrigues-Pousada C, Struhl K. Yap, a novel family of eight bZIP proteins in Saccharomyces cerevisiae with distinct biological functions. Mol Cell Biol. 1997;17:6982–6993. doi: 10.1128/mcb.17.12.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorner W, Durchschlag E, Martinez-Pastor MT, Estruch F, Ammerer G, Hamilton B, Ruis H, Schuller C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MJ, Hill HA. Chemistry of dioxygen. Meth Enzymol. 1984;105:3–22. doi: 10.1016/s0076-6879(84)05004-7. [DOI] [PubMed] [Google Scholar]
- Hardie DG. Roles of the AMP-activated/SNF1 protein kinase family in the response to cellular stress. Biochem Soc Symp. 1999;64:13–27. [PubMed] [Google Scholar]
- Hardie DG, Carling D. The AMP-activated protein kinase: fuel gauge of the mammalian cell? Eur J Biochem. 1997;246:259–273. doi: 10.1111/j.1432-1033.1997.00259.x. [DOI] [PubMed] [Google Scholar]
- Hohmann S, Mager WH. Yeast stress responses. New York, NY: Chapman & Hall,; 1997. [Google Scholar]
- Hounsa CG, Brandt EV, Thevelein J, Hohmann S, Prior BA. Role of trehalose in survival of Saccharomyces cerevisiae under osmotic stress. Microbiology. 1998;144:671–680. doi: 10.1099/00221287-144-3-671. [DOI] [PubMed] [Google Scholar]
- Huang D, Moffat J, Wilson WA, Moore L, Cheng C, Roach PJ, Andrews B. Cyclin partners determine Pho85 protein kinase substrate specificity in vitro and in vivo: control of glycogen biosynthesis by Pcl8 and Pcl10. Mol Cell Biol. 1998;18:3289–3299. doi: 10.1128/mcb.18.6.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JD, Estep PW, Tavazoie S, Church GM. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J Mol Biol. 2000;296:1205–1214. doi: 10.1006/jmbi.2000.3519. [DOI] [PubMed] [Google Scholar]
- Hwang PK, Tugendreich S, Fletterick RJ. Molecular analysis of GPH1, the gene encoding glycogen phosphorylase in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:1659–1666. doi: 10.1128/mcb.9.4.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson DJ. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast. 1998;14:1511–1527. doi: 10.1002/(SICI)1097-0061(199812)14:16<1511::AID-YEA356>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Jamsa E, Simonen M, Makarow M. Selective retention of secretory proteins in the yeast endoplasmic reticulum by treatment of cells with a reducing agent. Yeast. 1994;10:355–370. doi: 10.1002/yea.320100308. [DOI] [PubMed] [Google Scholar]
- Klein C, Struhl K. Protein kinase A mediates growth-regulated expression of yeast ribosomal protein genes by modulating RAP1 transcriptional activity. Mol Cell Biol. 1994;14:1920–1928. doi: 10.1128/mcb.14.3.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N, McEntee K. Identification of cis and trans components of a novel heat shock stress regulatory pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:248–256. doi: 10.1128/mcb.13.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Nierras CR, Warner JR. Transcriptional elements involved in the repression of ribosomal protein synthesis. Mol Cell Biol. 1999;19:5393–5404. doi: 10.1128/mcb.19.8.5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager WH, De Kruijff AJ. Stress-induced transcriptional activation. Microbiol Rev. 1995;59:506–531. doi: 10.1128/mr.59.3.506-531.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchase RB, Bounelis P, Brumley LM, Dey N, Browne B, Auger D, Fritz TA, Kulesza P, Bedwell DM. Phosphoglucomutase in Saccharomyces cerevisiae is a cytoplasmic glycoprotein and the acceptor for a Glc-phosphotransferase. J Biol Chem. 1993;268:8341–8349. [PubMed] [Google Scholar]
- Marchler G, Schuller C, Adam G, Ruis H. A Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO J. 1993;12:1997–2003. doi: 10.1002/j.1460-2075.1993.tb05849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Pastor MT, Marchler G, Schuller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (S.T.R.E) EMBO J. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- Moehle CM, Hinnebusch AG. Association of RAP1 binding sites with stringent control of ribosomal protein gene transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:2723–2735. doi: 10.1128/mcb.11.5.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskvina E, Schuller C, Maurer CT, Mager WH, Ruis H. A search in the genome of Saccharomyces cerevisiae for genes regulated via stress response elements. Yeast. 1998;14:1041–1050. doi: 10.1002/(SICI)1097-0061(199808)14:11<1041::AID-YEA296>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman-Silberberg FS, Bhattacharya S, Broach JR. Nutrient availability and the RAS/cyclic AMP pathway both induce expression of ribosomal protein genes in Saccharomyces cerevisiae but by different mechanisms. Mol Cell Biol. 1995;15:3187–3196. doi: 10.1128/mcb.15.6.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierras CR, Warner JR. Protein kinase C enables the regulatory circuit that connects membrane synthesis to ribosome synthesis in Saccharomyces cerevisiae. J Biol Chem. 1999;274:13235–13241. doi: 10.1074/jbc.274.19.13235. [DOI] [PubMed] [Google Scholar]
- Norbeck J, Blomberg A. Metabolic and regulatory changes associated with growth of Saccharomyces cerevisiae in 1.4 M NaCl. Evidence for osmotic induction of glycerol dissimilation via the dihydroxyacetone pathway. J Biol Chem. 1997;272:5544–5554. doi: 10.1074/jbc.272.9.5544. [DOI] [PubMed] [Google Scholar]
- Parrou JL, Enjalbert B, Plourde L, Bauche A, Gonzalez B, Francois J. Dynamic responses of reserve carbohydrate metabolism under carbon and nitrogen limitations in Saccharomyces cerevisiae. Yeast. 1999;15:191–203. doi: 10.1002/(SICI)1097-0061(199902)15:3<191::AID-YEA358>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Parrou JL, Teste MA, Francois J. Effects of various types of stress on the metabolism of reserve carbohydrates in Saccharomyces cerevisiae: genetic evidence for a stress-induced recycling of glycogen and trehalose. Microbiology. 1997;143:1891–1900. doi: 10.1099/00221287-143-6-1891. [DOI] [PubMed] [Google Scholar]
- Rep M, Krantz M, Thevelein JM, Hohmann S. The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J Biol Chem. 2000;275:8290–8300. doi: 10.1074/jbc.275.12.8290. [DOI] [PubMed] [Google Scholar]
- Rep M, Reiser V, Gartner U, Thevelein JM, Hohmann S, Ammerer G, Ruis H. Osmotic stress-induced gene expression in Saccharomyces cerevisiae requires Msn1p and the novel nuclear factor Hot1p. Mol Cell Biol. 1999;19:5474–5485. doi: 10.1128/mcb.19.8.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruis H, Schuller C. Stress signaling in yeast. Bioessays. 1995;17:959–965. doi: 10.1002/bies.950171109. [DOI] [PubMed] [Google Scholar]
- Schnell N, Krems B, Entian KD. The PAR1 (YAP1/SNQ3) gene of Saccharomyces cerevisiae, a c-jun homologue, is involved in oxygen metabolism. Curr Genet. 1992;21:269–273. doi: 10.1007/BF00351681. [DOI] [PubMed] [Google Scholar]
- Shalon D, Smith SJ, Brown PO. A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res. 1996;6:639–645. doi: 10.1101/gr.6.7.639. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. In: Fink GR, editor. Guide to Yeast Genetics and Molecular Biology. San Diego, CA: Academic Press; 1991. pp. 3–21. [Google Scholar]
- Singer MA, Lindquist S. Multiple effects of trehalose on protein folding in vitro and in vivo. Mol Cell. 1998;1:639–648. doi: 10.1016/s1097-2765(00)80064-7. [DOI] [PubMed] [Google Scholar]
- Smith A, Ward MP, Garrett S. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 1998;17:3556–3564. doi: 10.1093/emboj/17.13.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen DW, Rivers SL, Jamieson DJ. The role of the YAP1 and YAP2 genes in the regulation of the adaptive oxidative stress responses of Saccharomyces cerevisiae. Mol Microbiol. 1995;16:415–423. doi: 10.1111/j.1365-2958.1995.tb02407.x. [DOI] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Zahringer H, Burgert M, Holzer H, Nwaka S. Neutral trehalase Nth1p of Saccharomyces cerevisiae encoded by the NTH1 gene is a multiple stress responsive protein. FEBS Lett. 1997;412:615–620. doi: 10.1016/s0014-5793(97)00868-5. [DOI] [PubMed] [Google Scholar]