Abstract
One of several induced defense responses in plants is systemic acquired resistance (SAR), which is regulated by salicylic acid and in Arabidopsis by the NIM1/NPR1 protein. To identify additional components of the SAR pathway or other genes that regulate SAR-independent resistance, we performed genetic suppressor screens of mutagenized nim1-1 seedlings, which are highly susceptible to infection by Peronospora parasitica. We isolated the son1 (suppressor of nim1-1) mutant, which shows full restoration of pathogen resistance without the induction of SAR-associated genes and expresses resistance when combined with a salicylate hydroxylase (nahG) transgene. These features indicate that son1-mediated resistance is distinct from SAR. Resistance is effective against both the virulent oomycete Peronospora and the bacterial pathogen Pseudomonas syringae pv tomato strain DC3000. We cloned SON1 and found it to encode a novel protein containing an F-box motif, an element found within the specificity determinant in the E3 ubiquitin-ligase complex. We propose the existence of a novel defense response that is independent of SAR and negatively regulated in Arabidopsis by SON1 through the ubiquitin-proteosome pathway.
INTRODUCTION
Plants use a number of pathogen-induced defense pathways to prevent or control disease, which together constitute the general or innate immune system. Identification of several key regulators of plant immunity has revealed a complex network of interacting defense pathways (Feys and Parker, 2000). However, the means by which these pathways are coordinated or how they intersect are largely unknown. The most intensively studied pathogen-induced defense response is systemic acquired resistance (SAR), which provides to the plant broad-spectrum resistance against not only an initial pathogen but also against subsequent infection by a variety of other viral, fungal, and bacterial pathogens (Ryals et al., 1996; Delaney, 1997; Sticher et al., 1997).
The naturally occurring compound salicylic acid (SA) is a crucial signaling compound for the induction of SAR. Application to plants of SA or its functional analog 2,6-dichloroisonicotinic acid (INA) effectively induces SAR (Kessmann et al., 1994), enabling the facile manipulation of the SAR pathway for molecular and genetic studies. SA accumulation was shown to be essential for the induction of SAR in studies of transgenic tobacco plants that express nahG, a bacterial salicylate hydroxylase gene whose product catalyzes the conversion of SA into the inactive compound catechol; NahG plants showed neither SA accumulation nor induction of SAR (Gaffney et al., 1993). NahG plants also are hypersusceptible to a wide range of pathogens and are significantly compromised in their expression of several examples of resistance gene–mediated defense (Delaney et al., 1994).
Exogenous application of SA or INA to plants triggers the onset of disease resistance coincident with the induction of a large number of defense response genes, including many pathogenesis-related (PR) genes, which are used commonly as molecular markers to monitor SAR activation (Ward et al., 1991; Yalpani et al., 1991; Uknes et al., 1992). Despite the tight correlation between SAR activation and the high-level expression of many PR genes, transgenic plants that express these PR genes display only modest resistance against pathogens (Alexander et al., 1993), indicating that the concerted expression of several PR genes may be required to produce robust resistance or that other important resistance effectors have yet to be discovered.
Using SA or its functional analogs, several groups performed mutant screens to identify additional components in the SAR pathway. Two of these groups used transgenic plants that harbored PR gene promoter–reporter fusions and isolated mutants unable to induce the reporter after treatment with INA or SA (Cao et al., 1994; Shah et al., 1997). In our screen, we focused on the induced resistance phenotype and identified mutants unable to express INA- or SA-induced pathogen resistance (Delaney et al., 1995). The rationale for seeking loss of pathogen resistance as the mutant phenotype in our screen was based on the belief that this could provide access to a wider range of mutants than had we monitored PR gene expression as the terminal trait.
Despite these differences in design, the approximately 11 mutants isolated in these screens were allelic, having mutations in the same gene called NIM1/NPR1/SAI1 (Cao et al., 1994; Delaney et al., 1995; Shah et al., 1997). Plants carrying mutations in the NIM1/NPR1/SAI1 gene (henceforth, NIM1) are nonresponsive to SA accumulation and therefore are impaired in downstream transduction of the SA signal, resulting in a lack of PR gene expression, SAR, and other NIM1-dependent responses. A clue to the mechanism by which NIM1 functions was provided by the cloning of the gene by two groups, which showed the predicted protein product to contain ankyrin repeat domains and sequence similarity to the IκB family of mammalian transcriptional regulators (Cao et al., 1997; Ryals et al., 1997).
In addition to regulating SAR, NIM1 has been shown to be required for the activation of a SAR-independent defense pathway called induced systemic resistance (ISR) (Pieterse et al., 1996, 1998) that is induced upon exposure of Arabidopsis roots to certain nonpathogenic Pseudomonas fluorescens strains. Because the response does not require SA accumulation and is not associated with SAR-linked PR gene expression, ISR is distinct from SAR. Also unlike SAR, induction of ISR depends on jasmonic acid and ethylene signaling (Knoester et al., 1999; van Wees et al., 1999; Pieterse et al., 2000). How NIM1 modulates the expression of both SAR and ISR is unclear, but it is becoming apparent that NIM1 plays a central role in multiple defense signaling pathways.
To reveal additional regulators of the plant immune response associated with NIM1, we screened for genetic suppressor mutations that ameliorate the highly pathogen-susceptible nim1-1 mutant phenotype. We screened for plants resistant to a highly virulent strain of Peronospora parasitica, which gave us access to a range of types of mutations that could produce resistance in a nim1-1 mutant background. We identified five son (suppressor of nim1-1) mutants based on their pathogen-resistant phenotype. One of these, son1, exhibited heightened resistance without the attendant induction of SAR-associated PR genes or PDF1.2, a marker for a jasmonic acid–dependent inducible defense response (Epple et al., 1997; Manners et al., 1998). Resistance in son1 also was expressed in the presence of nahG, indicating that it does not require SA accumulation.
Because son1 mutants exhibit resistance in a nim1-1 background, do not accumulate PR or PDF1.2 gene transcripts, and are resistant independent of SA accumulation, pathogen resistance in son1 plants is unlike other previously characterized defense mechanisms. The son1 mutation is recessive, indicating that SON1 negatively regulates a novel SAR- and NIM1-independent defense response. Cloning of SON1 revealed that it encodes a novel protein containing an F-box, an element found within the specificity-determining component of the E3 ubiquitin-ligase complex (Bai et al., 1996), implicating the ubiquitin-proteosome pathway in SON1-regulated disease resistance.
RESULTS
nim1-1 Suppressor Mutant Screen and Genetic Analysis of son1
Homozygous Wassilewskija (Ws) nim1-1 kanamycin-resistant seeds were mutagenized using ethyl methanesulfonate and grown to mature plants, which were harvested to yield M2 seeds. Two-week-old M2 seedlings were screened for resistance against the highly virulent Peronospora isolate Emwa1. Approximately 95,000 M2 plants were screened to identify individuals that exhibited resistance to Peronospora infection, and five son mutants (son1 to son5) were isolated. M3 seeds derived from each son mutant were grown on agar medium containing kanamycin to verify that the drug resistance trait was fixed, ensuring that the mutants were not false positives resulting from contamination with wild-type seeds or pollen (data not shown).
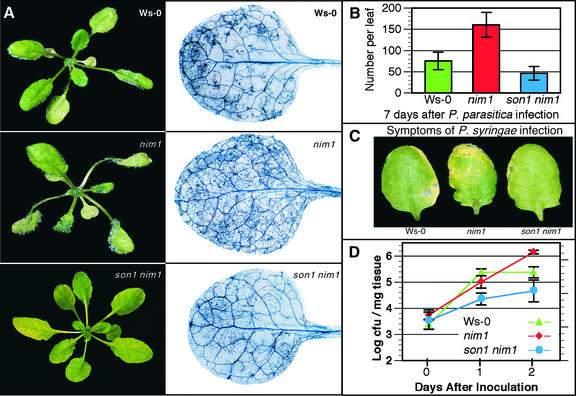
The son1 mutant showed a high level of resistance to Peronospora, with little evidence of downy mildew disease or hyphal development (Figure 1A). Quantification of conidiophore production showed that son1 plants had one-quarter the number of these reproductive structures compared with nim1-1, and ∼40% fewer than Ws-0 wild type (Figure 1B).
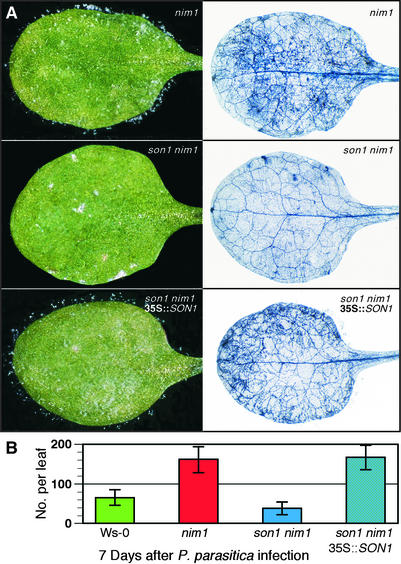
Figure 1.
Resistance of son1 nim1-1 Plants to Peronospora and Pst DC3000.
(A) The normally virulent Peronospora isolate Emwa1 showed moderate to heavy sporulation on wild-type Ws-0 and nim1-1 plants but failed to grow on son1 nim1-1 plants. Photographs were taken 7 days after inoculation with 5 × 104 conidiospores/mL (left column). Examination by lactophenol trypan blue staining of infected leaves 7 days after inoculation revealed little hyphal development in son1 nim1-1 leaves compared with the extensive growth seen in Ws-0 and nim1-1 leaves (right column). Host genotypes are indicated at top right in each panel.
(B) To quantify resistance, production of conidiophores was examined 7 days after inoculation with Peronospora Emwa1. Plants carrying the son1 mutation were resistant to this pathogen.
(C) The virulent Pst DC3000 failed to elicit disease symptoms in son1 nim1-1 plants. Three days after dip inoculation with a suspension of Pst DC3000, leaves from son1 nim1-1 mutants displayed fewer and less chlorotic lesions compared with leaves from infected wild-type Ws-0 and nim1-1 plants.
(D) Measurements of bacterial growth 24 and 48 h after inoculation show less bacterial replication in son1 nim1-1 plants than in wild-type or nim1-1 plants. cfu, colony-forming units.
Resistance of son1 to Pseudomonas syringae pv tomato
To determine whether disease resistance in son1 plants also was effective against a pathogen distinct from Peronospora, son1 mutants were infected with the virulent bacterial pathogen Pseudomonas syringae pv tomato strain DC3000 (Pst DC3000). Three days after infection, son1 mutants displayed less chlorosis and fewer disease symptoms than either wild-type or nim1-1 plants (Figure 1C). Bacterial growth also was restricted in son1 mutants compared with wild-type and nim1-1 plants (Figure 1D), indicating that son1-mediated resistance was active against Pst DC3000.
Genetic Analysis of the son1 Mutant
To determine the pattern of inheritance for the son1 mutation, son1 nim1-1 plants were backcrossed to SON1 nim1-1, and the resulting F1 and F2 progeny were tested for susceptibility to Peronospora Emwa1. Because all F1 plants produced were susceptible to the test pathogen, we concluded that son1-mediated resistance was inherited as a recessive trait. Analysis of 120 2-week-old F2 seedlings showed that 29 were resistant to Peronospora infection (∼3:1 susceptible:resistant plants; χ2 = 0.044, P > 0.5), supporting the conclusion that son1 is recessive and indicating that a single locus is responsible for the son1 phenotype. In crosses between son1 nim1-1 and wild-type Ws-0 plants, ∼25% of the F2 progeny were resistant to Peronospora, indicating that resistance was expressed independent of the nim1-1 mutation (data not shown).
To determine whether son1-mediated resistance was influenced by the presence of specific nim1 alleles, son1 nim1-1 plants were crossed to nim1-2 and nim1-4 plants. Allele-specific PCR was used to identify F2 seedlings homozygous for each of the introgressed nim1 alleles, and at least five such lines advanced to produce F3 seeds. Because each F2 plant had a 75% probability of carrying at least one copy of the son1 mutation, we were confident that, by analyzing five F3 populations, at least one would segregate son1 homozygous F3 progeny. In both the nim1-2 and nim1-4 homozygous F3 lines, we found resistant son1 nim1-2 and son1 nim1-4 plants, showing that son1-mediated resistance is not allele specific, because it was expressed in the presence of two other nim1 alleles (data not shown).
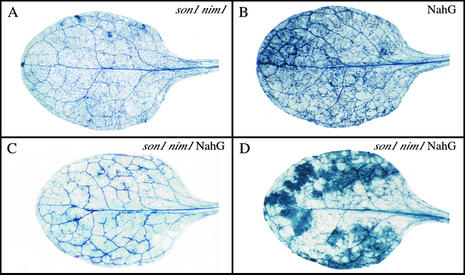
To determine whether son1-mediated resistance is dependent on SA accumulation, son1 nim1-1 plants were crossed to NahG transgenic plants. F2 son1 nim1-1 NahG homozygous lines were identified using PCR-based markers and the hygromycin marker linked to the NahG transgene. Mutant allele–specific primers were used to identify nim1-1 homozygotes (Lo et al., 1992), whereas derived CAPS were used to detect son1 homozygotes among F2 plants (Neff et al., 1998). F3 seeds from son1 nim1-1 F2 plants then were plated on Murashige and Skoog (1962) medium containing hygromycin (20 μg/mL) to screen for NahG homozygous lines. Several son1 nim1-1 NahG homozygous lines were identified and found to be resistant to Peronospora infection, showing that resistance is not dependent on SA (Figure 2). In some cases, inoculation triggered the formation of necrotic lesions on son1 nim1-1 NahG leaves (Figure 2D).
Figure 2.
Resistance of son1 nim1-1 NahG Plants to Peronospora.
Leaves were collected 7 days after inoculation with Peronospora isolate Emwa1 and stained with lactophenol trypan blue to visualize the extent of hyphal development. Leaves from son1 NahG plants ([C] and [D]) were as resistant as leaves from son1 nim1-1 (A) to pathogen infection and sometimes displayed extensive necrotic lesions (D). Host genotypes are indicated at top right in each panel.
Molecular Phenotype of son1 Plants
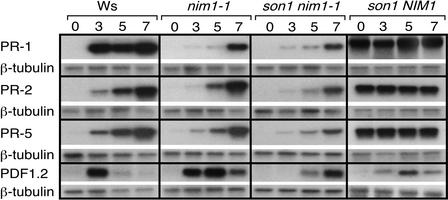
To determine whether resistance in son1 plants was accompanied by expression of PR or jasmonate-responsive genes, leaves from wild-type (Ws-0), nim1, son1 nim1, and son1 NIM1 plants were collected before and 3, 5, and 7 days after Peronospora Emwa1 inoculation and used for RNA extraction. RNA gel blots were probed with radiolabeled cDNA probes corresponding to the Arabidopsis PR-1, PR-2, PR-5, and PDF1.2 genes (Figure 3). The amount of PR-1 mRNA accumulation in the son1 nim1-1 double mutant after Peronospora infection was significantly less than the levels seen in wild type Ws-0 and slightly less than or equal to the amount observed in nim1-1. Similarly, PR-2 and PR-5 levels were reduced in son1 relative to nim1-1 and wild-type plants.
Figure 3.
Molecular Phenotype of son1 nim1-1 and son1 Plants after Inoculation with Peronospora.
Wild-type (Ws-0), nim1-1, son1 nim1-1, and son1 NIM1 plants were inoculated with the virulent Peronospora isolate Emwa1, and RNA accumulation was assessed 0, 3, 5, and 7 days later. RNA gel blots were probed with the SAR genes PR-1, PR-2, and PR-5 or with the jasmonate-responsive gene PDF1.2. Replicate blots were probed with the β-tubulin gene as a loading control.
These data indicate that resistance in son1 nim1-1 plants is not linked to the induction of SAR genes. Because son1 mutants display heightened resistance to Peronospora infection but do not show induction of PR gene expression, we tested whether the expression of the jasmonic acid–responsive defensin gene PDF1.2 was increased in son1 nim1-1 plants after inoculation with Peronospora. We detected no increase in PDF1.2 expression in son1 nim1-1 mutants after this treatment (Figure 3). Examination of the molecular phenotype of son1 plants that contain a functional NIM1 gene revealed an interesting constitutive increase in PR-1, PR-2, and PR-5 expression but not in PDF1.2 expression (Figure 3). This finding suggests that the son1 mutation activates both a SAR-independent defense response in nim1-1 mutants and SAR in NIM1 plants.
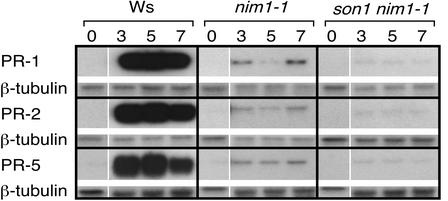
We also examined the accumulation of PR gene mRNAs after treatment with INA, an analog of SA. Wild-type plants showed the expected strong PR gene induction response to this treatment, whereas nim1-1 plants showed a much reduced response, and son1 nim1-1 plants were even less responsive than nim1-1 (Figure 4).
Figure 4.
PR Gene Expression in son1 Plants after INA Treatment.
Wild-type (Ws-0), nim1-1, and son1 nim1-1 plants were treated with the SAR-inducing chemical INA and examined for RNA accumulation 3, 5, and 7 days later. Pretreatment (time 0) samples are the same as those in Figure 3, in that Emwa1 and INA experiments were run simultaneously. SAR gene probes PR-1, PR-2, and PR-5 were tested on RNA gel blots. To assess equal loading, a β-tubulin gene probe was used on replicate blots.
Map-Based Cloning of SON1
The son1 mutant was isolated in a screen of Ws nim1-1 plants. Therefore, to map the SON1 gene, we performed crosses between son1-1 nim1-1 and Columbia (Col) npr1-2 plants. The son1 mutant phenotype was identified in F2 plants by their resistance to the Peronospora isolate Emco5, which is virulent in both Ws-0 and Col-0 accessions. The npr1-2 allele was included in the cross to reduce the possibility that the segregation of nim1-1 would complicate the identification of son1 F2 progeny.
The F2 mapping population was phenotyped for Emco5 resistance on 2-week-old plants, and resistant individuals were examined for segregation of PCR-based molecular markers. For the initial mapping, 48 F2 plants resistant to Emco5 infection were tested with codominant cleaved amplified polymorphic sequence (CAPS) markers from each of the five chromosomes (Konieczny and Ausubel, 1993). The THY1 marker on chromosome 2 showed cosegregation with the son1 phenotype.
Further analysis of this region showed the son1 mutation to be localized between THY1 and PHYB markers, which are ∼1030 kb apart (Figure 5A). To narrow the region in which SON1 was located, the Cereon Genomics single nucleotide polymorphisms (SNP) database was used to design SNP markers between THY1 and PHYB (Neff et al., 1998). Although the database is composed of SNPs between the Col-0 and Landsberg erecta accessions, we found that approximately one-third of the Col/Landsberg erecta polymorphisms tested also exist between Col-0 and Ws-0. After screening 948 F2 plants, SON1 was localized to a 41-kb region that contained 10 open reading frames (ORFs) (Figure 5C). We obtained and compared the DNA sequences of these 10 ORFs from son1 and wild-type Ws-0 plants, which revealed a single base pair polymorphism in ORF4 that causes an Arg-to-Gln change in the amino acid sequence of the predicted gene.
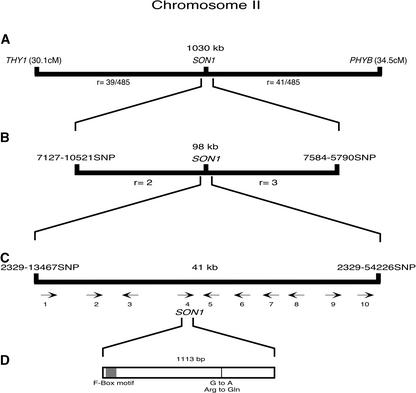
Figure 5.
Map-Based Cloning of SON1.
The initial mapping of SON1 using cleaved amplified polymorphic sequence markers located the gene in the interval between THY1 and PHYB on chromosome 2 (A). Additional PCR-based molecular markers (SNPs) were generated between THY1 and PHYB and used to narrow the region in which SON1 was located to 98 kb (B) and then 41 kb (C), which contains 10 putative open reading frames (the numbered arrows in [C] correspond to the 10 ORFs). Sequence comparison of the 10 ORFs from wild-type Ws-0 and son1 mutant plants revealed a single G-to-A transition in ORF4 (370 amino acids) that causes an Arg-to-Gln substitution at amino acid 257 in the predicted protein sequence (D). In (A), denominators indicate the number of F2 plants examined that showed the son1 phenotype. In (B) and (C), numbers ending in SNP indicate SNP molecular markers. cM, centimorgan; r, number of recombinant chromosomes identified in an F2 mapping population.
To confirm that ORF4 corresponded to the SON1 gene, Agrobacterium tumefaciens–mediated transformation was used to introduce into son1 plants a cDNA that corresponds to ORF4 under the control of the 35S promoter of Cauliflower mosaic virus. Thirty-three primary transgenic lines were obtained and found to have lost resistance to Peronospora isolate Emwa1 and Pst DC3000, indicating that ORF4 is equivalent to SON1 (Figure 6).
Figure 6.
Introduction of Wild-Type SON1 Complements the son1 Mutant.
A cDNA corresponding to ORF4 was cloned under the control of the 35S promoter of Cauliflower mosaic virus in a binary vector for transformation into son1 nim1-1 mutants. Thirty-three independent transgenic T1 lines tested showed complementation of the son1 mutation after inoculation with Peronospora isolate Emwa1.
(A) Complementation was demonstrated by the heavy Peronospora sporulation observed on son1 nim1-1 mutants that expressed ORF4, comparable to that observed on nim1-1 plants. Rows are as follows: top, nim1 is susceptible to Emwa1; middle, son1 nim1 is resistant; bottom, complementation of son1 nim1-1 with SON1 makes the transformed plants again susceptible to Peronospora. Leaves in the right column were stained with lactophenol trypan blue and then cleared to show hyphal development.
(B) Quantification of conidiophore production (ordinate) confirms that resistant son1 nim1 plants regained susceptibility after transformation with the wild-type SON1 gene.
Analysis of SON1 Structure
The SON1 gene has an uninterrupted ORF of 1113 bp, which encodes a novel 370–amino acid protein (Figure 7A). The N-terminal region of SON1 contains an F-box domain, which is a conserved 40– to 50–amino acid motif found within F-box proteins that are part of the E3 ubiquitin-ligase complex and are involved in recognition of both the E2 and substrate for ubiquitination (Bai et al., 1996). Approximately half of the known F-box proteins contain a C-terminal protein–protein interaction domain, such as a Leu-rich repeat, WD-40, or Kelch region, that is believed to play a role in substrate recognition (Takahashi et al., 1985; van der Voorn and Ploegh, 1992; Neer et al., 1994; Adams et al., 2000; Andrade et al., 2001; Kobe and Kajava, 2001). By contrast, SON1 and the other half of the known F-box proteins do not contain recognizable interaction domains at their C-terminal ends.
Figure 7.
SON1 Primary Structure and F-Box Motif.
(A) SON1 encodes a novel protein containing an F-box motif at its N-terminal end (underlined). The G-to-A missense mutation in son1-1 plants causes an Arg-to-Gln substitution at amino acid 257 in the protein (asterisk).
(B) The 40 to 50 amino acids that constitute the conserved F-box motif from SON1 are aligned with comparable regions from F-box–containing proteins from human (SKP2 and Cyclin F), yeast (CDC4), and Arabidopsis (TIR1, FKF1, UFO, and COI1); see text for literature citations. Black blocks indicate residues identical to the SON1 sequence, and gray blocks indicate similar amino acids. The consensus sequence at the bottom was generated using ClustalW on the aligned sequences.
However, the son1-1 mutation occurs in the carboxyl third of the protein, a G-to-A transition that results in an Arg-to-Gln substitution at amino acid 257, suggesting that this region is important to SON1 function (Figure 7A). Although SON1 is unlike other described proteins, its F-box region is highly similar to those of other F-box proteins from Arabidopsis, yeast, and human (Figure 7B), suggesting that SON1 also functions in conjunction with the E3 ubiquitin-ligase complex. Analysis of SON1 using reverse position-specific BLAST (http://www.ncbi.nlm.nih.gov) indicates that the putative F-box in this protein is supported by an expected value of 1 e-06, as related to the conserved F-box domain Smart00256 (LPDEILEEILSKLPPKDLLRLRKVSRKWRSLIDSHDFWFKL).
Accumulation of SON1 mRNA in Wild-Type, nim1-1, and son1 nim1 Plants
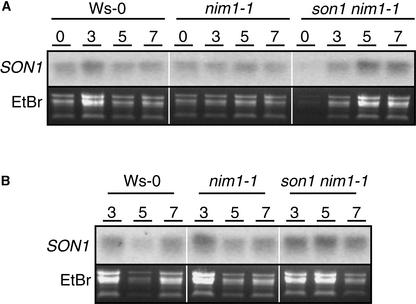
To determine if SON1 was regulated transcriptionally in plants after exposure to a pathogen or treatment with INA, we examined wild-type, nim1, and son1 nim1 plants after these treatments (Figure 8). We observed no significant change in SON1 RNA accumulation in any of the genotypes after inoculation with Peronospora or INA treatment, but we did observe a low constitutive level of expression in all plants examined (Figure 8).
Figure 8.
SON1 mRNA Accumulation in Ws-0, nim1-1, and son1 nim1-1 after Peronospora Infection or INA Treatment.
Wild-type (Ws-0), nim1-1, and son1 nim1-1 plants were inoculated with Peronospora isolate Emwa1 (A) or treated with INA (B) and examined for SON1 mRNA accumulation 3, 5, and 7 days later using RNA gel blot hybridization. To assess loading, the ethidium bromide (EtBr)–stained RNA gel was photographed before being transferred to the membrane.
DISCUSSION
In a screen to identify genetic suppressor mutations of the nim1-1 mutation and thereby reveal regulators of disease resistance, we isolated son1, a mutant that shows robust resistance to infection by both an oomycete and a bacterial pathogen and that appears to express a novel form of defense response. Resistance mediated by the recessive son1 mutation is not attributable to SAR, because son1 nim1-1 double mutants did not show significant accumulation of PR gene transcripts after pathogen exposure or in response to INA treatment, as would occur in wild-type plants or in nim1-1 plants carrying suppressors that restore function to the NIM1 pathway. Furthermore, SA accumulation is not required for the expression of son1-mediated resistance, because son1 nim1-1 NahG plants were as resistant to Peronospora and Pst DC3000 as son1 nim1 plants.
The jasmonic acid–induced defensin gene PDF1.2 also was not induced in son1 nim1-1 plants or in son1 plants, indicating that the mutation does not activate jasmonic acid signaling pathways. These unique characteristics distinguish son1 from other known nim1/npr1 suppressors, including sni1, ssi1, ssi2, cpr5, and cpr6. Plants carrying sni1 npr1-1 mutations show strong inducibility of PR genes after INA treatment (Li et al., 1999), whereas ssi1 npr1-5 and ssi2 npr1-5 mutants are dwarfed, develop necrotic lesions, and show constitutive expression of PR genes in the absence of a functional NIM1 gene (Shah et al., 1999, 2001). Unlike son1, ssi1 is a dominant mutation and is accompanied by constitutive expression of PDF1.2.
In the presence or absence of NIM1, the cpr5 and cpr6 mutants also show constitutive expression of PR and PDF1.2 genes and may express both SAR and SAR-independent defense responses (Bowling et al., 1997; Clarke et al., 1998). In son1 nim1-1 plants, on the other hand, the lack of any known defense-related gene induction indicates that this mutant expresses a NIM1-independent disease resistance mechanism unlike that observed in ssi or cpr mutants. In addition to the distinguishing phenotypic characteristics of son1 plants, the mapping and cloning of SON1 shows it to be distinct from any other cloned nim1/npr1 suppressor.
The constitutive expression of PR genes observed in son1 NIM1 plants was dependent on a functional NIM1, suggesting that in the absence of elicitation, SON1 acts to repress NIM1-dependent PR gene expression. Furthermore, the expression in son1 nim1-1 plants of SA- and PR gene–independent resistance indicates that SON1 acts as a negative regulator of a SAR-independent resistance (SIR) mechanism. Therefore, SON1 appears to have a dual role in repressing both SAR and SIR systems. Because constitutive expression of PR genes is not observed in son1 nim1-1 plants with respect to SAR, nim1 is epistatic to son1, indicating that the influence of SON1 on the SAR pathway occurs upstream of NIM1 or at NIM1 directly.
The dual role postulated for SON1 in SAR and SIR may be accomplished mechanistically by SON1 acting to repress the regulatory components required for both SAR and SIR or a regulator common to both defense systems. Alternatively, the pathogen-resistant phenotype of son1 plants may be a secondary effect of the mutation rather than the result of a direct activation of a SON1-regulated defense pathway. Indirect activation of defenses has been observed in plant mutants with defects in chlorophyll biosynthesis or catabolism, and a variety of other metabolic perturbations have been implicated in the induction of plant defense (Delaney, 1997; Glazebrook, 2001, and references therein).
To understand how SON1 may regulate two distinct defense response pathways, we cloned SON1 in a map-based approach using molecular markers and the published sequence of the Arabidopsis genome. SON1 is a 1.1-kb intronless gene that encodes a novel 370–amino acid protein that contains an N-terminal F-box motif. SON1 is like approximately half of the known F-box proteins in that it lacks a recognizable C-terminal protein–protein interaction domain. This region in SON1, though, is likely to be important to its function, because the son1-1 mutation causes a substitution in this part of the predicted protein.
Work with the yeast Saccharomyces cerevisiae has provided the greatest understanding of F-box protein function. In that species, these proteins have been shown to play a role as specificity factors in the SCF (Skp1, Cdc53/Cullin, F-box receptor) E3 ubiquitin-ligase complex that regulates the accumulation of specific cellular proteins through their polyubiquitination and proteolysis (Skowyra et al., 1997; Kaiser et al., 1998; Deshaies, 1999; Winston et al., 1999; Kipreos and Pagano, 2000). The F-box in these proteins has been shown to interact with Skp1, which helps form the E3 complex, whereas the C-terminal region of the F-box protein binds to specific substrate proteins, which consequently are targeted for polyubiquitination (for review, see Craig and Tyers, 1999).
The targeting specificity is conferred by the C-terminal end of the F-box protein, which often contains a recognizable protein–protein interaction region in the form of Leu-rich, WD-40, or kelch repeats (Craig and Tyers, 1999). Thus, F-box proteins have three roles: to interact with other components of the E3 complex; to mediate binding to E2; and to recruit specific substrates to the complex, leading to their ubiquitination and ultimate degradation (Patton et al., 1998; Xiao and Jang, 2000; Andrade et al., 2001). Because the family of F-box proteins is large, this mechanism provides cells with a rapid and irreversible system for the targeted destruction of a wide range of proteins, including many with regulatory functions.
In Arabidopsis, SON1 is the seventh F-box protein revealed to have a known function, although ∼337 F-box proteins are predicted based on genomic analysis of this plant (Arabidopsis Genome Initiative, 2000). The seven F-box proteins isolated in Arabidopsis to date have been found to regulate a diverse range of cellular functions, which include auxin responses by TIR1 (Ruegger et al., 1998), floral development by UFO (Samach et al., 1999), leaf senescence by ORE1 (Woo et al., 2001), circadian rhythms by ZTL1 and FKF1 (Nelson et al., 2000; Somers et al., 2000), wound- and jasmonate-regulated gene expression by COI1 (Xie et al., 1998), and regulation of SAR and a form of SIR by SON1 (this work). The large number of other F-box proteins encoded in the Arabidopsis genome suggests further diversification of F-box–regulated functions in plants.
Yeast two-hybrid studies also have shown that TIR1, UFO, ORE9, and COI1 are able to interact with AtASK1, the Arabidopsis homolog of the yeast SKP1 component of the SCF complex, supporting the involvement of ubiquitination-dependent proteolysis as an essential regulatory mechanism in plant signal transduction pathways (del Pozo and Estelle, 1999; Gray et al., 1999; Samach et al., 1999; Woo et al., 2001).
We suggest that SON1 functions as a component of an SCF ubiquitin ligase complex and interacts specifically with one or more substrate proteins, thus controlling their stability through ubiquitination. Therefore, learning the identity of the target substrate for SON1 is of great interest, because it is likely to act as a positive regulator of both the SAR and SIR pathways. This assumption is based on the induction of these defense responses in son1 plants and on the presumed role of SON1 in promoting the degradation of its substrate.
Based on the recognition that SON1 is an F-box protein, a possible mechanism for how the son1 mutation activates two distinct defense responses is that SON1 targets for degradation different positive regulators of SAR and SIR or a single factor common to both pathways. Thus, in son1 plants, the SON1 substrate(s) accumulates, activating SIR in nim1-1 as well as SAR in NIM1 plants. Candidates for such a regulatory factor would be proteins that displace negative transcriptional regulators of PR genes and genes whose expression may be associated with son1-mediated SIR. Alternatively, if NIM1 also regulates the SIR mechanism activated in son1 plants, as it does in SAR, the target of SON1 may be NIM1 itself.
The mechanism by which SON1 activity is regulated is unknown, but regulation of F-box proteins may occur at a number of levels. Although we found no evidence for pathogen- or INA-induced changes in SON1 mRNA accumulation, transcriptional and post-transcriptional regulation of F-box proteins has been observed in a few cases (Kornitzer and Ciechanover, 2000). However, a more important regulatory mechanism for F-box activity is in the phosphorylation of the F-box substrate protein, a modification that has been shown in many cases to be required for targeting by the SCF complex (Kornitzer and Ciechanover, 2000). Therefore, specific targeting mediated by substrate phosphorylation may be a general rule that governs this interaction. If this rule applies to SON1-controlled responses, then SON1 substrate protein(s) also may require phosphorylation before degradation.
The isolation and characterization of the son1 mutant demonstrates the existence of an effective, broad-spectrum defense system independent from known defense responses that are mediated by SA or jasmonic acid accumulation. The phenotype of son1 plants indicates that SON1 acts as a negative regulator of this defense response and, unexpectedly, also acts to negatively regulate SAR from a position upstream of or at NIM1. Cloning of the SON1 gene showed that it encodes an F-box protein, suggesting that the gene product plays a role in the selective ubiquitination and degradation of one or more target proteins. Thus, the protein(s) targeted by SON1 may act as a positive regulator of SAR and SIR. Greater understanding of the mechanism of SON1 action, and elucidation of its targets, will contribute to the engineering of disease resistance in agricultural crop species.
METHODS
Plants and Growth Conditions
Arabidopsis thaliana accession Wassilewskija (Ws-0) was obtained from the ABRC (Ohio State University, Columbus). Ws nim1-1 plants were described previously (Delaney et al., 1995), and the Ws NahG line (Molina et al., 1998) was provided by Syngenta (Research Triangle Park, NC). The Columbia (Col) npr1-2 line used for mapping was generously provided by Jane Glazebrook (Torrey Mesa Research Institute, San Diego, CA; Glazebrook et al., 1996). Plants were grown at 22°C in 14 h of light (∼150 μE provided by cool-white fluorescent lamps) at ∼60% RH on Cornell soil mix (Boodley and Sheldrake, 1977).
Genetic Suppressor Screen
Kanamycin-resistant nim1-1 seeds (Delaney et al., 1995) were mutagenized in a 0.3% solution of ethyl methanesulfonate in water for 17 h with gentle shaking. The seeds were washed extensively three times with 300 mL of sterilized water, vernalized overnight at 5°C, and sown on Cornell soil mix in 10 standard 21- × 11-inch plastic horticultural flats. The resulting M1 plants were partitioned into three groups per flat, and M2 seeds from each group were pooled upon harvest. To assess the efficiency of mutagenesis, slightly immature siliques from a sample of 100 M1 plants were dissected and scored for segregation of embryo-lethal (emb) mutations (Koncz et al., 1992). We observed ∼33% of the plants to exhibit segregation of the emb mutant phenotype.
Pathogen Inoculations
Peronospora parasitica isolates Emwa1 and Emco5 (Holub et al., 1994) were inoculated as a suspension of conidia (∼5 to 7 × 104 spores per mL of water) onto 2-week-old plants using a Preval spray mister (Precision Valve, Yonkers, NY). Inoculated plants were maintained at ∼100% RH in a Percival (Des Moines, IA) growth chamber at 18°C and 12-h-day/12-h-night cycles, as described previously (Donofrio and Delaney, 2001).
Pseudomonas syringae pv tomato (Pst) strain DC3000 was grown on King's medium B (KB; King et al., 1954) plates for 48 h at 30°C to provide a source of inoculum. Plates were washed with 10 mL of 10 mM MgCl2, which was transferred to a 250-mL flask containing 200 mL of 10 mM MgCl2 and adjusted to an OD600 of 0.05 by dilution with the MgCl2 solution. Two-week-old plants were dipped into the bacterial suspension containing 0.02% Silwet L-77 (OSI Specialties, Danbury, CT) according to Tornero and Dangl (2001). For each data point, four replicate samples consisting of pooled leaves from three identically treated plants (12 plants per treatment and time) were collected before and 24 and 48 h after inoculation to determine Pst DC3000 growth curves.
Collected leaf tissue from each sample was placed in preweighed 1.5-mL microcentrifuge tubes containing 1 mL of MgCl2 plus 0.02% Silwet. The Eppendorf tubes were reweighed with sample before shaking at 250 rpm at 28°C for 1 h. Dilutions (10−1 to 10−6) of each sample then were made using 10 mM MgCl2 in 96-well microtiter plates. Using a 96-well pin stamp, the dilutions were plated onto KB plates and left to incubate at 30°C. Colonies were counted 48 h later to determine the number of colony-forming units per milligram of leaf tissue.
RNA Gel Blot Analysis
RNA was extracted from harvested leaf tissue frozen in liquid nitrogen using a hot phenol/chloroform method followed by lithium chloride precipitation (Verwoerd et al., 1989), and samples were adjusted to 0.05 mg/mL ethidium bromide for loading onto a formaldehyde agarose gel and electrophoresis as described (Uknes et al., 1993). Gels were photographed under UV light to assess equal loading of the samples and then blotted onto Hybond N+ nylon membranes (Amersham Life Science, Arlington Heights, IL). Arabidopsis PR-1, PR-2, PR-5, PDF1.2, and SON1 cDNA clones were labeled with 32P by random priming using a commercial kit (Gibco Life Technologies/Invitrogen, Grand Island, NY). Hybridization of probe and subsequent washings were performed as described (Uknes et al., 1993).
2,6-Dichloroisonicotinic Acid Treatment
A 25% formulation of 2,6-dichloroisonicotinic acid (Syngenta, Basel, Switzerland) in wettable powder was dissolved in sterile water at 0.25 mg/mL (0.33 mM 2,6-dichloroisonicotinic acid) and spray misted onto 2-week-old plants. Leaf tissue from each plant was collected for RNA preparation and gel blot analysis at 0, 3, 5, and 7 days after treatment.
Fixation of Leaf Samples
Leaf samples were harvested and fixed in lactophenol trypan blue for 1 h before clearing with chloral hydrate (Uknes et al., 1993). Leaf samples were mounted on glass slides and photographed using a Leica MZ8 stereomicroscope (Wetzlar, Germany).
Map-Based Cloning of son1
To map SON1, the son1 nim1-1 mutant (derived from the Ws-0 accession) was crossed to npr1-2 in the Col-0 accession. Peronospora isolate Emco5, which is virulent on both Ws-0 and Col-0, was used to screen F2 progeny for the son1 resistant phenotype. DNA was extracted (Dellaporta et al., 1983) from frozen leaves of 48 F2 plants that were scored as resistant to Emco5. PCR then was performed using DNA from the plants with cleaved amplified polymorphic sequence (CAPS) markers (Konieczny and Ausubel, 1993; http://www.arabidopsis.org/) chosen from each chromosome and separated by an average distance of 5 to 8 centimorgan.
The CAPS marker THY1 on chromosome 2 was found to cosegregate with the son1 resistant phenotype, with six recombination breakpoints found between these loci among the 48 F2 plants tested. Additional CAPS markers surrounding THY1 were tested, and the son1 mutation was localized between CAPS markers THY1 and PHYB, 4.4 centimorgan or 1.02 Mb apart. DNA from an additional 485 F2 Emco5-resistant plants was tested with the THY1 and PHYB markers, and 33 additional recombinations were found using THY1 and 41 others found with PHYB.
To fine-map the location of SON1, single nucleotide polymorphism (SNP) markers were designed within the region flanked by THY1 and PHYB using the Cereon Genomics SNP database and sequence information from the Arabidopsis genome sequencing project (http://mips.gsf.de/proj/thal/db/index.html). Using two SNPs, 7127-10521SNP (forward primer, 5′-TCCTTTGACGTCTTTATGTG-3′; reverse primer, 5′-GCCTCCTGTTTACTAATGAT-3′; T/C polymorphism in Col-0 versus Ws-0) and 7584-5790SNP (forward primer, 5′-TAATTCCATGTGATGCTTGTG-3′; reverse primer, 5′-AGTTGATACAACTCTCATGAAC-3′; G/A polymorphism in Ws-0 versus Col-0), we were able to localize SON1 to a 98-kb region bounded by these markers.
The PCR protocol used for 7127-10521SNP and 7584-5790SNP was as follows: 95°C for 2 min, 95°C for 1 min, and 50°C for 2 min (for 30 cycles) followed by 72°C for 10 min. The 150-bp PCR product amplified using the 7127-10521SNP primers then was digested with the Fnu4HI restriction enzyme, which cleaves genomics DNA (gDNA) from Ws-0 plants but not Col-0 plants. The enzyme used to cut the 101-bp PCR product amplified with the 7584-5790SNP primers was AluI, which has a site in Col-0 gDNA but not in Ws-0. Both forward primers (7127-10521SNP and 7584-5790SNP) encompass the polymorphism and the resulting presence or absence of the restriction enzyme site. Of the 39 recombinations isolated using THY1, 2 remained at 7127-10521SNP, and of the 41 recombinations isolated using PHYB, 3 remained at 7584-5790SNP.
The interval between THY1 and PHYB was examined using four additional SNP markers found in the Cereon SNP database, and the closest two were used to fine-map SON1. With one, 2329-13467SNP, we were able to reduce the number of recombinations remaining to one at the left side of SON1, whereas using the 2329-54226SNP marker, the number of recombinations remaining was reduced to two on the right border of SON1. The narrower interval defined by the two markers encompassed 41 kb of genomic sequence and contained 10 open reading frames (ORFs).
The 2329-54226SNP marker (forward primer, 5′-CCAATTCATTGTTTTTGAACC-3′; reverse primer, 5′-GATGGAGAGATCAACGAGC-3′) reveals in the 152-bp amplicon an A/T polymorphism between Ws-0 and Col-0 that produces an MfeI site in the Ws-0 product that is absent in Col-0. The 2329-13467SNP marker (forward primer, 5′-TTTGCTCTAAGTTTCAACAG-3′; reverse primer, 5′-GCCGACGTACGTTAATCATTTG-3′) exploits a length polymorphism that causes a 180-bp product to be produced from Ws-0, whereas the product from Col-0 is 150 bp. The PCR conditions for both markers were identical to those described above except that the annealing temperature for 2329-13467SNP was 55°C instead of 50°C.
Each of the 10 ORFs flanked by markers around SON1 were examined by designing PCR primers to amplify each ORF from gDNA of the son1 mutant and Ws-0 wild-type plants. Comparisons between the sequences of each of the ORFs from son1 and wild-type plants revealed a single G-to-A base pair difference in ORF4 that produces an Arg-to-Gln substitution at amino acid 257 in the predicted protein sequence.
Complementation of son1 with the Wild-Type Gene
A cDNA of ORF4 was cloned under the control of the 35S promoter of Cauliflower mosaic virus into a modified pCAMBIA-1302 binary vector using NcoI and SpeI sites and transformed into son1 nim1 mutants using the floral dip method (Clough and Bent, 1998). T1 seeds were collected and plated onto Murashige and Skoog (1962) medium (Gibco Life Technologies/Invitrogen) containing 20 μg/mL hygromycin to select for primary transformants. Thirty-three independent lines were found to be resistant to hygromycin and were transferred to soil 1 week after germination. These 33 lines were tested in pathogen assays to assess the complementation of the son1 mutation.
At 2 weeks of age, all 33 lines were inoculated with Peronospora and incubated in Percival growth chambers as described above. All 33 lines were as susceptible to Peronospora infection as nim1-1 mutants and displayed increased sporulation compared with nontransformed son1 nim1 plants.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Accession Numbers
The GenBank accession number for the SON1 sequence is AF472589. The GenBank accession numbers for the sequences shown in Figure 7 are NP116026 (human SKP2), P41002 (human Cyclin F), CAA86341 (yeast CDC4), NP567135 (Arabidopsis TIR1), NP564919 (Arabidopsis FKF1), NP564368 (Arabidopsis UFO), and NP565919 (Arabidopsis COI1).
Acknowledgments
We thank Chang-Sik Oh and Erika Brutsaert for their assistance in mapping SON1. We appreciate Cristiana Argueso's careful reading of the manuscript. We gratefully recognize the support for this project and other work in the laboratory provided by the Cornell University Department of Plant Pathology and by grants to T.P.D. from the National Science Foundation CAREER program (Grant IBN-9722377) and the U.S. Department of Agriculture National Research Initiative Competitive Grants Program (Grant 9802134).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.001867.
References
- Adams, J., Kelso, R., and Cooley, L. (2000). The kelch repeat superfamily of proteins: Propellers of cell function. Trends Cell Biol. 10, 17–24. [DOI] [PubMed] [Google Scholar]
- Alexander, D., Goodman, R.M., Gut-Rella, M., Glascock, C., Weymann, K., Friedrich, L., Maddox, D., Ahl Goy, P., Luntz, T., Ward, E., and Ryals, J. (1993). Increased tolerance to two oomycete pathogens in transgenic tobacco expressing pathogenesis-related protein 1a. Proc. Natl. Acad. Sci. USA 90, 7327–7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade, M.A., Gonzalez-Guzman, M., Serrano, R., and Rodriguez, P.L. (2001). A combination of the F-box motif and kelch repeats defines a large Arabidopsis family of F-box proteins. Plant Mol. Biol. 46, 603–614. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Bai, C., Sen, P., Hofmann, K., Ma, L., Goebl, M., Harper, J.W., and Elledge, S.J. (1996). SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86, 263–274. [DOI] [PubMed] [Google Scholar]
- Boodley, J.W., and Sheldrake, R., Jr. (1977). Cornell peat-lite mixes for commercial plant growing. NY State Coll. Agric. Life Sci. Inf. Bull. 43, 1–8. [Google Scholar]
- Bowling, S.A., Clarke, J.D., Liu, Y., Klessig, D.F., and Dong, X. (1997). The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9, 1573–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Bowling, S.A., Gordon, A.S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Glazebrook, J., Clarke, J.D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63. [DOI] [PubMed] [Google Scholar]
- Clarke, J.D., Liu, Y., Klessig, D.F., and Dong, X. (1998). Uncoupling PR gene expression from NPR1 and bacterial resistance: Characterization of the dominant Arabidopsis cpr6-1 mutant. Plant Cell 10, 557–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Craig, K.L., and Tyers, M. (1999). The F-box: A new motif for ubiquitin dependent proteolysis in cell cycle regulation and signal transduction. Prog. Biophys. Mol. Biol. 72, 299–328. [DOI] [PubMed] [Google Scholar]
- Delaney, T.P. (1997). Genetic dissection of acquired resistance to disease. Plant Physiol. 106, 5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney, T.P., Friedrich, L., and Ryals, J.A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA 92, 6602–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney, T.P., Uknes, S., Vernooij, B., Friedrich, L., Weymann, K., Negrotto, D., Gaffney, T., Gut-Rella, M., Kessmann, H., Ward, E., and Ryals, J. (1994). A central role of salicylic acid in plant disease resistance. Science 266, 1247–1250. [DOI] [PubMed] [Google Scholar]
- Dellaporta, S.L., Wood, J., and Hicks, J.B. (1983). A plant DNA minipreparation: Version II. Plant Mol. Biol. Rep. 1, 19–21. [Google Scholar]
- del Pozo, J.C., and Estelle, M. (1999). Function of the ubiquitin-proteosome pathway in auxin response. Trends Plant Sci. 4, 107–112. [DOI] [PubMed] [Google Scholar]
- Deshaies, R.J. (1999). SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15, 435–467. [DOI] [PubMed] [Google Scholar]
- Donofrio, N.M., and Delaney, T.P. (2001). Abnormal callose response phenotype and hypersusceptibility to Peronospora parasitica in defense-compromised Arabidopsis nim1-1 and salicylate hydroxylase-expressing plants. Mol. Plant-Microbe Interact. 14, 439–450. [DOI] [PubMed] [Google Scholar]
- Epple, P., Apel, K., and Bohlmann, H. (1997). ESTs reveal a multigene family for plant defensins in Arabidopsis thaliana. FEBS Lett. 400, 168–172. [DOI] [PubMed] [Google Scholar]
- Feys, B.J., and Parker, J.E. (2000). Interplay of signaling pathways in plant disease resistance. Trends Genet. 16, 449–455. [DOI] [PubMed] [Google Scholar]
- Gaffney, T., Friedrich, L., Vernooij, B., Negrotto, D., Nye, G., Uknes, S., Ward, E., Kessmann, H., and Ryals, J. (1993). Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261, 754–756. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J. (2001). Genes controlling expression of defense responses in Arabidopsis: 2001 status. Curr. Opin. Plant Biol. 4, 301–308. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J., Rogers, E.E., and Ausubel, F.M. (1996). Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143, 973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., del Pozo, J.C., Walker, L., Hobbie, L., Risseeuw, E., Banks, T., Crosby, W.L., Yang, M., Ma, H., and Estelle, M. (1999). Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13, 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holub, E.B., Beynon, L.J., and Crute, I.R. (1994). Phenotypic and genotypic characterization of interactions between isolates of Peronospora parasitica and accessions of Arabidopsis thaliana. Mol. Plant-Microbe Interact. 7, 223–239. [Google Scholar]
- Kaiser, P., Sia, R.A., Bardes, E.G., Lew, D.J., and Reed, S.I. (1998). Cdc34 and the F-box protein Met30 are required for degradation of the Cdk-inhibitory kinase Swe1. Genes Dev. 12, 2587–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessmann, H., Staub, T., Hofmann, C., Maetzke, T., Herzog, J., Ward, E., Uknes, S., and Ryals, J. (1994). Induction of systemic acquired resistance in plants by chemicals. Annu. Rev. Phytopathol. 32, 439–459. [DOI] [PubMed] [Google Scholar]
- King, E.O., Ward, M.K., and Raney, D.E. (1954). Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44, 301–307. [PubMed] [Google Scholar]
- Kipreos, E.T., and Pagano, M. (2000). The F-box protein family. Genome Biol. 1, 3002.1–3002.7. [DOI] [PMC free article] [PubMed]
- Knoester, M., Pieterse, C.M., Bol, J.F., and Van Loon, L.C. (1999). Systemic resistance in Arabidopsis induced by rhizobacteria requires ethylene-dependent signaling at the site of application. Mol. Plant-Microbe Interact. 12, 720–727. [DOI] [PubMed] [Google Scholar]
- Kobe, B., and Kajava, A.V. (2001). The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11, 725–732. [DOI] [PubMed] [Google Scholar]
- Koncz, C., Chua, N.-H., and Schell, J. (1992). Methods in Arabidopsis Research. (London: World Scientific Publishing).
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Kornitzer, D., and Ciechanover, A. (2000). Modes of regulation of ubiquitin-mediated protein degradation. J. Cell Physiol. 182, 1–11. [DOI] [PubMed] [Google Scholar]
- Li, X., Zhang, Y., Clarke, J.D., Li, Y., and Dong, X. (1999). Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell 98, 329–339. [DOI] [PubMed] [Google Scholar]
- Lo, E.S., Lo, Y.M., Tse, C.H., and Fleming, K.A. (1992). Detection of hepatitis B pre-core mutant by allele specific polymerase chain reaction. J. Clin. Pathol. 45, 689–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manners, J.M., Penninckx, I.A., Vermaere, K., Kazan, K., Brown, R.L., Morgan, A., Maclean, D.J., Curtis, M.D., Cammue, B.P., and Broekaert, W.F. (1998). The promoter of the plant defensin gene PDF1.2 from Arabidopsis is systemically activated by fungal pathogens and responds to methyl jasmonate but not to salicylic acid. Plant Mol. Biol. 38, 1071–1080. [DOI] [PubMed] [Google Scholar]
- Molina, A., Hunt, M.D., and Ryals, J.A. (1998). Impaired fungicide activity in plants blocked in disease resistance signal transduction. Plant Cell 10, 1903–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Neer, E.J., Schmidt, C.J., Nambudripad, R., and Smith, T.F. (1994). The ancient regulatory-protein family of WD-repeat proteins. Nature 371, 297–300. [DOI] [PubMed] [Google Scholar]
- Neff, M.M., Neff, J.D., Chory, J., and Pepper, A.E. (1998). dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: Experimental applications in Arabidopsis thaliana genetics. Plant J. 14, 387–392. [DOI] [PubMed] [Google Scholar]
- Nelson, D.C., Lasswell, J., Rogg, L.E., Cohen, M.A., and Bartel, B. (2000). FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101, 331–340. [DOI] [PubMed] [Google Scholar]
- Patton, E.E., Willems, A.R., and Tyers, M. (1998). Combinatorial control in ubiquitin-dependent proteolysis: Don't Skp the F-box hypothesis. Trends Genet. 14, 236–243. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M., van Wees, S.C., Hoffland, E., van Pelt, J.A., and van Loon, L.C. (1996). Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression. Plant Cell 8, 1225–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M., van Wees, S.C., van Pelt, J.A., Knoester, M., Laan, R., Gerrits, H., Weisbeek, P.J., and van Loon, L.C. (1998). A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10, 1571–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M.J., Van Pelt, J.A., Ton, J., Parchmann, S., Mueller, M.J., Buchala, A.J., Metraux, J.P., and Van Loon, L.C. (2000). Rhizobacteria-mediated induced systemic resistance (ISR) in Arabidopsis requires sensitivity to jasmonate and ethylene but is not accompanied by an increase in their production. Physiol. Mol. Plant Pathol. 57, 123–134. [Google Scholar]
- Ruegger, M., Dewey, E., Gray, W.M., Hobbie, L., Turner, J., and Estelle, M. (1998). The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev. 12, 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals, J., Neuenschwander, U.H., Willits, M.G., Molina, A., Steiner, H.-Y., and Hunt, M.D. (1996). Systemic acquired resistance. Plant Cell 8, 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals, J., Weymann, K., Lawton, K.A., Friedrich, L., Ellis, D., Steiner, H.-Y., Johnson, J., Delaney, T.P., Jesse, T., Vos, P., and Uknes, S. (1997). The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor IκB. Plant Cell 9, 425–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach, A., Klenz, J.E., Kohalmi, S.E., Risseeuw, E., Haughn, G.W., and Crosby, W.L. (1999). The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant J. 20, 433–445. [DOI] [PubMed] [Google Scholar]
- Shah, J., Kachroo, P., and Klessig, D.F. (1999). The Arabidopsis ssi1 mutation restores pathogenesis-related gene expression in npr1 plants and renders defensin gene expression salicylic acid dependent. Plant Cell 11, 191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, J., Kachroo, P., Nandi, A., and Klessig, D.F. (2001). A recessive mutation in the Arabidopsis SSI2 gene confers SA- and NPR1-independent expression of PR genes and resistance against bacterial and oomycete pathogens. Plant J. 25, 563–574. [DOI] [PubMed] [Google Scholar]
- Shah, J., Tsui, F., and Klessig, D.F. (1997). Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana, identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol. Plant-Microbe Interact. 10, 69–78. [DOI] [PubMed] [Google Scholar]
- Skowyra, D., Craig, K.L., Tyers, M., Elledge, S.J., and Harper, J.W. (1997). F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91, 209–219. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Schultz, T.F., Milnamow, M., and Kay, S.A. (2000). ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101, 319–329. [DOI] [PubMed] [Google Scholar]
- Sticher, L., Mauch-Mani, B., and Metraux, J.P. (1997). Systemic acquired resistance. Annu. Rev. Phytopathol. 35, 235–270. [DOI] [PubMed] [Google Scholar]
- Takahashi, N., Takahashi, Y., and Putnam, F.W. (1985). Periodicity of leucine and tandem repetition of a 24-amino acid segment in the primary structure of leucine-rich alpha 2-glycoprotein of human serum. Proc. Natl. Acad. Sci. USA 82, 1906–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornero, P., and Dangl, J.L. (2001). A high-throughput method for quantifying growth of phytopathogenic bacteria in Arabidopsis thaliana. Plant J. 28, 475–481. [DOI] [PubMed] [Google Scholar]
- Uknes, S., Mauch, M.B., Moyer, M., Potter, S., Williams, S., Dincher, S., Chandler, D., Slusarenko, A., Ward, E., and Ryals, J. (1992). Acquired resistance in Arabidopsis. Plant Cell 4, 645–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uknes, S., Winter, A.M., Delaney, T., Vernooij, B., Morse, A., Friedrich, L., Nye, G., Potter, S., Ward, E., and Ryals, J. (1993). Biological induction of systemic acquired resistance in Arabidopsis. Mol. Plant-Microbe Interact. 6, 692–698. [Google Scholar]
- van der Voorn, L., and Ploegh, H.L. (1992). The WD-40 repeat. FEBS Lett. 307, 131–134. [DOI] [PubMed] [Google Scholar]
- van Wees, S.C.M., Luijendijk, M., Smoorenburg, I., van Loon, L.C., and Pieterse, C.M.J. (1999). Rhizobacteria-mediated induced systemic resistance (ISR) in Arabidopsis is not associated with a direct effect on expression of known defense-related genes but stimulates the expression of the jasmonate-inducible gene Atvsp upon challenge. Plant Mol. Biol. 41, 537–549. [DOI] [PubMed] [Google Scholar]
- Verwoerd, B., Dekker, M., and Hoekema, A. (1989). A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 17, 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, E.R., Uknes, S.J., Williams, S.C., Dincher, S.S., Wiederhold, D.L., Alexander, D.C., Ahl Goy, P., Métraux, J.P., and Ryals, J.A. (1991). Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3, 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston, J.T., Koepp, D.M., Zhu, C., Elledge, S.J., and Harper, J.W. (1999). A family of mammalian F-box proteins. Curr. Biol. 9, 1180–1182. [DOI] [PubMed] [Google Scholar]
- Woo, H.R., Chung, K.M., Park, J.H., Oh, S.A., Ahn, T., Hong, S.H., Jang, S.K., and Nam, H.G. (2001). ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13, 1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, W., and Jang, J. (2000). F-box proteins in Arabidopsis. Trends Plant Sci. 5, 454–457. [DOI] [PubMed] [Google Scholar]
- Xie, D.X., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. [DOI] [PubMed] [Google Scholar]
- Yalpani, N., Silverman, P., Wilson, T.M., Kleier, D.A., and Raskin, I. (1991). Salicylic acid is a systemic signal and an inducer of pathogenesis-related proteins in virus-infected tobacco. Plant Cell 3, 809–818. [DOI] [PMC free article] [PubMed] [Google Scholar]