Abstract
Phloem-mobile endogenous RNA is trafficked selectively into the shoot apex. In contrast, most viruses and long-distance post-transcriptional gene silencing (PTGS) signals are excluded from the shoot apex. These observations suggest the operation of an underlying regulatory mechanism. To examine this possibility, a potexvirus movement protein, known to modify cell-to-cell trafficking and PTGS, was expressed ectopically in transgenic plants. These plants were found to be compromised in their capacity to exclude both viral RNA and silencing signals from the shoot apex. The transgenic plants also displayed various degrees of abnormal leaf polarity depending on transgene expression level. Normal patterns of organ development were restored by either virus- or Agrobacterium tumefaciens–mediated induction of PTGS. This revealed the presence of an RNA signal surveillance system that acts to allow the selective entry of RNA into the shoot apex. We propose that this surveillance system regulates signaling and protects the shoot apex, in particular the cells that give rise to reproductive structures, from viral invasion.
INTRODUCTION
A fundamental difference between plant and animal development involves the establishment of the germline. Fate-mapping studies have shown that animals sequester their germline from somatic cells during embryogenesis, whereas in plants, germ cells arise from the shoot apical meristem after vegetative development has ceased (Eddy, 1975; Poethig et al., 1986; Jegla and Sussex, 1989; Irish and Sussex, 1992; Saffman and Lasko, 1999; Seydoux and Strome, 1999). Because viral infection could pose a challenge at any time in the life cycle of the plant, it would seem imperative that the shoot apex be protected against virus entry to ensure the integrity of the germline. Consistent with this hypothesis, most plant viruses cannot invade the shoot apex (Matthews, 1991), suggesting an underlying mechanism that protects this region of the plant from viral challenge.
An emerging paradigm in plant biology reflects the presence of mobile RNA species that act non-cell-autonomously to regulate developmental processes (Lucas et al., 2001). Within this context, post-transcriptional gene silencing (PTGS) appears to have evolved as a defensive mechanism to counter viral and transposon challenge (Jorgensen et al., 1998; Kooter et al., 1999; Vance and Vaucheret, 2001; Waterhouse et al., 2001). The vascular system, and specifically the phloem, serves to relay endogenous RNA species (Ruiz-Medrano et al., 1999; Xoconostle-Cázares et al., 1999; Kim et al., 2001) and sequence-specific PTGS signals (Palauqui et al., 1997; Voinnet et al., 1998, 2000) to distantly located tissues and/or organs. As in viral exclusion from the apex, the PTGS signal seems to be incapable of entering into and activating PTGS within the shoot apex (Beclin et al., 1998; Voinnet et al., 1998).
Viruses have evolved a range of countersurveillance mechanisms directed at interdicting the endogenous PTGS mechanism (Anandalakshmi et al., 1998, 2000; Brigneti et al., 1998; Li et al., 1999; Voinnet et al., 1999; Lucy et al., 2000; Guo and Ding, 2002). For example, the potyviral Helper Component–Proteinase has the capacity to suppress PTGS in ground (nonvascular) but not systemic (phloem) tissues (Kasschau and Carrington, 1998; Mallory et al., 2001). By contrast, the first protein encoded by the potexvirus triple gene block, the TGBp1, which functions as the viral movement protein (Lough et al., 1998), has little effect on PTGS in ground tissues but can inhibit the systemic transmission of the PTGS signal that can target RNA for degradation (Voinnet et al., 2000). A similar situation has been reported for the 2b protein of Cucumber mosaic virus (CMV2b) (Brigneti et al., 1998; Guo and Ding, 2002).
Although TGBp1 and CMV2b likely enhance the ability of these viruses to establish systemic infection, viral entry into the shoot apex remains blocked (Matthews, 1991), as is the movement of the systemic PTGS signal (Beclin et al., 1998; Voinnet et al., 1998). By contrast, the geminivirus Tomato golden mosaic virus and the potyvirus Pea seed–borne mosaic virus have the capacity to induce the propagation of a systemic PTGS signal that can exert its effects within the shoot apex (Jones et al., 1998; Peele et al., 2001). This observation is particularly intriguing, because both viruses were excluded from the shoot apex. Collectively, these findings suggest a bifurcation in the surveillance mechanism that detects the entry of viral infectious transcripts and PTGS signals during their delivery into the apex via the phloem.
Additional evidence in support of the hypothesis that plants evolved an RNA-based surveillance mechanism, located in the shoot apex, was provided by recent studies investigating the capacity of the phloem to mediate the long-distance delivery of RNA. A combination of heterografting experiments and in situ reverse transcriptase–mediated PCR analysis provided direct evidence that the phloem translocation stream contains a unique set of transcripts (Ruiz-Medrano et al., 1999). Furthermore, these studies provided evidence that specific RNA transcripts, derived from the stock, can be delivered to the scion apex, where they then exit the terminal phloem and move all the way into the shoot apical meristem (Ruiz-Medrano et al., 1999; Kim et al., 2001).
Analysis of a subset of these phloem-mobile transcripts demonstrated that only a fraction of the total number of transcripts examined were detected in the apex, suggesting the involvement of a selectivity filter (surveillance system) at the sites of unloading. A seminal finding from these studies was that the delivery of such phloem-mobile transcripts correlated with an alteration in lateral organ development within the scion apex (Kim et al., 2001). Such studies suggest that a perturbation to the putative RNA surveillance system in the apex may cause detectable alterations in plant (lateral organ) development.
In the present study, Nicotiana benthamiana ectopically expressing the White clover mosaic virus (WClMV) TGB1 was used to further our studies on the selective entry of RNA into the plant apex. We earlier demonstrated that efficient cell-to-cell movement of potexvirus WCIMV RNA requires the presence of TGBp1 to TGBp3 and coat protein (CP) (Lough et al., 1998, 2000). In the absence of TGBp2, TGBp3, and CP, TGBp1 can interact with plasmodesmata to induce an increase in size exclusion limit, but it is dysfunctional in terms of potentiating its own cell-to-cell transport (Lough et al., 1998, 2000).
The presence of “dysfunctional” TGBp1 in the apex resulted in a profound change in plant development. In the most extreme condition, named spikey, leaves failed to establish polarity about the adaxial (facing toward the apex)/abaxial (facing away from the apex) axis and developed as radially symmetric organs. Reestablishment of organ polarity, through the activation of PTGS, was used as an assay for penetration of the shoot apex by virus or the silencing signal. These studies demonstrated the operation of a zone of surveillance acting to regulate the entry of viral RNA and signaling molecules into the shoot apex. These results are discussed in terms of the evolution of a mechanism that prevents viral invasion of the shoot apex to protect the cells that ultimately give rise to reproductive structures.
RESULTS
TGB1 Overexpression Phenotype
Ectopic expression of WClMV TGB1 (Figure 1A) in N. benthamiana resulted in a profound alteration in leaf, shoot, and flower development (Figures 1B to 1D). The severity of the phenotype correlated directly with the level of TGB1 transcript accumulation (Figure 1E). A detailed descriptive analysis of the phenotype was undertaken to determine whether TGB1 expression perturbed the establishment of lateral organ polarity. The vegetative phenotypes ranged from slightly shortened plants with mildly epinastic leaves to dwarfed plants with bladeless lateral organs. The ectopic expression of TGB1 was correlated spatially and temporally with the earliest manifestation of the spikey phenotype.
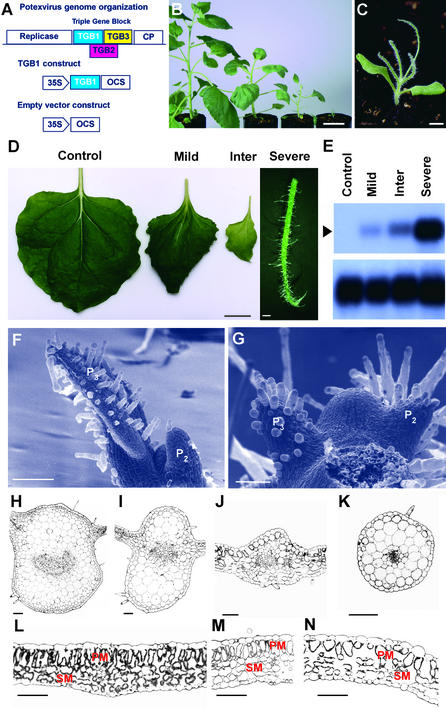
Figure 1.
Transgenic Expression of Potexvirus WCIMV TGB1 in N. benthamiana Alters Plant Form and Leaf Morphology.
(A) The potexvirus genome encodes a replicase, three overlapping open reading frames (TGB1 to TGB3), and a coat protein (CP). The TGB and CP act in concert to mediate the cell-to-cell movement of viral RNA. N. benthamiana plants were transformed with a binary vector designed to express either the TGB1 product or an empty cassette (control).
(B) Transgenic overexpression of TGB1 in N. benthamiana yielded a range of phenotypes that, relative to the control (left), included reduced internode length and altered leaf morphology.
(C) The most severe TGB1-induced phenotype, in which all true leaves were radially symmetric, was termed “spikey.”
(D) Expression of TGB1 resulted in both a net reduction in lamina size and a complete loss of polarity in spikey plants.
(E) The level of TGB1 transgene expression in N. benthamiana was correlated directly with phenotype severity. RNA gel blot analysis, using poly(A)+ RNA from control transgenic and TGB1-expressing plants displaying mild, intermediate, and severe phenotypes, was performed with a 32P-labeled TGB1-specific probe under high-stringency conditions (top). The position of the transcript of the expected size is indicated with an arrowhead. The membrane was stripped and reprobed with a 32P-labeled RbsC-specific probe (bottom).
(F) and (G) Scanning electron micrographs of control (F) and spikey (G) shoot apices and developing primordia (P2 and P3) illustrate an early defect in the development of spikey leaves. Spikey shoot apical meristems frequently were larger than those of control plants.
(H) to (K) Expression of the TGB1 transgene in N. benthamiana disrupts the establishment of leaf polarity. Transverse sections of control (H), mild (I), intermediate (J), and severe spikey (K) midribs show the positions of lamina and vascular tissue.
(L) to (N) Transverse sections of control (L), mild (M), and intermediate (N) lamina. Note the progressive loss of cell development in palisade parenchyma (PM) and spongy mesophyll (SM) layers and the concomitant increase in intercellular air spaces in mild to intermediate spikey leaves.
Bars = 5 cm in (B), 5 mm in (C), 1 cm in (D) (severe = 1 mm), 200 μm in (F) and (G), and 100 μm in (H) to (N).
Developing lateral organs were analyzed by scanning electron microscopy to determine the developmental stage of the earliest manifestation of the spikey phenotype. Lateral organs were initiated at P0, and their first physical appearance is defined as P1; younger or older organs are defined numerically according to this developmental series. Wild-type leaf primordia developed asymmetry about the adaxial/abaxial axes by P2 (Figure 1F). In contrast, spikey leaf primordia failed to initiate lamina at any stage and developed as radially symmetric organs (Figure 1G).
In TGB1 transgenic leaves, the lamina emerged from a more abaxial region of the midvein than in control plants (Figures 1H to 1J). When viewed in cross-section, the usually arc-shaped organization of the midvein vasculature was disorganized. The normally polarized pattern of tissue types in the lamina was perturbed in plants displaying a mild or intermediate phenotype (Figures 1L to 1N). The pattern of expression in the vasculature and shoot apex was consistent with the observed phenotypes (Figures 2A and 2B). With increasing levels of TGB1 RNA, leaf morphology was progressively more abnormal and internodal length was decreased (Figures 1B to 1D). In the most severe phenotype, lateral organs were radially symmetrical (Figure 1K), and these plants were named spikey (Figure 1C).
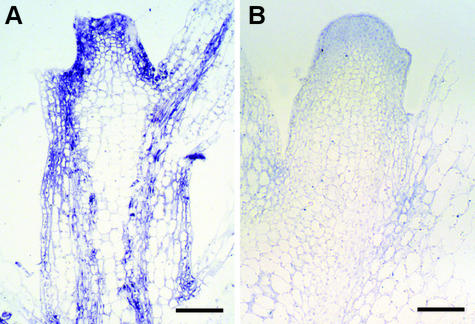
Figure 2.
WClMV TGB1 Ectopic Expression Pattern in Apical Tissues of N. benthamiana.
In situ hybridization with TGB1 probe illustrates high levels of TGB1 expression in the shoot apical meristem, young leaves, and vasculature of spikey plants (A) but none in control plants (B). Bars = 100 μm.
We next used a molecular marker for abaxial cells as an in situ hybridization probe to characterize cellular identity within spikey leaves. Primers were designed to the conserved YABBY domain common to all members of the Arabidopsis YABBY gene family (Bowman and Smyth, 1999; Siegfried et al., 1999). These primers were used to amplify a fragment of a homologous N. benthamiana gene. The deduced amino acid sequence displayed highest homology (83% over the C-terminal one-third of the protein) with FILAMENTOUS FLOWER (FIL), which was shown earlier to specify abaxial cell fate in Arabidopsis (Sawa et al., 1999; Siegfried et al., 1999).
In wild-type N. benthamiana, Nb-YABBY expression was detectable in the incipient leaf primordia P0. As primordia emerged from the shoot apical meristem, Nb-YABBY expression became restricted to the abaxial half of the leaf primordia (Figure 3A). In older leaves, expression decreased in the highly vacuolated cells of the abaxial midvein but remained high in abaxial portions of the lamina (Figure 3B). This pattern was consistent with that reported for FIL and other members of the YABBY family expressed in Arabidopsis (Sawa et al., 1999; Siegfried et al., 1999). In spikey apices, Nb-YABBY expression was similar to that in wild-type apices, although these expression domains were sometimes smaller, larger, or asymmetric relative to the wild-type pattern (Figure 3C).
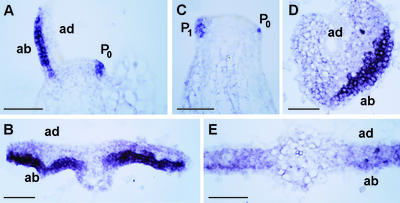
Figure 3.
Expression of TGB1 Results in Misexpression of a Molecular Marker for Organ Polarity.
Nb-YABBY, a molecular marker of abaxial leaf identity, is misexpressed in spikey leaves. Longitudinal sections of control (A) and spikey (C) apices show Nb-YABBY expression in shoot apical meristem and young leaves. (B), (D), and (E) show transverse sections through P3 and P4 leaves. Nb-YABBY is expressed throughout the abaxial half of the leaf primordia in control leaves (B), but in spikey leaves, it often is reduced or absent from abaxial regions (D) or is expressed at low levels throughout the adaxial and abaxial sides of the leaf (E). Adaxial (ad) and abaxial (ab) surfaces are oriented toward the top and bottom of the images, respectively, in (B), (D), and (E). Bars = 100 μm.
In spikey leaf primordia, Nb-YABBY expression often was reduced or absent from the abaxial domains of the leaf, especially toward the distal tip (Figure 3D). A low level of Nb-YABBY expression was detected throughout the abaxial and adaxial sides of spikey leaves (Figure 3E). Collectively, these morphological and anatomical studies, in conjunction with both the sporadic loss and the ectopic expression of Nb-YABBY in spikey leaves, are consistent with the concept that the ectopic expression of TGB1 in the meristem results in a defect in the establishment of lateral organ polarity.
Reversion of the Spikey Phenotype by Viral Infection
The potexvirus Potato virus X (PVX) lacks the capacity to induce gene silencing in the shoot apex (Ruiz et al., 1998; Ratcliff et al., 2001). Based on this observation, we predicted that although WClMV replication in spikey leaves would activate PTGS directed against both the virus and the TGB1 transgene, TGB1 expression in the shoot apex would remain unaffected. Consequently, the spikey phenotype would remain similarly unaffected during systemic infection by WClMV. Contrary to this prediction, the infection of spikey plants resulted in a reversion phenomenon that restored normal patterns of leaf development, whereas mock inoculation had no effect (Figure 4A). Normal plant architecture, floral morphology, and fertility also were restored in these reverted plants. Importantly, the progeny of reverted spikey plants again displayed the spikey phenotype.
Figure 4.
WClMV Inoculation Causes Reversion of the Spikey Phenotype.
(A) Inoculation of spikey cotyledons with WClMV resulted in the development of normal leaf and stem tissues (plant at right). In contrast, mock-inoculated plants retained the spikey phenotype (plant at left).
(B) Meristem cycling between spikey (s) and normal (n) habit during plant growth.
The observed lack of meiotic transmission of the reverted developmental state is consistent with the involvement of PTGS (Dorlhac de Borne et al., 1994). Support for this hypothesis was gained by performing ELISA on WClMV-infected control and reverted spikey leaves. A WClMV CP-specific antibody was used in these experiments, and they revealed an absence or a significant reduction in the level of WClMV in leaves that had developed on spikey plants subsequent to virus inoculation (data not shown). Over long periods, virus-infected spikey plants were observed to cycle between the initiation of normal and spikey leaves (Figure 4B). This cycling phenomenon suggested an oscillation of virus titer and TGB1 transgene RNA accumulation within the shoot apex, as was seen in earlier studies of virus-induced gene silencing of a green fluorescent protein (GFP) transgene in N. benthamiana (Ruiz et al., 1998).
Ectopic Expression of TGB1 Potentiates Viral Penetration into the Shoot Apex
The discovery that WClMV infection negated the effect(s) of TGBp1 on organ development suggested to us that, when expressed ectopically in the apex, this potexvirus movement protein could interfere with the proposed RNA surveillance system that protects the plant germline. To test this hypothesis, we examined the capacity of spikey plants to exclude virus from the shoot apex. Both control and spikey plants were inoculated with β-glucuronidase (GUS)–tagged (or GFP-tagged) PVX or wild-type WClMV. Systemically infected tissues were collected 5 to 10 days after inoculation and either stained histochemically for GUS activity or subjected to in situ hybridization using a PVX- or WClMV-specific probe. These studies clearly established that virus could be detected in the shoot apex of spikey but not control plants (Figures 5A to 5D and data not shown). Thus, in the presence of TGBp1, spikey plants are compromised in their ability to exclude virus from cells located within the shoot apex.
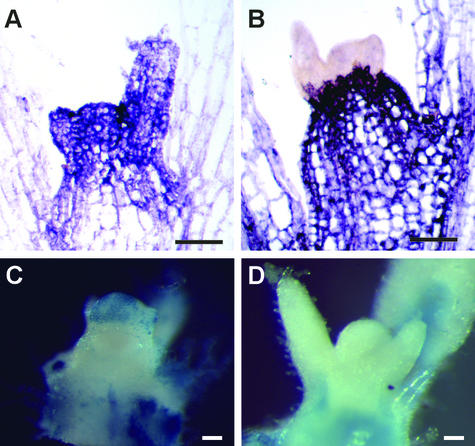
Figure 5.
Ectopic Expression of TGB1 Compromises the RNA Surveillance System of the Plant, Allowing Viral Entry into the Shoot Apex.
PVX infection domains identified in spikey and control plants 10 days after inoculation. In situ hybridization with PVX replicase probe indicates the presence of virus within the shoot apical meristem of spikey plants (A). In control plants (B), PVX was restricted to domains located beneath the shoot apex. PVX-GUS was detected histochemically in the shoot apex of spikey plants (C) but not control plants (D). Bars = 100 μm.
Reversion of the Spikey Phenotype Involves Targeted Degradation of TGB1 RNA
An alternative explanation for the reversion of the spikey meristem could involve the expression of viral products that sequester TGBp1. If this were the case, TGBp1 would no longer act to inhibit normal lateral organ development. To test this hypothesis, PVX was engineered to deliver a fragment of the WClMV TGB1 coding sequence (Figure 6A). Spikey plants inoculated with PVX.GFP failed to revert and retained the spikey phenotype, whereas those inoculated with PVX.GFP+W-TGB1 all reverted (Figure 6B). These experiments were repeated and consistently yielded the same results. Given the functional incompatibility between the PVX and WClMV TGB proteins (T.J. Lough, S.J. Emerson, R.L.S. Forster, and W.J. Lucas, unpublished data), these results provide support for the hypothesis that spikey plants revert as a result of the action of PTGS in the shoot apex rather than as a result of infection-mediated sequestration of TGBp1.
Figure 6.
A Viral Vector Incorporating WClMV TGB1 Causes Reversion of the Spikey Phenotype and Silences TGB1 Expression in the Apex.
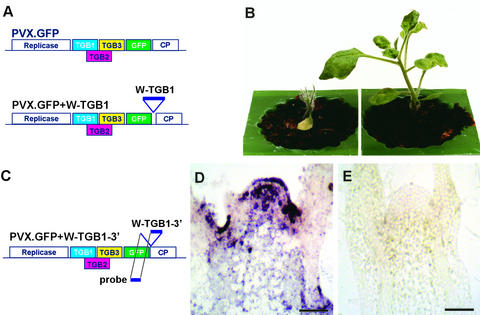
(A) The viral vector (PVX.GFP) (top) was engineered by insertion of a truncated, nontranslatable WClMV TGB1 coding sequence (incorporating WClMV TGB1 nucleotides 4056 to 4722) (PVX.GFP+W-TGB1; bottom).
(B) Plants inoculated with PVX.GFP retained the spikey phenotype (plant at left; representative of 50 plants inoculated), whereas inoculation with PVX.GFP+W-TGB1 resulted in the development of normal leaf and stem tissues, albeit with symptoms of PVX infection (plant at right; frequency of reversion was 49 of 50 plants inoculated).
(C) The viral vector (PVX.GFP) was engineered by insertion of a truncated, nontranslatable WClMV 3′ half TGB1 coding sequence (incorporating WClMV TGB1 nucleotides 4381 to 4705) (PVX.GFP+W-TGB-3′). The 5′ half of the TGB1 coding sequence was used as a probe for the in situ hybridization experiments described in (D) and (E).
(D) and (E) Longitudinal sections through the shoot apex of spikey plants illustrating the presence (D) and absence (E) of TGB1 transgene RNA after inoculation with PVX.GFP or PVX.GFP+W-TGB-3′. Bars = 100 μm.
To confirm that the loss of the spikey phenotype was associated with a reduction in TGB1 transgene transcript level in the shoot apex, an additional experiment was designed to distinguish transgene- and virus-encoded TGB1 transcripts. A fragment derived from the 3′ half of TGB1 was inserted into a heterologous viral vector (PVX.GFP+W-TGB1-3′), which was used to infect and subsequently revert spikey plants. The corresponding 5′ half of TGB1 was used as the probe for in situ localization experiments (Figure 6C). As anticipated, transgene transcripts were undetectable in the reverted apices, consistent with virus-induced gene silencing in the shoot apex (Figures 6D and 6E).
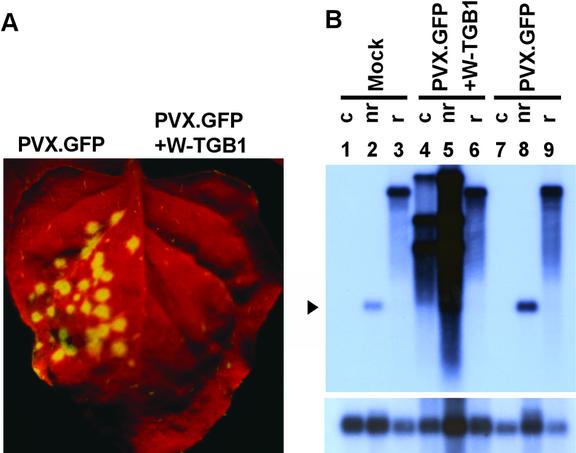
Earlier studies established that TGBp1 could suppress transgene-induced, but not virus-induced, local PTGS (Voinnet et al., 2000). In the present study, ectopic expression of TGB1 did not prevent viral induction of PTGS within the shoot apex. To further confirm the role played by PTGS in the reversion of spikey plants, we next sought to demonstrate that sequence-specific RNA degradation takes place in the reverted leaves. In contrast to the findings with PVX.GFP, infection could not be established with PVX.GFP+W-TGB1 (Figure 7A).
Figure 7.
Reverted Spikey Leaves Exhibit Sequence-Specific Degradation of WClMV TGB1.
(A) WClMV reverted spikey leaves were susceptible to PVX.GFP (left side of leaf) but resistant to infection by PVX.GFP+W-TGB1 (right side of leaf).
(B) RNA gel blot analysis performed using a WClMV TGB1-specific probe. RNA was extracted from control (c) N. benthamiana and WClMV reverted (r) spikey and nonreverted (nr) spikey leaves that were mock inoculated or inoculated with either PVX.GFP or PVX.GFP+W-TGB1. In mock-inoculation experiments, hybridization to WClMV TGB1 was not detected with control tissue (lane 1) but was observed with nonreverted spikey plants (lane 2). WClMV reverted spikey leaves revealed the presence of residual WClMV genomic RNA but the absence of WClMV TGB1 transcripts (lane 3). In control leaves of PVX.GFP+W-TGB1–inoculated tissues, the hybridization pattern expected for the presence of genomic and subgenomic PVX.GFP+W-TGB1 RNA was seen (lane 4); in nonreverted spikey leaves, note the presence of the same pattern of viral RNA species as in the control plus the transcript from the WClMV TGB1 transgene (lane 5); in WClMV reverted spikey leaves, only residual WClMV genomic RNA was detected (lane 6). In control leaves of PVX.GFP-inoculated tissues, no hybridization was detected (lane 7); in nonreverted spikey leaves, the probe detected the presence of transcripts from the WClMV TGB1 transgene (lane 8); in WClMV reverted spikey leaves, hybridization was confined to the residual WClMV genomic RNA (lane 9). As a loading control, membrane was stripped and reprobed with a 32P-labeled RbsC-specific probe (bottom). The arrowhead indicates the expected position of the WClMV TGB1 transcript.
RNA gel blot analysis using a WClMV TGB1-specific hybridization probe also was performed to further confirm the operation of PTGS directed against the WClMV TGB1 transgene (Figure 7B). No hybridization signal was detected against RNA extracted from wild-type N. benthamiana leaves (control [c], lane 1). As expected, the probe hybridized to TGB1 transgene transcripts extracted from nonreverted spikey leaves (nr, lane 2). Mock-inoculated WClMV-reverted spikey leaves revealed the presence of residual WClMV genomic RNA but an absence of WClMV TGB1 transgene transcripts (r, lane 3). This result was consistent with the operation of PTGS directed against the TGB1 transgene.
An additional set of hybridization experiments was conducted using RNA extracted from control tissue inoculated with PVX.GFP+W-TGB1. In this situation, we detected a hybridization pattern reflecting the presence of genomic and subgenomic viral RNA (Figure 7B, lane 4). Analysis of RNA extracted from nonreverted spikey leaves inoculated with PVX.GFP+W-TGB1 revealed the presence of the same viral RNA species plus the transcript from the WClMV TGB1 transgene (lane 5). By contrast, only residual WClMV genomic RNA was detected in PVX.GFP+W-TGB1–inoculated WClMV-reverted spikey tissues (lane 6). This result was consistent with the inability of PVX.GFP+W-TGB1 to establish infection in WClMV-reverted spikey leaves (Figure 7A), most likely as a result of the activation of PTGS directed against the WClMV TGB1 transgene.
Final proof of the involvement of PTGS was provided by infection studies using PVX.GFP. As expected, the probe did not hybridize to RNA extracted from control tissue inoculated with PVX.GFP (Figure 7B, lane 7). Detection of WClMV TGB1 transcripts in RNA obtained from PVX.GFP-infected nonreverted spikey leaves (lane 8) indicated that, because of the lack of homology between PVX.GFP and the WClMV TGB1 transgene, viral infection did not activate PTGS directed against the TGB1 transgene. Finally, only hybridization to residual WClMV genomic RNA was detected with RNA from PVX.GFP-inoculated WClMV-reverted spikey leaves (lane 9). Collectively, these data provide strong support for the hypotheses that, under the present experimental conditions, PTGS was induced in the shoot apex of spikey plants and that WClMV TGB1 sequence-specific RNA degradation was maintained in the subsequently initiated lateral organs.
To obviate the complications associated with viral infection in the shoot apex, we next used an Agrobacterium tumefaciens–mediated delivery system (Voinnet et al., 1998) to introduce double-stranded (ds) TGB1 (Figure 8A) to initiate a phloem-delivered systemic silencing signal. Reestablishment of leaf polarity by dsTGB1 confirmed that a phloem-mobile signal, initiated in the infiltrated cotyledons, could enter the spikey shoot apex to silence the TGB1 transgene (Figure 8B). As these dsTGB1-reverted spikey plants continued to grow, they exhibited a less severe (intermediate) spikey phenotype (data not shown). The efficacy of this delivery system was comparable to that reported for dsRNA-mediated silencing of transgenes in Arabidopsis plants (Chuang and Meyerowitz, 2000).
Figure 8.
Partial Reversion of the Spikey Phenotype by Transient Expression of dsTGB1.
(A) A binary vector designed to express an inverted repeat (linked by an intron) (Smith et al., 2000) to yield dsTGB1.
(B) Spikey plants were treated with either control (ds−) or dsTGB1 Agrobacterium. Approximately 50% of the dsTGB1-treated plants displayed normal patterns of development by 30 days after infiltration.
DISCUSSION
Recent studies on endogenous and viral RNA movement in plants support the concept of surveillance and selective RNA entry from the protophloem into the shoot apex (Ruiz-Medrano et al., 1999; Kim et al., 2001). Here, we provide direct evidence that ectopic expression of TGBp1 in the apex acts to compromise the capacity of the plant to control the entry of viral RNA into the shoot apex. To establish this point, we have used PTGS and reversion of a developmental phenotype to demonstrate the influence of virus on transgene expression within the shoot apex.
With a few notable exceptions (Ratcliff et al., 2001), plant viruses are excluded from the shoot apex (Matthews, 1991). One can speculate that this situation reflects the action of processes that protect the shoot apex, and hence the cell types that ultimately give rise to reproductive structures, from viral infection. It is important to note that spikey plants are infertile, whereas reverted plants produce viable seed. The geminivirus Tomato golden mosaic virus and the potyvirus Pea seed–borne mosaic virus are similarly excluded from the shoot apex, but unlike PVX, they induce the propagation of a systemic PTGS signal that exerts its effect in the shoot apex (Jones et al., 1998; Peele et al., 2001). These differences likely reflect the role played by TGB1 in blocking propagation of the systemic silencing signal into the shoot apex of N. benthamiana, rather than an inability of the potexviruses to induce PTGS within these specific tissues.
Reversion of the spikey phenotype is attributed to sequence-specific degradation of WClMV TGB1 transgene expression in the shoot apex. Small-nucleotide RNA fragments are associated with sequence-specific PTGS and RNA interference (Hamilton and Baulcombe, 1999; Zamore et al., 2000). Consistent with a role of PTGS in spikey reversion, small-nucleotide TGB1 antisense RNA fragments were detected in PVX.GFP+W-TGB1 reverted spikey leaves (data not shown). Future experiments will investigate the underlying events involved in the PTGS-mediated reversion of the spikey phenotype.
The molecular basis for the TGB1-mediated disruption of development in spikey plants remains to be elucidated. It is possible that the presence of TGBp1 in apical tissues causes a disruption in the cell-to-cell communication required to exert control over lateral organ development. Establishment of organ polarity likely requires the action of meristem-specific factors (Sussex, 1954, 1955; Snow and Snow, 1959) operating in a concentration-dependent manner to differentially activate PHABULOSA-like (PHB-like) genes (McConnell et al., 2001). PHB-like genes in turn suppress abaxial-promoting factors (Eshed et al., 2001). Gene products that promote abaxial cell fate in spikey plants likely include Nb-YABBY and KANADI family members (Eshed et al., 1999, 2001; Sawa et al., 1999; Siegfried et al., 1999).
Our in situ hybridization data clearly indicate that Nb-YABBY expression was relatively unaffected at the earliest stages of lateral organ development. However, Nb-YABBY expression was disrupted by P3, resulting in a patchy expression pattern. These data are consistent with TGBp1-mediated disruption in either PHB-like gene promotion of adaxial cell fate or the interaction between PHB-like genes and KANADI family members. As shown previously, TGBp1 on its own is dysfunctional at the level of trafficking through the plasmodesmata (Lough et al., 1998, 2000). However, ectopic expression of TGB1 in apical tissues could perturb the function of non-cell-autonomously acting genes involved in organ development.
Because viral movement proteins can be multifunctional, the ectopic expression of TGB1 also may act by influencing the translatability of genes required for the establishment of lateral organs. For example, the PVX TGBp1 has been proposed to bind the extreme 5′ end of the potexviral RNA to activate translatability (Atabekov et al., 2000). A link between PTGS and developmental regulation also can be made based on the phenotypic resemblance of spikey plants to the Arabidopsis mutant argonaute1 (ago1). AGO1 has been shown to be involved in both PTGS and the establishment of polarity in Arabidopsis (Bohmert et al., 1998; McConnell and Barton, 1998; Lynn et al., 1999; Fagard et al., 2000; Hammond et al., 2001). AGO1 belongs to a large gene family that includes a eukaryotic translation initiation factor (Fagard et al., 2000). It is tempting to speculate that AGO1 acts as a component of PTGS by mediating sequence-specific interference in translation and that ectopic expression of TGB1 disrupts this role. This could be achieved either by targeting transcripts for degradation or by regulating translation in a sequence-specific manner.
The present study also established that ectopic expression of TGB1 did not completely block the propagation of the systemic silencing signal after Agrobacterium-mediated dsRNA induction of PTGS in spikey cotyledons. The PVX TGBp1 was able to block the propagation of such systemic silencing signals (Voinnet et al., 2000), which appears to be in conflict with our current findings. Induction of PTGS directed against the TGB1 transgene by the dsRNA method would have resulted in a reduced level of TGBp1 within the infiltrated spikey cotyledons. Thus, we propose that a dynamic state was established between the reduced levels of TGBp1 and the block on systemic propagation of the TGB1-directed silencing signal. Partial reversion of the spikey phenotype was maintained over time, because the plants continued to grow, indicating that the systemic silencing signal had gained entry into the shoot apex. Clearly, although less efficient in the absence of viral infection, TGB1 appears to have acted to compromise the selective exclusion of the systemic silencing signal from the shoot apex.
Experimental evidence is accumulating in support of the concept that plants engage in the surveillance and selective entry of RNA into the shoot apex (Lucas et al., 2001). Figure 9 incorporates this concept into a model in which morphogenesis in the meristem depends on non-cell-autonomous communication to control the development of lateral organs. Leaf and flower development is orchestrated by signaling derived from both within the meristem (Sussex, 1954, 1955; Snow and Snow, 1959; Sessions et al., 2000; Eshed et al., 2001; McConnell et al., 2001) and via the phloem (Zeevaart, 1962; Zimmerman and Milburn, 1975; Voinnet and Baulcombe, 1997; Ruiz-Medrano et al., 1999; Kim et al., 2001). Transgene expression of WClMV TGB1 compromises the efficacy of the putative surveillance/selectivity system that controls the entry of signals into and exclusion of virus from the apex. Although the components responsible for regulating RNA delivery to the shoot apex remain to be defined (Lucas et al., 2001), this study provides an experimental foundation for their identification and characterization.
Figure 9.
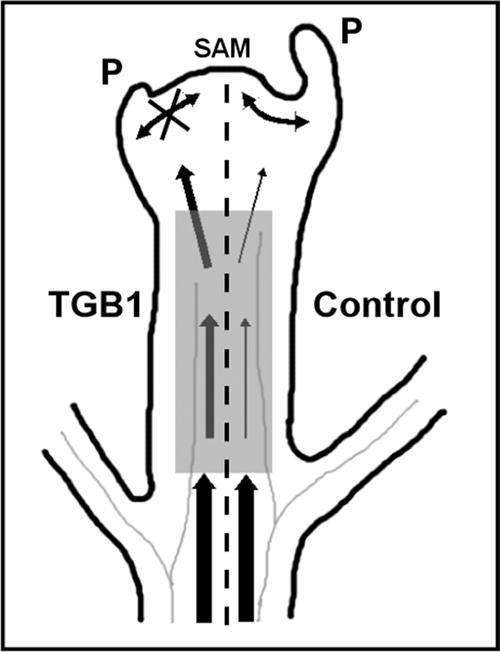
Effect of Potexvirus TGB1 on the Establishment of Leaf Polarity.
Model illustrating morphogenesis in the meristem and young leaf primordia involving non-cell-autonomous communication required for the development of normal lateral organs (right of the dashed line). Signaling that orchestrates development is derived from both within the meristem (double-headed arrows) and via the phloem (vertical arrows). WClMV TGB1 expression (left of the dashed line) compromises the selective entry of signals into and the exclusion of virus from the apex (the shaded box represents the zone of surveillance/selectivity). P, leaf primordia; SAM, shoot apical meristem.
METHODS
Cloning and RNA Gel Blot Analysis
All DNA manipulations were performed using standard techniques (Sambrook et al., 1988). Infectious White clover mosaic virus (WClMV) and Potato virus X (PVX; strain UK3) clones pWClMV-6 and pTXS.GFP-CP (green fluorescent protein–coat protein) have been described previously (Beck et al., 1990; Santa Cruz et al., 1996). The plasmid p806 was constructed by inserting a multiple cloning cassette (NsiI, SalI, and SmaI) into pTXS.GFP-CP downstream of the GFP terminator and upstream of a duplicated CP subgenomic promoter by a PCR-based mutagenesis procedure (kindly provided by Simon Santa Cruz, Scottish Crop Research Institute, Invergowrie, UK). This plasmid is referred to as pPVX.GFP throughout this study.
The plasmid pPVX.GFP+W-TGB1 was constructed by inserting WClMV TGB1 sequences, either nucleotides 4056 to 4722 or nucleotides 4381 to 4705, in the sense orientation as a PCR product flanked with XhoI sites into the compatible SalI site of the pPVX.GFP cloning cassette. All constructs were sequenced to confirm the nature of the engineered mutations. Nucleotide and amino acid sequences were analyzed using Genetics Computer Group (Madison, WI) version 10.0-UNIX programs (Devereux et al., 1984).
Infections established with the PVX or WClMV plasmids are referred to without the plasmid (p) prefix. Numbering of the nucleotide sequences of PVX and WClMV RNA is according to Huisman et al. (1988) and Beck et al. (1990), respectively. RNA gel blot analyses of uninoculated and inoculated leaves were performed using standard techniques (Sambrook et al., 1988; Beck et al., 1994). A randomly primed (RediprimeII; Amersham Pharmacia, Buckinghamshire, UK), 32P-labeled PCR product derived from WClMV TGB1 (nucleotides 4056 to 4722) was used as the hybridization probe.
In Situ Hybridization
In situ hybridizations were performed as described previously (Jackson et al., 1994). Nicotiana benthamiana apical tissues were excised from 2- to 4-week-old plants, fixed, dehydrated, and embedded in paraffin as described. PCR products derived from the TGB1 transgene (nucleotides 4056 to 4722), the 5′ half of the TGB1 transgene (nucleotides 3998 to 4380), WClMV replicase (nucleotides 3481 to 3951), and PVX replicase (nucleotides 3960 to 3301) were cloned into pGEM-EZ (Promega, Madison, WI). PCR fragments generated using external M13 forward and reverse primers allowed direct in vitro transcription of sense and antisense riboprobes from internal T7 or SP6 polymerase promoters.
Inoculation of PVX Transcripts
Transcripts were produced in vitro using T7 RNA polymerase, and plants were inoculated mechanically as described previously (Beck et al., 1990). Plants were maintained in a containment greenhouse, and GFP expression was detected with a hand-held, long-wavelength UV lamp (UVP, Upland, CA).
dsRNA TGB1-Mediated Reversion of Spikey Plants
The plasmid pRNA69 allows expression of sequences as an inverted repeat (linked by a YABBY5 intron). To facilitate Agrobacterium tumefaciens–mediated expression in plants, a NotI fragment incorporating the 35S promoter, the inverted repeat, and octopine synthase termination of transcription sequences was subcloned into the binary vector pART27 (Gleave, 1992). The plasmid pdsTGB1 was constructed by inserting WClMV TGB1 nucleotides 4056 to 4722 as a PCR product flanked with SalI-EcoRI or BamHI-NheI sites into either compatible XhoI-EcoRI or BamHI-XbaI sites, respectively.
The Agrobacterium inoculum for reversion experiments was derived from 40-mL cultures grown shaking for 36 h at 28°C in Luria-Bertani broth. These cultures were initiated from a starter culture grown from a single colony for 24 h. Bacteria were pelleted by centrifugation, washed in 10 mM MgCl2 at room temperature, and resuspended at an OD600 of 2.0 in 10 mM MgCl2 at room temperature. Spikey plants were treated with either control (pRNA69) or pdsTGB1 Agrobacterium. Cotyledons of 2- to 3-week-old seedlings were wounded mechanically using a hypodermic needle followed by infiltration of an Agrobacterium culture into the wounded tissues.
Fluorescence Microscopy Analysis
Visual detection of GFP fluorescence in whole plants was performed using a 100-W hand-held, long-wavelength UV Black Ray lamp (UVP). Photographs were taken with Kodak 100 ASA color slide film using an orange Hoya O (G) filter (Hoya, Tokyo, Japan). Analysis of the spatial distribution of GFP in plant tissues was performed 5 to 10 days after inoculation by epifluorescence microscopy (Leica MZFLIII stereomicroscope equipped with a DC200 digital camera; Wetzlar, Germany). Image analysis, display (adjustments in contrast, brightness, etc.), and preparation for plates were performed using Adobe Photoshop (Adobe Systems, Mountain View, CA).
Scanning Electron Microscopy
Two-week-old seedlings were dissected and fixed overnight in 3% glutaraldehyde in 0.1 M phosphate buffer, dehydrated in acetone, critical point dried in liquid CO2, and sputter coated with 25-nm gold using a super cool sputter coater (SCD-050; Bal-Tec, Balzers, Liechtenstein). Specimens were examined on a Cambridge 250 Mark III scanning electron microscope (Cambridge Instruments, Cambridge, UK) operated at 20 kV, and images were captured on 35-mm film.
Histology
Leaf tissue was fixed in 3% glutaraldehyde and 2% paraformaldehyde in 0.1 M phosphate buffer, infiltrated, and embedded in Procure 812 (ProSciTech, Kelso, Australia). Sections (1 μm) were cut, heat mounted, stained with 0.05% (w/v) toluidine blue, and photographed with an Axioplan microscope (Zeiss, Jena, Germany). Image analysis, display, and figure preparation were performed using Adobe Photoshop.
Plant Analysis
Histochemical assays for β-glucuronidase activity were performed as described by Jefferson (1989). Tissues were incubated in staining solution (50 mM sodium phosphate, pH 7.0, 0.2% Triton X-100, 2 mM potassium ferricyanide, 2 mM potassium ferrocyanide, and 0.5% X-GlcA-cyclohexylammonium salt [Duchefa, The Netherlands]) and incubated at 37°C for 12 h. Leaves excised from WClMV reverted plants (40 to 70 days after inoculation) were analyzed by sandwich ELISA (using WClMV CP-specific antiserum) to quantify virus accumulation using standard procedures (Clark and Adams, 1977). Original materials described in this article will be made available upon request.
Acknowledgments
Thanks are due to Brigitta Dudas for production of transgenic plants, Doug Hopcroft and Raymond Bennent for support with electron microscopy, Simon Santa Cruz for providing PVX.GFP constructs, Bruce Veit for assistance with in situ hybridization experiments, John Emery for pRNA69, and both Shou Wei Ding and Vicki Vance for providing information before publication. This work was supported by the Foundation of Research, Science, and Technology (Grant CO6816) and the National Science Foundation (Grant IBN 99-00539 to W.J.L. and Grant IBN 00-77984 to J.L.B.).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.001685.
References
- Anandalakshmi, R., Marathe, R., Ge, X., Herr, J.M.J., Mau, C., Mallory, A., Pruss, G., Bowman, L., and Vance, V.B. (2000). A calmodulin-related protein that suppresses posttranscriptional gene silencing in plants. Science 290, 142–144. [DOI] [PubMed] [Google Scholar]
- Anandalakshmi, R., Pruss, G.J., Ge, X., Marathe, R., Mallory, A.C., Smith, T.H., and Vance, V.B. (1998). A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA 95, 13079–13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atabekov, J.G., Rodionova, N.P., Karpova, O.V., Kozlovsky, S.V., and Poljakov, V.Y. (2000). The movement protein-triggered in situ conversion of potato virus X virion RNA from a nontranslatable into a translatable form. Virology 271, 259–263. [DOI] [PubMed] [Google Scholar]
- Beck, D.L., Forster, R.L., Bevan, M.W., Boxen, K.A., and Lowe, S.C. (1990). Infectious transcripts and nucleotide sequence of cloned cDNA of the potexvirus white clover mosaic virus. Virology 177, 152–158. [DOI] [PubMed] [Google Scholar]
- Beck, D.L., Van Dolleweerd, C.J., Lough, T.J., Balmori, E., Voot, D.M., Andersen, M.T., O'Brien, I.E., and Forster, R.L. (1994). Disruption of virus movement confers broad-spectrum resistance against systemic infection by plant viruses with a triple gene block. Proc. Natl. Acad. Sci. USA 91, 10310–10314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beclin, C., Berthome, R., Palauqui, J.C., Tepfer, M., and Vaucheret, H. (1998). Infection of tobacco or Arabidopsis plants by CMV counteracts systemic post-transcriptional silencing of nonviral (trans)genes. Virology 252, 313–317. [DOI] [PubMed] [Google Scholar]
- Bohmert, K., Camus, I., Bellini, C., Bouchez, D., Caboche, M., and Benning, C. (1998). AGO1 defines a novel locus of Arabidopsis controlling leaf development. EMBO J. 17, 170–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J.L., and Smyth, D.R. (1999). CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126, 2387–2396. [DOI] [PubMed] [Google Scholar]
- Brigneti, G., Voinnet, O., Li, W.X., Ji, L.H., Ding, S.W., and Baulcombe, D.C. (1998). Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17, 6739–6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chuang, C.F., and Meyerowitz, E.M. (2000). Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97, 4985–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, M.F., and Adams, A.N. (1977). Characteristics of the microplate method of enzyme-linked immunosorbent assay for the detection of plant viruses. J. Gen. Virol. 34, 475–483. [DOI] [PubMed] [Google Scholar]
- Devereux, J., Haeberli, P., and Smithies, O. (1984). A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12, 387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorlhac de Borne, F., Vincentz, M., Chupeau, Y., and Vaucheret, H. (1994). Co-suppression of nitrate reductase host genes and transgenes in transgenic tobacco plants. Mol. Gen. Genet. 243, 613–621. [DOI] [PubMed] [Google Scholar]
- Eddy, E.M. (1975). Germ plasm and the differentiation of the germ-cell line. Int. Rev. Cytol. 43, 229–280. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., Baum, S.F., and Bowman, J.L. (1999). Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99, 199–209. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., Baum, S.F., Perea, J.V., and Bowman, J.L. (2001). Establishment of polarity in lateral organs of plants. Curr. Biol. 11, 1251–1260. [DOI] [PubMed] [Google Scholar]
- Fagard, M., Boutet, S., Morel, J.B., Bellini, C., and Vaucheret, H. (2000). AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl. Acad. Sci. USA 97, 11650–11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave, A.P. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20, 1203–1207. [DOI] [PubMed] [Google Scholar]
- Guo, H.S., and Ding, S.W. (2002). A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J. 21, 398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, A.J., and Baulcombe, D.C. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286, 950–952. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M., Boettcher, S., Caudy, A.A., Kobayashi, R., and Hannon, G.J. (2001). Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293, 1146–1150. [DOI] [PubMed] [Google Scholar]
- Huisman, M.J.M., Linthorst, H.J., Bol, J.F., and Cornelissen, B.J.C. (1988). The complete nucleotide sequence of potato virus X and its homologies at the amino acid level with various plus-stranded RNA viruses. J. Gen. Virol. 69, 1789–1798. [DOI] [PubMed] [Google Scholar]
- Irish, V.F., and Sussex, I.M. (1992). A fate map of the Arabidopsis embryonic shoot apical meristem. Development 115, 745–753. [Google Scholar]
- Jackson, D., Veit, B., and Hake, S. (1994). Expression of maize KNOTTED-1 related genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120, 405–413. [Google Scholar]
- Jefferson, R.A. (1989). The GUS reporter gene system. Nature 342, 837–838. [DOI] [PubMed] [Google Scholar]
- Jegla, D.E., and Sussex, I.M. (1989). Cell lineage patterns in the shoot meristem of the sunflower embryo in the dry seed. Dev. Biol. 131, 215–225. [DOI] [PubMed] [Google Scholar]
- Jones, A.L., Thomas, C.L., and Maule, A.J. (1998). De novo methylation and co-suppression induced by a cytoplasmically replicating plant RNA virus. EMBO J. 17, 6385–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, R.A., Atkinson, R.G., Forster, R.L.S., and Lucas, W.J. (1998). An RNA-based information superhighway in plants. Science 279, 1486–1487. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D., and Carrington, J.C. (1998). A counterdefensive strategy of plant viruses: Suppression of posttranscriptional gene silencing. Cell 95, 461–470. [DOI] [PubMed] [Google Scholar]
- Kim, M., Canio, W., Kessler, S., and Sinha, N. (2001). Developmental changes due to long-distance movement of a homeobox fusion transcript in tomato. Science 293, 287–289. [DOI] [PubMed] [Google Scholar]
- Kooter, J.M., Matzke, M.A., and Meyer, P. (1999). Listening to the silent genes: Transgene silencing, gene regulation and pathogen control. Trends Plant Sci. 4, 340–347. [DOI] [PubMed] [Google Scholar]
- Li, H.W., Lucy, A.P., Guo, H.S., Li, W.X., Ji, L.H., Wong, S.M., and Ding, S.W. (1999). Strong host resistance targeted against a viral suppressor of the plant gene silencing defence mechanism. EMBO J. 18, 2683–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lough, T.J., Netzler, N.E., Emerson, S.J., Sutherland, P., Carr, F., Beck, D.L., Lucas, W.J., and Forster, R.L. (2000). Cell-to-cell movement of potexviruses: Evidence for a ribonucleoprotein complex involving the coat protein and first triple gene block protein. Mol. Plant-Microbe Interact. 13, 962–974. [DOI] [PubMed] [Google Scholar]
- Lough, T.J., Shash, K., Xoconostle-Cazares, B., Hofstra, K.R., Beck, D.L., Balmori, E., Forster, R.L.S., and Lucas, W.J. (1998). Molecular dissection of the mechanism by which potexvirus triple gene block proteins mediate cell-to-cell transport of infectious RNA. Mol. Plant-Microbe Interact. 11, 801–814. [Google Scholar]
- Lucas, W.J., Yoo, B.C., and Kragler, F. (2001). RNA as a long-distance information macromolecule in plants. Nat. Rev. Mol. Cell Biol. 2, 849–857. [DOI] [PubMed] [Google Scholar]
- Lucy, A.P., Guo, H.S., Li, W.X., and Ding, S.W. (2000). Suppression of post-transcriptional gene silencing by a plant viral protein localized in the nucleus. EMBO J. 19, 1672–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn, K., Fernandez, A., Aida, M., Sedbrook, J., Tasaka, M., Masson, P., and Barton, M.K. (1999). The PINHEAD/ZWILLE gene acts pleiotropically in Arabidopsis development and has overlapping functions with the ARGONAUTE1 gene. Development 126, 469–481. [DOI] [PubMed] [Google Scholar]
- Mallory, A.C., Ely, L., Smith, T.H., Marathe, R., Anandalakshmi, R., Fagard, M., Vaucheret, H., Pruss, G., Bowman, L., and Vance, V.B. (2001). HC-Pro suppression of transgene silencing eliminates the small RNAs but not transgene methylation or the mobile signal. Plant Cell 13, 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, R.E.F. (1991). Plant Virology, 3rd ed. (New York: Academic Press).
- McConnell, J.R., and Barton, M.K. (1998). Leaf polarity and meristem formation in Arabidopsis. Development 125, 2935–2942. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713. [DOI] [PubMed] [Google Scholar]
- Palauqui, J.C., Elmayan, T., Pollien, J.M., and Vaucheret, H. (1997). Systemic acquired silencing: Transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 16, 4738–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peele, C., Jordan, C.V., Muangsan, N., Turnage, M., Egelkrout, E., Eagle, P., Hanley-Bowdoin, L., and Robertson, D. (2001). Silencing of a meristematic gene using geminivirus-derived vectors. Plant J. 27, 357–366. [DOI] [PubMed] [Google Scholar]
- Poethig, R.S., Coe, E.H.J., and Johri, M.M. (1986). Cell lineage patterns in maize embryogenesis: A clonal analysis. Dev. Biol. 117, 392–404. [Google Scholar]
- Ratcliff, F., Martin-Hernandez, A.M., and Baulcombe, D.C. (2001). Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 25, 237–245. [DOI] [PubMed] [Google Scholar]
- Ruiz, M.T., Voinnet, O., and Baulcombe, D.C. (1998). Initiation and maintenance of virus-induced gene silencing. Plant Cell 10, 937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Medrano, R., Xoconostle-Cazares, B., and Lucas, W.J. (1999). Phloem long-distance transport of CmNACP mRNA: Implications for supracellular regulation in plants. Development 126, 4405–4419. [DOI] [PubMed] [Google Scholar]
- Saffman, E.E., and Lasko, P. (1999). Germline development in vertebrates and invertebrates. Cell. Mol. Life Sci. 55, 1141–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1988). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Santa Cruz, S., Chapman, S., Roberts, A.G., Roberts, I.M., Prior, D.A., and Oparka, K.J. (1996). Assembly and movement of a plant virus carrying a green fluorescent protein overcoat. Proc. Natl. Acad. Sci. USA 93, 6286–6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa, S., Watanabe, K., Goto, K., Liu, Y.G., Shibata, D., Kanaya, E., Morita, E.H., and Okada, K. (1999). FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes Dev. 13, 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions, A., Yanofsky, M.F., and Weigel, D. (2000). Cell-cell signaling and movement by the floral transcription factors LEAFY and APETALA1. Science 289, 779–782. [DOI] [PubMed] [Google Scholar]
- Seydoux, G., and Strome, S. (1999). Launching the germline in Caenorhabditis elegans: Regulation of gene expression in early germ cells. Development 126, 3275–3283. [DOI] [PubMed] [Google Scholar]
- Siegfried, K.R., Eshed, Y., Baum, S.F., Otsuga, D., Drews, G.N., and Bowman, J.L. (1999). Members of the YABBY gene family specify abaxial cell fate in Arabidopsis. Development 126, 4117–4128. [DOI] [PubMed] [Google Scholar]
- Smith, N.A., Singh, S.P., Wang, M.B., Stoutjesdijk, P.A., Green, A.G., and Waterhouse, P.M. (2000). Total silencing by intron-spliced hairpin RNAs. Nature 407, 319–320. [DOI] [PubMed] [Google Scholar]
- Snow, M., and Snow, R. (1959). The dorsiventrality of leaf primordia. New Phytol. 58, 188–207. [Google Scholar]
- Sussex, I.M. (1954). Experiments on the cause of dorsiventrality in leaves. Nature 174, 351–352. [DOI] [PubMed] [Google Scholar]
- Sussex, I.M. (1955). Morphogenesis in Solanum tuberosum L.: Experimental investigation of leaf dorsiventrality and orientation in the juvenile shoot. Phytomorphology 5, 286–300. [Google Scholar]
- Vance, V., and Vaucheret, H. (2001). RNA silencing in plants: Defense and counterdefense. Science 292, 2277–2280. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., and Baulcombe, D.C. (1997). Systemic signalling in gene silencing. Nature 389, 553. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., Lederer, C., and Baulcombe, D.C. (2000). A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103, 157–167. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., Pinto, Y.M., and Baulcombe, D.C. (1999). Suppression of gene silencing: A general strategy used by diverse DNA and RNA viruses of plants. Proc. Natl. Acad. Sci. USA 96, 14147–14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet, O., Vain, P., Angell, S., and Baulcombe, D.C. (1998). Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell 95, 177–187. [DOI] [PubMed] [Google Scholar]
- Waterhouse, P., Wang, M.-B., and Lough, T.J. (2001). Gene silencing: An adaptive defence against viruses. Nature 411, 834–842. [DOI] [PubMed] [Google Scholar]
- Xoconostle-Cázares, B., Xiang, Y., Ruiz-Medrano, R., Wang, H.L., Monzer, J., Yoo, B.C., McFarland, K.C., Franceschi, V.R., and Lucas, W.J. (1999). Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science 283, 94–98. [DOI] [PubMed] [Google Scholar]
- Zamore, P.D., Tuschl, T., Sharp, P.A., and Bartel, D.P. (2000). RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101, 25–33. [DOI] [PubMed] [Google Scholar]
- Zeevaart, Y.A.D. (1962). Physiology of flowering. Science 137, 723–731. [DOI] [PubMed] [Google Scholar]
- Zimmerman, H.M., and Milburn, J.A. (1975). Encyclopedia of Plant Physiology, New Series: Transport in Plants. I. Phloem Transport. (New York: Springer).