Abstract
Wild-type stomata are distributed nonrandomly, and their density is controlled by endogenous and exogenous factors. In the Arabidopsis mutant stomatal density and distribution1-1 (sdd1-1), the establishment of the stomatal pattern is disrupted, resulting in stomata clustering and twofold to fourfold increases in stomatal density. The SDD1 gene that encodes a subtilisin-like Ser protease is expressed strongly in stomatal precursor cells (meristemoids and guard mother cells), and the SDD1 promoter is controlled negatively by a feedback mechanism. The encoded protein is exported to the apoplast and probably is associated with the plasma membrane. SDD1 overexpression in the wild type leads to a phenotype opposite to that caused by the sdd1-1 mutation, with a twofold to threefold decrease in stomatal density and the formation of arrested stomata. While SDD1 overexpression was effective in the flp mutant, the tmm mutation acted epistatically. Thus, we propose that SDD1 generates an extracellular signal by meristemoids/guard mother cells and demonstrate that the function of SDD1 is dependent on TMM activity.
INTRODUCTION
During growth and development, all terrestrial plants form specialized epidermal structures called stomata. Stomata enable plants to adjust their gas exchange (i.e., H2O release and CO2 uptake) to suit surrounding environmental conditions by modulating the aperture of a pore delimited by two guard cells (GCs). However, regulating the density and distribution of stomata in the epidermis is as important as pore opening and closure in providing optimal gas flow.
Under natural growth conditions, stomata are distributed nonrandomly in almost all plant species (Willmer and Fricker, 1996). The presence of a stomata-free region surrounding each stoma is the major and universal principle of order in the established stomatal pattern (Sachs, 1991).
In Arabidopsis primary leaves, the bulk of stomatal complexes develop from single precursor cells (stomatal initials, also called meristemoid mother cells) via three rounds of asymmetric cell division (Berger and Altmann, 2000). The unequal division of a protodermal cell (stomatal initial) forms a primary meristemoid, a smaller, usually triangular daughter cell, and a neighboring cell. The meristemoid undergoes two more asymmetric divisions, finally resulting in the formation of a centrally located guard mother cell (GMC) surrounded by three neighboring cells. The GMC divides symmetrically into the two GCs, which then differentiate to acquire their unique structural and biochemical features (Pant and Kidwai, 1967; Larkin et al., 1997). The relevant cell types that occur during the different stages of stomatal development are shown in Figure 1. Thus, the cells belonging to a stomatal complex (GCs and the three neighboring cells) are in most cases clonally related derivatives of the stomatal initial (meristemoid mother cell).
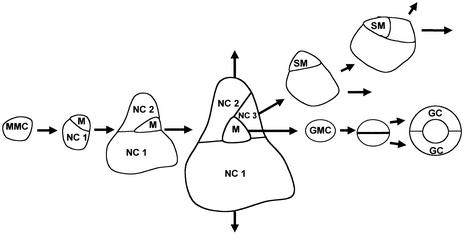
Figure 1.
Overview of Relevant Cell Types That Occur during the Different Stages of Stomatal Development.
A stomatal lineage is initiated by an unequal division of a stomatal initial, also called a meristemoid mother cell (MMC), creating a meristemoid (M). The meristemoid undergoes a series of additional (usually two) asymmetric divisions, after which it converts into a guard mother cell (GMC). The GMC surrounded by the clonally related neighboring cells (NC 1 to NC 3) divides symmetrically to produce two guard cells (GCs). Any neighboring cell (most frequently NC 3) also can divide asymmetrically to produce a satellite meristemoid (SM). SMs may undergo additional asymmetric divisions before they convert into GMCs and form satellite stomata. Reiteration of this process may result in the formation of even higher order stomatal complexes.
These findings, in accordance with cell lineage studies using transposon-induced sectors (Larkin et al., 1996; Serna and Fenoll, 2000; Serna et al., 2002), support the cell lineage hypothesis, which states that a series of asymmetric divisions leading to the formation of stomatal complexes establish the stomatal distribution pattern (Bünning, 1956). Consequently, the number and orientation of cell divisions that occur during stomatal complex formation are critical for correct stomatal pattern formation. Furthermore, patterning mistakes, such as adjacent meristemoids, have been observed. These may be corrected by either oriented asymmetric divisions or de/redifferentiation (Geisler et al., 2000) or by the formation of arrested stomata (Sachs et al., 1993; Sachs, 1994; Chin et al., 1995). Therefore, it is likely that cell–cell interactions play an additional important role in the formation of the final stomatal pattern.
In Brassicaceae, any of the neighboring cells of a primary stoma have the potential to divide asymmetrically to form a satellite meristemoid (SM), producing secondary, tertiary, or even higher order stomatal complexes (Pant and Kidwai, 1967). Like primary stomata, stomata of a higher order also are formed through a succession of asymmetric divisions, although these divisions vary from one to three in number (Berger and Altmann, 2000). The placement of the SM, the smaller product of the first asymmetric division, is opposite that of the previously formed meristemoid, GMC, or stomata and therefore plays an important role in establishing stomatal pattern (Berger and Altmann, 2000; Geisler et al., 2000). The SMs derive predominantly from the youngest (77%) neighboring cell and less frequently from the previously formed neighboring cell (Berger and Altmann, 2000).
The mutants stomatal density and distribution1-1 (sdd1-1), too many mouths (tmm), and four lips (flp), all of which are affected in stomatal differentiation and pattern formation, have been isolated in Arabidopsis (Yang and Sack, 1995; Berger and Altmann, 2000). The sdd1-1 mutant exhibits a twofold to fourfold increase in stomatal density in all aerial parts of the plant, and a fraction of the additional stomata occur in clusters (i.e., stomata placed in direct contact with each other). In the tmm mutant, the number of stomata is increased greatly, but in contrast to sdd1-1, the majority of additional stomata are arranged in large clusters predominantly in cotyledons and primary leaves. In contrast to sdd1-1 and tmm, the number of stomata is increased only slightly in flp, and the clusters that occur could be composed of even and odd numbers of GCs.
Analysis of serial dental resin imprints revealed a role of the SDD1 gene in the regulation of (1) the number of protodermal cells that form stomatal initials, (2) the number of asymmetric divisions of SMs, (3) the frequency with which neighboring cells undergo oriented asymmetric divisions to produce SMs, and (4) the orientation of the (first) asymmetric division of a neighboring cell (Berger and Altmann, 2000). These mechanisms are critical for controlling cell division patterns that lead to the differentiation of stomatal cells and their correct spacing. The orientation of asymmetric divisions and the frequency with which neighboring cells undergo asymmetric divisions that produce SMs also are controlled by TMM (Geisler et al., 2000), indicating that SDD1 and TMM are involved in similar mechanisms and may even act in the same signal transduction pathway. The appearance of clusters consisting of odd numbers of GCs in flp indicates a function of FLP in the control of GC/GMC identity (Larkin et al., 1997).
The SDD1 gene has been identified at the DNA level (Berger and Altmann, 2000). SDD1 encodes a 775–amino acid protein that shows homology with the subtilisin-like Ser proteases. In animals, proteases of this type activate precursors of hormones, growth factors, or receptors involved in the control of various developmental processes, including embryonic patterning via specific cleavage (Thacker et al., 1995; Cui et al., 1998; Steiner, 1998). In plants, little is known about possible subtilase substrates, but the high degree of similarity between the three catalytic domains and the substrate binding sites of animal and plant subtilases indicates that they share similar functions. Accordingly, SDD1 has been proposed to process factor(s) involved in the control of stomatal development (Berger and Altmann, 2000).
To further elucidate the molecular mechanisms that underlie the observed stomatal patterning processes, we describe the cellular and subcellular localization of SDD1, the activity of the SDD1 promoter in the wild type and in sdd1-1, and the effects of SDD1 overexpression in the wild type and in the mutants flp and tmm.
RESULTS
SDD1 Is Expressed Strongly in Meristemoids/GMCs
To investigate the cellular location of SDD1 mRNA in developing Arabidopsis seedlings and siliques, we performed RNA in situ hybridization experiments using an SDD1 antisense probe in 3-week-old Arabidopsis cv C24 plants and in developing siliques from mature Arabidopsis plants. The results showed that SDD1 was expressed predominantly in specialized cell types in the epidermis of developing leaves and siliques, whereas no SDD1 mRNA was detected in mature GCs (Figures 2A, 2C, 2D, and 2E). In paradermal sections of the epidermis, these specialized cell types were identified as meristemoids/GMCs (Ms/GMCs) according to their characteristic shape, as described in Zhao and Sack (1999) (Figure 2D). Additionally, weak expression of SDD1 mRNA was observed in the mesophyll cells of developing rosette leaves and leaf primordia and in all cell layers of the entire shoot apical meristem (Figures 2A and 2B).
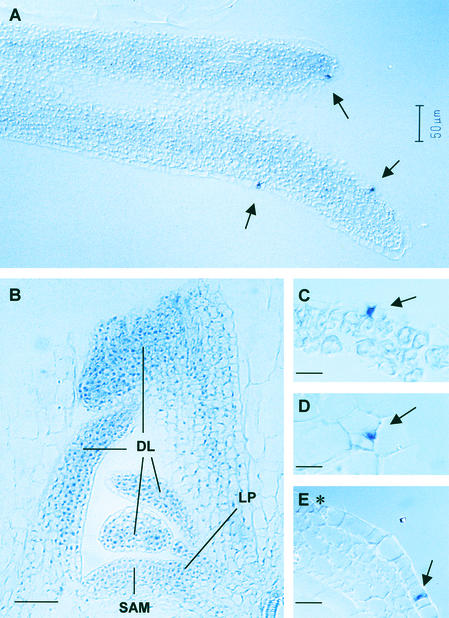
Figure 2.
In Situ Localization of SDD1 mRNA in Developing Rosette Leaves and Siliques of Arabidopsis.
Bright-field images of sections hybridized with digoxigenin-labeled antisense RNA probes of SDD1.
(A) to (C) Longitudinal sections through leaf primordia harvested 21 days after sowing. High levels of SDD1 mRNA are detectable in Ms/GMCs of the leaf epidermis (arrows in [A]), whereas low levels of SDD1 mRNA are present in mesophyll cells of leaf primordia and developing leaves as well as in the entire shoot apical meristem.
(D) Paradermal section through the leaf epidermis showing high levels of SDD1 mRNA in Ms/GMCs.
(E) Cross-sections through developing siliques showing high levels of SDD1 mRNA in Ms/GMCs of the silique epidermis but no hybridization of the SDD1 antisense probe to fully developed GCs.
Ms/GMCs are indicated by arrows, and fully developed GCs are indicated by asterisks. DL, developing leaf; LP, leaf primordia; SAM, shoot apical meristem. Bars = 50 μm in (B) and 10 μm in (C) to (E).
These observations were confirmed through analysis of four independent transgenic Arabidopsis wild-type cv C24 lines that harbored a fusion of the SDD1 promoter to the β-glucuronidase (GUS) reporter gene (pSDD1-GUS). The SDD1 promoter was particularly active during GC development. The highest level of GUS activity was detected in Ms/GMCs, whereas GUS activity decreased during GC formation and maturation. In fully developed leaves, almost no GUS staining was observed in mature stomata (Figures 3A and 3B).
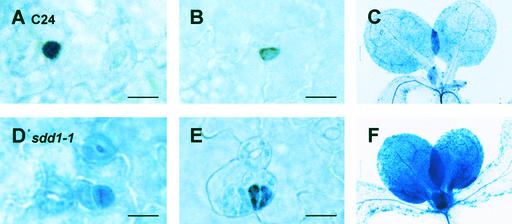
Figure 3.
GUS Staining Pattern in Developing Primary Leaves of 2-Week-Old Transgenic Arabidopsis Wild-Type and sdd1-1 Mutant Plants Carrying the pSDD1-GUS Transgene.
(A) to (C) Wild-type cv C24. β-Glucuronidase activity is strong and specific in the stomata precursor cells, the Ms and GMCs.
(D) to (F) Mutant sdd1-1. β-Glucuronidase activity is detected in dividing cells of developing leaves and leaf primordia; the activity is strongest in developing stomata.
Bars = 10 μm in (A), (B), (D), and (E) and 100 μm in (C) and (F).
The SDD1 Promoter Is Negatively Feedback Controlled
To investigate whether the SDD1 promoter is regulated by components of an SDD1-dependent signal transduction pathway, we compared the GUS staining pattern of transgenic Arabidopsis wild-type cv C24 and sdd1-1 mutant plants harboring the pSDD1-GUS construct. We selected four independent transgenic sdd1-1 mutant lines and compared the reporter gene activity with that in the wild type. In both the wild type and the sdd1-1 mutant, the SDD1 promoter was strongly active during GC development. The greatest GUS staining was observed in Ms/GMCs (Figures 3A, 3B, 3D, and 3E). In the sdd1-1 mutant background, however, strong GUS activity also was detected in the dividing cells of developing leaves and in leaf primordia (Figures 3C and 3F). The absence of repression of the SDD1 promoter in the sdd1-1 mutant, which lacks functional SDD1 protein, indicates that the SDD1 promoter is negatively feedback controlled by components of an SDD1-dependent signaling pathway.
The SDD1ct–Green Fluorescent Protein Fusion Protein Is Associated with the Plasma Membrane
SDD1 encodes a subtilisin-like Ser protease (Berger and Altmann, 2000). These proteases are expressed as preproprotein precursors, translocated via a signal (pre)peptide into the endomembrane system, and are activated through further cleavage of the propeptide (Siezen and Leunissen, 1997). Additional C- or N-terminal processing occurs in several subtilisins upon final maturation (Bogacheva, 1999; Janzik et al., 2000). The potentially complex processing of this protease poses difficulties in the design of translational green fluorescent protein (GFP) fusions to determine subcellular localization.
Because the exact processing sites within SDD1 were unknown, we created two different translational GFP fusions of SDD1. In the first construct (35S-SDD1-GFP), we fused the GFP C-terminal to the entire 2.3-kb coding region of SDD1 (Figure 4A). In the second construct (35S-SDD1ct-GFP), we fused the GFP C-terminal to a truncated 1.9-kb coding region of SDD1 lacking the region encoding the 121 C-terminal amino acids. This truncated SDD1 protein still harbors the common features of mature subtilisins: the catalytic triad, consisting of three highly conserved domains, and a fourth conserved domain, the substrate binding site (Dodson and Wlodawer, 1998) (Figure 4B). Both constructs were introduced into Arabidopsis wild-type cv C24.
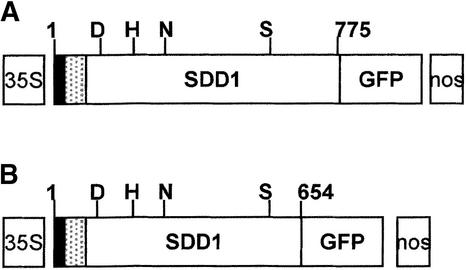
Figure 4.
Scheme of the Primary Structures of SDD1 Translational GFP Fusions.
The catalytic domain of mature SDD1 (white box) is preceded at the N terminus by a signal peptide and a propeptide (black and hatched boxes, respectively). The relative positions of the catalytically important Asp (D), His (H), Asn (N), and Ser (S) residues are indicated. nos, nopaline synthase terminator.
(A) GFP is fused to the C terminus of the entire SDD1 protein.
(B) C-terminally truncated version of the SDD1 protein fused to GFP.
Four independent transgenic lines overexpressing 35S-SDD1-GFP and 10 lines overexpressing 35S-SDD1ct-GFP were selected according to the results of protein gel blot analysis performed with an antibody raised against a synthetic SDD1 peptide (amino acids 586 to 599). In the 35S-SDD1-GFP–overexpressing lines, a minor 73-kD form and a predominant 63-kD form of the SDD1 protein were observed, whereas in all 35S-SDD1ct-GFP–overexpressing lines, an estimated 100-kD form of SDD1 was detected using the SDD1 peptide antibody (data not shown). The calculated molecular mass of the SDD1 protein without its prepropeptide is 73 kD, and the mass of the GFP protein is 27 kD. The expected molecular mass of the full-length SDD1-GFP fusion (derived from 35S-SDD1-GFP) after removal of the prepropeptide is 100 kD.
The predominant 63-kD form of SDD1 detected in the lines overexpressing 35S-SDD1-GFP probably represents a C-terminally processed form of the SDD1 protein. This assumption was confirmed using GFP antibodies that detected a 27-kD protein in these lines (data not shown). Accordingly, the GFP was liberated from the fusion. The 100-kD form of SDD1, which was observed in the lines overexpressing the SDD1ct-GFP fusion involving the C-terminally truncated SDD1 protein, is equivalent to the calculated molecular mass of the fusion protein without its prepeptide. This 100-kD protein also was detected using the anti-GFP antibody. These data demonstrate that in the 35S-SDD1ct-GFP lines, an SDD1ct-GFP fusion protein accumulated that lacked the prepeptide and thus probably was targeted to its correct destination.
In the 35S-SDD1ct-GFP–overexpressing lines, green fluorescence corresponding to GFP was localized to the plasma membrane (Figures 5A to 5D). Similar fluorescence patterns were observed when leaves were incubated for 1 to 12 h in a buffer at pH 7.0 in an attempt to neutralize the apoplast, because GFP is unstable at low pH (Haseloff et al., 1997) (Figures 5A and 5B). Plasmolysis with 1 M KNO3 confirmed the localization of the GFP fusion protein in association with the plasma membrane (Figure 5B). Because SDD1 has no predicted membrane-spanning domain, the 35S-SDD1ct-GFP fusion protein may be attached to the plasma membrane via additional factors.
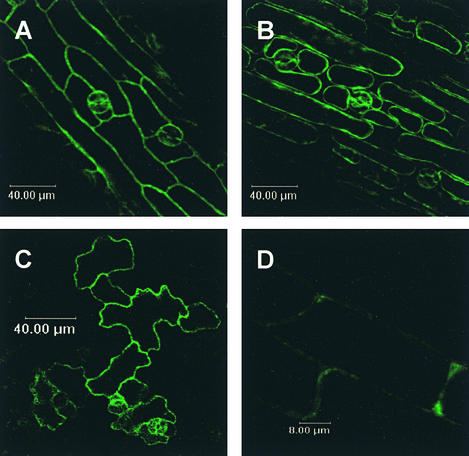
Figure 5.
Localization of 35S-SDD1ct-GFP in Developing Leaves of Arabidopsis.
The SDD1ct-GFP protein is localized to the plasma membrane.
(A), (B), and (D) Images from the midribs of developing leaves.
(C) Image from the leaf blade.
Leaves were taken from 21-day-old kanamycin-resistant plants of line SDD1ct-GFP #a22. In (A) and (B), the observed leaves were incubated in buffer (pH 7.0) for 12 h, and the leaf in (B) was further plasmolyzed thereafter with 1 M KNO3 for 5 min. In (C) and (D), the leaves were observed without any pretreatment.
Several attempts were made to use the SDD1 peptide antibody for immunolocalization of the SDD1 protein in tissue sections of young developing Arabidopsis leaves. Unfortunately, no antigen/antibody complexes were detected, indicating that the SDD1 peptide antibody is not suitable for immunolocalization. Thus, no independent proof of the localization of the native SDD1 protein is available.
SDD1 Overexpression Leads to a Twofold to Threefold Decrease in Stomatal Density and to the Formation of Arrested Stomata
To investigate the role of SDD1 during GC development and pattern formation, we prepared the 35S-SDD1-T7 construct (containing a chimeric gene created by the fusion of the 35S promoter of Cauliflower mosaic virus with the 2.3-kb open reading frame of SDD1 and a C-terminal T7 tag) and introduced it into Arabidopsis wild-type cv C24. The same construct was introduced into the sdd1-1 mutant as a control.
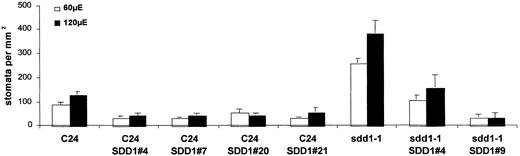
Stomatal density and differentiation were analyzed microscopically (the adaxial side of fully developed rosette leaves) in transformed plants selected for hygromycin resistance. Four independent transgenic wild-type lines and two transgenic sdd1-1 mutant lines overexpressing 35S-SDD1-T7 were selected according to this microscopic examination. The four transgenic wild-type lines exhibited a twofold to threefold reduction in stomatal density compared with the nontransgenic wild type (Figure 6). In transgenic line sdd1-1 SDD1#4, the stomatal density was reduced to wild-type levels, and in transgenic line sdd1-1 SDD1#9, a further twofold to threefold reduction of stomatal density was observed (Figure 6).
Figure 6.
Stomatal Density of Fully Developed Rosette Leaves of Transgenic Arabidopsis Plants Overexpressing SDD1 under Low- and High-Light Conditions.
Overexpression of SDD1 in the wild type and in the sdd1-1 mutant leads to a statistically significant decrease in stomatal density (Ryan-Einot-Gabriel-Welsch means test [P = 0.05]) that is independent of the light conditions used. The data represent average values of five individual plants ± sd.
With increasing light intensity, stomatal density increased in the wild type as well as in the sdd1-1 mutant and in most of the selected 35S-SDD1-T7–overexpressing lines, indicating that the adaptation of stomatal density to light is independent of the SDD1-controlled signal transduction pathway that negatively regulates stomatal density (Figure 6).
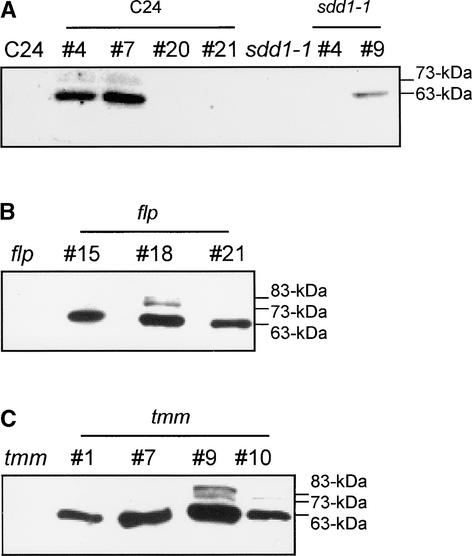
In the wild-type 35S-SDD1-T7–overexpressing lines C24 SDD1#4 and C24 SDD1#7 and in the transgenic line sdd1-1 SDD1#9, a minor 73-kD form and a predominant 63-kD form of SDD1 were detected via protein gel blot analysis using the SDD1 peptide antibody (Figure 7A). The fact that no SDD1 protein was detectable in the transgenic wild-type lines C24 SDD1#20 and C24 SDD1#21 but that a twofold to threefold decrease in stomatal density was observed indicates that minute amounts of additional SDD1 protein are sufficient to cause the developmental change. Using a T7 antibody against the C-terminal T7 tag fused to the SDD1 protein, no protein was detected in any of the transgenic lines (data not shown), supporting the results from the GFP fusion experiments that suggest that the SDD1 protein is C-terminally processed.
Figure 7.
Protein Blot Analysis of Transgenic Arabidopsis Plants Overexpressing SDD1.
(A) Analysis of transgenic wild-type and sdd1-1 mutant plants overexpressing SDD1.
(B) Analysis of transgenic flp mutant plants overexpressing SDD1.
(C) Analysis of transgenic tmm mutant plants overexpressing SDD1.
Total cellular protein was isolated from 21-day-old plants, and 40 μg of protein was separated using SDS-PAGE. The protein gel blot was probed with an antiserum (diluted 1:2000) raised against a peptide of SDD1.
For further analysis of stomatal density, stomatal index, and the differentiation status of the stomata, dental resin imprints of primary leaves of the transgenic wild-type lines C24 SDD1#4 and C24 SDD1#7 and the transgenic line sdd1-1 SDD1#9 were prepared and evaluated. A twofold to threefold reduction in stomatal density compared with the nontransgenic wild type was observed (Figures 8A and 8B). To determine whether stomatal density is affected by alterations in epidermal cell size and/or changes in the ratio of pavement cells to stomata, the stomatal indices were determined.
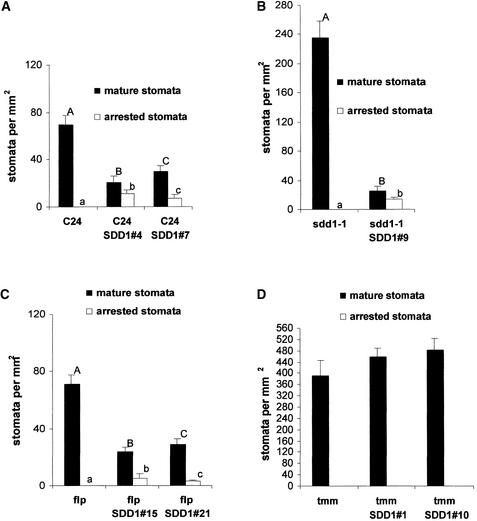
Figure 8.
Stomatal Density and Differentiation Status of Fully Developed Primary Leaves of Transgenic Arabidopsis Plants Overexpressing SDD1.
Density and differentiation status of the stomata were analyzed in nail polish copies prepared from dental resin imprints of the abaxial side of fully developed primary leaves.
(A) to (C) Overexpression of SDD1 in the wild type (A), the sdd1-1 mutant (B), and the flp mutant (C) leads to a twofold to threefold decrease in stomatal density and the formation of prematurely arrested stomata.
(D) Overexpression of SDD1 in the tmm mutant causes no phenotypic alterations.
The data represent average values of five individual plants ± sd. Letters in (A) to (C) indicate that the means are significantly different (Duncan means test [P = 0.05]).
The stomatal index shifted from 22.4% (on the abaxial surface of Arabidopsis wild-type cv C24 primary leaves) to 7.3 and 8.3% in the transgenic wild-type lines C24 SDD1#4 and C24 SDD1#7, respectively, and from 42.4% in the sdd1-1 mutant to 6.7% in the transgenic line sdd1-1 SDD1#9. Thus, in SDD1-overexpressing lines, the number of stomata was reduced as a result of a shift in the ratio of GCs to pavement cells. In addition to the twofold to threefold reduction in stomatal density, all three 35S-SDD1-T7–overexpressing lines exhibited the formation of arrested (at the M/GMC stage) stomata (Figures 9A to 9D). In the transgenic wild-type lines, 35% of C24 SDD1#4 and 19% of C24 SDD1#7 stomata were arrested at the M/GMC stage (Figure 8A). In the transgenic line sdd1-1 SDD1#9, 36% of the stomata were arrested (Figure 8B).
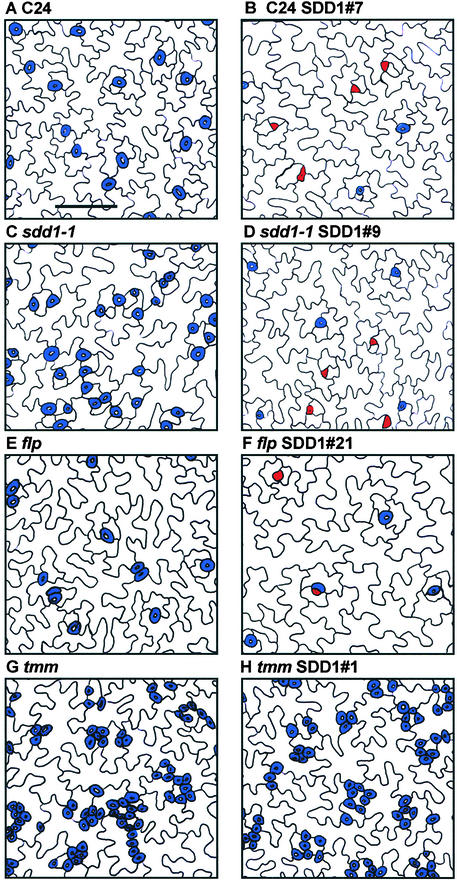
Figure 9.
Dental Resin Impressions of Fully Developed Primary Leaves of Transgenic Arabidopsis Plants Overexpressing SDD1.
Fully developed GCs are shown in blue, and stomata precursor cells (Ms and GMCs) are shown in red. Bar in (A) = 50 μm for (A) to (H).
(A) and (B) Impressions of a wild-type cv C24 primary leaf (A) and a primary leaf of a transgenic wild-type line (B) overexpressing SDD1.
(C) and (D) Impressions of a sdd1-1 mutant primary leaf (C) and a primary leaf of a transgenic sdd1-1 mutant line (D) overexpressing SDD1.
(E) and (F) Impressions of a flp mutant primary leaf (E) and a primary leaf of a transgenic flp mutant line (F) overexpressing SDD1.
(G) and (H) Impressions of a tmm mutant primary leaf (G) and a primary leaf of a transgenic tmm line (H) overexpressing SDD1.
The leaf size and the number of epidermis cells per leaf area of the analyzed primary leaves of the transgenic wild-type line C24 SDD1#4 did not differ from those of the nontransgenic wild type. The transgenic wild-type line C24 SDD1#7 had the same leaf size as the wild type, and the number of epidermis cells per leaf area was slightly higher compared with that in the wild type (Table 1).
Table 1.
Leaf Size and Epidermal Cell Number per mm2
| Line | Leaf Size (mm) (length/width) |
Epidermis Cells per mm2 |
|---|---|---|
| C24 | 6.18 ± 0.8/5.1 ± 1 | 240.24 ± 24.2 |
| C24 SDD1#4 | 5.26 ± 0.7/4.01 ± 0.5 | 247.03 ± 19.07 |
| C24 SDD1#7 | 5.7 ± 0.3/4.76 ± 0.3 | 324.72 ± 12.26 |
The data represent average values of five individual plants ± sd.
tmm but Not flp Acts Epistatically to SDD1
To determine whether the SDD1, TMM, and FLP genes are involved in common regulatory pathways, we monitored 35S-SDD1-T7 overexpression for alterations of stomatal characteristics in the flp and tmm mutants. We isolated three independent transgenic lines overexpressing 35S-SDD1-T7 in flp and four in tmm via protein gel blot analysis using the SDD1 peptide antibody (Figures 7B and 7C). Two transgenic lines were selected for each mutant and analyzed using dental resin imprints from primary leaves.
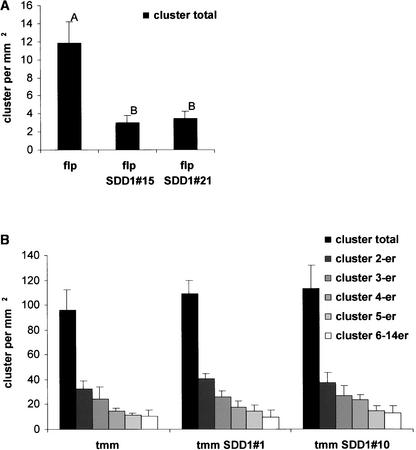
In the flp mutant, SDD1 overexpression led to the same effects as in the wild type and sdd1-1. The transgenic lines flp SDD1#15 and flp SDD1#21 showed twofold to threefold reductions in stomatal density. Nineteen percent and 11% of the stomata were arrested at the M/GMC stage in lines flp SDD1#15 and flp SDD1#21, respectively (Figures 8C, 9E, and 9F). The stomatal index shifted from 24.3% in the flp mutant to 10.2% in line flp SDD1#15 and 13.9% in line flp SDD1#21. Thus, as shown for wild-type cv C24 and sdd1-1, the ratio of GCs to pavement cells was altered. In addition, in lines flp SDD1#15 and flp SDD1#21, the fraction of stomata arranged in clusters was unchanged (Figure 10A). Accordingly, the effects of the loss of FLP function and increased SDD1 activity were additive.
Figure 10.
Clustering of Stomata in SDD1-Overexpressing flp and tmm Mutant Plants.
(A) Total cluster number in primary leaves of SDD1-overexpressing flp plants compared with nontransgenic flp plants.
(B) Phenotype of cluster size in primary leaves of SDD1-overexpressing tmm plants compared with nontransgenic tmm plants.
The data represent average values of five individual plants ± sd. Letters in (A) indicate that the means are significantly different (Duncan means test [P = 0.05]).
In sharp contrast to the effects of SDD1 overexpression in the wild type, in sdd1-1, or in flp, no differences in stomatal density or in average cluster size were observed between the tmm mutant and the transgenic lines tmm SDD1#1 and tmm SDD1#10 (Figures 8D and 10B). Furthermore, no arrested stomata occurred in these lines (Figures 9G and 9H). Thus, the tmm mutation acted completely epistatically to SDD1 overexpression.
DISCUSSION
Pattern formation is a fundamental aspect of development in all multicellular organisms. Stomata constitute a valuable plant system for the study of cell patterning because GCs are distributed nonrandomly in the epidermis, a two-dimensional tissue that can be examined easily.
In this study, we analyzed the localization and function of SDD1, a gene involved specifically in the regulation of stomatal differentiation and pattern formation (Berger and Altmann, 2000). SDD1 shows significant sequence similarity to subtilisin-like Ser proteases and has been proposed to act as a processing protease for which the (putative) substrate is unknown. Phenotypic analysis of the sdd1-1 mutant lacking functional SDD1 protein indicated that SDD1 is involved in several processes, including (1) the regulation of stomatal initial frequency in the developing epidermis, (2) the frequency of SM formation through asymmetric division of neighboring cells, (3) the number of asymmetric divisions of SMs, and (4) the orientation of the (first) asymmetric division of a neighboring cell (Berger and Altmann, 2000). This analysis demonstrated the involvement of SDD1 at several different stages of stomatal complex formation.
In the present study, SDD1 was shown to be expressed strongly in Ms/GMCs and weakly in developing leaf mesophyll cells and in the entire shoot apical meristem. Thus, neither protoderm cells nor neighboring cells, the developmental fates of which are affected by the action of SDD1, show detectable SDD1 expression. These findings can be explained readily by SDD1 involvement in signaling events between different cell types. According to this hypothesis, the SDD1-expressing mesophyll may exert control over protodermal cells, the developmental pathway of which is shifted from stomatal complex formation to differentiation into pavement cells. Thus, the loss of SDD1 activity and the concomitant lack of signal generation in the mesophyll would result in the observed increase in the fraction of protodermal cells that enter a stomatal cell lineage (from 35.3% in the wild type to 59.45% in the sdd1-1 mutant) (Berger and Altmann, 2000).
Similarly, the shift in the developmental fate of neighboring cells (from pavement cell to SM precursor) may be controlled by a signal created through an SDD1-dependent reaction in the Ms/GMCs, which show strong SDD1 expression. Within the limits of spatiotemporal expression analysis resolution (via in situ hybridization and reporter gene analysis), it appears that the timing of SDD1 expression coincides with the phase at which cell fate determination takes place in the majority of the neighboring cells. This is consistent with the proposed role of SDD1 in the generation of a signal that moves from Ms/GMCs to the neighboring cells and mediates cell-to-cell communication. This signal may either stimulate the development of neighboring cells into pavement cells or inhibit the conversion of neighboring cells into SM precursors.
Clearly, real-time high-resolution expression analysis will be necessary to determine the exact temporal order of events (the timing of SDD1 expression and developmental processes in the neighboring cells). Such an analysis also should reveal whether the role of SDD1 in regulating the number of asymmetric divisions in SMs involves cell-to-cell signaling or autonomous action. The proposed signaling from Ms/GMCs to neighboring cells is consistent with the observed role of SDD1 in the control of the orientation of asymmetric cell divisions in neighboring cells acting as satellite meristemoid mother cells.
The signal, probably generated in a SDD1-dependent manner in the Ms/GMCs, may provide a positional cue used by the neighboring cells for proper orientation of their (first) asymmetric division. Recent studies showing that cell fate is determined according to cell position and not cell ancestry indicate the likelihood that cell identity acquisition is dependent on cell–cell interactions (Jenik and Irish, 2000; Kidner et al., 2000) and support our hypothesis that the SDD1-dependent signal transduction pathway involves cell–cell interactions.
At present, the molecular nature of the proposed signal is unknown. According to the sequence similarity of SDD1 to eukaryotic subtilisin-like Ser proteases, it may act as a processing protease. In animal systems, these enzymes have been shown to exert their actions via specific cleavage of prohormones, growth factor precursors, or proreceptors involved in the regulation of various developmental processes (Thacker et al., 1995; Cui et al., 1998; Steiner, 1998). Plant subtilases are proposed to have a similar function (Berger and Altmann, 2000; Janzik et al., 2000). In view of this proposal, the signal in question may be a proteinaceous molecule activated through cleavage by SDD1 (Berger and Altmann, 2000). The putative extracellular but plasma membrane–associated localization of the SDD1 protein detected in this study is consistent with such a role. SDD1 may act as a processing protease that is exported into the apoplast, where it may interact with its substrates.
The necessity of precise temporal and spatial control of SDD1 expression was highlighted in this study by monitoring the activity of the SDD1 promoter in the wild type and in the sdd1-1 mutant and by the analysis of transgenic lines ectopically expressing SDD1 under the control of the 35S promoter of Cauliflower mosaic virus. In contrast to the situation in the wild type, in the sdd1-1 mutant, the SDD1 promoter was highly active not only in developing stomata but also in dividing cells of leaf primordia and developing leaves. In the wild type, the latter tissues exhibit only a transient weak expression of SDD1. Apparently, the presence of the SDD1 gene product in the wild type causes repression of the SDD1 promoter in the dividing cells of leaf primordia and developing leaves, except in Ms/GMCs. In other words, the SDD1 promoter is negatively feedback controlled by components of an SDD1-dependent signal transduction pathway, which may not be operative in the stomata precursor cells.
In transgenic plants overexpressing SDD1, this regulation is disrupted and SDD1 is expressed in all plant tissues. This deregulated SDD1 expression has dramatic consequences, causing a twofold to threefold reduction in stomatal density and the arrest of stomatal development at the M/GMC stage. It is likely that the reduction in stomatal density is caused by reduced SM production rather than by reduced initiation of stomatal cell lineages from protodermal cells.
This conclusion is based on the observation that leaf size and epidermal cell size are not changed considerably in the transgenic plants. Such changes would be expected if SDD1 overexpression caused reduced initiation of stomatal cell lineages, because the majority of epidermis cells (65 to 85% in leaves) are part of a stomatal lineage (Geisler et al., 2000). The reduced frequency of SM formation may be attributable to an enhanced production of the SDD1-dependent signal as a result of the ectopic overexpression of SDD1.
The observed arrest of stomatal precursors halted at the M/GMC stage is similar to the formation of arrested stomata in the monocots Tradescantia, Ruscus, and Aeonium and the dicots Anagallis and Pisum (Marks and Sachs, 1977; Sachs and Benouaiche, 1978; Kagan et al., 1992; Sachs et al., 1993; Chin et al., 1995). In Tradescantia and Pisum, arrested stomatal initials switch pathways and acquire the characteristics of epidermal cells (Kagan et al., 1992; Boetsch et al., 1995). In Arabidopsis as in Pisum, one of two adjacent meristemoids can arrest and further differentiate into an epidermal cell (Geisler et al., 2000). Whether the arrested stomatal initials observed in the SDD1-overexpressing plants acquired the characteristics of epidermal cells needs to be proven.
Furthermore, depending on when stomata arrest and their subsequent development, they might not be recognizable in mature organs. At present, it cannot be determined whether this response of the Ms/GMCs is attributable to the action of neighboring cells expressing SDD1 or extended expression in the Ms/GMCs themselves. The first possibility could be similar to the situation in which two Ms/GMCs, placed in direct contact with each other, results in one of the two arresting and dedifferentiating (Geisler et al., 2000). The other possibility is the occurrence of a sharp decrease in SDD1 expression upon GMC maturation to allow the completion of GC differentiation.
In any case, these observations highlight the importance of the SDD1 gene product during stomatal development and pattern formation. However, overexpression of SDD1 leads to a reduction in stomatal density but does not prevent the formation of stomata. Similarly, the fraction of protodermal cells that form stomatal initials is increased in sdd1-1, yet not all protodermal cells in sdd1-1 develop into stomatal initials. Thus, additional unknown independent signal transduction pathways may be operative in the control of protodermal cell fate.
The response of plants to SDD1 overexpression provided a means of studying epistatic relations between SDD1 and other known stomatal patterning genes such as FLP and TMM or mutant alleles thereof. To determine whether the SDD1, TMM, and FLP genes are involved in the same regulatory pathway, we examined the consequences of SDD1 overexpression in the tmm and flp mutants. This analysis revealed that SDD1 acts in a pathway independent of FLP. By contrast, none of the changes triggered in primary leaves and rosette leaves of the wild type occurred upon SDD1 overexpression in the tmm mutant. This epistatic relationship clearly demonstrates the necessity of TMM for SDD1 action and places the gene products of these two genes in the same signaling pathway.
However, according to the proposed function of SDD1 as a processing protease, the order of action of the two gene products cannot be deduced at present. Thus, TMM may be necessary for the synthesis of the substrate of SDD1 or may be involved in the perception/transduction of the SDD1-dependent signal. In any case, additional components are likely part of the proposed signaling pathway.
Precedence for signaling pathways involving extracellular proteinaceous signaling molecules (peptides) is provided by plant–pathogen interactions and by cell-to-cell communication in the shoot apical meristem. The bacterial elicitor flagellin and 15– to 22–amino acid peptides corresponding to the highly conserved N-terminal region of the Pseudomonas syringae pv tabaci flagellin have been shown to trigger defense responses in plants (Felix et al., 1999). The FLS2 Leu-rich repeat receptor-like kinase (LRR-RLK) of Arabidopsis is involved in this response (Gómez-Gómez and Boller, 2000) and has been shown to bind the active flagellin22 peptide (Gómez-Gómez et al., 2001). Like other LRR-RLKs (Stone et al., 1994; Braun et al., 1997; Trotochaud et al., 1999), FLS2 is regulated by the kinase-associated protein phosphatase. The CLAVATA (CLV) receptor complex provides an even more striking example of a mobile polypeptide. This complex is composed of the CLV1 LRR-RLK, which is linked to CLV2, a similar receptor-like protein that lacks a cytoplasmic signaling domain (Clark et al., 1997; Jeong et al., 1999). CLV3 encodes a small 76–amino acid polypeptide that is probably secreted and that acts as an essential ligand of CLV1 (Fletcher et al., 1999; Trotochaud et al., 2000). Additional components of the CLV complex are the aforementioned kinase-associated protein phosphatase and a Rho GTPase-related protein (Trotochaud et al., 1999). The CLV3 gene is expressed in the L1 and L2 layers, and the CLV3 polypeptide is proposed to diffuse to the CLV1-expressing cells in the L3 layer of the meristem (Clark et al., 1997; Fletcher et al., 1999). Because the CLV genes are key regulators of shoot apical meristem development and control the promotion of cells into differentiation, it is tempting to speculate that similar proteins may be operative in the SDD1-dependent signaling events involved in the control of stomatal development.
METHODS
Plant Material and Growth Conditions
tmm and flp seeds from a trichomeless gl1 mutant background of Arabidopsis thaliana (Columbia ecotype) were kindly provided by Fred D. Sack, Department of Plant Biology, Ohio State University, Columbus. The sdd1-1 mutant is in the Arabidopsis cv C24 background, which also carries a gl1 mutation (Berger and Altmann, 2000).
Plants cultivated in growth chambers were grown in standard soil (Einheitserde GS90; Gebrüder Patzer, Sinntal-Jossa, Germany) under a long-day light regime (16 h of fluorescent light [80 or 120 μmol·m−2·s−1] at 20°C and 60% RH/8 h of dark at 16°C and 75% RH). In tissue culture, seedlings up to 3 weeks old were grown in half-concentrated Murashige and Skoog (1962) medium supplemented with 1% Suc and solidified with 0.7% agar under a 16-h day (140 μmol·m−2·s−1, 22°C)/8-h night (22°C) regime.
Dental Resin Imprints
Arabidopsis wild-type cv C24, mutants sdd1-1, tmm, and flp, and the 35S-SDD1-T7–overexpressing lines were germinated and grown under 80 μmol·m−2·s−1 fluorescent light in soil. Dental resin imprints (Kagan et al., 1992; Berger and Altmann, 2000) were taken from the abaxial surfaces of fully developed primary leaves 1 and 2. Nail polish copies prepared from the dental resin imprints were analyzed by light microscopy and used to determine stomatal density and stomatal index. Data evaluation was based on dental resin imprints from five individual plants. For each plant, five separate fields of 0.31 mm2 were analyzed.
Statistics
The effect of SDD1 overexpression on the number of mature stomata, arrested stomata, and total clusters was tested by analysis of variance (SAS release 8.1; SAS Institute, Cary, NC). Where the effect of SDD1 on the dependent variable was significant (P < 0.05), means were compared by the Duncan means test (P = 0.05) or by the Ryan-Einot-Gabriel-Welsch means test (P = 0.05).
In Situ Hybridization
In situ hybridization was performed with 3-week-old Arabidopsis cv C24 seedlings grown in soil or in synthetic medium as described previously (http://www.edu/genetics/CATG/barton/protocols.html). A 388-bp BamHI-HindIII internal fragment of 35S-SDD1 (Berger and Altmann, 2000) was subcloned in pBluescript KS− (Stratagene), yielding pSDD1. This plasmid was linearized with XbaI, and antisense RNA was transcribed using T3 RNA polymerase. Sense control probes were synthesized using T7 RNA polymerase on pSDD1 linearized with SalI.
Construction of the SDD1 Promoter Fusion to the β-Glucuronidase Reporter Gene and Histochemical Localization of β-Glucuronidase Activity
To generate pSDD1–β-glucuronidase (GUS), the SDD1 promoter was inserted into pBI101.3 (Jefferson et al., 1987). The SDD1 promoter fragment ranged from position 60,961 to position 62,092 on BAC clone F20D22 and was amplified via Pfu DNA polymerase (Stratagene) with primers provided with additional XbaI and SmaI sites from BAC clone F20D22. After digestion of the PCR product and the vector with XbaI and SmaI, the SDD1 promoter fragment was inserted upstream of the β-glucuronidase (uidA) gene in pBI101.3. The construct was introduced into Arabidopsis cv C24 and the sdd1-1 mutant by Agrobacterium tumefaciens–mediated transformation (Bechtold et al., 1993). Transgenic lines transformed with these construct were selected using kanamycin.
5-Bromo-4-chloro-3 indolyl β-d-glucuronide was used to determine the localization of the enzyme activity of the GUS protein. Tissue samples were incubated at 37°C in GUS buffer (100 mM sodium phosphate buffer, pH 7.2, 0.1% Triton X-100, 2 mM K3[Fe(CN)6], and 0.5 mg/mL 5-bromo-4-chloro-3 indolyl β-d-glucuronide) for 2 to 15 h. After detection of the blue color, chlorophyll was extracted with 80% ethanol for 24 h.
Generation of SDD1 Translational Green Fluorescent Protein Fusions and Analysis of Transgenic Plants
To generate 35S-SDD1-green fluorescent protein (GFP), a fragment amplified with primers 5′-GCGTCGACATGGAACCCAAACCTT-TCTTTC-3′ (forward) and 5′-CTAGACTAGTACGTTAGTCTTCAA-GGTTAC-3′ (reverse) from BAC F20D22, which covered the 2328-bp SDD1 coding region and was provided with SalI linker sequences at the 5′ end and with SpeI linker sequences at the 3′ end, was inserted into the SalI- and SpeI-digested vector 35S-10H-GFP-JFH1 (Hong et al., 1999).
A second construct, called 35S-SDD1ct-GFP, was created by ligation of a fragment amplified with primers 5′-GCGTCGACATGGAACCCAAACCTTTCTTTC-3′ (forward) and 5′-GACTAGTACGCTCAC-GTTCTTATGAGTGATTGCTA-3′ (reverse) from the same BAC (F20D22) and provided with SalI linker sequences at the 5′ end and with SpeI linker sequences at the 3′ end into the SalI- and SpeI-digested vector 35S-10H-GFP-JFH1. In this construct, the SDD1 coding region is C-terminally truncated and terminates at position 658 of the deduced amino acid sequence. The constructs were introduced into Arabidopsis cv C24 by Agrobacterium-mediated transformation (Bechtold et al., 1993). Transgenic lines transformed with these constructs were selected using kanamycin.
Subcellular localization of 35S-SDD1ct-GFP in transformed Arabidopsis plants was examined using the Leica DM IRB confocal laser scanning microscope equipped with the Leica TCS SPII true confocal scanner and a HC PLAPO ×20/0.70 immersion objective (Leica, Wetzlar, Germany). Image acquisition and processing were performed using Leica confocal software. To visualize GFP in the apoplast, the leaves were incubated in 20 mM Pipes-KOH, pH 7.0, for 1 to 12 h on half-concentrated Murashige and Skoog (1962) medium supplemented with 1% Suc and solidified with 0.7% agar. For plasmolysis, the leaves were soaked in 1 M KNO3 for 5 min.
Generation of SDD1-Overexpressing Plants
The construct 35S-SDD1-T7 was created by ligation of a fragment amplified with primers 5′-ATGGAACCCAAACCTTTCTTT-3′ (forward) and 5′-ACCCGGTCTAGAACCCATTTGCTGTCCACCAGTCATGCT-AGCCATGTTAGTCTTCAAGGTTAC-3′ (reverse) from BAC F20D22, which carries the 2.3-kb SDD1 coding region, into the SmaI- and XbaI-digested vector pBluescript KS− (Stratagene) and subcloned into the Asp718- and XbaI-digested binary vector pBinAR-Hyg (Höfgen and Willmitzer, 1990). The reverse primer was provided with a T7 tag (underlined). The construct was introduced into Arabidopsis wild-type cv C24 and the mutants sdd1-1, tmm, and flp using Agrobacterium-mediated plant transformation (Bechtold et al., 1993). Transgenic lines transformed with this construct were selected using hygromycin.
Preparation of Protein Extracts and Immunoblotting
Total cellular protein extracts for use directly in immunoblotting experiments were prepared by homogenizing plant leaf material in a protein extraction buffer (20 mM Tris, pH 7.5, 10% [v/v] glycerol, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, and 0.5% Triton X-100). The samples were clarified in a microfuge, and the supernatant was collected. Protein concentration was measured by the method of Bradford (1976), and samples were stored at −20°C until further use.
Immunoblotting was conducted using standard laboratory procedures (Sambrook et al., 1989). An affinity-purified peptide antibody against the epitope DLYDRQGKAIKDGNK of SDD1 (generated by Eurogentec, Seraing, Belgium) was used at a dilution of 1:2000 in combination with the goat anti-rabbit IgG/peroxidase conjugate diluted 1:50,000 (Peribo Science, Bonn, Germany).
Acknowledgments
We thank F. Sack for providing the tmm and flp seeds, L. Bartetzko and H. Kulka for plant care, C. Schönberg and M. Preller for technical support, J. Bergstein for photographic work, M. McKenzie for critical reading of the manuscript, and Karin Köhl for the statistical analysis of the data. This work was supported by Grant Al 1-3 from the Deutsche Forschungsgemeintschaft.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.001016.
References
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris 316, 1194–1199. [Google Scholar]
- Berger, D., and Altmann, T. (2000). A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev. 14, 1119–1131. [PMC free article] [PubMed] [Google Scholar]
- Boetsch, J., Chin, J., and Croxdale, J. (1995). Arrest of stomatal initials in Tradescantia is linked to the proximity of neighbouring stomata and results in the arrested initials acquiring properties of epidermal cells. Dev. Biol. 168, 28–38. [DOI] [PubMed] [Google Scholar]
- Bogacheva, A.M. (1999). Plant subtilisins. Biochemistry 3, 287–293. [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–252. [DOI] [PubMed] [Google Scholar]
- Braun, D.M., Stone, J.M., and Walker, J.C. (1997). Interaction of the maize and Arabidopsis kinase interaction domains with a subset of receptor-like protein kinases: Implications for transmembrane signalling in plants. Plant J. 12, 83–95. [DOI] [PubMed] [Google Scholar]
- Bünning, E. (1956). General processes of differentiation. In The Growth of Leaves, F.L. Milthorpe, ed (London: Butterworths Scientific Publications), pp. 18–30.
- Chin, J.C., Wan, Y., Smith, J., and Croxdale, J. (1995). Linear aggregations of stomata and epidermis cells in Tradescantia leaves: Evidence for their group patterning as a function of the cell cycle. Dev. Biol. 168, 39–46. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Williams, R.W., and Meyerowitz, E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585. [DOI] [PubMed] [Google Scholar]
- Cui, Y., Jean, F., Thomas, G., and Christian, J.L. (1998). BMP-4 is proteolytically activated by furin and/or PC6 during vertebrate embryonic development. EMBO J. 16, 4735–4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson, G., and Wlodawer, A. (1998). Catalytic triads and their relatives. Trends Biochem. Sci. 23, 347–352. [DOI] [PubMed] [Google Scholar]
- Felix, G., Duran, J.D., Volko, S., and Boller, T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18, 265–276. [DOI] [PubMed] [Google Scholar]
- Fletcher, J.C., Brand, U., Running, M.P., Simon, R., and Meyerowitz, E.M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911–1914. [DOI] [PubMed] [Google Scholar]
- Geisler, M., Nadeau, J., and Sack, F.D. (2000). Oriented asymmetric divisions that generate the stomatal spacing pattern in Arabidopsis are disrupted by too many mouths mutation. Plant Cell 12, 2075–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez, L., Bauer, Z., and Boller, T. (2001). Both the extracellular leucine-rich repeat and the kinase activity of FLS2 are required for flagellin binding and signalling in Arabidopsis. Plant Cell 13, 1155–1163. [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez, L., and Boller, T. (2000). FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 5, 1003–1011. [DOI] [PubMed] [Google Scholar]
- Haseloff, J., Siemering, K., Prasher, D., and Hodge, S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94, 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfgen, R., and Willmitzer, L. (1990). Biochemical and genetic analysis of different patatin isoforms expressed in various organs of potato Solanum tuberosum. Plant Sci. 66, 221–230. [Google Scholar]
- Hong, B., Ichida, A., Wang, Y., Scott Gens, J., Pickard, B.G., and Harper, J.F. (1999). Identification of a calmodulin-regulated Ca2+-ATPase in the endoplasmic reticulum. Plant Physiol. 119, 1165–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzik, I., Macheroux, P., Amrhein, N., and Schaller, A. (2000). LeSBT1, a subtilase from tomato plants. J. Biol. Chem. 7, 5193–5199. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 13, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenik, P.D., and Irish, V.F. (2000). Regulation of cell proliferation patterns by homeotic genes during Arabidopsis floral development. Development 127, 1267–1276. [DOI] [PubMed] [Google Scholar]
- Jeong, S., Trotochaud, A.E., and Clark, S.E. (1999). The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11, 1925–1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan, M.L., Novoplansky, N., and Sachs, T. (1992). Variable cell lineages form the functional pea epidermis. Ann. Bot. 69, 303–312. [Google Scholar]
- Kidner, C., Sundaresan, V., Roberts, K., and Dolan, L. (2000). Clonal analysis of the Arabidopsis root confirms that position, not lineage, determines cell fate. Planta 211, 191–199. [DOI] [PubMed] [Google Scholar]
- Larkin, J.C., Marks, M.D., Nadeau, J., and Sack, F.D. (1997). Epidermal cell fate and patterning in leaves. Plant Cell 9, 1109–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin, J.C., Young, N., Prigge, M., and Marks, M.D. (1996). The control of trichome spacing and number in Arabidopsis. Development 122, 997–1005. [DOI] [PubMed] [Google Scholar]
- Marks, M.G., and Sachs, T. (1977). The determination of stomata pattern and frequency in Anagallis. Bot. Gaz. 138, 385–392. [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Pant, D.D., and Kidwai, P.F. (1967). Development of stomata in some Cruciferae. Ann. Bot. 31, 513–521. [Google Scholar]
- Sachs, T. (1991). Pattern formation in plant tissues. In Developmental and Cell Biology Series, P.W. Barlow, D. Bray, P.B. Green, and J.M.W. Slack, eds (Cambridge, UK: Cambridge University Press), pp. 107–109.
- Sachs, T. (1994). Both cell lineages and cell interactions contribute to stomatal patterning. Int. J. Plant Sci. 155, 245–247. [Google Scholar]
- Sachs, T., and Benouaiche, P. (1978). A control of stomata maturation in Aeonium. Isr. J. Bot. 27, 47–53. [Google Scholar]
- Sachs, T., Novoplansky, N., and Kagan, M. (1993). Variable development and cellular patterning in the epidermis of Ruscus. Ann. Bot. 71, 237–243. [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Serna, L., and Fenoll, C. (2000). Stomatal development and patterning in Arabidopsis leaves. Physiol. Plant. 109, 351–358. [Google Scholar]
- Serna, L., Torres-Contreras, J., and Fenoll, C. (2002). Clonal analysis of stomatal development and patterning in Arabidopsis leaves. Dev. Biol. 241, 24–33. [DOI] [PubMed] [Google Scholar]
- Siezen, R.J., and Leunissen, J.A.M. (1997). Subtilases: The superfamily of subtilisin-like serine proteases. Protein Sci. 6, 501–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner, D.F. (1998). The proprotein convertases. Curr. Opin. Chem. Biol. 2, 31–39. [DOI] [PubMed] [Google Scholar]
- Stone, J.M., Collinge, M.A., Smith, R.D., Horn, M.A., and Walker, J.C. (1994). Interaction of a protein phosphatase with an Arabidopsis serine-threonine receptor kinase. Science 266, 793–795. [DOI] [PubMed] [Google Scholar]
- Thacker, C., Peters, K., Stayko, M., and Rose, A.M. (1995). The bli-4 locus of Caenorhabditis elegans encodes structurally distinct kex2/subtilisin-like endoproteases essential for early development and adult morphology. Genes Dev. 9, 956–971. [DOI] [PubMed] [Google Scholar]
- Trotochaud, A.E., Hao, T., Wu, G., Yang, Z., and Clark, S.E. (1999). The CLAVATA1 receptor-like kinase requires CLAVATA3 for its assembly into a signalling complex that includes KAPP and a Rho-related protein. Plant Cell 11, 393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotochaud, A.E., Jeong, S., and Clark, S.E. (2000). CLAVATA3, a multimeric ligand for the CLAVATA1 receptor-kinase. Science 289, 613–617. [DOI] [PubMed] [Google Scholar]
- Willmer, C., and Fricker, M. (1996). Stomata. In Topics in Plant Functional Biology, Vol. 2, M. Black and B. Charlwood, eds (London: Chapman and Hall), pp. 95–125.
- Yang, M., and Sack, F.D. (1995). The too many mouths and four lips mutations affect stomatal production in Arabidopsis. Plant Cell 7, 2227–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, L., and Sack, F.D. (1999). Ultrastructure of stomatal development in Arabidopsis (Brassicaceae) leaves. Am. J. Bot. 86, 929–939. [PubMed] [Google Scholar]