Abstract
Chloroplast DNA (cpDNA) is packed into discrete structures called chloroplast nucleoids (cp-nucleoids). The structure of cpDNA is thought to be important for its maintenance and regulation. In bacteria and mitochondria, histone-like proteins (such as HU and Abf2, respectively) are abundant and play important roles in DNA organization. However, a primary structural protein has yet to be found in cp-nucleoids. Here, we identified an abundant DNA binding protein from isolated cp-nucleoids of the primitive red alga Cyanidioschyzon merolae. The purified protein had sequence homology with the bacterial histone-like protein HU, and it complemented HU-lacking Escherichia coli mutants. The protein, called HC (histone-like protein of chloroplast), was encoded by a single gene (CmhupA) in the C. merolae chloroplast genome. Using immunofluorescence and immunoelectron microscopy, we demonstrated that HC was distributed uniformly throughout the entire cp-nucleoid. The protein was expressed constitutively throughout the cell and the chloroplast division cycle, and it was able to condense DNA. These results indicate that HC, a bacteria-derived histone-like protein, primarily organizes cpDNA into the nucleoid.
INTRODUCTION
It is believed that chloroplasts and mitochondria arose from prokaryotic endosymbionts during eukaryotic evolution. According to this concept, chloroplasts originated from cyanobacteria (Margulis, 1970; Gray, 1992) and therefore have their own DNA. The chloroplast genome (cp-genome) often is divided into highly condensed, discrete structures called chloroplast nucleoids (cp-nucleoids) (Hewson and Wolstenholme, 1969; Kuroiwa et al., 1981; Kuroiwa, 1982; Yurina et al., 1995).
The cp-nucleoids are mainly classified into three types according to differences in their shape, size, and distribution (Kuroiwa et al., 1981; Kuroiwa, 1991; Gillham, 1994). The first type, the cp-nucleoids of primitive red algae, are located centrally in the chloroplast and are surrounded by thylakoids. Bacteria, including the chloroplast ancestor cyanobacteria, also have nucleoids that are restricted to the center. The second type is a large, ring-shaped cp-nucleoid found within the girdle lamellae (e.g., brown algae). In green plants and many green algal groups, the third type of cp-nucleoids are small, uniformly dispersed nucleoids found in the matrix between the thylakoids. Proteinase or trypsin treatment of ring-shaped cp-nucleoids isolated from brown algae suggests the presence of proteins that assemble chloroplast DNA (cpDNA) into cp-nucleoids (Kuroiwa and Suzuki, 1981), and it is now known that cp-nucleoids contain a large number of proteins (Nemoto et al., 1991; Yurina et al., 1995).
Accordingly, it is essential to investigate nucleoid proteins to understand the function of cp-nucleoids. The organization of cpDNA may offer a key to understanding the mechanism of gene expression in the cp-genome. However, little is known about the structures or mechanisms of action of proteins that bind to and organize cpDNA topology (Nemoto et al., 1991; Liu and Rose, 1992; Sato et al., 1993; Itoh et al., 1997).
The fundamental unit of chromatin in the cell nucleus is the nucleosome. Within the nucleosome, DNA is wrapped approximately twice around the histone core (Wolffe, 1998). The bacterial genome also is assembled into a nucleoid structure, and the abundant histone-like protein HU is involved in assembly (Rouviere-Yaniv and Gros, 1975; Drlica and Rouviere-Yaniv, 1987; Oberto and Rouviere-Yaniv, 2001). In mitochondria, Abf2 is an abundant basic protein that supercoils DNA (Diffley and Stillman, 1991). In chloroplasts, three proteins have been isolated from the cp-nucleoids: a 70-kD protein from pea (Sato et al., 1997), CND41 from tobacco (Nakano et al., 1997), and plastid-envelope DNA binding (PEND) protein from pea (Sato et al., 1993).
The 70-kD pea protein was dissociated selectively from nucleoids by salt washing and was identified as sulfite reductase (Sato et al., 2001). CND41 is a 41-kD DNA binding protein (Nakano et al., 1997) that is expressed abundantly in tobacco chloroplasts. The 130-kD PEND protein binds to DNA and was isolated from the envelope membrane of developing pea chloroplasts (Sato et al., 1998). Using nucleotide sequence analysis of the cryptomonad Guillardia theta (formerly Cryptomonas), Wang and Liu (1991) found a chloroplast gene (hlpA) that encodes a protein similar to the bacterial histone-like protein HU.
However, at present, there is no evidence to suggest that these proteins localize in vivo at cp-nucleoids or that they are involved in the organization or maintenance of the chloroplast nucleoid structure. Nucleoids are dispersed throughout the chloroplasts in higher plants such as pea and tobacco. As a result, the cp-nucleoid fraction is easily contaminated with fragments from cell nuclei during nucleoid isolation. Moreover, it is possible for basic proteins to contaminate isolated cp-nucleoids because DNA is negatively charged. Uncontaminated nucleoid fractions may be obtained from organisms with a simple structure that have cp-nucleoids concentrated at the center of chloroplasts.
Here, we report a DNA binding protein purified from the chloroplasts of the primitive red alga Cyanidioschyzon merolae. C. merolae is ideal for both cp-nucleoid protein isolation and the study of cpDNA interactions for the following reasons: (1) cp-nucleoids are restricted to the center of chloroplasts and resemble bacterial nucleoids; (2) mitotic and organelle division cycles can be highly synchronized by light/dark cycles (Suzuki et al., 1994); (3) the complete nucleotide sequence of its mitochondrial (Ohta et al., 1998) and chloroplast (Ohta et al., 1999) genomes have been determined; and (4) a method for isolating chloroplasts has been established (Miyagishima et al., 1999).
We isolated an abundant cpDNA binding protein from C. merolae. The protein was very similar to the bacterial histone-like protein HU and was named HC (histone-like protein of chloroplast). We demonstrate that the HC localized within cp-nucleoids is a cpDNA binding protein. HC has DNA-compacting activity and complements the deletion of HU in Escherichia coli.
RESULTS
Identification of DNA Binding Proteins in cp-Nucleoids
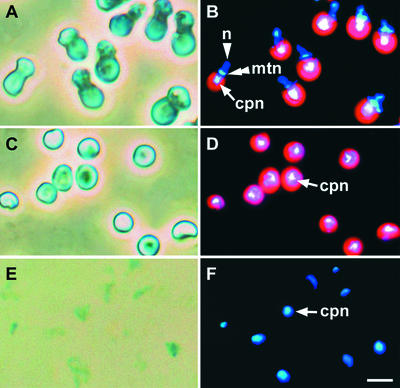
To analyze the cpDNA binding protein, we isolated cp-nucleoids from C. merolae. Phase-contrast and 4′,6-diamidino-2-phenylindole (DAPI)–stained images of whole cells, isolated chloroplasts, and cp-nucleoids from C. merolae are shown in Figure 1. Whole cells contained a single chloroplast, mitochondrion, and nucleus per cell, and cp-nucleoids were concentrated at the center of chloroplasts (Figures 1A and 1B). Nonsynchronous cells were disrupted in a French pressure cell after hypotonic treatment and layered on Percoll gradients. The intact chloroplast fraction was collected by centrifugation.
Figure 1.
Photomicrographs of cp-Nucleoid Subfraction Preparation.
(A), (C), and (E) Phase-contrast images of whole cells (A), isolated chloroplasts (C), and cp-nucleoid subfractions (E).
(B), (D), and (F) DAPI-stained DNA fluorescence images of whole cells (B), isolated chloroplasts (D), and cp-nucleoid subfractions (F).
Blue indicates DNA and red indicates the chlorophyll autofluorescence on the thylakoid. cpn, chloroplast nucleoids; mtn, mitochondrial nucleoids; n, nuclei. Bar in (F) = 3 μm for (A) to (F).
Observation of the isolated chloroplasts (Figures 1C and 1D) showed that cp-nucleoids maintained the same compact profiles seen in whole cells (Figure 1B) and were not contaminated by mitochondria or nuclei. The cp-nucleoids were isolated from this fraction by solubilizing the membrane with the nonionic detergent Nonidet P-40 and removing the solubilized components by centrifugation. Morphologically, the isolated cp-nucleoids (Figures 1E and 1F) remained intact and identical to those observed in whole cells (Figure 1B) and in isolated chloroplasts (Figure 1D). The combination of phase-contrast and DAPI-fluorescence microscopy showed that most of the chloroplast membrane had been removed (cf. Figures 1C and 1D with 1E and 1F).
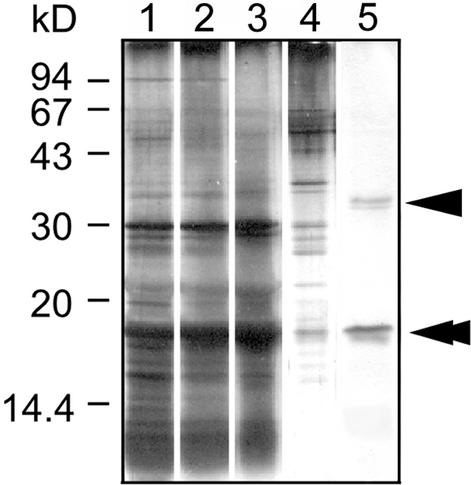
Next, to isolate DNA binding proteins involved in packaging cpDNA into nucleoid structures, we treated isolated cp-nucleoids with DNaseI. The proteins that dissociated from the cp-nucleoids were subjected to DNA-cellulose column chromatography and then to SDS-PAGE analysis. Figure 2 shows the protein patterns in the different fractions. After elution with start buffer containing 300 mM NaCl, several proteins were detected (lane 4). Proteins that bound DNA more strongly, with molecular masses of 35 kD (arrowhead) and 17 kD (double arrowhead), were eluted with 1 M NaCl (lane 5). The 17-kD protein was more abundant than the 35-kD protein, and two-dimensional gel electrophoresis of the cp-nucleoids revealed one spot with a molecular mass of 17 kD and a basic pI of 10.0 (data not shown). These results suggest that the 17-kD DNA binding protein is very basic, is abundant in cp-nucleoids, and binds DNA strongly.
Figure 2.
DNA-Cellulose Chromatography of C. merolae cp-Nucleoid Proteins.
Proteins were analyzed by SDS-PAGE using a 15% gel. Protein bands were visualized by silver staining. Lane 1, isolated cp-nucleoids; lane 2, supernatant of cp-nucleoid subfraction after DNaseI treatment; lane 3, flow-through fraction; lane 4, fraction eluted with 300 mM NaCl; lane 5, fraction eluted with 1 M NaCl. Arrowhead and double arrowhead indicate the 35- and 17-kD proteins, respectively.
Identification of the 17-kD Protein Gene
To identify the gene encoding the 17-kD protein, its N-terminal sequence was determined. The first 12 N-terminal residues were MDKTELISAVAE. The program FASTA (Pearson and Lipman, 1988) showed that this sequence was similar to part of HU, a bacterial histone-like protein (Rouviere-Yaniv and Gros, 1975). The 17-kD protein was similar to HU in many ways, including molecular mass, basic pI, and N-terminal sequence. This sequence also is similar to that of HlpA, the gene of which (hlpA) was found in the G. theta cp-genome (Wang and Liu, 1991).
Because G. theta is a cryptomonad alga with chloroplasts that are thought to have originated from red algae through secondary endosymbiosis, we expected to find the gene encoding the 17-kD protein in the cp-genome of C. merolae. Therefore, we examined the C. merolae cp-genome (Ohta et al., 1999) and found a gene that encoded the first 12 N-terminal residues of the 17-kD protein. Consequently, the gene was named CmhupA (hupA gene of Cyanidioschyzon merolae), and the protein encoded by CmhupA was named HC (histone-like protein of chloroplast).
The amino acid sequence of HC is shown in Figure 3. It encodes a protein with 113 amino acid residues, a molecular mass of 12.6 kD, and a calculated pI of 9.8. The molecular mass of HC was estimated to be 17 kD by SDS-PAGE analysis, and not 12.6 kD as deduced from the amino acid sequence (see above). Because the protein is very basic (estimated pI of 9.8), the band likely was shifted upward on SDS-PAGE, as is that of histone (Weber et al., 1972). Significant sequence similarity exists between HC and other HU-like proteins between residues 52 and 77, which corresponds to the DNA binding long arm of HU-like proteins (Tanaka et al., 1984). At the complete sequence level, 28 to 40% of the HC protein amino acid residues are identical to those in other HU-like proteins. If the comparison is limited to the region between residues 52 and 77, the sequence is 42 to 54% identical to those of other HU-like proteins.
Figure 3.
Comparison of HC Protein with Other HU-Like Proteins.
The amino acid sequence of HC is aligned with the sequences of other HU-like proteins from E. coli, Bacillus subtilis, Synechocystis sp strain PCC6803, Anabaena sp strain PCC7120, G. theta, and Toxoplasma gondii. The common amino acids appear in white type on a black background. Boxed sequences represent regions that form a long DNA binding arm (Tanaka et al., 1984).
There is no CmhupA homolog in the mitochondrial genome (Ohta et al., 1998) or in the nuclear genome, which was sequenced recently (our unpublished data). These results show that CmhupA is a single-copy gene that is encoded only by the cp-genome.
Complementation of HU-Lacking E. coli Mutants
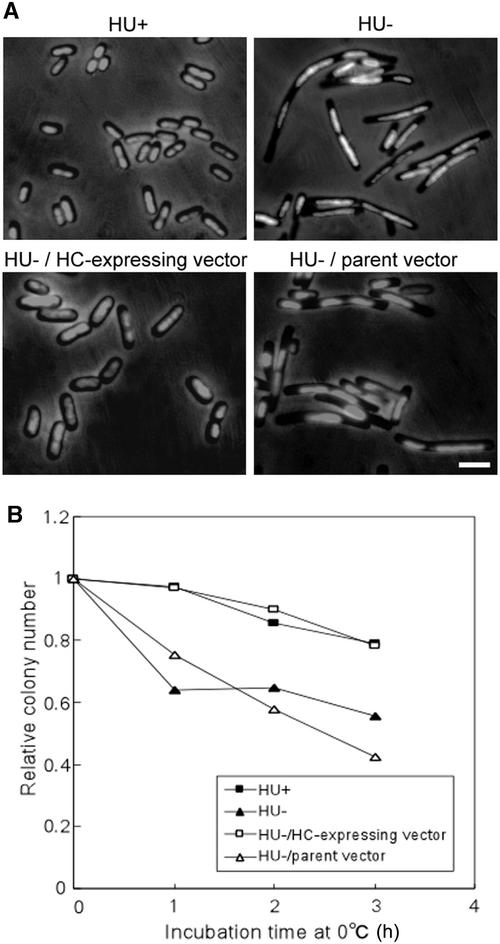
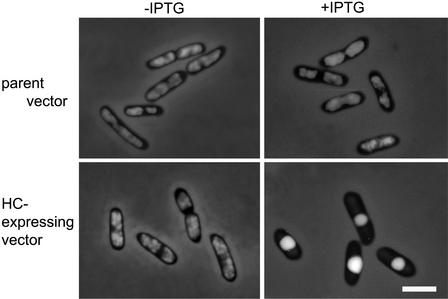
The HC protein sequence was similar to that of HU. We investigated whether HC was able to functionally complement E. coli mutants lacking HU. The E. coli HU protein is a heterodimer of HU-1 and HU-2, which are encoded by hupA and hupB, respectively. We constructed a hupA hupB double mutation by transducing the hupA::Cm mutation into the hupB derivative using P1 transduction (Miller, 1972). The double mutant exhibited filamentous morphology and a cold-sensitive phenotype, consistent with a previous report (Wada et al., 1988; Huisman et al., 1989).
The double mutants were transformed by an HC-expressing vector, and HC was expressed directly in the E. coli double mutants. Morphologically, they exhibited the wild-type phenotype, and the cold-sensitive phenotype was rescued (Figure 4). Furthermore, they are viable. In all cases, the double mutants carrying the parent vector retained the double mutant phenotypes. These results indicate that HC has not only sequence similarity to HU but also is functionally similar to HU.
Figure 4.
Complementation of HU-Lacking E. coli Mutants by HC.
(A) Effect of HC on the cell morphology of HU-lacking E. coli mutants. HU+, wild-type strain (JR1669); HU−, mutants lacking HU (JR1672); HU−/HC-expressing vector, pQE50-CmhupA mutants lacking HU carrying HC-expressing vector; HU−/parent vector, pQE50 mutants lacking HU carrying parent vector. Strains were grown in Luria-Bertani (LB) medium containing an appropriate antibiotic at 37°C. HU−/pQE50-CmhupA and HU−/pQE50 were grown in the presence of the inducer isopropylthio-β-galactoside (IPTG). Cells were fixed in 0.5% glutaraldehyde, stained with 1.0 μg/mL DAPI, and visualized by phase-contrast and fluorescence microscopy. Bar = 3 μm for all panels.
(B) Effect of HC on the cold-sensitive growth phenotype of HU-lacking E. coli mutants. Strains were grown to log phase in LB medium containing appropriate antibiotic and IPTG. They were transferred to 0°C for various periods and plated on LB agar containing appropriate antibiotic and IPTG. The plates were incubated at 37°C overnight, and the number of colonies was scored. The colony number was set at 1.0 immediately before transfer.
Localization and Expression of HC
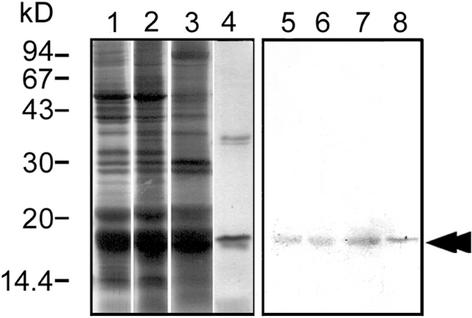
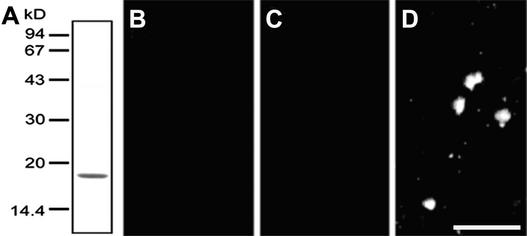
To analyze HC localization, we prepared an antibody against 6×His-tagged HC expressed in E. coli. Immunoblot analysis showed that this antibody reacted with a single band in a whole cell lysate, which had the same mobility as HC. The antibody also reacted with the same band in isolated chloroplasts and cp-nucleoids (Figure 5). This result indicates that HC is present in cp-nucleoids.
Figure 5.
Immunoblot Analysis with Anti-HC Antibody.
Proteins were separated by SDS-PAGE using a 15% gel. At left (lanes 1 to 4) are protein bands visualized by silver staining; at right (lanes 5 to 8) are immunoblots analyzed with anti-HC antibody. Lanes 1 and 5, whole cells; lanes 2 and 6, isolated chloroplasts; lanes 3 and 7, isolated cp-nucleoids; lanes 4 and 8, DNA-cellulose 1 M NaCl eluate; double arrowhead indicates the HC protein.
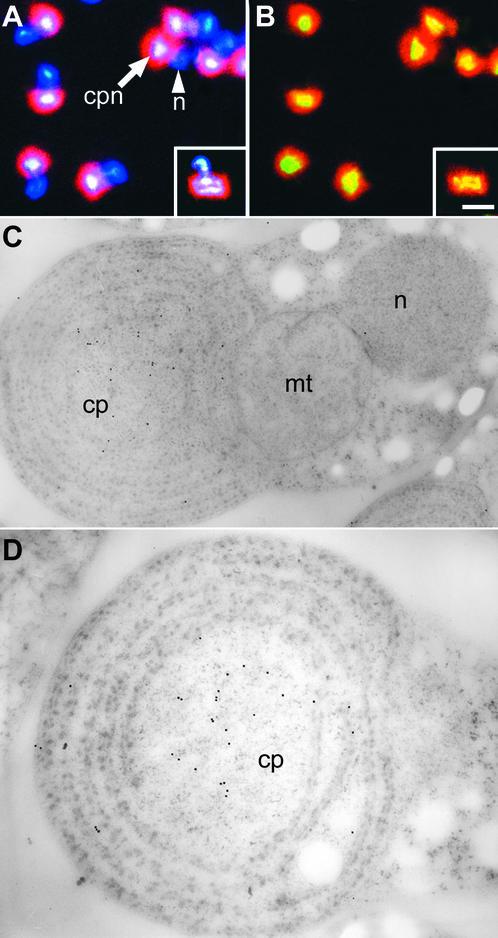
Immunofluorescence with anti-HC antibody was used to localize HC in vivo (Figures 6A and 6B). HC was located uniformly throughout the cp-nucleoid but was not seen in cell nuclei or in mitochondrial nucleoids (Figures 1B, 6A, and 6B). Fluorescein isothiocyanate fluorescence was seen clearly at exactly the same position in the chloroplast where the nucleoid was stained with DAPI fluorescence (Figures 6A and 6B). The use of anti-HC antibodies and immunoelectron microscopy allowed more precise localization of HC in cp-nucleoids at the ultrastructural level (Figures 6C and 6D).
Figure 6.
Cellular Immunolocalization of HC with Anti-HC Antibody.
(A) and (B) Fluorescence immunolocalization of HC to cp-nucleoids.
(A) DAPI staining of the same cells shown in (B).
(B) HC localization with a fluorescein isothiocyanate–labeled secondary antibody.
A dividing cell is shown in the inset. Yellow indicates the localization of HC. cpn, chloroplast nucleoids; n, nucleus. Bar in (B) = 1 μm for (A) and (B).
(C) and (D) HC localization to cp-nucleoids by immunoelectron microscopy. Cells are shown at two different magnifications after staining with anti-HupA antibodies and gold-labeled secondary antibodies. Gold particles are present at the cp-nucleoid. cp, chloroplast; mt, mitochondrion; n, nucleus. Bars = 0.5 μm.
Gold particles were observed in the chloroplast but not in the nucleus or the mitochondrion (Figure 6C), supporting the results of the immunofluorescence study (Figures 6A and 6B). Under higher magnification, gold particles were observed in the central part of the chloroplast where the cp-nucleoid was located (Figure 6D). Together, these results clearly demonstrate that HC is localized in cp-nucleoids (Figures 5 and 6).
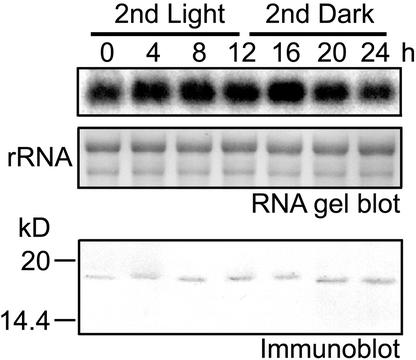
Next, we studied the relationship between the HC expression pattern and the chloroplast division cycle. Cell division and chloroplast division were synchronized by subjecting them to a 12-h-light/12-h-dark cycle. cpDNA replicates during the second light period, and chloroplasts divide during the second dark period (Itoh et al., 1997). RNA gel blot analysis was performed to investigate whether CmhupA transcription was correlated with the chloroplast division cycle (Figure 7).
Figure 7.
Time-Course Study of Cellular HC Level.
The cells were synchronized by a 12-h-light/12-h-dark cycle at 45°C. cpDNA replicated during the second light period, and most chloroplasts divided in the middle of the second dark period. Top and middle gels show RNA gel blot analysis using CmhupA probe. rRNA bands were visualized by methylene blue staining. Total RNA (5 μg/lane) was used. Bottom gel shows immunoblot analysis using anti-HC antibody. Whole cell lysates (10 μg protein/lane) were separated by SDS-PAGE on 15% gels.
Bands were detected at all time points, and signal intensities were constant. This result indicates that CmhupA was transcribed regardless of cpDNA replication. Immunoblot analysis showed that HC was expressed constitutively in the cell (Figure 7). When dividing chloroplasts were observed by immunofluorescence, HC was localized in the cp-nucleoids, and its fluorescence was approximately the same intensity as that of nondividing chloroplasts (Figures 6A and 6B, inset).
DNA Condensation Assay in Vitro and in E. coli
HC was the more abundant of the cpDNA binding proteins (Figure 2), was similar to the bacterial histone-like protein HU (Figures 3 and 4), and was localized within cp-nucleoids (Figure 6). Judging from these results, it seemed likely that HC is a structural protein involved in the organization of cpDNA into nucleoids. To examine the DNA-packing activity of HC, DNA-compacting assays were performed (Figures 8 and 9).
Figure 8.
Condensation of cpDNA by HC.
(A) Purified HC was separated by SDS-PAGE on a 15% gel, and the protein band was stained with Coomassie brilliant blue. C. merolae cpDNA was mixed with purified HC and dialyzed against isolation buffer. The mixture was stained with DAPI and examined by fluorescence microscopy.
(B) Control experiment without HC.
(C) Control experiment with BSA instead of HC.
(D) Mixture of cpDNA and HC.
Bar in (D) = 1 μm for (B) to (D).
Figure 9.
Fluorescence and Phase-Contrast Micrographs of the Effect of HC Protein on E. coli Nucleoid Structure.
E. coli cells carrying pQE50 (parent vector) or pQE50-CmhupA (HC-expressing vector) were fixed in 0.5% glutaraldehyde, stained with 1.0 μg/mL DAPI, and visualized by phase-contrast and fluorescence microscopy. Left column shows cells before induction (−IPTG), and right column shows cells after induction (+IPTG). Bar = 3 μm for all panels.
First, we confirmed that the purified HC was not contaminated with other proteins (Figure 8A). The purified HC was mixed with C. merolae cpDNA and dialyzed against isolation buffer. A number of small particles similar to nucleoids formed, although they were not the same size as cp-nucleoids in vivo (Figure 8D). In control experiments, neither cpDNA alone nor cpDNA plus BSA generated nucleoid-like particles (Figures 8B and 8C). This result suggests that HC can condense DNA into small particles without the help of other proteins.
To clarify the effect of HC on E. coli DNA, we expressed HC in E. coli (Figure 9). In the absence of the inducer IPTG, entire cells were stained uniformly. In HC-expressing cells, the nucleoid was condensed extensively, whereas no such change was observed in control cells. When we observed E. coli cells that were stained with the vital nucleic acid stain SYTO 11 to identify possible artifacts associated with fixation, only E. coli nucleoids expressing HC were condensed (data not shown). This result indicates that excess HC condenses nucleoids more compactly in E. coli.
DISCUSSION
cpDNA is arranged into protein/DNA complexes that are similar to bacteria and are called cp-nucleoids or cp-nuclei (Kuroiwa, 1991). Proteins bound to DNA may determine the structure of the nucleoid and influence DNA function as a template for either transcription or replication (Kuroiwa, 1991).
In this study, we used a newly developed method to purify DNA binding protein from cp-nucleoids that were isolated from C. merolae (Figures 1 and 2). Seventeen- and 35-kD proteins were eluted with the high-salt fraction, and the 17-kD protein was more abundant than the 35-kD protein. These results suggest that the 17-kD protein is the more abundant DNA binding protein in cp-nucleoids and that it binds DNA strongly.
We determined the N-terminal sequence of the 17-kD protein and performed a homology search. The sequence was completely different from those of other DNA binding proteins that have been isolated from cp-nucleoids (CND41 from tobacco [Nakano et al., 1997], PEND from pea [Sato et al., 1993], and a 70-kD protein from pea [Sato et al., 1997]). The 17-kD protein had high sequence homology with the bacterial histone-like protein HU and was named HC (Figure 3). HC is similar to HU in many ways (molecular mass and basic pI); moreover, HU is a major component of bacterial chromatin (60,000 monomers per E. coli cell) (Schmid, 1990).
Immunogold staining with anti-HU antibodies revealed the distribution of HU in E. coli cells, and the antibodies were observed primarily at the nucleoid periphery (Dürrenberger et al., 1988). In contrast, fluorescence microscopy of cells containing fluorescein-labeled HU showed that HU was distributed uniformly throughout the E. coli nucleoid (Shellman and Pettijohn, 1991). A subsequent study using immunofluorescence microscopy reported the same result (Azam et al., 2000).
In our study, both immunofluorescence and immunoelectron microscopy showed that HC was distributed uniformly throughout the entire cp-nucleoid (Figure 6). This observation is consistent with the localization of HU within the nucleoids, indicating that the distribution also is conserved between HC and bacterial HU. Furthermore, HC was expressed constitutively within the cells and located uniformly throughout the cp-nucleoids during chloroplast division (Figures 6 and 7). These results suggest that HC localizes within cp-nucleoids, that the DNA binding ability of HC is not restricted to any specific region of the nucleoid, and that HC plays an essential role in the maintenance of cp-nucleoids.
It is known that bacterial HU contributes to the compaction of DNA into nucleoid structures (Rouviere-Yaniv et al., 1979; Broyles and Pettijohn, 1986). In our DNA condensation assay, HC condensed DNA without the help of other proteins (Figure 8). Barry et al. (1992) confirmed that the nucleoids are condensed when Hc1 (a chlamydial histone H1 homolog) is expressed in E. coli. In the same way, excess expression of HC resulted in extremely condensed E. coli nucleoids (Figure 9).
These results suggest that HC is an important protein in the organization of cpDNA into nucleoids. However, HC organized DNA into a smaller size than the cp-nucleoids observed in vivo (Figure 8). This finding suggests that other proteins, such as the 35-kD protein eluted with 1 M NaCl (Figure 2), are necessary to organize DNA into natural cp-nucleoids. More detailed study is needed to clarify this point.
E. coli mutants lacking HU exhibited phenotypes that were different from that of the wild-type strain, such as filamentous and cold-sensitive phenotypes (Figure 4) (Wada et al., 1988; Huisman et al., 1989). These phenotypes are thought to occur because the absence of HU leads to disturbances in chromosome partitioning and cell division by an unknown mechanism (Wada et al., 1988). HC rescued these phenotypes in HU-lacking E. coli mutants (Figure 4), suggesting that HC is functionally very similar to E. coli HU. HU protein also participates in a number of cellular mechanisms, such as modulating the expression of specific genes (Manna and Gowrishankar, 1994; Wu and Datta, 1995), DNA melting at the initiation of replication (Skarstad et al., 1990; Kano et al., 1991), DNA breaking/rejoining in transposition and inversion reactions (Haykinson and Johnson, 1993; Lavoie and Chaconas, 1993), and homologous recombination (Kano and Imamoto, 1990; Dri et al., 1992). HC likely serves a fundamental biological function in chloroplasts, just as HU does in bacteria.
The primary structure of HU is highly conserved among bacterial species, including cyanobacteria, which are thought to be the ancestors of chloroplasts. In this study, the HU homolog HC also was found in the cp-genome of the primitive red algae C. merolae, and HC organized the cpDNA into nucleoids. A HU-like protein gene (hlpA) has been found in the G. theta cp-genome (Wang and Liu, 1991). G. theta is a cryptomonad alga, and its chloroplasts are thought to have originated from red algae through secondary endosymbiosis. The HlpA protein binds DNA in a sequence-independent manner and can bend DNA in vitro (Wu and Liu, 1997).
The HU homolog also has been found in several sequencing projects of apicomplexan parasites, including Toxoplasma gondii, Plasmodium yoelii, and Plasmodium berghei. The amino acid sequence of the HU homolog from T. gondii is aligned in Figure 3. These parasites have been shown to contain a vestigial nonphotosynthetic plastid, the apicoplast, which may have been derived from a red algal ancestor by secondary endosymbiosis (Fast et al., 2001). Therefore, bacterial HU seems to be highly conserved in red lineage plants (red algae and algae harbor red algal plastids).
No HU homolog has been detected in any of the chloroplast genomes of green algae or land plants. The immunological detection of an HU homolog in spinach chloroplast has been reported (Briat et al., 1984), as has the presence of a histone-like protein in pea chloroplast nucleoids (Yurina et al., 1995). However, the HU-like protein gene is not found in the cp-genome or the nuclear genome of Arabidopsis. The HU sequence may be too short to detect traces of the protein. It is possible that an HU homolog remains in the green lineage, but the sequence may have changed. It is certain that an HU-like protein was involved in the organization or maintenance of cp-nucleoid structure at an early stage of evolution.
METHODS
Culture Conditions
Cyanidioschyzon merolae was synchronized according to the method described by Suzuki et al. (1994). Cells were cultured in Allen's medium (Allen, 1959) at pH 2.5, and flasks were shaken under continuous light (40 W/m2) at 40°C. The cells were subcultured to <107 cells/mL and then synchronized by subjecting them to a 12-h-light/12-h-dark cycle at 45°C in aerated medium. After the second cycle, cells multiplied synchronously. During the light period, cells were in interphase, whereas during the dark period, the chloroplast, mitochondrion, and nucleus divided (in that order) before cytokinesis. Most chloroplasts divided in the middle of the second dark period.
Isolation of Chloroplast Nucleoids
Chloroplasts were isolated from nonsynchronous cells according to the method described by Miyagishima et al. (1999), with some modification. At the first hour of the first dark period, cells were collected by centrifugation and suspended in isolation buffer (20 mM Tris-HCl, pH 7.6, 5 mM KCl, 5 mM EDTA, and 1.2 mM spermidine) containing 5 mM MgCl2 and 180 mM Suc. Then, the cells were lysed in a French pressure cell (SLM Amico, Rochester, NY) at 2000 p.s.i., and 100 μg/mL DNaseI (DN-25; Sigma) was added. The lysate was incubated for 1 h at 4°C and then layered on three-step gradients of 80, 60, and 40% Percoll in isolation buffer containing 300 mM Suc.
Lysates were centrifuged for 50 min at 28,000g, and a band of intact chloroplasts was harvested from the 60 to 80% Percoll interface. Chloroplasts were washed with isolation buffer containing 300 mM Suc and suspended at a concentration of 1 mg protein/mL in isolation buffer containing 2% (v/v) Nonidet P-40. The lysed chloroplasts were incubated at room temperature for 15 min and then centrifuged at 18,500g for 15 min. The resulting pellet was resuspended in isolation buffer and 2% Nonidet P-40, incubated at room temperature for 15 min, and centrifuged at 18,500g for 15 min.
The final nucleoid pellet was suspended in isolation buffer and stored at 4°C. Cells, chloroplasts, and chloroplast nucleoids were fixed in 0.5% glutaraldehyde (distilled grade; TAAB, Aldermaston, UK), stained with 1.0 μg/mL 4′,6-diamidino-2-phenylindole (DAPI), and pressed between a microscope slide and a cover slip. The samples were observed with a fluorescence microscope (BHS-RFC; Olympus, Tokyo, Japan) using a UV excitation beam at 334 and 365 nm.
Identification of the DNA Binding Protein in Chloroplast Nucleoids
The nucleoid pellet was suspended in isolation buffer containing complete protease inhibitors (Roche, Mannheim, Germany), and 0.25 mg of a 1:1000 volume of 25 mg/mL DNaseI (D4527; Sigma) (final concentration of 25 μg/mL) and a 1:200 volume of 2 M MgCl2 (final concentration of 10 mM) were added. The suspension was incubated at 4°C for 1 h and then centrifuged at 100,000g for 30 min. The supernatant was dialyzed overnight at 4°C with start buffer (20 mM Tris-HCl, pH 7.6, 5 mM EDTA, 50 mM NaCl, 0.5 mM DTT, and complete protease inhibitors [Roche]) and loaded onto a native DNA-cellulose column (Amersham Pharmacia) that was washed with start buffer before use.
Proteins were eluted with start buffer containing 300 and 1000 mM NaCl. Proteins eluted with start buffer containing 1000 mM NaCl were mixed with one-third volume of 4 × Laemmli sample buffer (50 mM Tris, pH 6.8, 10% glycerol, 2% SDS, 5% 2-mercaptoethanol, and 0.01% bromphenol blue) and separated by SDS-PAGE on 15% polyacrylamide gels at 150 V for 1 h. Separated proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford, MA) for N-terminal sequencing or visualized by silver staining. The N-terminal sequence of the 17-kD protein was determined using a HP G1005A protein sequencing system (Hewlett Packard, Palo Alto, CA).
Plasmids
For plasmid pQE30-CmhupA, the coding sequence of CmhupA from C. merolae total DNA was amplified by PCR with primers SphI-HHU (5′-ACATGCATGCATGGACAAAACAGAACTTATA-3′) and PstI-HHU (5′-AACTGCAGTTATTCTTTTTTGTCATCTTG-3′). The underlined sequences are the SphI and PstI restriction sites, respectively. The DNA fragment was digested with SphI and PstI and inserted into vector pQE-30 (Qiagen, Hilden, Germany). For plasmid pQE50-CmhupA, which expresses HC directly (nontagged), the CmhupA coding sequence was amplified from C. merolae total DNA by PCR with primers SphI-HHU and PstI-HHU. The DNA fragment was digested with SphI and PstI and inserted into vector pQE-50 (Qiagen). The DNA sequence was confirmed.
RNA Gel Blot Analysis
Total RNA was isolated from C. merolae cells using a previously described method (Ohta et al., 1994). Aliquots (5 μg) of total RNA were electrophoresed on a 1.4% agarose gel for 2.5 h at 70 V in 10 mM sodium phosphate buffer, pH 7.0, and transferred to a nylon membrane by capillary blotting. The CmhupA probe was labeled with α-32P-dCTP, and hybridization, washing, and detection were performed as described above except that the hybridization buffer contained 50% formamide and washing was performed at 65°C (high-stringency conditions).
Complementation Analysis of HU-Lacking Escherichia coli Mutants Using HC
The single hupA and hupB Escherichia coli mutants (strains JR1670 and JR1671, respectively) were kindly provided by Josette Rouviere-Yaniv (Institut de Biolgie Physico-Chimique, Paris, France). The genotypes of JR1670 and JR1671 are EchupA::Cm and EchupB::Km, respectively. The double mutant (strain JR1672) was constructed by P1 transduction (Miller, 1972) starting from the single EchupA and EchupB mutants. Bacteria were grown in Luria-Bertani (LB) medium (10 g of tryptone peptone, 5 g of yeast extract, and 5 g of NaCl per liter). Kanamycin (50 μg/mL) and chloramphenicol (12.5 μg/mL) were included in the medium when appropriate.
The E. coli hupA hupB double mutant (JR1672) was transformed with plasmid pQE50-CmhupA. Transformed strains were grown in LB medium supplemented with 2% Glc, 50 μg/mL kanamycin, 12.5 μg/mL chloramphenicol, and 50 μg/mL ampicillin until they reached an optical density of 0.4 at 600 nm, at which time isopropylthio-β-galactoside (IPTG; final concentration of 2 mM) was added. Cells were fixed in 0.5% glutaraldehyde, stained with 1.0% DAPI, and examined by fluorescence and phase-contrast microscopy.
To detect the effect of cold shock, strains JR1669 (wild type), JR1672 (double mutant), JR1672/pQE50, and JR1672/pQE50-CmhupA were grown to log phase in LB medium containing 2 mM IPTG and the appropriate antibiotics, transferred to 0°C for 0, 1, 2, and 3 h, and plated on LB agar. JR1672/pQE50 and JR1672/pQE50-CmhupA were grown in LB medium in the presence of the inducer IPTG. The plates were incubated at 37°C overnight, and the number of colonies was scored. Colony number immediately before the transfer was set at 1.0.
Antibody Preparation and Immunoblot Analysis
Plasmid pQE30-CmhupA was used to express recombinant HC as a 6× His-tagged protein. This protein was purified using HisTrap (Amersham Pharmacia), and 1.5 mg of purified protein was used to raise polyclonal rabbit antisera. Anti-HC antibodies were purified from serum by protein G affinity chromatography (MAbTrap Kit; Amersham Pharmacia).
Proteins were separated by SDS-PAGE using 15% gel as described above and then transferred to a PVDF membrane (Millipore) in buffer (25 mM Tris, 190 mM Gly, 20% methanol, and 0.03% SDS) for 2 h at 100 V. The PVDF membrane was blocked for 1 h at room temperature in TBS (0.2 M Tris-HCl, pH 7.5, and 5 M NaCl) containing 3% gelatin. Blocked membranes were probed with rabbit polyclonal anti-HC antibody (1:10,000 in TBS containing 0.05% Tween 20) for 1 h at room temperature. Membranes then were washed with 1 × TBS containing 0.05% Tween 20 and exposed to an anti-rabbit IgG alkaline phosphatase conjugate (1:1000 dilution). Bands were resolved by incubation with 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium.
Immunofluorescence and Immunoelectron Microscopy Using Anti-HC Antibody
Immunofluorescence staining of HC and fluorescence microscopy were performed as described previously (Tanaka, 1991) with minor modifications. The primary antibody was anti-HC at a dilution of 1:1000 (v/v) in blocking buffer (PBS, pH 7.4, and 5% [w/v] BSA), and the secondary antibody was goat anti-rabbit IgG conjugated with fluorescein isothiocyanate (Biosource, Camarillo, CA) at a dilution of 1:100 in blocking buffer.
Immunogold labeling was performed as described previously (Miyagishima et al., 2001) with minor modifications. Cells were fixed by rapid freezing/freeze substitution fixation (in 1% glutaraldehyde dissolved in dry acetone). Anti-HC antibody (1:50 dilution) and goat anti-rabbit IgG conjugated with 10-nm colloidal gold (1:50 dilution; Zymed, San Francisco, CA) were used as primary and secondary antibodies, respectively. Finally, samples were stained with uranyl acetate and examined with a JEM-1200EX electron microscope (JEOL).
DNA Condensation Assay in Vitro
Using vector pGEX6P-1 (Amersham Pharmacia), HC was expressed in E. coli as a fusion protein with glutathione S-transferase (GST). The GST-fused HC protein was retained selectively on glutathione-Sepharose 4B columns. HC was released from the column by cleaving GST from GST-HC using Precision protease (Amersham Pharmacia). C. merolae chloroplast DNA was prepared as described above. The chloroplast DNA (1.0 μg) and HC (2.0 μg) were mixed and dialyzed against isolation buffer at 4°C for 5 h. The mixture was stained with 1.0 μg/mL DAPI and examined by fluorescence microscopy.
DNA Condensation Assay in E. coli
HC was expressed directly (nontagged) in E. coli (XL1-Blue) using vector pQE50 (Qiagen). The resulting expression plasmid was named pQE50-CmhupA. For observation, cells were cultured for 2 h after the addition of IPTG (final concentration of 2 mM). Then, cells were fixed in 0.5% glutaraldehyde and stained with 1.0 μg/mL DAPI.
Accession Numbers
The accession number for the nucleotide sequence of CmhupA is AB071964. The accession numbers for the sequences of HU-like proteins shown in Figure 3 are P02342 (E. coli), X52418-1 (B. subtilis), P73418 (Synechocystis sp strain PCC6803), P05514 (Anabaena sp strain PCC7120), M76547 (G. theta), and BM175970 (T. gondii). The accession numbers for other organisms mentioned in this article are BM166958 (P. yoelii) and BF295990 (P. berghei).
Acknowledgments
We thank Josette Rouviere-Yaniv (Institut de Biolgie Physico-Chimique, Paris, France) for providing the E. coli strains (JR1669, JR1670, and JR1671) and Tatsushi Mogi (University of Tokyo, Tokyo, Japan) for the kind gift of P1 phage. This work was supported by Grants 12440222 and 13206011 to T.K. from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and by a grant from the Program for the Promotion of Basic Research Activities for Innovative Bioscience.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.002717.
References
- Allen, M.B. (1959). Studies with Cyanidium caldarium, an anomalously pigmented chlorophyte. Arch. Microbiol. 32, 270–277. [DOI] [PubMed] [Google Scholar]
- Azam, T.A., Hiraga, S., and Ishihama, A. (2000). Two types of localization of the DNA-binding proteins within the Escherichia coli nucleoid. Genes Cells 5, 613–626. [DOI] [PubMed] [Google Scholar]
- Barry, C.E., III, Hayes, S.F., and Hackstadt, T. (1992). Nucleoid condensation in Escherichia coli that express a chlamydial histone homolog. Science 256, 377–379. [DOI] [PubMed] [Google Scholar]
- Briat, J.F., Letoffe, S., Mache, R., and Rouviere-Yaniv, J. (1984). Similarity between the bacterial histone-like protein HU and a protein from spinach chloroplast. FEBS Lett. 172, 75–79. [Google Scholar]
- Broyles, S., and Pettijohn, D.E. (1986). Interaction of the Escherichia coli HU protein with DNA evidence for formation of nucleosome-like structures with altered DNA helical pitch. J. Mol. Biol. 187, 47–60. [DOI] [PubMed] [Google Scholar]
- Diffley, J.F.X., and Stillman, B. (1991). A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc. Natl. Acad. Sci. USA 88, 7864–7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dri, A.M., Moreau, P.L., and Rouviere-Yaniv, J. (1992). Role of the histone-like proteins OsmZ and HU in homologous recombination. Gene 120, 11–16. [DOI] [PubMed] [Google Scholar]
- Drlica, K., and Rouviere-Yaniv, J. (1987). Histone like proteins of bacteria. Microbiol. Rev. 51, 301–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürrenberger, M., Bjornsti, M.A., Uetz, T., Hobot, J.A., and Kellenberger, E. (1988). Intracellular localization of the histone like protein HU in Escherichia coli. J. Bacteriol. 170, 4757–4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fast, N.M., Kissinger, J.C., Roos, D.S., and Keeling, P.J. (2001). Nuclear-encoded, plastid-targeted genes suggest a single common origin for apicomplexan and dinoflagellate plastids. Mol. Biol. Evol. 18, 418–426. [DOI] [PubMed] [Google Scholar]
- Gillham, N.W. (1994). Cytological organization and replication of organelle genomes. In Organelle Genes and Genomes. (New York: Oxford University Press), pp. 110–126.
- Gray, M.W. (1992). The endosymbiont hypothesis revisited. Int. Rev. Cytol. 141, 233–357. [DOI] [PubMed] [Google Scholar]
- Haykinson, M.J., and Johnson, R.C. (1993). DNA looping and the helical repeat in vitro and in vivo: Effect of HU protein and enhancer location on Hin invertasome assembly. EMBO J. 12, 2503–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewson, S., and Wolstenholme, R. (1969). Mitochondria and chloroplast: Nucleic acids and the problem of biogenesis (genetics and biology). In Handbook of Molecular Cytology, A. Lima-de-Faria, ed (Amsterdam: North-Holland), pp. 972–1046.
- Huisman, O., Faelen, M., Girard, D., Jaffe, A., Toussaint, A., and Rouviere-Yaniv, J. (1989). Multiple defects in Escherichia coli mutants lacking HU protein. J. Bacteriol. 171, 3704–3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, R., Takahashi, H., Toda, K., Kuroiwa, H., and Kuroiwa, T. (1997). DNA gyrase involvement in chloroplast-nucleoid division in Cyanidioschyzon merolae. Eur. J. Cell Biol. 73, 252–258. [PubMed] [Google Scholar]
- Kano, Y., and Imamoto, F. (1990). Requirement of integration host factor (IHF) for growth of Escherichia coli deficient in HU protein. Gene 89, 133–137. [DOI] [PubMed] [Google Scholar]
- Kano, Y., Ogawa, T., Ogura, S., Hiraga, T., Okazaki, T., and Imamoto, F. (1991). Participation of the histone-like protein HU and of IHF in minichromosomal maintenance in Escherichia coli. Gene 103, 25–30. [DOI] [PubMed] [Google Scholar]
- Kuroiwa, T. (1982). Mitochondrial nuclei. Int. Rev. Cytol. 75, 1–59. [DOI] [PubMed] [Google Scholar]
- Kuroiwa, T. (1991). The replication, differentiation, and inheritance of plastids with emphasis on the concept of organelle nuclei. Int. Rev. Cytol. 128, 1–62. [Google Scholar]
- Kuroiwa, T., and Suzuki, T. (1981). Circular nucleoids isolated from chloroplast in a brown alga Ectocarpus indicus. Exp. Cell Res. 134, 457–462. [DOI] [PubMed] [Google Scholar]
- Kuroiwa, T., Suzuki, T., Ogawa, K., and Kawano, S. (1981). The chloroplast nucleus: Distribution, number, size, and shape, and a model for the multiplication of the chloroplast genome during chloroplast development. Plant Cell Physiol. 22, 381–396. [Google Scholar]
- Lavoie, B.D., and Chaconas, G. (1993). Site-specific HU binding in the Mu transposome: Conversion of a sequence-independent DNA-binding protein into a chemical nuclease. Genes Dev. 7, 2510–2519. [DOI] [PubMed] [Google Scholar]
- Liu, J.-W., and Rose, R.J. (1992). The spinach chloroplast chromosome is bound to the thylakoid membrane in the region of the inverted repeat. Biochem. Biophys. Res. Commun. 184, 993–1000. [DOI] [PubMed] [Google Scholar]
- Manna, D., and Gowrishankar, J. (1994). Evidence for involvement of protein HU and RpoS in transcription of the osmoresponsive proU operon in Escherichia coli. J. Bacteriol. 176, 5378–5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis, L. (1970). Origin of Eukaryotic Cells. (New Haven, CT: Yale University Press).
- Miller, J.H. (1972). Experiments in Molecular Genetics. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 201–205.
- Miyagishima, S., Itoh, R., Aita, S., Kuroiwa, H., and Kuroiwa, T. (1999). Isolation of dividing chloroplasts with intact plastid-dividing rings from a synchronous culture of the unicellular red alga Cyanidioschyzon merolae. Planta 209, 371–375. [DOI] [PubMed] [Google Scholar]
- Miyagishima, S., Takahara, M., Mori, T., Kuroiwa, H., Higashiyama, T., and Kuroiwa, T. (2001). Plastid division is driven by a complex mechanism that involves differential transition of the bacterial and eukaryotic division rings. Plant Cell 13, 2257–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano, T., Murakami, S., Shoji, T., Yoshida, S., Yamada, Y., and Sato, F. (1997). A novel protein with DNA binding activity from tobacco chloroplast nuclei. Plant Cell 9, 1673–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto, Y., Kawano, S., Nagata, T., and Kuroiwa, T. (1991). Studies on plastid-nuclei (nucleoids) in Nicotiana tabacum L. IV. Association of chloroplast-DNA with proteins at several specific sites in isolated chloroplast-nuclei. Plant Cell Physiol. 32, 131–141. [Google Scholar]
- Oberto, J., and Rouviere-Yaniv, J. (2001). Does the parallel evolution pattern between the replication-segregation proteins and HU have a biological significance? Biochimie 83, 61–66. [DOI] [PubMed] [Google Scholar]
- Ohta, N., Sato, N., Kawano, S., and Kuroiwa, T. (1994). The trpA gene on the plastid genome of Cyanidium caldarium strain RK-1. Curr. Genet. 25, 357–361. [DOI] [PubMed] [Google Scholar]
- Ohta, N., Sato, N., and Kuroiwa, T. (1998). Structure and organization of the mitochondrial genome of the unicellular red alga Cyanidioschyzon merolae deduced from the complete nucleotide sequence. Nucleic Acids Res. 26, 5190–5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, N., Sato, N., and Kuroiwa, T. (1999). The organellar genomes of Cyanidioschyzon merolae. In Enigmatic Microorganisms and Extreme Environments, J. Seckbach, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 139–149.
- Pearson, W.R., and Lipman, D.J. (1988). Improved tools for biological sequence analysis. Proc. Natl. Acad. Sci. USA 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouviere-Yaniv, J., and Gros, F. (1975). Characterization of a novel, low-molecular-weight DNA-binding protein from Escherichia coli. Proc. Natl. Acad. Sci. USA 72, 3428–3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouviere-Yaniv, J., Yaniv, M., and Germond, J.E. (1979). E. coli DNA binding protein HU forms nucleosomelike structure with circular double-stranded DNA. Cell 17, 265–274. [DOI] [PubMed] [Google Scholar]
- Sato, N., Albrieux, C., Joyard, J., Douce, R., and Kuroiwa, T. (1993). Detection and characterization of a plastid envelope DNA-binding protein which may anchor plastid nucleoids. EMBO J. 12, 555–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, N., Misumi, O., Shinada, Y., Sasaki, M., and Yoine, M. (1997). Dynamics of localization and protein composition of plastid nucleoids in light-grown pea seedlings. Protoplasma 200, 163–173. [Google Scholar]
- Sato, N., Nakayama, M., and Hase, T. (2001). The 70-kD major DNA-binding protein of the chloroplast nucleoid is sulfite reductase. FEBS Lett. 487, 347–350. [DOI] [PubMed] [Google Scholar]
- Sato, N., Oshima, K., Watanabe, A., Ohta, N., Nishiyama, Y., Joyard, J., and Douce, R. (1998). Molecular characterization of the PEND protein, a novel bZIP protein present in the envelope membrane that is the site of nucleoid replication in developing plastid. Plant Cell 10, 859–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, M.B. (1990). More than just “Histone-like” proteins. Cell 63, 451–453. [DOI] [PubMed] [Google Scholar]
- Shellman, V.L., and Pettijohn, D.E. (1991). Introduction of proteins into living bacterial cells: Distribution of labeled HU protein in Escherichia coli. J. Bacteriol. 173, 3047–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skarstad, K., Baker, T.A., and Kornberg, A. (1990). Strand separation required for initiation of replication at the chromosome origin of E. coli is facilitated by a distant RNA-DNA hybrid. EMBO J. 9, 2341–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, K., Ehara, T., Osafune, T., Kuroiwa, H., Kawano, S., and Kuroiwa, T. (1994). Behavior of mitochondria, chloroplasts and their nuclei during the mitotic cycle in the ultramicroalga Cyanidioschyzon merolae. Eur. J. Cell Biol. 63, 280–288. [PubMed] [Google Scholar]
- Tanaka, I. (1991). Microtubule-determined plastid distribution during microsporogenesis in Lilium longiflorum. J. Cell Sci. 99, 21–31. [Google Scholar]
- Tanaka, I., Appelt, K., Dijk, J., White, S., and Wilson, K. (1984). 3-Å resolution structure of a protein with histone-like properties in prokaryotes. Nature 310, 376–381. [DOI] [PubMed] [Google Scholar]
- Wada, M., Kano, Y., Ogawa, T., Okazaki, T., and Imamoto, F. (1988). Construction and characterization of the deletion mutant of hupA and hupB genes in Escherichia coli. J. Mol. Biol. 204, 581–591. [DOI] [PubMed] [Google Scholar]
- Wang, S., and Liu, X.-Q. (1991). The plastid genome of Cryptomonas phi encodes an hsp70-like protein, a histone-like protein, and an acyl carrier protein. Proc. Natl. Acad. Sci. USA 88, 10783–10787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, K., Pringle, J.R., and Osborn, M. (1972). Measurement of molecular weights by electrophoresis on SDS-acrylamide gel. Methods Enzymol. 26, 3–27. [DOI] [PubMed] [Google Scholar]
- Wolffe, A. (1998). Chromatin structure. In Chromatin: Structure and Function, A. Wolffe, ed (San Diego: Academic Press), pp. 7–172.
- Wu, H., and Liu, X.-Q. (1997). DNA binding and bending by a chloroplast-encoded HU-like protein overexpressed in Escherichia coli. Plant Mol. Biol. 34, 339–343. [DOI] [PubMed] [Google Scholar]
- Wu, Y., and Datta, P. (1995). Influence of DNA topology on expression of the tdc operon in Escherichia coli K-12. Mol. Gen. Genet. 247, 764–767. [DOI] [PubMed] [Google Scholar]
- Yurina, N.P., Belkina, G.G., Karapetyan, N.V., and Odintsova, M.S. (1995). Nucleoids of pea chloroplasts: Microscopic and chemical characterization. Occurrence of histone-like proteins. Biochem. Mol. Biol. Int. 36, 145–154. [PubMed] [Google Scholar]