Abstract
The regulatory mechanism of centrosome function is crucial to the accurate transmission of chromosomes to the daughter cells in mitosis. Recent findings on the posttranslational modifications of many centrosomal proteins led us to speculate that these modifications might be involved in centrosome behavior. Poly(ADP-ribose) polymerase 1 (PARP-1) catalyzes poly(ADP-ribosyl)ation to various proteins. We show here that PARP-1 localizes to centrosomes and catalyzes poly(ADP-ribosyl)ation of centrosomal proteins. Moreover, centrosome hyperamplification is frequently observed with PARP inhibitor, as well as in PARP-1-null cells. Thus, it is possible that chromosomal instability known in PARP-1-null cells can be attributed to the centrosomal dysfunction. P53 tumor suppressor protein has been also shown to be localized at centrosomes and to be involved in the regulation of centrosome duplication and monitoring of the chromosomal stability. We found that centrosomal p53 is poly(ADP-ribosyl)ated in vivo and centrosomal PARP-1 directly catalyzes poly(ADP-ribosyl)ation of p53 in vitro. These results indicate that PARP-1 and PARP-1-mediated poly(ADP-ribosyl)ation of centrosomal proteins are involved in the regulation of centrosome function.
The centrosome functions as a major microtubule organizing center in animal cells and plays vital roles during mitosis as a core unit of spindle poles, including the assembly of bipolar mitotic spindles and determination of the plane where the cleavage furrow is introduced (for reviews, see references 6 and 27). Since each daughter cell receives only one centrosome, the centrosome must duplicate once during each cell cycle. Thus, centrosome duplication must take place in coordination with other cell cycle events, including DNA duplication. In mammalian somatic cells, centrosome duplication begins near the G1/S boundary of the cell cycle and is completed in G2 phase (61, 63). Abrogation of the regulatory mechanisms that ensure the coordinated progression of centrosome duplication and other cell cycle events, including DNA duplication, and that prevent reduplication of the duplicated centrosome within the same cell cycle results in hyperamplification of centrosomes (7, 57). This, in turn, leads to increased frequency of defective (multipolar) mitotic spindles and unbalanced segregation of chromosomes into daughter cells as observed in cancer cells (11, 28, 47, 57).
Recently, it has been reported that some of the centrosomal proteins undergo various posttranslational modifications, including kinases such as Aurora A, Plks, and Nek2 (17, 19, 31); phosphorylation of NPM/B23 and Mps1p by CDK2 (15, 45); and ubiqutination complex (SCF complex) such as Skp1, Skp2, and Cul1 (16, 39, 69). These modifications could affect the properties of the proteins. For example, NPM/B23 is associated with unduplicated centrosomes but not with duplicated centrosomes and dissociates from centrosomes upon phosphorylation by CDK2/cyclin E (45). Furthermore, several studies have reported that tumor suppressor protein p53 is localized to centrosome (4, 8, 36) and changes the regulatory activity of centrosome duplication with mutations of p53 phosphorylation sites (58, 59). Thus, these studies suggest that the modifications of centrosomal proteins are important for centrosome (centriole) behavior.
Poly(ADP-ribosyl)ation is known to be one of the major posttranslational modifications. Poly(ADP-ribose) polymerase 1 (PARP-1; EC 2.4.2.30) catalyzes the formation of long-branched poly(ADP-ribose) polymers on glutamic acid, aspartic acid, and lysine residues of target proteins with NAD+ as a substrate (42, 56). It has been reported that poly(ADP-ribose) glycohydrolase (PARG) rapidly hydrolyzes the polymer of poly(ADP-ribose) from the poly(ADP-ribosyl)ated proteins to produce free ADP-ribose residues (13, 33). Recently, a quite large family of PARP enzymes have been identified and characterized (PARP-1, PARP-2, PARP-3, Tankyrase-1, Tankyrase-2, and vault PARP). Many proteins that are poly(ADP-ribosyl)ated by PARP-1 have been identified, including PARP-1 itself (43), histones (26), lamins (1), topoisomerases (25), DNA polymerases (44, 70), c-Fos (2), and p53 tumor suppressor protein (68). Since the attachment of the negatively charged polymer changes the properties of the acceptor protein (40, 46), PARP-1 could be involved in a variety of cellular events, including modulation of chromatin structure, DNA synthesis, DNA repair, gene transcription, and cell cycle regulation (13). In particular, the studies with PARP inhibitors have shown that PARP-1 plays an important role in maintenance of genome integrity (10, 34, 35). More recently, it has been shown that cells derived from PARP-1-deficient mice exhibit chromosomal instability and increased frequency of aneuploidy (12, 14, 38, 48, 53, 60, 62, 66), although the mechanism is not clear. PARP-1 was originally described as a nuclear protein (9, 52), but we have recently found that PARP-1 can also be localized to the centrosome of cancer cell lines (22). Centrosomal localization of PARP-1, as well as chromosome instability in PARP-1-deficient (PARP-1−/−) cells, suggest that PARP-1 and/or poly(ADP-ribosyl)ation may also function as a regulator of centrosomes, and thus loss or reduction of PARP-1 may induce chromosome instability (aneuploidy) through altering either centrosome function and/or centrosome copy number.
P53 has been shown to physically interact with PARP-1, to be poly(ADP-ribosyl)ated by PARP-1 (30, 64, 67), and to show changes of its property (30). These observations led to an attractive hypothesis that PARP-1 and PARP-1-mediated poly(ADP-ribosyl)ation may control centrosome duplication partly by influencing p53 activity at the centrosomes.
We examined here the potential role of PARP-1 and/or PARP-1-mediated poly(ADP-ribosyl)ation in the maintenance of proper numbers of centrosomes. We found that extensive hyperamplification of centrosomes occurred both in cells treated with PARP inhibitor and in PARP-1−/− cells, indicating that PARP-1-mediated poly(ADP-ribosyl)ation is important for maintaining the proper copy number of centrosomes. Consistently, PARP-1−/− cells exhibit the deregulation of the centrosome duplication cycle and the chromosomal instability, including the aneuploidy (hypoploidy and hyperploidy). We further found that several centrosomal components were poly(ADP-ribosyl)ated, one of which was p53. These observations raise the possibility that PARP-1 and/or poly(ADP-ribosyl)ation may regulate the centrosome function.
MATERIALS AND METHODS
Cells.
PARP-1+/+ and PARP-1−/− primary mouse embryonic fibroblasts (PMEFs) were prepared from day 13.5 embryos derived from heterozygous crosses as described previously (60). PARP-1+/+ (F20) and PARP-1−/− (PARP-1 knocked out; clones A11 and A12) mouse embryonic fibroblasts (MEFs) were immortalized by standard 3T3 protocol (66). Cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum (FBS), penicillin (100 U/ml), and streptomycin (100 μg/ml) in an atmosphere containing 10% CO2 at 37°C.
Indirect immunofluorescence.
Cells were first extracted briefly with extraction buffer (0.75% Triton X-100, 5 mM PIPES, 2 mM EGTA [pH 6.7]) to eliminate free proteins and proteins loosely associated with organelles (45) and then were fixed with 10% formalin and 10% methanol for 20 min at 25°C, washed with phosphate-buffered saline (PBS), and permeabilized with 1% NP-40 in PBS for 5 min at 25°C. Cells were then incubated with blocking solution (10% normal goat serum in PBS) for 1 h and subjected to immunostaining. For coimmunostaining of γ-tubulin and PARP-1 or poly(ADP-ribose), cells were probed with anti-γ-tubulin monoclonal (Sigma) and anti-PARP-1 polyclonal (which does not cross-react with other PARP family members; Upstate Biotechnology) antibodies or with anti-γ-tubulin polyclonal (18) and anti-poly(ADP-ribose) monoclonal (23) antibodies for 1 h at 25°C. Cells were probed with anti-γ-tubulin polyclonal and anti-α-tubulin monoclonal (DM1A; Sigma) antibodies for 1 h at 25°C. The antibody-antigen complexes were detected with fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin G (IgG) and rhodamine-conjugated goat anti-mouse IgG antibodies by incubation for 1 h at 25°C. The samples were counterstained with DAPI (4′,6′-diamidino-2-phenylindole).
Electron microscopy.
Cells were fixed and processed for transmission electron microscopy as previously described (45). In brief, cells were fixed with 2.5% glutaraldehyde and 1% osmium tetroxide, embedded, and polymerized in epoxy resin. Thin-sectioned cells were stained with uranyl acetate and lead citrate and examined by using a Hitachi H7000 electron microscope.
Flow cytometry.
Cells were treated with trypsin and resuspended in 100 μl of a solution containing 250 mM sucrose, 40 mM sodium citrate (pH 7.6), and 5% dimethyl sulfoxide. Cells were then incubated in a solution containing 3.4 mM sodium citrate, 0.1% NP-40, 1.5 mM spermine tetrahydrochloride, and 0.5 mM Tris-HCl (pH 7.6) for 10 min, followed by RNase A (0.1 mg/ml) treatment for 10 min. Nuclei were stained for 15 min with propidium iodide (0.42 mg/ml), filtered through a 37-μm-pore-size nylon mesh, and analyzed with a dual-laser flow cytometer (FACScan; Becton Dickinson).
BrdU incorporation assay.
The assay was performed by using the bromodeoxyuridine (BrdU) labeling kit (Boehringer Mannheim). Briefly, cells were serum starved for 36 h and then treated with fresh medium containing 20% FBS and BrdU. At each point, cells were extracted and fixed in 70% ethanol in 50 mM glycine (pH 2.0) for 20 min at −20°C and then probed with anti-BrdU monoclonal antibody for 30 min at 37°C, followed by incubation with fluorescein isothiocyanate-conjugated sheep anti-mouse IgG.
Centrosome isolation and immunoblot analysis.
Centrosomes were prepared from PARP-1+/+ and PARP-1−/− MEFs as described previously (22, 37, 45). Exponentially growing cells were incubated with culture medium containing 1 μg of cytochalasin D/ml and 0.2 μM nocodazole for 1 h at 37°C to depolymerize the actin and microtubule filaments. Then, 1 × 107 to 3 × 107 cells were harvested by trypsinization and lysed in a solution containing 1 mM HEPES (pH 7.2), 0.5% NP-40, 0.5 mM MgCl2, and 0.1% 2-mercaptoethanol with proteinase inhibitors (EDTA-free proteinase inhibitor cocktail; Roche Diagnostics, Mannheim, Germany) and phosphatase inhibitors (50 mM sodium fluoride and 1 mM sodium orthovanadate). Swollen nuclei and chromatin aggregates were removed by centrifugation at 2,500 × g for 10 min, and the supernatant was filtered through a 37-μm nylon mesh. The supernatant was treated with 2 U of DNase I (Roche)/ml in 10 mM HEPES (pH 7.2) for 30 min on ice. The lysate was then underlaid with 60% sucrose solution (60% [wt/wt] sucrose in 10 mM PIPES [pH 7.2]-0.1% Triton X-100-0.1% 2-mercaptoethanol). Centrosomes were sedimented into the sucrose cushion by centrifugation at 10,000 × g for 30 min at 4°C in a Beckman L8.50B ultracentrifuge equipped with an SW50.1 rotor. After centrifugation by using a discontinuous gradient consisting of 500 μl of 70% sucrose, 300 μl of 50% sucrose, and 300 μl of 40% sucrose solutions, aliquots of 200 μl were collected from the bottom as fractions 1 to 7. Each fraction was diluted with 1 ml of 10 mM PIPES (pH 7.2), centrifuged for 10 min at 15,000 rpm, and then denatured in sodium dodecyl sulfate (SDS) sample buffer (62.5 mM Tris-HCl [pH 6.8], 10% glycerol, 2% SDS, 1.4% 2-mercaptoethanol, 0.001% bromophenol blue). Proteins from isolated centrosomes were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and electrotransferred onto an Immobilon-P membrane (Millipore). The membranes were incubated with the primary and the secondary antibodies in Tris-buffered saline with 0.05% Tween 20 for 60 min and then were washed in the same medium. The primary antibodies for these experiments included mouse monoclonal antibody for poly(ADP-ribose) (23, 40) and rabbit polyclonal antibody for γ-tubulin. The bound primary antibody was detected with alkaline phosphatase-conjugated secondary antibody and visualized with NBT/BCIP (Roche).
Immunodepletion of p53 from the enriched centrosome fractions.
Centrosomes purified by sucrose gradient fractionation were denatured in 10 mM PIPES buffer containing 0.5% SDS at 95°C for 10 min and diluted with PBS to 0.05% SDS. The samples were subjected to immunoprecipitation with anti-p53 mouse monoclonal antibody (DO-1; Santa Cruz and Pab421; Biomol) or normal mouse IgG (Santa Cruz). The immunocomplexes were precipitated with protein G-agarose beads (Amersham Pharmacia). Since the precipitated abundant IgG inhibits the detection of modified p53 by some cross-reaction with either rabbit polyclonal or mouse monoclonal antibodies against anti-poly(ADP-ribose) on a Western blot, immunodepleted proteins in the supernatants were resolved by SDS-PAGE.
Preparation of anti-poly(ADP-ribose) antibody affinity column and detection of poly(ADP-ribosyl)ated p53 in the centrosome.
10H-2 cells producing monoclonal anti-poly(ADP-ribose) antibody were incubated in original medium (23). Produced antibody was purified by HiTrap protein G HP column (Amersham Pharmacia Biotech) and coupled in a HiTrap NHS-activated HP column (Amersham Pharmacia Biotech) according to the manufacturer's instructions. The heat-denatured centrosome fraction (0.5% SDS at 95°C and diluted with PBS to 0.05% SDS) was applied to the anti-poly(ADP-ribose) antibody affinity column and eluted by elution buffer of pH 5.0 and pH 3.0. Each fraction was resolved by SDS-PAGE and immunoblotted with anti-p53 antibody (DO-1 and Pab421) or normal mouse IgG.
In vitro poly(ADP-ribosyl)ation assay with enriched centrosome fractions.
Centrosomes were resuspended in the in vitro poly(ADP-ribosyl)ation reaction buffer (50 mM Tris-HCl, 2 mM dithiothreitol, 20 mM MgCl2, 0.5% NP-40) and incubated with 20 μM NAD+ and 12.5 μCi of [32P]NAD for 20 min at 25°C. After the reaction, the samples were subjected to immunoprecipitation with anti-p53 antibody (DO-1 and Pab421), resolved by SDS-PAGE, and analyzed with a BAS Analyzer (Fuji Film).
RESULTS
PARP-1 and poly(ADP-ribose) localize to the centrosomes.
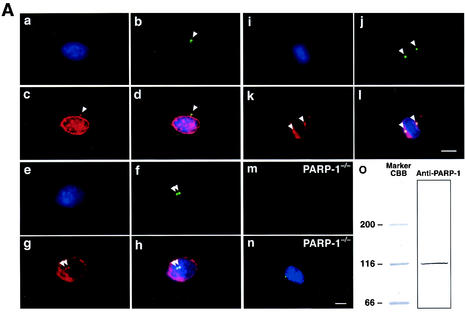
We have previously shown that PARP-1 localizes to the centrosomes, by immunocytochemical (antibodies F-2 and F-1-23) and biochemical analysis, in human osteosarcoma cells (SaOs2) and glioma cells (U-251) (22). We extended this observation to MEFs. Cells were then coimmunostained for PARP-1 and γ-tubulin, a major centrosomal protein (reviewed in reference 21). In wild-type MEFs, centrosomal colocalization of PARP-1 and γ-tubulin was observed in virtually all cells examined: PARP-1 localized to both unduplicated and duplicated centrosomes during interphase and to spindle poles during mitosis (representative immunostaining images are shown in Fig. 1Aa to l). The PARP-1 antibody (Upstate Biotechnology) detected a band at 116 kDa, which corresponded to the molecular mass of PARP-1 (Fig. 1Ao). In addition, PARP-1−/− MEFs were not stained by this antibody (Fig. 1Am and n). Thus, PARP-1 antibody did not detect other PARP family proteins with different molecular masses. Therefore, consistent with our previous findings, PARP-1 associates with the centrosomes throughout the cell cycle, and centrosomal association of PARP-1 is not specific to a particular species or cell type.
FIG. 1.
(A) PARP-1 physically associates with centrosomes throughout the cell cycle. Exponentially growing wild-type MEFs were coimmunostained with anti-γ-tubulin monoclonal antibodies (panels b, f, and j, green) and anti-PARP-1 polyclonal antibodies (panels c, g, and k, red). Cells were also counterstained with DAPI (panels a, e, and i, blue). Panels d, h, and l show the overlay images. Panels a to d show a cell with unduplicated centrosome. Panels e to h show a cell with duplicated centrosomes. Panels i to l show a mitotic cell. Arrowheads point to centrosomes. Panels m and n show that negative immunostaining of PARP-1−/− MEFs by anti-PARP-1 antibody. Exponentially growing PARP-1−/− MEFs were coimmunostained with anti-PARP-1 polyclonal (panel m, red) and anti-γ-tubulin polyclonal (panel n, green) antibodies. Cells were also counterstained with DAPI (panel n, blue). Scale bar, 10 μm. Wild-type MEFs were lysed in RIPA buffer, resolved by SDS-PAGE, and immunoblotted with anti-PARP-1 antibody (panel o). (B) Centrosomal proteins are poly(ADP-ribosyl)ated. Exponentially growing wild-type MEFs were coimmunostained with anti-γ-tubulin polyclonal (panels b, f, and j, red) and anti-poly(ADP-ribose) monoclonal (panels c, g, and k, green) antibodies. Cells were also counterstained with DAPI (panels a, e, and i, blue). Panels d, h, and l show the overlay images. Panels a to d show a cell with unduplicated centrosome. Panels e to h show a cell with duplicated centrosomes. Panels I to l show a mitotic cell. The arrowheads point to centrosomes. Panels m and n show the immunostaining of PARP-1−/− MEFs by anti-poly(ADP-ribose) antibody. Exponentially growing PARP-1−/− MEFs were coimmunostained with anti-poly(ADP-ribose) monoclonal (panel m, green) and anti-γ-tubulin polyclonal (panel n, red) antibodies. Cells were also counterstained with DAPI (panel n, blue). Scale bar, 10 μm.
PARP catalyzes polymerization of ADP-ribose residues of NAD+, adding long-branched chains of poly(ADP-ribose) to a variety of proteins (9, 13, 52). Centrosomal localization of PARP-1 suggests that at least some of the centrosomal components may be poly(ADP-ribosyl)ated. To test this, exponentially growing wild-type MEFs were coimmunostained for poly(ADP-ribose) and γ-tubulin. Anti-poly(ADP-ribose) antibody-reactive signals were readily observed in both unduplicated and duplicated centrosomes during interphase and at spindle poles during mitosis in MEFs (representative images are shown in Fig. 1Ba to l). In addition, the immunostaining for anti-poly(ADP-ribose) antibody in PARP-1−/− MEFs showed much weaker staining than in wild-type MEFs (compare Fig. 1Bc and m) and indicated that PARP-1-mediated poly(ADP-ribosyl)ation of the centrosome is the most common type in the PARP family. These observations suggest that at least some of centrosomal components are poly(ADP-ribosyl)ated.
Suppression of poly(ADP-ribosyl)ation results in centrosome hyperamplification.
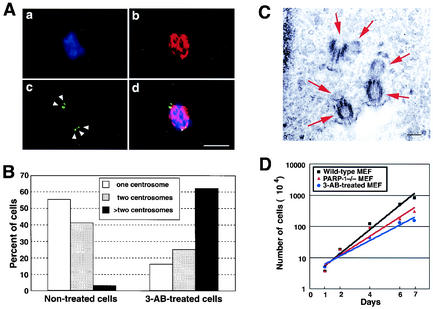
To examine whether poly(ADP-ribosyl)ation is involved in centrosome regulation, wild-type MEFs were cultured for a week in the presence of 3-aminobenzamide (3-AB), a potent inhibitor of poly(ADP-ribosyl)ation (29, 35, 51). Cells were then coimmunostained for γ-tubulin and α-tubulin. Extensive centrosome hyperamplification was observed in ∼60% of the 3-AB-treated cells (Fig. 2A and B) compared to the untreated cells, among which <5% had hyperamplified centrosomes (Fig. 2B). To rule out the possibility that the dot signals detected by anti-γ-tubulin antibody are fragmented centrosomes, we used electron microscopy to confirm the existence of a pair of centrioles in each hyperamplified centrosome (Fig. 2C). Each cell examined contained more than four centrioles. Representative images from one such cell, in which we found three centrosomes, showed that each centrosome contained two centrioles, as shown in Fig. 2C. These data suggest that the multiple anti-γ-tubulin antibody-reactive signals represent intact centrosomes. Moreover, it is possible that the hyperamplified centrosomes were functionally intact, since they serve as spindle poles during mitosis (Fig. 2A). As a negative control, we used 3-aminobenzoic acid, a 3-AB homologue that does not inhibit PARP activity, and found that the 3-aminobenzoic acid-treated cells do not change the number of cells and centrosomes (data not shown). Thus, inhibition of poly(ADP-ribosyl)ation results in centrosome hyperamplification.
FIG. 2.
Inhibition of PARP induces abnormal centrosome amplification. (A) Representative immunostaining of abnormally amplified centrosomes in 3-AB-treated MEFs. Wild-type MEFs were cultured in the presence of 7 mM 3-AB for 1 week. Cells were coimmunostained for anti-γ-tubulin polyclonal (panel c, green) and anti-α-tubulin monoclonal (panel b, red) antibodies. Cells were also counterstained with DAPI (panel a, blue). Panel d shows the overlay images. (B) Statistical analysis of centrosome hyperamplification in 3-AB-treated MEFs. The number of centrosomes per cell (i.e., 1, 2, or >2 centrosomes) was determined by coimmunostaining of α-tubulin and γ-tubulin of cells that were cold treated and briefly extracted before fixation. For the analysis, >200 cells were examined. (C) Electron micrographs of thin sections in 3-AB-treated cells. The arrows indicate the centriole. Scale bar, 0.2 μm. (D) Kinetic analysis of cell growth. Wild-type MEFs, PARP-1−/− MEFs, and 3-AB-treated cells were treated with trypsin, negatively stained with 0.4% trypan blue, and counted on a hemacytometer.
Additionally, we performed the kinetic analysis of cell growth in 3-AB-treated MEFs and PARP-1−/− MEFs. The number of 3-AB-treated cells and PARP-1−/− MEFs increased but at a lower rate compared to the PARP-1+/+ MEFs (Fig. 2D). However, the fluorescence-activated cell sorting (FACS) analysis and morphology indicated that 3-AB-treated cells and PARP-1−/− MEFs included the multinuclei and/or single larger nucleus but not dead cells (Fig. 4C and unpublished observations). Thus, the decreased growth rate and cell number indicates the failure of mitosis, including abnormal centrosome behavior.
FIG. 4.
(A) Uncoupling of initiation of DNA and centrosome duplication in PARP-1−/− MEFs. PARP-1+/+ and PARP-1−/− MEFs were serum starved for 36 h, followed by serum stimulation with the medium containing 20% FBS and BrdU. At indicated time points, cells were briefly extracted, fixed, and coimmunostained with anti-BrdU monoclonal and anti-γ-tubulin polyclonal antibodies. The number of centrosomes per cell and BrdU incorporation were scored by fluorescence microscopy. For each immunostaining, >200 cells were examined. (B) Centrosome reduplication assay. PARP-1+/+ and PARP-1−/− MEFs were incubated for 36 h with Aph, stained with an antibody against γ-tubulin, and analyzed by immunofluorescence microscopy. The number of centrosomes was then counted. (C) Flow cytometric analysis of PARP-1−/− MEFs. Nuclei were prepared and stained with propidium iodide for flow cytometric analysis. In addition to the two major peaks of nuclei at 2N and 4N that appeared in the DNA histograms of PARP-1+/+ MEFs, PARP-1−/− MEFs exhibit peaks at 1N, 3N, and 8N (arrows).
Loss of PARP-1 results in centrosome hyperamplification.
To confirm this finding and further test whether PARP-1 plays a role in either centrosomal function or numeral homeostasis of centrosomes, we examined MEFs derived from PARP-1−/− mice. Exponentially growing PARP-1−/− MEFs (A11) (Fig. 3Ac and d) and PARP-1+/+ MEFs (F20) (Fig. 3Aa and b) were immunostained for γ-tubulin. Statistical analysis showed that ∼35% of PARP-1−/− MEFs contained abnormal numbers (≥3) of centrosomes (Fig. 3B) compared to PARP-1+/+ MEFs, among which <10% had hyperamplified centrosomes. In addition, other immortalized PARP-1−/− MEFs (A12) showed the centrosome hyperamplification (data not shown). Abnormally amplified centrosomes seen in PARP-1−/− MEFs are not due to fragmentation of centrosomes, since we detected a pair of centrioles for each case of hyperamplified centrosomes detected by electron microscopy (data not shown). Thus, loss of PARP-1 results in centrosome hyperamplification.
FIG. 3.
Centrosome hyperamplification in PARP-1−/− MEFs. (A) Exponentially growing PARP-1−/− and PARP-1+/+ MEFs were immunostained with anti-γ-tubulin polyclonal antibody (red). Cells were also counterstained with DAPI (blue). Panels a and b are PARP-1+/+ MEFs, and panels c and d are PARP-1−/− MEFs. Panels a and c show mitotic cells, whereas panels b and d show interphase cells. The arrowheads point to centrosomes, and an arrow indicates aggregated centrosomes. Scale bar, 10 μm. (B) Statistical analysis of centrosome hyperamplification in PARP-1−/− MEFs. The number of centrosomes per cell (i.e., 1, 2, or >2 centrosomes) was determined by immunostaining of γ-tubulin. For the analysis, >200 cells were examined. Bars represent the average ± the standard error calculated from three independent measurements. (C) PARP-1+/+ and PARP-1−/− primary MEFs were immunostained with anti-γ-tubulin polyclonal (green) and/or α-tubulin monoclonal (red) antibodies. Cells were counterstainined with DAPI (blue). Representative images in panels a and b show the mitotic cells. Scale bar, 10 μm. (D) Quantification of centrosome hyperamplification in PARP-1−/− PMEFs. The number of centrosomes per cell (1, 2, or >2 centrosomes) was determined by immunostaining of γ-tubulin. For the analysis, >200 cells were examined.
To extend this observation, we prepared PMEFs from E13.5 PARP-1+/+ and PARP-1−/− embryos. These PARP-1+/+ and PARP-1−/− PMEFs were immunostained for γ-tubulin and/or α-tubulin. Representative images are shown in Fig. 3C. Quantification of all images showed that ca. 27 to 32% of the PARP-1−/− PMEFs and ca. 0 to 3% of the PARP-1+/+ PMEFs contained an abnormal number (≥3) of centrosomes (Fig. 3D). These results confirmed that the centrosome hyperamplification occurred in PARP-1−/− PMEFs before immortalization.
Abrogation of the regulatory mechanisms that ensure the coordinated progression of centrosome duplication and other cell cycle events, including DNA duplication, and that prevent reduplication of duplicated centrosome within the same cell cycle results in the hyperamplification of centrosomes (7, 27). We examined whether the centrosome duplication cycle was deregulated in PARP-1−/− MEFs, especially with respect to coordination between the initiation of centrosome and DNA duplication. Cells were serum starved for 36 h and then serum stimulated in the medium containing BrdU to monitor S-phase entry. At every 4 h for a period of 16 h, BrdU incorporation and the number of centrosomes per cell were scored (Fig. 4A). Uncoupling of centrosome and DNA duplication cycle was evident in PARP-1−/− MEFs. For instance, the percent increase in the number of cells with ≥2 centrosomes during the initial 8 h of serum stimulation [denoted as RCEN(0 to 8 h)] was 24.8% (3.1%/h) in PARP-1−/− MEFs, which is significantly higher than 6.4% (0.8%/h) in PARP-1+/+ MEFs. However, during this period the increases in BrdU incorporation [denoted as RBU(0 to 8 h)] were similar for PARP-1−/− and PARP-1+/+ MEFs: 10.4% (1.3%/h) for PARP-1+/+ MEFs and 12% (1.5%/h) for PARP-1−/− MEFs. The coordination of centrosome and DNA duplication was significantly uncoupled in PARP-1−/− MEFs.
To confirm these data, we examined the centrosome profiles of cells arrested at the G1/S boundary (by aphidicolin [Aph] or hydroxyurea treatment for 20 to 60 h) (3), and the number of centrosomes per cell was analyzed. Wild-type and PARP-1−/− MEFs were treated with Aph for 36 h, immunostained for centrosomes, and assayed for the number of centrosomes (Fig. 4B and Fig. 3B). In PARP-1−/− MEFs, there was a substantial decrease in the number of cells with one centrosome (from 37 to 10%) and an increase in the number of cells with more than two centrosomes (from 37 to 59%). In contrast, there was no change in the centrosome profile in wild-type MEFs before and after Aph treatment. The centrosome should not undergo reduplication when the cell cycle is halted to S phase by Aph treatment. However, in PARP-1−/− MEFs, the centrosomes underwent reduplication in S-phase-arrested cells, suggesting that the loss of PARP-1 might cause the uncoupling of DNA and centrosome duplication.
Since hyperamplified centrosomes lead to an increased frequency of defective (multipolar) mitotic spindles and unbalanced segregation of chromosomes into daughter cells, we examined them by FACS. Flow cytometric analysis of the PARP-1−/− MEFs typically showed two peaks (2N and 4N; 70%) and a readily recognizable population of hypoploid (1N and 3N; 17%) and hyperploid (8N; 13%) cells (Fig. 4C). Additionally, we analyzed the chromosomal instability in another PARP-1−/− MEF (A12) by FACS. Although PARP-1−/− MEF A12 did not show the same chromosomal abnormality formed in PARP-1−/− MEF A11 (1N, 3N, and 8N), ∼20% of the cell showed the hyperploidy and hypoploidy, which could be caused by the centrosome abnormality (data not shown). These hypo- and hyperploid cells were not observed in PARP-1+/+ MEFs. Thus, the loss of PARP-1 causes chromosomal instability, including hypoploidy (1N and 3N) and hyperploidy (8N) with hyperamplified centrosomes.
Loss of PARP-1 results in hypo-poly(ADP-ribosyl)ation of centrosomes.
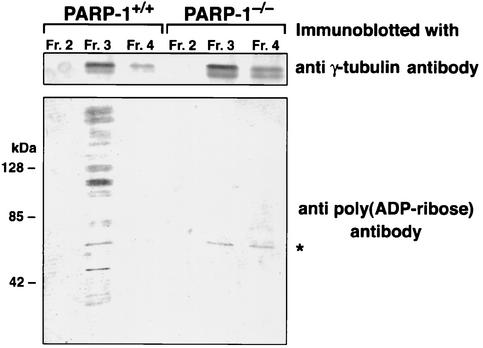
The findings that PARP-1 localizes to the centrosomes and that centrosomes can be detected by anti-poly(ADP-ribose) antibody (Fig. 1) strongly suggest that poly(ADP-ribosyl)ation of centrosomes are catalyzed by PARP-1. If this is the case, the absence of reduced poly(ADP-ribosyl)ation should be observed in the centrosomes of PARP-1−/− MEFs. To test this, centrosomes were isolated from PARP-1+/+ and PARP-1−/− MEFs by discontinuous sucrose gradient fractionation (see Materials and Methods). The fraction enriched for centrosomes were subjected to immunoblot analysis with anti-γ-tubulin antibody (Fig. 5, upper panel) and with anti-poly(ADP-ribose) antibody (lower panel). Poly(ADP-ribosyl)ation was abundantly detected in the centrosome fraction of PARP-1+/+ MEFs, whereas it was barely detectable in that of PARP-1−/− MEFs. One band that appeared in PARP-1−/− MEFs, which also appeared in PARP-1+/+ MEFs, may be a poly(ADP-ribosyl)ated protein catalyzed by another PARP family member (indicated by asterisk). Since PARP-1 or poly(ADP-ribose) is known to localize abundantly on chromosomes, we tested the possibility that the centrosome fraction may be contaminated with DNA-protein complexes by immunoblot analysis with anti-histone H1 antibody. We performed DNase I digestion during the preparation of centrosomes (see Materials and Methods) and did not detect histones in the centrosome fraction, thus excluding the possibility of chromosome contamination (data not shown). We therefore concluded that some centrosomal proteins are poly(ADP-ribosyl)ated by PARP-1 and that the loss of PARP-1 results in the reduction of poly(ADP-ribosyl)ation of centrosomal components.
FIG. 5.
Hypo-poly(ADP-ribosyl)ation of centrosome in PARP-1−/− MEFs. The cell homogenates derived from exponentially growing PARP-1+/+ and PARP-1−/− MEFs (∼2 × 107 cells) were subjected to a discontinuous sucrose gradient fractionation. The fractions were subjected to immunoblot analysis for γ-tubulin (upper panels) to identify the centrosomal fractions. The fraction most enriched for centrosomes (fraction 3) and neighboring fractions (fractions 2 and 4) were subjected to immunoblot analysis for poly(ADP-ribose) (lower panels). The asterisk indicates a nonspecific band or a protein poly(ADP-ribosyl)ated by other PARP family.
Identification of p53 as one of the poly(ADP-ribosyl)ated centrosomal proteins.
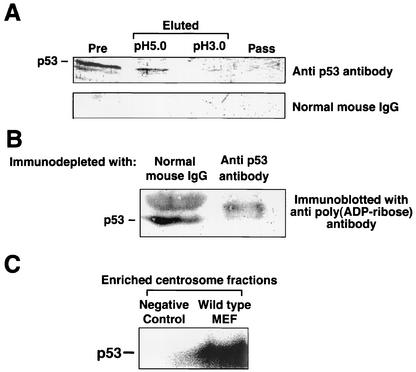
p53 has been shown to localize at centrosomes and to control centrosome duplication (18, 59). Moreover, p53 has been shown to be one of the targets of PARP-1 (30, 67). For these reasons, we examined whether p53 could be a target of poly(ADP-ribosyl)ation by PARP-1 at centrosomes. Centrosomes were purified from PARP-1+/+ MEFs by sucrose gradient fractionations. The centrosomes were then resuspended in PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] buffer containing 0.5% SDS, heat-denatured, and diluted with buffer to 0.05% SDS. The heat-denatured centrosome fraction derived from wild-type MEFs was passed through the anti-poly(ADP-ribose) antibody affinity column. The pass-through fraction and two eluates, by successive elutions at pH 5.0 and pH 3.0, were analyzed by immunoblotting with anti-p53 antibody. p53 was not detected in the pass-through fraction, whereas readily detectable levels of p53 were present in the pH 5.0 eluates and, to a lesser degree, in the pH 3.0 eluates (Fig. 6A).
FIG. 6.
p53 is poly(ADP-ribosyl)ated in the centrosome in vivo and in vitro. (A) Detection of poly(ADP-ribosyl)ated p53 in centrosome by using an affinity column of anti-poly(ADP-ribose) monoclonal antibody. The centrosome fraction of wild-type MEFs were heat denatured in 10 mM PIPES buffer containing 0.5% SDS at 95°C for 10 min and then diluted with PBS to 0.05% SDS. The denatured centrosomes were applied onto the column of anti-poly(ADP-ribose) antibody. The samples before appliaction to the column (lane 1 [Pre]), the eluates (lane 2 [eluted at pH 5.0] and lane 3 [eluted at pH 3.0]), and a pass-through fraction (lane 4 [Pass]) were resolved by SDS-PAGE and immunoblotted with anti-p53 monoclonal antibody (upper panel) or normal mouse IgG (lower panel). (B) Detection of poly(ADP-ribosyl)ated p53 in centrosomes. The centrosomes isolated from wild-type MEFs were heat denatured as described for panel A and subjected to immunodepletion with normal mouse IgG or anti-p53 monoclonal antibody. The supernatants after precipitation of the antibody-antigen complexes were immunoblotted with anti-poly(ADP-ribose) antibody. (C) In vitro poly(ADP-ribosyl)ation of p53 in the isolated centrosomes. The enriched centrosome fractions were resuspended in the in vitro poly(ADP-ribosyl)ation reaction buffer and were incubated with [32P]NAD. A negative control was subjected to analysis with the enriched centrosome fractions derived from PARP-1−/− MEFs. After the reaction, the centrosomes were heat denatured as described for panel A and subjected to immunoprecipitation with anti-p53 antibody. The immunoprecipitates were resolved by SDS-PAGE and subjected to phospho-imaging analysis.
In other experiments, the enriched and denatured centrosomes were immunodepleted by anti-p53 monoclonal antibody and analyzed by immunoblotting with anti-poly(ADP-ribose) antibody. When the centrosomes were immunodepleted with the control mouse IgG, there was a clear band at a molecular mass of ∼53 kDa, which was absent in centrosomes immunodepleted for p53 (Fig. 6B). The nature of a big band seen just above the p53 protein is not clear, but it might be a protein band modified by a longer chain of poly(ADP-ribose). These results confirm that p53 is one of the poly(ADP-ribosyl)ated centrosomal proteins.
We next examined whether centrosomal PARP-1 directly catalyzes poly(ADP-ribosyl)ation of p53. The centrosome fractions were resuspended in the in vitro poly(ADP-ribosyl)ation reaction buffer, incubated with [32P]NAD, heat denatured in the presence of 0.5% SDS, diluted with the buffer to 0.05% SDS, and then immunoprecipitated with anti-p53 antibody. The immunoprecipitates were resolved by SDS-PAGE, and radiolabeled proteins were analyzed by a phospho-imaging analyzer. [32P]NAD-labeled p53 was detected in the centrosome fraction prepared from wild-type MEFs, whereas no detectable label from p53 was observed in the negative control with the centrosome fraction prepared from PARP-1−/− MEFs (Fig. 6C). Thus, centrosomal PARP-1 can catalyze poly(ADP-ribosyl)ation of p53 in vitro, strongly suggesting that p53 is one of the targets of poly(ADP-ribosyl)ation by PARP-1 in the centrosome.
DISCUSSION
Involvement of PAPR-1 in centrosome behavior.
We have previously reported that PARP-1 is localized at centrosomes throughout the cell cycle (22), suggesting that PARP-1 and PARP-1-mediated poly(ADP-ribosyl)ation may play some roles in centrosome behavior. Here, we provide the evidence that PARP-1-mediated poly(ADP-ribosyl)ation is important for numeral homeostasis of centrosomes: treatment of normal cells with 3-AB, a potent inhibitor of poly(ADP-ribosyl)ation (29, 35, 51), induced abnormally amplified centrosomes. Consistent with the inhibitor studies, we found that PARP-1-null cells exhibited a high degree of centrosome abnormality. This conclusion has been supported by analysis of immortalized and freshly isolated PMEFs.
Since each daughter cell inherits one centrosome, the centrosome must duplicate prior to the subsequent mitosis and do so only once in each cell cycle. Centriole, the core component of centrosome, initiates duplication near the G1/S boundary of the cell cycle, and centrosome duplication is completed in the G2 phase. Thus, the centrosome duplication cycle proceeds in a precise coordination with other cell cycle events, including DNA duplication (reviewed in reference 63). Abrogation of the regulation that coordinates centrosome and DNA duplication will likely increase the frequency of centrosome hyperamplification (7). A close examination of PARP-1−/− cells revealed that the initiation of centrosome and DNA duplication is uncoupled in significant numbers of cells. Centrosomes initiate duplication much earlier than S-phase entry and reduplication at S-phase arrest in PARP-1−/− MEFs. Thus, loss of PARP-1 causes the uncoupling of the centrosome and DNA duplication, suggesting the occurrence of centrosome hyperamplification.
A quite large family of PARP enzymes and poly(ADP-ribosyl)ated proteins have been identified and characterized over the past 3 years. Especially, it has been reported that PARP-1 is localized to the centromere on the chromosomes and interacts with CENPA, CENPB, and Bub3 (49); that vault PARP (193 kDa) is localized to the nucleus during interphase or the mitotic spindle during metaphase (24); and that tankyrase (142 kDa) is localized to the telomere, nuclear envelope, nuclear pore complex, and pericentrimatrix (55). Furthermore, it has been reported that purified chicken NAD-arginine ADP-ribosyltransferase mono-ADP-ribosylates tubulin from bovine brain, and this leads to inhibition of self-assembly and rapid depolymerization (50). It is not yet known whether tubulin and/or microtubule proteins are poly(ADP-ribosyl)ated by PARP family proteins in mammalian cells. Together, these reports suggest that poly(ADP-ribosyl)ation might be involved in the chromosome stability through the mitotic machinery. In the present study, immunostaining and immunoblotting with enriched centrosome fractions in PARP-1−/− MEFs by anti-poly(ADP-ribose) antibody (Fig. 1Bm and 5) indicated that PAPR-1 is a major enzyme responsible for poly(ADP-ribosyl)ation of proteins in the centrosome.
Loss of PARP-1 and poly(ADP-ribosyl)ation could cause the chromosomal instability through centrosome hyperamplification.
Centrosome amplification causes various types of chromosomal instability. (i) Hyperamplified centrosomes with tripolar mitotic spindles would show a typical chromosomal instability patterns (1N, 3N, and 8N). (ii) Hyperamplified centrosomes with pseudobipolar (clustering) mitotic spindles would cause aneuploidy (changes of a few numbers of chromosomes). In addition, hyperamplified centrosomes might cause other chromosomal or genomic instabilities (structural chromosome abnormalities, leading to chromosome breakage and micronucleus-multinucleus formation). It is well known that cells treated with a potent PARP inhibitor (3-AB) show genomic instability (34, 35, 65). However, other researchers recently reported that a new PARP inhibitor, GPI6150, did not cause tetraploidy (54). However, whether this drug causes other types of chromosomal instability is not known. We detected here that ∼60% of the cells treated with 3-AB showed the centrosome hyperamplification (Fig. 2B), with ∼15% of the cells showing hyperploidy (not tetraploidy) on FACS analysis (data not shown). Although it is still unknown whether 3-AB-mediated loss of poly(ADP-ribosyl)ation caused only tetraploidization, it is possible that the cells treated with 3-AB could cause the chromosomal and/or genomic instability through the centrosome abnormality. Several groups of researchers have recently developed PARP-1 knockout mice (41, 48, 53, 60). Each PARP-1−/− mouse demonstrated chromosomal instability, including aneuploidy (hyperploidy and/or tetraploidy) (12, 14, 38, 48, 53, 60, 62, 66). Recent studies have shown that centrosome hyperamplification leads to the formation of multipolar spindles and unequal segregation of chromosomes to daughter cells (18, 57) and is a major factor contributing to chromosome instability (11, 28, 47). FACS analysis revealed that a profile of PARP-1+/+ cells showed a typical pattern characterized by two major peaks of 2N and 4N. In contrast, in addition to these two major peaks, ∼30% of PARP-1−/− cells showed a typical chromosomal instability, which is either hypoploidy (1N and 3N; 17%) or hyperploidy (8N; 13%). Although the cells with hyperploidy (8N; 13%) might be produced by errors of cytokinesis and/or abnormal centrosome hyperamplification, the cells with hypoploidy (1N and 3N; 17%) might be produced by hyperamplified centrosome but not from errors of cytokinesis. Thus, the loss of PARP-1 causes chromosomal instability, including hyperploidy and hypoploidy, possibly through centrosome hyperamplification. However, it was reported that another immortalized PARP-1−/− cells (i.e., A12) did not show the chromosomal instability pattern as 1N, 3N, and 8N (20; unpublished data). This means the centrosome abnormality could cause many patterns of chromosome distribution, including aneuploidy and/or typical 1N, 3N, and 8N populations. For the present, it is known that the chromosomal instability is caused by abnormalities of centrosome duplication, microtubule dynamics, chromosome condensation, kinetochore assembly, and/or chromatid cohesion. Furthermore, the immortalization process might select certain cell populations with various chromosomal instability patterns because specific cells, which contain more proliferation genes, could be selected by this process. In the present study, we detected the centrosome hyperamplification in both immortalized and primary PARP-1−/− MEFs. Thus, PARP-1 is important for the centrosome regulation and a part of the chromosomal instabilities could be caused by the centrosome abnormality through the loss of PARP-1.
At present, the mechanism of abnormal hyperamplification of centrosomes in PARP-1−/− MEFs or in the presence of PARP inhibitor is still unknown. Recently, Borel et al. showed that centrosome amplification occurs in two steps: (i) failure to arrest at a G1 tetraploidy checkpoint after failure to segregate the genome in mitosis and (ii) clustering of centrosomes at a single spindle pole in subsequent tetraploid or aneuploid mitosis (5). Moreover, Meraldi et al. indicated that a major route to centrosome amplification is through defects in cell division with HeLa cells (32). Based on our data, it is plausible that there might be some fractions of PARP-1−/− cells with hyperamplified centrosomes due to defects in cytokinesis. However, as shown in Fig. 4A, centrosomes initiate duplication much earlier than S-phase entry in serum-stimulated PARP-1−/− MEFs, and in Fig. 4B ca. 60% of the PARP-1−/− MEFs have hyperamplified centrosomes, whereas <10% of the PARP-1+/+ MEFs have hyperamplified centrosomes when DNA synthesis was stopped by Aph. This indicates that the hyperamplification of the centrosomes in PARP-1−/− MEFs might be caused not only by defects of cytokinesis after DNA synthesis but also by the uncoupling of centrosome duplication and DNA synthesis. Therefore, as a further study, it is important to clarify the mechanism of centrosome hyperamplification and its significance for tumor biology in general.
p53 is a target for poly(ADP-ribosyl)ation by PARP-1 in the centrosome.
Poly(ADP-ribosyl)ation catalyzed by PARP-1 is one of the major posttranslational modifications of proteins, which would greatly affect the properties and functions of the target proteins. We found that some of centrosomal components were poly(ADP-ribosyl)ated when the centrosomes isolated from PARP-1+/+ cells were examined. More importantly, almost no poly(ADP-ribosyl)ation was detected in the centrosomes prepared from PARP-1−/− cells. Together with the finding that PARP-1 localizes to the centrosomes, these observations suggest that PARP-1 posttranslationally modifies the proteins that associate with centrosomes. Indeed, we identified p53 as one of the proteins to be modified in vivo. Consistent with this we found, by using the in vitro poly(ADP-ribosyl)ation assay of the isolated centrosomes, that centrosomal PARP-1 can catalyze poly(ADP-ribosyl)ation of p53 at the centrosome. Since p53 has been shown to control the centrosome duplication cycle through its physical association with centrosomes (18, 58), our findings put forward an attractive hypothesis: that p53 may be one of the primary targets of PARP-1 and that poly(ADP-ribosyl)ation of p53 in the centrosome may regulate the centrosome function through p53 activity at the centrosome. This in turn controls the proper progression of the centrosome duplication cycle.
Role of reversible poly(ADP-ribosyl)ation in the centrosome.
Recent findings suggest that the modifications of centrosomal proteins are important for centrosome (centriole) behavior. We found that some of the centrosomal proteins were poly(ADP-ribosyl)ated in vivo. Many proteins involved in centrosome behavior, as well as the cell cycle, may need such dynamic changes through posttranslational modifications. Thus, it is possible that such dynamic structural or functional changes of proteins associated with centrosomes through reversible poly(ADP-ribosyl)ation by PARP-1 and PARG may be required for proper centrosome behavior. Since we showed that, for centrosome duplication, PARP-1-mediated poly(ADP-ribosyl)ation of some centrosome proteins is important, PARG-mediated hydrolysis of poly(ADP-ribose) bound to proteins may also be important for centrosome properties. Indeed, immunoblot analysis of centrosomes isolated from wild-type cells with anti-poly(ADP-ribose) antibody detected several distinct bands in addition to p53. Thus, identification of these poly(ADP-ribosyl)ated centrosomal component(s) and clarification of possible shuttling between the centrosome and nucleus will further our understanding not only of the role of PARP-1 and poly(ADP-ribosyl)ation for the centrosome structure and function but also of the regulation of centrosome duplication in general.
Since PARP-1 is known to be activated by binding to either single- or double-stranded DNA breaks after exposure to DNA-damaging agents, it is of great interest to examine what triggers PARP-1-mediated poly(ADP-ribosyl)ation in centrosomes. Indeed, centrosomal PARP-1 has poly(ADP-ribosyl)ation activity in the absence of DNA (Fig. 6C). The present observations suggest an attractive hypothesis in which the poly(ADP-ribosyl)ation activity of centrosomal PARP-1 may be triggered by some posttranslational modification or interaction with certain proteins. Identification of the trigger(s) will certainly advance our understanding of the involvement of PARP-1 and poly(ADP-ribosyl)ation in centrosome behavior.
Acknowledgments
We thank L. Haddad, Y. Nogi, S. Ohashi, A. Kabayama, N. Sugae, and Y. Ohno for technical assistance.
This work was supported in part by Grants-in-Aid for Cancer Research from the Ministry of Health, Labour, and Welfare (Japan), the Japan Society for the Promotion of Sciences (Japan), and the National Institutes of Health (CA90522 and CA90925). W.-M. Tong and Z.-Q. Wang are supported by the Association for International Cancer Research (United Kingdom) and by the Association pour la Recherche contre le Cancer (France).
REFERENCES
- 1.Adolph, K. W., and M. K. Song. 1985. Decrease in ADP-ribosylation of HeLa non-histone proteins from interphase to metaphase. Biochemistry 24:345-352. [DOI] [PubMed] [Google Scholar]
- 2.Amstad, P. A., G. Krupitza, and P. A. Cerutti. 1992. Mechanism of c-fos induction by active oxygen. Cancer Res. 52:3952-3960. [PubMed] [Google Scholar]
- 3.Balczon, R., L. Bao, W. E. Zimmer, K. Brown, R. P. Zinkowski, and B. R. Brinkley. 1995. Dissociation of centrosome replication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. J. Cell Biol. 130:105-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blair Zajdel, M. E., and G. E. Blair. 1988. The intracellular distribution of the transformation-associated protein p53 in adenovirus-transformed rodent cells. Oncogene 2:579-584. [PubMed] [Google Scholar]
- 5.Borel, F., O. D. Lohez, F. B. Lacroix, and R. L. Margolis. 2002. Multiple centrosomes arise from tetraploidy checkpoint failure and mitotic centrosome clusters in p53 and RB pocket protein-compromised cells. Proc. Natl. Acad. Sci. USA 99:9819-9824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bornens, M. 1992. Structure and functions of isolated centrosomes, p. 1-43. In V. I. Kaknins (ed.), The centrosome. Academic Press, Inc., New York, N.Y.
- 7.Brinkley, B. R., and T. M. Goepfert. 1998. Supernumerary centrosomes and cancer: Boveri's hypothesis resurrected. Cell Motil. Cytoskeleton 41:281-288. [DOI] [PubMed] [Google Scholar]
- 8.Brown, C. R., S. J. Doxsey, E. White, and W. J. Welch. 1994. Both viral (adenovirus E1B) and cellular (hsp 70, p53) components interact with centrosomes. J. Cell Physiol. 160:47-60. [DOI] [PubMed] [Google Scholar]
- 9.Bürkle, A. 2001. Poly(ADP-ribosyl)ation, a DNA damage-driven protein modification and regulator of genomic instability. Cancer Lett. 163:1-5. [DOI] [PubMed] [Google Scholar]
- 10.Bürkle, A., R. Heilbronn, and H. zur Hausen. 1990. Potentiation of carcinogen-induced methotrexate resistance and dihydrofolate reductase gene amplification by inhibitors of poly(adenosine diphosphate-ribose) polymerase. Cancer Res. 50:5756-5760. [PubMed] [Google Scholar]
- 11.Carroll, P. E., M. Okuda, H. F. Horn, P. Biddinger, P. J. Stambrook, L. L. Gleich, Y. Q. Li, P. Tarapore, and K. Fukasawa. 1999. Centrosome hyperamplification in human cancer: chromosome instability induced by p53 mutation and/or Mdm2 overexpression. Oncogene 18:1935-1944. [DOI] [PubMed] [Google Scholar]
- 12.d'Adda di Fagagna, F., M. P. Hande, W. M. Tong, P. M. Lansdorp, Z. Q. Wang, and S. P. Jackson. 1999. Functions of poly(ADP-ribose) polymerase in controlling telomere length and chromosomal stability. Nat. Genet. 23:76-80. [DOI] [PubMed] [Google Scholar]
- 13.D'Amours, D., S. Desnoyers, I. D'Silva, and G. G. Poirier. 1999. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 342:249-268. [PMC free article] [PubMed] [Google Scholar]
- 14.de Murcia, J. M., C. Niedergang, C. Trucco, M. Ricoul, B. Dutrillaux, M. Mark, F. J. Oliver, M. Masson, A. Dierich, M. LeMeur, C. Walztinger, P. Chambon, and G. de Murcia. 1997. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc. Natl. Acad. Sci. USA 94:7303-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisk, H. A., and M. Winey. 2001. The mouse Mps1p-like kinase regulates centrosome duplication. Cell 106:95-104. [DOI] [PubMed] [Google Scholar]
- 16.Freed, E., K. R. Lacey, P. Huie, S. A. Lyapina, R. J. Deshaies, T. Stearns, and P. K. Jackson. 1999. Components of an SCF ubiquitin ligase localize to the centrosome and regulate the centrosome duplication cycle. Genes Dev. 13:2242-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fry, A. M., P. Meraldi, and E. A. Nigg. 1998. A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J. 17:470-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukasawa, K., T. Choi, R. Kuriyama, S. Rulong, and G. F. Vande Woude. 1996. Abnormal centrosome amplification in the absence of p53. Science 271:1744-1747. [DOI] [PubMed] [Google Scholar]
- 19.Goepfert, T. M., and B. R. Brinkley. 2000. The centrosome-associated Aurora/Ipl-like kinase family. Curr. Top. Dev. Biol. 49:331-342. [DOI] [PubMed] [Google Scholar]
- 20.Halappanavar, S. S., Y. L. Rhun, S. Mounir, L. M. Martins, J. Huot, W. C. Earnshaw, and G. M. Shah. 1999. Survival and proliferation of cells expressing caspase-uncleavable poly(ADP-ribose) polymerase in response to death-inducing DNA damage by an alkylating agent. J. Biol. Chem. 274:37097-37104. [DOI] [PubMed] [Google Scholar]
- 21.Joshi, H. C. 1994. Microtubule organizing centers and gamma-tubulin. Curr. Opin. Cell Biol. 6:54-62. [DOI] [PubMed] [Google Scholar]
- 22.Kanai, M., M. Uchida, S. Hanai, N. Uematsu, K. Uchida, and M. Miwa. 2000. Poly(ADP-ribose) polymerase localizes to the centrosomes and chromosomes. Biochem. Biophys. Res. Commun. 278:385-389. [DOI] [PubMed] [Google Scholar]
- 23.Kawamitsu, H., H. Hoshino, H. Okada, M. Miwa, H. Momoi, and T. Sugimura. 1984. Monoclonal antibodies to poly(adenosine diphosphate ribose) recognize different structures. Biochemistry 23:3771-3777. [DOI] [PubMed] [Google Scholar]
- 24.Kickhoefer, V. A., A. C. Siva, N. L. Kedersha, E. M. Inman, C. Ruland, M. Streuli, and L. H. Rome. 1999. The 193-kD vault protein, VPARP, is a novel poly(ADP-ribose) polymerase. J. Cell Biol. 146:917-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krupitza, G., and P. Cerutti. 1989. ADP-ribosylation of ADPR-transferase and topoisomerase I in intact mouse epidermal cells JB6. Biochemistry 28:2034-2040. [DOI] [PubMed] [Google Scholar]
- 26.Krupitza, G., and P. Cerutti. 1989. Poly(ADP-ribosylation) of histones in intact human keratinocytes. Biochemistry 28:4054-4060. [DOI] [PubMed] [Google Scholar]
- 27.Lange, B. M. H., and K. Gull. 1996. Structure and function of the centriole in animal cells; progress and questions. Trends Cell. Biol. 6:348-352. [DOI] [PubMed] [Google Scholar]
- 28.Lingle, W. L., W. H. Lutz, J. N. Ingle, N. J. Maihle, and J. L. Salisbury. 1998. Centrosome hypertrophy in human breast tumors: implications for genomic stability and cell polarity. Proc. Natl. Acad. Sci. USA 95:2950-2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacLaren, R. A., W. W. Au, and M. S. Legator. 1989. The effect of 3-aminobenzamide on X-ray induction of chromosome aberrations in Down syndrome lymphocytes. Mutat. Res. 222:1-7. [DOI] [PubMed] [Google Scholar]
- 30.Malanga, M., J. M. Pleschke, H. E. Kleczkowska, and F. R. Althaus. 1998. Poly(ADP-ribose) binds to specific domains of p53 and alters its DNA binding functions. J. Biol. Chem. 273:11839-11843. [DOI] [PubMed] [Google Scholar]
- 31.Mayor, T., Y. D. Stierhof, K. Tanaka, A. M. Fry, and E. A. Nigg. 2000. The centrosomal protein C-Nap1 is required for cell cycle-regulated centrosome cohesion. J. Cell Biol. 151:837-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meraldi, P., R. Honda, and E. A. Nigg. 2002. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53−/− cells. EMBO J. 21:483-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miwa, M., and T. Sugimura. 1971. Splitting of the ribose-ribose linkage of poly(adenosine diphosphate-ribose) by a calf thymus extract. J. Biol. Chem. 246:6362-6364. [PubMed] [Google Scholar]
- 34.Morgan, W. F., and J. E. Cleaver. 1982. 3-Aminobenzamide synergistically increases sister-chromatid exchanges in cells exposed to methyl methanesulfonate but not to ultraviolet light. Mutat. Res. 104:361-366. [DOI] [PubMed] [Google Scholar]
- 35.Morgan, W. F., and J. E. Cleaver. 1983. Effect of 3-aminobenzamide on the rate of ligation during repair of alkylated DNA in human fibroblasts. Cancer Res. 43:3104-3107. [PubMed] [Google Scholar]
- 36.Morris, V. B., J. Brammall, J. Noble, and R. Reddel. 2000. p53 localizes to the centrosomes and spindles of mitotic cells in the embryonic chick epiblast, human cell lines, and a human primary culture: an immunofluorescence study. Exp. Cell Res. 256:122-130. [DOI] [PubMed] [Google Scholar]
- 37.Moudjou, M., and M. Bornens. 1994. Method of centrosome isolation from cultured animal cells. Academic Press, Inc., San Diego, Calif.
- 38.Muiras, M. L., and A. Bürkle. 2000. Defending genomic stability over life span: a proposed role for PARP-1. Exp. Gerontol. 35:703-709. [DOI] [PubMed] [Google Scholar]
- 39.Nakayama, K., H. Nagahama, Y. A. Minamishima, M. Matsumoto, I. Nakamichi, K. Kitagawa, M. Shirane, R. Tsunematsu, T. Tsukiyama, N. Ishida, M. Kitagawa, and S. Hatakeyama. 2000. Targeted disruption of Skp2 results in accumulation of cyclin E and p27Kip1, polyploidy, and centrosome overduplication. EMBO J. 19:2069-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nozaki, T., M. Masutani, T. Akagawa, T. Sugimura, and H. Esumi. 1994. Noncovalent interaction between poly(ADP-ribose) and cellular proteins: an application of a poly(ADP-ribose)-Western blotting method to detect poly(ADP-ribose) binding on protein blotted filter. Biochem. Biophys. Res. Commun. 198:45-51. [DOI] [PubMed] [Google Scholar]
- 41.Nozaki, T., H. Fujihara, N. Kamada, O. Ueda, T. Takato, H. Nakagama, and T. Sugimura. 2001. Hyperploidy of embryonic fibroblasts derived from Parp-1 knockout mouse. Proc. Japan Acad. 77:121-124. [Google Scholar]
- 42.Ogata, N., K. Ueda, H. Kagamiyama, and O. Hayaishi. 1980. ADP-ribosylation of histone H1: identification of glutamic acid residues 2, 14, and the COOH-terminal lysine residue as modification sites. J. Biol. Chem. 255:7616-7620. [PubMed] [Google Scholar]
- 43.Ogata, N., K. Ueda, M. Kawaichi, and O. Hayaishi. 1981. Poly(ADP-ribose) synthetase, a main acceptor of poly(ADP-ribose) in isolated nuclei. J. Biol. Chem. 256:4135-4137. [PubMed] [Google Scholar]
- 44.Ohashi, Y., A. Itaya, Y. Tanaka, K. Yoshihara, T. Kamiya, and A. Matsukage. 1986. Poly(ADP-ribosyl)ation of DNA polymerase beta in vitro. Biochem. Biophys. Res. Commun. 140:666-673. [DOI] [PubMed] [Google Scholar]
- 45.Okuda, M., H. F. Horn, P. Tarapore, Y. Tokuyama, A. G. Smulian, P. K. Chan, E. S. Knudsen, I. A. Hofmann, J. D. Snyder, K. E. Bove, and K. Fukasawa. 2000. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell 103:127-140. [DOI] [PubMed] [Google Scholar]
- 46.Panzeter, P. L., C. A. Realini, and F. R. Althaus. 1992. Noncovalent interactions of poly(adenosine diphosphate ribose) with histones. Biochemistry 31:1379-1385. [DOI] [PubMed] [Google Scholar]
- 47.Pihan, G. A., A. Purohit, J. Wallace, H. Knecht, B. Woda, P. Quesenberry, and S. J. Doxsey. 1998. Centrosome defects and genetic instability in malignant tumors. Cancer Res. 58:3974-3985. [PubMed] [Google Scholar]
- 48.Samper, E., F. A. Goytisolo, J. Ménissier-de Murcia, E. Gonzalez-Suarez, J. C. Cigudosa, G. de Murcia, and M. A. Blasco. 2001. Normal telomere length and chromosomal end capping in poly(ADP-ribose) polymerase-deficient mice and primary cells despite increased chromosomal instability. J. Cell Biol. 154:49-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saxena, A., R. Saffery, L. H. Wong, P. Kalitsis, and K. H. Choo. 2002. Centromere proteins Cenpa, Cenpb, and Bub3 interact with poly(ADP-ribose) polymerase-1 protein and are poly(ADP-ribosyl)ated. J. Biol. Chem. 277:26921-26926. [DOI] [PubMed] [Google Scholar]
- 50.Scaife, R. M., L. Wilson, and D. L. Purich. 1992. Microtubule protein ADP-ribosylation in vitro leads to assembly inhibition and rapid depolymerization. Biochemistry 31:310-316. [DOI] [PubMed] [Google Scholar]
- 51.Schwartz, J. L., W. F. Morgan, L. N. Kapp, and S. Wolff. 1983. Effects of 3-aminobenzamide on DNA synthesis and cell cycle progression in Chinese hamster ovary cells. Exp. Cell Res. 143:377-382. [DOI] [PubMed] [Google Scholar]
- 52.Shall, S., and G. de Murcia. 2000. Poly(ADP-ribose) polymerase-1: what have we learned from the deficient mouse model? Mutat. Res. 460:1-15. [DOI] [PubMed] [Google Scholar]
- 53.Simbulan-Rosenthal, C. M., B. R. Haddad, D. S. Rosenthal, Z. Weaver, A. Coleman, R. Luo, H. M. Young, Z. Q. Wang, T. Ried, and M. E. Smulson. 1999. Chromosomal aberrations in PARP−/− mice: genome stabilization in immortalized cells by reintroduction of poly(ADP-ribose) polymerase cDNA. Proc. Natl. Acad. Sci. USA 96:13191-13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simbulan-Rosenthal, C. M., D. S. Rosenthal, R. Luo, J. H. Li, J. Zhang, and M. E. Smulson. 2001. Inhibition of poly(ADP-ribose) polymerase activity is insufficient to induce tetraploidy. Nucleic Acids Res. 29:841-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith, S., and T. de Lange. 1999. Cell cycle dependent localization of the telomeric PARP, tankyrase, to nuclear pore complexes and centrosomes. J. Cell Sci. 112:3649-3656. [DOI] [PubMed] [Google Scholar]
- 56.Suzuki, H., P. Quesada, B. Farina, and E. Leone. 1986. In vitro poly(ADP-ribosyl)ation of seminal ribonuclease. J. Biol. Chem. 261:6048-6055. [PubMed] [Google Scholar]
- 57.Tarapore, P., and K. Fukasawa. 2000. p53 mutation and mitotic infidelity. Cancer Investig. 18:148-155. [DOI] [PubMed] [Google Scholar]
- 58.Tarapore, P., H. F. Horn, Y. Tokuyama, and K. Fukasawa. 2001. Direct regulation of the centrosome duplication cycle by the p53-p21Waf1/Cip1 pathway. Oncogene 20:3173-3184. [DOI] [PubMed] [Google Scholar]
- 59.Tarapore, P., Y. Tokuyama, H. F. Horn, and K. Fukasawa. 2001. Difference in the centrosome duplication regulatory activity among p53 ‘hot spot' mutants: potential role of Ser 315 phosphorylation-dependent centrosome binding of p53. Oncogene 20:6851-6863. [DOI] [PubMed] [Google Scholar]
- 60.Tong, W. M., M. P. Hande, P. M. Lansdorp, and Z. Q. Wang. 2001. DNA strand break-sensing molecule poly(ADP-ribose) polymerase cooperates with p53 in telomere function, chromosome stability, and tumor suppression. Mol. Cell. Biol. 21:4046-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tournier, F., and M. Bornens. 1994. Cell cycle regulation of centrosome function. Wiley-Liss, New York, N.Y.
- 62.Trucco, C., F. J. Oliver, G. de Murcia, and J. Ménissier-de Murcia. 1998. DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res. 26:2644-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vandre, D. D., and G. G. Borisy. 1989. The centrosome cycle in animal cells. Academic Press, Inc., San Diego, Calif.
- 64.Vaziri, H., M. D. West, R. C. Allsopp, T. S. Davison, Y. S. Wu, C. H. Arrowsmith, G. G. Poirier, and S. Benchimol. 1997. ATM-dependent telomere loss in aging human diploid fibroblasts and DNA damage lead to the post-translational activation of p53 protein involving poly(ADP-ribose) polymerase. EMBO J. 16:6018-6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Waldman, A. S., and B. C. Waldman. 1991. Stimulation of intrachromosomal homologous recombination in mammalian cells by an inhibitor of poly(ADP-ribosylation). Nucleic Acids Res. 19:5943-5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang, Z. Q., L. Stingl, C. Morrison, M. Jantsch, M. Los, K. Schulze-Osthoff, and E. F. Wagner. 1997. PARP is important for genomic stability but dispensable in apoptosis. Genes Dev. 11:2347-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wesierska-Gadek, J., A. Bugajska-Schretter, and C. Cerni. 1996. ADP-ribosylation of p53 tumor suppressor protein: mutant but not wild-type p53 is modified. J. Cell. Biochem. 62:90-101. [DOI] [PubMed] [Google Scholar]
- 68.Wesierska-Gadek, J., G. Schmid, and C. Cerni. 1996. ADP-ribosylation of wild-type p53 in vitro: binding of p53 protein to specific p53 consensus sequence prevents its modification. Biochem. Biophys. Res. Commun. 224:96-102. [DOI] [PubMed] [Google Scholar]
- 69.Wojcik, E. J., D. M. Glover, and T. S. Hays. 2000. The SCF ubiquitin ligase protein slimb regulates centrosome duplication in Drosophila. Curr. Biol. 10:1131-1134. [DOI] [PubMed] [Google Scholar]
- 70.Yoshihara, K., A. Itaya, Y. Tanaka, Y. Ohashi, K. Ito, H. Teraoka, K. Tsukada, A. Matsukage, and T. Kamiya. 1985. Inhibition of DNA polymerase alpha, DNA polymerase beta, terminal deoxynucleotidyl transferase, and DNA ligase II by poly(ADP-ribosyl)ation reaction in vitro. Biochem. Biophys. Res. Commun. 128:61-67. [DOI] [PubMed] [Google Scholar]