Abstract
The HR6A and -B genes, homologues of the yeast Rad6 gene, encode ubiquitin-conjugating enzymes that are required for postreplication repair of DNA and damage-induced mutagenesis. Using surface plasmon resonance, we show here that HR6 protein (referred as Rad6) physically interacts with p53. Analysis of proteins coimmunoprecipitated with Rad6 antibody from metabolically labeled normal MCF10A human breast epithelial cells not only confirmed Rad6-p53 interactions in vivo but also demonstrated for the first time that exposure of MCF10A cells to cisplatin or adriamycin (ADR) induces recruitment of p14ARF into Rad6-p53 complexes. Further analysis of ADR-induced p53 response showed that stable Rad6-p53-p14ARF complex formation is associated with a parallel increase and decrease in monoubiquitinated and polyubiquitinated p53, respectively, and arrest in G2/M phase of the cell cycle. Interestingly, the ADR-induced suppression of p53 polyubiquitination correlated with a corresponding decline in intact Hdm2 protein levels. Treatment of MCF10A cells with MG132, a 26S proteasome inhibitor, effectively stabilized monoubiquitinated p53 and rescued ADR-induced downregulation of Hdm2. These data suggest that ADR-induced degradation of Hdm2 occurs via the ubiquitin-proteasome pathway. Rad6 is present in both the cytoplasmic and nuclear compartments of normal MCF10A cells, although in response to DNA damage it is predominantly found in the nucleus colocalizing with ubiquitinated p53, whereas Hdm2 is undetectable. Consistent with in vivo data, results from in vitro ubiquitination assays show that Rad6 mediates addition of one (mono-) to two (multimono-) ubiquitin molecules on p53 and that inclusion of Mdm2 is essential for its polyubiquitination. The data presented in the present study suggest that Rad6-p53-p14ARF complex formation and p53 ubiquitin modification are important damage-induced responses that perhaps determine the fidelity of DNA postreplication repair.
Postreplication DNA repair is believed to function during and after DNA synthesis when unrepaired lesions in the template strand induce stalling of the DNA replication assembly, thereby causing gaps in the newly synthesized strand. The postreplication repair system is composed of two separate processes, error-free and error-prone, that are jointly regulated by the Rad6 gene and are designed to promote the completion of DNA synthesis (30). Error-prone recovery entails a mutagenic bypass of damaged sites by a specialized DNA polymerase dedicated to translesion synthesis (30), whereas error-free recovery involves bypass by template switching and/or gap filling by recombination, although the mechanism of error-free repair has not been fully understood (30, 31).
The Rad6 gene encodes a 17-kDa protein that belongs to a group of ubiquitin-conjugating enzymes (E2) that covalently adds ubiquitin to specific lysine residues of a substrate protein (24, 53). All functions performed by Rad6 appear to result from ubiquitination since replacement of the conserved Cys88 with serine produces a totally null phenotype (55, 56). Mutations in Rad6 confer extreme sensitivity toward a variety of DNA-damaging agents but are defective in damage-induced mutagenesis (43). Rad6 is highly conserved among eukaryotes. Two closely related human DNA repair genes, HHR6A and HHR6B (human homologues of yeast Rad6), encode ubiquitin-conjugating enzymes and complement the DNA repair and UV mutagenesis defects of the Saccharomyces cerevisiae rad6 mutant (26). HHR6A and HHR6B share 95% identical amino acid residues and are localized on human chromosome Xq24-q25 and 5q23-q31, respectively (27). In S. cerevisiae, error-free and error-prone lesion bypass require Rad6 and Rad18 genes (8, 41, 58, 59). Rad18 is a zinc finger protein with single-stranded DNA-binding activity and forms a complex with Rad6 (3-5). Recently, a human homolog of Rad18 has been identified that encodes for a 54-kDa protein and forms stable protein complexes with both HHR6A and HHR6B when coexpressed in yeast (61). However, proteins relevant to DNA repair that are ubiquitinated by Rad6 remain unknown.
Proteasomal degradation of key regulatory proteins control physiological events involving cell cycle, differentiation, DNA repair, apoptosis, and immune responses (18). Ubiquitination of a protein involves three separate enzymatic activities designated E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzyme), and E3 (ubiquitin ligase [9, 18, 19]). Ubiquitin is first activated through a covalent thiol ester linkage to E1. The activated ubiquitin is transferred to one of the several different ubiquitin-conjugating enzymes (E2) in an ATP-dependent manner. E2 then functions in combination with an E3 ligase to transfer ubiquitin to the target protein. The final ubiquitin transfer results in an isopeptide bond between the carboxyl-terminal Gly of ubiquitin and the ɛ-amino group of a Lys residue on the target protein. Whereas monoubiquitination may serve as targeting or localization signal (52), further conjugation of ubiquitin, usually to Lys48 of the previous ubiquitin moiety, results in the formation of polyubiquitin chains that labels the substrate for selective degradation by the 26S proteasome (10, 12, 14). Different E2s can function with a given E3. Thus, the formation of different E2-E3 complexes may provide additional levels of substrate specificity (29).
The tumor suppressor p53 is a latent and highly labile transcription factor that is mutated in 50% of human tumors mainly by missense mutation in the DNA-binding region. It plays a central role in maintaining genomic integrity by coordinating cell cycle (25), DNA repair (16), and programmed cell death in response to DNA damage (32, 49). In normal cells, p53 is present at very low levels; however, in response to DNA damage, wild-type p53 accumulates in the nucleus and coordinates a change in the balance of gene expression leading to growth arrest or apoptosis, events that prevent the growth or survival of damaged cells. Signals arising from cellular stresses trigger a complex series of regulatory events in the p53 pathway that lead to increased stability of p53 and activation of its biochemical functions (15). The stability and half-life of p53 are tightly regulated by Mdm2 and the ubiquitin-proteasome pathway (2, 16, 28). Binding of Mdm2 to the amino terminus of p53 (amino acid residues 19 to 26) represses p53 transcriptional activity (38, 39), promotes ubiquitination of p53 by acting as the E3 ubiquitin ligase, and targets p53 to the cytoplasm for 26S proteasome-dependent degradation (17). Disruption of p53-Mdm2 complexes is a pivotal event during the induction of p53 and is sufficient to invoke p53-mediated gene expression and cell cycle arrest (48). The increase in p53 is thought to result from p14ARF binding to the Mdm2, which interferes with p53-Mdm2 complex formation and proteasome degradation by inhibiting the E3 ubiquitin ligase activity of Mdm2 (23, 28, 35).
Here we show that Rad6 functions in cells after exposure to DNA-damaging agents by forming supramolecular complexes with p53 and p14ARF that correlate with p53 stability. Adriamycin (ADR)-induced p53 response in normal MCF10A human breast epithelial cells is accompanied by an increase in monoubiquitinated p53 and a simultaneous decrease in p53 polyubiquitination that is coincident with Hdm2 downregulation via the ubiquitin-proteasome pathway. The stable Rad6-p53 interaction observed is not unique to MCF10A cells since it is also seen after ADR treatment, albeit only temporarily, in metastatic MDA-MB-231 human breast cancer cells that express mutant p53. Results from in vitro ubiquitination assays show that Rad6 mediates limited addition of ubiquitin molecules on p53, and inclusion of Mdm2 is essential for extending the ubiquitin chains. The data presented in the present study suggest that (i) p53 ubiquitination is an important and regulated damage-induced modification and (ii) the accuracy of postreplication DNA repair process may be determined by the stability of supramolecular complexes formed between Rad6 and proteins of the p53 pathway.
MATERIALS AND METHODS
Cell lines, cell culture, and drug treatment.
MCF10A cells are normal-behaving pseudodiploid cells that lack tumorigenicity in nude mice and are unable to support anchorage-independent growth (50). The metastatic human breast cancer cell line MDA-MB-231 harbors an R280K mutation in p53 (T. Soussi, p53 mutation database [http://p53.curie.fr/]) and was purchased from the American Type Culture Collection (Manassas, Va.). MCF10A cells were maintained in Dulbecco modified Eagle medium (DMEM)-F-12 medium supplemented with 2.5% horse serum, 0.02 μg of epidermal growth factor, 0.5 μg of hydrocortisone/ml, 10 μg of insulin/ml, 0.1 μg of cholera toxin/ml, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. MDA-MB-231 cells were grown in DMEM supplemented with 10% fetal calf serum. Cells were treated with ADR (inhibition concentration [IC50] = 0.1 μg/ml for MCF10A or 0.5 μg/ml for MDA-MB-231) for 1 h, rinsed, and replaced with fresh medium to allow recovery for 0 to 72 h. For p53 or Hdm2 ubiquitination, the proteasome inhibitor MG132 (Calbiochem, San Diego, Calif.) was added to a final concentration of 25 μM to the culture media of untreated or ADR-treated cells 3 h before lysates were prepared.
Metabolic labeling and immunoprecipitation.
Control or cisplatin-treated (5 μg/ml for 6 h) MCF10A cells (5 × 106 cells per 100-mm dish) were incubated in methionine-free DMEM-2% dialyzed fetal bovine serum supplemented with 100 μCi of [l-35S]-methionine (specific activity, 1,083 Ci/mmol; NEN Life Science Products). Cells were labeled for 3 h, after which the monolayers were gently rinsed twice with phosphate-buffered saline and lysed with lysis buffer (10 mM Tris-HCl, pH 7.5; 150 mM NaCl; 1% Triton X-100; 1 mM phenylmethylsulfonyl fluoride; 5 μg of leupeptin, aprotonin, pepstatin, and chymostatin/ml; 1 mM sodium orthovanadate). Aliquots of lysates containing equivalent amounts of [35S]methionine incorporated into trichloroacetic acid-insoluble material (107 cpm) were immunoprecipitated by incubating them overnight with Rad6 antibody or nonimmune rabbit immunoglobulin G (IgG). Immune complexes were pelleted by incubating them with protein A/G-agarose and washed several times with lysis buffer. Rad6-immunodepleted lysates and total cellular lysates were subjected to immunoprecipitation with p53 Ab421 antibody, and immune complexes similarly recovered with protein A/G-agarose. Bound proteins were solubilized in sodium dodecyl sulfate (SDS) buffer and subjected to SDS-polyacrylamide gel electrophoresis (PAGE). Gels were either processed for Western blot analysis with polyclonal p53 CM-1 antibody or for fluorography.
Antibodies, immunoprecipitation, and Western blot analysis.
Control or ADR-treated MCF10A cells harvested after 0-, 2-, 4-, 8-, 24-, 48-, or 72-h recovery periods were lysed as described above, and aliquots of lysates containing 100 μg of protein were then either subjected to Western blot analysis of Rad6, Rad18, p53, p14ARF, Hdm2, and β-actin or to immunoprecipitation with Rad6 antibody. Immune complexes were pelleted with protein A/G-agarose, washed, and subjected to SDS-PAGE after solubilization in SDS sample buffer under reducing or nonreducing conditions. For reprecipitation experiments, immune complexes resulting from immunoprecipitation with Rad6 antibody were boiled in 100 μl of 50 mM Tris-HCl (pH 7.5)-1% SDS-5 mM dithiothreitol (DTT), diluted 10-fold in lysis buffer, and reprecipitated with antibodies to p53, p14ARF, Mdm2, or Rad18. p53, p14ARF, Hdm2, or Rad18 proteins coprecipitated with Rad6 antibody were detected by Western blot analysis with pAb421, p14ARF, Mdm2, or Rad18 antibody, respectively. To examine effects of the 26S proteasome inhibitor MG132 on p53 ubiquitination, cell extracts were immunoprecipitated with p53 pAb421 antibody, followed by Western blot analysis with p53 CM-1, and antibodies that specifically recognize ubiquitin-protein conjugates (FK2) or polyubiquitinated proteins (FK1) but not free ubiquitin. Effects of MG132 on Hdm2 ubiquitination were determined by Western blot analysis with Mdm2 antibody. Protein bands were visualized after reaction with appropriate anti-rabbit IgG, anti-mouse IgG, or anti-mouse IgM coupled to horseradish peroxidase by using ECL kit (Amersham, Arlington Heights, Ill.). The relative steady-state levels of HHR6A/HHR6B, Rad18, p14ARF, p53, or Hdm2 to β-actin bands were quantitated with a scanner-densitometer (model 300A densitometer; Molecular Dynamics, Sunnyvale, Calif.). Antibody to Rad6 was generated by multiple immunizations of New Zealand White rabbits with a synthetic peptide (K plus amino acid residues 131 to 152; accession no. NM_009458) that is conserved 100% in mouse and human HR6B and 91% in human HR6A (46). Since the antibody is unable to distinguish between HR6A and -B forms of Rad6, the 17-kDa proteins detected by this antibody are referred to as Rad6. p53 CM-1 polyclonal antibody was purchased from Novocastra Laboratories, Ltd. (Newcastle upon Tyne, United Kingdom); p53 pAb421 and pAb1801 antibodies recognize epitope on amino acids 372 to 382 and amino acids 46 to 55, respectively; Mdm2 antibody recognizes an epitope in the N terminus of Mdm2 and was purchased from Oncogene Science (Cambridge, Mass.). Other antibodies used were specific for human Rad18 (Imgenex, San Diego, Calif.), ubiquitin (Zymed Labs, San Francisco, Calif.), ubiquitin-protein conjugates (FK2) and polyubiquitin-protein conjugates FK1 (Affiniti Research Products, Ltd., Mamhead Castle, United Kingdom), p14ARF, and β-actin (Oncogene Science).
Immunofluorescence microscopy.
Control or ADR-treated MCF10A cells were grown on coverslips and fixed in methanol-acetone (1:1 [vol/vol]) at −20°C. Cells were preincubated with 2% horse serum-phosphate-buffered saline and then incubated with anti-Rad6 antibody. To assess colocalization and/or regulation by ADR of Rad6, p53, or Hdm2 expression, cells were stained with a 1:1 mixture of Rad6 and p53 pAb421 antibodies, pAb421 and ubiquitin antibodies, or Rad6 and Mdm2 antibodies. Cells were washed, and Rad6, ubiquitin, p53, or Hdm2 was detected with fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG or Texas red-conjugated goat anti-mouse IgG. Nonbinding mouse or rabbit IgG was used as a control in all double-labeling experiments. All images were collected on a Olympus BX-4 fluorescence microscope equipped with Sony high-resolution/sensitivity charge-coupled device video camera.
Surface plasmon resonance assay.
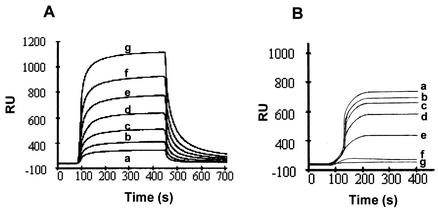
p53-Rad6 interaction studies were carried out by using a BIAcore 3000 surface plasmon resonance instrument (BIAcoreAB, Uppsala, Sweden). All of the experiments were carried out at 25°C by using a constant flow rate (5 μl/min) of running buffer. Glutathione S-transferase (GST)-tagged p53, a gift from H. Yasuda (School of Life Science, Tokyo University of Pharmacy and Life Science, Tokyo, Japan) was affinity purified on a glutathione-conjugated Sepharose 4B column and covalently attached to CM5 sensor chip surface by the standard amine coupling procedure. Reactive sites remaining on the surface were blocked by reaction with ethanolamine. Various concentrations (50 to 1,000 nM) of human recombinant Rad6 (UbcH2; BostonBiochem, Boston, Mass.) prepared in 35-μl portions were sequentially injected into 20 mM HEPES-150 mM NaCl-3.4 mM EDTA-0.05% Tween 20 (pH 7.4) through the flow cell. Between experiments, the sensor chips were regenerated by washing them with two pulses of 20 mM HCl, followed by an EXTRACLEAN procedure that was done as recommended by the manufacturer. For competition assays, GST-p53 was immobilized on the sensor chip, and Rad6 (400 nM) was injected, followed by the immediate injection of various dilutions of Rad6 antibody or corresponding normal rabbit serum (1:500, 1:2,000, 1:5,000, or 1:10,000) by the coinjection mode. In some experiments, anti-Rad6 antibody (1:500) or bovine serum albumin (BSA; 400 nM) was injected over the chip containing immobilized GST-p53 in the absence of Rad6. Sensorgrams were prepared and globally fit by using nonlinear least-squares analysis and numerical integration of the differential rate equations by using the SPRevolution software package. Sensorgrams were generated after subtraction of the signal due to nonspecific binding of Rad6 to the control chip. Duplicate injections were made and the response units reported are the average of two injections.
Ubiquitination assay.
Reactions were performed in the presence or absence of 0.5 μg of affinity-purified GST-Mdm2 (generous gift from H. Yasuda) in mixtures containing 100 ng of E1, 0.5 μg of Rad6 (BostonBiochem), 0.5 μg of purified GST-p53, and 10 μg of ubiquitin in 20 mM Tris-HCl (pH 7.6)-50 mM NaCl-4 mM ATP-10 mM MgCl2-0.2 mM DTT for 2 h at 30°C. Reactions were also performed in the absence of Rad6 to determine the specific effects of Rad6 on p53 ubiquitination. Reactions were terminated and resolved by SDS-PAGE on 6% or 8 to 16% gradient gels under reducing and nonreducing conditions. Ubiquitinated p53 products were visualized by Western blot analysis with p53 CM-1-, ubiquitin-, or polyubiquitin-specific antibodies.
Flow cytometry.
Control or ADR-treated MCF10A and MDA-MB-231 cells were trypsinized into single cell suspension, fixed in ice-cold 70% ethanol, and stored at 4°C until required. Before analysis, cells were resuspended in 100 μg of RNase A (Promega Corp., Madison, Wis.)/ml, 40 μg of propidium iodide/ml, and phosphate-buffered saline. Analysis was performed on a Becton Dickinson fluorescence-activated cell sorter (FACScan).
Deubiquitinating enzyme assay.
Deubiquitinating enzyme activity in control and ADR-treated MCF10A cells exposed to MG132 was measured according to the method of Dang et al. (13). Briefly, aliquots of cell extracts containing 25 μg of protein were incubated in assay buffer (50 mM HEPES-0.5 mM EDTA [pH 7.8] containing 0.1 mg ovalbumin/ml and 1 mM DTT) at room temperature for 30 min to allow DTT-mediated activation of isopeptidase T and UCH-L3 prior to the addition of predetermined concentration (25 nM) of Ub-AMC (BostonBiochem). Reaction progress was monitored by measuring the increase in fluorescence emission at 460 nm (λex = 380 nm) that accompanies cleavage of AMC from Ub-AMC by using a Hitachi 2000 fluorescence spectrophotometer. The specificity of ubiquitin-hydrolyzing activity measured in cell extracts was confirmed by preincubating reactions with a 10-fold excess of ubiquitin aldehyde, a ubiquitin hydrolase inhibitor (BostonBiochem).
RESULTS
Treatment with a DNA-damaging agent stabilizes de novo interaction of Rad6 with p53 and p14ARF proteins.
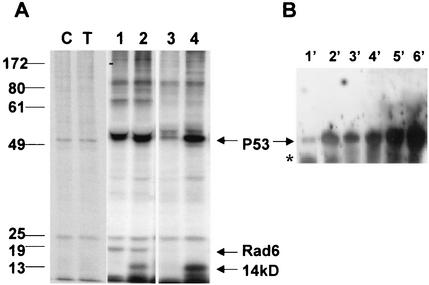
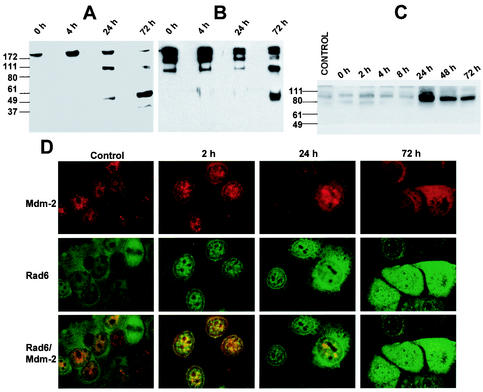
To determine the effects of DNA damage on de novo expression of Rad6, normal MCF10A cells treated with cisplatin (5 μg/ml) or untreated were metabolically labeled with [35S]methionine and analyzed for Rad6 expression by immunoprecipitation with anti-Rad6 antibody. Control immunoprecipitations were performed with equivalent amounts of normal rabbit IgG. Whereas nonimmune IgG did not precipitate 17-kDa proteins from control or cisplatin-treated lysates of MCF10A cells (Fig. 1A, lanes C and T), immunoprecipitation with Rad6 antibody caused similar levels of de novo-synthesized Rad6 to immunoprecipitate from both control and cisplatin-treated MCF10A cells (Fig. 1A, lanes 1 and 2). Besides immunoprecipitating Rad6, the Rad6 antibody effectively coprecipitated a protein doublet of ∼53 kDa from both control and treated cells (Fig. 1A, lanes 1 and 2). Since functional interactions between p53 and several key proteins involved in DNA repair have been reported (7, 37, 60), we determined the identity of the 53-kDa protein to be p53 by subjecting the proteins immunoprecipitated by Rad6 antibody (Fig. 1A, lanes 1 and 2) and those reimmunoprecipitated from Rad6-immunodepleted supernatants by p53 pAb421 antibody (Fig. 1A, lanes 3 and 4) to Western blot analysis with p53 CM-1 antibody (Fig. 1B, lanes 1′ to 4′). The relative levels of p53 recovered from Rad6 immunoprecipitates (Fig. 1B, lanes 1′ and 2′) versus Rad6-immunodepleted supernatants (Fig. 1B, lanes 3′ and 4′) were determined by comparing total p53 levels detected from corresponding samples without prior immunoprecipitation with Rad6 antibody (Fig. 1B, lanes 5′ and 6′). The data from Fig. 1B show that, whereas in the control cells ca. 80% of p53 was present in Rad6-immunodepleted supernatants, >50% of p53 was found to be associated with Rad6 after exposure to the drug. That a physical interaction, perhaps regulated by DNA-damaging agent, occurs between Rad6 and p53 is further confirmed by coprecipitation of a 14-kDa protein, identified to be p14ARF by Western blot analysis (data not shown and Fig. 5B), by both Rad6 and p53 antibodies from only cisplatin-treated MCF10A cells (Fig. 1A, lanes 2 and 4).
FIG. 1.
De novo interaction between Rad6 and p53. Exponentially growing MCF10A cells were either untreated or treated with 5 μg of cisplatin/ml for 6 h and then incubated in methionine-free DMEM supplemented with 100 μCi of [35S]methionine for 3 h. Equivalent amounts of [35S]methionine-labeled trichloroacetic acid-precipitable proteins were immunoprecipitated with anti-Rad6 antibody or normal IgG, and immune complexes were pelleted by incubating with protein A/G-agarose. Rad6-immunodepleted lysates were subjected to a second immunoprecipitation with p53 pAb421 antibody, and immune complexes were similarly recovered with protein A/G-agarose. Bound proteins were resolved by SDS-PAGE, followed by fluorography (A) or Western blot analysis with p53 CM-1 antibody (B). (A) Lanes C and T, control and cisplatin-treated cell extracts, respectively, precipitated with normal IgG; lanes 1 and 2, control and cisplatin-treated cell extracts, respectively, immunoprecipitated with anti-Rad6 antibody; lanes 3 and 4, immunoprecipitation of Rad6-depleted supernatants from control and cisplatin-treated cells, respectively, with pAb421 antibody. (B) Lanes 1′ to 4′, p53 CM-1 reactivity of corresponding lanes 1 to 4 of panel A; lanes 5′ and 6′, p53 CM-1 reactivity of pAb421 immunoprecipitated control and cisplatin-treated cell extracts, respectively. Positions of Rad6, p14ARF, and p53 are indicated. Note the presence of p14ARF coprecipitating with both Rad6 (lane 2) and p53 (lane 4) antibodies only in cisplatin-treated cells.
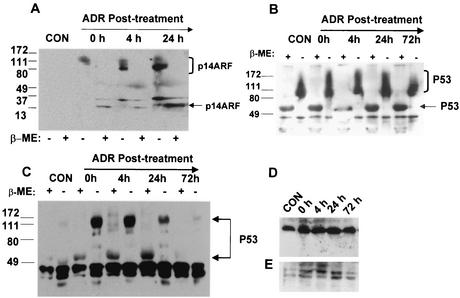
FIG. 5.
Effect of ADR on Rad6-p53-p14ARF interactions. MCF10A (A and B) or MDA-MB-231 (C, D, and E) cells were treated with ADR for 1 h, and cultures were rinsed and incubated with drug-free medium as described in Materials and Methods. At the indicated periods of recovery, aliquots of cell extracts containing equivalent amounts of total protein were subjected to immunoprecipitation with anti-Rad6 antibody (A to D) or immunoblotted with pAb421 without prior immunoprecipitation (E). Immune complexes were resolved by SDS-PAGE under reducing and nonreducing conditions as indicated and then analyzed by immunoblotting with p14ARF (A), pAb421 (B and C), or Rad6 (D) antibody. The positions of p14ARF and p53 in immune complexes are indicated.
Regulation of Rad6, p53, p14ARF, Hdm2, and Rad18 protein levels by ADR treatment.
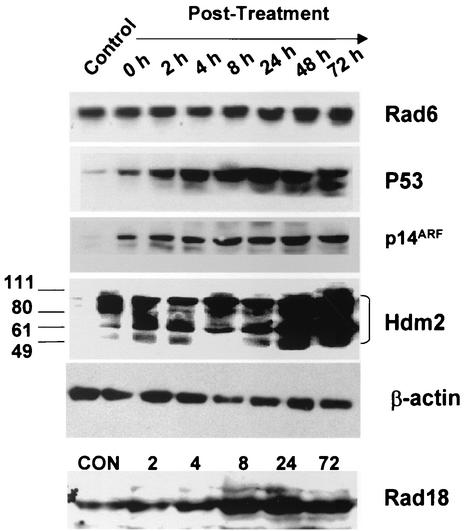
Since results from Fig. 1 demonstrated a potential de novo interaction regulated by cisplatin between Rad6, p53, and p14ARF, we examined the regulation of Rad6, Rad18, p53, p14ARF, and Hdm2 steady-state levels in normal MCF10A cells after exposure to ADR, a potent anthracycline and topoisomerase II inhibitor that is one of the most widely used cancer chemotherapeutic drugs. MCF10A cells were either left untreated or treated with predetermined IC50 dose of ADR (0.1 μg/ml) for 1 h; cells were rinsed, and cultures were incubated with fresh drug-free medium. Cell lysates were prepared from untreated samples and at 0-, 2-, 4-, 8-, 24-, 48-, and 72-h recovery periods after exposure to the drug and then analyzed for Rad6, Rad18, p53, p14ARF, and Hdm2 steady-state levels relative to β-actin by Western blot analysis. As shown in Fig. 2, an increase in Rad6 levels was evident at 8 h of recovery and steadily increased by 24 to 72 h posttreatment to approximately two- to fivefold-higher levels relative to the untreated control lysates. Steady-state levels of Rad18 protein exhibited a similar regulation profile by ADR treatment and reached approximately four- to sixfold-higher levels relative to the untreated control cells by 8 to 72 h of treatment (Fig. 2). Both p14ARF and p53 exhibited dramatic and steady increases that were evident immediately after exposure to ADR. Levels of p14ARF were enhanced 5-fold relative to untreated control lysates at 0 h of recovery and steadily increased to ∼15-fold by 48 to 72 h posttreatment (Fig. 2). The p53 expression pattern after ADR exposure mirrored the p14ARF profile; however, levels of p53 were upregulated and were maintained at ca. 25- to 40-fold-higher levels relative to the controls in the period from 4 to 72 h after recovery from drug exposure. Induction of p53 was accompanied by a simultaneous increase in the appearance of p53 as a doublet band, a finding that may indicate a posttranslational modification of p53 (Fig. 2). Although the steady-state levels of Hdm2 exhibited a modest increase after drug treatment, maintenance of higher levels of intact Hdm2 is probably impaired by the simultaneous accumulation of several lower-molecular-weight Mdm2-immunoreactive proteins (Fig. 2). These data suggest that ADR significantly enhances Rad6, Rad18, p14ARF, and p53 proteins with only modest effects on Hdm2 levels.
FIG. 2.
Effects of ADR on steady-state levels of Rad6, Rad18, p53, p14ARF, and Hdm2 proteins. MCF10A cells were treated with ADR (0.1 μg/ml) for 1 h, and the cultures were washed and replaced with fresh drug-free medium to allow for recovery. Cell lysates were prepared from control (untreated) and ADR-treated cells at the indicated time periods of recovery. Aliquots of cell lysates containing 100 μg of total protein were subjected to SDS-PAGE and Western blot analysis. Immunoblots were reacted with antibodies to Rad6, p53 pAb421, p14ARF, Mdm2, Rad18, or β-actin as discussed in Materials and Methods.
Intact p53, p14ARF, and Hdm2 proteins are physically complexed with Rad6.
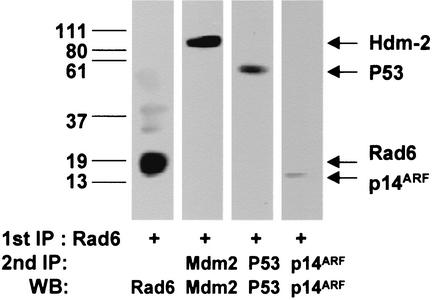
Since our data from Fig. 1 and 2 have demonstrated physical interactions between de novo-synthesized Rad6, p53, and p14ARF in cisplatin-treated cells and similar ADR-mediated inductory effects on Rad6, p14ARF, and p53 proteins, respectively, we investigated whether Rad6 exists in vivo as part of a supramolecular complex with proteins of the p53 pathway in ADR-treated MCF10A cells. MCF10A lysates prepared at 8 h of recovery after ADR treatment were subjected to immunoprecipitation with Rad6 antibody. Lysates prepared at 8 h posttreatment showed that Rad6, p53, p14ARF, Hdm2, and Rad18 are all upregulated (Fig. 2). Rad6 immunoprecipitates were washed, boiled, and diluted 10-fold prior to reprecipitation with p53 pAb421, p14ARF, Mdm2, or Rad18 antibodies. The results (Fig. 3) indicated the presence of immunoprecipitable Hdm2, p53, and p14ARF in Rad6-immunoprecipitated proteins from ADR-treated MCF10A cells. A similar analysis of control MCF10A lysates failed to reveal detectable amounts of p53, p14ARF, or Hdm2 in Rad6-immunoprecipitated proteins (data not shown). These data suggest the presence of a weak interaction between Rad6 and proteins of the p53 pathway in untreated MCF10A cells and that drug-induced effects on p53, p14ARF, and Hdm2 levels perhaps stabilize their interaction with Rad6. Rad18 was not detectable in Rad6 immunoprecipitates from both control and ADR-treated MCF10A lysates, and the majority of Rad18 was detected in Rad6-immunodepleted supernatants (data not shown). Amino acid residues 141 to 149 at the carboxyl terminus of Rad6 are essential for Rad18 binding (5); thus, immunoprecipitation with the Rad6 antibody, which recognizes an epitope on amino acids 138 to 152, could occlude Rad6 interaction with Rad18.
FIG. 3.
Rad6 interacts with p53, p14ARF, and Hdm2. ADR-treated MCF10A cell extracts (200 μg of protein) prepared at 8 h of recovery were subjected to immunoprecipitation with anti-Rad6 antibody. Immune complexes were pelleted with protein A/G-agarose, boiled, and reprecipitated with antibodies to p53 pAb421, p14ARF, or Mdm2 as described in Materials and Methods. Coprecipitating p53, p14ARF, or Hdm2 were detected by Western blot analysis with antibodies to p53 (pAb1801), p14ARF, or Mdm2. The positions of Hdm2, p53, and p14ARF that coprecipitated with Rad6 are indicated.
Rad6-p53 interactions by surface plasmon resonance assay.
Our data from metabolic-labeling and immunoprecipitation experiments (Fig. 1 and 3) demonstrated the existence of physical interactions between endogenous Rad6 and p53 proteins. Surface plasmon resonance was used to verify whether there is a direct interaction between p53 and Rad6 proteins or whether this association is dependent on the presence of extraneous cellular factors. As shown in Fig. 4A, Rad6 showed a dose-dependent increase in binding to immobilized GST-p53, suggesting that Rad6-p53 interaction occurred independently of cellular factors. The kon was determined from the association phase of binding of Rad6 to immobilized p53 by using the sensorgrams obtained with different concentrations of the soluble component and was 2.6 × 105 M−1 s−1. Similarly, koff was calculated from the dissociation phase and was 9.4 × 10−3 s−1. These data suggest that Rad6 complexes readily with p53, and the apparent Kd (koff/kon) value 3.6 × 10−8 M is in the range of strong interactions. In order to further determine the specificity of the Rad6-p53 interaction, the effects of various amounts of anti-Rad6 antiserum or the corresponding normal rabbit serum were tested on binding of Rad6 (400 nM) to immobilized GST-p53. As shown in Fig. 4B, coinjection of Rad6 antibody caused a dose-dependent inhibition in the binding of Rad6 to immobilized GST-p53. Whereas the injection of Rad6 antibody at 1:10,000, 1:5,000, 1:2,000, or 1:500 produced 7, 15.5, 25.2, or 44% inhibition of Rad6 binding to GST-p53, respectively (Fig. 4B), similar coinjection of normal rabbit serum failed to alter Rad6 binding to immobilized GST-p53 (data not shown). Direct injection of Rad6 antibody (1:500) or BSA into immobilized p53 failed to elicit binding, further confirming the specificity of binding reactions (Fig. 4B).
FIG. 4.
Rad6 binds to p53 in vitro. (A) Affinity-purified p53-GST was immobilized on a BIAcore 3000 sensor chip, and various concentrations of Rad6 protein were injected over the chip surface. Curves a to g represent binding curves obtained with 50, 100, 200, 300, 400, 600, and 1,000 nM concentrations, respectively. (B) GST-p53 immobilized chip was injected with 400 nM Rad6; this step was immediately followed by injection of various dilutions of anti-Rad6 antibody by the coinjection mode. Curve a represents Rad6 binding in the absence of antibody; curves b to e represent Rad6 binding in the presence of 1:10,000, 1:,5000, 1:2,000, and 1:500 dilutions of anti-Rad6 antibody; curves f and g represent the binding of 400 nM BSA or a 1:500 dilution of anti-Rad6 antibody alone, to immobilized GST-p53 chip.
Normal breast epithelial cells exhibit ADR-mediated stabilization of Rad6 complexed p53.
Since our data from Fig. 3 have shown a direct interaction between Rad6 and proteins of the p53 pathway, we examined the effects of ADR on the stability of molecular complexes formed between Rad6 and p53 in normal MCF10A and metastatic MDA-MB-231 breast cells. MDA-MB-231 and MCF10A cells were chosen since they express mutant (T. Soussi, [http://p53.curie.fr/]) and wild-type (47) p53, respectively. MCF10A cell lysates prepared from untreated cells and at 0, 4, 24, and 72 h in the recovery period after ADR treatment were subjected to immunoprecipitation with Rad6 antibody, and immune complexes resolved by SDS-PAGE were assessed by Western blot analysis with antibodies to p53 or p14ARF. Complex formation was tested under both nonreducing and reducing conditions to determine whether variations in size and immunoreactivity to specific proteins are detectable. Consistent with ADR-induced regulation of p14ARF steady-state levels (Fig. 2), the results (Fig. 5A) showed that no Rad6-immunoprecipitable p14ARF was detectable in the control MCF10A cell lysates. However, upon exposure to the drug, Rad6-immunoprecipitable p14ARF was detectable immediately after treatment and was localized to an ∼150-kDa band. By 4 h posttreatment, p14ARF immunoreactivity was observed as a broad band spanning ca. 100 to 150 kDa, and by 24 h the majority of p14ARF immunoreactivity was increasingly localized to the 100-kDa band (Fig. 5A). Analysis of the corresponding immunoprecipitates in the presence of β-mercaptoethanol not only confirmed the presence of p14ARF in Rad6-immunoprecipitable complexes but also corroborated DNA damage-induced effects on p14ARF recruitment and formation of Rad6-p14ARF complexes observed in Fig. 1.
Similar analysis of Rad6-p53 interaction in MCF10A cell extracts from untreated control and 0, 4, 24, and 72 h after ADR treatment revealed the presence of Rad6-immunoprecipitable pAb421-immunoreactive p53 both in control and ADR-treated cells (Fig. 5B). These data suggest that Rad6-p53 interaction in MCF10A cells, unlike that observed with p14ARF, is not contingent upon exposure to drug. Rad6-immunoprecipitable p53 was found to be present as a broad smear spanning ca. 100 to 150 kDa (Fig. 5B). Analysis of the corresponding immunoprecipitates under reducing conditions confirmed the presence of p53 in Rad6-p53 (Fig. 5B).
Although equivalent amounts of antibody and total cellular proteins were included in each immunoprecipitation, it is interesting that the sizes of molecular complexes formed not only reflect a supershift caused by antibody reaction but also the stability of interactions between Rad6, p53, and p14ARF. This is evident from regulation of Rad6-p53 complex formation observed in ADR-treated metastatic MDA-MB-231 breast cancer cells (Fig. 5C). Analysis of Rad6-p53 interaction in metastatic MDA-MB-231 cell extracts from untreated controls and 0, 4, 24, and 72 h after ADR treatment, revealed the presence of detectable Rad6-immunoprecipitable pAb421-immunoreactive p53 in an ∼160-kDa band only in drug-exposed cells (Fig. 5C). These data suggest that, unlike in normal MCF10A cells, exposure to the DNA-damaging drug is required to enhance and/or stabilize Rad6-p53 interaction in metastatic MDA-MB-231 cells (Fig. 5C). Interestingly, unlike in MCF10A cells, in which stable complexes of Rad6-p53 were detectable at least up to 72 h after ADR treatment, a 75% decrease in Rad6-immunoprecipitable p53 was observed at 24 h posttreatment, and by 72 h negligible p53 reactivity was seen under nonreducing conditions (Fig. 5C). The presence of p53 in Rad6-immunoprecipitable complexes was confirmed not only by the immunoreactivity of the complex with p53 antibody but also by derivation under reducing conditions of p53-immunoreactive 53-kDa band in amounts that were proportional to that present in complexes under nonreducing conditions (Fig. 5C). The reduction in the amounts of Rad6-p53 complexes observed at 24 and 72 h after ADR treatment was not due to inefficient immunoprecipitation by Rad6 antibody, since Western blot analysis of corresponding Rad6 immunoprecipitates showed significant amounts of Rad6 in all samples (Fig. 5D). In contrast, Western analysis of p53 steady-state levels showed p53 induction in ADR-treated samples at 0, 4, and 24 h and significant reduction at 72 h posttreatment (Fig. 5E). These data indicate that, although ADR-induced p53 response is associated with an upregulation in interaction between Rad6 and p53 in metastatic MDA-MB-231 cells, prolonged maintenance of Rad6-complexed p53 in metastatic MDA-MB-231 cells is impaired, in contrast to the situation in normal MCF10A cells.
ADR induces Hdm2 degradation via the ubiquitin-proteasome pathway.
Since the results of Fig. 2 showed ADR to exert dramatic inductory effects on Rad6, p53, and p14ARF accumulation and modest stimulatory and/or degradation-inducing effects on Hdm2 in MCF10A cells, we investigated whether ADR-induced decline in Hdm2 occurs via the ubiquitin-proteasome pathway. Lysates of control and ADR-treated MCF10A cells were subjected to immunoprecipitation with Mdm2 antibody, and immune complexes resolved by SDS-PAGE under nonreducing conditions were analyzed by Western blotting with antibodies to ubiquitin or Mdm2. The results (Fig. 6A) demonstrated the presence of a prominent Mdm2-immunoreactive band at ∼172 kDa during early periods of recovery (0 to 4 h) after ADR treatment that was not detectable in control cells (data not shown). At 24 h, a reduction in the signal of the 172-kDa band and the emergence of Mdm2-immunoreactive 110- and 55-kDa bands were observed. By 72 h, the intensities of the 172- and 110-kDa bands decreased >90% and were replaced by a proportional increase in a band at 55 kDa and a smaller band at 40 kDa. When the same blot was stripped and reprobed with antiubiquitin antibody, a similar pattern of immunostaining was observed (Fig. 6B). Intense ubiquitin-immunoreactive Mdm2-immunoprecipitable bands were observed at ∼172 and 150 kDa during 0 to 4 h of recovery, followed by decreases in the 172- and 150-kDa bands at 24 h and subsequent increases in bands at ∼110 and 55 kDa at 72 h posttreatment (Fig. 6B). These data suggest that ADR treatment may facilitate Hdm2 degradation via the ubiquitination pathway.
FIG. 6.
ADR treatment induces Hdm2 degradation. MCF10A cells were treated with ADR for 1 h, and cultures were rinsed and incubated with drug-free medium. Cell extracts prepared at indicated periods of recovery were subjected to immunoprecipitation with Mdm2 antibody, and immune complexes resolved by SDS-PAGE under nonreducing conditions were analyzed by Western blot analysis with anti-Mdm2 (A) or antiubiquitin (B) antibody. (C) Steady-state levels of Hdm2 were measured by Western blot analysis of extracts prepared at the indicated times from control and ADR-treated MCF10A cells pretreated with MG132 to stabilize Hdm2. (D) Distribution and colocalization of Rad6 and Hdm2 were visualized in control and ADR-treated MCF10A cells at 2, 24, and 72 h of recovery by immunofluorescence staining with FITC-conjugated or Texas red-conjugated secondary antibody for Rad6 and Mdm2, respectively.
To obtain further evidence that ADR-induced Hdm2 ubiquitination and degradation occur via the 26S proteasome, MCF10A cells were treated with ADR prior to treatment with MG132, a 26S proteasome inhibitor, and lysates were analyzed for Hdm2 steady-state levels by Western blot analysis with Mdm2 antibody. The results of Fig. 6C show that exposure of ADR-treated cells to MG132 resulted in the accumulation of >25-fold-higher levels of intact Hdm2 by 24 h compared to those in untreated control samples or during early periods of recovery. These data suggest that the ADR-induced degradation of Hdm2 in MCF10A cells occurs at least in part via the ubiquitin-proteasome pathway since the decrease in intact Hdm2 levels induced by ADR (Fig. 6A and B) can be rescued by treatment with a proteasome inhibitor (Fig. 6C).
To determine whether the stabilization of Rad6-p53 complex formation observed in MCF10A cells parallels a corresponding decay of Hdm2, we examined the distribution of Hdm2 and Rad6 by immunofluorescence microscopy in control and ADR-treated MCF10A cells. Hdm2 was localized in the nucleus and excluded from nucleoli in untreated MCF10A cells (Fig. 6D). After exposure to ADR, i.e., at 2 h of recovery, Hdm2 levels were significantly elevated and Hdm2 immunoreactivity was localized both to nuclear bodies and nucleoli (Fig. 6D). Consistent with immunoblotting experiments (Fig. 2), a 60% decline in Hdm2 immunoreactivity was observed in the nuclei at 24 h posttreatment, and by 72 h >90% of nuclei exhibited only diffuse staining in the nucleoplasm and substantial staining in the cytoplasm (Fig. 6D). Immunofluorescence localization of Rad6 in control MCF10A cells revealed diffuse staining in the cytoplasmic and nuclear compartments. However, treatment with ADR induced a preferential redistribution of Rad6 from the cytoplasm to the nucleus that was reflected by detection of elevated levels of Rad6 in the nucleus at least until 72 h posttreatment (Fig. 6D). These results suggest that ADR exerts opposing effects on the stability of Rad6 and Hdm2 in MCF10A cells.
ADR-stabilized p53 is ubiquitinated and colocalizes with Rad6 in the nucleus.
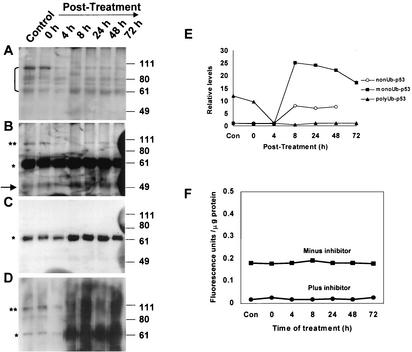
Since treatment of MCF10A cells with ADR enhances both the steady-state levels of p53 and Rad6 and prolongs the stability of Rad6-p53 complexes, we investigated the effects of ADR on in vivo p53 ubiquitination status. Lysates prepared from MCF10A cells treated with ADR prior to treatment with MG132 were immunoprecipitated with pAb421 antibody. Immune complexes were resolved by SDS-PAGE and subjected to Western blot analysis with antibodies specific to polyubiquitinated protein conjugates (Fig. 7A), p53 CM-1 (Fig. 7B and C), or ubiquitin-protein conjugates (Fig. 7D). Analysis of p53 with CM-1 antibody in control and ADR-treated MCF10A cells exposed to MG132 revealed that the majority of p53 immunoprecipitated with the p53 antibody was ubiquitinated since overexposure of the blots was necessary to visualize the presence of normal nonubiquitinated p53 (Fig. 7B and C). Quantitation of relative intensities of nonubiquitinated p53 showed that samples at 8, 24, 48, and 72 h after ADR treatment contained ∼8-fold-higher levels of p53 compared to control and earlier periods of post-ADR treatment (Fig. 7B). Short-time exposure revealed that ca. 15- to 25-fold-higher levels of monoubiquitinated p53 were present in ADR-treated samples at 8, 24, 48, and 72 h posttreatment compared to controls or at 0 and 4 h after ADR treatment (Fig. 7C). In addition to the 62-kDa p53 immunoreactive band, a prominent p53-immunoreactive band with a molecular size of ∼100 kDa was also detected, the latter probably comprising p53 molecules carrying five molecules of ubiquitin (Fig. 7B). The levels of polyubiquitinated p53 in control and at 0 h after ADR treatment were ∼8-fold higher than in samples at 4 and 8 h posttreatment and ∼5-fold higher than in samples at 24, 48, and 72 h posttreatment (Fig. 7B).
FIG. 7.
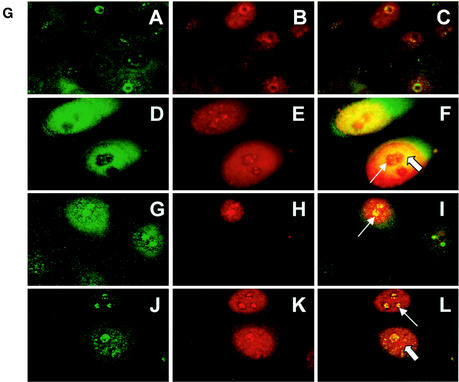
ADR enhances monoubiquitination of p53. Control and ADR-treated cultures were pretreated with MG132, and extracts prepared at the indicated time points were subjected to immunoprecipitation with p53 pAb421 antibody. Immune complexes were subjected to SDS-PAGE and analyzed by Western blotting with polyubiquitin-specific FK1 (A), p53 CM-1 (B and C), or ubiquitin-protein conjugate-specific FK2 (D) antibodies. Panels B and C show an overexposure and a brief exposure, respectively. The positions of unubiquitinated (→), monoubiquitinated (✽), and polyubiquitinated (✽✽) p53 are indicated. (E) Graphic representation of relative levels of nonubiquitinated and monoubiquitinated p53 versus polyubiquitinated p53 detected by p53 CM-1 or polyubiquitin-specific FK1 antibodies, respectively. (F) Deubiquitinating enzyme activity in control and ADR-treated cells. (G) Colocalization of Rad6 and p53 was visualized in control MCF10A cells (subpanels A to C) and at 72 h of recovery post-ADR treatment of MCF10A cells (subpanels D to F) by immunofluorescence staining with FITC-conjugated or Texas red-conjugated secondary antibody for Rad6 and p53, respectively. (Subpanels G to I) Colocalization of Rad6 and ubiquitin were visualized in ADR-treated MCF10A cells with FITC-conjugated or Texas red-conjugated secondary antibodies for Rad6 and ubiquitin, respectively. (Subpanels J to L) Colocalization of ubiquitin and p53 were visualized in ADR-treated MCF10A cells with FITC-conjugated or Texas red-conjugated secondary antibodies for ubiquitin and p53, respectively. Note the colocalization of Rad6 and p53, Rad6 and ubiquitin, or p53 and ubiquitin in the nucleoli (thick arrows) and nucleoplasm (thin arrows).
Reprobing the blots with polyubiquitinated protein-specific FK1 antibody further substantiated that the 100-kDa p53-immunoreactive band indeed represented polyubiquitinated p53 and was most prominent in control samples and in samples at 0 h post-ADR treatment (Fig. 7A). It is interesting that although nonubiquitinated p53 was not detected with polyubiquitin-specific antibody, faint bands corresponding to 62, 70, 80, and 90 kDa were detected in most samples. It is not clear whether these bands represent nonspecific immunoreactivity to p53 molecules with limited ubiquitination or multimonoubiquitination. Densitometric analysis of the FK1-reactive 100-kDa band indicated its presence at ∼10-fold-higher levels in control samples and in samples at 0 h post-ADR treatment compared to samples at 4, 8, 24, 48, and 72 h posttreatment. Immunoblotting with FK2, an antibody that recognizes all ubiquitin-protein conjugates, showed the presence of >50-fold-higher levels of monoubiquitinated p53 in ADR-treated samples at 8, 24, 48, and 72 h posttreatment compared to those in control samples or in samples at 0 and 4 h post-ADR treatment (Fig. 7D). The relative levels of unubiquitinated p53, monoubiquitinated p53, and polyubiquitinated p53 detected with p53 CM-1 and FK1 antibodies, respectively, are graphically summarized in Fig. 7E. These data indicate that p53 is polyubiquitinated in control samples and during the initial periods of ADR treatment. However, the drug-induced response is accompanied by a decrease in polyubiquitinated p53 that is coupled with a dramatic and concomitant increase in the levels of monoubiquitinated p53.
In order to determine whether the increase or decrease in monoubiquitinated p53 versus polyubiquitinated forms, respectively, reflected an increase in deubiquitinating enzyme activity in ADR-treated cells, we measured the ubiquitin-hydrolyzing activity in control and ADR-treated cells at 0, 4, 8, 24, 48, and 72 h of recovery according to the assay described by Dang et al. (13). The results in Fig. 7F show that there was no significant difference in Ub-AMC-hydrolyzing activity between control and ADR-treated samples that can account for the high levels of polyubiquitinated p53 or monoubiquitinated p53 in control and ADR-treated samples, respectively. Inclusion of ubiquitin hydrolase inhibitor, ubiquitin aldehyde, abolished the Ub-AMC-hydrolyzing capacity of the extracts by >90%, thus confirming the specificity of ubiquitin hydrolase (Fig. 7F). These data suggest that alterations in the ratio of monoubiquitinated p53 to its polyubiquitinated forms is not a result of an increase in deubiquitinating activity but rather is due to alterations in Hdm2 E3 ligase activity that is required for the polyubiquitination of p53.
To determine the cellular localization of Rad6 and p53 and to confirm whether the Rad6 complexed p53 is indeed ubiquitinated, we probed control and ADR-treated (at 72 h of recovery) MCF10A cells with antibodies to Rad6, p53, or ubiquitin. Whereas negligible immunoreactivity to p53 was observed in untreated cells (Fig. 7GB), exposure to ADR caused a dramatic appearance of pAb421-immunoreactive p53 in the nucleoplasm and nucleoli of MCF10A cells (Fig. 7GE and GK). Similarly, treatment with ADR induced preferential accumulation of Rad6 in the nuclei of MCF10A cells (Fig. 7GD and GG and Fig. 6D) compared to untreated cells that displayed diffuse Rad6 staining in the cytoplasm and nucleus (Fig. 7GA and Fig. 6D). Double immunofluorescence labeling and image-merging experiments demonstrated the colocalization of Rad6 with p53 (Fig. 7GF), p53 with ubiquitin (Fig. 7GL) and Rad6 with ubiquitin (Fig. 7GI) in the nucleoplasm and nucleoli of ADR-treated cells when Hdm2 was undetectable (Fig. 6D). These data not only confirm the results from coprecipitation studies but also provide further evidence for ADR-induced effects on p53 ubiquitination, colocalization of p53 with Rad6, and the stability of Rad6-p53 complexes.
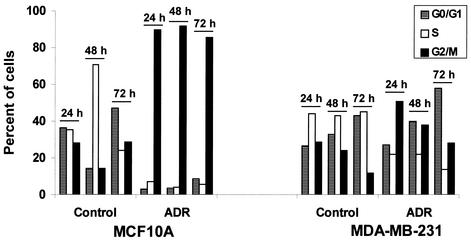
We next examined the effects of ADR on cell cycle progression in MCF10A and MDA-MB-231 cells since both cell lines show Rad6-p53 complex formation but with distinctly different stabilities. Treatment of MCF10A cells with 0.1 μg of ADR/ml induced G2/M cell cycle arrest by 24 h in ∼90% of cells that persisted at least up to 72 h of treatment, whereas similar analysis of untreated MCF10A cells at corresponding time points revealed normal cell cycle progression (Fig. 8). Similar flow cytometric analysis of MDA-MB-231 cells revealed that 51% of the cells arrested in G2/M at 24 h, and at 72 h a majority of the cells (58%) were found to arrest in G0/G1 compared to only 28% in the G2/M phase (Fig. 8).
FIG. 8.
ADR treatment induces differential effects on cell cycle arrest in MCF10A and MDA-MB-231 cells. Cell cycle progression was analyzed in control or ADR-treated MCF10A and MDA-MB-231 cells at 24, 48, and 72 h by using a FACScan.
Demonstration in vitro of Rad6- and Mdm2-mediated effects on p53 ubiquitination.
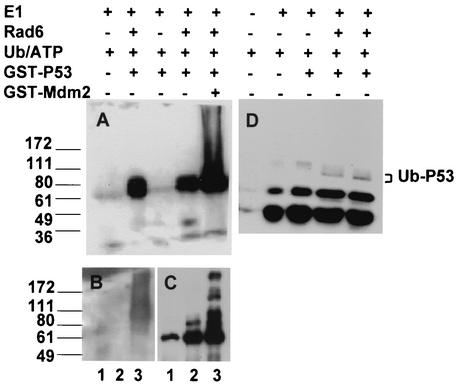
Since our data showed that (i) a significant amount of p53 is associated with Rad6 in ADR-exposed MCF10A cells, (ii) ADR induces physical colocalization of Rad6 with p53 in MCF10A nuclei, (iii) a substantial amount of the p53 present in the nuclei of ADR treated MCF10A cells is ubiquitinated under conditions when Hdm2 is undetectable, and (iv) the majority of the posttranslationally stabilized p53 in ADR-treated MCF10A cells is indeed monoubiquitinated, we tested the effects of Rad6 singly or in combination with Mdm2 on p53 ubiquitination in a cell-free system. Recombinant Rad6 was incubated with GST-p53 in the presence or absence of GST-Mdm2 as indicated (Fig. 9). Under nonreducing conditions, E1-Ub thioesters are detectable both in the presence and in the absence of p53, and E1 per se has no effect on p53 ubiquitination (Fig. 9D). Interestingly, although thioester intermediates of E1-Rad6 are readily detectable in reactions lacking p53 (data not shown), E1-Rad6 thioester intermediates were not observed in reactions containing p53. These data suggest that inclusion of a suitable ubiquitination substrate such as p53 may promote rapid transfer of activated ubiquitin to the substrate. The molecular sizes of ubiquitinated p53 correspond with the presence of one or two ubiquitin molecules (Fig. 9A). The effects of Rad6 on p53 ubiquitination are specific since assay mixtures not containing Rad6 are unable to mediate transfer of ubiquitin to p53. Inclusion of GST-Mdm2 into reactions containing E1 and Rad6 induced polyubiquitination of p53, as confirmed by its immunoreactivity to ubiquitin (Fig. 9A), polyubiquitin (Fig. 9B), and p53 CM-1 (Fig. 9C) antibodies. These data suggest that, whereas Rad6 mediates restrictive ubiquitination of p53, Mdm2 functions by extending ubiquitin chains.
FIG. 9.
Effects of Rad6 and Mdm2 on p53 ubiquitination. GST-p53 was incubated with ATP, ubiquitin, and E1 in the absence or presence of Rad6 or in the presence of both Rad6 and GST-Mdm2 as indicated. Reactions were terminated after 2 h at 30°C and then analyzed by SDS-PAGE under reducing (A to and C) or nonreducing (D) conditions and by Western blot analysis with ubiquitin (A and D), polyubiquitin (B), or p53 CM-1 (C) antibody. The positions of ubiquitinated p53 are indicated. The commercial preparation of ubiquitin contained significant levels of diubiquitin (D). The reactions and panels B and C contained E1, Rad6, Mdm2, ubiquitin, ATP, and p53 (lane 3); lacked Mdm2 (lane 2); or lacked Rad6 and Mdm2 (lane 1). Note the formation of fewer ubiquitin- or p53-immunoreactive bands in the presence of Rad6 as opposed to the formation of ubiquitin (A)-, polyubiquitin (B)-, or p53 CM-1 (C)-immunoreactive bands extending to the top of the gel in reactions containing Rad6 and Mdm2.
DISCUSSION
The catalytic activity of Rad6 as a ubiquitin-conjugating enzyme is essential for its functions in DNA repair since Rad6 with a defective catalytic site confers hypersensitivity to a variety of DNA-damaging agents (36, 42, 56). However, proteins relevant to DNA repair that are ubiquitinated by Rad6 besides histone H2b have remained unknown until recently. While this study was in review, Hoege et al. (20) demonstrated that proliferating cell nuclear antigen (PCNA), a DNA polymerase involved in DNA replication and repair, is a substrate of Rad6-mediated monoubiquitination and that damage-induced PCNA ubiquitination is essential for DNA repair. The results from the present study provide evidence for the first time that Rad6 may function in vivo during DNA damage-induced response by undergoing stable associations with proteins of the p53 pathway that ultimately result in restrictive ubiquitination of p53. Based on coimmunoprecipitation, surface plasmon resonance, and GST pull-down assays (unpublished data), we have demonstrated that Rad6 physically interacts with p53 and that this interaction is not dependent on Rad6's ubiquitination status. Thus, Rad6 can exist in complexes with p53 in the cells both before and after treatment with ADR. However, since ADR treatment induces quantitative and/or qualitative effects on Rad6, p53, p14ARF, and Mdm2 proteins, the levels and stability of Rad6-p53 complexes formed in vivo may be subject to regulation by ADR-induced posttranslational modifications. The results presented here are also more relevant to the clinical situation since the markers of drug-induced response are measured at various times of recovery or repair after a single exposure to the damaging agent rather than at different time points of treatment.
Rad6 is known to attach ubiquitin directly to a substrate protein either with or without the help of a ubiquitin ligase (54). Results from in vitro ubiquitination assays clearly show that p53 ubiquitination is a regulated process, wherein Rad6 mediates limited addition of ubiquitin moieties to p53 and further addition of ubiquitin molecules for extension of ubiquitin chains is dependent upon the presence of Mdm2-mediated E3 ligase activity. The physiological relevance of these in vitro data is underscored by results from in vivo experiments that demonstrated that the majority of ADR-stabilized p53 in MCF10A cells is not only monoubiquitinated and physically complexed with Rad6 but that the increase or decrease in levels of mono- versus polyubiquitinated p53 species, respectively, correlates with the decline in intact Hdm2 levels. Thus, p53 protein degradation, but not its monoubiquitination, is impaired during the ADR-induced p53 response. Unlike other ubiquitin-conjugating enzymes, such as UbcH5 (22) and UbcH7 (29), that efficiently ubiquitinate p53 in vitro, Rad6 expression is inducible by a variety of DNA-damaging agents. These results suggest a unique role for Rad6 in p53 ubiquitin modification and DNA damage signaling that are distinct from other known ubiquitin-conjugating enzymes that participate in the generation of degradation signal required for recognition by the 26S proteasome (21).
It is interesting that the chemotherapeutic drug ADR influences the stability of p53 by (i) enhancing p53 monoubiquitination, (ii) interfering with the cocompartmentalization of p53 and Mdm2 (62), and (iii) inhibiting p53 polyubiquitination that is required for nuclear export by Mdm2 and degradation by the proteasome (57). Thus, DNA damage-induced p53 ubiquitin modification may serve specific function(s) in the recovery process rather than simply be a tag for directing proteasome degradation. These findings are consistent with previous reports that have described ubiquitin modifications of histone H2B (45), p53 (34), and histone H1 (40) that do not lead directly to degradation. Since Hdm2 is not detectable and since high levels of ubiquitinated p53 are found to colocalize with Rad6 in the nuclei of ADR-treated MCF10A cells, it remains to be determined how ubiquitinated p53 is regulated in damaged cells.
Although expression of p14ARF is not linked to DNA damage (51), our data show that ADR exerts similar inductory effects on p14ARF protein expression as on p53 and Rad6. Results from our de novo metabolic-labeling studies clearly demonstrate that p14ARF recruitment into Rad6-p53 complexes is dependent on the presence of cisplatin-induced damage. Since Hdm2 is degraded at least in part via the ubiquitin-proteasome pathway in ADR-treated MCF10A cells, it is not clear whether p14ARF functions in p53 stabilization (44) by inhibiting E3 ubiquitin ligase activity of Hdm2 (23) or by facilitating Hdm2 destabilization, as proposed by Zhang et al. (63). ADR-induced downregulation of Mdm2 via mechanisms independent of the proteasome has been reported (33); however, these results conflict with the present data, indicating variations in anthracycline-induced responses that may be attributable to cell-specific differences. In this regard, it can be noted that proteasome-mediated proteolysis of Mdm2 has been reported to play an important role in etoposide-induced early downregulation of Mdm2 in HL-60 cells since Mdm2 levels could be partially restored by proteasome inhibitors lactacystin and LLnL (11). Although the precise nature and regulation of interactions between Rad6, p53, p14ARF, and Hdm2 remain to be determined, our findings from both coimmunoprecipitation (endogenous Rad6 and p53 proteins) and plasmon resonance (recombinant Rad6 and GST-p53) studies suggest that Rad6 interacts directly with p53 in the absence of Hdm2 or p14ARF and that the stability of Rad6-p53 complexes in vivo are perhaps subject to regulation by drug-mediated effects on p14ARF and Hdm2 levels.
Although the region(s) of Rad6 molecule interacting with p53 is currently being determined, the present study represents the first demonstration of an in vivo correlation between ADR-induced p53 response, p53 ubiquitination status, p14ARF recruitment, Hdm2 decay, and stabilization of interactions between Rad6, a ubiquitin-conjugating enzyme and DNA repair protein, and p53. Thus, the longevity and functionality of the DNA damage-induced p53 response may be subject to regulation by the levels and activity of Rad6, proteins of the p53 pathway, and/or by the mutational status of p53. In this context, it is interesting that, in contrast to normal MCF10A cells that express wild-type p53 and exhibit stable Rad6-p53 complex formation and prolonged G2/M arrest, metastatic MDA-MB-231 human breast cancer cells express mutant R280K p53, form complexes with Rad6 with transient stability, and exhibit short-lived G2/M arrest. DNA damage has been believed to stabilize p53 by inducing posttranslational modifications such as phosphorylation, acetylation, sumoylation, or ubiquitination (1). Although phosphorylation of p53 is likely to be an important determinant of p53 stability, phosphorylation of amino and carboxy terminus sites of p53 is not absolutely required for p53 stabilization in response to certain DNA-damaging agents. In fact, p53 mutated in all potential amino and carboxy terminus phosphorylation sites remained susceptible to stabilization induced by UV irradiation and actinomycin D (2, 6). These data suggest that stabilization of p53 in response to various DNA damaging agents is not likely to occur through a single pathway but instead may involve multiple mechanisms that are unique to the cell and individual DNA-damaging agents. These mechanisms may include regulation of Hdm2 expression and stability, interaction of Hdm2 with enzymes of the ubiquitin system, regulation of Hdm2 E3 ligase activity, p53-Hdm2-p14ARF complex formation, Rad6-p53-p14ARF-Hdm2 complex formation, p53 ubiquitination, nuclear export of p53, and degradation of ubiquitinated p53 by the proteasome. Based on our data, it appears that the chemotherapeutic drug ADR elicits DNA damage-induced p53 response by influencing several of these steps in normal MCF10A and metastatic MDA-MB-231 breast cells.
Rad18 has been proposed to act to load or target Rad6 to sites of DNA damage where its ubiquitin-conjugating activity could subsequently modify the stalled DNA replication complex (3). In the yeast, complex formation between Rad6 and Rad18 is believed to be important during error-free and error-prone lesion bypass; however, the mechanism by which this complex participates in lesion bypass processes remains to be determined. Residues 141 to 149 at the carboxyl terminus of Rad6 are essential for Rad18 binding (5); thus, immunoprecipitation with a Rad6 antibody directed to the carboxyl end appears to have impeded Rad6-Rad18 binding. Since ADR exerts similar inductory effects on Rad6 and Rad18, studies utilizing different Rad6 antibodies are necessary to determine whether Rad18 is a significant component of the supramolecular complexes formed between Rad6 and proteins of the p53 pathway.
In summary, our findings suggest that ubiquitin modification mediated by Rad6 may be an important regulator of p53 activity. RNAi experiments are under way to confirm whether Rad6 is a major regulator of p53 ubiquitination in drug-treated cells. Although very little is known about the mechanisms regulating error-free versus error-prone postreplication repair, our data suggest that maintenance of stable Rad6-p53 interactions and monoubiquitinated p53 resulting in the prolonged presence of the DNA damage sensor, may be one such mechanism. Thus, alterations in Rad6 levels and/or activity or mutations in p53 that can potentially disrupt or influence the stability of Rad6-p53 interactions can lead to transitory G2 arrest and increased promiscuous repair. Modulation of this interaction during DNA damage-induced cellular response, such as by Rad6-mediated p53 monoubiquitination, may serve to regulate and guarantee the fidelity of postreplication repair.
Acknowledgments
This work was supported by U.S. Army Medical Research and Materiel Command grant DAMD17-99-I-9443 to M.P.V.S.
We thank K. Shavorskaya (BIAcore Facilities, Swedish University of Agricultural Sciences, Uppsala, Sweden) for performing biomolecular interaction experiments, A. Iakovenko (Max-Planck Institute for Molecular Biology, Germany) for calculating dissociation constant, H. Yasuda (School of Life Science, Tokyo University of Pharmacy and Life Science, Tokyo, Japan) for generously providing GST-p53 and GST-Mdm2 fusion proteins, and Gloria Heppner for critically reading the manuscript. We thank Noelle Kondrat for technical assistance and two anonymous reviewers for suggesting experiments shown in Fig. 7A and F.
REFERENCES
- 1.Appella, E., and C. W. Anderson. 2001. Post-translational modifications and activation of p53 by genotoxic stresses. Eur. J. Biochem. 268:2764-2772. [DOI] [PubMed] [Google Scholar]
- 2.Ashcroft, M., and K. H. Vousden. 1999. Regulation of p53 stability. Oncogene 18:7637-7643. [DOI] [PubMed] [Google Scholar]
- 3.Bailly, V., J. Lamb, P. Sung, S. Prakash, and L. Prakash. 1994. Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 8:811-820. [DOI] [PubMed] [Google Scholar]
- 4.Bailly, V., S. Lauder, S. Prakash, and L. Prakash. 1997. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J. Biol. Chem. 272:23360-23365. [DOI] [PubMed] [Google Scholar]
- 5.Bailly, V., S. Prakash, and L. Prakash. 1997. Domains required for dimerization of yeast Rad6 ubiquitin-conjugating enzyme and Rad18 DNA binding protein. Mol. Cell. Biol. 17:4536-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blattner, C., E. Tobiasch, M. Litfen, H. J. Rahmsdorf, and P. Herrlich. 1999. DNA damage induced p53 stabilization: no indication for an involvement of p53 phosphorylation. Oncogene 8:1723-1732. [DOI] [PubMed] [Google Scholar]
- 7.Bucchop, S., M. K. Gibson, X. W. Wang, P. Wagner, H. W. Sturzbecher, and C. C. Harris. 1997. Interaction of p53 with the human Rad51 protein. Nucleic Acids Res. 25:3868-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cassier-Chauvat, C., and F. Fabre. 1991. A similar defect in UV-induced mutagenesis conferred by the rad6 and rad18 mutations of Saccharomyces cerevisiae. Mutat. Res. 254:247-253. [DOI] [PubMed] [Google Scholar]
- 9.Ciechanover, A. 1994. The ubiquitin-proteasome proteolytic pathway. Cell 79:13-22. [DOI] [PubMed] [Google Scholar]
- 10.Chau, V., J. W. Tobias, A. Bachmair, D. Marriott, D. J. Ecker, D. K. Gonda, and A. Varshavsky. 1989. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243:1576-1583. [DOI] [PubMed] [Google Scholar]
- 11.Cho, J. W., J. C. Park, J. C. Lee, T. K. Kwon, J. W. Park, W. K. Baek, S. I. Suh, and M. H. Suh. 2001. The levels of Mdm2 protein are decreased by a proteasome-mediated proteolysis prior to caspase-3 dependent pRb and PARP cleavages. J. Korean Med. Sci. 16:135-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coux, O., K. Tanaka, and A. L. Goldberg. 1996. Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 65:801-847. [DOI] [PubMed] [Google Scholar]
- 13.Dang, L. C., F. D. Melandri, and R. L. Stein. 1998. Kinetic and mechanistic studies on the hydrolysis of ubiquitin C-terminal 7-amido-4-methylcoumarin by deubiquitinating enzymes. Biochemistry 37:1868-1879. [DOI] [PubMed] [Google Scholar]
- 14.Finley, D., S. Sadis, B. P. Monia, P. Boucher, D. J. Ecker, S. T. Crooke, and V. Chau. 1994. Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol. Cell. Biol. 14:5501-5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottleib, T. M., and C. J. Sherr. 1996. p53 in growth control and neoplasia. Biochim. Biophys. Acta 1287:77-102. [DOI] [PubMed] [Google Scholar]
- 16.Haffner, R., and M. Oren. 1995. Biochemical properties and biological effects of p53. Curr. Opin. Genet. Dev. 5:84-90. [DOI] [PubMed] [Google Scholar]
- 17.Haupt, Y., R. Maya, A. Kazaz, and M. Oren. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296-299. [DOI] [PubMed] [Google Scholar]
- 18.Hershko, A., and A. Ceichanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 19.Hochstrasser, M. 1995. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr. Opin. Cell. Biol. 7:215-223. [DOI] [PubMed] [Google Scholar]
- 20.Hoege, C., B. Pfander, G. L. Moldovan, G. Pyrowolakis, and S. Jentsch. 2002. Rad6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419:135-141. [DOI] [PubMed] [Google Scholar]
- 21.Hoffman, R. M., and C. M. Pickart. 1999. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96:645-653. [DOI] [PubMed] [Google Scholar]
- 22.Honda, R., H. Tanaka, and H. Yasuda. 1997. Oncoprotein Mdm2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420:25-27. [DOI] [PubMed] [Google Scholar]
- 23.Honda, R., and H. Yasuda. 1999. Association of p19(ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 18:22-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jentsch, S., J. P. McGrath, and A. Varshavsky. 1987. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature 329:131-134. [DOI] [PubMed] [Google Scholar]
- 25.Kastan, M. B., O. Onyekwere, D. Sidransky, B. Vogelstein, and R. W. Craig. 1991. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 51:6304-6311. [PubMed] [Google Scholar]
- 26.Koken, M. H., P. Reynolds, I. Jaspers-Dekker, L. Prakash, S. Prakash, D. Bootsma, and J. H. Hoeijmakers. 1991. Structural and functional conservation of two human homologs of the yeast DNA repair gene RAD6. Proc. Natl. Acad. Sci. USA 88:8865-8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koken, M. H., E. M. Smit, I. Jaspers-Dekker, B. A. Oostra, A. Hagemeijer, D. Bootsma, and J. H. Hoeijmakers. 1992. Localization of two human homologs, HHR6A and HHR6B, of the yeast DNA repair gene RAD6 to chromosomes Xq24-q25 and 5q23-q31. Genomics 12:447-453-1992. [DOI] [PubMed] [Google Scholar]
- 28.Kubbutat, M. H., S. N. Jones, and K. H. Vousden. 1997. Regulation of p53 stability by Mdm2. Nature 387:299-303. [DOI] [PubMed] [Google Scholar]
- 29.Kumar, S., W. H. Kao, and P. M. Howley. 1997. Physical interaction between specific E2 and Hect E3 enzymes determines functional cooperativity. J. Biol. Chem. 272:13548-13554. [DOI] [PubMed] [Google Scholar]
- 30.Lawrence, C. 1994. The RAD6 DNA repair pathway in Saccharomyces cerevisiae: what does it do, and how does it do it? Bioessays 16:253-258. [DOI] [PubMed] [Google Scholar]
- 31.Lawrence, C. W., and D. C. Hinkle. 1996. DNA polymerase zeta and the control of DNA damage induced mutagenesis in eukaryotes. Cancer Surveys 28:21-31. [PubMed] [Google Scholar]
- 32.Levine, A. J. 1997. p53, the cellular gatekeeper for growth and division. Cell 88:323-331. [DOI] [PubMed] [Google Scholar]
- 33.Ma, Y., R. Yuan, Q. Meng, I. D. Goldberg, E. M. Rosen, and S. Fan. 2000. P53-independent downregulation of Mdm2 in human cancer cells treated with adriamycin. Mol. Cell. Biol. Res. Commun. 3:122-128. [DOI] [PubMed] [Google Scholar]
- 34.Maki, C. G., and P. M. Howley. 1997. Ubiquitination of p53 and p21 is differentially affected by ionizing and UV radiation. Mol. Cell. Biol. 17:355-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Midgley, C. A., J. M. Desterro, M. K. Saville, S. Howard, A. Sparks, R. T. Hay, and D. P. Lane. 2000. An N-terminal p14ARF peptide blocks Mdm2-dependent ubiquitination in vitro and can activate p53 in vivo. Oncogene 19:2312-2323. [DOI] [PubMed] [Google Scholar]
- 36.Montelone, S. Prakash, and L. Prakash. 1981. Recombination and mutagenesis in rad6 mutants of Saccharomyces cerevisiae: evidence for multiple functions of the RAD6 gene. Mol. Gen. Genet. 184:410-415. [DOI] [PubMed] [Google Scholar]
- 37.Offer, H., M. Milyavsky, N. Erez, D. Matas, I. Zurer, C. C. Harris, and V. Rotter. 2001. Structural and functional involvement of p53 in BER in vitro and in vivo. Oncogene 20:581-589. [DOI] [PubMed] [Google Scholar]
- 38.Oliner, J. D., K. W. Kinzler, P. S. Meltzer, D. L. George, and B. Vogelstein. 1992. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 358:80-83. [DOI] [PubMed] [Google Scholar]
- 39.Oliner, J. D., J. A. Pietenpol, S. Thialingam, J. Gyuris, K. W. Kinzler, and B. Vogelstein. 1993. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature 362:857-860. [DOI] [PubMed] [Google Scholar]
- 40.Pham, A. D., and F. Sauer. 2000. Ubiquitin-activating/conjugating activity of TAFII250, a mediator of activation of gene expression in Drosophila. Science 289:2357-2360. [DOI] [PubMed] [Google Scholar]
- 41.Prakash, L. 1981. Characterization of postreplication repair in the yeast Saccharomyces cerevisiae and effects of the rad6, rad18, rev3, and rad52 mutations. Mol. Gen. Genet. 184:471-478. [DOI] [PubMed] [Google Scholar]
- 42.Prakash, S., P. Sung, and L. Prakash. 1990. Structure and function of RAD3, RAD6, and other DNA repair genes of Saccharomyces cerevisiae, p. 275-292. In P. R. Strauss and S. H. Wilson (ed.), The eukaryotic nucleus. Molecular structure and macromolecular assemblies, vol. 1. Telford Press, Caldwell, N.J.
- 43.Prakash, S., P. Sung, and L. Prakash. 1993. DNA repair genes and proteins of Saccharomyces cerevisiae. Annu. Rev. Genet. 27:33-70. [DOI] [PubMed] [Google Scholar]
- 44.Prives, C. 1998. Signaling to p53: breaking the MDM2-p53 circuit. Cell 95:5-8. [DOI] [PubMed] [Google Scholar]
- 45.Robzyk, K., J. Recht, and M. A. Osley. 2000. Rad6-dependent ubiquitination of histone H2B in yeast. Science 287:501-504. [DOI] [PubMed] [Google Scholar]
- 46.Shekhar, P. V. M., A. Lyakhovich, H. Heng, D. W. Visscher, and N. Kondrat. 2002. Rad6 overexpression induces multinucleation, centrosome amplification, abnormal mitosis, aneuploidy, and transformation. Cancer Res. 62:2115-2124. [PubMed] [Google Scholar]
- 47.Shekhar, P. V. M., R. Welte, R., J. K. Christman, H. Wang, and J. Werdell. 1997. Altered p53 conformation: a novel mechanism of wild type p53 functional inactivation in a model for early human breast cancer. Int. J. Oncol. 11:1087-1094. [DOI] [PubMed] [Google Scholar]
- 48.Shieh, S. Y., M. Ikeda, Y. Taya, and C. Prives. 1997. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 91:325-334. [DOI] [PubMed] [Google Scholar]
- 49.Smith, M. L., and A. J. Fornace, Jr. 1996. The two faces of tumor suppressor p53. Am. J. Pathol. 148:1019-1022. [PMC free article] [PubMed] [Google Scholar]
- 50.Soule, H. D., T. M. Maloney, S. R. Wolman, W. D., Peterson, Jr., R. Brenz, C. M. McGrath, J. Russo, R. J. Pauley, R. F. Jones, and S. C. Brooks. 1990. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 50:6075-6086. [PubMed] [Google Scholar]
- 51.Stott, F., S. Bates, M. C. James, B. B. McConnell, M. Starborg, S. Brookes, I. Palmero, K. Ryan, E. Hara, K. H. Vousden, and G. Peters. 1998. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 17:5001-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strous, G. J., and R. Govers. 1999. The ubiquitin-proteasome system and endocytosis. J. Cell Sci. 112:1417-1423. [DOI] [PubMed] [Google Scholar]
- 53.Sung, P., E. Berleth, C. Pickart, S. Prakash, and L. Prakash. 1991. Yeast RAD6 encoded ubiquitin-conjugating enzyme mediates protein degradation dependent on the N-end-recognizing E3 enzyme. EMBO J. 10:2187-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sung, P., S. Prakash, and L. Prakash. 1988. The RAD6 protein of Saccharomyces cerevisiae polyubiquitinates histones, and its acidic domain mediates this activity. Genes Dev. 2:1476-1485. [DOI] [PubMed] [Google Scholar]
- 55.Sung, P., S. Prakash, and L. Prakash. 1990. Mutation of cysteine 88 in Saccharomyces cerevisiae RAD6 protein abolishes its ubiquitin-conjugating activity and its various biological functions. Proc. Natl. Acad. Sci. USA 87:2695-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sung, P., S. Prakash, and L. Prakash. 1991. Stable ester conjugate between the Saccharomyces cerevisiae RAD6 protein and ubiquitin has no biological activity. J. Mol. Biol. 221:745-749. [DOI] [PubMed] [Google Scholar]
- 57.Tao, W., and A. J. Levine. 1999. P19(ARF) stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc. Natl. Acad. Sci. USA 96:3077-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Torres-Ramos, C. A., S. Prakash, and L. Prakash. 2002. Requirement of RAD5 and MMS2 for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 22:2419-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Torres-Ramos, C. A., B. L. Yoder, P. M. Burgers, S. Prakash, and L. Prakash. 1996. Requirement of proliferating cell nuclear antigen in RAD6-dependent postreplicational DNA repair. Proc. Natl. Acad. Sci. USA 93:9676-9681. [DOI] [PMC free article] [PubMed]
- 60.Wang, X. W., A. Tseng, N. A. Ellis, E. A. Spillare, S. P. Linke, A. I. Robles, H. Seker, Q. Yang, P. Hu, S. Beresten, N. A. Bemmels, S. Garfield, and C. C. Harris. 2001. Functional interaction of p53 and BLM DNA helicase in apoptosis. J. Biol. Chem. 276:32948-32955. [DOI] [PubMed] [Google Scholar]
- 61.Xin, H., W. Lin, W. Sumanasekera, Y. Zhang, X. Wu, and Z. Wang. 2000. The human RAD18 gene product interacts with HHR6A and HHR6B. Nucleic Acids Res. 28:2847-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xirodimas, D. P., C. W. Stephen, D. P. Lane. 2001. Cocompartmentalization of p53 and Mdm2 is a major determinant for Mdm2-mediated degradation of p53. Exp. Cell Res. 270:66-77. [DOI] [PubMed] [Google Scholar]
- 63.Zhang, Y., Y. Xiong, and W. G. Yarbrough. 1998. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 92:725-734. [DOI] [PubMed] [Google Scholar]