Abstract
A strong epithelial specific enhancer drives transcription of the human papillomavirus type 18 (HPV18) oncogenes. Its activity depends on the formation of a higher-order nucleoprotein complex (enhanceosome) involving the sequence-specific JunB/Fra2 transcription factor and the HMG-I(Y) architectural protein. Here we show that proteins from HeLa cell nuclear extract cover almost all of the HPV18 enhancer sequences and that it contains seven binding sites for the purified HMG-I(Y) protein, providing evidence for a tight nucleoprotein structure. Binding of HMG-I(Y) and the AP1 heterodimer from HeLa nuclear extract to overlapping sites of the core enhanceosome is cooperative. The integrity of this specific HMG-I(Y) binding site is as essential as the AP1 binding site for the enhancer function, indicating the fundamental role played by this architectural protein. We demonstrate that the CBP/p300 coactivator is recruited by the HPV18 enhanceosome and that it is limiting for transcriptional activation, since it is sequestered by the adenovirus E1A protein and by the JunB/Fra2 positive factor in excess. We show the involvement of JunB and p300 in vivo in the HPV18 transcription by chromatin immunoprecipitation of HPV18 sequences in HeLa cells.
The high-mobility-group (HMG) proteins are nonhistone components of the chromatin that act as architectural factors in the assembly of specific DNA-protein complexes. The HMG-I(Y) subfamily member is characterized by AT hook motifs, basic domains of the protein that mediate binding to the minor groove of the DNA to AT-rich stretches longer than 3 bp (43). HMG-I(Y) is involved in the regulation of a wide range of cellular genes such as the beta interferon (IFN-β) (56), E-selectin (53), interleukin-2 receptor α-chain (25), and interleukin-4 (28) genes the CD44 receptor gene in smooth muscle (18), and the rhodopsin gene (14) but is also involved in the regulation of viral genes, for viruses such as human immunodeficiency virus (23), herpes simplex virus type 1 (19, 40), JC papovavirus (31), and human papillomavirus type 18 (HPV18) (11). By allosteric changes induced in the DNA, HMG-I(Y) cooperates with transcription factors to promote formation of higher-order nucleoprotein complexes called enhanceosomes (56).
Enhanceosomes are synergistic multiprotein complexes allowing high levels of tissue-specific transcription (13). In these complexes, bending of the DNA and stereospecific alignment of factors are absolutely required, to form a new interaction surface that efficiently recruits coactivators, such as the CREB binding protein (CBP), for the IFN-β enhanceosome, or CIITA, a B-lymphocyte specific coactivator, for the major histocompatibility complex class II enhanceosome (1, 27, 35). CBP was first identified by its ability to bind to and coactivate cyclic AMP response element binding protein (CREB) (5, 29). CBP, as well as its closely related p300 protein, were shown to interact with the adenovirus E1A protein (32; Z. Arany, W. R. Sellers, D. M. Livingston, and R. Eckner, Letter, Cell 77:799-800, 1994). At present, a large number of cellular or viral transcription factors have been shown to interact with CBP, which notably can functionally, as well as physically, interact with JunB and c-Jun (30), whereas the third zinc finger domain of CBP (C/H3) interacts with the basal transcriptional machinery (29). By linking these two classes of proteins, CBP integrates a large number of signaling pathways involved in cell growth, transformation, and development (22, 24).
One of the major functions of CBP in activating transcription is acetylation and remodeling of the chromatin, either by its intrinsic histone acetyltransferase activity (2, 9, 39), or via its interaction with other histone acetyltransferases such as P/CAF (55), ACTR (15), or SRC-1 (48). In contrast, in the IFN-β enhanceosome, recruitment of CBP is preceded by that of P/CAF, which acetylates the histones and the HMG-I(Y) architectural protein to allow assembly of the enhanceosome (1, 37). Following this step, CBP is mobilized to orderly recruit all the components involved in transcriptional activation: the RNA polymerase II, the SWI/SNF chromatin remodeling complex, and finally the TFIID complex (1). This role of integration site for interaction between upstream transcription factors and the basal machinery may be a characteristic trait of enhanceosomes.
Papillomaviruses are small double-stranded DNA viruses associated with benign proliferative lesions of the epidermis, such as skin warts, but also associated with cervical carcinomas. HPV18 is associated with more advanced genital neoplasia than other high-risk HPV types. This correlates with higher levels of activity of the early promoter that directs transcription of the E6 and E7 oncogenes (4, 44). We have shown that the HPV18 early promoter is controlled by a strong cell-specific enhanceosome (11).
Footprinting experiments and functional analysis indicate that the saturating occupancy of factors of the HPV18 enhanceosome correlates both with binding of transcription or architectural factors, such as HMG-I(Y), and with structural features. The HPV18 enhanceosome is architecturally and functionally based on the central AP1 binding site binding specifically the JunB/Fra-2 heterodimer (11). In the present study, we show that cooperative binding of the HMG-I(Y) protein with the JunB/Fra2 heterodimer to a combined AP1/HMG-I(Y) binding site constitute the core of the enhanceosome. Site-directed mutagenesis indicates that the architectural factor is essential for the HPV18 transcription. We also show that the CBP/p300 coactivator is recruited by the HPV18 enhanceosome and is required in vivo for transcription of the HPV18 sequences integrated in the HeLa cell genome.
MATERIALS AND METHODS
Plasmids and transfections.
A thymidine kinase-chloramphenicol acetyltransferase (tk-CAT) reporter plasmid was used to clone the enhancer fragment (nucleotides 7510 to 7740 of the HPV18 genome) upstream of the minimal tk promoter (11). Point mutations or deletions were obtained by a PCR-based directed mutagenesis. Fragments A, C, and E were obtained by PCR amplification between nucleotides 7566 and 7740, 7510 and 7720, and 7584 and 7740, respectively. Fragments B, D, and F were subjected to deletion of nucleotides 7541 and 7566, 7655 and 7713, and 7626 and 7630, respectively, but the stereoalignment was maintained by addition of unrelated nucleotides. Fragment G was mutated in the AP1 binding site (TTAGTCATTTTCC→ggAGTCATTTTCC [binding site is underlined]). Mutation of the HMG-I(Y) binding site adjacent to the AP1 binding site was done in the four-T tract downstream from the AP1 consensus, with 2T-Mut corresponding to mutation of the first two Ts (TTAGTCATTTTCC→TTAGTCAggTTCC) and 4T-Mut corresponding to mutation of all the Ts (TTAGTCATTTTCC→TTAGTCAggggCC).
The collagenase reporter plasmid contains the −517 to 140 promoter region (3). pCMV 12S E1A expression vectors and its versions with deletions were a gift of Y. Shi (26, 47), the cytomegalovirus (CMV) expression plasmids for the AP1 fusion proteins are described elsewhere (7), the CMV-CBP expression plasmid was a gift of A. Harel-Bellan (2), and the E6 expression plasmid was provided by M. Scheffner (45).
Transfection experiments were done in HeLa cells by the calcium phosphate coprecipitation method. Cells were harvested 40 h after transfection. All transfections were done in the presence of CMV β-galactosidase and CMV green fluorescent protein (GFP)-C1 expression plasmid to standardize the transfection efficiencies.
Nuclear extracts and purified protein.
Nuclear extracts were prepared from HeLa cells according to the method of Dignam et al. (17). Protein concentrations were determined by the method of Bradford.
The glutathione S-transferase (GST)-JunB-Fra-2 fusion protein was constructed by cloning the JunB/Fra-2 fusion in the pGEX4T-1 plasmid (Pharmacia Biotech). Transformed BL21 strain was induced by 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 2 h at 30°C. They were then lysed by sonication in A250 (25 mM Tris [pH 7.5], 15 mM MgCl2, 15 mM EGTA, 10% glycerol, 0.3% Triton, 250 mM NaCl, 1 mM NaCl, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride). The supernatant was incubated with glutathione-agarose beads overnight at 4°C. Following three washes with phosphate-buffered saline and two washes with 50 mM Tris, proteins were eluted with 10 mM glutathione in 50 mM Tris.
DNase I footprinting assays.
In the context of the 230-bp enhancer cloned in the tk-CAT plasmid, unique restriction sites upstream (HindIII) and downstream (BstBI) of the enhancer were used. Fragments were labeled at the HindIII or BstBI restriction sites to perform footprinting, respectively, with the bottom or the top strand of DNA. A total of 3.3 pmol of the DNA probe was labeled by Klenow filling in the presence of [α-32P]dATP. Footprinting experiments with purified proteins were done with probes prepared by PCR amplification of the enhancer sequences with primers end-labeled by the T4 polynucleotide kinase in the presence of [γ-32P]dATP. The two probes contain DNA sequences from nucleotide 7510 to 7674 (upper strand) or from nucleotide 7740 to 7566 (bottom strand).
Binding reactions mixtures containing DNA (0.04 pmol) and nuclear extracts (24 or 50 μg) or purified proteins [0.5 to 1 μg of HMG-I(Y) or 2 to 3 μg of GST-JunB/Fra-2], were done in a 10 mM HEPES (pH 7.9) buffer supplemented with 4 mM spermidine, 4 mM MgCl2, 30 mM KCl, 10% glycerol, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, and 0.1 mM EDTA. To prevent nonspecific DNA binding, a preincubation of 10 min was done with 1 μg of poly(dI-dC) for nuclear extract or 400 ng of poly(dC-dG) for purified proteins, before the addition of the labeled probes. After addition of DNase I, tubes were transferred at 37°C and incubated for 2 min. Reactions were stopped by addition of 25 μl of a stop solution (50 mM EDTA, tRNA [0.1 mg/ml], 0.2% sodium dodecyl sulfate). Proteins were then digested with 0.5 μg of proteinase K (45 min at 42°C), and the DNA was phenol extracted, ethanol precipitated, denatured, and loaded on a 7% polyacrylamide sequencing gel. Chemical sequences were done by the Maxam and Gilbert method.
Gel shift assays.
Probes containing the extended AP1 binding site from HPV18 were obtained by hybridization of the following oligonucleotides: 5′-CTAGACCACCTGGTATTAGTCATTTTCCTGTCT-3′ and 5′-CTAGAGACAGGAAAATGACTAATACCAGGTGGT-3′ for the wild type, 5′-CTAGACCACCTGGTATTAGTCAggTTCCTGTCT-3′ and 5′-CTAGAGACAGGAAccTGACTAATACCAGGTGGT-3′ for the mutated downstream T tract (2T-Mut), or 5′-CTAGACCACCTGGTATTAGTCAggggCCTGTCT-3′ and 5′-CTAGAGACAGGccccTGACTAATACCAGGTGGT-3′ (4T-Mut) and 5′-CTAGACCACCTGGTATTACTTATTTTCCTGTCT-3′ and 5′-CTAGAGACAGGAAAATAAGTAATACCAGGTGGT-3′ for the AP1 mut. The TRE-containing probe was obtained by hybridization of the two oligonucleotides: 5′-AGCTAGCTGACTCAGATGTCCT-3′ and 5′-AGCTAGGACATCTGAGTCAGCT-3′. (In the sequences shown in this paragraph, lowercase letters are mutated sequences and underlined letters indicate AP1 binding sites.)
Purified proteins were preincubated for 30 min with 0.2 μg of poly(dC-dG) and nuclear extract with 0.1 to 1 μg of poly(dI-dC), before addition of 0.04 pmol of the hybridized oligonucleotides labeled by Klenow filling in the presence of [α-32P]dATP. After 5 min of incubation, samples were loaded on a 5% native polyacrylamide gel. For competition experiments, excess cold DNA was added to the binding reaction mixtures s just before the addition of labeled probes. For supershift experiments with specific antibodies, 2 μl of undiluted antiserum was added to the binding reactions and incubated 15 min at room temperature before addition of the labeled probes.
Immunodetection of p53.
Immunofluorescence was done on HeLa cells grown on coverslips cotransfected with E1A, the HPV18 E6, or the control CMV expression plasmids, in the presence of a fixed amount of the GFP expression plasmid. At 40 h after transfection they were rinsed with phosphate-buffered saline and then fixed in 4% paraformaldehyde. Following rehydration, cells were permeabilized with 0.1% Triton and incubated with the 1801 monoclonal antibody against p53 purchased from Santa Cruz Biotechnology, and this was followed by an anti-mouse antibody coupled to Texas red and DAPI (4′,6′-diamidino-2-phenylindole(0.15 μg/ml). Western blots were done with total cellular extracts of transfected HeLa cells harvested 40 h posttransfection. Ten micrograms of proteins was loaded on a 10% acrylamide gel, transferred to nitrocellulose membranes, and revealed using the 1801 primary antibody against p53 and the ECL kit (Boehringer-Mannheim).
Semiquantitative RT-PCR.
Total RNA from HeLa cells grown in 10-cm-diameter petri dishes, either transfected with 4 μg of the E1A expression plasmid or not, was extracted by the guanidinium thiocyanate method and digested with DNase I. Reverse transcription (RT) and PCR amplification were done on various dilutions of total cDNA with the following primers: 5′-TGCCTGCGGTGCCAGAAACCG-3′ and 5′-ATGGCGCGCTTTGAGGATCCA-3′ for the HPV18 E6 gene and (5′-GACCTGACAGACTACCTCAT-3′ and 5′-AGACAGCACTGTGTTGGCAT-3′ for β-actin.
Chromatin immunoprecipitation.
HeLa cells grown to 80% confluency in 10-cm-diameter petri dishes, either transfected by 10 μg of p300 expression vector or not, were cross-linked in 1% formaldehyde for 30 min at 37°C. Reactions were stopped by addition of glycine (0.1 M) for 10 min at room temperature. These cells were pelleted and resuspended in 210 μl of HNB (0.5 M sucrose, 15 mM Tris [pH 8], 0.25 mM EDTA, 0.125 mM EGTA, 0.1% NP-40) and sonicated. Immunoprecipitations were done with rabbit polyclonal anti-JunB (11) and with a rabbit polyclonal anti-p300 from Santa Cruz (sc:8981) or with preimmune antiserum for controls. Cell lysates, completed with 10% calf serum and 500 μg of sonicated salmon sperm DNA, were precleared with 10 μg of protein A-Sepharose (Amersham Biosciences). Immunoprecipitates were washed three times with 1 ml of HNB (260 mM KCl), resuspended in 100 μl of Tris (pH 8)-0.1% sodium dodecyl sulfate at 65°C for 10 min, and digested with proteinase K overnight. DNA was extracted and PCR was amplified using the primers specific either for the HPV18 LCR (5′-GGCGCCGCCTCTTTGGCG-3′ and 5′-GTATGTGCTGCCCAACC-3′) or for the β-actin gene (5′-CCTGAGCGCAAGTAC-3′ and 5′-GCGGTGGACGATGG-3′). Amplified products were analyzed on an agarose gel.
RESULTS
DNase I footprinting and functional analysis of the HPV18 enhancer.
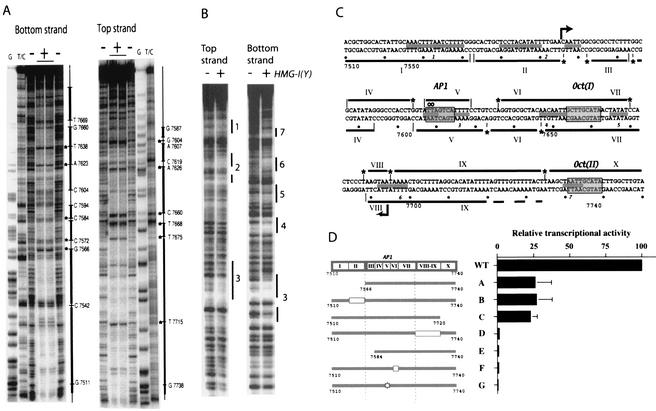
Transcription of the HPV18 oncoproteins E6 and E7 is positively regulated by a cell-specific enhancer contained within the noncoding region of the viral genome. This enhancer has been located to a 230-bp fragment containing binding sites for cellular factors, among which an essential AP1 binding site was identified (12, 20, 33, 38, 51). To characterize this fragment, we performed DNase I footprinting experiments with an extract containing highly concentrated nuclear proteins from the HeLa cervical carcinoma cell line, in which the rate of HPV18 transcription is high. These experiments delineated a total of 10 protected regions (I to X) (Fig. 1A and C), the 5′ and 3′ ends being extensively covered, as well as the central 100-bp core fragment spreading from nucleotide 7566 to 7674. These results support the idea that transcription of HPV18 depends on formation of a three-dimensional structure shaped around a core fragment that cooperates with factors binding to the ends of the 230-bp fragment (11). This in vitro footprinting pattern is similar to the dimethyl sulfate reactivity pattern obtained in vivo with the integrated HPV18 genome in HeLa cells (10).
FIG. 1.
DNase I footprinting experiments and functional analysis of the complete HPV18 enhancer. (A) DNase I digestion of the DNA probe in the presence (+) or absence (−) of 50 μg of HeLa cells nuclear extracts. The G or T/C lanes contain chemical sequencing reactions. Bold lines to the right of each gel indicate protected regions, and stars represent hypersensitive sites. (B) Footprints obtained with the purified HMG-I(Y). Protected regions are shown by lines and numbered from the 5′ end of the enhancer. (C) Summary of the footprints observed on the HPV18 enhancer. Areas protected by the HeLa cells nuclear extract are depicted by bold lines above or under the respective DNA strand and numbered from I to X. HMG-I(Y) protections are schematized by gray boxes between the strands and numbered from 1 to 7. Stars represent hypersensitive sites. Arrows delineate the core enhancer sequence (from nucleotide 7566 to 7674). Empty circles indicate point mutations in footprint V in the AP1 binding site. The AP1 and the two putative Oct binding sites are boxed. (D) The activity of various mutant fragments of the enhancer resulting from point mutations (star), or from deletions (boxes) cloned in front of the tk-CAT reporter, was tested by transfection of 2 μg of reporter plasmids in HeLa cells. Activities are given relative to the wild-type enhancer activity, which was set at 100%. WT, wild type. The values are the means from at least three independent experiments with standard deviations (error bars).
We have previously defined a core enhancer sequence (between the arrows in Fig. 1C) which can form a multiprotein complex in gel shift assays, in the presence of HeLa nuclear extract (11). This core enhancer sequence retained only 10% of the total activity of the enhanceosome, indicating that interactions with surrounding sequences are required for full enhancer activity. To decipher other important sequences in the 230-bp HPV18 enhancer, we introduced point mutations and deletions (summarized in Fig. 1D). Point mutations in the AP1 binding site are shown in fragment G. Three deletion mutants, D, E, and F, showed complete loss of function, while three others, A, B, and C, were affected in their transcriptional activity. Interestingly, some of these sequences do not match known binding sites for transcription factors (41), and we speculate that their function could be essentially conformational. In this regard, footprints VIII and IX as well as footprint I and II for example, are composed of stretches of AT-rich sequences that may play a structural role. On the other hand, we demonstrated by point mutations that the Oct binding site, contained within footprint X, is an important sequence and may account for the decrease in transcriptional activity exhibited by fragment C (data not shown).
These experiments suggested that some of the footprints observed on the 230-bp sequence of the HPV18 enhancer may represent bona fide recognition sites for binding factors (as footprint III, V, VII, or X for instance), whereas other protected sequences may result from wrapping of the DNA around proteins in a tight three-dimensional nucleoprotein.
HMG-I(Y) is known to bind to AT-rich sequences in the minor groove of the DNA α-helix. We observed that the HMG-I(Y) purified protein covered at least seven AT-rich sequences along the HPV18 enhancer (sequences 1 to 7) (Fig. 1B and gray boxes between the two strands in Fig. 1C). Many of these AT tracts are long enough to be bound by two or three of the protein AT hooks, giving rise to high affinity-binding sites (34). As in previously described enhanceosomes, HMG-I(Y) could facilitate assembly of the HPV18 enhanceosome by inducing allosteric changes in the DNA that facilitate binding of transcription factors. However, more work is needed to decipher the role of HMG binding to these different sites and the structural and functional relationship between the architectural protein and transcription factors.
HMG-I(Y) and JunB/Fra-2 bind together to adjacent sequences.
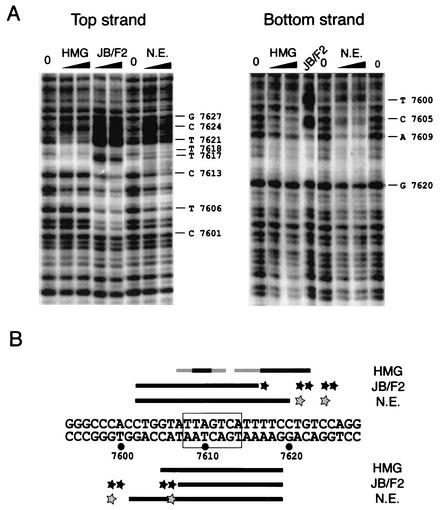
Within the core enhancer sequence, footprint V, obtained with the HeLa nuclear extract, matched the binding site for AP1 (Fig. 1A and C). Interestingly, the purified HMG-I(Y) protein also binds to this region, footprint 3 (Fig. 1B), covering the two AT-rich sequences surrounding the core of the AP1 binding site (Fig. 1B and C). We therefore studied the relationship between the binding of AP1 and HMG-I(Y) to these sequences in DNase I footprinting experiments with purified bacterially expressed HMG-I(Y) and GST-JunB-Fra-2 fusion proteins (Fig. 2A). This fusion protein allowed studies with a predefined heterodimer that is functionally identical to the association of the two partners (7). Sequences protected by HMG-I(Y) around the AP1 binding site corresponded to a four-T tract downstream of the AP1 binding site and AT-rich sequences upstream and within the core sequence itself (Fig. 2B). Sequences protected by the JunB/Fra-2 fusion protein encompass the core recognition site with strong hypersensitive sites on both sides. Interestingly, the pattern of DNase protection, observed with the HeLa cell nuclear extract, is similar to combined patterns of the two purified proteins, suggesting that, in vivo, the two proteins may bind together to their adjacent sites.
FIG. 2.
Footprint on the AP1 binding site is a combination of the HMG-I(Y) and JunB/Fra-2 footprints. (A) The DNA probes used in these experiments are PCR fragments amplified between nucleotides 7510 and 7674 for the upper strand or from nucleotide 7740 to 7566 of the HPV18 genome for the bottom strand, end labeled by the T4 polynucleotide kinase. DNase I digestions of the DNA probes were done in the absence (0) or in the presence of 500 ng or 1 μg of the HMG-I(Y) purified protein, 2 or 3 μg of the GST-JunB-Fra-2 fusion protein, and 24 or 36 μg of HeLa cells nuclear extract. (B) Summary of the footprints observed on the AP1 region. The areas of protection are depicted by bold lines above or under the respective DNA strand. Stars represent hypersensitive sites. Black symbols correspond to strong protection or hypersensitivity, while gray symbols depict signals of lower intensity. N.E., nuclear extract.
Binding of HMG-I(Y) adjacent to AP1 is functionally essential.
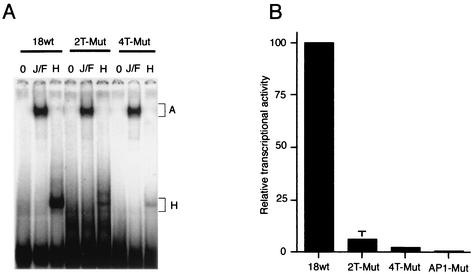
Footprinting experiments indicated that HMG-I(Y) and AP1 could bind to adjacent sites in the HPV18 core enhanceosome. We studied the binding of each purified proteins by gel shift assays on a 26-bp probe containing the AP1 binding site and its AT-rich surrounding regions (nucleotides 7598 to 7624). Using the wild-type probe, shifted complexes corresponding to the binding of the purified JunB/Fra-2 fusion (A) or HMG-I(Y) (H) proteins were observed (Fig. 3A). Specificity of the HMG-I(Y) binding to this probe was demonstrated by mutation of the four-T tract, downstream of the AP1 binding site (nucleotides 7615 to 7618). Mutation of the two closer Ts (2T-Mut: TTTT→GGTT) almost completely abolished HMG-I(Y) binding, as well as mutation of the four Ts (4T-Mut: TTTT→GGGG) (Fig. 3A). Therefore, HMG-I(Y) binds specifically to the AT-rich region downstream of the AP1 binding site. In contrast, these mutations did not alter binding of the JunB/Fra-2 heterodimer, as shown in Fig. 3A.
FIG. 3.
The HMG-I(Y) binding site adjacent to the AP1 binding site is functionally important. (A) Electrophoretic mobility shift assays were done with 100 ng of purified HMG-I(Y) protein and 1 μg of the GST-JunB-Fra-2 fusion protein, with oligonucleotide probes containing 10 additional nucleotides upstream and downstream of the AP1 consensus site, either wild-type (wt) or mutated in the four-T tract downstream of AP1 (2T-Mut and 4T-Mut). The AP1 shift is indicated as A while the HMG-I(Y) shift is labeled as H. (B) Transient-transfection experiments were done in HeLa cells with the HPV18 enhancer-tk-CAT reporter plasmid either wild-type (wt) or containing mutations in the four-T tract or in the AP1 binding site as indicated. Error bars, standard deviations.
We examined the functional involvement of this HMG-I(Y) binding site in the HPV18 transcriptional activity by introducing identical mutations of the four-T tract downstream of the AP1 binding site, in the context of the complete HPV18 enhancer, placed upstream of the minimal tk promoter and the CAT reporter gene. These mutated sequences were transfected in HeLa cells and their activities were compared to the wild-type enhancer, and to a mutant of the AP1 core binding site. We observed that both mutants (two-T or four-T substitutions) of the HMG-I(Y) binding site, drastically reduced the activity of the enhancer, the activity of the four-T mutant being almost as reduced as the activity of the AP1 core mutant (Fig. 3B). These results suggest that the HMG-I(Y) binding to this four-T tract is as crucial as the AP1 binding for HPV18 transcription.
HMG-I(Y) and JunB/Fra2 from HeLa nuclear extract bind cooperatively to the HPV18 enhanceosome.
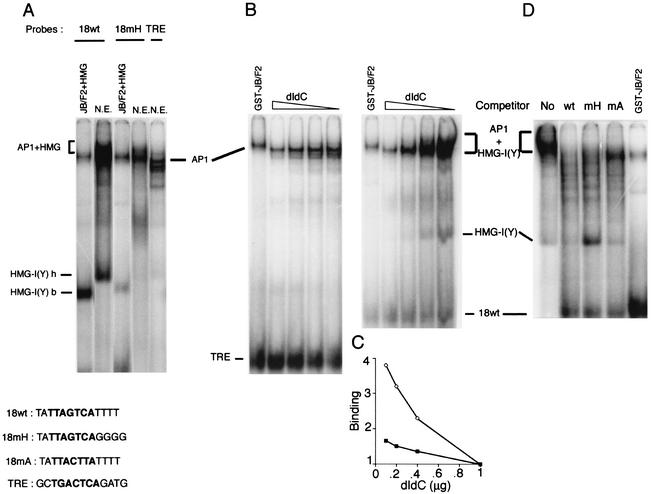
We have previously shown that HMG-I(Y) and JunB/Fra2 could interact in vitro at the protein-protein level. To decipher whether this interaction could induce cooperative binding to the overlapping sites on the HPV18 enhancer, gel shift binding assays were done with the two purified proteins at different relative concentrations. Unexpectedly, we could neither detect the appearance of a double complex containing the AP1 and HMG-I(Y) proteins, nor any cooperative induction of AP1 binding (Fig. 4A). In contrast, when using HeLa nuclear extract, we observed a strong increase in the apparent AP1 binding on the chimerical HPV18 sequence (18wt), when compared to the collagenase AP1 binding site (TRE) (Fig. 4A). As noted in Fig. 4A, the two complexes formed with the bacterial HMG-I(Y) protein (b) and the protein from nuclear extract (h) migrate differently, indicating possible posttranslational modifications. Formation of the complex from nuclear extract was strongly reduced with the HPV18 probe mutated in the four-T tract, downstream of the AP1 binding site (18mH), suggesting that the binding of HMG-I(Y) is required for this higher affinity (Fig. 4A). This apparent increase in AP1 binding to the wild-type probe, compared to the mutated or TRE probes, is about twofold. Competition experiments with increasing amounts of nonspecific poly(dI-dC) DNA or with excess of the specific nonlabeled probes indicated that the complex obtained with the HeLa nuclear extract was indeed composed of HMG-I(Y) and AP1 (Fig. 4B and D). HMG-I(Y) binds to the AT-rich tract with not very high sequence specificity and is therefore easily competed with nonspecific poly(dI-dC). We show here that increasing concentrations of poly(dI-dC) induces an increasing competition of the two complexes bound to the HPV18 probe, while the competitive effect was much less visible on the collagenase probe (Fig. 4B). At the highest concentration of poly(dI-dC) (1 μg), only binding of AP1 was detectable at identical levels on the two probes (Fig. 4C). At the lowest dI-dC concentration (0.1 μg), the increase of the slower-migrating complex on the HPV18 wild-type sequence, was about threefold, when compared to the TRE probe, indicating a cooperative binding of the two proteins to the HPV18 sequence (Fig. 4C).
FIG. 4.
AP1 and HMG-I(Y) bind cooperatively to the HPV18 probe from HeLa nuclear extract. (A) Binding of purified GST-JunB/Fra-2 (JB/F2) and HMG-I(Y) and HeLa nuclear extract (N.E.) to labeled probes containing the HPV18 AP1 binding site either wild type (18 wt) or mutated in the downstream four-T tract (18 mH) or the TRE, containing an AP1 binding site from the collagenase promoter. Sequences of the various probes are given. A cooperative complex only forms on the HPV18 wt probe with HeLa cell nuclear extract. AP1 and HMG-I(Y) binding from bacterially expressed HMG-I(Y) (labeled with a b) or HeLa nuclear extract (labeled with an h) is shown. (B) The AP1/HMG-I(Y) complex is competed by poly(dI-dC). Increasing amounts (0.1, 0.2, 0.4, and 1 μg) of the poly(dI-dC) nonspecific competitor were added to the binding reactions before addition of the labeled specific probes, TRE or 18 wt, as indicated. (C) Complexes formed on the two probes at various poly(dI-dC) concentrations are quantified by ImageQuant and plotted. (D) Specific competition of the AP1 and HMG-I(Y) binding on the HPV18 wt probe. An 80-fold excess of nonlabeled probes containing the HPV18 AP1 binding site, either wild-type or mutated in the HMG-I(Y) binding site (mH) or in the AP1 binding site (mA), whose sequences are given, were added to the binding reactions before addition of the labeled 18 wt probe.
Excess unlabeled HPV18 wild-type probe competed for all specific binding, while excess probe mutated in the AP1 core sequence (mA), competed for the HMG-I(Y) binding and the slower-migrating complex, only leaving binding of AP1. Finally, excess of a probe mutated in the downstream AT-tract (mH) competed with the totality of the slower-migrating complex and induced a concomitant increase in HMG-I(Y) binding (Fig. 4D). In conclusion, these competition experiments demonstrated that the slower-migrating complex contains both AP1 and HMG-I(Y) bound to two specific overlapping sequences.
Overexpression of JunB/Fra-2 titrates the CBP coactivator.
We have previously shown that the AP1 binding site is specifically bound by the JunB/Fra-2 heterodimer and that mutations of the binding site that prevent interaction with the protein totally abolished transcriptional activity of the HPV18 enhancer. However, we found that overexpression of the JunB/Fra-2 heterodimer did not increase the HPV18 transcription, but instead, repressed it more than 10-fold, while it could activate more than 25-fold the collagenase promoter (Fig. 5A). Overexpression of the other AP1 fusion proteins also led to repression of the HPV18 transcription, although it appeared milder than with JunB/Fra2 (two- to threefold), while they all activated efficiently the collagenase promoter (7) (data not shown). These experiments suggested that excess of the JunB/Fra-2 heterodimer titrated an essential limiting factor(s) for the formation of the HPV18 enhanceosome. Such limiting factors could be coactivators that interact directly with the interface formed by DNA bound proteins.
FIG. 5.
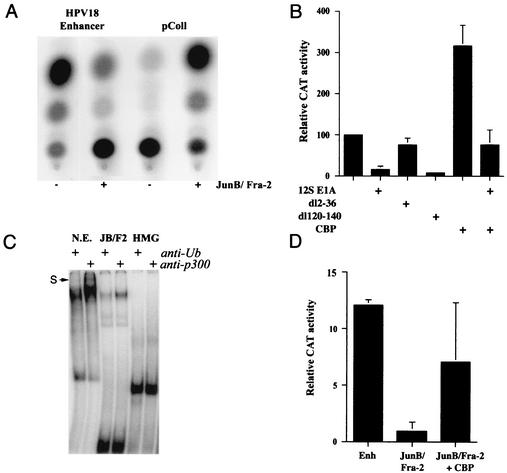
The CBP coactivator is required for the activity of the HPV18 enhancer in transfected cells. (A) Overexpression of JunB/Fra-2 leads to a strong repression of the enhancer. Representative CAT assays of transient-transfection experiments done in HeLa cells with 2 μg of the HPV18 enhancer-tk-CAT, or collagenase promoter reporter plasmid (pColl), in the absence (−) or in the presence (+) of 0.5 μg of plasmid expressing the JunB-Fra-2 fusion protein. Repression of the HPV18 enhancer is 10-fold, while activation of the collagenase promoter is 25-fold. (B) The adenovirus E1A protein represses the HPV18 enhancer via its CBP/p300 binding domain and this repression is relieved by excess of CBP. HeLa cells were cotransfected with 2 μg of the HPV18 enhancer-tk-CAT reporter plasmid and 8 μg of vector expressing CBP and/or 50 ng of vectors expressing either the wild-type (WT) 12S E1A, a deletion mutant form (dl2-36) which does not bind to CBP/p300, or a deletion mutant form (dl120-140) which does not bind to pRb. Transfections were done at least three times, and activities were standardized with a β-galactosidase expression vector. Error bars, standard deviations. (C) Supershift experiments with anti-p300 antibody. Binding reactions were preincubated with a rabbit anti-p300 antibody or a rabbit antiubiquitin antibody (Santa Cruz) as a negative control. The supershifted complex is shown by an arrow (S) and is observed with the HeLa nuclear extract and not with the purified JunB/Fra2 and HMG-I(Y) proteins. (D) A partial relief of the inhibition induced by JunB/Fra-2 was observed by cotransfecting 8 μg of the CBP expression plasmid, with 1 μg of the JunB/Fra-2 fusion protein expression plasmid. CAT assays are the mean of at least three independent transfection experiments. Error bars, standard deviations.
The CBP/p300 coactivators have been shown to play an essential role in transcriptional regulation of enhanceosomes, notably of the IFN-β gene. In addition, it has previously been shown to interact at the protein-protein level with AP1 (30) and HMG-I(Y) (36). We examined the putative role of CBP in the HPV18 enhanceosome by transfecting expression vectors of the adenovirus E1A protein, which is known to target the CBP/p300 coactivators specifically (49). Indeed, expression of wild-type 12S E1A protein represses CBP/p300-mediated transcriptional activation, by sequestering the coactivators. We found that the HPV18 enhancer activity was greatly reduced by expression of the wild-type 12S E1A protein and by a deletion mutant that fails to bind to pRb (dl120-140, Fig. 5B). In contrast, a mutant with the CBP/p300 interaction domain deleted (dl2-36) could no longer repress HPV18 transcription (Fig. 5B). The specificity of this repression was assessed by control experiments indicating that the three E1A constructs could activate the adenovirus E2 promoter while they did not affect activity of the tk promoter (not shown). Direct overexpression of CBP activated HPV18 transcription about threefold (Fig. 5B). In addition, CBP overexpression could relieve almost completely the repression mediated by 12S E1A protein, indicating that, indeed, E1A-mediated repression of the enhancer was due to sequestering of CBP/p300.
Taken together, these results suggested that CBP/p300 plays a critical role in the activity of the HPV18 enhanceosome. We postulated that the coactivator interacts with the interface created by the multiple proteins bound to the HPV18 enhanceosome. We checked whether CBP/p300 was present in the slower-migrating complex formed on the HPV18 sequence, in gel shift experiments. Specific antibodies raised against p300 induced a supershift, specifically with the complex formed on the HPV18 probe, indicating that CBP/p300 interacts in vitro with this complex (Fig. 5C). We also examined whether the transcriptional repression, observed with excess JunB/Fra-2, was due to the sequestering of the CBP/p300 coactivators, as demonstrated for the adenovirus E1A protein. We cotransfected the JunB/Fra-2 heterodimer with an excess of CBP expression plasmid and observed a partial relief of the repression by the JunB/Fra-2 heterodimer (Fig. 5D). This partial relief of transcriptional repression could indicate that other limiting factor(s) might be involved.
CBP/p300 is a coactivator of the HPV18 enhanceosome in vivo.
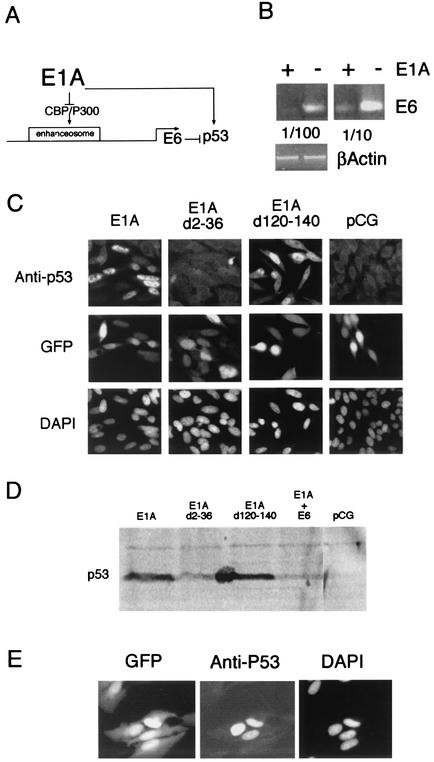
HeLa cells contain HPV18 sequences integrated in the cellular genome, including the E6 and E7 open reading frames under the control of the viral upstream regulatory region (46). In these cells, the E6 and E7 transforming functions, which induce the degradation of p53 and pRb, respectively, are actively transcribed due to the HPV18 enhancer. In order to check whether these integrated sequences were regulated similarly to the transfected sequences, we studied the effect of E1A transfection on endogenous E6 expression in HeLa cells. In transfected cells, E1A efficiently repressed the HPV18 enhancer through CBP/p300 sequestering (Fig. 5B). Repression of endogenous HPV18 sequences should lead to transcriptional repression of E6, which, in turn, will induce stabilization of the p53 protein (Fig. 6A). Indeed, overexpression of E1A, which was cotransfected with a GFP expression plasmid in HeLa cells, induced repression of the endogenous E6 transcription, as detected by semiquantitative PCR (Fig. 6B). This transcriptional repression led to a high degree of stabilization of the p53 protein, as detected both by immunofluorescence and Western blotting (Fig. 6C and D). This induction was dependent on CBP/p300 binding since the E1A mutant dl2-36, which cannot bind to the coactivators, could not induce p53, whereas the mutant dl120-140, impaired in its binding to pRb, has the same effect as the wild-type E1A (Fig. 6C and D). Cotransfection of a vector expressing HPV18 E6 with E1A counterbalanced the effect of E1A alone by reducing the level of p53 to its basal level in HeLa cells (Fig. 6D).
FIG. 6.
The CBP/p300 coactivator is required for the activity of the HPV18 enhancer in vivo. (A) Model for E1A repression of the HPV18 enhanceosome in vivo. Transfection of E1A in HeLa cells, containing the HPV18 regulatory region and E6 and E7 ORF integrated in their genome, should induce p53 stabilization through transcriptional repression of E6. (B) Semiquantitative RT-PCR done on HPV18 E6/E7 RNA extracted from HeLa cells either transfected with E1A or not. (C) p53 is stabilized in the nuclei of HeLa cells transfected by 2 μg of the wild-type 12S E1A or by the dl120-140 mutant, but not by the dl2-36 E1A mutant. p53 was revealed by immunostaining of fixed HeLa cells with Texas red anti-mouse antibody against an anti-p53 monoclonal antibody, GFP staining indicated the successfully transfected cells and DAPI labeled the nuclei of all the cells present in the fields. (D) Western blot analysis revealing that p53 is stabilized in HeLa cells transfected by 2 μg of the wild type or dl120-140 12S E1A, but not by the dl2-36 E1A mutant. Cotransfection of HPV18 E6 with E1A reduced p53 to its background level shown in control HeLa cells transfected with the empty vector (pCG). (E) HeLa cells were cotransfected with 2 μg of the JunB/Fra-2 and 0.5 μg of the GFP expression plasmids. Positively transfected cells are labeled by a Texas red anti-mouse antibody against the anti-p53 antibody. Nuclei are revealed by DAPI staining.
We checked whether activation of p53 also occurred in the presence of excess JunB/Fra2 transfected in HeLa cells, assuming that the transcriptional repression observed in this case was due to a mechanism similar to the repression observed with E1A. As shown in Fig. 6E, cells that overexpress JunB/Fra2, revealed by cotransfected GFP, expressed a strongly stabilized p53 in their nuclei. This result is in agreement with the idea that excess JunB/Fra2 sequester the CBP coactivator similarly to E1A.
In vivo interaction of JunB and p300 with the HPV18 chromatin.
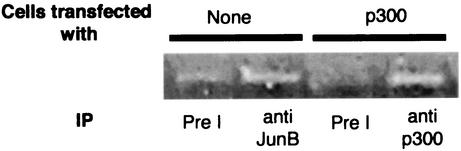
The preceding experiments indicated that CBP/p300 is involved in the control of the endogenous HPV18 transcription, and suggested that the regulation of the HPV18 sequence in vivo correlates with data obtained in cotransfection experiments. We therefore decided to directly check the involvement of the coactivator in vivo by chromatin immunoprecipitation. HeLa cells were either mock transfected or transfected with excess p300 expression plasmid. After cross-linking of the chromatin and immunoprecipitation with specific antibodies raised against the JunB protein or the p300 coactivator, DNA of the HPV18 regulatory region was PCR amplified and analyzed. Amplification of a 3′ part of the actin gene was used as a negative control (not shown) as well as immunoprecipitation with preimmune antibodies (Fig. 7). A more efficient amplification of the HPV18 DNA was observed with the anti-JunB antibody than with preimmune antibody in HeLa cells, indicating that this Jun protein is specifically recruited by the HPV18 chromatin in vivo. An efficient amplification was also observed with an anti-p300 antibody, in p300 overexpressing cells, thus demonstrating recruitment of the coactivator in vivo (Fig. 7).
FIG. 7.
JunB and p300 are able to bind the HPV-18 LCR in vivo. The agarose gel shows the PCR-amplified 200-bp DNA fragments obtained with the LCR specific primers after immunoprecipitation (IP). The immunoprecipitations were done with the JunB and p300 antiserum. Negative controls with rabbit preimmune serum (Pre I) were done for each chromatin preparation. Chromatin immunoprecipitations were done with HeLa cells either not transfected or transfected with p300 expression vector as indicated.
DISCUSSION
Papillomaviruses infect only keratinocytes of the squamous and mucous epithelia. The viral tropism is mainly based on the specificity of the viral gene expression that is restricted to keratinocytes and cervical carcinoma cell lines for all genital HPVs. Although the general organization does not appear well conserved, a common structure in the enhancers of all genital mucosal viruses may activate transcription of the E6 and E7 transforming genes (38). One model system to study HPV transcription is based on the cervical carcinoma cell lines established from genital lesions, in which the HPV early genes E6 and E7 are transcribed (46). We chose to use HeLa cells to investigate the mode of action of the HPV18 transcriptional enhancer since, in these cells, the enhancer can activate transcription of a heterologous promoter more than a hundredfold. The molecular basis of this strong transcriptional activity was unclear until we demonstrated that the HPV18 transcription is controlled by a three-dimensional nucleoprotein structure resembling the previously described enhanceosomes. Two major elements of this structure are the JunB/Fra-2 and HMG-I(Y) factors (11).
Here, we show by footprinting experiments, that proteins in a HeLa nuclear extract could bind most of the sequences of the 230-bp HPV18 enhancer. Other studies of the HPV18 regulatory region, by in vivo dimethyl sulfate analysis of the viral endogenous genome in HeLa cells, have also shown that the enhancer is densely occupied by proteins (10). In fact, our functional data indicated that this saturating occupancy of factors might correspond both to binding of transcription factors and to structural features that is in agreement with our working model. Moreover, preliminary footprinting experiments indicated that a purified HMG-I(Y) protein could bind to several regions along the enhancer sequences. Architectural proteins have been shown to play an essential role in the formation of the three-dimensional structure of enhanceosomes. Indeed, architectural factors, either from the HMG family, or not (LEF-1 for example), produce topological changes in the DNA (21, 52, 56) and even looped structures, as shown for the β-globin promoter (6). In addition, HMG-I(Y) has been shown to increase binding affinities of several transcription factors (28, 31, 23, 40, 56), and we can therefore speculate that it plays such a role in the HPV18 enhanceosome, although more work is needed to determine the other factors involved.
The functional role of the HMG-I(Y) binding site adjacent to AP1 was investigated by directed mutagenesis. We observed that this sequence is absolutely required for transcriptional activity of the enhancer. We showed that indeed, the presence of the architectural factor strongly increased AP1 binding to the enhancer in vitro. However, this effect seems to require either posttranslational modifications of HMG-I(Y), or intervening of an additional factor, since it was only detected with the proteins from HeLa nuclear extract. Acetylation of HMG-I(Y) by P/CAF and CBP have been shown to play a critical role in the formation of the IFN-β enhanceosome. In a recent work, acetylation of HMG-I(Y) lysine-71 by P/CAF was shown to potentiate stabilization of the IFN-β enhanceosome, while in contrast, acetylation of HMG-I(Y) on lysine-65 by the CBP coactivator, which is recruited later by the enhanceosome, rather led to disruption of the three-dimensional structure (37). Such a transcriptional switch is certainly not required for the HPV18 enhancer, whose activity is constitutive in HeLa cells. However, we showed here that it recruits the CBP/p300 coactivator that is essential for its transcriptional activity in vitro and in vivo. Acetylation of HMG-I(Y) by this coactivator may therefore play a role in the HPV18 enhancer constitution as well as acetylation by P/CAF. More work is needed to answer these questions.
Besides its involvement in regulating gene transcription, HMG-I(Y) has frequently been associated with both neoplastic transformation of cells and metastatic tumor progression (54), notably in cervical cancers (8). HMG-I(Y), particularly the HMG-Y isoform, is now considered to be a proto-oncogene, being able, when overexpressed, to induce both primary and metastatic tumors (42, 54). Comparative array analysis of transcription profiles of cells expressing HMG-I(Y) demonstrated modulation of a wide range of genes involved in signal transduction, cell proliferation, tumor initiation, invasion, migration, induction of angiogenesis, and colonization, these genes being often themselves involved in the transformation and/or tumor progression processes (42). Strikingly, HMG-I(Y) induced tumors have undergone an epithelial-mesenchymal transition, due to an upregulation of specific genes (type I, III, and IV collagens and vimentin, etc.). In our system, HMG-I(Y) is strongly involved in the HPV18 early promoter directing transcription of the E6 and E7 oncogenes. Increased expression of HMG-I(Y) could therefore represent a crucial step in the conversion of HPV associated lesions to cervical carcinomas.
HMG-I(Y) is required for the formation of enhanceosomes structures to form a novel activating surface that optimally recruits the transcriptional machinery (1, 50, 56). A major putative target of this surface is the CBP-Pol II holoenzyme, whose recruitment is essential for assembly of the transcriptional preinitiation complex. In this paper, we show that the CBP/p300 coactivator was recruited by the HPV18 enhanceosome in transient transfection and in vivo. We show that transfection of adenovirus E1A inhibits the endogenous E6 transcription by sequestering the CBP/p300 coactivator, and thereby induces stabilization of p53. We previously described that the outcome of the viral E2 protein overexpression in HeLa cells was identical, leading to p53 stabilization, although E2 represses the endogenous HPV18 E6 promoter through a completely different mechanism (16). Therefore, these experiments indicate that CBP/p300 positively controls the HPV18 integrated sequences as well as transfected sequences. Immunoprecipitation analysis of the HPV18 chromatin from HeLa cells gives an additional strong indication of the ability of the endogenous viral sequences to recruit JunB and p300 specifically.
These results reinforced the idea that the HPV18 transcription is highly synergistic, being stabilized by its interaction with the CBP/p300 coactivator. Moreover, we found that while overexpression of the JunB/Fra-2 heterodimer in HeLa cells could efficiently activate the collagenase promoter, it drastically reduced the activity of the HPV18 enhancer. An interpretation of these results is that excess AP1 could titrate limiting amounts of other crucial factors. The CBP coactivator appears limiting, since it activates the HPV18 transcription when overexpressed in cotransfection experiments. In addition, its overexpression could partially relieve inhibition by excess of AP1 or of the adenovirus E1A protein, indicating that it may directly interact with both proteins and could therefore be one of the limiting factors involved in the formation of the HPV18 enhanceosome.
This study also provides new insights into our understanding of enhanceosomes. While the HPV18 enhanceosome shares many characteristics with structures previously described, it exhibits several novel features; it is considerably longer (230 bp), compared with the 57 or 75 bp of IFN-β and T-cell receptor α enhancers, and exhibits a new type of association between AP1 and the HMG-I(Y) architectural factor. In addition, it is a strong constitutive cell-specific enhancer with a keratinocyte-specific AP1 heterodimer, JunB/Fra-2, that may constitute a model for keratinocyte-specific enhanceosomes.
Acknowledgments
We thank Pascale Debey and Martine Chebrout for the kind gift of the HMG-I(Y) purified protein; Yang Shi for the E1A expression plasmids; Annick Harel-Bellan for the CBP expression plasmids, Latifa Bakiri for the AP1 expression vectors, and Dominique Lallemand for the JunB antibodies. We are very grateful to Caroline Demeret and Jonathan Weitzman for critical reading of the manuscript.
This work was supported by the Association pour la Recherche contre le Cancer (ARC). I.B. was the recipient of a fellowship from the Ligue Nationale contre le cancer.
REFERENCES
- 1.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 2.Ait-Si-Ali, S., A. Polesskaya, S. Filleur, R. Ferreira, A. Duquet, P. Robin, A. Vervish, D. Trouche, F. Cabon, and A. Harel-Bellan. 2000. CBP/p300 histone acetyl-transferase activity is important for the G1/S transition. Oncogene 19:2430-2437. [DOI] [PubMed] [Google Scholar]
- 3.Angel, P., I. Baumann, B. Stein, H. Delius, H. J. Rahmsdorf, and P. Herrlich. 1987. 12-O-Tetradecanoyl-phorbol-13-acetate induction of the human collagenase gene is mediated by an inducible enhancer element located in the 5′-flanking region. Mol. Cell. Biol. 7:2256-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arends, M. J., Y. K. Donaldson, E. Duvall, A. H. Wyllie, and C. C. Bird. 1993. Human papillomavirus type 18 associates with more advanced cervical neoplasia than human papillomavirus type 16. Hum. Pathol. 24:432-437. [DOI] [PubMed] [Google Scholar]
- 5.Arias, J., A. S. Alberts, P. Brindle, F. X. Claret, T. Smeal, M. Karin, J. Feramisco, and M. Montminy. 1994. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature 370:226-229. [DOI] [PubMed] [Google Scholar]
- 6.Bagga, R., S. Michalowski, R. Sabnis, J. D. Griffith, and B. M. Emerson. 2000. HMG I/Y regulates long-range enhancer-dependent transcription on DNA and chromatin by changes in DNA topology. Nucleic Acids Res. 28:2541-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bakiri, L., K. Matsuo, M. Wisniewska, E. F. Wagner, and M. Yaniv. 2002. Promoter specificity and biological activity of tethered AP-1 dimers. Mol. Cell. Biol. 22:4952-4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bandiera, A., D. Bonifacio, G. Manfioletti, F. Mantovani, A. Rustighi, F. Zanconati, A. Fusco, L. Di Bonito, and V. Giancotti. 1998. Expression of HMGI(Y) proteins in squamous intraepithelial and invasive lesions of the uterine cervix. Cancer Res. 58:426-431. [PubMed] [Google Scholar]
- 9.Bannister, A. J., and T. Kouzarides. 1996. The CBP co-activator is a histone acetyltransferase. Nature 384:641-643. [DOI] [PubMed] [Google Scholar]
- 10.Bednarek, P. H., B. J. Lee, S. Gandhi, E. Lee, and B. Phillips. 1998. Novel binding sites for regulatory factors in the human papillomavirus type 18 enhancer and promoter identified by in vivo footprinting. J. Virol. 72:708-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouallaga, I., S. Massicard, M. Yaniv, and F. Thierry. 2000. An enhanceosome containing the JunB/Fra-2 heterodimer and the HMG-I(Y) architectural protein controls HPV18 transcription. EMBO Rep. 1:422-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butz, K., and F. Hoppe-Seyler. 1993. Transcriptional control of human papillomavirus (HPV) oncogene expression: composition of the HPV type 18 upstream regulatory region. J. Virol. 67:6476-6486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carey, M. 1998. The enhanceosome and transcriptional synergy. Cell 92:5-8. [DOI] [PubMed] [Google Scholar]
- 14.Chau, K. Y., N. Munshi, A. Keane-Myers, K. W. Cheung-Chau, A. K. Tai, G. Manfioletti, C. K. Dorey, D. Thanos, D. J. Zack, and S. J. Ono. 2000. The architectural transcription factor high mobility group I(Y) participates in photoreceptor-specific gene expression. J. Neurosci. 20:7317-7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, H., R. J. Lin, R. L. Schiltz, D. Chakravarti, A. Nash, L. Nagy, M. L. Privalsky, Y. Nakatani, and R. M. Evans. 1997. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell 90:569-580. [DOI] [PubMed] [Google Scholar]
- 16.Desaintes, C., C. Demeret, S. Goyat, M. Yaniv, and F. Thierry. 1997. Expression of the papillomavirus E2 protein in HeLa cells leads to apoptosis. EMBO J. 16:504-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foster, L. C., P. Wiesel, G. S. Huggins, R. Panares, M. T. Chin, A. Pellacani, and M. A. Perrella. 2000. Role of activating protein-1 and high mobility group-I(Y) protein in the induction of CD44 gene expression by interleukin-1beta in vascular smooth muscle cells. FASEB J. 14:368-378. [DOI] [PubMed] [Google Scholar]
- 19.French, S. W., M. C. Schmidt, and J. C. Glorioso. 1996. Involvement of a high-mobility-group protein in the transcriptional activity of herpes simplex virus latency-active promoter 2. Mol. Cell. Biol. 16:5393-5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Carranca, A., F. Thierry, and M. Yaniv. 1988. Interplay of viral and cellular proteins along the long control region of human papillomavirus type 18. J. Virol. 62:4321-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giese, K., C. Kingsley, J. R. Kirshner, and R. Grosschedl. 1995. Assembly and function of a TCR alpha enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 9:995-1008. [DOI] [PubMed] [Google Scholar]
- 22.Goodman, R. H., and S. Smolik. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14:1553-1577. [PubMed] [Google Scholar]
- 23.Henderson, A., M. Bunce, N. Siddon, R. Reeves, and D. J. Tremethick. 2000. High-mobility-group protein I can modulate binding of transcription factors to the U5 region of the human immunodeficiency virus type 1 proviral promoter. J. Virol. 74:10523-10534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janknecht, R., and T. Hunter. 1996. Transcription. A growing coactivator network. Nature 383:22-23. [DOI] [PubMed] [Google Scholar]
- 25.John, S., R. B. Reeves, J. X. Lin, R. Child, J. M. Leiden, C. B. Thompson, and W. J. Leonard. 1995. Regulation of cell-type-specific interleukin-2 receptor alpha-chain gene expression: potential role of physical interactions between Elf-1, HMG-I(Y), and NF-κB family proteins. Mol. Cell. Biol. 15:1786-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kannabiran, C., G. F. Morris, C. Labrie, and M. B. Mathews. 1993. The adenovirus E1A 12S product displays functional redundancy in activating the human proliferating cell nuclear antigen promoter. J. Virol. 67:507-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, T. K., T. H. Kim, and T. Maniatis. 1998. Efficient recruitment of TFIIB and CBP-RNA polymerase II holoenzyme by an interferon-beta enhanceosome in vitro. Proc. Natl. Acad. Sci. USA 95:12191-12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klein-Hessling, S., G. Schneider, A. Heinfling, S. Chuvpilo, and E. Serfling. 1996. HMG I(Y) interferes with the DNA binding of NF-AT factors and the induction of the interleukin 4 promoter in T cells. Proc. Natl. Acad. Sci. USA 93:15311-15316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwok, R. P., J. R. Lundblad, J. C. Chrivia, J. P. Richards, H. P. Bachinger, R. G. Brennan, S. G. Roberts, M. R. Green, and R. H. Goodman. 1994. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature 370:223-226. [DOI] [PubMed] [Google Scholar]
- 30.Lee, J. S., R. H. See, T. Deng, and Y. Shi. 1996. Adenovirus E1A downregulates cJun- and JunB-mediated transcription by targeting their coactivator p300. Mol. Cell. Biol. 16:4312-4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leger, H., E. Sock, K. Renner, F. Grummt, and M. Wegner. 1995. Functional interaction between the POU domain protein Tst-1/Oct-6 and the high-mobility-group protein HMG-I/Y. Mol. Cell. Biol. 15:3738-3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lundblad, J. R., R. P. Kwok, M. E. Laurance, M. L. Harter, and R. H. Goodman. 1995. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature 374:85-88. [DOI] [PubMed] [Google Scholar]
- 33.Mack, D. H., and L. A. Laimins. 1991. A keratinocyte-specific transcription factor, KRF-1, interacts with AP1 to activate expression of human papillomavirus type 18 in squamous epithelial cells. Proc. Natl. Acad. Sci. USA 88:9102-9106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maher, J. F., and D. Nathans. 1996. Multivalent DNA-binding properties of the HMG-1 proteins. Proc. Natl. Acad. Sci. USA 93:6716-6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masternak, K., A. Muhlethaler-Mottet, J. Villard, M. Zufferey, V. Steimle, and W. Reith. 2000. CIITA is a transcriptional coactivator that is recruited to MHC class II promoters by multiple synergistic interactions with an enhanceosome complex. Genes Dev. 14:1156-1166. [PMC free article] [PubMed] [Google Scholar]
- 36.Merika, M., A. J. Williams, G. Chen, T. Collins, and D. Thanos. 1998. Recruitment of CBP/p300 by the IFN beta enhanceosome is required for synergistic activation of transcription. Mol. Cell 1:277-287. [DOI] [PubMed] [Google Scholar]
- 37.Munshi, N., T. Agalioti, S. Lomvardas, M. Merika, G. Chen, and D. Thanos. 2001. Coordination of a transcriptional switch by HMGI(Y) acetylation. Science 293:1133-1136. [DOI] [PubMed] [Google Scholar]
- 38.O'Connor, M., S. Chan, and H. U. Bernard. 1995. Transcription factor binding sites in the long control region of genital HPVs, p. 21-40. In G. Myers, H. U. Bernard, H. Delius, K. Baker, J. Icenogle, A. Halpern, and C. Wheeler (ed.), Human papillomaviruses, 1995 compendium, part III. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 39.Ogryzko, V. V., R. L. Schiltz, V. Russanova, B. H. Howard, and Y. Nakatani. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87:953-959. [DOI] [PubMed] [Google Scholar]
- 40.Panagiotidis, C. A., and S. J. Silverstein. 1999. The host-cell architectural protein HMG I(Y) modulates binding of herpes simplex virus type 1 ICP4 to its cognate promoter. Virology 256:64-74. [DOI] [PubMed] [Google Scholar]
- 41.Quandt, K., K. Frech, H. Karas, E. Wingender, and T. Werner. 1995. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23:4878-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reeves, R., D. D. Edberg, and Y. Li. 2001. Architectural transcription factor HMGI(Y) promotes tumor progression and mesenchymal transition of human epithelial cells. Mol. Cell. Biol. 21:575-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reeves, R., and M. S. Nissen. 1990. The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J. Biol. Chem. 265:8573-8582. [PubMed] [Google Scholar]
- 44.Romanczuk, H., L. L. Villa, R. Schlegel, and P. M. Howley. 1991. The viral transcriptional regulatory region upstream of the E6 and E7 genes is a major determinant of the differential immortalization activities of human papillomavirus types 16 and 18. J. Virol. 65:2739-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129-1136. [DOI] [PubMed] [Google Scholar]
- 46.Schwarz, E., U. K. Freese, L. Gissmann, W. Mayer, B. Roggenbuck, A. Stremlau, and H. zur Hausen. 1985. Structure and transcription of human papillomavirus sequences in cervical carcinoma cells. Nature 314:111-114. [DOI] [PubMed] [Google Scholar]
- 47.Shi, Y., E. Seto, L. S. Chang, and T. Shenk. 1991. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell 67:377-388. [DOI] [PubMed] [Google Scholar]
- 48.Spencer, T. E., G. Jenster, M. M. Burcin, C. D. Allis, J. Zhou, C. A. Mizzen, N. J. McKenna, S. A. Onate, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 1997. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature 389:194-198. [DOI] [PubMed] [Google Scholar]
- 49.Stein, R. W., M. Corrigan, P. Yaciuk, J. Whelan, and E. Moran. 1990. Analysis of E1A-mediated growth regulation functions: binding of the 300-kilodalton cellular product correlates with E1A enhancer repression function and DNA synthesis-inducing activity. J. Virol. 64:4421-4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thanos, D., and T. Maniatis. 1992. The high mobility group protein HMG I(Y) is required for NF-kappa B-dependent virus induction of the human IFN-beta gene. Cell 71:777-789. [DOI] [PubMed] [Google Scholar]
- 51.Thierry, F., G. Spyrou, M. Yaniv, and P. M. Howley. 1992. Two AP1 sites binding JunB are essential for HPV18 transcription in keratinocytes. J. Virol. 66:3740-3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Werner, M. H., and S. K. Burley. 1997. Architectural transcription factors: proteins that remodel DNA. Cell 88:733-736. [DOI] [PubMed] [Google Scholar]
- 53.Whitley, M. Z., D. Thanos, M. A. Read, T. Maniatis, and T. Collins. 1994. A striking similarity in the organization of the E-selectin and beta interferon gene promoters. Mol. Cell. Biol. 14:6464-6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wood, L. J., J. F. Maher, T. E. Bunton, and L. M. Resar. 2000. The oncogenic properties of the HMG-I gene family. Cancer Res. 60:4256-4261. [PubMed] [Google Scholar]
- 55.Yang, X. J., V. V. Ogryzko, J. Nishikawa, B. H. Howard, and Y. Nakatani. 1996. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382:319-324. [DOI] [PubMed] [Google Scholar]
- 56.Yie, J., M. Merika, N. Munshi, G. Chen, and D. Thanos. 1999. The role of HMG I(Y) in the assembly and function of the IFN-beta enhanceosome. EMBO J. 18:3074-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]