Abstract
Although oncogenic Ras commonly contributes to the development of cancer, in normal primary cells it induces cell cycle arrest rather than transformation. Here we analyze the additional genetic changes required for Ras to promote cell cycle progression rather than arrest. We show that loss of p53 is sufficient for oncogenic Ras to stimulate proliferation in the absence of extrinsic mitogens in attached cells. However, surprisingly, we find that p53 loss is not sufficient for Ras to overcome anchorage dependence or contact inhibition. In contrast, expression of simian virus 40 (SV40) large T antigen (LT) allows Ras to overcome these additional cell cycle controls. Mutational analysis of SV40 LT shows that this action of SV40 LT depends on its ability to inactivate the retinoblastoma (Rb) family of proteins, in concert with the loss of p53. Importantly, we show that inactivation of the Rb family of proteins can be mimicked by loss of the cyclin-dependent kinase inhibitor p16INK4A. p16INK4A is commonly lost in human tumors, but its contribution to the transformed phenotype is unknown. We demonstrate here a role for p16INK4A in the loss of cell cycle controls required for tumorigenesis and show how accumulating genetic changes cooperate and contribute to the transformed phenotype.
The transformation of a normal cell to a cancer cell is associated with a disruption of normal cellular controls that tightly regulate when and where a cell may divide and survive. Many of the genetic changes responsible for the generation of a cancer cell have now been identified. What remains less clear is how each of these changes contributes to the phenotype of the tumor. The Ras oncogene is mutated in a high proportion of many human tumor types. In vitro, Ras-transformed cells have lost many normal checkpoint controls. These include loss of contact inhibition and loss of the requirements for mitogens and anchorage in order to proliferate. Each of these properties could be envisaged to contribute to the loss of proliferative controls seen in Ras-induced tumors.
Normal cells appear to have intrinsic mechanisms that protect against oncogenic activation. In the case of the Ras oncogene, it appears that most normal human and rodent cells have checkpoints that render oncogenic Ras signals inhibitory to the cell cycle. Signals through the mitogen-activated protein kinase pathway via Raf result in a G1 cell cycle arrest as a consequence of the induction of cyclin-dependent kinase inhibitors (CDKIs) (38, 39, 57, 73). In fibroblasts this proliferative arrest is associated with a senescent phenotype (57). For Ras to signal positively to the cell cycle, further genetic changes are required that act to abrogate the inhibitory signal to the cell cycle and act synergistically with Ras to promote proliferation.
p53 and INK4A play important roles in mediating the signals that constrain the cell cycle in response to hyperproliferative signals from Ras and, furthermore, are the most frequently inactivated tumor suppressor genes in human cancer (27, 28). In rodent fibroblasts, Schwann cells, and keratinocytes Ras-induced G1 arrest is dependent on p53, and cells lacking functional p53 no longer undergo a cell cycle arrest in response to Ras (39, 52, 57). The INK4A locus encodes two unrelated proteins: p19ARF and p16INK4A. p16INK4A is a specific inhibitor of the cyclin D-dependent kinases CDK4 and CDK6 (56) and antagonizes their ability to phosphorylate the retinoblastoma (Rb) family of proteins and so prevent exit from the G1 phase of the cell cycle (59). p19ARF antagonizes the function of the p53 negative regulator MDM2, thereby stabilizing p53 (33, 61, 72). In addition, p19ARF has functions that are independent of p53 (9, 67). Both 19ARF and p16INK4A are induced in response to oncogenic Ras (57; I. Palmero, C. Pantoja, and M. Serrano, Letter, Nature 395:125-126, 1998). p19ARF is required to mediate the induction of p53 and the resultant cell cycle arrest. p16INK4A−/− cells still arrest in response to Ras, making the role of p16INK4A in cellular transformation less clear (35, 58).
In the present study we used primary rat Schwann cells to identify additional genetic changes that enable Ras to signal positively to the cell cycle and attribute a role to each change in conferring the various properties characteristic of Ras transformed cells. We find that loss of the inhibitory signal from Ras by inactivation of p53, while permitting mitogen-independent proliferation, is insufficient for Ras to confer loss of contact inhibition and anchorage-independent growth. This requires, in addition, inactivation of the Rb family of proteins, a function conferred by loss of the tumor suppressor p16INK4A.
MATERIALS AND METHODS
Cell culture and BrdU incorporation.
Schwann cells were purified from the sciatic nerves of 7-day-old rats as described previously (11) and were maintained in culture as described elsewhere (39). Cells were routinely cultured in Dulbecco modified Eagle medium (DMEM) with 1.5 mg of glucose/ml, supplemented with 3% fetal calf serum, 1 μM Forskolin (Calbiochem), and glial growth factor (20 ng/ml; R&D Systems) on poly-l-lysine (Sigma)-coated plasticware. For culture in defined medium, cells were first seeded in DMEM containing 0.2% serum. After 12 h, the cells were washed twice with defined medium, which consisted of DMEM (plus 1.5 mg of glucose/ml) with B/S supplement (6), and then cultured in B/S-supplemented medium for 48 h. To determine the proportion of cells entering S phase, bromodeoxyuridine (BrdU; 10 μM) was added to the culture medium 12 h prior to fixation, and incorporated BrdU was detected by immunostaining with an anti-BrdU antibody (Boehringer Mannheim) and Cy3-labeled secondary antibody (Jackson Laboratories). Labeled nuclei were visualized by fluorescence microscopy, and an average of 300 nuclei were counted per assay point.
Retroviral vectors and retroviral transduction.
The Val12 mutant H-Ras cDNA in the retrovirus LXSN was a kind gift of Pablo Rodriguez-Viciana. The simian virus 40 (SV40) large T antigen (LT) cDNA (a kind gift from James DeCaprio) and the human p53175 cDNA were subcloned into pBabePuro. The N-terminal SV40 LT cDNAs TN136 WT, J domain mutant TN136 5110, and Rb-binding domain mutant TN136 K1 (as described elsewhere [60]) were a generous gift from Jim Pipas) were subcloned into pBabeHygro. The vector pWZL Hygro was used to express the cdk4R24C mutant. The primers 5′-CCGCCATCGATGGCATGGAGTCCTCTGCAGATAGACT-3′ and 5′-AACCCAAGCTTGGGCCTGTATCGGGGTACGACCGAAA-3′ were used to amplify rat p16INK4A, which was subcloned into the HindIII/ClaI sites of the retroviral vector pLHCX (Clontech) that utilizes a cytomegalovirus promoter. High-titer retroviral stocks were produced by transient transfection of Phoenix cells. To achieve 100% infectivity in order to avoid selection pressures, subconfluent cells were infected three times in succession in a 36-h period. Cells were split 1:3 into the relevant drug selection. In order to avoid the accumulation of any additional genetic changes, all experiments were performed on cells in fewer than four passages after drug selection.
Focus and soft agar assays.
For focus assays, 5 × 105 Schwann cells per 9-cm culture plate were incubated with diluted RasLXSN or LXSN viral supernatants. At 48 h after infection the cells were treated with trypsin and replated onto two 9-cm culture plates. Cells on one plate were allowed to reach confluence, whereas cells on the duplicate plate were cultured in the presence of 0.4 mg of G418/ml to select for infected cells. Foci and drug-resistant colonies of cells were stained with methylene blue 2 to 3 weeks later. For soft agar assays, 4 × 103 or 8 × 103 cells were suspended in 4 ml of normal culture medium containing 0.5% (wt/vol) low-melting-point agarose (FMC) at 37°C and then plated onto 6-cm dishes coated with normal culture medium containing 0.9% (wt/vol) low-melting-point agarose. Cultures were fed twice weekly, and colonies of cells were stained by the addition of 1 ml of neutral red solution (0.015% [wt/vol] in phosphate-buffered saline [Sigma]).
Western blots.
Cells were lysed in buffer A (1% NP-40, 50 mM Tris-HCl [pH 8], 150 mM NaCl, 20 mM NaF, 1 mM Na3VO4, 1× Complete protease inhibitor cocktail [Boehringer Mannheim]). The concentration of the lysates was determined by using the Bio-Rad protein assay reagents. Protein lysates (30 μg) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto Immobilon-P membrane (Millipore). The following antibodies were used: p16INK4A (Ab7; Neomarkers) and tubulin (Sigma). The p19ARF antiserum was raised against the peptide NH2-RRGPQPHPGPGDDDGQRQSGSSPC-COOH of the rat amino acid sequence by AbCam. The antisera were affinity purified by using the peptide coupled to CH Sepharose. Cyclin D1 (no. 450; Santa Cruz), cdk4 (no. 260; Santa Cruz), cdk6 (Ab3; Neomarkers), cdc25A (no. 7389; Santa Cruz), myc (Upstate Biotech), and Ras (Y13-259 [a gift from Alan Hall]) were obtained as indicated. Immunoreactive proteins were visualized by using the chemiluminescence detection reagent ECL Plus (Amersham Pharmacia)
Semiquantitative reverse transcription-PCR (RT-PCR).
RNA was isolated from cells by the method of Chomczynski and Sacchi (13), and cDNA was synthesized by using Superscript preamplification reagents (Life Technologies). A 350-bp product spanning exons 1a and 2 of the rat INK4A locus was amplified with the following primers: sense, CTCCGAGAGGAAGGCGAACTCG; and antisense, GGTGCAGTACTACCAGAGTG. A 350-bp product corresponding to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was amplified with the sense primer AAAAGGGTCATCATCTCCGC and the antisense primer GGATGCAGGGATGATGTTCT. Amplifications were carried out in the presence of [α-32P]dCTP, products were resolved by polyacrylamide gel electrophoresis, visualized, and quantified by using a Bio-Rad molecular imager. The number of cycles were limited so that the amplification was linear, with between 25 and 27 cycles for p16INK4a and between 16 and 18 cycles for GAPDH.
Spin-down immunofluorescence.
Colonies were sucked out of the agar by using a pipette into ice-cold phosphate-buffered saline. Cells were immediately spun at 4°C onto poly-d-lysine-coated coverslips by using a cyclospin centrifuge. The cells were immediately fixed in 4% paraformaldehyde and then stained for Rb phosphorylation with the Rb (Ser-795) antibody (Cell Signaling Technology) according to the manufacturer's instructions, with the addition of an amplification step using the TSA fluorescence systems kit (Molecular Probes).
Schwann cell transfections.
A total of 5 × 104 cells in six-well dishes were transfected by using the Fugene reagent according to the manufacturer's instructions. Cells were transfected with 0.5 μg of the green fluorescent protein (GFP)-expressing vector pBirdCME (65) and 0.1 μg of pcDNA3 Rb(NPC−) (12) or control vector. At 30 h after transfection, the cells were labeled for 14 h with BrdU and then fixed and immunostained.
RESULTS
Loss of p53 function is sufficient for Ras-induced proliferation in the absence of mitogens.
Primary rat Schwann cells are an ideal cell type to study the contribution of cooperating oncogenes to the transformed phenotype since they can be expanded indefinitely in culture while maintaining cell cycle checkpoints normally lost during the immortalization process (43). Oncogenic Ras expression in normal Schwann cells results in a cell cycle arrest that we have previously shown is dependent on functional p53 (39). We wanted to establish whether the loss of p53 was sufficient for Ras to stimulate proliferation independently of extrinsic factors or whether further genetic changes were required. To do this, we studied the proliferation of normal Schwann cells expressing oncogenic Ras and a dominant-negative mutant of p53 (dnp53) or SV40 LT (see Materials and Methods for details of the constructs). LT, a multifunctional viral protein, has previously been shown to cooperate with Ras in transforming primary Schwann cells (49). Early-passage rat Schwann cells were infected with a high-titer retroviral vector, pBabePuro, expressing either dnp53 or wild-type SV40 LT. Large pools of early-passage, puromycin-resistant Schwann cells expressing either a dnp53 (NSdnp53) or wild-type SV40 LT (NSLT) were then infected with an oncogenic H-RasV12-expressing retroviral vector, Ras LXSN. G418-resistant cells expressing Ras, together with dnp53 or SV40 LT (NSdnp53Ras and NSLTRas, respectively) or the empty vector LXSN, together with mutant p53 or SV40 LT (NSdnp53LXSN and NSLTLXSN, respectively), were expanded in culture. In all of the infections described, the infectivity rate was close to 100%, so that each infection was equivalent to only a single passage of the cells. Minimal expansion of selected cells was used in order to minimize the possibility of the selection of additional genetic changes. Western blotting showed that the NSdn53Ras and NSLTRas cells expressed similar levels of Ras protein (data not shown). Both populations of Ras-expressing cells showed a dramatic change in morphology similar to that observed in Schwann cells when an estrogen-inducible Raf is activated (39) or when injected with Ras protein (49) (Fig. 1A).
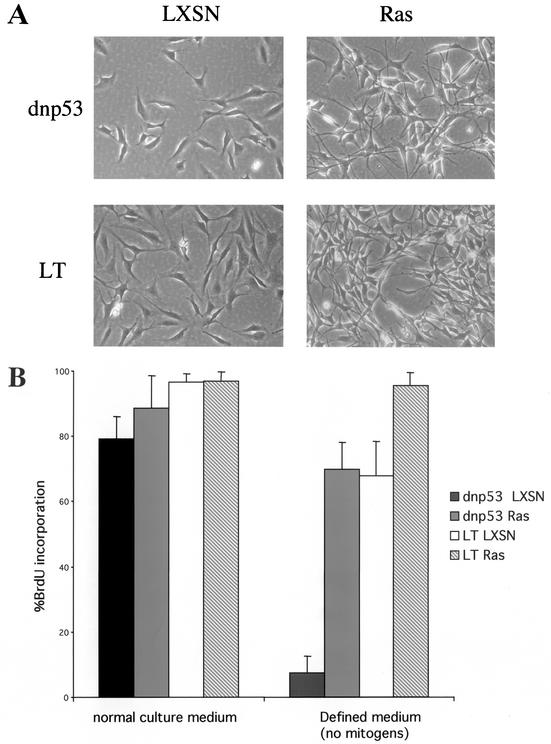
FIG. 1.
Loss of p53 function is sufficient for Ras induced proliferation in the absence of mitogens. (A) Phase-contrast images of pools of Schwann cells expressing either a dominant-negative mutant of p53 (dnp53) or the SV40 LT, together with HRasVal12 (Ras) or the empty retroviral vector LXSN. (B) Asynchronous populations of Schwann cells were cultured in normal culture medium or defined medium containing no mitogen for 40 h. The percentage of cells entering S phase was determined by measuring BrdU incorporation 12 h prior to fixing and staining. The error bars indicate the standard deviations of triplicate assays.
We initially assessed the ability of Ras in the different genetic backgrounds to confer independence from extrinsic mitogens. These experiments can be performed in the complete absence of mitogen since Schwann cells survive well in a defined medium. We found that cells expressing Ras and dnp53 were able to enter S phase efficiently in the absence of mitogen (Fig. 1B). Cells expressing dnp53 alone required mitogens for proliferation, thus demonstrating that Ras is required to confer mitogen independence. These results show that only one further genetic change, mutation of p53, is required for Ras to stimulate proliferation in the absence of mitogen.
Loss of p53 is not sufficient for Ras to overcome contact inhibition or for anchorage-independent proliferation.
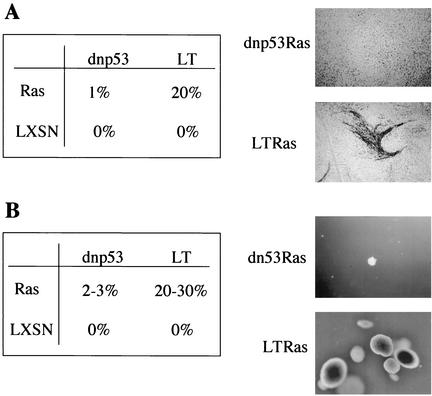
Ras was first identified as an oncogene by virtue of its ability to overcome cell-cell contact inhibition of proliferation to form transformed foci of cells within a confluent monolayer of immortal fibroblasts. To establish whether loss of p53 function is sufficient for Ras expressing cells to become insensitive to contact inhibition, Ras was expressed at low frequency within monolayers of NSdnp53 or NSLT cells. Interestingly, although Ras efficiently formed foci within monolayers of cells expressing LT (ca. 20% of infected cells), only a few foci were formed in the monolayers of the cells expressing dnp53 (<1%) (Fig. 2A). Thus, LT abrogates additional cell cycle controls to render them insensitive to contact inhibition. However, loss of these controls in the absence of Ras is not sufficient to overcome contact inhibition since we never observed any foci in either dnp53- or LT-expressing cells.
FIG. 2.
Loss of p53 is not sufficient for Ras to overcome contact inhibition or for anchorage-independent proliferation. (A) Percentage of Ras or LXSN-infected Schwann cells, expressing dnp53 or SV40 LT (LT), which proliferated to form foci in a confluent monolayer after 2 weeks, as visualized by staining with methylene blue. A representative focus of SV40 LT cells (LT) expressing Ras is shown. (B) Percentage of Schwann cells expressing dnp53 or LT, together with Ras or the empty vector LXSN, which proliferated to form colonies in soft agar. Pools of cells (4 × 103) were seeded into soft agar. After 2 weeks the colonies were photographed and counted. Representative colonies of cells expressing Ras, together with dnp53 or LT, are shown. Assays for panels A and B were carried out in duplicate and are representative of three independent experiments.
Most normal cells in culture require attachment to a substratum for proliferation. Cells that have acquired the ability to proliferate independently of anchorage in vitro usually have the ability to form tumors in vivo. We therefore examined the ability of NSdnp53Ras and NSLTRas cells to proliferate in the absence of anchorage. NSLTRas cells efficiently formed colonies in soft agar with 20 to 30% of the seeded cells proliferating to form smooth, regular-shaped colonies of more than a thousand cells in a period of 2 weeks (Fig. 2B). NSLT cells, in contrast, did not proliferate and remained as single refractive cells even after extended periods in suspension (over 3 weeks), demonstrating the strict requirement for a Ras signal for anchorage-independent proliferation. Interestingly, only a small proportion (2 to 3%) of NSdnp53Ras cells were capable of forming colonies, and these tended to be smaller and irregular in shape (Fig. 2B). Thus, as for the loss of contact inhibition, Ras requires signals provided by LT expression, in addition to inactivation of p53, in order for the cells to proliferate in the absence of anchorage.
Inactivation of both p53 and Rb family required for Ras-dependent, anchorage-independent proliferation.
SV40 LT binds many proteins that play important roles in controlling entry into and progression through the cell cycle. The N terminus of SV40 LT binds and inactivates members of the Rb family, Rb, p107, and p130 (46), whereas the large C-terminal region of LT sequesters p53 (41) and the transcriptional coactivator p300 (2, 21, 37). To define the domain of LT that cooperates with Ras, we used a retroviral construct (pBabeHygro) expressing just the N-terminal 136 amino acids of SV40 LT (N-LT), which is truncated immediately after the Rb-binding domain. The vector expressing the N terminus of LT was introduced into early-passage Schwann cells that did or did not express dnp53. These cells were subsequently infected with the Ras LXSN retroviral vector. Pools of G418-resistant cells expressing Ras or the empty vector were then seeded at low density in soft agar.
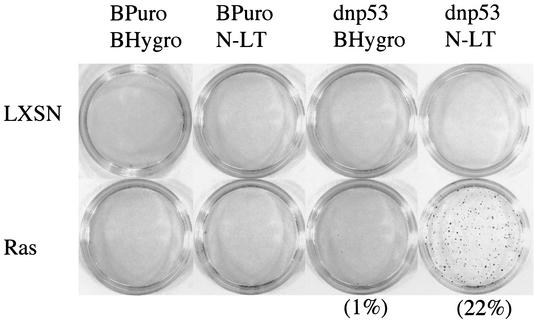
Cells expressing Ras, together with mutant p53 and the N terminus of LT, formed colonies in soft agar as efficiently as the RasLT cells (Fig. 3), demonstrating that the N terminus of LT contained a domain that was sufficient to confer anchorage-independent proliferation to these cells. Cells expressing Ras with just the N terminus of LT were unable to proliferate independently of anchorage and remained as single cells, thus demonstrating the requirement for both loss of p53 and expression of N-LT. These results show that the loss of anchorage independence requires additional genetic changes other than activation of Ras and loss of p53, and these are conferred by the N terminus of LT.
FIG. 3.
The N terminus of LT is responsible for abrogating the additional checkpoints that enables Ras to be proliferative in the absence of anchorage. Pools of drug selected Schwann cells (4 × 103), expressing dnp53 and the N terminus of LT together or on their own with Ras or the empty retroviral vector LXSN, were seeded in soft agar. After 2 weeks the colonies were stained with neutral red prior to being counted and photographed. Assays were carried out in duplicate, and a representative result of three independent experiments is shown.
Importance of inactivation of the Rb family for anchorage-independent proliferation.
Within the N-terminal 136 amino acids of SV40 LT are two domains that are important for inactivating the Rb family of tumor suppressor proteins. Between residues 103 to 107 is the LXCXE motif that binds the pocket containing proteins Rb and its two family members, p107 and p130 (18, 20, 22, 70). Nearer the N terminus is a region that is homologous to the J domain found in cellular DnaJ or Hsp40 molecular chaperone proteins. This domain has been shown to disrupt Rb family-E2F complexes (64, 71) and regulate the specific inactivation of p107 and p130 (17, 62, 63). We therefore wanted to determine whether these domains were required for anchorage-independent proliferation. Retroviral constructs expressing the wild-type form of the N terminus of LT and two mutants, one with a defective J domain and the other with a defective LXCXE motif, were introduced into early-passage NSdnp53 cells and then infected with the retroviral vector RasLXSN. Western blotting of lysates prepared from the N-LT-expressing cells demonstrated that all of the N-LT constructs were expressed at equivalent levels (data not shown). Pools of G418-resistant cells expressing Ras or the vector LXSN were then seeded at low density in soft agar.
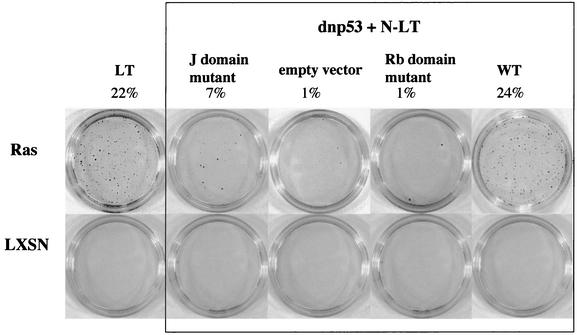
The results showed that cells that expressed N-LT with a defective LXCXE motif were unable to proliferate in soft agar, clearly demonstrating that inactivation of the Rb family was necessary for anchorage-independent proliferation (Fig. 4). Moreover, for the N terminus to confer anchorage independence efficiently, an intact J domain was required, indicating a specific role for p107 and p130. Therefore, efficient inactivation of the Rb family, rather than just Rb, appears to be critical, in addition to p53 loss and Ras activation, for cells to proliferate independently of anchorage. The finding that inactivation of the Rb family alone is insufficient to allow Ras-induced anchorage-independent proliferation is in agreement with the findings of Peeper et al. (47), who showed that the loss of Rb/p107 was sufficient to overcome the Ras-induced cell cycle arrest but was insufficient to promote anchorage-independent proliferation. In contrast to their report, however, we find that p53 loss alone is insufficient to promote efficient anchorage-independent proliferation but requires, in addition, inactivation of the Rb family.
FIG. 4.
Inactivation of both p53 and the Rb family are required for Ras-dependent, anchorage-independent proliferation. Pools of drug selected Schwann cells (4 × 103) expressing dnp53 with wild type (WT) or mutants of the N terminus of LT (N-LT), together with Ras or the empty vector LXSN, were seeded in agarose. After 2 weeks the colonies were stained with neutral red prior to being counted and photographed. Assays were carried out in duplicate, and a representative result of three independent assays is shown.
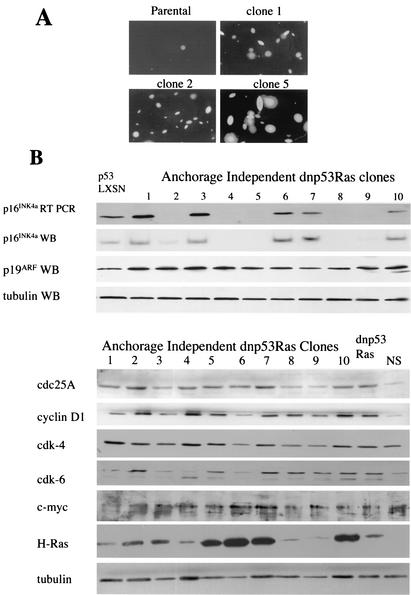
Spontaneous acquisition of anchorage-independent proliferation is associated with frequent loss of p16INK4A expression.
We were interested as to whether the small proportion of NSdnp53Ras cells that formed colonies in soft agar had acquired an additional genetic change that permitted anchorage-independent growth or whether these cells had some ability to proliferate in soft agar albeit at a much lower frequency. In order to test this, single colonies of NSdnp53Ras cells were picked from soft agar, dissociated, and expanded in culture. Cells from these clones were then seeded at low density in soft agar. Of the 10 clones examined, all 10 proliferated in soft agar with high efficiency (Fig. 5A). This indicated that the NSdnp53Ras cells had acquired an additional genetic change, reiterating the finding that Ras activation and loss of p53 are not sufficient for anchorage-independent proliferation.
FIG. 5.
Spontaneous acquisition of anchorage-independent proliferation is associated with frequent loss of p16INK4A expression. (A) Colonies of cells expressing dnp53 and Ras that proliferated in soft agar were picked, dissociated, and expanded in culture. A total of 4 × 103 cells were reseeded in agarose. Two weeks later the colonies were photographed. The parental polyclonal dnp53/Ras-expressing cells are shown adjacent to three clones. Assays were carried out in duplicate, and a representative of three independent assays is shown. (B) Immunoblots of lysates prepared from 10 dnp53/Ras-expressing clones that had acquired the ability to proliferate efficiently in soft agar. p16INK4A mRNA levels were determined by semiquantitative RT-PCR.
Analysis of the protein levels of CDKIs expressed by the 10 clones showed that 5 of the 10 clones had lost expression of p16INK4A (Fig. 5B). This is in contrast to clones of normal Schwann cells (43) or to 10 clones of NSdnp53LXSN cells, expanded as controls, all of which continue to express p16INK4A (data not shown). The loss of p16INK4A expression was mirrored by a reduction in mRNA levels, as determined by semiquantitative RT-PCR (Fig. 5B). This effect would appear to be specific, since we did not detect any loss of p19ARF expression, a protein that is encoded by the same locus (Fig. 5B). Loss of p16INK4A would be predicted to result in the activation of cyclin D-cdk complexes, resulting in the phosphorylation and subsequent inactivation of Rb, p107, and p130 (19, 25, 30). We therefore screened the p16INK4A-positive clones for changes in the expression of other proteins that act on this pathway and that have been shown to cooperate with Ras and a loss of p53 in animal models of tumor formation (3). We analyzed the expression of cyclin D1, cdc25A, cdk-4, cdk-6, and Myc. In addition, we analyzed the levels of Ras protein in the clones to address whether high levels of Ras expression were able to substitute for p16INK4A loss. Our results showed that, although the levels of cyclin D1 and cdc25A were higher in the dnp53Ras cells compared to the normal Schwann cells, the proteins were not expressed to higher levels in the p16INK4A-expressing clones (Fig. 5B). Likewise, we did not find overexpression of cdk-4, cdk-6, or Myc in any of the clones. Ras expression did vary considerably between the clones, but the expression levels did not correlate with the p16INK4A status. It therefore remains unclear what additional genetic change is responsible for anchorage independence in half of the clones.
Our LT mutational analysis showed that inactivation of the Rb family of proteins is required for anchorage-independent growth. This would imply that loss of p16INK4A and subsequent deregulation of cdk4/6 activity is sufficient to inactivate the Rb family constraint of anchorage-independent proliferation. This strongly indicates that loss of p16INK4A, together with loss of p53 function, is likely to be sufficient to overcome the additional checkpoint to enable Ras to be proliferative in the absence of anchorage.
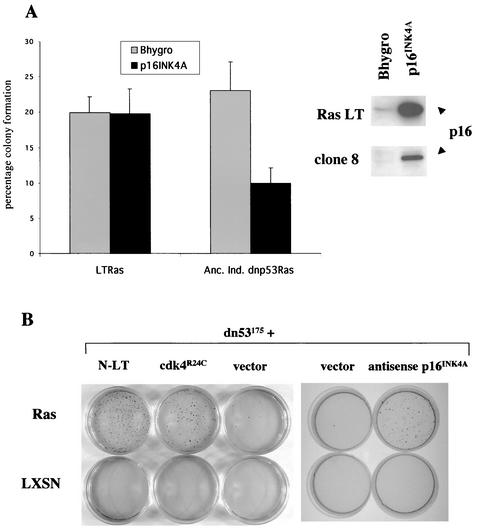
p16INK4A loss confers anchorage independence.
Loss of p16INK4A expression as the result of promoter methylation is a major mechanism of tumor suppressor gene silencing (50). We therefore examined whether promoter methylation was responsible for the loss of p16INK4A expression in the NSdnp53Ras clones. Treatment of the dnp53Ras clones that grew in soft agar with the demethylating agent 5-aza-deoxycytidine resulted in reexpression of p16INK4A in all cases (data not shown), demonstrating that promoter methylation was responsible for silencing p16INK4A expression in these cells. These cells would no longer proliferate in the absence of anchorage; however, this effect was nonspecific since 5-aza-deoxycytidine treatment of RasLT cells also inhibited their proliferation. To test whether reexpression of p16INK4A could inhibit proliferation of the NSdnp53Ras clones, we infected anchorage-independent NSdnp53Ras cells that had lost p16INK4a expression and NSLTRas cells with a retroviral vector expressing wild-type rat p16INK4A or with the control empty vector. Pools of hygromycin-resistant cells expressing wild-type p16INK4A or the empty retroviral vector were expanded in culture and subsequently seeded in soft agar. Reexpression of p16INK4A greatly reduced the ability of anchorage-independent dnp53Ras cells to proliferate in soft agar, whereas p16INK4A had no effect in NSLTRas cells despite the levels being higher (Fig. 6A). To verify that loss of p16INK4A expression is capable of cooperating with Ras and dnp53 to confer anchorage independence, we took two approaches: (i) expression of antisense p16INK4A by an approach similar to one described elsewhere (9) and (ii) expression of a tumor-derived mutant of cdk4 that is insensitive to inhibition by p16INK4A. The cdk4R24C mutant can no longer bind p16INK4A but can still associate and form active complexes with cyclin D (4, 68), thereby mimicking the loss of p16INK4A expression. Early-passage NSdnp53 cells were infected with a retroviral vector expressing the p16INK4A-insensitive cdk4 mutant (cdk4R24C) or a high-expressing retroviral vector expressing antisense p16INK4A (α/sp16) and the corresponding control vectors. Pools of hygromycin-resistant cells were subsequently infected with RasLXSN or LXSN. Pools of G418-resistant cells expressing Ras or the empty vector were then seeded in soft agar. A substantial proportion of the dnp53cdkR24C Ras and dnp53αsp16Ras cells proliferated to form colonies in soft agar compared to dnp53Ras-expressing cells (Fig. 6B). Thus, p16INK4A loss or deregulation of cdk4 activity cooperate with loss of p53 function to enable Ras to overcome the dependence on anchorage for proliferation.
FIG. 6.
p16INK4A loss is responsible for conferring anchorage independence. (A) Pools of LTRas and anchorage-independent dnp53Ras Schwann clones that no longer expressed endogenous p16INK4A, expressing either wild-type rat p16INK4A or the empty vector pWZLhygro, were seeded in soft agar. The graph is representative of two independent experiments showing the average percentage of cells seeded that proliferated to form colonies in soft agar. The error bars represent the standard deviations of triplicate assays. The levels of p16INK4A as determined by immunoblotting are shown in the inset panel. (B) Populations of drug-selected Schwann cells, expressing dnp53 with the p16INK4A-insensitive mutant of cdk4 (cdk4R24C), antisense p16INK4A, or the N terminus of LT, together with Ras or the empty vector LXSN, were seeded in agarose. After 2 weeks the colonies were stained with neutral red prior to being counted and photographed.
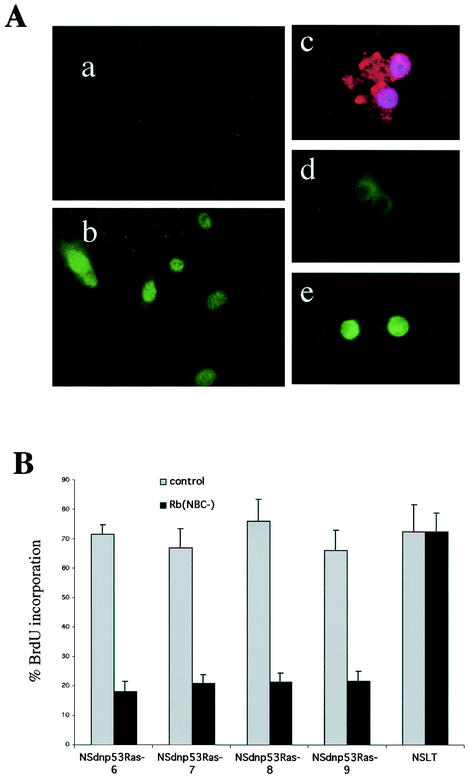
Our results predict that the dnp53Ras cells lacking p16INK4A expression should phosphorylate Rb in the absence of anchorage and should maintain a requirement for Rb inactivation in order to proliferate. To test for Rb phosphorylation in the absence of anchorage, we made use of a phospho-specific antibody that specifically recognizes Rb phosphorylated on residue Ser-795. To confirm the specificity of this antibody, we stained quiescent normal Schwann cells and Schwann cells 16 h after refeeding with mitogen (Fig. 7Aa and b). To analyze the Rb phosphorylation status of unattached cells, the cells were pipetted out of the agarose and spun onto coverslips, immediately fixed, and then immunostained. The parental pools of dnp53Ras cells that are unable to proliferate in soft agar were consistently negative for Rb staining. In contrast, we found that the majority of the dnp53Ras clones spun out of soft agar were positive for Rb staining. (An example of negative and positive staining is shown in Fig. 7Ad and e). This finding was irrespective of the p16INK4A status of the cells, suggesting that the clones that maintain p16INK4A expression also have the capacity to inactivate Rb in the absence of anchorage signals. To demonstrate the Rb dependence of these cells, we transfected into the clones a mutant of Rb that cannot be phosphorylated [Rb(NPC−) (12)]. Transfected cells were monitored by cotransfection with a GFP-expressing vector. We then analyzed the ability of these cells to enter S phase by monitoring BrdU incorporation. The dnp53Ras clones irrespective of their p16INK4A status were inhibited in their ability to enter S phase by the expression of the nonphosphorylatable form of Rb, thus demonstrating that Rb inactivation is necessary for these cells to proliferate. This was in contrast to Schwann cells expressing LT, which were insensitive to the effects of Rb(NPC−) expression (Fig. 7B).
FIG. 7.
Anchorage-independent Ras/dnp53 cells phosphorylate Rb in suspension and maintain Rb dependency for proliferation. (A) Antibody phospho-Rb (Ser-795) recognizes Rb in proliferating (b) but not in quiescent (a) rat Schwann cells. Cells from growing colonies in soft agar were removed and spun onto coverslips and immediately fixed. The cells were immunostained for either a cytoplasmic protein S100 (c) or phospho-Rb (d and e, which show examples of cells negative and positive for phospho-Rb staining, respectively). (B) BrdU incorporation of cells after transfection with a nonphosphorylatable form of Rb (12) or control vector. Four NSdnp53Ras clones are shown. Clones 6 and 7 are p16INK4A positive; clones 8 and 9 are p16INK4A negative. NSLT are Schwann cells expressing SV40 LT. Transfected cells were monitored by determining GFP expression. The error bars indicate the standard deviations of triplicate assays.
DISCUSSION
Many normal cell types respond to oncogenic Ras signaling with a cell cycle arrest (49, 52, 57). This inhibitory signal appears to be the result of strong constitutive activation of signaling pathways that normally mediate mitogenic signaling and as such has been proposed to act as a tumor suppressor mechanism. Certainly, normal cells are resistant in vitro to oncogenic Ras, requiring additional genetic changes provided by so-called cooperating genes, in order for Ras to be transforming. One property of a Ras cooperating gene is that it must provide a function able to block or overcome the inhibitory signal from Ras. Signals from oncogenic Ras are then able to act positively on the cell cycle.
Rat Schwann cells can be cultured extensively in culture, maintaining cell cycle checkpoints normally lost upon immortalization. These cells have a strict requirement for mitogens and anchorage to proliferate and maintain contact inhibition for long periods of time. Chromosomal analysis of late-passage cells suggests they are relatively genetically stable (43), and the rate of spontaneous transformation appears to be very low since the background rates in anchorage independence and focus formation assays are always zero. These cells thus offer an ideal cell system for analyzing how genetic changes can contribute to loss of normal cell cycle checkpoint controls.
We have previously shown in rat Schwann cells that the inhibitory signal from activated Ras requires functional p53 (39). Here we show that cells expressing Ras and dnp53 are able to proliferate in the absence of mitogen. Therefore, in cells with defective p53 signaling, oncogenic Ras is able to substitute for a mitogenic signal to the cell cycle. These cells, however, maintain some extrinsic regulation of the cell cycle since Ras/dnp53 cells are unable to proliferate in the absence of anchorage and are unable to form colonies within monolayers of contact-inhibited cells. This is in contrast to Ras/LT cells that proliferate with high efficiency in soft agar and within monolayers. Deletion analysis of LT showed that the amino terminus conferred anchorage independence but only when coexpressed with Ras and dnp53. This would indicate that additional genetic changes are required for anchorage independence compared to mitogen independence. This idea is reinforced by the finding that, although LT expression is sufficient for mitogen independence (Fig. 1B), additional signals from Ras are required for anchorage-free proliferation and to overcome contact inhibition.
Mutational analysis of LT has demonstrated an absolute requirement for the LXCXE domain for anchorage-independent proliferation. The LXCXE domain binds to a motif within the conserved pocket domain of the Rb family members, Rb, p107, and p130. Members of the Rb family bind and negatively regulate the E2F family of transcription factors that control the transcription of key cell cycle regulatory genes. LT binding to the Rb family results in the release of E2Fs and the subsequent deregulation of E2F-responsive genes (19). In addition, p107 and p130 negatively regulate the cell cycle by binding and inhibiting cyclin E-cdk2 and cyclin A-ckd2 complexes via a CDKI-like domain that is absent in Rb (10, 14, 69). The ability to bind cyclin E and A complexes has been shown to be independent from their role as suppressors of E2F-mediated transcription since mutants of p130, which can bind E2Fs but are unable to bind cyclin E-cdk2 and cyclin A-cdk2 complexes, are defective at preventing entry into S phase (29). Therefore, for Ras signals to be proliferative in the absence of anchorage may involve the removal of E2F-dependent and -independent growth suppressor functions of the Rb family.
For LT to efficiently confer anchorage independence requires a functional J domain, indicating a role for p107 and p130 (63). The J domain is required for LT to transform wild-type and Rb−/− mouse embryonic fibroblasts (MEFs) but is dispensable for transformation of p107−/−/p130−/− MEFs (62). Consistent with a role for p107 and p130, E2F4 and E2F5 that primarily associate with p107 and p130 have been shown to be required for pocket protein-induced cell cycle arrest (24).
The ability of cells expressing dnp53 and activated Ras to spontaneously acquire anchorage independence is associated with the acquisition of additional genetic changes. This loss is specific to p16INK4A since we did not observe a loss of expression of p19ARF, which is encoded by the same locus (34, 36). The mechanism of p16INK4A loss is likely to involve silencing of the p16INK4A promoter by methylation since treatment of these cells with a demethylating agent resulted in the restoration of p16INK4A expression. We have not been able to determine the genetic change responsible for anchorage independence in the clones that maintain p16INK4A expression; however, since these clones also show Rb phosphorylation in the absence of an anchorage signal, a dysregulation of this pathway is probably involved.
The ability of antisense p16INK4A and a p16INK4A-insensitive mutant of cdk4 to confer Ras-dependent anchorage-independent proliferation, in conjunction with a loss of p53 function, confirms that the loss of p16INK4A is sufficient to confer anchorage independence to these cells. This is substantiated by the finding that reintroduction of p16INK4A into Ras/dnp53 cells that have lost p16INK4A inhibits anchorage-independent proliferation. As expected, we find that these clones phosphorylate Rb in the absence of anchorage and maintain a requirement for Rb phosphorylation in order to proliferate. Until recently, it had been thought that Rb was responsible for mediating the growth-inhibitory effects of p16INK4A (40, 44). Recently, however, it has been established that p107 and p130 also play important roles in mediating p16INK4A inhibitory signals. p107−/−/p130−/− MEFs are defective in their response to the growth-inhibitory effects of p16INK4A (7), although they respond normally to other types of G1 arrest (31, 32). Similarly, MEFs lacking both E2F4 and E2F5, with which p107 and p130 primarily associate, are less sensitive to p16INK4A (24). Our findings indicate that for efficient anchorage-independent proliferation, inactivation of p107/p130 are required. This can be achieved either by expression of the N terminus of LT or by loss of p16INK4A expression. This demonstrates that in terms of regulating the anchorage dependence of proliferation, p16 loss and inactivation of the Rb family appear to be genetically equivalent. This result is in contrast to those of previous studies analyzing the growth properties of Rb−/−, p107−/−, p130−/− (TKO) cells and p16−/− cells. Whereas TKO cells are immortal and show a reduced requirement for mitogens (16, 54), p16−/− cells appear to retain normal growth controls (35, 58). Here we demonstrate a role of Rb family loss in conferring anchorage independence and that p16INK4A loss has the same effect. These results therefore identify how p16INK4A inactivation can contribute to the loss of extrinsic growth requirements associated with cellular transformation.
The mechanisms by which integrin and mitogenic signals cooperate to promote cell proliferation are still poorly understood. In many immortalized cell lines, mitogen and integrin signals cooperate to achieve sustained activation of the ERK pathway, which has been shown to be sufficient for anchorage-independent proliferation (15, 51). Studies in primary cells, however, have shown that the situation is more complex and have identified Rac as an important mediator of integrin signaling to the cell cycle (45). Rac signaling appears to cooperate with activation of the ERK pathway to induce cyclin D1 levels. However, cyclin D1 expression is insufficient to promote proliferation, indicating that other signaling pathways, as yet unidentified, are important for cell cycle progression. The complexity of the signals required for a cell to enter S phase is reflected by our findings that indicate that multiple genetic changes are required to overcome anchorage dependency. These studies also reemphasize the inadequacies of using immortalized cell lines to study cell cycle checkpoints since many of the genetic changes involved in the immortalization process appear to mediate or inactivate these checkpoints.
Removal of anchorage has also been reported to result in the induction of CDKIs and the loss of a signal required for cyclin A transcription (23, 26). LT expression has been reported to confer insensitivity to CDKIs, and LT-expressing Schwann cells have high levels of cyclin A (39, 42). However, LT cells are unable to proliferate in the absence of anchorage. This would suggest an additional checkpoint that is insensitive to LT expression but can be overcome by Ras. Further studies are required to investigate the mechanisms responsible.
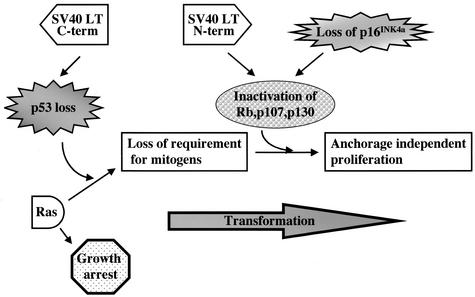
We have demonstrated how the accumulation of specific genetic defects leads to a progressive independence from extrinsic signals (Fig. 8). Although Ras signals alone cause a cell cycle arrest, loss of p53 abolishes the arrest and signals from Ras confer mitogen independence to the cells. However, these cells maintain anchorage dependence and are susceptible to contact inhibition. Loss of the Rb family or p16INK4A, together with p53 loss, enables Ras signals to drive proliferation in the absence of anchorage or the presence of contact inhibition. These results would appear to have direct relevance to human carcinogenesis. The genetic defects required for Ras to transform primary cells in vitro are paralleled by the spectrum of genetic defects found in tumors that have a high incidence of Ras mutations. In pancreatic carcinomas, in which the incidence of Ras mutations is nearly 100% (1), mutation or loss of both p16INK4A and p53 are commonly found (8, 53). The frequency of p53 mutations has been found to be more than 60% (5, 48), whereas mutation or loss of expression of p16INK4A, as a result of promoter methylation, has been found in >95% of the primary tumors (55). In contrast, Rb loss is much less frequent, reflecting our findings that Rb loss alone is not sufficient to confer full cellular transformation. Intriguingly, a murine pancreatic tumor progression model mirrors our in vitro findings (66). In these mice, tumor progression involves activation of Ras, loss of p53, and loss of INK4A. Our results demonstrate how these genetic changes can cooperate to result in cells that can proliferate independently of both extrinsic mitogenic and anchorage signals.
FIG. 8.
Contribution of the inactivation of checkpoints to Ras transformation. Model of how the progressive loss of checkpoints allows Ras to act positively on the cell cycle. Loss of p53 activity causes Ras to be proliferative in the absence of mitogens. Inactivation of the Rb family, by loss of p16INK4A or by inactivation by the N terminus of SV40 LT, renders the cells capable of proliferation independently of anchorage and insensitive to contact inhibition.
Acknowledgments
We thank Nicole Mathon, Michel Cayouette, and Martin Raff for helpful discussions. We are grateful to Sibylle Mittnacht for providing the Rb(NPC−) construct.
This work was funded by a project grant from Cancer Research UK that was sponsored by the Neurofibromatosis Association of the United Kingdom. A.C.L. is a Cancer Research UK Senior Research Fellow.
REFERENCES
- 1.Almoguera, C., D. Shibata, K. Forrester, J. Martin, N. Arnheim, and M. Perucho. 1988. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell 53:549-554. [DOI] [PubMed] [Google Scholar]
- 2.Avantaggiati, M. L., M. Carbone, A. Graessmann, Y. Nakatani, B. Howard, and A. S. Levine. 1996. The SV40 large T antigen and adenovirus E1a oncoproteins interact with distinct isoforms of the transcriptional co-activator, p300. EMBO J. 15:2236-2248. [PMC free article] [PubMed] [Google Scholar]
- 3.Bardeesy, N., B. C. Bastian, A. Hezel, D. Pinkel, R. A. DePinho, and L. Chin. 2001. Dual inactivation of RB and p53 pathways in RAS-induced melanomas. Mol. Cell. Biol. 21:2144-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartkova, J., J. Lukas, P. Guldberg, J. Alsner, A. F. Kirkin, J. Zeuthen, and J. Bartek. 1996. The p16-cyclin D/Cdk4-pRb pathway as a functional unit frequently altered in melanoma pathogenesis. Cancer Res. 56:5475-5483. [PubMed] [Google Scholar]
- 5.Barton, C. M., S. L. Staddon, C. M. Hughes, P. A. Hall, C. O'Sullivan, G. Kloppel, B. Theis, R. C. Russell, J. Neoptolemos, R. C. Williamson, et al. 1991. Abnormalities of the p53 tumour suppressor gene in human pancreatic cancer. Br. J. Cancer 64:1076-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bottenstein, J. E., and G. H. Sato. 1980. Fibronectin and polylysine requirement for proliferation of neuroblastoma cells in defined medium. Exp. Cell Res. 129:361-366. [DOI] [PubMed] [Google Scholar]
- 7.Bruce, J. L., R. K. Hurford, Jr., M. Classon, J. Koh, and N. Dyson. 2000. Requirements for cell cycle arrest by p16INK4a. Mol. Cell 6:737-742. [DOI] [PubMed] [Google Scholar]
- 8.Caldas, C., S. A. Hahn, C. L. da, M. S. Redston, M. Schutte, A. B. Seymour, C. L. Weinstein, R. H. Hruban, C. J. Yeo, and S. E. Kern. 1994. Frequent somatic mutations and homozygous deletions of the p16 (MTS1) gene in pancreatic adenocarcinoma. Nat. Genet. 8:27-32. (Erratum, 8:410.) [DOI] [PubMed]
- 9.Carnero, A., J. D. Hudson, C. M. Price, and D. H. Beach. 2000. p16INK4A and p19ARF act in overlapping pathways in cellular immortalization. Nat. Cell Biol. 2:148-155. [DOI] [PubMed] [Google Scholar]
- 10.Castano, E., Y. Kleyner, and B. D. Dynlacht. 1998. Dual cyclin-binding domains are required for p107 to function as a kinase inhibitor. Mol. Cell. Biol. 18:5380-5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng, L., M. Khan, and A. W. Mudge. 1995. Calcitonin gene-related peptide promotes Schwann cell proliferation. J. Cell Biol. 129:789-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chew, Y. P., M. Ellis, S. Wilkie, and S. Mittnacht. 1998. pRB phosphorylation mutants reveal role of pRB in regulating S phase completion by a mechanism independent of E2F. Oncogene 17:2177-2186. [DOI] [PubMed] [Google Scholar]
- 13.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 14.Coats, S., P. Whyte, M. L. Fero, S. Lacy, G. Chung, E. Randel, E. Firpo, and J. M. Roberts. 1999. A new pathway for mitogen-dependent cdk2 regulation uncovered in p27Kip1-deficient cells. Curr. Biol. 9:163-173. [DOI] [PubMed] [Google Scholar]
- 15.Cowley, S., H. Paterson, P. Kemp, and C. J. Marshall. 1994. Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell 77:841-852. [DOI] [PubMed] [Google Scholar]
- 16.Dannenberg, J. H., A. van Rossum, L. Schuijff, and H. te Riele. 2000. Ablation of the retinoblastoma gene family deregulates G1 control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev. 14:3051-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeCaprio, J. A. 1999. The role of the J domain of SV40 large T in cellular transformation. Biologicals 27:23-28. [DOI] [PubMed] [Google Scholar]
- 18.DeCaprio, J. A., J. W. Ludlow, J. Figge, J. Y. Shew, C. M. Huang, W. H. Lee, E. Marsilio, E. Paucha, and D. M. Livingston. 1988. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell 54:275-283. [DOI] [PubMed] [Google Scholar]
- 19.Dyson, N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12:2245-2262. [DOI] [PubMed] [Google Scholar]
- 20.Dyson, N., R. Bernards, S. H. Friend, L. R. Gooding, J. A. Hassell, E. O. Major, J. M. Pipas, T. Vandyke, and E. Harlow. 1990. Large T antigens of many polyomaviruses are able to form complexes with the retinoblastoma protein. J. Virol. 64:1353-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eckner, R., J. W. Ludlow, N. L. Lill, E. Oldread, Z. Arany, N. Modjtahedi, J. A. DeCaprio, D. M. Livingston, and J. A. Morgan. 1996. Association of p300 and CBP with simian virus 40 large T antigen. Mol. Cell. Biol. 16:3454-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ewen, M. E., Y. G. Xing, J. B. Lawrence, and D. M. Livingston. 1991. Molecular cloning, chromosomal mapping, and expression of the cDNA for p107, a retinoblastoma gene product-related protein. Cell 66:1155-1164. [DOI] [PubMed] [Google Scholar]
- 23.Fang, F., G. Orend, N. Watanabe, T. Hunter, and E. Ruoslahti. 1996. Dependence of cyclin E-CDK2 kinase activity on cell anchorage. Science 271:499-502. [DOI] [PubMed] [Google Scholar]
- 24.Gaubatz, S., G. J. Lindeman, S. Ishida, L. Jakoi, J. R. Nevins, D. M. Livingston, and R. E. Rempel. 2000. E2F4 and E2F5 play an essential role in pocket protein-mediated G1 control. Mol. Cell 6:729-735. [DOI] [PubMed] [Google Scholar]
- 25.Grana, X., J. Garriga, and X. Mayol. 1998. Role of the retinoblastoma protein family, pRB, p107, and p130 in the negative control of cell growth. Oncogene 17:3365-3383. [DOI] [PubMed] [Google Scholar]
- 26.Guadagno, T. M., M. Ohtsubo, J. M. Roberts, and R. K. Assoian. 1993. A link between cyclin A expression and adhesion-dependent cell cycle progression. Science 262:1572-1575. (Erratum, 263:455, 1994.) [DOI] [PubMed]
- 27.Hainaut, P., T. Soussi, B. Shomer, M. Hollstein, M. Greenblatt, E. Hovig, C. C. Harris, and R. Montesano. 1997. Database of p53 gene somatic mutations in human tumors and cell lines: updated compilation and future prospects. Nucleic Acids Res. 25:151-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall, M., and G. Peters. 1996. Genetic alterations of cyclins, cyclin-dependent kinases, and Cdk inhibitors in human cancer. Adv. Cancer Res. 68:67-108. [DOI] [PubMed] [Google Scholar]
- 29.Hansen, K., T. Farkas, J. Lukas, K. Holm, L. Ronnstrand, and J. Bartek. 2001. Phosphorylation-dependent and -independent functions of p130 cooperate to evoke a sustained G1 block. EMBO J. 20:422-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harbour, J. W., and D. C. Dean. 2000. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 14:2393-2409. [DOI] [PubMed]
- 31.Harrington, E. A., J. L. Bruce, E. Harlow, and N. Dyson. 1998. pRB plays an essential role in cell cycle arrest induced by DNA damage. Proc. Natl. Acad. Sci. USA 95:11945-11950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hurford, R. K., Jr., D. Cobrinik, M. H. Lee, and N. Dyson. 1997. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 11:1447-1463. [DOI] [PubMed] [Google Scholar]
- 33.Kamijo, T., J. D. Weber, G. Zambetti, F. Zindy, M. F. Roussel, and C. J. Sherr. 1998. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc. Natl. Acad. Sci. USA 95:8292-8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamijo, T., F. Zindy, M. F. Roussel, D. E. Quelle, J. R. Downing, R. A. Ashmun, G. Grosveld, and C. J. Sherr. 1997. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 91:649-659. [DOI] [PubMed] [Google Scholar]
- 35.Krimpenfort, P., K. C. Quon, W. J. Mooi, A. Loonstra, and A. Berns. 2001. Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature 413:83-86. [DOI] [PubMed] [Google Scholar]
- 36.Latres, E., M. Malumbres, R. Sotillo, J. Martin, S. Ortega, J. Martin-Caballero, J. M. Flores, C. Cordon-Cardo, and M. Barbacid. 2000. Limited overlapping roles of p15INK4b and p18INK4c cell cycle inhibitors in proliferation and tumorigenesis. EMBO J. 19:3496-3506. [DOI] [PMC free article] [PubMed]
- 37.Lill, N. L., M. J. Tevethia, R. Eckner, D. M. Livingston, and N. Modjtahedi. 1997. p300 family members associate with the carboxyl terminus of simian virus 40 large tumor antigen. J. Virol. 71:129-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin, A. W., M. Barradas, J. C. Stone, L. van Aelst, M. Serrano, and S. W. Lowe. 1998. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev. 12:3008-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lloyd, A. C., F. Obermuller, S. Staddon, C. F. Barth, M. McMahon, and H. Land. 1997. Cooperating oncogenes converge to regulate cyclin/cdk complexes. Genes Dev. 11:663-677. [DOI] [PubMed] [Google Scholar]
- 40.Lukas, J., D. Parry, L. Aagaard, D. J. Mann, J. Bartkova, M. Strauss, G. Peters, and J. Bartek. 1995. Retinoblastoma-protein-dependent cell-cycle inhibition by the tumour suppressor p16. Nature 375:503-506. [DOI] [PubMed] [Google Scholar]
- 41.Manfredi, J. J., and C. Prives. 1994. The transforming activity of simian virus 40 large tumor antigen. Biochim. Biophys. Acta 1198:65-83. [DOI] [PubMed] [Google Scholar]
- 42.Mann, D. J., and N. C. Jones. 1996. E2F-1 but not E2F-4 can overcome p16-induced G1 cell-cycle arrest. Curr. Biol. 6:474-483. [DOI] [PubMed] [Google Scholar]
- 43.Mathon, N. F., D. S. Malcolm, M. C. Harrisingh, L. Cheng, and A. C. Lloyd. 2001. Lack of replicative senescence in normal rodent glia. Science 291:872-875. [DOI] [PubMed] [Google Scholar]
- 44.Merlino, G., and L. J. Helman. 1999. Rhabdomyosarcoma: working out the pathways. Oncogene 18:5340-5348. [DOI] [PubMed] [Google Scholar]
- 45.Mettouchi, A., S. Klein, W. Guo, M. Lopez-Lago, E. Lemichez, J. K. Westwick, and F. G. Giancotti. 2001. Integrin-specific activation of Rac controls progression through the G1 phase of the cell cycle. Mol. Cell 8:115-127. [DOI] [PubMed] [Google Scholar]
- 46.Moran, E. 1993. DNA tumor virus transforming proteins and the cell cycle. Curr. Opin. Genet. Dev. 3:63-70. [DOI] [PubMed] [Google Scholar]
- 47.Peeper, D. S., J. H. Dannenberg, S. Douma, H. te Riele, and R. Bernards. 2001. Escape from premature senescence is not sufficient for oncogenic transformation by Ras. Nat. Cell Biol. 3:198-203. [DOI] [PubMed] [Google Scholar]
- 48.Redston, M. S., C. Caldas, A. B. Seymour, R. H. Hruban, C. L. Da, C. J. Yeo, and S. E. Kern. 1994. p53 mutations in pancreatic carcinoma and evidence of common involvement of homocopolymer tracts in DNA microdeletions. Cancer Res. 54:3025-3033. [PubMed] [Google Scholar]
- 49.Ridley, A. J., H. F. Paterson, M. Noble, and H. Land. 1988. Ras-mediated cell cycle arrest is altered by nuclear oncogenes to induce Schwann cell transformation. EMBO J. 7:1635-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rocco, J. W., and D. Sidransky. 2001. p16(MTS-1/CDKN2/INK4a) in cancer progression. Exp. Cell Res. 264:42-55. [DOI] [PubMed] [Google Scholar]
- 51.Roovers, K., and R. K. Assoian. 2000. Integrating the MAP kinase signal into the G1 phase cell cycle machinery. Bioessays 22:818-826. [DOI] [PubMed] [Google Scholar]
- 52.Roper, E., W. Weinberg, F. M. Watt, and H. Land. 2001. p19ARF-independent induction of p53 and cell cycle arrest by Raf in murine keratinocytes. EMBO Rep. 2:145-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rozenblum, E., M. Schutte, M. Goggins, S. A. Hahn, S. Panzer, M. Zahurak, S. N. Goodman, T. A. Sohn, R. H. Hruban, C. J. Yeo, and S. E. Kern. 1997. Tumor-suppressive pathways in pancreatic carcinoma. Cancer Res. 57:1731-1734. [PubMed] [Google Scholar]
- 54.Sage, J., G. J. Mulligan, L. D. Attardi, A. Miller, S. Chen, B. Williams, E. Theodorou, and T. Jacks. 2000. Targeted disruption of the three Rb-related genes leads to loss of G1 control and immortalization. Genes Dev. 14:3037-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schutte, M., R. H. Hruban, J. Geradts, R. Maynard, W. Hilgers, S. K. Rabindran, C. A. Moskaluk, S. A. Hahn, I. Schwarte-Waldhoff, W. Schmiegel, S. B. Baylin, S. E. Kern, and J. G. Herman. 1997. Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer Res. 57:3126-3130. [PubMed] [Google Scholar]
- 56.Serrano, M., G. J. Hannon, and D. Beach. 1993. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature 366:704-707. [DOI] [PubMed] [Google Scholar]
- 57.Serrano, M., A. W. Lin, M. E. McCurrach, D. Beach, and S. W. Lowe. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88:593-602. [DOI] [PubMed] [Google Scholar]
- 58.Sharpless, N. E., N. Bardeesy, K. H. Lee, D. Carrasco, D. H. Castrillon, A. J. Aguirre, E. A. Wu, J. W. Horner, and R. A. DePinho. 2001. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature 413:86-91. [DOI] [PubMed] [Google Scholar]
- 59.Sherr, C. J., and J. M. Roberts. 1999. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13:1501-1512. [DOI] [PubMed] [Google Scholar]
- 60.Srinivasan, A., A. J. McClellan, J. Vartikar, I. Marks, P. Cantalupo, Y. Li, P. Whyte, K. Rundell, J. L. Brodsky, and J. M. Pipas. 1997. The amino-terminal transforming region of simian virus 40 large T and small t antigens functions as a J domain. Mol. Cell. Biol. 17:4761-4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stott, F. J., S. Bates, M. C. James, B. B. McConnell, M. Starborg, S. Brookes, I. Palmero, K. Ryan, E. Hara, K. H. Vousden, and G. Peters. 1998. The alternative product from the human CDKN2A locus, p14ARF, participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 17:5001-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stubdal, H., J. Zalvide, K. S. Campbell, C. Schweitzer, T. M. Roberts, and J. A. DeCaprio. 1997. Inactivation of pRB-related proteins p130 and p107 mediated by the J domain of simian virus 40 large T antigen. Mol. Cell. Biol. 17:4979-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stubdal, H., J. Zalvide, and J. A. DeCaprio. 1996. Simian virus 40 large T antigen alters the phosphorylation state of the RB-related proteins p130 and p107. J. Virol. 70:2781-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sullivan, C. S., P. Cantalupo, and J. M. Pipas. 2000. The molecular chaperone activity of simian virus 40 large T antigen is required to disrupt Rb-E2F family complexes by an ATP-dependent mechanism. Mol. Cell. Biol. 20:6233-6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tang, D. G., Y. M. Tokumoto, J. A. Apperly, A. C. Lloyd, and M. C. Raff. 2001. Lack of replicative senescence in cultured rat oligodendrocyte precursor cells. Science 291:868-871. [DOI] [PubMed] [Google Scholar]
- 66.Wagner, M., F. R. Greten, C. K. Weber, S. Koschnick, T. Mattfeldt, W. Deppert, H. Kern, G. Adler, and R. M. Schmid. 2001. A murine tumor progression model for pancreatic cancer recapitulating the genetic alterations of the human disease. Genes Dev. 15:286-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weber, J. D., J. R. Jeffers, J. E. Rehg, D. H. Randle, G. Lozano, M. F. Roussel, C. J. Sherr, and G. P. Zambetti. 2000. p53-independent functions of the p19ARF tumor suppressor. Genes Dev. 14:2358-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wolfel, T., M. Hauer, J. Schneider, M. Serrano, C. Wolfel, E. Klehmann-Hieb, E. De Plaen, T. Hankeln, K. H. Meyer zum Buschenfelde, and D. Beach. 1995. A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science 269:1281-1284. [DOI] [PubMed] [Google Scholar]
- 69.Woo, M. S., I. Sanchez, and B. D. Dynlacht. 1997. p130 and p107 use a conserved domain to inhibit cellular cyclin-dependent kinase activity. Mol. Cell. Biol. 17:3566-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zalvide, J., and J. A. DeCaprio. 1995. Role of pRb-related proteins in simian virus 40 large-T-antigen-mediated transformation. Mol. Cell. Biol. 15:5800-5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zalvide, J., H. Stubdal, and J. A. DeCaprio. 1998. The J domain of simian virus 40 large T antigen is required to functionally inactivate RB family proteins. Mol. Cell. Biol. 18:1408-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang, Y., Y. Xiong, and W. G. Yarbrough. 1998. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 92:725-734. [DOI] [PubMed] [Google Scholar]
- 73.Zhu, J., D. Woods, M. McMahon, and J. M. Bishop. 1998. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev. 12:2997-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]