Abstract
Antioxidant enzymes are critical in oxidative stress responses. Radioresistant variants isolated from MCF-7 human carcinoma cells following fractionated ionizing radiation (MCF+FIR cells) or overexpression of manganese superoxide dismutase (MCF+SOD cells) demonstrated dose-modifying factors at 10% isosurvival of 1.8 and 2.3, respectively. MCF+FIR and MCF-7 cells (exposed to single-dose radiation) demonstrated 5- to 10-fold increases in MnSOD activity, mRNA, and immunoreactive protein. Radioresistance in MCF+FIR and MCF+SOD cells was reduced following expression of antisense MnSOD. DNA microarray analysis and immunoblotting identified p21, Myc, 14-3-3 zeta, cyclin A, cyclin B1, and GADD153 as genes constitutively overexpressed (2- to 10-fold) in both MCF+FIR and MCF+SOD cells. Radiation-induced expression of these six genes was suppressed in fibroblasts from Sod2 knockout mice (−/−) as well as in MCF+FIR and MCF+SOD cells expressing antisense MnSOD. Inhibiting NF-κB transcriptional activity in MCF+FIR cells, by using mutant IκBα, inhibited radioresistance as well as reducing steady-state levels of MnSOD, 14-3-3 zeta, GADD153, cyclin A, and cyclin B1 mRNA. In contrast, mutant IκBα was unable to inhibit radioresistance or reduce 14-3-3 zeta, GADD153, cyclin A, and cyclin B1 mRNAs in MCF+SOD cells, where MnSOD overexpression was independent of NF-κB. These results support the hypothesis that NF-κB is capable of regulating the expression of MnSOD, which in turn is capable of increasing the expression of genes that participate in radiation-induced adaptive responses.
Chronic exposure of cells to ionizing radiation (IR) induces an adaptive response that results in enhanced tolerance to the subsequent cytotoxicity of IR. The fate of irradiated cells is believed to be controlled by the network of signaling elements that lead to different modes of cell death or survival. Many stress-responsive genes are inducible by IR (20), but only a fraction of radiation-inducible genes are believed to play a key role in the stress-tolerance phenotype, i.e., elements in cell cycle checkpoints, apoptosis, and DNA repair (36, 45).
Reactive oxygen species (ROS) are produced by the metabolism of O2 in all aerobic cells (1) and are essential for normal cellular signaling functions. However, excessive ROS formation such as that seen during IR can cause cell death, and intrinsic antioxidant enzymes (AEs) are induced to protect cells from ROS-induced lethality (21). ROS have been shown to activate preapoptosis signaling elements such as JNK (66). MnSOD(SOD2, manganese-containing superoxide dismutase) is a nuclear-encoded mitochondrial enzyme that scavenges superoxide radicals in the mitochondrial matrix and is thought to be an important determinant of sensitivity to ROS-induced cytotoxicity. Using MnSOD knockout mice (Sod2−/−), MnSOD was shown to be essential in protecting against ROS-induced injury during O2 metabolism (57). MnSOD is also essential in resistance to tumor necrosis factor (TNF)-induced apoptosis (46, 59) and is involved in MnCl2-induced apoptosis (55). The protective effect of MnSOD against normal tissue damage caused by radiation is highlighted by in vivo experiments showing induction of MnSOD following radiation in the heart and gut (42, 49) and the finding that overexpression of MnSOD in normal mouse epithelial tissues protected them from radiation injury (18). Endogenous SOD activity is also involved in the resistance of Wilms' tumors to chemotherapeutic agents (21) and in protecting human leukemic and cancer cells from radiation (63). Further evidence suggests that radiation-induced esophagitis is linked with activation of a group of cytokines that are responsive to MnSOD (19). Also observed in cells overexpressing MnSOD were significant phenotypic changes, including cell differentiation (48) and alterations in Jun-associated transcription factors (30). Despite this plethora of evidence suggesting that MnSOD is associated with protection from free radical-mediated radiation damage as well as causing alterations in gene expression, the relationship between MnSOD, IR-inducible genes, and radioprotection remains obscure.
Expression of endogenous MnSOD has been found to be reduced in many human cancer cells and transformed cell lines (32, 39, 64). Expression of MnSOD causes significant alterations in the malignant phenotype as well as inhibition of tumor growth in vivo (32, 64, 68). In many human tumor cells, MnSOD immunoreactive protein and activity are barely detectable in contrast to other AEs (CuZnSOD, CAT, and GPx) (40). Therefore, induction of endogenous MnSOD by different stresses could contribute to adaptive responses seen in some tumor cells treated with fractionated radiation (FIR). It also follows that a fraction of genes in the radiation-induced gene expression profiles in tumor cells could represent genes that are regulated by MnSOD induction. Identifying MnSOD-responsive genes could therefore yield crucial insights into the genes involved in radioresistant phenotypes.
NF-κB is a stress-responsive transcription factor made up of five Rel family members (p65/RelA, p50, p52, c-Rel, and RelB [6, 7]) that has been suggested to be actively involved in ROS-induced apoptosis in some model systems (7, 22, 44). With links to the expression of about 150 target genes, NF-κB is also thought to prevent apoptosis via activation of antiapoptosis genes in other model systems (7). Overexpression of MnSOD is also believed to modulate NF-κB activity (8, 37), and NF-κB binding sequences were found in the MnSOD promoter (17, 56). NF-κB is also closely linked to MnSOD expression induced by phorbol myristate acetate, cytokine (31), and serum starvation (53) leading to antiapoptotic responses to TNF (10, 15).
To determine in the present study if genes governing the FIR-induced resistant phenotype in human cancer cells were regulated via alterations in NF-κB-mediated MnSOD expression, gene expression profiles of 1,176 genes in MCF+FIR and MCF+SOD cells were compared to that in wild-type MCF-7. Increased expression of proteins p21, Myc, 14-3-3 zeta, cyclin A, cyclin B1, and GADD153 were detected in the genes up-regulated in both radioresistant MCF+FIR and MCF+SOD cell lines, relative to that in MCF-7. The involvement of MnSOD in the FIR-induced alterations in gene expression was confirmed by a lack of radiation responsiveness of these genes in Sod2 knockout mouse cells. Moreover, conditional expression of antisense MnSOD inhibited the up-regulation of these genes in MCF+FIR and MCF+SOD cells, as well as inhibiting the radiation-resistant phenotype demonstrated by these cell lines. Finally, blocking NF-κB transcriptional activity using mutant IκB expression in MCF+FIR cells inhibited radioresistance as well as inhibiting the transcription of MnSOD, 14-3-3 zeta, cyclin A, cyclin B1, and GADD153. These results provide strong evidence that MnSOD regulates the expression of at least a subset of the genes involved with the induction of the radioresistant phenotype seen in MCF-7 cells following fractionated radiation exposure. These results also suggest that a pathway involving NF-κB-inducible genes in combination with alterations in the metabolism of superoxide can mediate radiation-inducible adaptive responses.
MATERIALS AND METHODS
DNA constructs.
pTet-On and pTRE vectors were purchased from Clontech Laboratories, Inc. (Palo Alto, Calif.). The pTRE-antiSOD plasmid was constructed by inserting a previously described full-length human MnSOD cDNA in the reverse orientation into pTRE (32). The antisense MnSOD orientation was confirmed by DNA sequencing. NF-κB- and AP-1-controlled luciferase reporters were the same as described before (35). I2E (intronic enhancer element of the human SOD2 gene)-controlled luciferase reporters were kindly provided by Daret St. Clair at the University of Kentucky (31). The pcDNA3 vector with a neomycin resistance gene was obtained from Invitrogen Co. (Carlsbad, Calif.).
Cell lines isolated following exposure to FIR or MnSOD overexpression.
MCF-7 human breast adenocarcinoma cells (starting at passage 168) were purchased from the American Type Culture Collection and utilized because previously well-characterized MnSOD overexpressors were available in this cell line (32). MCF+FIR cells were obtained from MCF-7 cells by exposure to FIR with a total dose of 60 Gy of γ-irradiation (2 Gy per fraction, five times per week for 6 weeks). Radiation was delivered at room temperature at a rate of 46 cGy/min (Theratron-80 S/N 140 Co-60 unit; Atomic Energy of Canada Limited). MCF-7 cells treated with sham FIR were maintained as control cells. Experiments were performed with the MCF+FIR cells within 7 to 10 passages after FIR. MCF+SOD cells were obtained by stably overexpressing MnSOD cDNA in MCF-7 cells, which caused a 5.4-fold increase in MnSOD activity (32). Fibroblasts isolated from Sod2 knockout mice (−/−), heterozygotes (+/−), and wild-type controls (+/+) were previously shown to have the expected levels of MnSOD expression (26). Radioresistance was measured by clonogenic survival following exposure to IR. Irradiated cells were trypsinized and cloned in a 37°C incubator with 5% CO2 for 14 to 21 days. Clones containing more than 50 cells were scored as survivors, and the data were normalized to the appropriate sham-irradiated control group. Dose-modifying factors (DMFs) were calculated using the following expression: DMF = (dose to reach the specified survival in resistant cells)/(dose to reach the same survival in the control).
Establishment of cell lines that conditionally expressed antisense MnSOD.
Tetracycline (Tet)-inducible MnSOD antisense transfectants were established using the same procedure used to produce Tet-controlled antisense MnSOD in MCF+SOD cells (MCF+SOD+antiSOD) (34). Briefly, MCF+FIR cells in 100-mm-diameter dishes were transfected with 15 μg of tTA regulator, 15 μg of pTRE-antiSOD plasmids, 2 μg of pcDNA3 as a selection marker, 40 μg of Lipofectamine, and 5 μg of Plus reagent in 6 ml of serum-free Dulbecco's modified Eagle's medium (DMEM). Transfectants were selected with relevant antibiotics, and pooled clones were maintained under noninducing conditions for two passages. All the transfectants were maintained in 10% fetal bovine serum (FBS)-DMEM under noninducing conditions. Transfectants with the Tet-controlled luciferase gene (34) demonstrated no detectable transgene expression under noninducing conditions (without doxycycline [DOX]). An ∼24-fold increase in luciferase activity was induced by 0.1 μg of DOX/ml. No cytotoxic effects of DOX (0.1 to 1.0 μg/ml) were observed.
Establishment of mutant IκB transfectants.
Stable transfectants were obtained using a previously described method (13) with Lipofectamine reagent (Life Technologies, Inc., Gaithersburg, Md.). Briefly, 5 × 106 cells grown in 100-mm-diameter cell culture dishes were transfected with 15 μg of mutant IκB plasmid controlled by a cytomegalovirus promoter (provided by Nancy Rice at the National Cancer Institute, National Institutes of Health [NIH]), 2 μg of hygromycin marker pCEP4, and 40 μg of Lipofectamine in 6 ml of serum-reduced OPTI-EMEM (Life Technologies, Inc.). pcDNA3 in place of mutant IκB was transfected as vector control. Cells were transfected for 72 h, trypsinized, and cultured in the selecting medium with 80 μg of hygromycin B/ml for 21 days. The selected mutant IκB (MCF+FIR+mIκB or MCF+SOD+mIκB) and vector control clones were pooled and passaged with 20 μg of hygromycin B/ml. The transfected cells were cultured for at least two passages in hygromycin B-free medium before experiments.
SOD activity analysis using native gel analysis.
Cells were rinsed three times with 10 ml of ice-cold phosphate-buffered saline (2.7 mM KCl, 1.5 mM KH2PO4, 8 mM NaHPO4, and 136.9 mM NaCl; pH 7.0) and scraped into phosphate buffer using a sterile rubber policeman. The cell suspension was then sonicated on ice using a 550 Sonic Dismembrator (Fisher Scientific Co., Tustin, Calif.), and the samples were stored at −80°C before measurements. Equal protein, as determined by the Bradford protein assay (Bio-Rad, Hercules, Calif.), was loaded into each lane. Proteins were size separated in 12% polyacrylamide gels with 5% stacking gels following 1 h of preelectrophoresis, and SOD activity was visualized by the nitroblue tetrazolium method (9).
DNA microarray analysis.
Total RNA was extracted using TRIzol reagent (Life Technologies, Inc.). After confirmation of the RNA integrity on an agarose gel, total RNA was digested using RNase-free DNase I for 20 min, extracted with phenol-chloroform, and then precipitated with 2.5 volumes of ethanol. poly(A)+ RNA was isolated using the Oligotex RNA kit (QIAGEN Inc., Valencia, Calif.). Gene expression was analyzed using the Atlas human cancer cDNA expression array filter (1,176 genes) from Clontech Laboratories, Inc. To prepare for hybridization, 1 μg of poly(A)+ RNA was transcribed with nucleotides containing [α-32P]dATP, and the labeled cDNA was purified, denatured, and added to 5 ml of ExpressHyb hybridization solution (Clontech). The final probe with a concentration of 106 cpm/ml was freshly applied to an array membrane that was prehybridized in the ExpressHyb hybridization solution (Clontech) for 30 min. Hybridization was allowed to proceed overnight at 68°C in a roller bottle. The filters were then stringently washed four times with agitation for 20 min in 200 ml of prewarmed (68°C) solution 1 (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 1% sodium dodecyl sulfate [SDS]) and twice with solution 2 (0.1× SSC, 0.5% SDS) before exposing them to X-ray film overnight at −80°C. The filters were also exposed to a phosphor screen overnight and scanned using a Storm 840 PhosphorImager (Molecular Dynamics, Inc., Sunnyvale, Calif.), and signals of paired genes were quantified with ImageQuant software. Results represent the quantitation of one hybridization with two sets of fluorescence-labeled RNA probes, and changes less than twofold were not considered for further study.
Immunoblotting techniques.
Wild-type MCF-7, MCF+FIR, and MCF+SOD cells were cultured in 10% FBS-DMEM, and total protein extracts were prepared. The Tet-antisense MnSOD transfectants were incubated with or without transgene-inducing conditions (DOX at 0.1 μg/ml) in 10% FBS-DMEM, cells were trypsinized, and total proteins were analyzed 24 h after gene induction. Equal quantities of sample protein were mixed with 50 μl of loading buffer, heated at 70°C for 10 min, size separated in 12.5% acrylamide SDS-polyacrylamide gel electrophoresis, and transferred to nitrocellulose membranes (Bio-Rad). The membranes were then blocked at room temperature for 2 h in blocking solution (Pierce Co., Rockford, Ill.), washed with 0.01% Tween-phosphate-buffered saline, and incubated overnight at 4°C with rabbit antiserum to human kidney MnSOD or human placental CuZnSOD as described elsewhere (41, 47) at a dilution of 1:200. Antibodies to p21, Myc, 14-3-3 zeta, cyclin A, cyclin B1, GADD153, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were purchased from Santa Cruz Biotechnology (Santa Cruz, Calif.). The blots were then incubated with horseradish peroxidase-conjugated secondary antibody at a dilution of 1:1,000. Protein bands were visualized using the ECL Plus detection system (Amersham Life Science, Arlington Heights, Ill.).
Reverse transcription-PCR (RT-PCR).
Total RNA was isolated using TRIzol reagent (Life Technologies, Inc.) as described above. The cDNA was synthesized from 2 μg of total RNA using Moloney murine leukemia virus reverse transcriptase and oligo(dT). For PCR, 5 μl of cDNA products was mixed with PCR buffer (Promega, Madison, Wis.) and 0.1 μM concentrations of human or mouse PCR primers synthesized according to the sequence in GenBank. The primer sequences utilized in these studies are listed in Table 1.
TABLE 1.
DNA primers
| GenBank no. | Gene name | Primers |
|---|---|---|
| U24173 | Mouse p21 | 5′-GTCCAATCCTGGTGATGTCC (forward) |
| 5′-CAGGGCAGAGGAAGTACTGG (reverse) | ||
| NM_009741 | Mouse Bcl-2 alpha | 5′-ATGATAACCGGGAGATCGTG (forward) |
| 5′-GACGGTAGCGACGAGAGAAG (reverse) | ||
| D87660 | Mouse 14-3-3 zeta | 5′-AGCAGGCAGAGCGATATGAT (forward) |
| 5′-CTTTCTGGTTGCGAAGCATT (reverse) | ||
| BC013718 | Mouse GADD153 | 5′-CAGAGTTCTATGGCCCAGGA (forward) |
| 5′-ATGGTGCTGGGTACACTTCC (reverse) | ||
| NM_008183 | Mouse glutathione S-transferase theta 2 | 5′-AGTTGGCCATGGTTTGCTAC (forward) |
| 5′-GCAAAGATTGGCTTGGAGAG (reverse) | ||
| D13866 | Mouse α-catenin | 5′-GTTTTGGCTGCATCTGTTGA (forward) |
| 5′-TTTGGCTGCCATAATGTTCA (reverse) | ||
| X51688 | Mouse cyclin A | 5′-CTGGACCCAGAAAACCATTG (forward) |
| 5′-CCTCTCAGCACTGACATGGA (reverse) | ||
| V00568 | Mouse c-Myc | 5′-CTCCTGGCAAAAGGTCAGAG (forward) |
| 5′-TTTCCGCAACAAGTCCTCTT (reverse) | ||
| X64713 | Mouse cyclin B1 | 5′-GGCTAACGGAAGTTGTCGAA (forward) |
| 5′-AGACTTGGGGGCAAATTCTT (reverse) | ||
| NM_008084 | Mouse GAPDH | 5′-GTGTTCCTACCCCCAATGTG (forward) |
| 5′-CTTGCTCAGTGTCCTTGCTG (reverse) | ||
| XM_034952 | Human 14-3-3 zeta | 5′-AAAAACGGAAGGTGCTGAGA (forward) |
| 5′-TGCTTGTTGTGACTGATCCA (reverse) | ||
| S40706 | Human GADD153 | 5′-AGCAGAGGTCACAAGCACCT (forward) |
| 5′-TTCATGCTTGGTGCAGATTC (reverse) | ||
| M13267 | Human CuZnSOD | 5′-GAAGGTGTGGGGAAGCATTA (forward) |
| 5′-ACCTTTGCCCAAGTCATCTG (reverse) | ||
| Y00985 | Human MnSOD | 5′-CCTGAACATCAACGAGGAGAAG (forward) |
| 5′-CTCCCAGTTGATTACATTCCAA (reverse) | ||
| X51688 | Human cyclin A | 5′-CTGGACCCAGAAAACCATTG (forward) |
| 5′-CCTCTCAGCACTGACATGGA (reverse) | ||
| NM_078467 | Human p21 | 5′-GCGACTGTGATGCGCTAAT (forward) |
| 5′-GGCGTTTGGAGTGGTAGAAA (reverse) | ||
| X00364 | Human c-Myc | 5′-CCAGCGAGGATATCTGGAAG (forward) |
| 5′-AGGTACAAGCTGGAGGTGGA (reverse) | ||
| M25753 | Human cyclin B1 | 5′-TGTGGATGCAGAAGATGGAG (forward) |
| 5′-AAACATGGCAGTGACACCAA (reverse) | ||
| AF261085 | Human GAPDH | 5′-ATCCCATCACCATCTTCCAG (forward) |
| 5′-GCCATCACGCCACAGTTTCC (reverse) |
Reporter transfection and luciferase assay.
Cells in 12-well plates were cotransfected with 0.3 μg of AP-1, NF-κB, or I2E luciferase reporters and 0.2 μg of β-galactosidase reporters. Cells were transfected for 3 h, and luciferase activity was measured at different times following radiation. For control of reporter transfection efficiency, an aliquot of the same cell lysate was used for measurement of β-galactosidase activity, and luciferase activity was normalized to β-galactosidase activity.
Northern blot analysis.
Nylon filters containing 20 μg of total RNA were hybridized with radioactively labeled oligonucleotides at 42°C for 20 h in 50% formamide, 5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 10× Denhardt's solution, 2% SDS, and 100 μg of denatured herring sperm DNA/ml. Filters were washed with 0.1× SSC, 0.1% SDS for 2 h at 50°C and processed for autoradiography. Gene expression levels were estimated by densitometry.
RESULTS
Radioresistance induced by MnSOD overexpression and FIR.
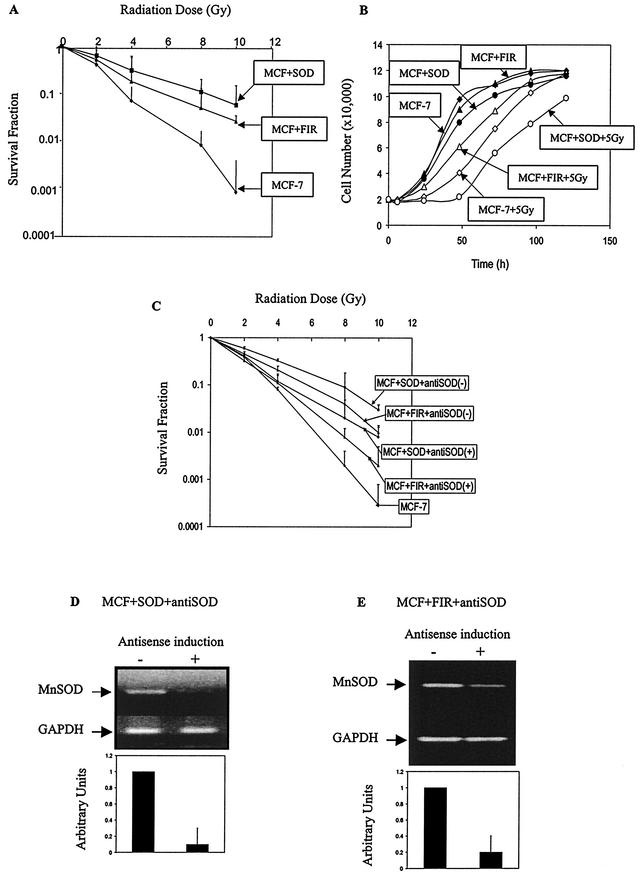
To determine the radioresistant phenotypes in MCF+FIR and MCF+SOD cells, clonogenic survival was determined, and the DMFs at 10% isosurvival were found to be 1.8 and 2.3 for MCF+FIR and MCF+SOD cells, respectively (Fig. 1A). These results are in agreement with previous reports of MnSOD-mediated radioprotection (50, 58). A longer (∼10-h) growth delay (following a single 5-Gy dose) was also observed when MCF-7 cells were compared to MCF+SOD cells (Fig. 1B). In contrast, MCF+FIR cells demonstrated a shorter (∼11-h) IR-induced growth delay relative to parental MCF-7 cells (Fig. 1B). These results clearly indicate that both MCF+SOD and MCF+FIR cells demonstrate a radioresistant phenotype as well as alterations in growth following single-dose radiation exposure, but they suggest that differences in the mechanisms governing growth delays exist.
FIG.1.
Radioresistance and MnSOD expression. (A) Radioresistance induced by FIR or MnSOD overexpression. Clonogenic survival of parental MCF-7 cells, MCF+FIR, and MCF+SOD cells was measured following exposure to γ-irradiation. Plating efficiencies for MCF-7, MCF+SOD, and MCF+FIR cells were 0.19, 0.08, and 0.28, respectively. Results were normalized to nonirradiated cells (data represent mean ± 1 standard deviation [SD] of three independently irradiated cultures, each plated into at least two cloning dishes that were counted). (B) Cell growth following IR (5 Gy) in MCF+FIR and MCF+SOD cells. Cell number was plotted as a function of time. (C) Conditional expression of antisense MnSOD sensitizes both MCF+SOD and MCF+FIR cells to radiation. Tet-inducible constructs containing a full-length antisense MnSOD cDNA were transfected into MCF+SOD (MCF+SOD+antiSOD) and MCF+FIR (MCF+FIR+antiSOD) cells, and stable transfectants were selected. Clonogenic survival following exposure to IR was measured with (+) or without (−) DOX induction of the antisense constructs (data represent mean ± 1 SD of three independently irradiated cultures, each plated into at least two cloning dishes that were counted). (D) Inhibition of MnSOD expression in MCF+SOD+antiSOD and MCF+FIR+antiSOD cells treated in the presence of DOX. MnSOD expression was measured with RT-PCR following 24 h with (+) or without (−) antisense-inducing conditions. Primers for GAPDH were included as internal controls (three experiments were performed, and a single representative analysis is shown). Densitometry analysis is shown in arbitrary units normalized to the GAPDH loading control (errors represent ±1 SD of three separate densitometric analyses done on the single representative set of samples shown above).
Conditional expression of antisense MnSOD sensitized both MCF+SOD and MCF+FIR cells to IR.
To determine if a causal relationship between MnSOD expression and radiosensitivity exists, MCF+SOD and MCF+FIR cells were transfected with MnSOD antisense constructs under the control of the Tet-on promoter (34). Antisense expression and decreases in MnSOD immunoreactive protein had been confirmed previously (34). Antisense MnSOD expression was induced 6 h before and during radiation, and clonogenic survival was determined. The results (Fig. 1C) show that expression of antisense MnSOD inhibited radioresistance, as shown by DMFs of 1.2 and 1.3 for MCF+FIR+antiSOD and MCF+SOD+antiSOD cells, respectively, at 10% isosurvival (relative to MCF-7). Figure 1D confirms reduced expression of MnSOD mRNA in both MCF+SOD+antiSOD and MCF+FIR+antiSOD cell lines under antisense-inducing conditions (25). These observations support the hypothesis that MnSOD expression regulates a portion of the radioresistant phenotype.
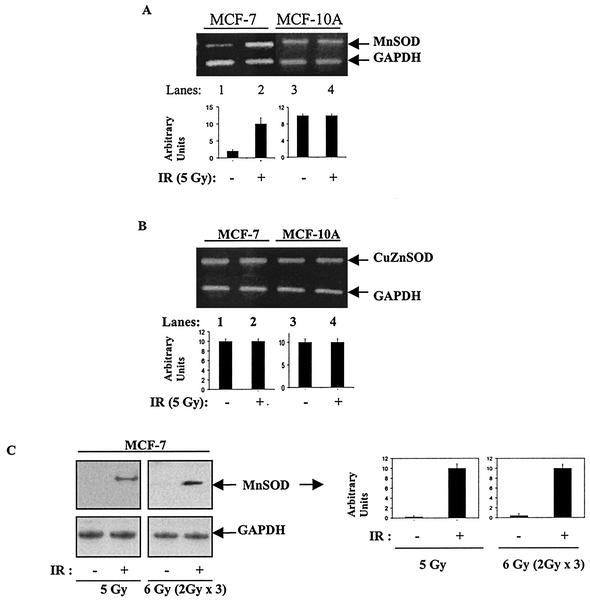
Endogenous MnSOD expression in wild-type MCF-7 cells exposed to IR.
By using the MnSOD/GAPDH ratio, Fig. 2A demonstrates that a single dose of radiation (5 Gy) induced about a fivefold increase in the steady-state level of MnSOD transcripts in wild-type MCF-7 cells but not in nonmalignant human breast epithelial cells (MCF-10A) that constitutively demonstrated a fivefold-greater level of endogenous MnSOD mRNA (Fig. 2A). In contrast, the cytosolic CuZnSOD, which is expressed at similar basal levels in both MCF-7 and MCF-10A cells, did not show any alteration following irradiation (Fig. 2B). Furthermore, MnSOD immunoreactive protein was induced significantly in MCF-7 cells by a single dose of 5 Gy or FIR (Fig. 2C). These results indicate that the induction of MnSOD by radiation is more pronounced in MCF-7 cells, relative to MCF-10A cells, and the induction appears specific for the mitochondrial SOD (MnSOD) versus the cytosolic SOD (CuZnSOD).
FIG. 2.
IR-induced MnSOD expression in MCF-7 cells. (A) IR induced MnSOD expression in MCF-7 but not in MCF-10A cells. MCF-7 and MCF-10A cells were irradiated with 5 Gy and harvested using trypsin 12 h after radiation (+) or sham treatment (−). MnSOD expression was detected by RT-PCR using 5 μg of total RNA. PCR fragments were enhanced for 23 cycles, and relative levels of MnSOD transcripts were estimated by densitometric analysis. (B) Expression of CuZnSOD was not changed in irradiated MCF-7 cells. CuZnSOD mRNA was measured by RT-PCR. PCR fragments were enhanced for 21 cycles with 5 μg of total RNA. The expression levels were estimated using densitometry. (C) Immunoreactive MnSOD protein induced by single or fractionated exposure to irradiation. Wild-type MCF-7 cells were irradiated at room temperature with a single dose of 5 Gy or three doses of 2 Gy FIR. Twenty micrograms of protein from sham-irradiated (−) and irradiated (+) MCF-7 cells 24 h after irradiation was run per lane, and immunoreactive protein levels were detected by Western blotting using rabbit antiserum to human MnSOD and GAPDH as the loading control. In all cases, the quantitation was done using densitometry and presented as arbitrary units after normalization to GAPDH (three experiments were done in each case and one representative image is shown; the errors represent ± 1 standard deviation of three separate densitometric analyses done on the single representative image).
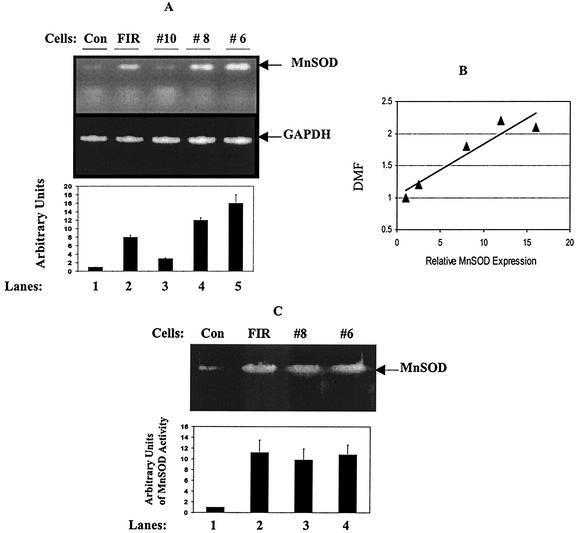
Increased MnSOD in radioresistant clones isolated from MCF+FIR cells.
Figure 3 shows that, compared to MCF-7 cells (Fig. 3A, lane 1), steady-state levels of MnSOD transcripts were increased in MCF+FIR cells (lane 2) as well as in two radioresistant clones (clones 8 and 6; lanes 4 and 5), but not in a relatively radiosensitive clone (clone 10; lane 3) isolated from the MCF+FIR population. The MnSOD mRNA levels were estimated by densitometry and normalized to GAPDH (Fig. 3A, lower panel), and radioresistance correlated to MnSOD expression levels (Fig. 3B) (linear regression r = 0.95; P < 0.05). Enzyme activity detected by native gel analysis (Fig. 3C) further showed that endogenous MnSOD activity was increased in the MCF+FIR cells (Fig. 3C, lane 2) and in radioresistant MCF+FIR clones (lanes 3 and 4). Since FIR could induce gene mutations and/or deletions leading to alterations in the activity of the MnSOD protein, the MnSOD cDNA sequence from MCF+FIR cells was determined. No mutations or deletions in the coding region were detected (data not shown). These results suggest that the endogenous MnSOD protein in MCF+FIR cells has normal structure and activity.
FIG. 3.
MnSOD expression in radioresistant clones of MCF+FIR cells. (A) Increased MnSOD expression in MCF+FIR cells and clones isolated from MCF+FIR cells. MnSOD expression levels in MCF-7 (Con; lane 1), MCF+FIR (FIR; lane 2), and three clones isolated from MCF+FIR cells demonstrating different radiosensitivities (DMF = 1.2, 2.2, and 2.1 for clone 10, 8, and 6, respectively; lanes 3 to 5) were measured by RT-PCR. Expression levels were estimated by densitometry and normalized to GAPDH (lower panel; data represent the mean ± 1 standard deviation [SD] of three RT-PCR measurements of a single set of samples, and a representative image is shown). (B) Correlation analysis of MnSOD expression and radioresistance. DMFs of wild-type MCF-7 cells, MCF+FIR cells, and three individual clones (6, 8, and 10) isolated from MCF+FIR cells were determined by clonogenic survival and correlated to the MnSOD/GAPDH mRNA ratio (linear regression r = 0.95; P < 0.05). (C) MnSOD activity gel analysis. MnSOD activity of MCF-7 cells (Con; lane 1), MCF+FIR cells (lane 2), and two radioresistant clones from MCF+FIR cells (clones 8 and 6; lanes 3 and 4) was measured by native gel assays (image represents one of two separate analyses). The MnSOD activity was estimated by densitometry and expressed in arbitrary units relative to the control (lower panel; data represent the mean ± 1 SD of three densitometric analyses of the single image that is shown).
Overlapping gene expression profiles in MCF+SOD and MCF+FIR cells.
To analyze common genes induced in both MCF+FIR and MCF+SOD cells, gene expression profiles were analyzed using Clontech Atlas human cancer cDNA expression cDNA microarrays for 1,176 genes as described previously (34). Since MnSOD overexpression clearly induced radioresistance in MCF-7 cells and ionizing radiation induced endogenous MnSOD as well as radioresistance in these same cells, the goal was to identify genes altered in MCF+SOD profiles that were required for radioresistance in MCF+FIR cells. Each gene was analyzed in two resistant cell lines (clone 8 from MCF+FIR and MCF+SOD). The results from MCF+FIR clone 8 were compared to those for revertant clone 10, and the results from MCF+SOD cells were compared to those from the parental MCF-7 cells. The genes found to be regulated in a similar fashion in both MCF+FIR clone 8 and MCF+SOD cells are listed in Tables 2 and 3. Sixteen genes demonstrated reduced expression (Table 2) and 32 genes demonstrated increased expression (Table 3) in both resistant phenotypes (grouped by gene functions), relative to control MCF-7 cells. A cluster of six genes that also contained NF-κB consensus sequences in their promoter regions was increased in both resistant phenotypes. These included p21, Myc, cyclin A, cyclin B1, 14-3-3 zeta, and GADD153. These six genes were chosen for further study because NF-κB has been found to be activated by exposure of human keratinocytes to FIR, and activation of NF-κB has been shown to mediate a portion of the resistant phenotype demonstrated by FIR-treated cells (13).
TABLE 2.
Down-regulated genes in both MCF+FIR and MCF+SOD cells
| GenBank no. | Gene name | Gene functiona | Foldb change in:
|
|
|---|---|---|---|---|
| MCF+FIR/control | MCF+SOD/MCF-7c | |||
| M32315 | TNF receptor 2 precursor | Apoptosis | 4.5 | 4.6 |
| U69611 | TNF-α converting enzyme | Apoptosis | 4.4 | 4.0 |
| Y09392 | WSL-LR (Apo-3) | Apoptosis | 3.9 | 3.0 |
| L13689 | DNA-binding protein BMI-1 | Signal transduction | 4.2 | 2.4 |
| X60188 | Extracellular signal-regulated kinase 1 | Signal transduction | 2.4 | 2.9 |
| M10901 | Glucocorticoid receptor alpha | Transcription factor | 3.6 | 3.5 |
| Q00421 | GA-binding protein beta-2 chain | Transcription factor | 2.0 | 3.5 |
| D12763 | ST2 protein precursor | Cytokine | 3.6 | 2.1 |
| M16961 | Alpha-2-HS-glycoprotein precursor | Insulin tyrosine kinase inhibitor | 4.9 | 4.3 |
| L10240 | Basin precursor | Collagenase | 4.3 | 5.2 |
| M24283 | Intercellular adhesion molecule 1 precursor | Cell adhesion | 3.5 | 3.2 |
| M14091 | Thyroxine-binding globulin (TBG) precursor | TBG function | 3.1 | 3.7 |
| M59911 | Integrin alpha-3 | Adhesion molecules | 2.2 | 3.5 |
| M96995 | Growth factor receptor-bound protein 2 | T4-binding globulin | 2.1 | 3.2 |
| X53795 | Inducible membrane protein R2; CD82 | Tumor suppressor antigen | 4.4 | 9.8 |
| L12693 | Cellular nucleic acid-binding protein | Unknown | 3.7 | 9.1 |
Gene function was updated by MedLine database searches.
Results represent the quantitation of one hybridization of two sets of fluorescence-labeled RNA probes, not including values with changes of less than twofold.
MCF+FIR profiles were obtained by normalizing gene expression levels of MCF+FIR radioresistant clone 8 to the level of the revertant MCF+FIR clone 10 (shown in Fig. 3A); MCF+SOD profiles were obtained by normalizing expression levels of the MCF+SOD cell line to the level in wild-type MCF-7 cells.
TABLE 3.
Up-regulated genes in both MCF+FIR and MCF+SOD cells
| GenBank no. | Gene name | Functiona | Foldb change in:
|
|
|---|---|---|---|---|
| MCF+FIR/controlc | MCF+SOD/MCF-7c | |||
| L25081 | Transforming protein RHOC | Apoptosis, proliferation, carcinogenesis | 5.8 | 3.2 |
| M86400 | 14-3-3 zeta | Apoptosis | 5.1 | 3.2 |
| M14745 | BCL-2-alpha | Apoptosis | 3.1 | 3.3 |
| Q99537 | BRCA1, RHO7, and VATI | Apoptosis | 2.2 | 3.4 |
| AI376462 | Lupus KU autoantigen protein P86 | Apoptosis, radioresistance | 2.0 | 6.4 |
| V00568 | MYC proto-oncogene protein | Antiapoptosis | 5.5 | 3.3 |
| S40706 | GADD 153 (growth arrest and DNA-damage-inducible protein) | DNA damage response | 5.4 | 9.1 |
| Cell cycle control | ||||
| U09579 | p21 (waf1, CDKNIA) | Cell cycle regulator | 4.6 | 5.5 |
| M25753 | Cyclin B1 | Cell cycle control | 3.3 | 2.1 |
| X51688 | Cyclin A | Cell cycle control | 3.3 | 6.4 |
| X12530 | B-lymphocyte antigen CD20 | Cell cycle antigen | 3.5 | 7.7 |
| Y00371 | Heat-shock cognate 71-kDa protein | Stress response | 8.3 | 3.6 |
| M14091 | Thyroxin-binding globulin precursor | Stress response | 3.4 | 2.5 |
| L38503 | Glutathione S-transferase theta 2 | Stress response | 3.2 | 6.2 |
| Q64347 | Zinc finger protein 101 | Transcription activation | 4.2 | 2.1 |
| X66899 | RNA-binding protein EWS | Transcription activation | 3.4 | 5.5 |
| G29507 | General transcription factor | Transcription factor | 4.1 | 3.8 |
| M31523 | Transcription factor 3 | Transcription factor | 3.2 | 6.3 |
| Q00421 | GA-binding protein beta-2 chain | Transcription factor | 3.2 | 12.2 |
| M55422 | Krueppel-related zinc finger protein | Nuclear receptors | 8.3 | 5.6 |
| S72008 | CDC10 protein homolog | Growth regulator | 4.4 | 6.2 |
| M63488 | Replication protein A | DNA replication | 4.4 | 8.8 |
| J04088 | DNA topoisomerase II alpha isozyme | DNA replication | 4.4 | 8.5 |
| X06745 | RNA1 DNA polymerase alpha | DNA synthesis | 3.9 | 4.2 |
| U57342 | Myeloid leukemia factor 2 | Cell differentiation | 18.9 | 8.9 |
| M27396 | Asparagine synthetase | Cell proliferation | 6.3 | 7.3 |
| AA629900 | NAD-dependent methylenetetrahydrofolate dehydrogenase | Cell proliferation | 5.3 | 3.7 |
| H38136 | TGF-βd-inducible early growth response 2 | Cell proliferation | 3.2 | 4.5 |
| D13748 | Eukaryotic initiation factor 4A-I | Cell proliferation | 4.1 | 7.7 |
| L07594 | TGF-β receptor type III precursor | Cell proliferation, differentiation | 3.3 | 7.4 |
| X60221 | ATP synthase B chain, mitochondrial precursor | ATP synthesis | 3.2 | 6.4 |
| AF026816 | Putative oncogene protein | Oncogene | 12.4 | 6.4 |
Gene function was updated by MedLine database searches.
Results represent the quantitation of one hybridization of two sets of fluorescence-labeled RNA probes, not including values with changes of less than twofold.
MCF+FIR profiles were obtained by normalizing gene expression levels of MCF+FIR radioresistant clone 8 to the level of the revertant MCF+FIR clone 10; MCF+SOD profiles were obtained by normalizing expression levels of the MCF+SOD cell line to the level of wild-type MCF-7 cells.
TGF-β, transforming growth factor β.
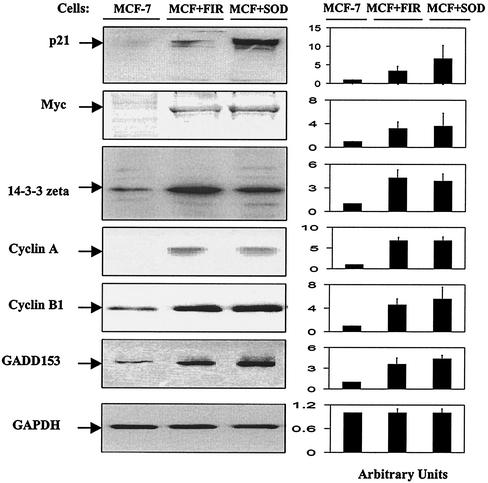
To confirm the DNA microarray results, expression of immunoreactive proteins corresponding to the six genes containing NF-κB binding sites in their gene promoters and up-regulated in both MCF+FIR clone 8 and MCF+SOD cells was analyzed. Western blotting showed that immunoreactive proteins corresponding to these proteins increased in both MCF+FIR and MCF+SOD cells (Fig. 4). Compared to basal levels in wild-type MCF-7 cells, immunoreactive protein corresponding to p21, c-Myc, 14-3-3- zeta, cyclin A, cyclin B1, and GADD153 showed the largest increases in the radioresistant cells (2- to 6-fold), and 14-3-3 zeta was increased to a more modest extent (1.5- to 2.0-fold). These results show that the six genes found to be up-regulated in the microarray analysis were also increased at the level of immunoreactive protein.
FIG. 4.
Western blotting for p21, Myc, 14-3-3 zeta, cyclin A, cyclin B1, and GADD153. Immunoreactive protein levels for p21, Myc, 14-3-3 zeta, cyclin A, cyclin B1, and GADD153 were analyzed by immunoblotting. Aliquots of protein (20 μg/lane) from MCF-7, MCF+SOD, and MCF+FIR cells were separated using SDS-12.5% polyacrylamide gel electrophoresis and transferred to membranes, and membranes were incubated with antibodies and visualized using an ECL Plus detection system with GAPDH as the loading control. (The blots were run with three independently harvested sets of samples, and a single representative set of samples probed with the different antibodies is shown.) Expression levels on the blots that are shown were determined by densitometry and normalized to control MCF-7 cells (right panel; the data represent the mean ± 1 standard deviation of three densitometric analyses of the single set of blots which are shown).
Radiation inducibility of p21, Myc, 14-3-3 zeta, cyclin A, cyclin B1, and GADD153 in fibroblasts from Sod2 −/− mice.
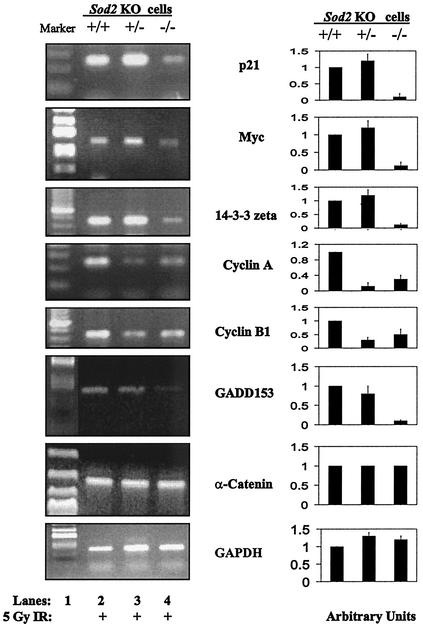
To further confirm the causal link between alterations in MnSOD expression and regulation of signaling elements following radiation, expression was analyzed in fibroblasts isolated from Sod2 (MnSOD) knockout mice with the Sod2 genotypes −/−, +/−, and +/+. These cells demonstrated the expected levels of MnSOD expression, with the −/− animals lacking MnSOD and the +/− animals having approximately 50% of control levels of MnSOD mRNA (26). Mouse PCR primers for p21, c-Myc, 14-3-3 zeta, cyclin A, cyclin B1, and GADD153 were synthesized, and mouse GAPDH and α-catenin were included as controls. Levels of transcripts corresponding to α-catenin and GAPDH were detected but not altered in any of the Sod2 knockout variants. In contrast, the transcripts of all six genes of interest were reduced (60 to 90%) in Sod2 −/− cells, relative to Sod2+/+ cells 24 h following 5-Gy irradiation (Fig. 5). In the case of the Sod2 heterozygotes (+/−), cyclin A and cyclin B1 were reduced relative to levels in the +/+ animals following radiation. Without radiation, all genes were hardly detectable by RT-PCR (data not shown). These results strongly suggest that this group of stress-responsive genes does not respond to IR in cells lacking MnSOD, which supports the hypothesis that MnSOD expression was in part responsible for induction of radiation-responsive genes, including p21, GADD153, cyclin A, cyclin B1, c-Myc, and 14-3-3 zeta.
FIG. 5.
Reduced expression of radiation-responsive genes in fibroblasts derived from Sod2 (MnSOD) knockout mice. Expression of p21, Myc, 14-3-3 zeta, cyclin A, cyclin B1, and GADD153 as well as two control genes (α-catenin and GAPDH) was measured by RT-PCR 24 h following 5-Gy irradiation in fibroblasts from wild-type (+/+), homozygous (−/−), and heterozygous (+/−) Sod2 knockout mice. PCR primers were synthesized according to sequence data from GenBank. Expression levels were measured using densitometry and normalized to +/+ cells (right panel; the data represent the mean ± 1 standard deviation of three densitometric analyses of the single set of PCR analyses which are shown).
Down-regulation of p21, c-Myc, 14-3-3 zeta, cyclin A, cyclin B1, and GADD153 and radiosensitization in MCF+FIR cells following conditional expression of antisense MnSOD.
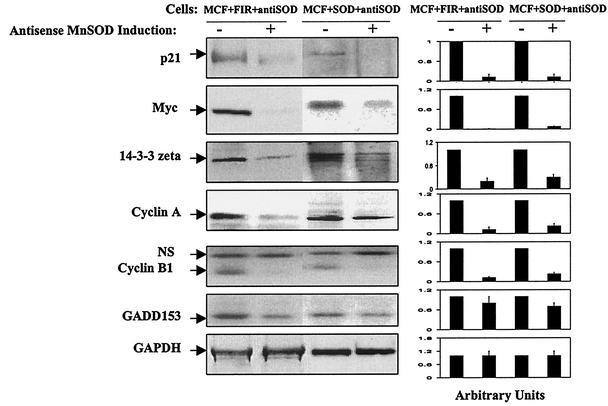
The causal relationship between MnSOD overexpression and expression of p21, c-Myc, 14-3-3 zeta, cyclin A, cyclin B1, and GADD153 in radioresistant cells was assessed by conditional overexpression of antisense MnSOD using the same cell lines and conditions shown in Fig. 1. In general, levels of immunoreactive protein corresponding to these genes were significantly reduced (70 to 90%) under enforced expression of antisense MnSOD in both MCF+FIR and MCF+SOD cells (Fig. 6). Compared to the immunoreactive protein levels found under non-antisense-inducing conditions, p21, c-Myc, 14-3-3 zeta, cyclin A, cyclin B1, and GADD153 were reduced 80 to 95% in MCF+FIR+antiSOD and MCF+SOD+antiSOD cells under antisense-inducing conditions (Fig. 6). These results further support the hypothesis that key signaling elements such as 14-3-3 zeta, cyclin A, and cyclin B1 are up-regulated in MCF+FIR cells due to MnSOD expression.
FIG. 6.
Down-regulation of radiation-responsive genes by conditional expression of antisense MnSOD in MCF+FIR and MCF+SOD cells. Immunoreactive protein corresponding to p21, Myc, 14-3-3 zeta, cyclin A, cyclin B1, and GADD153 was monitored in controls and 12 h after antisense-inducing conditions in MCF+SOD+antiSOD and MCF+FIR+antiSOD cells (NS = nonspecific band). GAPDH was included as the loading control, and each experiment was done three times with one representative blot, using each antibody shown. Expression levels were measured using densitometry and normalized to the levels seen in the absence of DOX (right panel; the data represent the mean ± 1 standard deviation of three densitometric analyses of the single set of blots which are shown).
Inhibition of MnSOD expression in MCF+FIR cells by mutant IκB.
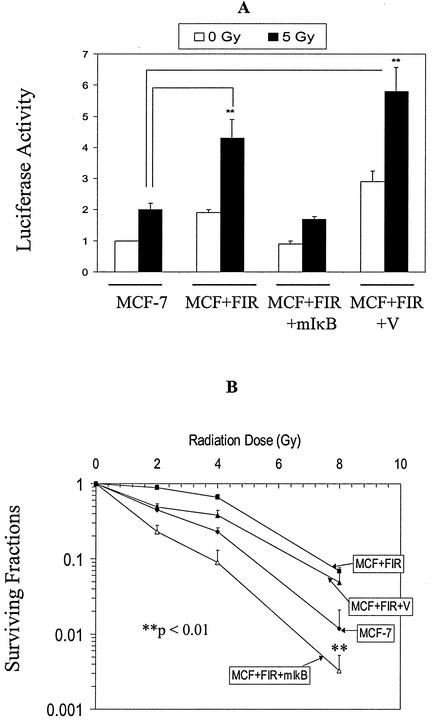
NF-κB is a well-documented redox-sensitive transcription factor that is regulated by IκB. Figure 7 shows that the mutant IκB transfectants (MCF+FIR+mIκB) were radiosensitized, relative to MCF+FIR cells, and that NF-κB-driven reporter gene activity was also suppressed in the cells exposed to 5 Gy. These results strongly suggest that NF-κB-mediated transcriptional activity could contribute to the radioresistant phenotype seen in MCF+FIR cells.
FIG. 7.
Inhibition of NF-κB by mutant IκB sensitizes MCF+FIR cells to radiation. (A) Basal and 5-Gy-induced NF-κB-driven luciferase reporter activity was inhibited in MCF+FIR cells by mutant IκB transfection. A luciferase reporter containing NF-κB sites was cotransfected with a β-galactosidase reporter into MCF-7, MCF+FIR, MCF+FIR+mIkB, and vector control (MCF+FIR+V) cells, and luciferase activity was measured at 24 h after 5 Gy of IR (data represent mean ± 1 standard deviation of three separate experiments. **, irradiated groups that are significantly different from irradiated MCF-7 results (paired Student's t test, P < 0.05). (B) Radiosensitivity was increased in MCF+FIR+mIκB cells. Parental MCF-7, MCF+FIR, and MCF+FIR+mIκB cells and MCF+FIR+V cells were exposed to a range of IR doses, and clonogenic survival was determined. The surviving fraction was normalized to nonirradiated cells from each group (data represent mean ± 1 standard deviation of three separate experiments) **, the irradiated group, MCF+FIR+mIκB, is significantly different from the irradiated control, MCF+FIR+V, at the 8-Gy dose (paired Student's t test, P < 0.01).
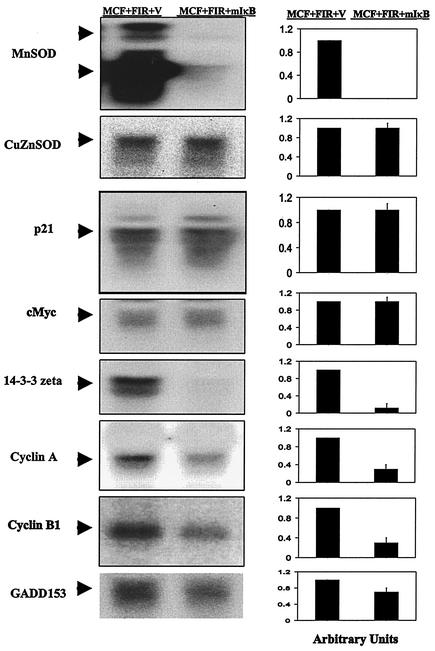
Down-regulation of MnSOD-responsive genes in MCF+FIR+mIκB cells.
Figure 8 demonstrates that mutant IκB expression dramatically inhibited the increased steady-state levels of MnSOD RNA in MCF+FIR cells, relative to the control, without affecting CuZnSOD mRNA levels. In addition, the increased expression of 14-3-3 zeta, cyclin A, cyclin B1, and GADD153 was inhibited in MCF+FIR+mIκB cells (Fig. 8) relative to the vector control. These results together with the data shown in Fig. 4 to 6 strongly suggest that at least a portion of the signaling elements involved in cell cycle control and antiapoptosis genes in MCF+FIR cells are very sensitive to MnSOD expression, which in turn is controlled by NF-κB-mediated transcription.
FIG. 8.
Down-regulation of MnSOD-responsive genes in MCF+FIR+mIκB cells. Transcription of 14-3-3 zeta, cyclin A, cyclin B1, GADD153, p21, and c-Myc as well as MnSOD and CuZnSOD were analyzed by Northern blotting of total RNA isolated from MCF+FIR+V and MCF+FIR+mIκB cells. Oligo probes were synthesized using RT-PCR with the human primers listed in Table 1. The MnSOD and 14-3-3 zeta lanes were overexposed to show the mRNA levels in the cells containing the mutant IκB construct. The blots represent one set of representative results obtained from three experiments. The right panel data represent the mean ± 1 standard deviation of three densitometric analyses of the single set of blots that is shown.
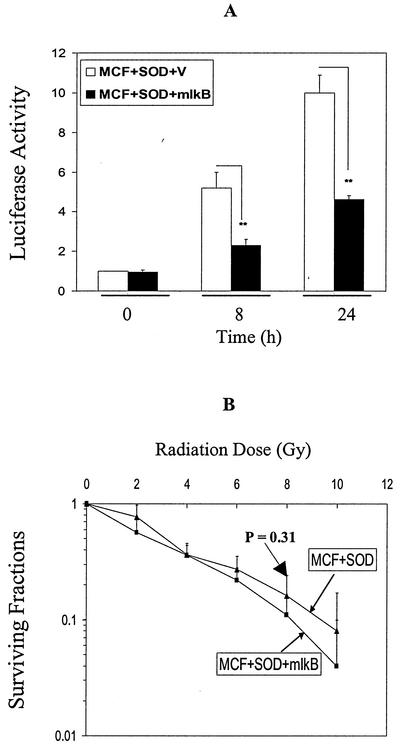
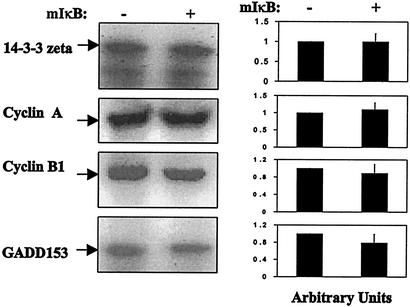
Inhibition of NF-κB by mutant IκB does not sensitize MCF+SOD cells to radiation.
The MCF+SOD cell line overexpresses MnSOD that is not under the control of NF-κB, but still demonstrates a radioresistant phenotype (Fig. 1). To determine if MnSOD-mediated radioresistance seen in MCF+SOD cells was under the control of NF-κB, MCF+SOD cells were transfected with mutant IκB. Luciferase reporter activity controlled by the human SOD2 gene did show inhibition due to mutant IκB transfection in the MCF+SOD+mIκB cells (Fig. 9A), demonstrating that the mutant IκB was capable of inhibiting radiation-induced activation of an NF-κB-containing reporter construct from the SOD2 gene. In addition, there was no significant reduction in clonogenic survival in MCF+SOD+mIκB versus MCF+SOD+V cells (Fig. 9B). Figure 10 further indicates that expression of 14-3-3 zeta, cyclin A, cyclin B1, and GADD153 was not inhibited in MCF+SOD cells overexpressing mutant IκB. These results demonstrate that when MnSOD is overexpressed in an NF-κB-independent fashion, radioresistance and the expression of 14-3-3 zeta, cyclin A, cyclin B, and GADD153 are unaffected by mutant IκB overexpression. These results, combined with the results in Fig. 1, 7, and 8, support the hypothesis that MnSOD regulates genes that govern radioresistance and, in MCF+FIR cells, NF-κB regulates the expression of MnSOD.
FIG. 9.
Inhibition of NF-κB by mutant IκB did not sensitize MCF+SOD cells to radiation. (A) The activity of the I2E luciferase reporter that contains the intronic enhancer element from the SOD2 gene was inhibited in irradiated MCF+SOD+mIκB cells. Five grays induced I2E (containing an NF-κB site) luciferase reporter activity at 8 and 24 h following IR, and this IR-induced activation was inhibited in MCF+SOD cells by mutant IκB transfection. I2E reporters were cotransfected with the β-galactosidase reporter into MCF+SOD+mIκB and vector control MCF+SOD+V cells, and luciferase activity was measured at 0 (without IR), 8, and 24 h after 5-Gy IR (data represent mean ± 1 standard deviation [SD] of five separate experiments). **, irradiated groups that are significantly different from their irradiated vector control (paired Student's t test, P < 0.05). (B) MCF+SOD+mIκB cells were not radiosensitized. MCF+SOD+mIκB and MCF+SOD+V cells were exposed to a range of IR doses, and clonogenic survival was obtained after IR treatments. Surviving fractions were normalized to nonirradiated cells (data represent mean ± 1 SD of three independently irradiated cultures, each plated into at least two cloning dishes that were counted; P > 0.05, paired t test).
FIG. 10.
MnSOD-responsive genes were not inhibited in MCF+SOD+mIκB cells. Transcription of 14-3-3 zeta, cyclin A, cyclin B1, and GADD153 was analyzed by Northern blotting with RNA isolated from MCF+SOD+V and MCF+SOD+mIκB cells. The data that are shown are one representative image from one set of samples that was run three times. The data in the right panel represent the mean ± 1 standard deviation of three densitometric analyses of the single set of blots that is shown.
DISCUSSION
The present study provides the first evidence that a group of signaling proteins in the expression profile of radioresistant MCF-7 cells are regulated as a result of alterations in MnSOD expression. Indirect evidence in support of this conclusion includes the fact that MCF-7 cells treated with FIR or overexpressing MnSOD showed significant radioresistance, and endogenous MnSOD was induced in irradiated wild-type MCF-7 cells as well as in radioresistant MCF+FIR populations. Further support was obtained when expression of stress proteins induced in both MCF+FIR and MCF+SOD cells was also found to be reduced in cells from Sod2 knockout mice following irradiation. A causal relationship between MnSOD expression, increased radiation resistance, and increased expression of stress-responsive genes (such as cyclin B1, cyclin A, GADD153, and 14-3-3 zeta) was further suggested by experiments showing that conditional expression of MnSOD antisense mRNA down-regulated the stress-responsive genes and significantly radiosensitized both MCF+FIR and MCF+SOD cells. In addition, inhibition of NF-κB-mediated transcriptional activity with mutant IκB radiosensitized MCF+FIR cells as well as reducing the transcription of MnSOD, 14-3-3 zeta, cyclin B1, cyclin A, and GADD153. Finally, expression of IκB in MCF+SOD cells where MnSOD was not controlled by NF-κB failed to induce radiosensitization or alter expression of the stress-responsive genes. Thus, a pathway of NF-κB → MnSOD → stress effector genes is proposed for radiation-induced adaptive tolerance in MCF-7 cells exposed to FIR.
Identifying gene targets for radiosensitization and/or chemosensitization is an important strategy in improving anticancer treatments (14, 23, 38). Although the exact roles of many IR-responsive genes remain unclear (3, 4, 29), most are believed to participate in cell cycle regulation, apoptotic responses, and DNA repair (2, 36). Amundson et al. reported an alteration rate of 3.87% (48 of 1,238 genes) in a DNA microarray analysis in MCF-7 cells following single-dose IR (3). The present report, using comparative microarray analysis of gene expression profiles between MCF-7, MCF+FIR, and MCF+SOD cells, showed alterations in the expression of 4.6% of genes (54 of 1,176) that overlapped both IR-resistant phenotypes. The number of alterations in gene expression seen in both radioresistant phenotypes (MCF+FIR and MCF+SOD, relative to MCF-7) appears to be similar to those seen following a single IR exposure (3).
The present results provide evidence for MnSOD expression participating in FIR-induced radioresistance. The present results also demonstrate that MnSOD expression is induced by single or multiple doses of IR (Fig. 2 and 3). In contrast, IR did not alter the expression of the cytoplasmic form of superoxide dismutase (CuZnSOD) (Fig. 2 and 8). Although the mechanism by which MnSOD impacts FIR-induced adaptive responses is currently unknown, MnSOD overexpression has been suggested by other studies to alter the intracellular redox environment via changes in mitochondrial hydroperoxide production (33), initiating downstream signaling cascades. Lack of IR-responsive genes in Sod2−/+ and Sod2−/− mouse embryo fibroblasts (Fig. 5) and inhibition of IR-responsive gene expression by antisense MnSOD further support the notion that MnSOD may alter radioresistance via gene regulation. However, MnSOD-mediated induction of radioresistance may be species and cell line dependent, which is evidenced by studies using CHO cells that overexpress MnSOD activity and demonstrate no alterations in radiosensitivity as determined by clonogenic survival (data not shown).
The stress-responsive transcription factor NF-κB is believed to play a key role in IR-induced adaptive responses and, thus, may have possible applications in cancer therapy. However, inhibition of NF-κB by overexpressing IκB did not affect radiosensitivity in PC3 prostate cancer cells and HD-MyZ Hodgkin's lymphoma cells (43), but it did increase the radiosensitivity of HeLa cells (11) and virus-transformed human keratinocytes (13). This apparent paradox may be due to the fact that a variety of signaling functions regulated by these transcription factors are dependent upon the metabolic state of these different cell lines at the time of treatment. Recent results have demonstrated that pro- and antiapoptotic responses can be activated by NF-κB-mediated MnSOD expression (10). Therefore, key genes regulated by these transcription factors that might impact the metabolic state of the cell need to be identified.
Most transcription factors are known to be redox sensitive (24, 28), and several redox-sensitive transcription factors are believed to contribute to MnSOD expression (30, 37). Levels of MnSOD also appear to regulate the activity of several transcription factors, including the basal DNA-binding activity and transcriptional activation caused by NF-κB and AP-1, but not SP-1 (8). Sequence analysis has confirmed the binding site for NF-κB in the promoter region of MnSOD (56) and in the control region of the second intron of SOD2 (61). Because IR activates NF-κB (25) and NF-κB is actively involved in expression of MnSOD (27, 52, 61), the present study results suggest that FIR-induced MnSOD expression is mediated via NF-κB activation. This is evidenced by the fact that IR-induced luciferase activity (controlled by the element in the second intron of the human SOD2 gene [61]) and radioresistance was inhibited in MCF+FIR cells transfected with mutant IκB (Fig. 7 and 8). Additionally, NF-κB activation is suggested to be involved in antiapoptotic pathways activated following exposure to anticancer agents (16, 54). Inhibition of NF-κB DNA-binding activity by indomethacin increases radiosensitivity (11), and mutant IκB partially reversed the radioresistant phenotype of human keratinocytes (13). Together, these results support the hypothesis that radiation-induced NF-κB contributes to the increased expression of MnSOD, which in turn alters signaling pathways leading to the activation of radioresistance genes. This signaling loop in irradiated cells may be important to understanding the variety of radiation-induced adaptive responses that have been observed. The present results, indicating that 14-3-3 zeta, cyclin A, cyclin B1, and GADD153 are potentially regulated by NF-κB through its induction of MnSOD following FIR, further narrow the candidate genes in the signaling network(s) leading to radiation-induced adaptive responses.
Many signaling cascades are believed to contain redundancies in the pathways leading to the induction of an adaptive response(s) to radiation and oxidative stress. In addition, H2O2 represents the product of MnSOD catalytic activity and may act as a signaling molecule in a variety of biological responses (51). Currently, genes signaling radiation-induced adaptive responses have been grouped into three general categories: arrest of cell cycle progression, DNA repair, and apoptosis. Proteins with signaling functions, including Raf-1, p21, GADD45, 14-3-3, Bax, Fas/APO1, KILLER/DR5, PIG3, Tsp1, and IGF-BP3 (5), are among the genes represented in radiation-induced gene expression profiles. Microarray analysis of the expression profiles in MCF+FIR and MCF+SOD cells demonstrated a group of stress-responsive genes overexpressed in both cell lines. These results, coupled with immunoblot analysis results, demonstrated overexpression of at least six genes (p21, Myc, 14-3-3 zeta, cyclin A, cyclin B1, and GADD153) that appear to be responsive to MnSOD expression. Endogenous MnSOD was strikingly induced in MCF+FIR cells and, in addition, mutant IκB did not affect the MnSOD-responsive genes in MCF+SOD cells where MnSOD was not controlled by NF-κB. Therefore, these results support the hypothesis that a subset of IR-induced stress-responsive genes might be the result of NF-κB-induced MnSOD expression leading to alterations in the intracellular redox environment and activation of transcription factors that control the stress-responsive signaling pathways. This speculation is supported by recent results suggesting that MnSOD induces expression of matrix metalloproteinase-2 via alterations in intracellular ROS metabolism (67).
The stress-signaling proteins responsive to MnSOD expression have been correlated to the signaling pathways governing radiation responses and radioresistance. We have shown that enforced expression of antisense cyclin B1 in MCF+FIR cells significantly increases radiosensitization (35). Cyclin B1 was found to be up-regulated in both MCF+FIR and MCF+SOD cells (Table 3), and the protein level was down-regulated following antisense MnSOD expression in both MCF+FIR and MCF+SOD cells during the same time that radiosensitization was occurring (Fig. 1 and 6). Therefore, we hypothesize that cyclin B1 may also play a specific role in radioresistance that is dependent upon the expression of MnSOD.
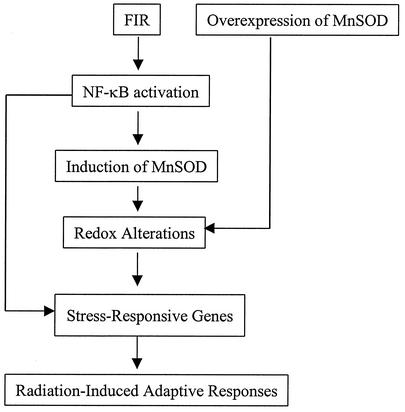
Another significant protein that was found to be regulated by MnSOD expression in the present study is 14-3-3 zeta (Fig. 8). 14-3-3 zeta was found to be down-regulated in MCF+FIR cells by antisense MnSOD and mutant IκB. 14-3-3 proteins are phosphoserine-binding molecules (62) with a primary function of inhibiting apoptosis (12, 60, 65). Recently, c-Rel/NF-κB-controlled MnSOD was found to play a key role in antiapoptosis (10), and E2F1 as well as c-Myc appear to potentiate apoptosis through inhibition of NF-κB activity that facilitates MnSOD-mediated ROS elimination (53). Therefore, our present data together with previous results support the hypothesis that the mitochondrial AE, MnSOD, may function to regulate specific stress-responsive genes, especially in tumor cells that acquire radioresistance following FIR. A pathway leading from NF-κB to MnSOD to effector genes (with antiapoptotic functions) is therefore a possible contributor to radiation-induced adaptive responses (shown schematically in Fig. 11).
FIG. 11.
Hypothesized model of stress-responsive gene regulation activated by NF-κB and MnSOD in MCF+FIR cells. FIR is hypothesized to induce NF-κB activation that up-regulates the expression of endogenous SOD2 (MnSOD), which in turn up-regulates gene expression associated with acquisition of resistance. Among the FIR-induced genes (some directly controlled by NF-κB), a group of cell cycle and antiapoptotic proteins appear to be sensitive to MnSOD expression and could potentially represent some of the genes participating in the signaling network leading to radiation-induced cellular adaptive responses.
In conclusion, a subset of signaling proteins from FIR-induced gene expression profiles have been shown to be regulated via MnSOD activation. Constitutive increases in these genes were confirmed in both radioresistant MCF+FIR and MCF+SOD cells, and the expression of these genes was inhibited in Sod2 knockout cells and in cell lines that conditionally overexpressed antisense MnSOD. Blocking NF-κB-mediated transcription by using mutant IκB transfection produced a similar inhibition of MnSOD and MnSOD-responsive genes in MCF+FIR cells. Thus, activation of the pathway involving NF-κB and MnSOD leading to increased expression of effector genes appears to be an important signaling network in radiation-induced adaptive responses.
Acknowledgments
We thank William Dewey at the University of California San Francisco for insightful discussion, Daret St. Clair for providing EI2 luciferase reporters and suggestions, Shiuan Chen for providing immortalized MCF-10A cells, Tom LeBon for critical reading of the manuscript, and Vicki Boore and Kellie Bodeker for manuscript preparation.
This work was supported by an International Union Against Cancer Fellowship (G.G.), a Beckman Fellowship of the Beckman Research Institute (T.W.), the City of Hope Graduate School Research Program (J.O.), an Intramural Research Award from the Beckman Research Institute of City of Hope (J.J.L.), NIH P01 CA66081 (L.W.O.), NIH RO1 HL51469 (D.R.S.), and NIH T32CA78586 (C.W.).
REFERENCES
- 1.Adler, V., Z. Yin, K. D. Tew, and Z. Ronai. 1999. Role of redox potential and reactive oxygen species in stress signaling. Oncogene 18:6104-6111. [DOI] [PubMed] [Google Scholar]
- 2.Almasan, A. 2000. Cellular commitment to radiation-induced apoptosis. Radiat. Res. 153:347-350. [DOI] [PubMed] [Google Scholar]
- 3.Amundson, S. A., M. Bittner, Y. Chen, J. Trent, P. Meltzer, and A. J. Fornace, Jr. 1999. Fluorescent cDNA microarray hybridization reveals complexity and heterogeneity of cellular genotoxic stress responses. Oncogene 18:3666-3672. [DOI] [PubMed] [Google Scholar]
- 4.Amundson, S. A., K. T. Do, and A. J. Fornace, Jr. 1999. Induction of stress genes by low doses of gamma rays. Radiat. Res. 152:225-231. [PubMed] [Google Scholar]
- 5.Amundson, S. A., T. G. Myers, and A. J. Fornace, Jr. 1998. Roles for p53 in growth arrest and apoptosis: putting on the brakes after genotoxic stress. Oncogene 17:3287-3299. [DOI] [PubMed] [Google Scholar]
- 6.Baeuerle, P. A., and D. Baltimore. 1996. NF-κB: ten years after. Cell 87:13-20. [DOI] [PubMed] [Google Scholar]
- 7.Barkett, M., and T. D. Gilmore. 1999. Control of apoptosis by Rel/NF-kappaB transcription factors. Oncogene 18:6910-6924. [DOI] [PubMed] [Google Scholar]
- 8.Basu, S., K. R. Rosenzweig, M. Youmell, and B. D. Price. 1998. The DNA-dependent protein kinase participates in the activation of NF kappa B following DNA damage. Biochem. Biophys. Res. Commun. 247:79-83. [DOI] [PubMed] [Google Scholar]
- 9.Beauchamp, C., and I. Fridovich. 1971. Superoxide dismutase: improved assays and an assay applicable to polyacrylamide gel. Anal. Biochem. 44:276-287. [DOI] [PubMed] [Google Scholar]
- 10.Bernard, D., D. Monte, B. Vandenbunder, and C. Abbadie. 2002. The c-Rel transcription factor can both induce and inhibit apoptosis in the same cells via the upregulation of MnSOD. Oncogene 21:4392-4402. [DOI] [PubMed] [Google Scholar]
- 11.Bradbury, C. M., S. Markovina, S. J. Wei, L. M. Rene, I. Zoberi, N. Horikoshi, and D. Gius. 2001. Indomethacin-induced radiosensitization and inhibition of ionizing radiation-induced NF-kappaB activation in HeLa cells occur via a mechanism involving p38 MAP kinase. Cancer Res. 61:7689-7696. [PubMed] [Google Scholar]
- 12.Brunet, A., A. Bonni, M. J. Zigmond, M. Z. Lin, P. Juo, L. S. Hu, M. J. Anderson, K. C. Arden, J. Blenis, and M. E. Greenberg. 1999. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96:857-868. [DOI] [PubMed] [Google Scholar]
- 13.Chen, X., B. Shen, L. Xia, A. Khaletzkiy, D. Chu, J. Y. Wong, and J. J. Li. 2002. Activation of nuclear factor kappaB in radioresistance of TP53-inactive human keratinocytes. Cancer Res. 62:1213-1221. [PubMed] [Google Scholar]
- 14.Coleman, C. N., and M. A. Stevenson. 1996. Biologic basis for radiation oncology. Oncology 10:399-411. [PubMed] [Google Scholar]
- 15.Delhalle, S., V. Deregowski, V. Benoit, M. P. Merville, and V. Bours. 2002. NF-kappaB-dependent MnSOD expression protects adenocarcinoma cells from TNF-alpha-induced apoptosis. Oncogene 21:3917-3924. [DOI] [PubMed] [Google Scholar]
- 16.De Smaele, E., F. Zazzeroni, S. Papa, D. U. Nguyen, R. Jin, J. Jones, R. Cong, and G. Franzoso. 2001. Induction of gadd45beta by NF-kappaB downregulates pro-apoptotic JNK signalling. Nature 414:308-313. [DOI] [PubMed] [Google Scholar]
- 17.Duttaroy, A., T. Parkes, P. Emtage, K. Kirby, G. L. Boulianne, X. Wang, A. J. Hilliker, and J. P. Phillips. 1997. The manganese superoxide dismutase gene of Drosophila: structure, expression, and evidence for regulation by MAP kinase. DNA Cell Biol. 16:391-399. [DOI] [PubMed] [Google Scholar]
- 18.Epperly, M. W., S. Defilippi, C. Sikora, J. Gretton, A. Kalend, and J. S. Greenberger. 2000. Intratracheal injection of manganese superoxide dismutase (MnSOD) plasmid/liposomes protects normal lung but not orthotopic tumors from irradiation. Gene Ther. 7:1011-1018. [DOI] [PubMed] [Google Scholar]
- 19.Epperly, M. W., J. A. Gretton, S. J. DeFilippi, J. S. Greenberger, C. A. Sikora, D. Liggitt, and G. Koe. 2001. Modulation of radiation-induced cytokine elevation associated with esophagitis and esophageal stricture by manganese superoxide dismutase-plasmid/liposome (SOD2-PL) gene therapy. Radiat. Res. 155:2-14. [DOI] [PubMed] [Google Scholar]
- 20.Fornace, A. J., Jr., S. A. Amundson, M. Bittner, T. G. Myers, P. Meltzer, J. N. Weinsten, and J. Trent. 1999. The complexity of radiation stress responses: analysis by informatics and functional genomics approaches. Gene Expr. 7:387-400. [PMC free article] [PubMed] [Google Scholar]
- 21.Gajewska, J., M. Szczypka, T. Izbicki, T. Klepacka, and T. Laskowska-Klita. 1996. Antioxidant and glutathione-associated enzymes in Wilms' tumour after chemotherapy. J. Cancer Res. Clin. Oncol. 122:483-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Granville, D. J., C. M. Carthy, H. Jiang, J. G. Levy, B. M. McManus, J. Y. Matroule, J. Piette, and D. W. Hunt. 2000. Nuclear factor-kappaB activation by the photochemotherapeutic agent verteporfin. Blood 95:256-262. [PubMed] [Google Scholar]
- 23.Hahn, S. M., A. Russo, J. A. Cook, and J. B. Mitchell. 1999. A multidrug-resistant breast cancer cell line induced by weekly exposure to doxorubicin. Int. J. Oncol. 14:273-279. [DOI] [PubMed] [Google Scholar]
- 24.Harris, A. L. 2002. Hypoxia—a key regulatory factor in tumour growth. Nat. Rev. Cancer 2:38-47. [DOI] [PubMed] [Google Scholar]
- 25.Herscher, L. L., J. A. Cook, R. Pacelli, H. I. Pass, A. Russo, and J. B. Mitchell. 1999. Principles of chemoradiation: theoretical and practical considerations. Oncology 13:11-22. [PubMed] [Google Scholar]
- 26.Huang, T. T., M. Yasunami, E. J. Carlson, A. M. Gillespie, A. G. Reaume, E. K. Hoffman, P. H. Chan, R. W. Scott, and C. J. Epstein. 1997. Superoxide-mediated cytotoxicity in superoxide dismutase-deficient fetal fibroblasts. Arch. Biochem. Biophys. 344:424-432. [DOI] [PubMed] [Google Scholar]
- 27.Jones, P. L., D. Ping, and J. M. Boss. 1997. Tumor necrosis factor alpha and interleukin-1β regulate the murine manganese superoxide dismutase gene through a complex intronic enhancer involving C/EBP-β and NF-κB. Mol. Cell. Biol. 17:6970-6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamata, H., and H. Hirata. 1999. Redox regulation of cellular signalling. Cell Signal 11:1-14. [DOI] [PubMed] [Google Scholar]
- 29.Keyse, S. M. 1993. The induction of gene expression in mammalian cells by radiation. Semin. Cancer Biol. 4:119-128. [PubMed] [Google Scholar]
- 30.Kiningham, K. K., and D. K. St. Clair. 1997. Overexpression of manganese superoxide dismutase selectively modulates the activity of Jun-associated transcription factors in fibrosarcoma cells. Cancer Res. 57:5265-5271. [PubMed] [Google Scholar]
- 31.Kiningham, K. K., Y. Xu, C. Daosukho, B. Popova, and D. K. St. Clair. 2001. Nuclear factor kappaB-dependent mechanisms coordinate the synergistic effect of PMA and cytokines on the induction of superoxide dismutase 2. Biochem. J. 353:147-156. [PMC free article] [PubMed] [Google Scholar]
- 32.Li, J. J., L. W. Oberley, D. St. Clair, L. A. Ridnour, and T. D. Oberley. 1995. Phenotypic changes induced in human breast cancer cells by overexpression of manganese-containing superoxide dismutase cDNA. Oncogene 10:1989-2000. [PubMed] [Google Scholar]
- 33.Li, S., T. Yan, J. Q. Yang, T. D. Oberley, and L. W. Oberley. 2000. The role of cellular glutathione peroxidase redox regulation in the suppression of tumor cell growth by manganese superoxide dismutase. Cancer Res. 60:3927-3939. [PubMed] [Google Scholar]
- 34.Li, Z., A. Khaletskiy, J. Wang, J. Y. Wong, L. W. Oberley, and J. J. Li. 2001. Genes regulated in human breast cancer cells overexpressing manganese-containing superoxide dismutase. Free Radic. Biol. Med. 30:260-267. [DOI] [PubMed] [Google Scholar]
- 35.Li, Z., L. Xia, M. L. Lee, A. Khaletskiy, J. Wang, J. Y. C. Wong, and J. J. Li. 2001. Effector genes altered in MCF-7 human breast cancer cells after exposure to fractionated ionizing radiation. Radiat. Res. 155:543-553. [DOI] [PubMed] [Google Scholar]
- 36.Maity, A., W. G. McKenna, and R. J. Muschel. 1994. The molecular basis for cell cycle delays following ionizing radiation: a review. Radiother. Oncol. 31:1-13. [DOI] [PubMed] [Google Scholar]
- 37.Manna, S. K., H. J. Zhang, T. Yan, L. W. Oberley, and B. B. Aggarwal. 1998. Overexpression of manganese superoxide dismutase suppresses tumor necrosis factor-induced apoptosis and activation of nuclear transcription factor-B and activated protein-1. J. Biol. Chem. 273:13245-13254. [DOI] [PubMed] [Google Scholar]
- 38.Mitchell, J. B., A. Russo, J. A. Cook, and E. Glatstein. 1989. Tumor cell drug and radiation resistance: does an interrelationship exist? Cancer Treat. Res. 48:189-203. [DOI] [PubMed] [Google Scholar]
- 39.Oberley, L. W. 2001. Anticancer therapy by overexpression of superoxide dismutase. Antioxid. Redox Signal 3:461-472. [DOI] [PubMed] [Google Scholar]
- 40.Oberley, L. W., and T. D. Oberley. 1994. Reactive oxygen species in the aetiology of cancer, p. 47-63. In C. Ioannides and D. F. C. Lewis (ed.), Drugs diet and disease. Ellis Horwoord, Hemel Hemstead, United Kingdom.
- 41.Oberley, L. W., L. A. Ridnour, E. Sierra-Rivera, T. D. Oberley, and D. L. Guernsey. 1989. Superoxide dismutase activities of differentiating clones from an immortal cell line. J. Cell. Physiol. 138:50-60. [DOI] [PubMed] [Google Scholar]
- 42.Oberley, L. W., D. K. St. Clair, A. P. Autor, and T. D. Oberley. 1987. Increase in manganese superoxide dismutase activity in the mouse heart after X-irradiation. Arch. Biochem. Biophys. 254:69-80. [DOI] [PubMed] [Google Scholar]
- 43.Pajonk, F., K. Pajonk, and W. H. McBride. 1999. Inhibition of NF-kappaB, clonogenicity, and radiosensitivity of human cancer cells. J. Natl. Cancer Inst. 91:1956-1960. [DOI] [PubMed] [Google Scholar]
- 44.Ravi, R., G. C. Bedi, L. W. Engstrom, Q. Zeng, B. Mookerjee, C. Gelinas, E. J. Fuchs, and A. Bedi. 2001. Regulation of death receptor expression and TRAIL/Apo2L-induced apoptosis by NF-kappaB. Nat. Cell Biol. 3:409-416. [DOI] [PubMed] [Google Scholar]
- 45.Schmidt-Ullrich, R. K., J. N. Contessa, P. Dent, R. B. Mikkelsen, K. Valerie, D. B. Reardon, G. Bowers, and P. S. Lin. 1999. Molecular mechanisms of radiation-induced accelerated repopulation. Radiat. Oncol. Investig. 7:321-330. [DOI] [PubMed] [Google Scholar]
- 46.Siemankowski, L. M., J. Morreale, and M. M. Briehl. 1999. Antioxidant defenses in the TNF-treated MCF-7 cells: selective increase in MnSOD. Free Radic. Biol. Med. 26:919-924. [DOI] [PubMed] [Google Scholar]
- 47.Spitz, D. R., J. H. Elwell, Y. Sun, L. W. Oberley, T. D. Oberley, S. J. Sullivan, and R. J. Roberts. 1990. Oxygen toxicity in control and H2O2-resistant Chinese hamster fibroblast cell lines. Arch. Biochem. Biophys. 279:249-260. [DOI] [PubMed] [Google Scholar]
- 48.St. Clair, D. K., T. D. Oberley, K. E. Muse, and W. H. St. Clair. 1994. Expression of manganese superoxide dismutase promotes cellular differentiation. Free Radic. Biol. Med. 16:275-282. [DOI] [PubMed] [Google Scholar]
- 49.Summers, R. W., B. V. Maves, R. D. Reeves, L. J. Arjes, and L. W. Oberley. 1989. Irradiation increases superoxide dismutase in rat intestinal smooth muscle. Free Radic. Biol. Med. 6:261-270. [DOI] [PubMed] [Google Scholar]
- 50.Sun, J., Y. Chen, M. Li, and Z. Ge. 1998. Role of antioxidant enzymes on ionizing radiation resistance. Free Radic. Biol. Med. 24:586-593. [DOI] [PubMed] [Google Scholar]
- 51.Sundaresan, M., Z. X. Yu, V. J. Ferrans, K. Irani, and T. Finkel. 1995. Requirement for generation of H2O2 for platelet-derived growth factor signal transduction. Science 270:296-299. [DOI] [PubMed] [Google Scholar]
- 52.Tan, Y., X. Sun, M. Xu, X. Tan, A. Sasson, B. Rashidi, Q. Han, X. Wang, Z. An, F. X. Sun, and R. M. Hoffman. 1999. Efficacy of recombinant methioninase in combination with cisplatin on human colon tumors in nude mice. Clin. Cancer Res. 5:2157-2163. [PubMed] [Google Scholar]
- 53.Tanaka, H., I. Matsumura, S. Ezoe, Y. Satoh, T. Sakamaki, C. Albanese, T. Machii, R. G. Pestell, and Y. Kanakura. 2002. E2F1 and c-Myc potentiate apoptosis through inhibition of NF-kappaB activity that facilitates MnSOD-mediated ROS elimination. Mol. Cell 9:1017-1029. [DOI] [PubMed] [Google Scholar]
- 54.Tang, G., Y. Minemoto, B. Dibling, N. H. Purcell, Z. Li, M. Karin, and A. Lin. 2001. Inhibition of JNK activation through NF-kappaB target genes. Nature 414:313-317. [DOI] [PubMed] [Google Scholar]
- 55.Thongphasuk, J., L. W. Oberley, and T. D. Oberley. 1999. Induction of superoxide dismutase and cytotoxicity by manganese in human breast cancer cells. Arch. Biochem. Biophys. 365:317-327. [DOI] [PubMed] [Google Scholar]
- 56.Wan, X. S., M. N. Devalaraja, and D. K. St. Clair. 1994. Molecular structure and organization of the human manganese superoxide dismutase gene. DNA Cell Biol. 13:1127-1136. [DOI] [PubMed] [Google Scholar]
- 57.Williams, M. D., H. Van Remmen, C. C. Conrad, T. T. Huang, C. J. Epstein, and A. Richardson. 1998. Increased oxidative damage is correlated to altered mitochondrial function in heterozygous manganese superoxide dismutase knockout mice. J. Biol. Chem. 273:28510-28515. [DOI] [PubMed] [Google Scholar]
- 58.Wong, G. H. 1995. Protective roles of cytokines against radiation: induction of mitochondrial MnSOD. Biochim. Biophys. Acta 1271:205-209. [DOI] [PubMed] [Google Scholar]
- 59.Wong, G. H., J. H. Elwell, L. W. Oberley, and D. V. Goeddel. 1989. Manganous superoxide dismutase is essential for cellular resistance to cytotoxicity of tumor necrosis factor. Cell 58:923-931. [DOI] [PubMed] [Google Scholar]
- 60.Xing, H., S. Zhang, C. Weinheimer, A. Kovacs, and A. J. Muslin. 2000. 14-3-3 proteins block apoptosis and differentially regulate MAPK cascades. EMBO J. 19:349-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu, Y., K. K. Kiningham, M. N. Devalaraja, C. C. Yeh, H. Majima, E. J. Kasarskis, and D. K. St. Clair. 1999. An intronic NF-kappaB element is essential for induction of the human manganese superoxide dismutase gene by tumor necrosis factor-alpha and interleukin-1beta. DNA Cell Biol. 18:709-722. [DOI] [PubMed] [Google Scholar]
- 62.Yaffe, M. B., K. Rittinger, S. Volinia, P. R. Caron, A. Aitken, H. Leffers, S. J. Gamblin, S. J. Smerdon, and L. C. Cantley. 1997. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell 91:961-971. [DOI] [PubMed] [Google Scholar]
- 63.Yamaguchi, S., S. Sakurada, and M. Nagumo. 1994. Role of intracellular SOD in protecting human leukemic and cancer cells against superoxide and radiation. Free Radic. Biol. Med. 17:389-395. [DOI] [PubMed] [Google Scholar]
- 64.Yan, T., L. W. Oberley, W. Zhong, and D. K. St. Clair. 1996. Manganese-containing superoxide dismutase overexpression causes phenotypic reversion in SV-40 transformed human lung fibroblasts. Cancer Res. 56:2864-2871. [PubMed] [Google Scholar]
- 65.Zha, J., H. Harada, E. Yang, J. Jockel, and S. J. Korsmeyer. 1996. Serine phosphorylation of death agonist BAD in response to survival factor results in binding to 14-3-3 not BCL-X(L). Cell 87:619-628. [DOI] [PubMed] [Google Scholar]
- 66.Zhang, H., R. L. Zheng, Z. Q. Wei, W. J. Li, Q. X. Gao, W. Q. Chen, Z. H. Wang, J. He, J. P. Liang, G. W. Han, T. Huang, Q. Li, H. M. Xie, S. M. Zhang, and X. C. Cai. 1998. Effects of preexposure of mouse testis with low-dose (16)O8+ ions or 60Co gamma-rays on sperm shape abnormalities, lipid peroxidation and superoxide dismutase (SOD) activity induced by subsequent high-dose irradiation. Int. J. Radiat. Biol. 73:163-167. [DOI] [PubMed] [Google Scholar]
- 67.Zhang, H. J., W. Zhao, S. Venkataraman, M. E. Robbins, G. R. Buettner, K. C. Kregel, and L. W. Oberley. 2002. Activation of matrix metalloproteinase-2 by overexpression of manganese superoxide dismutase in human breast cancer MCF-7 cells involves reactive oxygen species. J. Biol. Chem. 277:20919-20926. [DOI] [PubMed] [Google Scholar]
- 68.Zhong, W., L. W. Oberley, T. D. Oberley, T. Yan, F. E. Domann, and D. K. St. Clair. 1996. Inhibition of cell growth and sensitization to oxidative damage by overexpression of manganese superoxide dismutase in rat glioma cells. Cell Growth Differ. 7:1175-1186. [PubMed] [Google Scholar]