Abstract
Members of the SREBP family of transcription factors control cholesterol and lipid homeostasis and play important roles during adipocyte differentiation. The transcriptional activity of SREBPs is dependent on the coactivators p300 and CBP. We now present evidence that SREBPs are acetylated by the intrinsic acetyltransferase activity of p300 and CBP. In SREBP1a, the acetylated lysine residue resides in the DNA-binding domain of the protein. Coexpression with p300 dramatically increases the expression of both SREBP1a and SREBP2, and this effect is dependent on the acetyltransferase activity of p300, indicating that acetylation of SREBPs regulates their stability. Indeed, acetylation or mutation of the acetylated lysine residue in SREBP1a stabilizes the protein. We demonstrate that the acetylated residue in SREBP1a is also targeted by ubiquitination and that acetylation inhibits this process. Thus, our studies define acetylation-dependent stabilization of transcription factors as a novel mechanism for coactivators to regulate gene expression.
Members of the sterol regulatory element binding (SREBP) family of transcription factors control cholesterol and lipid metabolism and play critical roles during adipocyte differentiation (7, 12, 31, 37). In addition, SREBP1c is an important regulator of insulin-dependent gene expression (16, 43). The genes srebp1 and srebp2 encode three different SREBP proteins, each with a molecular mass of approximately 125 kDa (13). The SREBP2 gene encodes a single protein that has approximately 50% identity with SREBP1. As a result of alternative splicing and use of alternative promoters, the SREBP1 gene encodes two proteins, SREBP1a and SREBP1c (also known as ADD-1), that differ in the length of the amino-terminal transactivation domain.
Newly synthesized SREBPs are inserted into the nuclear and endoplasmic reticulum membranes and are transcriptionally inactive. When cellular sterol concentrations are lowered, the N-terminal domain of SREBP is released from membranes by a two-step proteolytic cascade (35, 39). This transcriptionally active fragment of SREBP is translocated to the nucleus and binds to the promoters of SREBP target genes. It has been demonstrated that transcriptional activation of SREBP target genes requires interactions between SREBP and other transcription factors, including Sp1 and NF-Y (15, 40). In addition, SREBPs interact with various transcriptional coactivators, such as CBP, p300, and ARC/DRIP (14, 26, 27). The interactions between SREBP and CBP and p300 involve N-terminal domains in all three proteins, and these interactions are required for the transcriptional activity of SREBPs. Both CBP and p300 have intrinsic histone acetyltransferase (HAT) activity, and it has been demonstrated that activation of SREBPs results in hyperacetylation of histone H3 at SREBP target promoters in vivo (5).
Proteins with intrinsic HAT activity act as transcriptional coactivators by acetylating histones and thereby induce an open chromatin conformation, allowing the transcriptional machinery access to promoters (38). The best-characterized HATs are p300, CBP, and P/CAF (38). These proteins interact with a large number of transcription factors, such as SREBPs, as well as components of the basic transcriptional machinery. Thus, it is believed that HATs are central integrators of various signaling pathways in the nucleus. Since the observation that p53 is a direct target for acetylation by the coactivators p300 and P/CAF (19), a number of transcription factors and nuclear proteins have been found to be modified in this manner (1, 23, 45). The functional consequences of acetylation are diverse and include increased DNA binding (p53 [19], GATA-1 [6], and MyoD [34, 41]), decreased DNA binding [HMGI(Y) (25)], increased stability (E2F1 [24] and Smad7 [17]), inhibition of nuclear export (importin-α [2] and HNF4 [44]), and changes in protein-protein interactions (T-cell factor [47], ACTR [10], and erythroid Krüppel-like factor [49]).
In this paper, we demonstrate that SREBP1a and SREBP2 are direct targets of the coactivators p300 and CBP and identify the acetylated lysine residue in SREBP-1a. Coexpression with p300 dramatically increases the expression of both SREBP1a and SREBP2, and this effect is dependent on the acetyltransferase activity of p300, indicating that acetylation of SREBPs regulates their stability. We demonstrate that the acetylated residue in SREBP1a is also targeted by ubiquitination. Thus, our data suggest that competition between ubiquitination and acetylation of overlapping lysine residues regulates the expression of SREBPs and may constitute a novel mechanism to regulate protein stability.
MATERIALS AND METHODS
Cell culture.
All tissue culture media and antibiotics were obtained from Invitrogen and Sigma. Human embryonic kidney epithelial 293T, HepG2, and Cos7 cells were from the American Type Culture Collection. All cells were maintained at 37°C in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum, 50 U of penicillin/ml, and 50 μg of streptomycin/ml, in 5% CO2.
Reagents and antibodies.
[14C]acetyl-coenzyme A (CoA; 51.6 mCi/mmol) was from ICN. Peroxidase-conjugated sheep anti-mouse and donkey anti-rabbit immunoglobulin G were from Amersham-Pharmacia Biotech. Acetyl-CoA, ubiquitin, and an anti-Flag antibody (M5) were from Sigma. Rabbit antihemagglutinin (anti-HA; Y-11), anti-p300 (N-15), monoclonal anti-Myc (9E10), antiubiquitin (P4D1), anti-SREBP1a (2A4), and anti-Gal4 DNA-binding domain (RK5C1) antibodies were from Santa Cruz Biotechnology. Monoclonal anti-HA (12CA5) antibodies were from Roche. Anti-acetyl lysine antibodies were from Cell Signaling Technology and Upstate Biotechnology. Protein A was obtained from Zymed. Tetramethyl rhodamine isocyanate-conjugated anti-mouse antibodies were from DAKO.
Plasmids and DNA transfections.
The expression vectors for Flag-tagged SREBP1a (amino acids 2 to 490) and SREBP2 (amino acids 2 to 485) in the mammalian expression vector pcDNA3 (Invitrogen) have been described earlier (14). Enhanced green fluorescent protein (EGFP)-tagged SREBP1a was generated by subcloning the EcoRI fragment from pcDNA3-Flag-SREBP1a into pEGFP-C2 (Clontech). Glutathione S-transferase (GST)-tagged SREBP1a constructs were generated by cloning the corresponding PCR fragments into pGEX4T-1 (Amersham-Pharmacia Biotech). SREBP1a point mutants were generated by site-directed mutagenesis (QuickChange; Stratagene). The expression vectors for HA-tagged p300 and Flag-tagged P/CAF were kind gifts from T. Kouzarides (Wellcome/CRC Institute, Cambridge, United Kingdom). Expression vectors for Gal4-tagged p300 and CBP in pVR1012 (Vical) were from N. Perkins (University of Dundee, Dundee, United Kingdom). Expression vectors for Flag-tagged p300 (wild type and ΔHAT1 and ΔHAT2 mutants) were kind gifts from Y. Nakatani (Dana-Farber Cancer Institute, Boston, Mass.). The expression vector for Myc-tagged SOCS3 was a kind gift from A. Yoshimura (Kyushu University, Kyushu, Japan). The 3-hydroxy-3-methylglutaryl (HMG)-CoA synthase promoter-reporter construct (SYNSRE-luc) has been described earlier (14). Transient transfections were performed with the MBS transfection kit (Stratagene).
Immunoprecipitations and immunoblotting.
Cells were lysed in ice-cold lysis buffer (50 mM HEPES [pH 7.2], 150 mM NaCl, 1 mM EDTA, 20 mM NaF, 2 mM sodium orthovanadate, 1% [wt/vol] Triton X-100, 10% [wt/vol] glycerol, 1 mM phenylmethylsulfonyl fluoride, 10 mM sodium butyrate, 1% aprotinin) and cleared by centrifugation. Immunoprecipitations were carried out by adding the appropriate antibodies plus protein A- or protein G-Sepharose beads, followed by incubation at 4°C. The immunoprecipitates were washed extensively, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to nitrocellulose membranes (Millipore). The membranes were incubated with the appropriate antibodies, washed, and incubated with horseradish peroxidase-coupled anti-rabbit or anti-mouse immunoglobulin G. After being washed, the blots were visualized by using Western blotting chemiluminescence luminol reagent (Santa Cruz Biotechnology).
Generation of recombinant proteins.
GST-SREBP1a fusion proteins and His-tagged SREBP1a and SREBP2 were expressed in Escherichia coli BL21 and purified according to standard protocols. Recombinant p300 was a kind gift from J. Boyes (ICR, London, United Kingdom).
Northern blot analysis and quantitative real-time PCR.
Total RNA was isolated from transiently transfected 293T cells with Trizol reagent (Invitrogen). Ten-microgram aliquots of total RNA were fractionated by electrophoresis on 1% agarose-formaldehyde gels, transferred to nylon membranes, and cross-linked with UV light. [32P]dCTP-radiolabeled cDNA probes were generated by random priming (Amersham-Pharmacia Biotech). Hybridization and quantitation using phosphorimage analysis (Fuji) were as described previously (15). Total RNA from transiently transfected 293T cells was extracted as described above and reverse transcribed with Super Script RNase H− reverse transcriptase (Invitrogen). cDNA was analyzed for the expression of human low-density lipoprotein receptor (LDLR) and HMG-CoA reductase. Gene-specific PCR products were continuously measured by means of an ABI PRISM 7700 sequence detection system (Applied Biosystems) during 40 cycles by using SYBR green PCR master mixture (Applied Biosystems). The level of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA in each sample was used for normalization. The gene-specific primers were designed by using Primer Express software (Applied Biosystems). The following primer pairs were used in the study: LDLR, 5′-CTGGACCGGAGCGAGTACAC-3′ and 5′-TGGGTGCTGCAGATCATTCTC-3′; HMG-CoA reductase, 5′-CATGATTCACAACAGGTCGAAGA-3′ and 5′-TCAGAACTGTCGGGCTATTCAG-3′; GAPDH, 5′-CCCATGTTCGTCATGGGTGT-3′ and 5′-TGGTCATGAGTCCTTCCACGATA-3′.
Protein-DNA interactions.
Cell lysates from transiently transfected 293T cells were precleared with streptavidin-agarose (Sigma) and subsequently used in oligonucleotide precipitation (DNAP) assays as described previously (30). The biotinylated double-stranded DNA was composed of two copies of the sterol regulatory element 1 (SRE-1) sequence (5′-ATCACCCCAC-3′) from the LDLR promoter. DNA-bound proteins were precipitated with streptavidin-agarose for 30 min at 4°C, washed, and detected by Western blot analysis.
The double-stranded oligonucleotide used in the electrophoretic mobility shift assay (EMSA) corresponds to a single copy of the SRE-1 (see above) from the LDLR promoter. The radiolabeled probe was incubated with total cell lysates from transfected 293T cells, in vitro-translated Flag-tagged SREBP1a, or recombinant polyhistidine-tagged SREBP1a as described earlier (15). In some cases, the SREBP1a protein was acetylated prior to the EMSA.
In vitro acetylation assay.
SREBP proteins (1 μg) were incubated in the absence or presence of p300 (50 ng) or HA-tagged p300 immunoprecipitated from transfected 293T cells as described earlier (17). Acetylated proteins were separated by SDS-PAGE. The gels were dried and analyzed by phosphorimage analysis.
In vitro ubiquitination assay.
GST-SREBP1a was used as the substrate in in vitro ubiquitination reactions involving 20 mM HEPES (pH 7.5), 5 mM MgCl2, 2 mM dithiothreitol, 2 mM ATP, 5 μg of ubiquitin, and 20 μM MG132 in the absence or presence of 5 μl of crude rabbit reticulocyte lysate (Promega) as described previously (17). In some cases, GST-SREBP1a was acetylated prior to the ubiquitination reaction by using recombinant p300 and unlabeled acetyl-CoA. Reaction mixtures were incubated for 1 h at 30°C. GST-SREBP1a was captured on glutathione beads and washed extensively in order to remove p300. The samples were resolved by SDS-PAGE and transferred to nitrocellulose membranes, and the ubiquitination of SREBP1a was determined by Western blotting.
Determination of protein half-life.
293T cells were transfected with Flag-tagged SREBP1a, either the wild type or K333Q, in the absence or presence of p300-HA. Thirty-six hours after transfection, cells were treated with cycloheximide (100 μM) to stop protein synthesis and incubated for various times. Total cell lysates were prepared, and the proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes. SREBP1a was visualized by Western blotting using anti-Flag antibodies, followed by quantitation with a charge-coupled device camera (Fuji) and image analysis software (Aida Image Analyzer, version 3.10).
Luciferase and β-galactosidase assays.
HepG2 cells were transiently transfected with the SYNSRE-luc promoter-reporter gene in the absence or presence of expression vectors for SREBP1a, either the wild type or a mutant. After 36 h, cells were lysed and luciferase activities in duplicate samples were determined as described by the manufacturer (Promega). The pCH110 vector carrying the β-galactosidase gene under the control of the simian virus 40 promoter (Amersham-Pharmacia Biotech) was used as an internal control for transfection efficiency. Luciferase values (relative light units) were calculated by dividing the luciferase activity by the β-galactosidase activity.
Immunohistochemistry.
Cells were grown on coverslips and fixed in 2% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min at room temperature. The cells were subsequently permeabilized in 0.2% Triton X-100 in PBS, washed in PBS, and incubated in 10 mM glycine in PBS. Primary and secondary antibodies were diluted in PBS containing 5% fetal bovine serum. Cells were incubated with primary antibodies followed by secondary antibodies for 1 h with a washing step in between. Nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole). The coverslips were mounted on glass object slides by the use of Fluoromount-G (Southern Biotechnology Associates). Cells were photographed with a Hamamatsu ORCA charge-coupled device digital camera by using the QED imaging system software with a Zeiss Axioplan2 microscope.
Analysis of cholesterol biosynthesis.
293T cells were transfected with either the vector alone (pCDNA3), SREBP1a (wild type), or SREBP1a (K333Q). Thirty-six hours after transfection, cells were placed in fresh media supplemented with [14C]acetate (10 μCi/ml) and incubated for 2 h. The cells were washed and collected by centrifugation. The pelleted cells were hydrolyzed in 60% (wt/vol) KOH in methanol. Nonpolar lipids were extracted in hexane and resolved by thin-layer chromatography (Silica Gel 60; Merck). The radioactive products were identified by comparison with unlabeled standards, which were visualized with iodine vapor. The plates were exposed to X-ray film at −80°C.
RESULTS
p300 recruits SREBP1a to nuclear speckles.
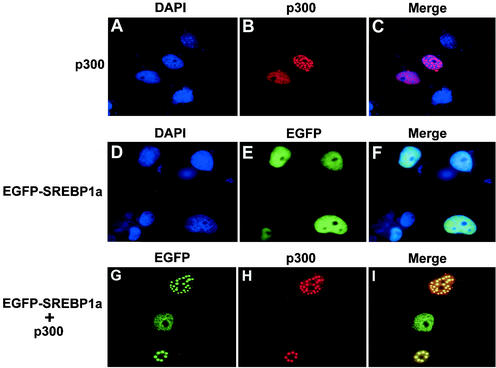
Earlier studies have demonstrated that SREBPs interact with the acetyltransferases CBP and p300 and that these interactions are important for their transcriptional activity. To determine if SREBPs interact with p300 in vivo, Cos7 cells were transfected with EGFP-SREBP1a and p300-HA, either alone or in combination (Fig. 1). p300 accumulated in nuclear speckles in the absence of SREBP1a expression (Fig. 1A to C). In agreement with earlier studies (28), EGFP-SREBP1a showed a uniform nuclear staining in the absence of p300 expression (Fig. 1D to F). Interestingly, p300 and EGFP-SREBP1a colocalized in nuclear speckles when expressed together, suggesting that SREBP1a is recruited into these structures through its interaction with p300 (Fig. 1G to I). Similar results were obtained with EGFP-SREBP2 and with Flag-tagged SREBP1a and SREBP2 (data not shown).
FIG. 1.
SREBP1a interacts with the coactivator p300. Cos7 cells were transfected with EGFP-SREBP1a and HA-tagged p300, either alone or together, and the subcellular localization of the transfected proteins was determined as described in Materials and Methods. (A to C) p300 (red) is localized to nuclear speckles in cells transfected with p300-HA alone. (D to F) EGFP-SREBP1a (green) shows a uniform nuclear staining in the absence of p300 expression. (G to I) EGFP-SREBP1a (green) is recruited into nuclear speckles when coexpressed with p300-HA (red).
SREBPs are acetylated in vitro.
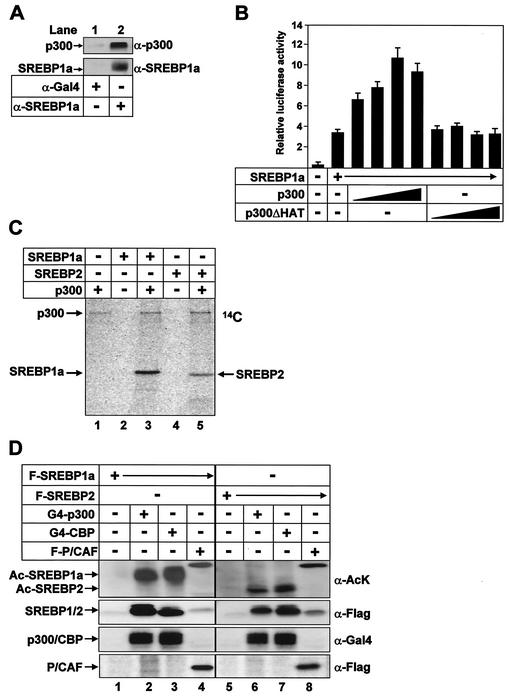
To determine whether endogenous SREBP1a and p300 form a complex in vivo, we used anti-SREBP1a antibodies to immunoprecipitate SREBP1a from HeLa cell lysates. p300 was detected in the SREBP1a immunoprecipitates, indicating that SREBP1a and p300 interact in vivo (Fig. 2A). In contrast, no p300 was detected when an unrelated antibody was used in the immunoprecipitation. A growing number of transcription factors have been found to be acetylated as a result of their interactions with CBP and p300. The acetyltransferase p300 enhanced the transcriptional activity of SREBP1a, and this effect was dependent on the acetyltransferase activity of p300 (Fig. 2B), suggesting that SREBPs could be direct targets of the acetyltransferase activity of p300. Indeed, both SREBP1a and SREBP2 were substrates of the acetyltransferase activity of p300 in vitro (Fig. 2C). Similar results were obtained with CBP immunoprecipitated from 293T cells transfected with HA-tagged CBP (data not shown). When expressed in 293T cells, p300 and CBP induced a robust acetylation of SREBP1a and SREBP2, whereas P/CAF failed to acetylate either protein (Fig. 2D). P/CAF was able to acetylate p53, both in vivo and in vitro (data not shown), and it was autoacetylated (Fig. 2D, lanes 4 and 8), verifying its activity.
FIG. 2.
SREBPs are substrates for p300 and CBP. (A) Lysates from HeLa cells grown in lipoprotein-deficient media were immunoprecipitated with anti-Gal4 (lane 1) or anti-SREBP1a (lane 2) antibodies. The immunoprecipitates were washed, and the proteins were separated by SDS-PAGE. Coimmunoprecipitated p300 was detected by Western blotting using anti-p300 antibodies. The amount of mature SREBP1a in the immunoprecipitates was determined with anti-SREBP1a antibodies. (B) HepG2 cells were transfected with SYNSRE-luc in the absence (lane 1) or presence (lanes 2 to 10) of Flag-tagged SREBP1a and in the absence or presence of p300 (25 to 250 ng), either the wild type (lanes 3 to 6) or the ΔHAT1 mutant (lanes 7 to 10). Thirty-six hours after transfection, the luciferase activity was measured. The data represent the averages ± standard deviations of three independent experiments performed in duplicate. (C) His-tagged SREBP1a and SREBP2 were subjected to in vitro acetylation in the absence (lanes 2 and 4) or presence (lanes 3 and 5) of p300 as described in Materials and Methods. After the proteins were separated by SDS-PAGE, the gel was dried and analyzed by phosphorimage analysis. (D) Flag-tagged SREBP1a (lanes 1 to 4) and SREBP2 (lanes 5 to 8) were expressed in 293T cells in the absence or presence of the indicated acetyltransferases. Following immunoprecipitation with anti-Flag antibodies, the samples were resolved by SDS-PAGE and the acetylation of the SREBPs was determined with anti-acetyl lysine antibodies (α-AcK). The levels of SREBPs in the immunoprecipitates were determined with anti-Flag antibodies (α-Flag). The levels of Gal4-p300, Gal4-CBP, and Flag-P/CAF in the cell lysates were determined by Western blotting.
The p300/CBP-binding motif in SREBP1a is important for its acetylation.
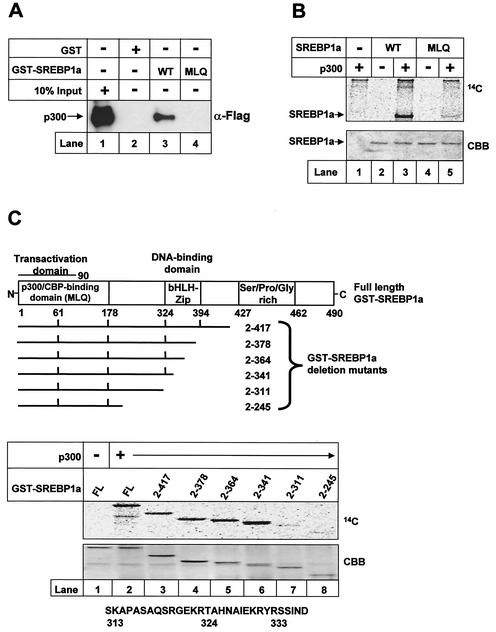
The N-terminal transactivation domain of SREBP1a contains a conserved protein-protein interaction motif (DIEDMLQ) that is also found in the CBP-binding domain of CREB (8). To test if this motif in SREBP1a is important for its interaction with p300, we created a GST-SREBP1a mutant in which the DIEDMLQ sequence was changed to DIEDAAA. Wild-type GST-SREBP1a was able to interact with p300 in GST pull-down assays (Fig. 3A, lane 3). However, the MLQ mutant was unable to interact with p300, indicating that this motif is part of a functional domain in SREBP1a that binds to p300 or CBP (p300/CBP-binding domain; Fig. 3A, lane 4).
FIG. 3.
The p300/CBP-binding motif in SREBP1a is important for its acetylation. (A) Flag-tagged p300 was expressed in 293T cells, and the cell lysates were incubated with GST alone (lane 2) or GST-SREBP1a, the wild type (WT; lane 3) and the MLQ mutant (lane 4), coupled to glutathione beads. The complexes were washed, and the proteins were separated by SDS-PAGE. The presence of p300 was detected by Western blotting with anti-Flag antibodies. Ten percent of the material used in the pull-down was loaded on the gel (lane 1). (B) Wild-type GST-SREBP1a and the MLQ mutant were subjected to in vitro acetylation in the absence (lanes 2 and 4) or presence (lanes 3 and 5) of p300 as described in Materials and Methods. After the proteins were separated by SDS-PAGE, the gel was dried and analyzed by phosphorimage analysis. The gel was stained with Coomassie brilliant blue to ensure that equal amounts of GST fusion proteins were used in the assay (CBB). (C) Schematic illustration of the GST-SREBP1a proteins used to map the acetylated residues in SREBP1a. Proteins consisting of GST fused to full-length SREBP1a (amino acids 2 to 490) and the indicated deletion mutants were subjected to in vitro acetylation by using p300 as described in Materials and Methods. After the proteins were separated by SDS-PAGE, the gel was dried and exposed to phosphorimage analysis (top). The gel was stained with Coomassie brilliant blue to ensure that equal amounts of GST fusion proteins were used in the assay (CBB). The core DNA-binding domain (amino acids 312 to 341) of human SREBP1a, including the potential acetylated lysine residues, is shown at the bottom. bHLH-Zip, basic helix-loop-helix zipper.
GST-SREBP1a, either the wild type or the MLQ mutant, was used in in vitro acetylation assays to test if the p300/CBP-binding motif was important for the acetylation of SREBP1a (Fig. 3B). Wild-type SREBP1a was a good substrate for recombinant p300 (Fig. 3B, lane 3). However, the acetylation of SREBP1a was lost when the MLQ sequence was mutated (Fig. 3B, lane 5), confirming the function of the p300/CBP-binding domain in SREBP1a.
SREBP1a is acetylated in its DNA-binding domain.
Full-length GST-SREBP1a was a good substrate for p300, and deletion of a major part of its C terminus (amino acids 342 to 490) did not affect the acetylation of SREBP1a (Fig. 3C, lanes 2 to 6). However, the acetylation of SREBP1a was lost when the core DNA-binding domain (amino acids 311 to 341) was removed, indicating that the acetylated lysine residue(s) resides in the DNA-binding domain of SREBP1a. All GST-SREBP1a fragments interacted with p300 (data not shown), consistent with the finding that this interaction is dependent on the N-terminal transactivation domain of SREBPs (14, 26).
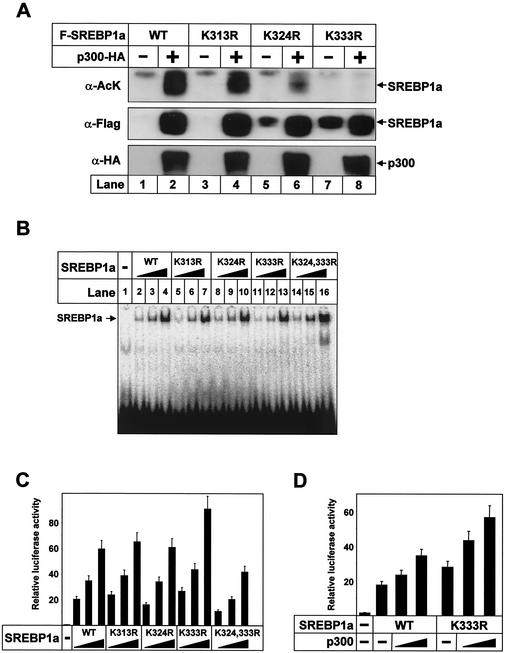
As illustrated in Fig. 3C, the sequence between amino acids 311 and 341 of SREBP1a contains three lysine residues that could be potential targets for acetylation (K313, K324, and K333). To determine which of these residues are acetylated in vivo, Flag-tagged SREBP1a, either the wild type or the lysine-to-arginine mutants indicated in Fig. 4, were expressed in 293T cells alone or together with p300-HA. Wild-type SREBP1a was acetylated following coexpression with p300, and mutation of K313 had only a limited effect on the acetylation of SREBP1a (Fig. 4A). However, mutation of K324 significantly decreased the acetylation of SREBP1a, and mutation of K333 completely prevented acetylation of SREBP1a (Fig. 4A, lanes 6 and 8). The K333R SREBP1a mutant was still able to interact with p300 in GST pull-down experiments, indicating that the lack of acetylation of this mutant was not due to effects on its interaction with p300 (data not shown).
FIG. 4.
SREBP1a is acetylated in its core DNA-binding domain by p300. (A) Flag-tagged wild-type (WT) SREBP1a and the indicated lysine mutants were expressed in 293T cells in the absence or presence of p300-HA. Following immunoprecipitation of SREBP1a, samples were resolved by SDS-PAGE and the acetylation of SREBP1a was detected with anti-acetyl lysine antibodies (α-AcK). The amount of SREBP1a in the immunoprecipitates was determined with anti-Flag antibodies (α-Flag). The levels of p300-HA in the cell lysates were determined by Western blotting (α-HA). (B) Increasing amounts of in vitro-translated Flag-SREBP1a, the wild type and the indicated mutants, were used in EMSAs with a 32P-labeled probe containing the SRE-1 sequence from the LDLR promoter. (C) Cos7 cells were transfected with SYNSRE-luc in the absence (lane 1) or presence (lanes 2 to 16) of increasing amounts (0.5 to 2.5 ng) of Flag-tagged SREBP1a, the wild type and the indicated mutants. Thirty-six hours after transfection, the luciferase activity was measured. The data represent the averages ± standard deviations (SD) of three independent experiments performed in duplicate. (D) HepG2 cells were transfected with SYNSRE-luc in the absence or presence of Flag-tagged SREBP1a, the wild type and the K333R mutant, in the absence or presence of p300 (25 or 100 ng). Thirty-six hours after transfection, the luciferase activity was measured. The data represent the averages ± SD of three independent experiments performed in duplicate.
Our initial results indicated that SREBP1a is acetylated on K324 and K333 in its DNA-binding domain, with K333 being the major site (Fig. 4A). Consequently, we wanted to determine if these residues were important for the DNA-binding or transcriptional activity of SREBP1a. Mutation of the potential acetylation sites in SREBP1a to arginines did not affect its DNA-binding or transcriptional activity, except that the K333R mutant displayed a slightly enhanced transcriptional activity (Fig. 4B and C). However, mutation of both lysine 324 and 333 (K324,333R) reduced the transcriptional activity of SREBP1a (Fig. 4C). This effect could not be explained by differences in DNA binding, since the DNA-binding activity of the double mutant was slightly enhanced (Fig. 4B). One possibility is that acetylation of both these residues affects the recruitment of additional coactivators, as was recently described for p53 (3).
The transcriptionally active fragments of SREBPs are highly unstable and are rapidly degraded, potentially through the ubiquitin-proteasome pathway. We were, therefore, intrigued to see that the expression of wild-type SREBP1a was dramatically enhanced following coexpression with p300 (compare lanes 1 and 2 in Fig. 4A). Interestingly, the expression of the lysine mutants, and especially K324R and K333R, was increased in the absence of p300 compared to that of wild-type SREBP1a, indicating that the acetylated lysine residues confer instability on SREBP1a and that acetylation prevents this process (compare lanes 1, 3, 5, and 7 in Fig. 4A).
Our results indicated that p300-mediated acetylation of K333 in SREBP1a stabilizes the protein (Fig. 4A). We therefore wanted to determine if p300 could further coactivate transcription from the K333R mutant. To do this, HepG2 cells were transfected with SREBP1a, either the wild type or the K333R mutant, and a promoter-reporter gene derived from the SREBP-responsive HMG-CoA synthase promoter. The transcriptional activity of both wild-type and K333R SREBP1a was enhanced by coexpressed p300, suggesting that p300 has additional activities as a coactivator of SREBP1a (Fig. 4D). It has been demonstrated that the transcriptional activity of SREBPs is dependent on interactions with p300 or CBP and that activation of SREBPs results in hyperacetylation of histone H3 at SREBP target promoters in vivo (5, 14, 26, 27). Therefore, it is not surprising that p300 is able to function as a coactivator of the stabilized form of SREBP1a also.
Acetylation enhances the expression of SREBPs.
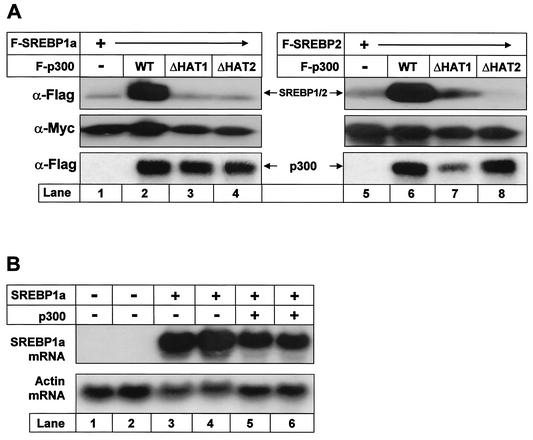
The expression of both SREBP1a and SREBP2 was enhanced in the presence of coexpressed p300 (Fig. 5A). This effect was dependent on the acetyltransferase activity of p300, since acetyltransferase-deficient forms of p300 failed to enhance the expression of SREBPs. The effect was specific, since p300 had no effect on the expression of a cotransfected nonacetylated control protein (Myc-tagged SOCS3) (Fig. 5A, middle). Expression of the transfected SREBP1a gene was high in the absence of p300, and no increase in the SREBP1a mRNA levels was observed following p300 expression, indicating that the effect of p300 on the expression of SREBPs is posttranscriptional (Fig. 5B).
FIG. 5.
Acetylation enhances the stability of SREBPs. (A) Flag-tagged wild-type (WT) SREBP1a (lanes 1 to 4) and SREBP2 (lanes 5 to 8) were expressed in 293T cells in the absence or presence of Flag-p300, either the wild type (lanes 2 and 6) or two different acetylation-deficient mutants (lanes 3, 4, 7, and 8). Thirty-six hours after transfection, cell lysates were prepared and resolved by SDS-PAGE. The levels of SREBPs were detected by Western blotting using anti-Flag antibodies (α-Flag). The levels of Flag-p300 in the cell lysates were determined by Western blotting (α-Flag). To ensure that equal amounts of protein were loaded in each well, the levels of Myc-SOCS3 in the samples were estimated by Western blotting using anti-Myc antibodies (α-Myc). (B) Flag-tagged SREBP1a was expressed in 293T cells in the absence (lanes 3 and 4) or presence (lanes 5 and 6) of p300-HA. Thirty-six hours posttransfection, RNA was extracted and resolved on agarose gels, and the mRNA levels for SREBP1a and actin were determined as described in Materials and Methods.
Acetylated SREBP1a binds DNA.
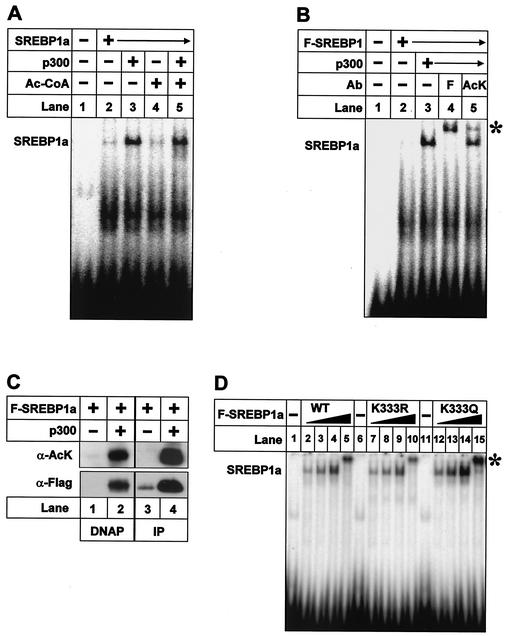
K324 and K333 are located close to the core DNA-binding domain of SREBP1a, and acetylation has been shown to affect the DNA-binding activity of many transcription factors. Therefore, we wanted to determine if the acetylated residues in SREBP1a are important for its DNA-binding activity and if acetylation affects this activity (Fig. 6). Interestingly, recombinant p300 enhanced the DNA-binding activity of purified SREBP1a in the absence of acetyl-CoA, and no further increase was observed when acetyl-CoA was included in the assay, indicating that the p300-mediated effect is unrelated to acetylation of SREBP1a (compare lanes 2, 3, and 5 in Fig. 6A). p300 enhanced the expression of SREBP1a when the two proteins were coexpressed in 293T cells (Fig. 2 to 5 and data not shown), resulting in a dramatic increase in total DNA-binding activity (compare lanes 2 and 3 in Fig. 6B). The fact that a portion of the DNA-bound SREBP1a was supershifted by the anti-acetyl lysine antibody indicated that acetylated SREBP1a is able to bind DNA (Fig. 6B, lane 5). The anti-acetyl lysine antibody did not shift DNA-protein complexes containing nonacetylated SREBP1a (data not shown). The major acetylation site in SREBP1a, i.e., K333, is located between two amino acids that directly contact DNA in the SREBP-DNA cocrystal structure (32). It is, therefore, unlikely that the anti-acetyl lysine antibody interacts with this lysine residue without affecting DNA binding. Rather, we hypothesize that the anti-acetyl lysine recognizes K324 in the SREBP1a-DNA complex. To confirm that acetylated SREBP1a can bind DNA, nuclear extracts were prepared from cells transfected as described above and the extracts were used for DNAP assays using a biotin-labeled oligonucleotide containing an SREBP binding site (Fig. 6C). We could detect only low levels of SREBP1a associated with the oligonucleotide in the absence of p300, and none of this SREBP1a was acetylated (Fig. 6C, lanes 1 and 3). The amount of DNA-bound SREBP1a increased drastically following coexpression with p300, and a large portion of the SREBP1a associated with DNA was acetylated (Fig. 6C, lanes 2 and 4), suggesting that acetylated SREBP1a is able to bind DNA. Taken together, our results indicate that acetylated SREBP1a can bind DNA and that acetylation of SREBP1a within its core DNA-binding domain does not affect its DNA-binding activity. To confirm these observations, we generated two constructs in which the major acetylation site in SREBP1a (K333) was mutated to either R (K333R) or Q (K333Q) to mimic nonacetylated and acetylated SREBP1a, respectively (48). The corresponding SREBP1a proteins were generated by in vitro translation and were subsequently used in EMSAs (Fig. 6D). Both K333R and K333Q were able to bind DNA, and we were unable to detect any significant difference in affinity for DNA between the mutants and wild-type SREBP1a, suggesting that acetylation of SREBP1a does not affect its ability to bind DNA.
FIG. 6.
Acetylated SREBP1a binds DNA. (A) Recombinant SREBP1a was used in EMSAs with a 32P-labeled probe containing the SRE-1 sequence from the LDLR promoter. Where indicated, SREBP1a was incubated with p300 (lane 3), acetyl-CoA (Ac-CoA; lane 4), or p300 plus acetyl-CoA (lane 5) prior to the EMSA. (B) Flag-tagged wild-type SREBP1a was expressed in 293T cells in the absence (lane 2) or presence of p300-HA (lanes 3 to 5). Thirty-six hours after transfection, nuclear extracts were prepared and used in EMSA with a 32P-labeled probe containing the SRE-1 sequence from the LDLR promoter. Where indicated, anti-Flag (F; lane 4) or anti-acetyl lysine (AcK; lane 5) antibodies were included in the assay. ∗, supershifted complexes. (C) Nuclear extracts were prepared from 293T cells transfected as in panel B and used in DNAP assays with the SRE-1 sequence from the LDLR promoter (lanes 1 and 2) or immunoprecipitated (IP) with anti-Flag antibodies (lanes 3 and 4). The levels and acetylation of SREBP1a were determined by Western blotting using anti-Flag (α-Flag) and anti-acetyl lysine (α-AcK) antibodies, respectively. (D) Increasing amounts of in vitro-translated Flag-SREBP1a, the wild type (WT) and the indicated mutants, were used in EMSAs with a 32P-labeled probe containing the SRE-1 sequence from the LDLR promoter. The SREBP1a-DNA complexes were supershifted with anti-Flag antibodies (lanes 5, 10, and 15; asterisk).
Acetylation stabilizes SREBP1a.
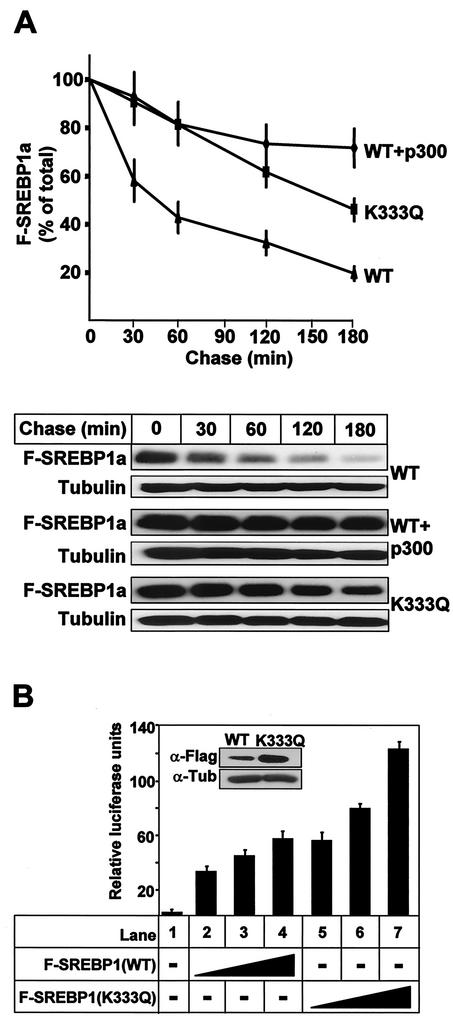
The results presented in Fig. 2 to 6 demonstrate that p300 is able to enhance the expression of SREBP1a and SREBP2 and that the acetyltransferase activity of p300 is required for this effect. In addition, we have demonstrated that mutation of the major acetylated residue in SREBP1a (K333) enhances its expression. Based on these observations, we hypothesized that the acetylated lysine residue in SREBP1a is important for its stability. To address this possibility, we determined the half-lives of wild-type and acetylation-deficient (K333Q) SREBP1a, both in the absence and presence of p300. As seen in Fig. 7A, wild-type SREBP1a was rapidly degraded in the absence of p300, while the K333Q mutant was relatively stable. Interestingly, the degradation of wild-type SREBP1a was prevented following coexpression with p300, suggesting that acetylation of K333 inhibits the degradation of SREBP1a.
FIG. 7.
Acetylation stabilizes SREBP1a. (A) 293T cells were transfected with Flag-SREBP1a, either the wild type (WT) or the K333Q mutant, in the absence or presence of p300-HA. Thirty-six hours after transfection, cells were treated with cycloheximide for the indicated times, and the levels of Flag-SREBP1a in the cell lysates were determined as described in Materials and Methods. The amount of SREBP1a at each time point is plotted as a percentage of the amount at the start of the chase and represents the average ± the standard error of the mean of three independent experiments (top). The results from one representative experiment are illustrated at the bottom. To ensure that equal amounts of protein were loaded in each well, the levels of tubulin in the samples were estimated by Western blotting using antitubulin antibodies. (B) HepG2 cells were transfected with SYNSRE-luc in the absence (lane 1) or presence (lanes 2 to 7) of increasing amounts of Flag-SREBP1a (1 to 5 ng), either the wild type (lanes 2 to 4) or K333Q (lanes 5 to 7). Thirty-six hours after transfection, the luciferase activity was measured. The data represent the averages ± standard deviations of three independent experiments per-formed in duplicate. The levels of Flag-SREBP1a in samples 4 (WT) and 7 (K333Q) were determined by Western blotting using anti-Flag antibodies (inset).
To confirm that the stabilized SREBP1a was active, HepG2 cells were transfected with the SREBP-responsive HMG-CoA synthase promoter-reporter gene in the absence or presence of Flag-tagged SREBP1a, either the wild type or the K333Q mutant. As illustrated in Fig. 7B, the expression of the reporter gene was induced following expression of wild-type SREBP1a. Interestingly, the stabilized form of SREBP1a was more efficient in inducing transcription from the HMG-CoA synthase promoter-reporter gene. In the transfected HepG2 cells, the level of the K333Q mutant was elevated compared to that of wild-type SREBP1a (Fig. 7B, inset), indicating that the enhanced SREBP-dependent transcription was due to the increased stability of the mutant form of SREBP1a.
Acetylation of SREBP1a prevents ubiquitination.
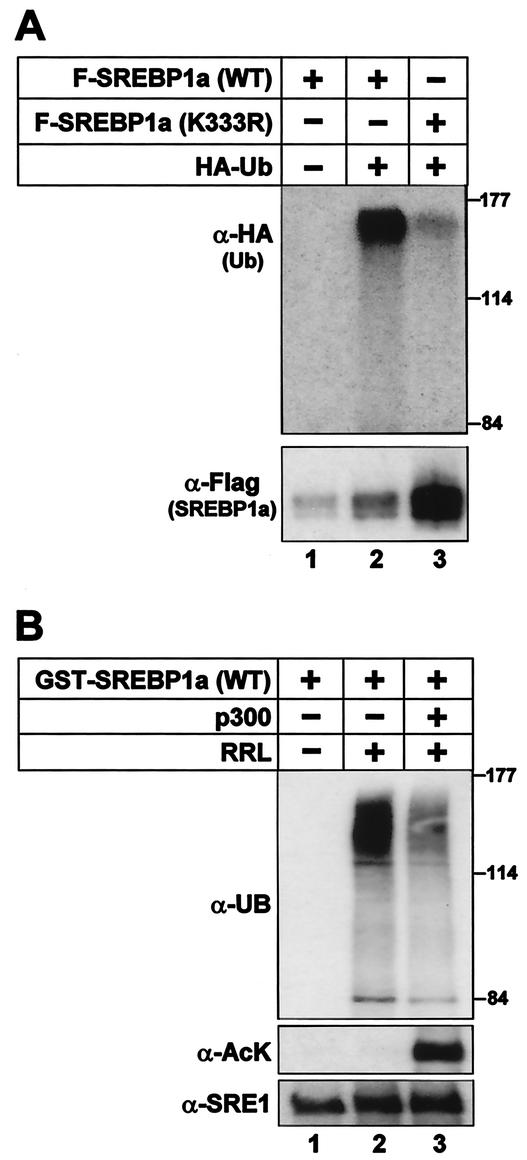
Our results indicate that acetylation of SREBPs enhances their stability, potentially by interfering with degradation. Acetylation of proteins involves the attachment of an acetyl group to the side chain of lysine residues. The side chain of lysine is also targeted by a second posttranslational modification, i.e., ubiquitination, and ubiquitinated proteins are recognized and degraded by the ubiquitin-proteasome pathway. We were able to detect ubiquitination of wild-type SREBP1a as a collection of high-molecular-weight bands following coexpression with HA-tagged ubiquitin, while no HA-tagged bands were observed in the absence of HA-ubiquitin (Fig. 8A). Mutation of K333 significantly reduced the ubiquitination of SREBP1a, suggesting that the acetylated lysine residue is also targeted by ubiquitination in vivo.
FIG. 8.
Acetylation of SREBP1a prevents its ubiquitination. (A) Flag-tagged wild-type SREBP1a and the K333R mutant were expressed in 293T cells in the absence or presence of HA-ubiquitin. Thirty-six hours after transfection, SREBP1a was immunoprecipitated from cell lysates and resolved by SDS-PAGE. The ubiquitination of SREBP1a was determined by Western blotting using anti-HA antibodies (α-HA). The migration of molecular mass standards (in kilodaltons) is indicated to the right. The levels of SREBP1a in the immunoprecipitates were determined by Western blotting using anti-Flag antibodies (α-Flag). (B) GST-SREBP1a was used as the substrate in in vitro ubiquitination reactions in the absence or presence of rabbit reticulocyte lysate (RRL). Where indicated, GST-SREBP1a was acetylated prior to the ubiquitination reaction (lane 3). The samples were resolved by SDS-PAGE, and the ubiquitination of SREBP1a was determined with antiubiquitin antibodies (α-UB). The migration of molecular mass standards (in kilodaltons) is indicated to the right. The amount of SREBP1a and its acetylation were determined with anti-SREBP1a (α-SRE1) and anti-acetyl lysine (α-AcK) antibodies, respectively.
To test if p300-mediated acetylation is able to affect the ubiquitination of SREBP1a, we used recombinant SREBP1a, either acetylated or nonacetylated, as the substrates in reconstituted in vitro ubiquitination assays. As illustrated in Fig. 8B, nonacetylated SREBP1a was ubiquitinated in the presence of rabbit reticulocyte lysate and acetylation of SREBP1a prevented its ubiquitination. These results suggest that K333 in SREBP1a is targeted by ubiquitination and that p300-mediated acetylation of the same lysine residue prevents this process.
Stabilization of SREBP1a affects sterol synthesis.
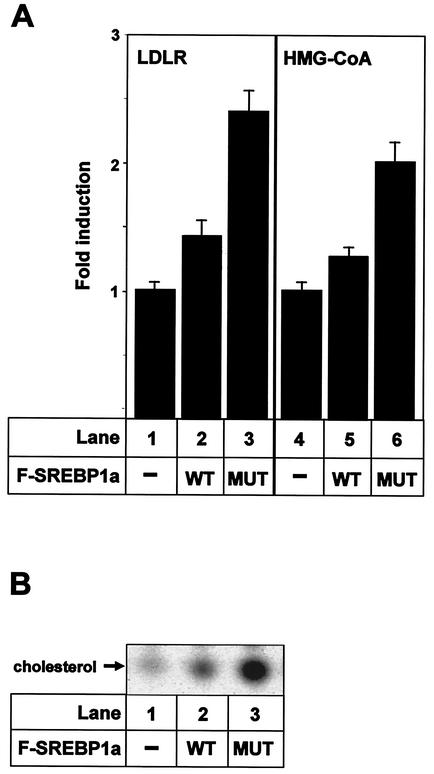
Theoretically, stabilization of SREBP1a would increase the expression of SREBP target genes and, thereby, would affect cholesterol metabolism. To test this hypothesis, 293T cells were transfected with SREBP1a, either the wild type or K333Q, and the mRNA levels for the LDLR and HMG-CoA reductase genes were determined by real-time PCR. The expression of both genes was higher in cells expressing the stabilized form of SREBP1a than in cells expressing wild-type SREBP1a (Fig. 9A). To determine if this enhanced expression of SREBP target genes could affect sterol metabolism, the incorporation of [14C]acetate into cholesterol in 293T cells transfected with SREBP1a, either the wild type or the K333Q mutant, was determined (Fig. 9B). Cholesterol synthesis in vector-transfected control cells was low and was induced in response to expression of wild-type SREBP1a. Interestingly, cholesterol synthesis was further enhanced in cells expressing the stabilized form of SREBP1a. Taken together, these results indicate that stabilization of transcriptionally active SREBP1a could affect the expression of SREBP target genes and lipid metabolism.
FIG. 9.
Stabilization of SREBP1a affects cholesterol metabolism. (A) 293T cells were transfected with the vector alone (−) or Flag-tagged wild-type SREBP1a (WT) or K333Q (MUT). Thirty-six hours after transfection, total RNA was extracted and used to determine the expression of the LDLR and HMG-CoA reductase genes by quantitative real-time PCR. The results are presented as the factors by which mRNA levels increased relative to the mRNA levels found in cells transfected with the vector alone. The results represent the averages ± standard deviations of one representative experiment performed in triplicate. (B) 293T cells were transfected with either the vector alone (−) or Flag-tagged wild-type SREBP1a or K333Q (MUT). Thirty-six hours after transfection, cells were placed in fresh media supplemented with [14C]acetate. Nonpolar lipids were extracted and resolved by thin-layer chromatography (TLC). Radioactive products were visualized after exposure of the TLC plates to X-ray film.
DISCUSSION
Here we present evidence that SREBPs interact with the transcriptional coactivator p300 in vivo, resulting in acetylation of the SREBPs. Acetylation of SREBP1a and SREBP2 enhances the stability of these transcription factors. In SREBP1a, we have mapped the acetylation to a specific lysine residue (K333) in its core DNA-binding domain. Acetylation or mutation of this residue stabilizes SREBP1a by preventing ubiquitination of the same lysine residue, indicating that competition between acetylation and ubiquitination controls the function of SREBP1a.
SREBPs are known to interact with CBP and p300, and these interactions are important for the transcriptional activity of this family of transcription factors. We now demonstrate that SREBP1a colocalizes with the acetyltransferase p300 in nuclear speckles in vivo (Fig. 1). A large number of nuclear proteins, including transcription factors, coactivators, and corepressors, have been localized to various subnuclear particles (11). Interestingly, it has been demonstrated that activation of Ras induces relocalization of p53 and CBP to PML bodies and that Ras-induced acetylation of p53 is lost in PML−/− fibroblasts (33). In addition, the transcription factor EVI1 colocalizes with CBP and P/CAF in nuclear speckles and is acetylated by both of these coactivators (9). Certain CBP-containing nuclear structures are rich in components of the 26S proteasome (4). It is, therefore, tempting to hypothesize that CBP- or p300-mediated acetylation of SREBP protects it from degradation within these structures. Further studies, using confocal microscopy, are necessary to characterize these structures and to determine if the subnuclear localization of SREBPs is of functional importance.
We found that SREBP1a and SREBP2 are acetylated by both CBP and p300, but not by P/CAF (Fig. 2). In SREBP1a, the acetylation was dependent on its N-terminal p300/CBP-binding domain and the acetylated residues were mapped to its core DNA-binding domain (Fig. 3). Further studies identified K333 as the major acetylation site (Fig. 4). Acetylation or mutation of K333 did not affect DNA binding. However, we observed that p300 enhanced the DNA-binding activity of SREBP1a in vitro in the absence of acetylation. It was recently demonstrated that p300 enhances the DNA-binding activity of Sp1, and this effect was independent of the acetyltransferase activity of p300 (46). Further studies are necessary to determine if p300 affects the DNA-binding activity of SREBPs in vivo and if this is of functional importance. The DNA-binding domain is also acetylated in SREBP2, although the targeted lysine residue(s) is not identical (unpublished data). Further studies are needed to identify the acetylated residues in SREBP2 and to determine if acetylation of these residues affects the function of SREBP2. Further studies are also necessary to determine if acetylation of K324 and K333 in SREBP1a has distinct functional effects.
We observed a significant increase in the expression of transfected SREBPs in the presence of cotransfected p300, and this effect was dependent on the acetyltransferase activity of p300 (Fig. 4 and 5). Interestingly, we observed a similar enhanced expression when the major acetylated residue in SREBP1a (K333) was mutated. These observations indicated that the acetylated residues in SREBP1a and SREBP2 are involved in regulating the stability of these proteins, potentially by inhibiting their degradation. Indeed, when we determined the half-life of SREBP1a, we found that the degradation of the wild-type protein was very rapid, while acetylation or mutation of K333 stabilized the protein (Fig. 7).
Transcriptionally active SREBP is rapidly turned over, especially when cells accumulate high levels of cholesterol. In addition, SREBPs are stabilized by proteasome inhibitors, and the expression of SREBP-regulated genes is induced in response to these inhibitors, indicating that the stability of SREBPs is regulated by the ubiquitin-proteasome pathway. Indeed, it was recently demonstrated that SREBP1a is ubiquitinated in vivo, although the lysine residues targeted by this modification were not identified (20). Thus, one interpretation of our results is that K333 confers instability on SREBP1a, potentially because this lysine residue is also targeted by ubiquitination. This hypothesis gained support from our observation that wild-type SREBP1a is ubiquitinated in vivo and that mutation of K333 decreased the ubiquitination of SREBP1a (Fig. 8). These observations were confirmed by our demonstration that p300-mediated acetylation of SREBP1a prevented its ubiquitination in vitro. It has been demonstrated that the expression levels of E2F1 are enhanced following coexpression with P/CAF (24). This enhanced expression was dependent on the HAT domain of P/CAF, indicating that acetylation of E2F1 is important for this effect. However, when the acetylated lysines in E2F1 were mutated, the mutant protein did not accumulate. Rather, and in contrast to the case for SREBP1a, the lysine-mutated version of E2F1 was rapidly degraded. Further studies are, therefore, necessary to determine if competition between ubiquitination and acetylation of common lysine residues is responsible for the stabilization of E2F1 in response to P/CAF expression. There have also been reports of a potential correlation between the acetylation and stability of p53 and Mdm2 (21, 22, 29, 36). It is, however, unclear if acetylation of these proteins is the only factor contributing to the observed effects, and the stabilization was not linked to individual lysine residues. The issue is further complicated by the fact that p53 is destabilized through its interaction with p300 and CBP under certain conditions (18). However, our results are similar to those reported for the inhibitory Smad7 protein (17). Acetylation or mutation of the acetylated lysine residues in Smad7 stabilized the protein and protected it from transforming growth factor β-induced degradation. Furthermore, it was demonstrated that the acetylated residues in Smad7 were targeted by ubiquitination and that acetylation of these residues prevented the subsequent ubiquitination of Smad7 by the ubiquitin ligase Smurf1.
The potential link between acetylation or deacetylation and ubiquitination received further support from the recent identification of the components of the murine histone deacetylase 6 (HDAC6) complex (42). It was demonstrated that a number of the proteins in this complex are involved in the ubiquitin-signaling pathway. For acetylated SREBPs to be degraded they need to be deacetylated and ubiquitinated. Interestingly, we have found that SREBP1a and SREBP2 interact with certain HDACs (unpublished data). Future studies will address whether acetylated SREBPs are substrates of HDACs and if deacetylation of SREBPs promotes their ubiquitination and degradation.
Our results suggest that acetylation regulates the stability and, thereby, the transcriptional activity of SREBPs. The most common treatment for elevated cholesterol levels in humans is a group of drugs called statins. These compounds block cholesterol synthesis and, therefore, activate SREBPs. Activation of SREBPs leads to an enhanced expression of the LDLR gene and, thereby, increased clearance of LDL from the circulation. We hypothesize that compounds that enhance the acetylation of SREBPs could increase the levels of transcriptionally active SREBP in cells. Such compounds could potentially be used to enhance the cholesterol-lowering activities of statins. This hypothesis is supported by the results presented in Fig. 9, where cells expressing a stabilized form of SREBP1a (K333Q) expressed higher levels of the LDLR and HMG-CoA reductase genes, well-established SREBP target genes. In addition, the synthesis of cholesterol in the cells expressing the stabilized form of SREBP1a was enhanced.
It is striking that SREBP1a undergoes two different modifications, which both target the same lysine residue. We propose that acetylation of specific lysines in SREBPs prevents subsequent ubiquitination of the same residues, thereby blocking proteasome-mediated degradation. Furthermore, our studies define acetylation-dependent stabilization of transcription factors as a novel mechanism for coactivators to regulate gene expression. The ubiquitin-proteasome pathway regulates a large number of nuclear proteins, and many of these are also acetylated. Therefore, it will be of the utmost importance to determine if competition between acetylation and ubiquitination is a general mechanism to regulate protein stability.
Acknowledgments
We are grateful to J. Boyes, N. Perkins, T. Kouzarides, A. Yoshimura, and Y. Nakatani for kindly providing expression vectors and reagents and C.-H. Heldin for stimulating discussions and suggestions. We also thank J.-B. Demoulin for assistance with real-time PCR analysis.
This work was in part supported by grants from the Swedish Research Council and the Novo Nordisk Foundation to J.E. V.G. is a recipient of a fellowship from the Wenner-Gren Foundations. J.E. is a Research Fellow of the Royal Swedish Academy of Sciences through a grant from the Knut and Alice Wallenberg Foundation.
REFERENCES
- 1.Bannister, A. J., and E. A. Miska. 2000. Regulation of gene expression by transcription factor acetylation. Cell. Mol. Life Sci. 57:1184-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bannister, A. J., E. A. Miska, D. Gorlich, and T. Kouzarides. 2000. Acetylation of importin-alpha nuclear import factors by CBP/p300. Curr. Biol. 10:467-470. [DOI] [PubMed] [Google Scholar]
- 3.Barlev, N. A., L. Liu, N. H. Chehab, K. Mansfield, K. G. Harris, T. D. Halazonetis, and S. L. Berger. 2001. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 8:1243-1254. [DOI] [PubMed] [Google Scholar]
- 4.Baumann, C. T., H. Ma, R. Wolford, J. C. Reyes, P. Maruvada, C. Lim, P. M. Yen, M. R. Stallcup, and G. L. Hager. 2001. The glucocorticoid receptor interacting protein 1 (GRIP1) localizes in discrete nuclear foci that associate with ND10 bodies and are enriched in components of the 26S proteasome. Mol. Endocrinol. 15:485-500. [DOI] [PubMed] [Google Scholar]
- 5.Bennett, M. K., and T. F. Osborne. 2000. Nutrient regulation of gene expression by the sterol regulatory element binding proteins: increased recruitment of gene-specific coregulatory factors and selective hyperacetylation of histone H3 in vivo. Proc. Natl. Acad. Sci. USA 97:6340-6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyes, J., P. Byfield, Y. Nakatani, and V. Ogryzko. 1998. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature 396:594-598. [DOI] [PubMed] [Google Scholar]
- 7.Brown, M. S., and J. L. Goldstein. 1999. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc. Natl. Acad. Sci. USA 96:11041-11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cardinaux, J. R., J. C. Notis, Q. Zhang, N. Vo, J. C. Craig, D. M. Fass, R. G. Brennan, and R. H. Goodman. 2000. Recruitment of CREB binding protein is sufficient for CREB-mediated gene activation. Mol. Cell. Biol. 20:1546-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakraborty, S., V. Senyuk, S. Sitailo, Y. Chi, and G. Nucifora. 2001. Interaction of EVI1 with cAMP-responsive element-binding protein (CBP) and p300/CBP-associated factor (P/CAF) results in reversible acetylation of EVI1 and in co-localization in nuclear speckles. J. Biol. Chem. 276:44936-44943. [DOI] [PubMed] [Google Scholar]
- 10.Chen, H., R. J. Lin, W. Xie, D. Wilpitz, and R. M. Evans. 1999. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell 98:675-686. [DOI] [PubMed] [Google Scholar]
- 11.Cockell, M., and S. M. Gasser. 1999. Nuclear compartments and gene regulation. Curr. Opin. Genet. Dev. 9:199-205. [DOI] [PubMed] [Google Scholar]
- 12.Edwards, P. A., and J. Ericsson. 1999. Sterols and isoprenoids: signaling molecules derived from the cholesterol biosynthetic pathway. Annu. Rev. Biochem. 68:157-185. [DOI] [PubMed] [Google Scholar]
- 13.Edwards, P. A., D. Tabor, H. R. Kast, and A. Venkateswaran. 2000. Regulation of gene expression by SREBP and SCAP. Biochim. Biophys. Acta 1529:103-113. [DOI] [PubMed] [Google Scholar]
- 14.Ericsson, J., and P. A. Edwards. 1998. CBP is required for sterol-regulated and sterol regulatory element-binding protein-regulated transcription. J. Biol. Chem. 273:17865-17870. [DOI] [PubMed] [Google Scholar]
- 15.Ericsson, J., S. M. Jackson, and P. A. Edwards. 1996. Synergistic binding of sterol regulatory element-binding protein and NF-Y to the farnesyl diphosphate synthase promoter is critical for sterol-regulated expression of the gene. J. Biol. Chem. 271:24359-24364. [DOI] [PubMed] [Google Scholar]
- 16.Foretz, M., C. Guichard, P. Ferre, and F. Foufelle. 1999. Sterol regulatory element binding protein-1c is a major mediator of insulin action on the hepatic expression of glucokinase and lipogenesis-related genes. Proc. Natl. Acad. Sci. USA 96:12737-12742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grönroos, E., U. Hellman, C. H. Heldin, and J. Ericsson. 2002. Control of Smad7 stability by competition between acetylation and ubiquitination. Mol. Cell 10:483-493. [DOI] [PubMed] [Google Scholar]
- 18.Grossman, S. R., M. Perez, A. L. Kung, M. Joseph, C. Mansur, Z. X. Xiao, S. Kumar, P. M. Howley, and D. M. Livingston. 1998. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol. Cell 2:405-415. [DOI] [PubMed] [Google Scholar]
- 19.Gu, W., and R. G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90:595-606. [DOI] [PubMed] [Google Scholar]
- 20.Hirano, Y., M. Yoshida, M. Shimizu, and R. Sato. 2001. Direct demonstration of rapid degradation of nuclear sterol regulatory element-binding proteins by the ubiquitin-proteasome pathway. J. Biol. Chem. 276:36431-36437. [DOI] [PubMed] [Google Scholar]
- 21.Ito, A., C. H. Lai, X. Zhao, S. Saito, M. H. Hamilton, E. Appella, and T. P. Yao. 2001. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 20:1331-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawai, H., L. Nie, D. Wiederschain, and Z. M. Yuan. 2001. Dual role of p300 in the regulation of p53 stability. J. Biol. Chem. 276:45928-45932. [DOI] [PubMed] [Google Scholar]
- 23.Kouzarides, T. 2000. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 19:1176-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez-Balbas, M. A., U. M. Bauer, S. J. Nielsen, A. Brehm, and T. Kouzarides. 2000. Regulation of E2F1 activity by acetylation. EMBO J. 19:662-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munshi, N., M. Merika, J. Yie, K. Senger, G. Chen, and D. Thanos. 1998. Acetylation of HMG I(Y) by CBP turns off IFN beta expression by disrupting the enhanceosome. Mol. Cell 2:457-467. [DOI] [PubMed] [Google Scholar]
- 26.Naar, A. M., P. A. Beaurang, K. M. Robinson, J. D. Oliner, D. Avizonis, S. Scheek, J. Zwicker, J. T. Kadonaga, and R. Tjian. 1998. Chromatin, TAFs, and a novel multiprotein coactivator are required for synergistic activation by Sp1 and SREBP-1a in vitro. Genes Dev. 12:3020-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naar, A. M., P. A. Beaurang, S. Zhou, S. Abraham, W. Solomon, and R. Tjian. 1999. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature 398:828-832. [DOI] [PubMed] [Google Scholar]
- 28.Nagoshi, E., and Y. Yoneda. 2001. Dimerization of sterol regulatory element-binding protein 2 via the helix-loop-helix-leucine zipper domain is a prerequisite for its nuclear localization mediated by importin β. Mol. Cell. Biol. 21:2779-2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamura, S., J. A. Roth, and T. Mukhopadhyay. 2000. Multiple lysine mutations in the C-terminal domain of p53 interfere with MDM2-dependent protein degradation and ubiquitination. Mol. Cell. Biol. 20:9391-9398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishihara, A., J. Hanai, T. Imamura, K. Miyazono, and M. Kawabata. 1999. E1A inhibits transforming growth factor-beta signaling through binding to Smad proteins. J. Biol. Chem. 274:28716-28723. [DOI] [PubMed] [Google Scholar]
- 31.Osborne, T. F. 2000. Sterol regulatory element-binding proteins (SREBPs): key regulators of nutritional homeostasis and insulin action. J. Biol. Chem. 275:32379-32382. [DOI] [PubMed] [Google Scholar]
- 32.Parraga, A., L. Bellsolell, A. R. Ferre-D'Amare, and S. K. Burley. 1998. Co-crystal structure of sterol regulatory element binding protein 1a at 2.3 Å resolution. Structure 6:661-672. [DOI] [PubMed] [Google Scholar]
- 33.Pearson, M., R. Carbone, C. Sebastiani, M. Cioce, M. Fagioli, S. Saito, Y. Higashimoto, E. Appella, S. Minucci, P. P. Pandolfi, and P. G. Pelicci. 2000. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature 406:207-210. [DOI] [PubMed] [Google Scholar]
- 34.Polesskaya, A., A. Duquet, I. Naguibneva, C. Weise, A. Vervisch, E. Bengal, F. Hucho, P. Robin, and A. Harel-Bellan. 2000. CREB-binding protein/p300 activates MyoD by acetylation. J. Biol. Chem. 275:34359-34364. [DOI] [PubMed] [Google Scholar]
- 35.Rawson, R. B., N. G. Zelenski, D. Nijhawan, J. Ye, J. Sakai, M. T. Hasan, T. Y. Chang, M. S. Brown, and J. L. Goldstein. 1997. Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol. Cell 1:47-57. [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez, M. S., J. M. P. Desterro, S. Lain, D. P. Lane, and R. T. Hay. 2000. Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol. Cell. Biol. 20:8458-8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosen, E. D., C. J. Walkey, P. Puigserver, and B. M. Spiegelman. 2000. Transcriptional regulation of adipogenesis. Genes Dev. 14:1293-1307. [PubMed] [Google Scholar]
- 38.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70:81-120. [DOI] [PubMed] [Google Scholar]
- 39.Sakai, J., R. B. Rawson, P. J. Espenshade, D. Cheng, A. C. Seegmiller, J. L. Goldstein, and M. S. Brown. 1998. Molecular identification of the sterol-regulated luminal protease that cleaves SREBPs and controls lipid composition of animal cells. Mol. Cell 2:505-514. [DOI] [PubMed] [Google Scholar]
- 40.Sanchez, H. B., L. Yieh, and T. F. Osborne. 1995. Cooperation by sterol regulatory element-binding protein and Sp1 in sterol regulation of low density lipoprotein receptor gene. J. Biol. Chem. 270:1161-1169. [DOI] [PubMed] [Google Scholar]
- 41.Sartorelli, V., P. L. Puri, Y. Hamamori, V. Ogryzko, G. Chung, Y. Nakatani, J. Y. Wang, and L. Kedes. 1999. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol. Cell 4:725-734. [DOI] [PubMed] [Google Scholar]
- 42.Seigneurin-Berny, D., A. Verdel, S. Curtet, C. Lemercier, J. Garin, S. Rousseaux, and S. Khochbin. 2001. Identification of components of the murine histone deacetylase 6 complex: link between acetylation and ubiquitination signaling pathways. Mol. Cell. Biol. 21:8035-8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shimomura, I., Y. Bashmakov, S. Ikemoto, J. D. Horton, M. S. Brown, and J. L. Goldstein. 1999. Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes. Proc. Natl. Acad. Sci. USA 96:13656-13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soutoglou, E., N. Katrakili, and I. Talianidis. 2000. Acetylation regulates transcription factor activity at multiple levels. Mol. Cell 5:745-751. [DOI] [PubMed] [Google Scholar]
- 45.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki, T., A. Kimura, R. Nagai, and M. Horikoshi. 2000. Regulation of interaction of the acetyltransferase region of p300 and the DNA-binding domain of Sp1 on and through DNA binding. Genes Cells 5:29-41. [DOI] [PubMed] [Google Scholar]
- 47.Waltzer, L., and M. Bienz. 1998. Drosophila CBP represses the transcription factor TCF to antagonize Wingless signalling. Nature 395:521-525. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, Q., H. Yao, N. Vo, and R. H. Goodman. 2000. Acetylation of adenovirus E1A regulates binding of the transcriptional corepressor CtBP. Proc. Natl. Acad. Sci. USA 97:14323-14328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, W., S. Kadam, B. M. Emerson, and J. J. Bieker. 2001. Site-specific acetylation by p300 or CREB binding protein regulates erythroid Kruppel-like factor transcriptional activity via its interaction with the SWI-SNF complex. Mol. Cell. Biol. 21:2413-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]