Abstract
The level of genomic DNA methylation plays an important role in development and disease. In order to establish an experimental system for the functional analysis of genome-wide hypermethylation, we overexpressed the mouse de novo methyltransferase Dnmt3a in Drosophila melanogaster. These flies showed severe developmental defects that could be linked to reduced rates of cell cycle progression and irregular chromosome condensation. In addition, hypermethylated chromosomes revealed elevated rates of histone H3-K9 methylation and a more restricted pattern of H3-S10 phosphorylation. The developmental and chromosomal defects induced by DNA hypermethylation could be rescued by mutant alleles of the histone H3-K9 methyltransferase gene Su(var)3-9. This mutation also resulted in a significantly decreased level of genomic DNA methylation. Our results thus uncover the molecular consequences of genomic hypermethylation and demonstrate a mutual interaction between DNA methylation and histone methylation.
Aberrant DNA methylation patterns are one of the most consistent hallmarks of cancer (31). Many of the epigenetic aberrations that occur during tumorigenesis can be related to ectopic hypermethylation (4). While the analysis by various experimental systems of reduced levels of DNA methylation has helped to define the role of DNA methylation, the functional consequences of DNA hypermethylation remain unclear. DNA methylation is catalyzed by a class of enzymes called DNA methyltransferases (7). During early mammalian embryogenesis, de novo methyltransferases catalyze the methylation of previously unmethylated cytosine residues. During later stages, DNA methylation patterns are copied postreplicatively from the parental strand to the daughter strand (29). This step requires a functionally specialized DNA methyltransferase that has been termed the maintenance methyltransferase. To date, there are four known mammalian DNA methyltransferase genes. Dnmt1 encodes an enzyme with maintenance methyltransferase activity (8, 22), while the closely related genes Dnmt3a and Dnmt3b encode de novo methyltransferases (26, 39, 48). The function of the fourth gene, Dnmt2, remains enigmatic (16, 49, 65).
Disruption of DNA methyltransferase activity proved that DNA methylation was essential for proper development of mice (35), frogs (59), and plants (18, 54). The analysis of corresponding DNA hypomethylation phenotypes identified a role for DNA methylation in the regulation of gene activity (28, 59), control of foreign DNA elements (42, 63), and genome stability (14). Similar conclusions have been reached from the analysis of ICF (immunodeficiency, centromeric instability, facial anomalies) syndrome patients. These patients lack the majority of DNMT3B methyltransferase activity (23, 48, 64) and show striking chromosome aberrations that have been attributed to the demethylation and concomitant decondensation of pericentromeric satellite DNA (57). Together, these results suggest that DNA methylation might influence yet undefined aspects of higher-order chromatin structure rather than encoding site-specific information about gene activity.
DNA methylation signals can be interpreted by methyl-DNA binding proteins (10). These proteins act as bifunctional molecules that specifically bind to methylated DNA and then recruit histone deacetylase complexes to their site of binding (46). This results in the establishment of chromatin structures that are characterized by reduced acetylation levels at the N-terminal tails of histones H3 and H4. Histone acetylation and deacetylation play important roles in epigenetic gene regulation, and methyl-DNA binding proteins provide a direct link between histone acetylation and DNA methylation.
In addition to histone acetylation, modifications such as phosphorylation and methylation of H3 have been described. Together, these modifications might constitute an epigenetic histone code (30, 62). Histone phosphorylation has long been held as a simple marker for chromosome condensation during mitosis, but more recent results also indicate a function in transcriptional regulation in Drosophila melanogaster (47). Histone methylation can have roles in gene activation (60) and repression (3, 34, 44), depending on the lysine residue being methylated. Recent work has led to an extensive characterization of H3-K9 methylation, which is associated with inactive regions in the genome such as constitutive (51) and facultative (11, 24, 50) heterochromatin. Furthermore, targeting of the mouse Suv39h histone methyltransferase gene has shown that H3-K9 methylation is required for the maintenance of genome stability. Mutant mice revealed various abnormalities in chromosome condensation and segregation (51). Interestingly, there are indications for a direct connection between H3-K9 methylation and DNA methylation. Loss of H3-K9 methylation in Neurospora crassa resulted in a dominant loss of DNA methylation (61). Similarly, DNA methylation in Arabidopsis thaliana has also been shown to be dependent on histone H3-K9 methylation (27). These results reinforced the notion of epigenetic cross talk between DNA methylation and histone modifications (6).
D. melanogaster contains a simple DNA methylation system that is active primarily during early embryonic development (37). In order to analyze the functional consequences of DNA methylation, we previously generated transgenic D. melanogaster strains that allow genomic hypermethylation by inducible overexpression of the murine Dnmt3a methyltransferase (39). This established a unique system with which to study the effects of genomic hypermethylation and demonstrated that the induction of ectopic de novo methylation resulted in developmental defects by an unknown mechanism (39). We have now determined the consequences of DNA hypermethylation at the cytological and molecular levels. We found that developmental defects are induced by DNA hypermethylation and are linked to delayed cell cycle progression. In this regard, chromosome analysis revealed a high incidence of structural abnormalities. In addition, hypermethylated chromosomes showed widespread misregulation of histone H3 methylation and phosphorylation. The DNA hypermethylation-induced phenotype could be partially rescued by mutations in the Su(var)3-9 histone H3 methyltransferase gene. Our results thus demonstrate a functional interaction between DNA methylation and epigenetic chromatin modifications in D. melanogaster.
MATERIALS AND METHODS
Construction of plasmids.
We generated plasmid pUAST-D3aΔc by removing parts of the C-terminal methyltransferase domain from Dnmt3a. To this end, we amplified the sequence encoding amino acids 520 to 656 by PCR with the primers 5′-GGAGTGTGCTTACCAGTATGACG-3′ and 5′-ATCGTCTAGATTAAATGTAGCGGTCCACTTGG-3′. The latter primer contains a novel XbaI restriction site and a stop codon. The PCR product was digested with KpnI and XbaI. We then exchanged the 3′ sequence of pUAST-Dnmt3a (39), encoding amino acids 531 to 908, for the digested PCR product encoding amino acids 531 to 656.
Fly stocks and generation of transgenic flies.
Fly stocks were maintained under standard conditions. UAS-D3aΔc strains were generated by P-element-mediated transformation with standard procedures and a w1118 strain as the host. Other strains used carried UAS-Dnmt3a (39), GAL4-1032 (39), GawB(69B) (12), Da-GAL4 (a kind gift from J. M. Dura), Su(var)3-906 (56), and Su(var)3-919 (a kind gift from G. Reuter).
Antibodies.
The following antibodies were used for immunofluorescence: rat antibromodeoxyuridine (α-BrdU; Harlan Sera Laboratories, 1:2); rabbit anti-5-methylcytosine (Megabase Research Products, 1:100); rabbit anti-H3 (Cell Signaling, 1:200); rabbit anti-Cid (25) (1:200); rabbit anti-acetylated H4 (Upstate Biotechnology, 1:200); rabbit anti-acetylated H3-K9 (Cell Signaling, 1:200); rabbit anti-4x-methylated H3-K9 (51) (1:2,000); and mouse anti-phosphorylated H3-S10 (Cell Signaling, 1:50). Secondary antibodies were indocarbocyanine (Cy3)-conjugated goat anti-rat immunoglobulin (Dianova; 1:500); Cy3-conjugated goat anti-rabbit immunoglobulin (Jackson Immuno Research, 1:500); Cy3-conjugated goat anti-mouse immunoglobulin (Jackson Immuno Research, 1:500); and fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin (Jackson Immuno Research; 1:500).
Western blotting.
Overexpression of transgenes was confirmed by Western analysis of protein extracts from third-instar larvae with standard procedures. For immunodetection, anti-Dnmt3a (a kind gift from En Li) was diluted 1:4,000, anti-4x-methylated H3-K9 (51) was diluted 1:10,000, and anti-H3 (Cell Signaling) was diluted 1:1,000.
Determination of cytosine methylation levels.
Genomic DNA was isolated from larval and pupal homogenates. Standard calf thymus DNA was purchased from Sigma. DNA samples were derivatized and analyzed by capillary electrophoresis as described previously (58). Samples from independent DNA preparations were measured at least three times, and the results were found to be strictly reproducible.
BrdU incorporation assays.
For in vitro incorporation assays, third-instar larvae were rinsed in phosphate-buffered saline (PBS), and brains were dissected in 0.7% NaCl. Whole brains were incubated in 10 mg of BrdU/PBS per ml at 25°C for 1.5 h. Brains were then fixed in 4% paraformaldehyde in PBS for 20 min and washed in PBTx (PBS plus 0.05% Triton X-100) for 10 min. DNA was denatured in fresh 2 M HCl for 30 min, and brains were washed for 5 min in PBTx and incubated with the primary antibody diluted in PBS overnight at 4°C. Brains were washed three times for 5 min each with PBTx and incubated with secondary antibody diluted in PBS containing 0.3% bovine serum albumin for 2 h at room temperature and washed three times for 5 min each. Nuclei were stained for 10 min with 4′,6′-diamidino-2-phenylindole (DAPI) (100 ng/ml in PBTx). After washing for 5 min in PBTx, brains were transferred to polylysine slides containing Mowiol. Several independent preparations were examined with a Leica DMR microscope, and the results were found to be strictly reproducible.
For in vivo incorporation assays, third-instar larvae were fed for 1 h with 50 mg of BrdU (Sigma) per ml in D. melanogaster instant food (Sigma). The food contained food coloring to visualize which larvae had ingested the drug. The brains were dissected, hypotonically swollen, fixed, and immunostained as described previously (36). The preparations were examined with a Leica DMR microscope.
Staining of salivary glands.
Third-instar salivary glands were dissected in PBS, permeabilized for 10 min in PBS-0.1% Triton X-100, stained with DAPI (100 ng/ml in PBS) for 10 min, and mounted in Fluoromount G (Southern Biotechnology).
Mitotic neuroblast chromosomes.
Brains of third-instar larvae were dissected in 1× EBR buffer (130 mM NaCl, 4.7 mM KCl, 1.9 mM CaCl2, 10 mM HEPES, pH 6.9) and hypotonically swollen for 3 min in 0.25× EBR. Brains were then fixed for 5 min in 4% paraformaldehyde-5% acetic acid-0.1% Triton X-100 in 0.1725× EBR on a coverslip and during this time were squashed repeatedly onto polylysine-treated slides. Slides were frozen in liquid nitrogen, and the coverslip was flicked off with a razor blade. Slides were subsequently washed for 2 min in PBS, permeablized three times for 10 min each in PBTx (0.1% Triton X-100 in PBS).
For 5-methylcytosine staining, we denatured the DNA for 30 min in 2 M HCl at room temperature and then neutralized samples in 100 mM Tris (pH 8.5). Samples were then blocked in PBS containing 3% bovine serum albumin for 1 h at room temperature. Slides were then washed for 5 min in PBTx and incubated for 16 h at 4°C with the primary antibody diluted in PBS containing 0.3% bovine serum albumin. Slides were washed six times for 5 min each in PBTx and incubated for 2 h at room temperature with the appropriate secondary antibody. Slides were again washed six times for 5 min each in PBTx. Chromosomes were stained for 10 min with DAPI (100 ng/ml in PBTx). After washing for 5 min in PBTx, slides were mounted in Fluoromount G (Southern Biotechnology). Several independent preparations were examined with a Leica DMR microscope, and the results were always found to be reproducible.
RESULTS
Characterization of DNA hypermethylation-induced phenotype in D. melanogaster.
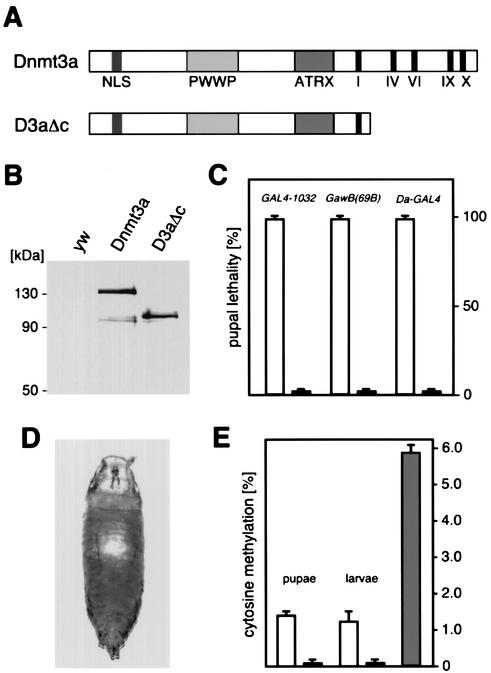
A transgenic Drosophila system that allows genomic hypermethylation by GAL4-inducible expression of the mouse de novo DNA methyltransferase Dnmt3a has been described previously (39). Overexpression with several different GAL4 drivers caused lethality and/or developmental defects. This suggested a hypermethylation phenotype that can potentially be linked to the biological function of DNA methylation. In order to validate the connection between the phenotype and DNA methylation, we generated a control Dnmt3a transgene that carries a deletion of the catalytic domain but retains the entire N-terminal domain of Dnmt3a (D3aΔc, Fig. 1A). Due to the removal of essential catalytic motifs, this protein has lost its ability to methylate DNA (also see Fig. 1E and Fig. 4A). The protein from this transgene was present at levels similar to that of transgenically overexpressed Dnmt3a (Fig. 1B). Overexpression of Dnmt3a invariably resulted in pupal lethality with the GAL4 lines tested (Fig. 1C). In contrast, overexpression of the catalytically inactive control protein did not affect fly development at any detectable rate (Fig. 1C). This demonstrated that the phenotype is caused by the DNA methylation activity rather than by other properties of the Dnmt3a protein.
FIG. 1.
Expression of mouse DNA methyltransferase Dnmt3a in D. melanogaster. (A) Dnmt3a constructs used for GAL4-dependent overexpression. The conserved catalytic motifs of the C-terminal catalytic domain are shown as black bars. NLS indicates the predicted nuclear localization signal. The PWWP domain has been implicated in DNA binding (52), and the ATRX domain has been implicated in histone deacetylase interactions (19). (B) Equal expression from all transgenes was confirmed by Western blotting. No protein was detectable in control crosses (yw). (C) Expression of Dnmt3a transgenes with various GAL4 lines that result in high-level and widespread transactivation. Expression of the full-length transgene caused lethality in all of the crosses analyzed (open bars), and no phenotype could be observed upon overexpression of the control construct (black bars). (D) Appearance of the DNA hypermethylation-induced phenotype after Da-GAL4 transactivation. (E) Genomic cytosine methylation levels were determined by capillary electrophoretic analysis. Dnmt3a overexpression with Da-GAL4 resulted in 1.4% cytosine methylation in early pupae and 1.2% cytosine methylation in third-instar larvae (open bars). Only background levels of cytosine methylation were detectable in control pupae and larvae overexpressing the D3aΔc control protein (black bars). Normal calf thymus DNA was included as an additional control (grey bar).
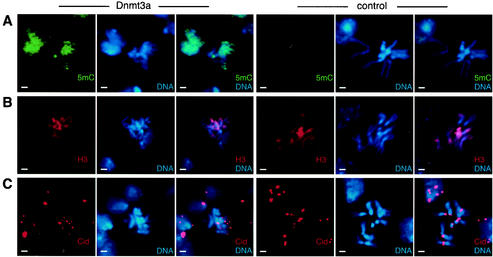
FIG. 4.
Mitotic chromosomes are methylated homogeneously and show defects in chromosome condensation. Larval neuroblast chromosome preparations were stained with various antibodies, as indicated. Bars, 2 μm. (A) Immunodetection of 5-methylcytosine (5mC) revealed widespread DNA methylation in Dnmt3a-overexpressing flies, while no DNA methylation was detectable in flies overexpressing the catalytically inactive control. (B) Immunodetection of histone H3 indicated that the distribution of nucleosomes was not affected by DNA hypermethylation. (C) The localization of the centromere identifier protein Cid was not altered by genomic hypermethylation. However, the pairwise staining pattern of the centromere was disrupted in Dnmt3a-overexpressing flies. Note the frequent overcondensation of hypermethylated mitotic chromosomes.
In order to further characterize the hypermethylation-induced phenotype, we focused on UAS-Dnmt3a induction by Daughterless-GAL4 (Da-GAL4). This GAL4 strain results in potent transgene activation during all stages of development and at various tissues, including salivary glands and larval brains (data not shown). As a consequence of Dnmt3a-induced hypermethylation, flies died as early, undifferentiated pupae (Fig. 1D). To determine the genomic methylation level in phenotypically affected pupae, we isolated genomic DNA from pupal homogenates and determined the cytosine methylation level by capillary electrophoresis. This revealed 1.4% genomic cytosine methylation (Fig. 1E). The preceding developmental stage (third-instar larvae) did not show any lethal effects from Da-GAL4-mediated Dnmt3a overexpression. Analysis of genomic DNA from these larvae revealed a slightly lower level of cytosine methylation (Fig. 1E). Control larvae and pupae overexpressing the catalytically inactive Dnmt3a protein showed only background levels of DNA methylation. Consistent with published data (20), standard calf thymus DNA revealed 5.6% cytosine methylation (Fig. 1E). The moderate cytosine methylation level observed in phenotypically affected Drosophila tissues is unlikely to have direct consequences on DNA structure or stability. Therefore we focused our analyses on cellular pathways that potentially interact with DNA methylation.
Genomic hypermethylation delays cell cycle progression.
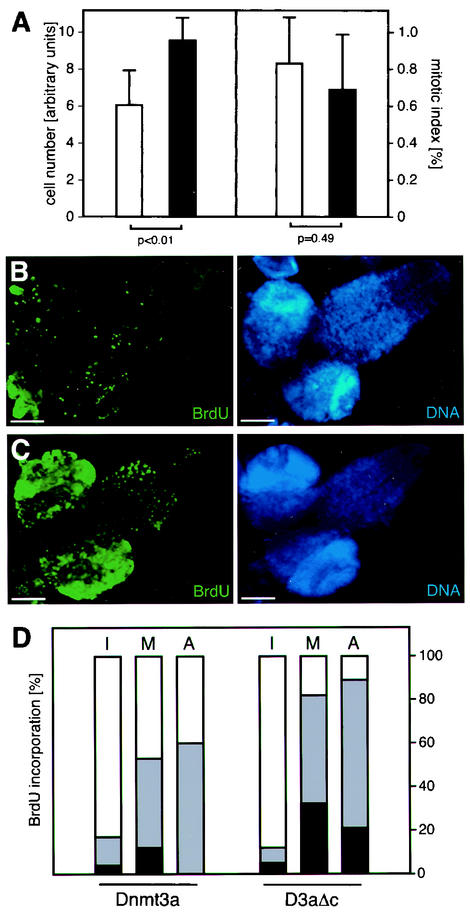
For a general characterization of the hypermethylation-induced phenotype, we analyzed the cell number and the mitotic index. Overexpression of Dnmt3a was induced by Da-GAL4, and imaginal discs as well as larval brains were isolated from third-instar larvae. Cells were counted from trypsinized wing discs, and the mitotic index was determined in larval neuroblast preparations. This revealed a significant reduction in cell numbers and a slightly elevated mitotic index in flies overexpressing Dnmt3a (Fig. 2A). These results prompted us to analyze the mitotic activity in higher detail. Isolated larval brains were incubated in a medium containing BrdU, and incorporation was visualized by immunostaining. This revealed dramatically reduced amounts of BrdU staining in brains from Dnmt3a-overexpressing flies compared to flies overexpressing the catalytically inactive control (Fig. 2B, compare to 2C). Thus, flies with ectopic DNA methylation showed a reduced rate of DNA synthesis.
FIG. 2.
Genomic hypermethylation induces a delay in cell cycle progression. (A) Imaginal discs and brains were isolated from third-instar larvae and used for the determination of cell numbers and the mitotic index, respectively. White bars represent results from flies overexpressing Dnmt3a, and black bars represent results from flies overexpressing the catalytically inactive protein. Overexpression of Dnmt3a caused a significant (P < 0.01, as determined by a standard t test) reduction in cell numbers and also resulted in a slightly increased mitotic index. (B) Isolated brains were cultured in vitro in the presence of BrdU. BrdU incorporation was visualized by immunodetection. Bars, 100 μm. Overexpression of Dnmt3a caused dramatically reduced BrdU incorporation. (C) Overexpression of the catalytically inactive protein had no effect on BrdU incorporation. (D) In vivo BrdU uptake into larval neuroblasts. Larvae were fed BrdU-containing medium for 1 h, followed by preparation of brain neuroblasts. Staining with a BrdU-specific antibody revealed reduced levels of BrdU incorporation in hypermethylated metaphase (M) and anaphase (A) chromosomes. Black fractions represent complete staining, grey fractions partial staining, and white fractions no staining. Because most of the interphase (I) nuclei were presumably in G1 phase, only a small percentage incorporated BrdU. The data are based on the examination of several hundred nuclei from several independent preparations.
In order to further analyze this effect under in vivo conditions, third-instar larvae were fed BrdU-containing medium for 1 h. Larval brains were then isolated, and neuroblast preparations were double stained with a BrdU-specific antibody and with DAPI to visualize DNA. This allowed us to determine the rate of BrdU incorporation and to distinguish between different stages of the cell cycle. Chromosomes at interphase, metaphase, or anaphase of the cell cycle can be easily distinguished by their visual appearance. Therefore, we were able to discriminate between cells that had different prerequisites with respect to BrdU incorporation. It was observed that the majority of interphase cells did not incorporate BrdU, indicating that these cells were in G1 phase. In contrast, cells which are in the mitotic cycle stained positively for BrdU. This pattern could be observed in both Dnmt3a-overexpressing flies and in flies overexpressing the catalytically inactive control protein (Fig. 2D). However, the fraction of labeled metaphase and anaphase chromosomes was significantly lower in hypermethylated neuroblasts compared to controls (Fig. 2D, compare left and right panels). These results are consistent with our in vitro BrdU incorporation assay and indicate a delay in cell cycle progression dependent on DNA methylation.
Genomic hypermethylation affects polytene chromosome structure.
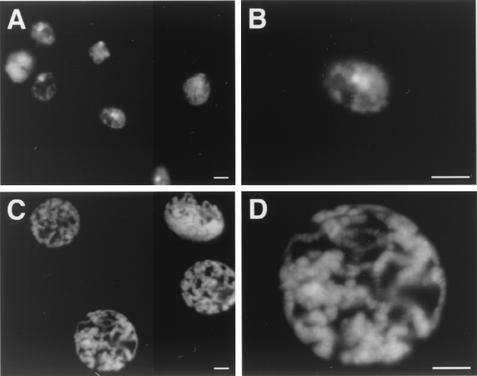
Impaired chromatin structure can conceivably account for the observed delay of the cell cycle. To explore this possibility, we analyzed polytene chromosomes from the salivary glands of third-instar larvae. Polytene chromosomes are generated by several rounds of DNA replication without mitosis. Because of their large size and their polytenization, they provide an excellent system with which to investigate the effect of DNA hypermethylation on chromatin structure. We induced protein expression with various GAL4 lines (including Da-GAL4) that result in strong transactivation in salivary glands. After dissection of third-instar larvae, we observed a significant size reduction of salivary glands overexpressing Dnmt3a compared to control glands overexpressing the catalytically inactive Dnmt3a derivative (data not shown).
Interestingly, we were unable to obtain polytene chromosome preparations from glands overexpressing Dnmt3a, whereas control glands showed polytene chromosomes of normal appearance. Therefore, we prepared whole-mount salivary glands from Dnmt3a-overexpressing and control larvae and stained them with DAPI. This revealed a highly unusual DNA organization in nuclei from Dnmt3a-overexpressing glands (Fig. 3A). While some nuclei showed rudimentary polytenization, the chromosomes frequently appeared underreplicated (Fig. 3A). In most cases, the DNA looked similar to interphase nuclei with no obvious polytenization (Fig. 3B). In contrast, nuclei from control glands overexpressing the catalytically inactive protein showed distinct polytene chromosomes with no detectable structural alterations (Fig. 3C and D). Therefore, we concluded that hypermethylation of salivary gland chromosomes affects their structural organization and interferes with their polytenization.
FIG. 3.
Ectopic de novo methylation affects polytene chromosome structure. (A) Overexpression of Dnmt3a resulted in loss of polytene chromosome organization in salivary glands. (B) Higher magnification of a single nucleus. Bars, 10 μm. (C) Overexpression of the catalytically inactive Dnmt3a protein had no effect. (D) Higher magnification of a single nucleus. Nuclei with hypermethylated DNA frequently appeared smaller and showed indications of DNA underreplication.
Genomic hypermethylation results in irregular chromosome condensation.
In order to characterize structural chromosome abnormalities in higher detail and to determine their relationship to DNA methylation, we stained mitotic chromosomes from larval neuroblasts with an antibody that specifically recognizes 5-methylcytosine. Significant levels of endogenous DNA methylation in D. melanogaster have been detected only during embryogenesis (21, 38). Thus, a low background methylation level is expected in third-instar larvae. 5-Methylcytosine staining of larval neuroblast preparations from transgenic fly strains demonstrated widespread chromosome staining in Dnmt3a-overexpressing flies (Fig. 4A). No staining was detectable in preparations from larval brains overexpressing the control protein (Fig. 4A). At the same time, about a third of the methylated mitotic chromosomes revealed structural changes (Fig. 4). No structural changes could be detected with unmethylated control chromosomes (Fig. 4, Fig. 5). Methylated chromosomes were frequently overcondensed to a variable degree, ranging from a visible compaction of chromosome arms to the appearance of highly condensed structures (Fig. 4). In some cases we also observed chromosome rearrangements and misaligned sister chromatids (data not shown). These results demonstrate a pronounced effect of DNA methylation on chromosome structure.
FIG. 5.
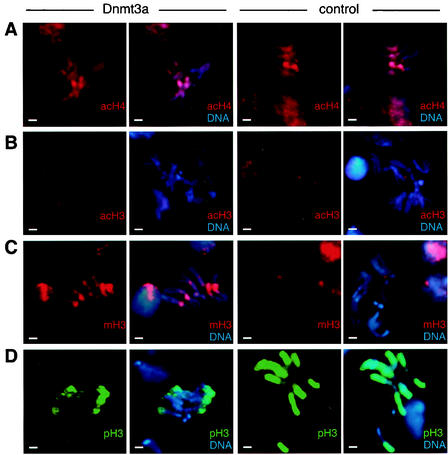
DNA hypermethylation causes changes in epigenetic histone modifications. (A) Immunodetection of acetylated histone H4 (acH4) on chromosome preparations from larval neuroblasts. There was no detectable difference between flies expressing full-length or deleted Dnmt3a protein. (B) Similarly, H3-K9 acetylation (acH3) was found to be very low on mitotic chromosomes and did not change upon DNA methylation. (C) Immunodetection of K9-methylated histone H3 (mH3). Chromosomes from Dnmt3a-overexpressing flies showed a strongly increased level of H3 methylation compared to controls. The majority of K9-methylated H3 staining was located in pericentromeric heterochromatin. (D) Phosphorylation of H3-S10 (pH3) was strongly reduced on chromosomes from Dnmt3a-overexpressing flies. Only the telomere-proximal regions of the chromosome arms remained stained. Control chromosomes showed widespread and homogeneous phosphorylation of H3. Bars, 2 μm.
In order to investigate the basal chromatin organization of hypermethylated chromosomes, we also stained larval neuroblast preparations with antibodies against structural chromatin components. Staining with a histone H3-specific antibody resulted in homogeneous labeling of mitotic chromosomes (Fig. 4B). The staining pattern of methylated chromosomes was indistinguishable from that of unmethylated controls (Fig. 4B), indicating a similar distribution of nucleosomes in both cases. Similarly, the distribution of heterochromatin protein 1 did not change upon chromosome hypermethylation (data not shown). Staining with an antibody against the centromere-specific H3 variant Cid (25) revealed a characteristic centromeric double-dot pattern for control chromosomes (Fig. 4C). The pairwise alignment of signals was frequently disturbed in hypermethylated chromosomes (Fig. 4C), which is probably a consequence of the structural chromosome changes described above. However, in all cases Cid was found strictly at the centromeres. Taken together, our results thus suggest substantial preservation of the overall chromatin organization in hypermethylated chromosomes.
Genomic hypermethylation influences epigenetic histone modifications.
As an alternative to gross structural chromatin changes, we also considered the possibility of DNA methylation-induced changes in histone modifications. Therefore, we stained mitotic chromosomes from larval neuroblasts with various antibodies against modified histone H3 and histone H4. We first analyzed histone acetylation, a chromatin modification that has frequently been found to be associated with DNA methylation (46). More specifically, acetylation of histone H4 has been demonstrated to be involved in various epigenetic phenomena (62). When we stained chromosome preparations from larval neuroblasts with an antibody that specifically recognizes acetylated H4, we were unable to detect any significant differences between methylated and unmethylated chromosomes (Fig. 5A). Similarly, H3-K9 acetylation levels were rather low and appeared not to change upon chromosome methylation (Fig. 5B). We therefore concluded that DNA hypermethylation has no detectable effect on the acetylation status of histones in mitotic chromosomes.
We then sought to determine the status of histone H3-K9 methylation. This modification has been shown to be a marker of constitutive (51) and facultative (11, 24, 50) heterochromatin. When we compared hypermethylated larval neuroblast chromosomes to unmethylated chromosomes, we observed a strong increase in histone methylation in response to DNA methylation. This increase was restricted to pericentromeric regions and was observed in numerous independent preparations with two independent antibodies (tetramethyl H3 [Fig. 5C] and dimethyl H3 [data not shown]). The changes in H3 methylation did not depend on structural chromosome abnormalities, since they were equally observed in structurally normal chromosomes.
Histone H3 phosphorylation is primarily detectable during metaphase and has been shown to be a marker of transcriptionally active chromosome regions in D. melanogaster (47). The level of H3-S10 phosphorylation appears to be inversely correlated with the level of H3-K9 methylation (53). Our previous results would therefore predict a reduced level of H3 phosphorylation on hypermethylated chromosomes. Indeed, many hypermethylated chromosomes showed an irregular staining pattern that was frequently restricted to regions close to the telomeres (Fig. 5D). This is in marked contrast to control chromosomes, which revealed intense and homogeneous staining of the euchromatic arms (Fig. 5D). We therefore concluded that DNA hypermethylation causes changes in histone phosphorylation.
Rescue of DNA hypermethylation-induced defects by mutations in the histone methyltransferase gene Su(var)3-9.
Several characteristics of the DNA hypermethylation phenotype in flies are reminiscent of the cellular phenotype induced by overexpression of the mammalian SUV39H1 H3-K9 methyltransferase (41). Therefore, we addressed the functional interaction between DNA methylation and histone methylation by inducing DNA hypermethylation in flies with a heterozygous null mutation for the histone H3-K9 methyltransferase gene Su(var)3-9 (56). To this end, we introduced Su(var)3-9 mutant alleles into our UAS-Dnmt3a strains and activated transgene expression with GAL4-1032. This revealed a partial but unambiguous rescue of the developmental defects compared to controls with wild-type Su(var)3-9 activity (Fig. 6A).
FIG. 6.
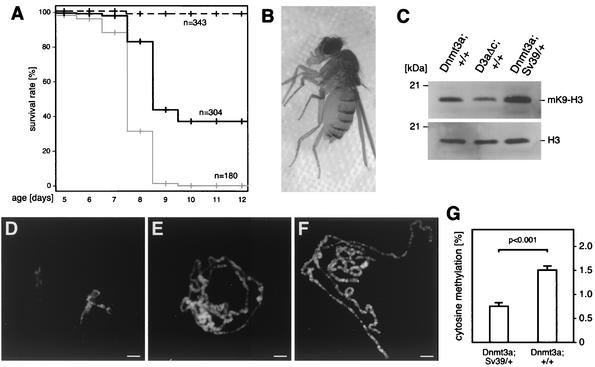
Rescue of DNA hypermethylation-induced phenotype by Su(var)3-9 mutations in H3-K9 methyltransferase. (A) Survival of flies was calculated by determining their stage of developmental arrest during pupal development. Flies overexpressing Dnmt3a in a wild-type Su(var)3-9 background are shown as a grey line, and flies overexpressing Dnmt3a in a heterozygous Su(var)3-9 null mutant background are shown as a black line. The mutant allele as such showed no detectable effect on fly development (dashed black line). For each experiment, we analyzed several independent crosses. (B) Rescued adult flies were viable and fertile and had a normal appearance. (C) Western analysis indicated larger amounts of methylated K9-H3 both in third-instar larvae overexpressing Dnmt3a in a wild-type background and in a heterozygous Su(var)3-9 mutant background. See text for details. (D to F) Rescue of the polytene chromosome phenotype. Bars, 10 μm. (D) No polytene chromosomes could be prepared from flies overexpressing Dnmt3a in a wild-type background. (E) The presence of a heterozygous mutant Su(var)3-9 allele resulted in the appearance of polytene chromosomes that were similar to wild-type chromosomes. (F) Overexpression of the catalytically inactive Dnmt3a protein combined with a heterozygous Su(var)3-9 mutation had no effect on polytene chromosome morphology. (G) Quantification of genomic DNA methylation levels in rescued (Dnmt3a; Sv39/+) and nonrescued (Dnmt3a; +/+) pupae. The reduction of H3-K9 methylation in rescued flies resulted in a significant decrease of the genomic DNA methylation level (P < 0.001, as determined by a standard t test).
While none of the flies overexpressing Dnmt3a in a wild-type Su(var)3-9 background survived the pupal stage, more than 30% of the rescued flies reached the adult stage (Fig. 6A). These flies had a normal appearance and were viable and fertile (Fig. 6B). Similar results were observed with two independent Su(var)3-9 alleles and an additional, independent Dnmt3a-overexpressing strain; homozygous Su(var)3-9 mutations increased the rescue effect only marginally, which is probably due to suppression of the Su(var)3-9 phenotype by Dnmt3a. Indeed, protein extracts from third-instar larvae overexpressing Dnmt3a in a heterozygous mutant Su(var)3-9 background showed slightly increased amounts of K9-methylated histone H3 (Fig. 6C). This might be due to the rescue activity of Dnmt3a and/or an altered activity of other H3-K9 methyltransferases, such as E(z) and Ash1 (5, 15, 43). Consistent with our mitotic chromosome analysis, protein extracts from whole third-instar larvae also showed an increased amount of K9-methylated histone H3 in flies overexpressing Dnmt3a in a wild-type Su(var)3-9 background (Fig. 6C, compare lane 1 and lane 2).
Finally, we also looked for a rescue of the salivary gland chromosome phenotype. Chromosome squashes were prepared from flies overexpressing either Dnmt3a or the control protein in a wild-type or heterozygous Su(var)3-9 mutant background. The results confirmed that no polytene chromosomes could be prepared from larvae overexpressing Dnmt3a in a wild-type background (Fig. 6D). In contrast, a large number of larvae overexpressing Dnmt3a in a heterozygous Su(var)3-9 mutant background showed polytene chromosomes that were similar to those of controls (Fig. 6E, compare to Fig. 6F). This chromosomal rescue effect confirmed our developmental rescue data. In order to analyze the effect of the Su(var)3-9 mutation on DNA methylation, we also prepared genomic DNA from rescued and nonrescued pupae and determined the global DNA methylation level by capillary electrophoretic analysis. This analysis revealed significantly reduced methylation level in rescued pupae (Fig. 6G). The dependence of DNA methylation on H3-K9 methylation is consistent with previous observations from other model organisms (27, 61), and it can also be observed for endogenous Drosophila DNA methylation (J. Marhold and F. Lyko, unpublished data). This effect is probably the underlying cause for the rescue activity of the Su(var)3-9 mutation, and this strongly suggests that the interaction between DNA methylation and H3-K9 methylation is mutual.
DISCUSSION
DNA hypermethylation has been frequently observed in tumors and also plays a role in several other diseases. However, attempts to generate experimental systems for ectopic DNA methylation have been largely unsuccessful. Slight hypermethylation of the mouse genome has recently been achieved by overexpression of the maintenance methyltransferase Dnmt1 in embryonic stem cells (9). However, transgenic mice could not be derived due to embryonic lethality, and central aspects of DNA methylation could not be addressed. We used a stringent inducible system (12) to improve control over experimental conditions. To this end, ectopic DNA methylation was induced by widespread overexpression of the mouse de novo methyltransferase Dnmt3a. As a consequence, flies died during the pupal stage of development. This stage might be particularly sensitive to DNA hypermethylation because cells are still actively dividing and are also subjected to differentiation processes.
Dnmt3a overexpression resulted in a maximum cytosine methylation level of 1.4%. Thus, the level of exogenous pupal methylation is considerably more than the 0.4% endogenous DNA methylation observed in D. melanogaster embryos (38). As judged from chromosome staining, DNA methylation appeared to be evenly distributed over the entire genome. Therefore, numerous loci should have been methylated during the majority of Drosophila development. While the effects of DNA hypermethylation on gene expression remain to be analyzed, our results strongly suggest an effect of DNA methylation on higher-order chromosome structure. In addition, DNA hypermethylation also induced a delay in cell cycle progression. Our observation of underreplicated polytene chromosomes in larval salivary glands is also in agreement with reduced DNA replication activity in tissues with hypermethylated genomic DNA.
As a consequence of genome hypermethylation, we detected frequent and consistent structural chromosome abnormalities. The majority of structurally deviant chromosomes appeared overcondensed, and a smaller number revealed chromosome rearrangements and sister chromatid cohesion defects (data not shown). Intriguingly, the overall structural changes appeared similar to those seen with certain cell cycle mutants of D. melanogaster (33, 36), which is consistent with our observation of delayed cell cycle progression. Lastly, a role of DNA methylation in the maintenance of structural chromosome integrity can also explain some of the mutations observed in mouse embryonic stem cells with a strongly demethylated genome (14).
The cellular pathways interacting with DNA methylation are still poorly understood. There is, however, mounting evidence for interactions between DNA methylation and covalent histone modifications. The first example was provided by vertebrate methyl-DNA binding proteins, which target histone deacetylases to methylated DNA (32, 45). Here, we did not detect any significant DNA methylation-dependent histone deacetylation in D. melanogaster. This is consistent with the fact that the Drosophila dMBD2/3 protein, which is associated with histone deacetylase activity and shows extensive homologies to mammalian methyl-DNA binding proteins, is expressed exclusively during embryogenesis (40). In addition, we limited our analysis to histone modifications of metaphase chromosomes. If a major fraction of deacetylated histones fails to be transmitted through mitosis, it would not have been detected in our experiments. It is remarkable that both the increased histone methylation and the more restricted phosphorylation were readily detectable on mitotic chromosomes. Thus, they are potentially able to act as independent epigenetic signals that can be stably transmitted through many cell generations. Mitotic stability has long been held as an exclusive hallmark of DNA methylation, but there is increasing evidence that specialized chromatin configurations can be similarly effective (13, 17).
The phenotype induced by DNA hypermethylation in flies appears strikingly similar to the defects caused by overexpression of the human SUV39H1 H3-K9 methyltransferase in cell lines (41). These cells revealed delayed cell cycle progression and irregular chromosome segregation and condensation, as well as abnormal histone H3 phosphorylation. For this reason, we addressed the possibility of a functional relationship between DNA methylation and histone methylation in D. melanogaster. Indeed, it has been shown recently that DNA methylation and H3-K9 methylation interact in N. crassa and in Arabidopsis thaliana (27, 61). Both reports concluded that DNA methylation depends on histone H3-K9 methylation. Both results also suggested that DNA methylation acts downstream of histone methylation.
Our results confirm a dependency of DNA methylation on histone methylation, but they also show that histone methylation can be induced by DNA methylation. This is an important distinction because a mutual relationship between DNA methylation and H3-K9 methylation implies a role for histone methylation in various stages of mammalian development and disease that had been characterized previously only through DNA methylation. The mechanistic basis for the induction of H3-K9 methylation by DNA methylation is still unknown but could conceivably involve methyl-DNA binding proteins. Apart from the Drosophila dMBD2/3 protein, which is associated with histone deacetylase activity (2), flies express several other proteins with a putative methyl-DNA binding domain (MBD) (1, 55) (data not shown). These proteins might either directly methylate histone H3 through a SET domain (the CG12196 gene contains both an MBD and a SET domain) or they might recruit the Su(var)3-9 histone methyltransferase to methylated DNA.
Acknowledgments
We thank Gunter Reuter, Bernard Mechler, and Margarete Heck for support and advice and En Li, Steve Henikoff, and Thomas Jenuwein for antibodies.
I.M.C. received a National Science Foundation Graduate Fellowship. The work of R.P. was supported by a grant from the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie. The work of F.L. was supported by an Emmy Noether fellowship from the Deutsche Forschungsgemeinschaft.
Footnotes
This paper is dedicated to Harald zur Hausen on the occasion of his retirement as head of the German Cancer Research Center, with gratitude and appreciation for 20 years of leadership.
REFERENCES
- 1.Adams, M. D., et al. 2000. The genome sequence of Drosophila melanogaster. Science 287:2185-2195. [DOI] [PubMed] [Google Scholar]
- 2.Ballestar, E., L. A. Pile, D. A. Wassarman, A. P. Wolffe, and P. A. Wade. 2001. A Drosophila MBD family member is a transcriptional corepressor associated with specific genes. Eur. J. Biochem. 268:5397-5406. [DOI] [PubMed] [Google Scholar]
- 3.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 4.Baylin, S. B., and J. G. Herman. 2000. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 16:168-174. [DOI] [PubMed] [Google Scholar]
- 5.Beisel, C., A. Imhof, J. Greene, E. Kremmer, and F. Sauer. 2002. Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature 419:857-862. [DOI] [PubMed] [Google Scholar]
- 6.Ben-Porath, I., and H. Cedar. 2001. Epigenetic crosstalk. Mol. Cell 8:933-935. [DOI] [PubMed] [Google Scholar]
- 7.Bestor, T. H. 2000. The DNA methyltransferases of mammals. Hum. Mol. Genet. 9:2395-2402. [DOI] [PubMed] [Google Scholar]
- 8.Bestor, T. H., and V. M. Ingram. 1983. Two DNA methyltransferases from murine erythroleukemia cells: purification, sequence specificity, and mode of interaction with DNA. Proc. Natl. Acad. Sci. USA 80:5559-5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Biniszkiewicz, D., J. Gribnau, B. Ramsahoye, F. Gaudet, K. Eggan, D. Humpherys, M. A. Mastrangelo, Z. Jun, J. Walter, and R. Jaenisch. 2002. Dnmt1 overexpression causes genomic hypermethylation, loss of imprinting, and embryonic lethality. Mol. Cell. Biol. 22:2124-2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bird, A. P., and A. P. Wolffe. 1999. Methylation-induced repression—belts, braces, and chromatin. Cell 99:451-454. [DOI] [PubMed] [Google Scholar]
- 11.Boggs, B. A., P. Cheung, E. Heard, D. L. Spector, A. C. Chinault, and C. D. Allis. 2001. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat. Genet. 30:73-76. [DOI] [PubMed] [Google Scholar]
- 12.Brand, A. H., and N. Perrimon. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401-415. [DOI] [PubMed] [Google Scholar]
- 13.Cavalli, G., and R. Paro. 1998. The Drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell 93:505-518. [DOI] [PubMed] [Google Scholar]
- 14.Chen, R. Z., U. Pettersson, C. Beard, L. Jackson-Grusby, and R. Jaenisch. 1998. DNA hypomethylation leads to elevated mutation rates. Nature 395:89-93. [DOI] [PubMed] [Google Scholar]
- 15.Czermin, B., R. Melfi, D. McCabe, V. Seitz, A. Imhof, and V. Pirrotta. 2002. Drosophila Enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal polycomb sites. Cell 111:185-196. [DOI] [PubMed] [Google Scholar]
- 16.Dong, A., J. A. Yoder, X. Zhang, L. Zhou, T. H. Bestor, and X. Cheng. 2001. Structure of human DNMT2, an enigmatic DNA methyltransferase homolog that displays denaturant-resistant binding to DNA. Nucleic Acids Res. 29:439-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ekwall, K., T. Olsson, B. M. Turner, G. Cranston, and R. C. Allshire. 1997. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell 91:1021-1032. [DOI] [PubMed] [Google Scholar]
- 18.Finnegan, E. J., W. J. Peacock, and E. S. Dennis. 1996. Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc. Natl. Acad. Sci. USA 93:8449-8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuks, F., W. A. Burgers, N. Godin, M. Kasai, and T. Kouzarides. 2001. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J. 20:2536-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gama-Sosa, M. A., R. M. Midgett, V. A. Slagel, S. Githens, K. C. Kuo, C. W. Gehrke, and M. Ehrlich. 1983. Tissue-specific differences in DNA methylation in various mammals. Biochim. Biophys. Acta 740:212-219. [DOI] [PubMed] [Google Scholar]
- 21.Gowher, H., O. Leismann, and A. Jeltsch. 2000. DNA of Drosophila melanogaster contains 5-methylcytosine. EMBO J. 19:6918-6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gruenbaum, Y., H. Cedar, and A. Razin. 1982. Substrate and sequence specificity of a eukaryotic DNA methylase. Nature 295:620-622. [DOI] [PubMed] [Google Scholar]
- 23.Hansen, R. S., C. Wijmenga, P. Luo, A. M. Stanek, T. K. Canfield, C. M. Weemaes, and S. M. Gartler. 1999. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc. Natl. Acad. Sci. USA 96:14412-14417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heard, E., C. Rougeulle, D. Arnaud, P. Avner, C. D. Allis, and D. L. Spector. 2001. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell 107:727-738. [DOI] [PubMed] [Google Scholar]
- 25.Henikoff, S., K. Ahmad, J. S. Platero, and B. van Steensel. 2000. Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl. Acad. Sci. USA 97:716-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsieh, C. L. 1999. In vivo activity of murine de novo methyltransferases Dnmt3a and Dnmt3b. Mol. Cell. Biol. 19:8211-8218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson, J. P., A. M. Lindroth, X. Cao, and S. E. Jacobsen. 2002. Control of CpNpG DNA methylation by the Kryptonite histone H3 methyltransferase. Nature 416:556-560. [DOI] [PubMed] [Google Scholar]
- 28.Jackson-Grusby, L., C. Beard, R. Possemato, M. Tudor, D. Fambrough, G. Csankovszki, J. Dausman, P. Lee, C. Wilson, E. Lander, and R. Jaenisch. 2001. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat. Genet. 27:31-39. [DOI] [PubMed] [Google Scholar]
- 29.Jaenisch, R. 1997. DNA methylation and imprinting: why bother? Trends Genet. 13:323-329. [DOI] [PubMed] [Google Scholar]
- 30.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 31.Jones, P. A., and S. B. Baylin. 2002. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 3:415-428. [DOI] [PubMed] [Google Scholar]
- 32.Jones, P. L., G. J. Veenstra, P. A. Wade, D. Vermaak, S. U. Kass, N. Landsberger, J. Strouboulis, and A. P. Wolffe. 1998. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 19:187-191. [DOI] [PubMed] [Google Scholar]
- 33.Krause, S. A., M. L. Loupart, S. Vass, S. Schoenfelder, S. Harrison, and M. M. Heck. 2001. Loss of cell cycle checkpoint control in Drosophila Rfc4 mutants. Mol. Cell. Biol. 21:5156-5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 35.Li, E., T. H. Bestor, and R. Jaenisch. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69:915-926. [DOI] [PubMed] [Google Scholar]
- 36.Loupart, M. L., S. A. Krause, and M. S. Heck. 2000. Aberrant replication timing induces defective chromosome condensation in Drosophila ORC2 mutants. Curr. Biol. 10:1547-1556. [DOI] [PubMed] [Google Scholar]
- 37.Lyko, F. 2001. DNA methylation learns to fly. Trends Genet. 17:169-172. [DOI] [PubMed] [Google Scholar]
- 38.Lyko, F., B. H. Ramsahoye, and R. Jaenisch. 2000. DNA methylation in Drosophila melanogaster. Nature 408:538-540. [DOI] [PubMed] [Google Scholar]
- 39.Lyko, F., B. H. Ramsahoye, H. Kashevsky, M. Tudor, M. A. Mastrangelo, T. L. Orr-Weaver, and R. Jaenisch. 1999. Mammalian (cytosine-5) methyltransferases cause genomic DNA methylation and lethality in Drosophila. Nat. Genet. 23:363-366. [DOI] [PubMed] [Google Scholar]
- 40.Marhold, J., M. Zbylut, D. H. Lankenau, M. Li, D. Gerlich, E. Ballestar, B. M. Mechler, and F. Lyko. 2002. Stage-specific chromosomal association of Drosophila dMBD2/3 during genome activation. Chromosoma 111:13-21. [DOI] [PubMed] [Google Scholar]
- 41.Melcher, M., M. Schmid, L. Aagaard, P. Selenko, G. Laible, and T. Jenuwein. 2000. Structure-function analysis of SUV39H1 reveals a dominant role in heterochromatin organization, chromosome segregation, and mitotic progression. Mol. Cell. Biol. 20:3728-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miura, A., S. Yonebayashi, K. Watanabe, T. Toyama, H. Shimada, and T. Kakutani. 2001. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 411:212-214. [DOI] [PubMed] [Google Scholar]
- 43.Muller, J., C. M. Hart, N. J. Francis, M. L. Vargas, A. Sengupta, B. Wild, E. L. Miller, M. B. O'Connor, R. E. Kingston, and J. A. Simon. 2002. Histone methyltransferase activity of a Drosophila polycomb group repressor complex. Cell 111:197-208. [DOI] [PubMed] [Google Scholar]
- 44.Nakayama, J., J. C. Rice, B. D. Strahl, C. D. Allis, and S. I. Grewal. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110-113. [DOI] [PubMed] [Google Scholar]
- 45.Nan, X., H. H. Ng, C. A. Johnson, C. D. Laherty, B. M. Turner, R. N. Eisenman, and A. Bird. 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393:386-389. [DOI] [PubMed] [Google Scholar]
- 46.Ng, H. H., and A. Bird. 1999. DNA methylation and chromatin modification. Curr. Opin. Genet. Dev. 9:158-163. [DOI] [PubMed] [Google Scholar]
- 47.Nowak, S. J., and V. G. Corces. 2000. Phosphorylation of histone H3 correlates with transcriptionally active loci. Genes Dev. 14:3003-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okano, M., D. W. Bell, D. A. Haber, and E. Li. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247-257. [DOI] [PubMed] [Google Scholar]
- 49.Okano, M., S. Xie, and E. Li. 1998. Dnmt2 is not required for de novo and maintenance methylation of viral DNA in embryonic stem cells. Nucleic Acids Res. 26:2536-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peters, A. H., J. E. Mermoud, D. O'Carroll, M. Pagani, D. Schweizer, N. Brockdorff, and T. Jenuwein. 2001. Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat. Genet. 30:77-80. [DOI] [PubMed] [Google Scholar]
- 51.Peters, A. H., D. O'Carroll, H. Scherthan, K. Mechtler, S. Sauer, C. Schofer, K. Weipoltshammer, M. Pagani, M. Lachner, A. Kohlmaier, S. Opravil, M. Doyle, M. Sibilia, and T. Jenuwein. 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107:323-337. [DOI] [PubMed] [Google Scholar]
- 52.Qiu, C., K. Sawada, X. Zhang, and X. Cheng. 2002. The PWWP domain of mammalian DNA methyltransferase Dnmt3b defines a new family of DNA-binding folds. Nat. Struct. Biol. 9:217-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rea, S., F. Eisenhaber, D. O'Carroll, B. D. Strahl, Z. W. Sun, M. Schmid, S. Opravil, K. Mechtler, C. P. Ponting, C. D. Allis, and T. Jenuwein. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593-599. [DOI] [PubMed] [Google Scholar]
- 54.Ronemus, M. J., M. Galbiati, C. Ticknor, J. Chen, and S. L. Dellaporta. 1996. Demethylation-induced developmental pleiotropy in Arabidopsis. Science 273:654-657. [DOI] [PubMed] [Google Scholar]
- 55.Rubin, G. M., L. Hong, P. Brokstein, M. Evans-Holm, E. Frise, M. Stapleton, and D. A. Harvey. 2000. A Drosophila complementary DNA resource. Science 287:2222-2224. [DOI] [PubMed] [Google Scholar]
- 56.Schotta, G., A. Ebert, V. Krauss, A. Fischer, J. Hoffmann, S. Rea, T. Jenuwein, R. Dorn, and G. Reuter. 2002. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 21:1121-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smeets, D. F., U. Moog, C. M. Weemaes, G. Vaes-Peeters, G. F. Merkx, J. P. Niehof, and G. Hamers. 1994. ICF syndrome: a new case and review of the literature. Hum. Genet. 94:240-246. [DOI] [PubMed] [Google Scholar]
- 58.Stach, D., O. J. Schmitz, S. Stilgenbauer, A. Benner, H. Dohner, M. Wiessler, and F. Lyko. Capillary electrophoretic analysis of genomic DNA methylation levels. Nucleic Acids Res., in press. [DOI] [PMC free article] [PubMed]
- 59.Stancheva, I., and R. R. Meehan. 2000. Transient depletion of xDnmt1 leads to premature gene activation in Xenopus embryos. Genes Dev. 14:313-327. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 60.Strahl, B. D., R. Ohba, R. G. Cook, and C. D. Allis. 1999. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc. Natl. Acad. Sci. USA 96:14967-14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tamaru, H., and E. U. Selker. 2001. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414:277-283. [DOI] [PubMed] [Google Scholar]
- 62.Turner, B. M. 2000. Histone acetylation and an epigenetic code. Bioessays 22:836-845. [DOI] [PubMed] [Google Scholar]
- 63.Walsh, C. P., J. R. Chaillet, and T. H. Bestor. 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20:116-117. [DOI] [PubMed] [Google Scholar]
- 64.Xu, G. L., T. H. Bestor, D. Bourc'his, C. L. Hsieh, N. Tommerup, M. Bugge, M. Hulten, X. Qu, J. J. Russo, and E. Viegas-Pequignot. 1999. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 402:187-191. [DOI] [PubMed] [Google Scholar]
- 65.Yoder, J. A., and T. H. Bestor. 1998. A candidate mammalian DNA methyltransferase related to pmt1p of fission yeast. Hum. Mol. Genet. 7:279-284. [DOI] [PubMed] [Google Scholar]