Abstract
A drastic reduction in BRCA1 gene expression is a characteristic feature of aggressive sporadic breast carcinoma. However, the mechanisms underlying BRCA1 downregulation in breast cancer are not well understood. Here we report that both in vitro and in vivo HMGA1b protein binds to and inhibits the activity of both human and mouse BRCA1 promoters. Consistently, murine embryonic stem (ES) cells with the Hmga1 gene deleted display higher Brca1 mRNA and protein levels than do wild-type ES cells. Stable transfection of MCF-7 cells with the HMGA1b cDNA results in a decrease of BRCA1 gene expression and in a lack of BRCA1 induction after estrogen treatment. Finally, we found an inverse correlation between HMGA1 and BRCA1 mRNA and protein expression in human mammary carcinoma cell lines and tissues. These data indicate that HMGA1 proteins are involved in transcriptional regulation of the BRCA1 gene, and their overexpression may have a role in BRCA1 downregulation observed in aggressive mammary carcinomas.
BRCA1 was isolated as the gene responsible for increased susceptibility to familial breast and ovarian cancer (31). Germ line mutations of BRCA1 have been detected in approximately 90% of familial breast and ovarian cancers and in approximately 50% of familial breast cancers alone. Full-length BRCA1 is a nuclear protein of 220 kDa and 1,863 amino acids. The BRCA1 gene is highly expressed in rapidly proliferating mammary epithelial cells during pregnancy and is downregulated during lactation (30, 39). BRCA1 has pleiotropic biological functions, possibly playing a role in transcriptional regulation, chromatin remodeling, DNA damage repair, cell cycle regulation, and checkpoint control (reviewed in reference 43).
Although BRCA1 mutations play a critical role in familial breast carcinomas, sporadic breast carcinomas rarely show mutations in the BRCA1 gene (21). However, reduced expression of BRCA1 has been frequently observed in sporadic breast carcinomas, and reduced expression of BRCA1 has shown a positive correlation with increased invasiveness of human breast carcinomas (49, 51).
Many studies have examined the downregulation of BRCA1 expression in advanced sporadic cancer, but the mechanisms for this remain poorly understood. Alterations of methylation patterns are rarely detected in the vicinity of the major transcription initiation site of the BRCA1 gene (9, 11, 16, 29, 36), and loss of heterozygosity is not related with BRCA1 mRNA and protein expression level (26, 40, 49). Therefore, different mechanisms may account for the inactivation of BRCA1 function in sporadic breast cancer.
HMGA1 is a structural gene that encodes a nonhistone chromatin protein. Two isoforms, HMGA1a and HMGA1b, are produced through an alternative splicing mechanism. Both isoforms are able to bind DNA in AT-rich regions and interact with various transcription factors to enhance or inhibit gene transcription by acting as architectural proteins (reviewed in reference 35). HMGA1 protein is abundant during embryogenesis (12, 53) but is absent or present only at low concentration in normal adult tissues.
Induction of HMGA1 expression occurs in several human malignant neoplasias, including thyroid (13, 15), colon (1, 14, 19), prostate (46), cervix (5), and pancreas (2) carcinomas. HMGA1 protein expression significantly correlates with parameters indicating a poor prognosis in patients with colon carcinomas (1, 14). Increased expression of the HMGA1 gene in mouse (33) and human (28) breast carcinomas has been demonstrated, with a direct correlation between HMGA1 protein levels and the metastatic phenotype of human breast cancer cell lines (28).
Inhibition of HMGA synthesis prevented the neoplastic transformation induced by the myeloproliferative sarcoma virus or by the Kirsten murine sarcoma virus, suggesting that HMGA overexpression plays a key role in the induction of the malignant phenotype (6). Moreover, suppression of HMGA1 protein synthesis by an adenovirus construct carrying the HMGA1b cDNA in antisense orientation led several carcinoma cell lines to death (41).
Here we report that HMGA1b protein directly binds to the BRCA1 promoter, resulting in the downregulation of BRCA1 promoter activity both in vitro and in vivo. Murine embryonic stem (ES) cells with the Hmga1 gene deleted have higher Brca1 mRNA and protein levels than do wild-type ES cells. Moreover, MCF-7 cells stably transfected with HMGA1 cDNA show decreased BRCA1 gene expression. Finally, we demonstrate an inverse correlation between HMGA1 and BRCA1 expression levels in human breast carcinoma cell lines and tissues, suggesting that downregulation of BRCA1 expression by HMGA1 may account for the low BRCA1 levels observed in aggressive mammary carcinomas.
MATERIALS AND METHODS
Expression vectors.
pCMV-β-Gal was purchased from Invitrogen. The pCEFL-hemagglutinin (HA)-tagged HMGA1b expression plasmid (20) and reporter constructs containing the BRCA1 promoter region (48) have been previously described. BRCA1-198 and -198Mut were generated by PCR using as the 5′primer the follow sequence: 5′-TCCTCTTCCGTCTCTTTCCTTTTACGTCA-3′ or TCCTCTTCCGTCTCTTTCCTTggACGTCA, respectively. The point mutations generated in the HMGA binding site of the BRCA1 promoter are shown in lowercase type. As 3′ primer, the region between base +5 and +36 was used as described previously (48). pCMV-YT, which contains the first 79 amino acids, including the three AT hook domains, was obtained by PCR amplification and subcloned into the pcDNA3 myc-His vector (Invitrogen). The mouse Brca1 promoter region −266/+45 was PCR amplified (based on the sequence published in reference 8 [GenBank accession no. AF080589]) and cloned in the pGL3 reporter vector (Promega).
Cell culture.
Tumor cell lines were obtained from American Type Culture Collection, and H-MEC cells were purchased from Clonetics. Cell media and reagents were obtained from GIBCO, except as noted. H-MEC cells were cultured in MGEM bullet kit medium (Clonetics) according to manufacturer instructions. All the experiments involving H-MEC cells were performed using cells between passages 9 and 10.
MCF-7 cells were maintained in minimum essential medium (MEM) supplemented with 10% fetal bovine serum, 2 mM sodium pyruvate, 2 mM glutamine, 100 nM insulin, 1× MEM nonessential amino acids, penicillin (100 IU/ml), and streptomycin (100 μg/ml). The other mammary carcinoma-derived cell lines were grown in Dulbecco's MEM, supplemented with 10% fetal bovine serum, 2 mM glutamine, penicillin (100 IU/ml), and streptomycin (100 μg/ml). Stably transfected cell lines MCF-7 CMV, MCF-7 YHA, and MCF-7 YT were selected for neomycin resistance in medium containing G418 (800 μg/ml) and maintained in medium containing G418 (200 μg/ml). Cells were incubated in a humidified atmosphere of 95% air and 5% CO2 at 37°C.
For estrogen induction experiments, MCF-7 cells were plated in supplemented MEM with no insulin and with 10% charcoal-stripped serum instead of fetal bovine serum. After 3 days, the medium was replaced with the same medium containing 10 nM (final concentration) 17-β-estradiol (E2). Cells were harvested at 0, 3, 6, 9, 11, 12, 18, 24, and 48 h and divided in three aliquots for flow cytometry analysis, RNA extraction, and protein extraction.
Culture of murine ES cells.
Murine ES cells AB2.1 have been described elsewhere (44). For targeted disruption of the Hmga1 gene, exons 6 and 7 were replaced with a Neo cassette (S. Battista et al., unpublished data). Double-knockout clones, also indicated as Hmga1−/−, were obtained by selecting Hmga1+/− clones with higher doses (6 mg/ml) of G418. ES cells were grown on a fibroblast feeder layer, as described elsewhere (44). Before transient transfection, fibroblasts were removed, by three passages on gelatin-coated plates. Leukemia inhibiting factor (100 U/ml) was added to maintain the pluripotent undifferentiated state.
For functional assays of mouse and human BRCA1 promoter ES cells were cultured in six-well plates at a density of 2 × 105 cells/well. Cells were transfected 48 h later using FuGENE 6 transfection reagent (Roche Diagnostic Corporation, Indianapolis, Ind.), according to the manufacturer's instructions. Wild-type or Hmga1−/− ES cells were transfected with a mixture of 500 ng of reporter construct, 1 μg each of the HMGA1b and HMGA1b/T constructs, and 0.5 μg of pCMV-β-Gal construct. The vector alone was transfected as a control. Cells were lysed and processed after 36 h as described below.
Transient-transfection experiments.
MCF-7, T47D, and H-MEC cells were transfected using FuGENE 6, using a total of 5 μg of DNA per transfection (0.5 μg of promoter construct and 0.5 μg of pCMV-β-Gal, plus a total of 4 μg of HMGA1b vectors and/or control cytomegalovirus vector). Transfections were optimized for six-well plates. Cells were harvested 48 h posttransfection, and lysates were analyzed for luciferase activity. Transfection efficiency was normalized using β-galactosidase activity. All assays were performed in triplicate and repeated in at least three independent experiments.
For the experiments involving E2 stimulation, 2 × 105 cells were plated in steroid-free medium (see above) and transfected 2 days later. Twenty-four hours posttransfection, E2 was added at a final concentration of 10 nM. Cells were then cultured for 24 h and processed as described above.
RNA isolation reverse transcription (RT)-PCR and Northern blot analysis.
Total RNA was extracted using either the RNAeasy kit (Qiagen) or TriReagent (Molecular Research Center, Cincinnati, Ohio) solution, according to the manufacturer's instructions. Northern blot hybridization was performed as described previously (20) using Hybond-XL membranes (Amersham Pharmacia BioTech). All cDNA probes were radiolabeled with [α-32P]dCTP (NEN) using a random prime synthesis kit II (Stratagene). The entire HMGA1b coding region was used as a molecular probe for the HMGA1 gene. A 3,035-bp EcoRI/BamHI fragment of human BRCA1 cDNA (47) was used as a probe for human BRCA1. A 335-bp PstI fragment of pS2 cDNA (obtained from the American Type Culture Collection) was used as a probe for human pS2, and a radiolabeled internal oligonucleotide (5′-CTC CTG ACA ACA TAA ATC AGG GAA CTG ACC-3′) was used as a specific probe for the mouse Brca1 cDNA. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression was used as an internal control.
Protein extraction, Western blotting, and antibodies.
Cells were washed twice in ice-cold phosphate-buffered saline (PBS) and lysed in NP-40 lysis buffer (2) supplemented with Complete protease inhibitors cocktail (Roche Diagnostic Corporation). Total proteins were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes (Bio-Rad). Membranes were blocked with 5% nonfat milk and incubated with the appropriate antibodies. Anti-HMGA1 antibodies were previously described (20) or purchased from Santa Cruz Biotechnology. Antibodies against human BRCA1 included Ab1 mouse monoclonal antibody (Oncogene Science), rabbit polyclonal antibodies raised against epitopes between amino acids 768 and 793 and amino acids 1847 and 1863 of the human BRCA1 protein (PharMingen), and K-18 rabbit polyclonal antibody (Santa Cruz Biotechnology). Anti mouse-BRCA1 M-20, anti-HA epitope Y-11 rabbit polyclonal, anti-myc 9E10 mouse monoclonal antibody, and antitubulin mouse monoclonal antibody were purchased from Santa Cruz Biotechnology. Bound antibodies were detected by the appropriate secondary antibodies and revealed with an enhanced chemiluminescence system (Amersham Pharmacia Biotech).
Flow cytometry.
Cells were analyzed for DNA content as described previously (3). After the cells were collected, washed in PBS, and fixed in ethanol, the DNA was stained with propidium iodide (50 μg/ml) and the cells were analyzed using a FACScan flow cytometer (Becton Dickinson). Cell cycle data were analyzed with the XLII system program (Becton Dickinson).
Production of recombinant proteins.
The full-length or the truncated versions of HMGA1 cDNAs were cloned in the pET2c vector (Novagen). BL21/DE3 cells transformed with each vector were grown in Luria-Bertani medium, induced with IPTG (isopropyl-β-d-thiogalactopyranoside, and washed in PBS. The cells were then sonicated and purified using the His-Trap purification kit (Amersham Pharmacia Biotech) following the manufacturer instructions as previously described (4).
DNase I footprinting assays.
A 242-bp (−208 to + 36) fragment representing a portion of the human BRCA1 promoter radiolabeled on either the top strand or the bottom strand, was incubated with recombinant HMGA1b protein and digested with DNase I, using the DNase footprinting kit (Promega). The optimal enzyme concentration and digestion time were determined empirically. Single-stranded DNA cleavage products were loaded onto a 6% polyacrylamide sequencing gel. Electrophoresis was performed at room temperature for roughly 2 h at 45 W, after which the gels were dried and autoradiographed.
DNA binding assays.
Electrophoretic mobility shift assays (EMSA) were performed essentially as described previously (4, 48). The oligonucleotides used in the assays corresponded to the region of nucleotides (nt) −209 to −169 and the region of nt −198 to −177 of the human BRCA1 gene. Five to 100 ng of recombinant proteins was incubated in the presence of radiolabeled oligonucleotide, with 0.5 μg of poly(dC-dG) (Sigma) serving as a nonspecific competitor. In some experiments a 50- or 100-fold excess of specific or nonspecific unlabeled competitor oligonucleotides was added. The DNA-protein complexes were resolved on 5% nondenaturing acrylamide gels and were visualized by exposure to autoradiographic films.
Formaldehyde cross-linked chromatin preparation.
Subconfluent plates of wild-type and Hmga1−/− ES cells and of MCF-7 cells transfected with YHA6 or YHA7 were formaldehyde cross-linked for 10 min at room temperature by adding 0.1 volume of cross-linking solution (11% formaldehyde, 50 mM HEPES [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA). Cross-linking was stopped by the addition of glycine to a final concentration of 125 mM. Cells were washed twice with ice-cold PBS and then harvested in lysis buffer (10 mM Tris-HCl [pH 8.0], 0.25% Triton X-100, 10 mM EDTA, 0.5 mM EGTA, 10 mM sodium butyrate, 100 mM sodium chloride, 20 mM β-glycerophosphate, 100 μM sodium orthovanadate, protease inhibitors Complete) and left on ice for 10 min. Cells were centrifuged at 800 × g for 4 min at 4°C. Nuclei were washed in wash buffer (10 mM Tris-HCl [pH 8.0], 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM sodium butyrate, 20 mM β-glycerophosphate, 100 μM sodium orthovanadate, protease inhibitors Complete), resuspended in sonication buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA, 0.5 mM EGTA, 100 mM NaCl, 10 mM sodium butyrate, 20 mM β-glycerophosphate, 100 μM sodium orthovanadate, protease inhibitors Complete), and sonicated. SDS was added to a final concentration of 1%, and the chromatin solutions were rotated at room temperature for 1 h. Insoluble material was removed, and the soluble supernatant chromatin was used for immunoprecipitation as described below.
ChIP.
Chromatin immunoprecipitation (ChIP) experiments were carried out as described previously (10, 32) with a few modifications. Cross-linked released chromatin fractions were adjusted to RIPA buffer (10 mM Tris-HCl [pH 8.0], 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate, 1 mM EDTA, 0.5 mM EGTA, 140 mM NaCl, 10 mM sodium butyrate, 20 mM β-glycerophosphate, 100 μM sodium orthovanadate, protease inhibitors Complete). For immunoprecipitation, chromatin was mixed with anti-HMGA1, anti-HA, or normal rabbit immunoglobulin G (IgG) and incubated with rotation for 2 h at 4°C. Protein A/G-Sepharose beads (Santa Cruz Biotechnology) were added, and the mixture was incubated with rotation at 4°C for a further 15 h. The beads were harvested by centrifugation and then washed sequentially with (i) RIPA buffer; (ii) RIPA, 500 mM NaCl; (iii) RIPA, 1 M NaCl; (iv) 0.25 M LiCl, 10 mM Tris-HCl (pH 8.0), 1 mM EDTA, 1 mM EGTA, 1% NP-40, 1% sodium deoxycholate, 20 mM β-glycerophosphate, 10 mM sodium butyrate, 100 μM sodium orthovanadate, and protease inhibitors Complete; and (v) Tris-EDTA (TE) (pH 7.5), 10 mM sodium butyrate, 20 mM β-glycerophosphate, and protease inhibitors Complete. The antibody-bound chromatin was eluted from the beads with 200 μl of elution buffer (Tris-EDTA [pH 7.5], 10 mM sodium butyrate, 20 mM β-glycerophosphate, 30 mM NaCl) containing 1.5% SDS, and this was followed by elution with 200 μl of elution buffer containing 0.5% SDS. Formaldehyde cross-links were reversed, and the DNA was ethanol precipitated, resuspended in water, and treated with RNase A (50 μg/ml), and this was followed by proteinase K treatment (100 μg/ml). The DNAs were extracted with phenol-chloroform and chloroform, precipitated with ethanol, and dissolved in water. Input DNA and immunoprecipitated DNAs were analyzed by PCR for the presence of BRCA1 promoter sequences.
PCR analysis of immunoprecipitated DNA.
All PCRs were performed using a Perkin-Elmer GeneAmp 2400 thermal cycler and Takara Taq DNA polymerase. The primers used to amplify the human BRCA1 promoter were (forward) 5′-ACG CGT TAG AGG CTA GAG GGC AGG-3′ and (reverse) 5′-CTC GAG GGA AGT CTC AGC GAG CTC-3′. The primers used to amplify the mouse Brca1 promoter were (forward) 5′-ACG CGT TCT TTC TCT TCC GTC TCT-3′ and (reverse) 5′-CTC GAG CCG AGG ACG GAG AAG CCC CAA-3′. PCR products were resolved on agarose gels, transferred to nylon membrane, and hybridized with the −202 to +36 region of human BRCA1 promoter or the −236 to +44 region of the mouse Brca1 promoter.
RESULTS
HMGA1 proteins bind to the BRCA1 promoter region.
The positive regulatory region of BRCA1 promoter encompasses nt −202 to −177, and in this region has been located the binding site of some yet-unidentified proteins that may positively regulate the activity of BRCA1 promoter (48). This region is flanked upstream and downstream by AT-rich sequences which represent a preferential binding site for the HMGA proteins.
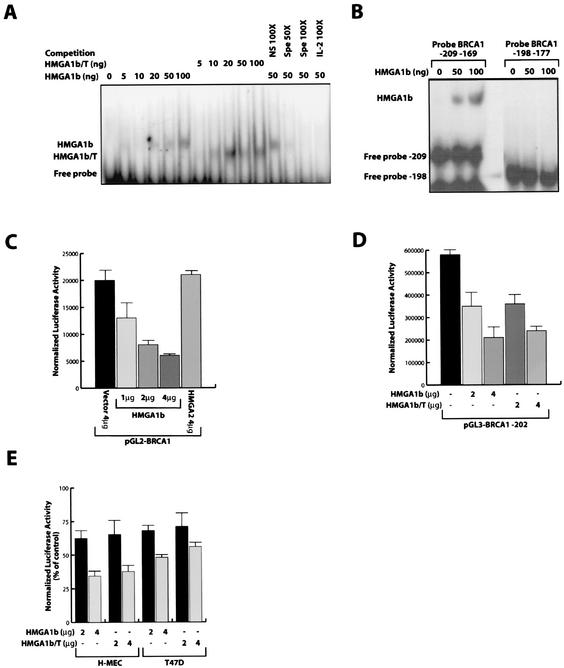
To investigate whether the HMGA1 proteins were able to bind the BRCA1 promoter, an EMSA was performed using a 42-bp oligonucleotide spanning nt −209 to −169 of the human BRCA1 promoter, encompassing the positive regulatory region (48). As shown in Fig. 1A, recombinant HMGA1b protein can directly bind to this region. Binding specificity is demonstrated by competition experiments showing loss of binding with the addition of a 100-fold molar excess of a specific, unlabeled oligonucleotide, but not with the addition of an unrelated, unlabeled oligonucleotide at the same concentration (Fig. 2A, compare lane 12 with lanes 13 and 14). Moreover, the addition of a 100-fold molar excess of an unlabeled oligonucleotide encompassing the CD-28 responsive element of the interleukin-2 (IL-2) promoter region (4) that contain an HMGA binding site, was able to displace the HMGA1 binding. To verify that the binding to the BRCA1 promoter was due to the ability of HMGA1b protein to bind the DNA we used a truncated HMGA1b protein, which retains only the three AT hook domains and lacks the carboxyl-terminal tail (HMGA1b/T). HMGA1b/T recombinant protein displayed almost the same binding capacity as the wild-type protein (Fig. 1A), demonstrating that the presence of the DNA binding domains is important while the presence of the carboxyl-terminal tail is not. Deletion of the putative HMGA1 binding site (using an oligonucleotide spanning nt −198 to −177 of human BRCA1 promoter) in the 42-bp oligonucleotide resulted in loss of binding activity for both the wild type and the truncated recombinant HMGA1b proteins (Fig. 1B).
FIG. 1.
HMGA1 protein binds to and downregulates the human BRCA1 promoter. (A) EMSA performed with a radiolabeled oligonucleotide spanning from nt −209 to −169 of the human BRCA1 promoter incubated with increasing amounts of recombinant HMGA1b or HMGA1b/T proteins as indicated. To assess the specificity of the binding, 50 ng of recombinant protein was incubated in the presence of a 100-fold excess of unlabeled nonspecific oligonucleotide (lane 12), in the presence of a 50- to 100-fold excess of the nt −209 to −169 unlabeled oligonucleotide as specific competitor (lane 13 and 14), or a 100-fold excess of the IL-2 CD28RE unlabeled oligonucleotide (lane 15). (B) EMSA performed with a radiolabeled oligonucleotide spanning from nt −209 to −169 and from nt −198 to −177 of the human BRCA1 promoter incubated with increasing amounts of recombinant HMGA1b protein as indicated. (C) Effect of increasing amounts (as indicated) of the HMGA1b expression vector on the human BRCA1 promoter region (pGL2-BRCA1) transfected in the MCF-7 cells. (Error bar, standard deviation.) (D) Effect of HMGA1b and HMGA1b/T constructs on the activity of the BRCA1 minimal promoter region (pGL3-BRCA1-202) transfected in the MCF-7 cells. (Error bar, standard deviation). (E) Effect of HMGA1b and HMGA1b/T constructs on the activity of the pGL3-BRCA1-202 reporter vector transfected in the H-MEC and in the T47D cell lines.
FIG. 2.
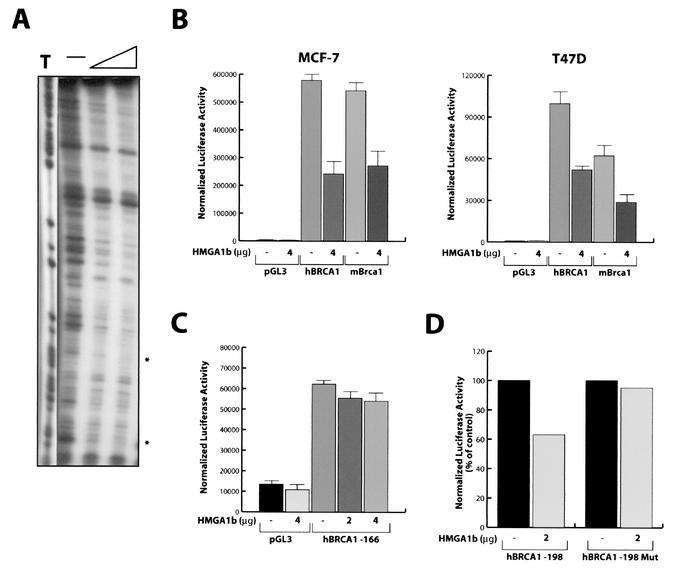
HMGA1 protein interferes with the activity of mouse Brca1 promoter. (A) DNase I footprinting assay of human BRCA1 promoter fragment nt −209 to +45. The asterisks indicate the regions of binding between the human BRCA1 promoter and the HMGA1b recombinant protein flanking the positive regulatory region. (B) Effect of HMGA1b and HMGA1b/T overexpression on the activity of human and mouse BRCA1 minimal promoter regions transfected in the MCF-7 and in the T47D cell lines. (C) Effect of HMGA1b overexpression on the activity of human BRCA1 promoter mutant −166, which lacks the putative HMGA1 binding sites, transfected in the MCF-7 cells. (D) Effect of HMGA1b overexpression on the activity of human BRCA1-198 and BRCA1-198Mut, in which T −177 and −178 were replaced with G in MCF-7 cells. (B to D) Error bar, standard deviation.
HMGA1 proteins inhibit BRCA1 promoter activity.
To define the functional consequences of the interaction between the HMGA1 protein and the BRCA1 promoter, we cotransfected a luciferase reporter construct regulated by the human BRCA1 promoter (pGL2-BRCA1) (48) in MCF-7 cells with increasing amounts of an HMGA1b expression vector (HA-HMGA1b). The expression of HMGA1b protein in MCF-7 cells resulted in a three- to fourfold decrease of the activity of BRCA1 promoter in the luciferase reporter construct. This effect was dose dependent (Fig. 1C) and was not observed in cells transfected with the control vector pGL3 (data not shown). No decrease in BRCA1 promoter activity was obtained when the cells were transfected with a construct expressing HMGA2 (Myc-HMGA2), the other member of the HMGA family (Fig. 1C). To confirm the specificity of the HMGA1 activity on the BRCA1 promoter we also analyzed the activity of the IL-15 receptor α promoter in MCF-7, and we obtained a little stimulation rather than inhibition of the activity of this promoter, transfecting 2 to 4 μg of HMGA1b expression vector together with the pGL3 IL-15 receptor α reporter vector (data not shown).
Plasmid pGL3-202 is a reporter vector containing nt −202 through +36 of the BRCA1 gene, a region that retains full promoter activity (48). HMGA1b overexpression resulted in threefold reduction of the BRCA1 promoter activity on plasmid pGL3-202 in MCF-7 cells (Fig. 1D). The truncated HMGA1b/T (Myc-HMGA1b/T) protein was as active as the wild-type protein in inhibiting pGL3-202 activity (Fig. 1D). This finding demonstrates again that the presence of the DNA binding domains in the HMGA1b protein is sufficient to obtain the donwregulation of BRCA1 promoter. These data were further confirmed using the H-MEC and T47D cell lines that express low levels of HMGA1 protein (Table 1). In both the cell lines overexpression of either HMGA1b or HMGA1b/T protein resulted in a strong reduction in the activity of the pGL3-202 construct (Fig. 1E). Interestingly, the T47D cell line has been reported to have a p53 mutation (42), suggesting that downregulation of the BRCA1 promoter by HMGA1b is independent from p53 activity.
TABLE 1.
Relationship between HMGA1 expression, BRCA1 expression, and cell cycle distribution in normal mammary and mammary carcinoma-derived cell lines
| Cell line | Expressiona of:
|
Cell cycle distributionb (%)
|
|||
|---|---|---|---|---|---|
| HMGA1 | BRCA1 | G1 | S | G2/M | |
| H-MEC | − | ++++++ | 72 | 18 | 5 |
| MCF-7 | + | +++ | 70 | 18 | 11 |
| T47D | − | ++++ | 79 | 9 | 11 |
| HBL-100 | + | ++++ | 70 | 13 | 14 |
| MDA-MB-453 | + | + | 67 | 22 | 8 |
| BT-549 | ++ | ++++ | 58 | 16 | 20 |
| BT-20 | ++ | ++ | 57 | 18 | 19 |
| MDA-MB-436 | +++ | + | 59 | 16 | 20 |
| SK-BR-3 | +++ | + | 73 | 11 | 7 |
| MDA-MB-231 | ++++ | + | 51 | 23 | 14 |
| MDA-MB-361 | ++ | ++ | NT | NT | NT |
The expression of HMGA1 and BRCA1 is indicated in arbitrary units and was evaluated by Western blot analysis and normalized using the tubulin expression levels. The results reported are the means of four different independent experiments carried out using three different anti-BRCA1 antibodies and two different anti-HMGA1 antibodies. Symbols: −, no expression; +, low expression; ++, moderate expression; +++, high expression; ++++, very high expression.
The cell cycle distribution observed in the different cell lines represents the value obtained in a single experiment and reflects the distribution observed in two other independent assays. In each case the cell lines were cultured to 70 to 80% of confluence, and harvested 15 to 18 h after the addition of fresh medium. NT, not tested.
HMGA1b protein binds to and inhibits the mouse Brca1 promoter.
To define the DNA sequences in the BRCA1 promoter interacting with HMGA1b, we performed a DNase I footprinting assay using the purified HMGA1b protein and the BRCA1promoter region nt −202 to +43. The recombinant HMGA1b protein protected the region spanning the positive regulatory site between nt −198 and −166 (highlighted by asterisk in Fig. 2A).
A comparative analysis of human and murine BRCA1 promoters revealed that the pyrimidine-rich region, first identified as a positive regulatory region in the human BRCA1 promoter (48), is highly conserved at nearly the same position of the mouse Brca1 promoter (data not shown). Experiments using the mouse promoter region nt −266 to +45 cloned in the pGL3 luciferase reporter vector indicated that the negative effect of HMGA1b overexpression on this vector was comparable to the inhibition exerted on the pGL3-202 construct containing the human BRCA1 promoter, as tested in two different mammary carcinoma-derived cell lines (Fig. 2B). Interestingly, deletion of the putative HMGA1 binding sites from the BRCA1 promoter impaired the HMGA1 activity on the human BRCA1 promoter. In fact, we generated a deletion mutant of BRCA1 promoter that starts from base −166, in the pGL3 reporter vector. This fragment lacks the putative HMGA1 binding sites. Transfection of increasing amount of HMGA1 vector, together with the pGL3-166 BRCA1 promoter, did not significantly affect the luciferase activity of this vector when compared with the activity obtained transfecting the pGL3-166 BRCA1 promoter with a control expression vector (Fig. 2C). Finally, we generated a point mutation in the putative binding site for the HMGA1 protein present in the BRCA1 promoter spanning between base −185 and −176 by replacing thymidines −177 and −178 with two guanines. To this aim we inserted by PCR these point mutations in the pGL3-198 construct that displays an activity similar to that obtained by transfecting the pGL3-202 construct (48). Overexpression of HMGA1 was able to reduce the activity of the wild-type pGL3-198 but it completely failed in its inhibitory effect if the HMGA1 binding site of this construct was mutated (Fig. 2D).
Constitutive expression of HMGA1b in MCF-7 cells results in endogenous BRCA1 downregulation.
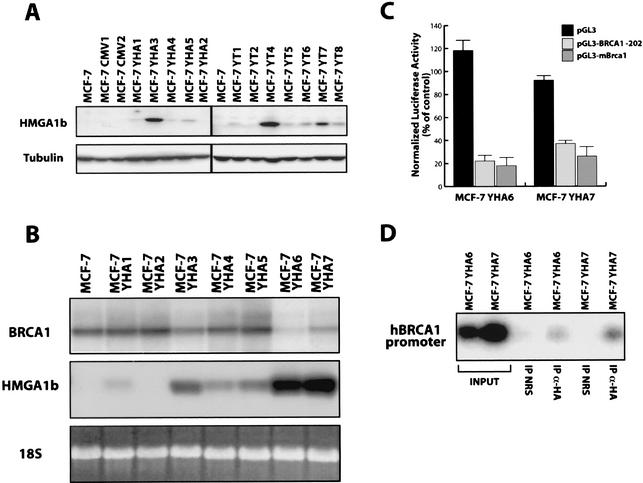
To better understand the role of the HMGA1b-mediated BRCA1 down regulation in breast cancer cell lines, we generated MCF-7 cell lines stable transfected with the full-length or truncated HMGA1b constructs. As control MCF-7 cells were transfected with the vector alone (MCF-7 CMV). Several clones were isolated and analyzed for HMGA1 protein expression (Fig. 3A). Since the effects of the full-length (YHA) or the truncated (YT) HMGA1b constructs were comparable, only the results obtained with two different full-length MCF-7 YHA clones (YHA6 and YHA7) are presented here.
FIG. 3.
Impaired expression of BRCA1 in MCF-7 Y-HA cells. (A) Expression of HMGA1b and HMGA1b/T proteins in MCF-7 cells stable transfected with the full-length (YHA) or the truncated HMGA1b (YT) expression vectors. (B) Northern blot analysis of BRCA1 (upper panel) and HMGA1 (middle panel) expression in MCF-7 cells and in seven MCF-7 YHA clones expressing different levels of HMGA1b mRNA. Levels of 18S rRNA were used to monitor RNA amounts in each lane (lower panel). (C) Activity of the human and mouse BRCA1 minimal promoter regions transfected in MCF-7 YHA6 and YHA7 cell lines. The pGL3 vector was transfected as a control. The activity is expressed as a percentage of the activity exhibited in the MCF-7 cells. (D) The ChIP assay was performed on MCF-7 YHA6 and YHA7 cells. The purified DNA was used as a template for PCRs with primers that amplify the human BRCA1 promoter region comprised between base −202 and +36.
Examination of BRCA1 mRNA levels in several different MCF-7 HMGA1b clones showed an inverse relation between HMGA1b and BRCA1 expression. Northern blot analysis demonstrated that overexpression of HMGA1b resulted in downregulation of BRCA1 mRNA in a dose-dependent manner (Fig. 3B). Similarly, transient-transfection experiments with the human and mouse BRCA1 promoter regions cloned in the pGL3 vector (described above) revealed a 70 to 80% decrease in BRCA1 promoter activity in both MCF-7 YHA6 and MCF-7 YHA7 transfectants compared to parental MCF-7 cells (Fig. 3C). No effect was observed on the basal activity of the pGL3 vector, indicating the specificity of the effect on the BRCA1 promoter. Taken together, these results suggest that the inhibition of BRCA1 expression in MCF-7 YHA clones is due to the direct action of the HMGA1b protein on the endogenous BRCA1 promoter.
ChIP experiments confirmed that the exogenous HMGA1b protein was able to bind in vivo the human BRCA1 promoter. Chromatin prepared as previously described (10, 32) from MCF-7 YHA6 and YHA7 cells was immunoprecipitated with an anti-HA polyclonal rabbit antibody or with normal rabbit IgG. As shown in Fig. 3D the −202 to +36 region of the human BRCA1 promoter was amplified only from the DNA recovered with the anti-HA antibody and not from the DNA recovered using the normal rabbit IgG (Fig. 3D, compare lanes 3 and 5 with lanes 6 and 7).
Overexpression of HMGA1b proteins in MCF-7 cells increases the S phase population.
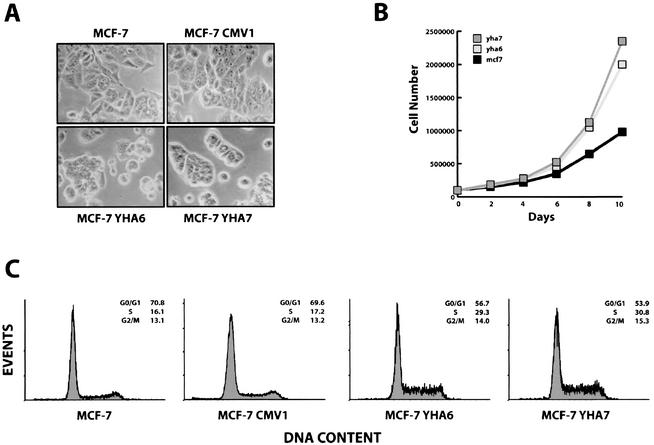
HMGA1b-transfected MCF-7 cells showed a change in morphology, with the acquisition of a round-like shape, and were less adherent to the plate; by contrast, no changes in morphology were noticed in MCF-7 cells transfected with the vector alone (MCF-7 CMV) (Fig. 4A). Since an increased growth rate in the MCF-7 YHA clones compared to control cells was observed (Fig. 4B), we used flow cytometry to analyze the DNA content of the different cell lines under exponential growth conditions. Parental MCF-7 cells and vector-transfected MCF-7 cells showed 16% of cells in S phase when analyzed in exponential phase of growth (Fig. 4C, first and second panels). Under the same culture conditions, 28 to 30% of MCF-7 cells transfected with HMGA1b were in S phase, with a corresponding reduction of the G0/G1 population (Fig. 4C, third and fourth panels). These results indicate that overexpression of HMGA1b protein in MCF-7 cells is associated with an increased proliferation rate and a less-differentiated phenotype. It is worthwhile to observe that the differences in BRCA1 expression could not be ascribed to a decreased number of cells in S phase in MCF-7 cell clones, since the percentage of cells in S phase was higher in HMGA1 transfected cells. It is known, in fact, that BRCA1 is a cell-cycle-regulated protein that is expressed at highest levels during the S phase of the cell cycle (39). Therefore, the flow cytometry analysis confirms that the downregulation of BRCA1 expression is not related to a decreased proliferation rate.
FIG. 4.
Increased growth ability in MCF-7 overexpressing the HMGA1b protein. (A) Light microscopy photograph of parental MCF-7 cells (MCF-7) and cells transfected with the backbone vector (MCF-7 CMV1), or with the HMGA1b-HA tagged construct (YHA6 and YHA7). (B) Monolayer growth curves compare the growth abilities of MCF-7, MCF-7 YHA6, and MCF-7 YHA7 cell lines. (C) Flow cytometry analysis of MCF-7 cells and its derivative clones CMV-1, YHA6, and YHA7. A representative experiment is reported.
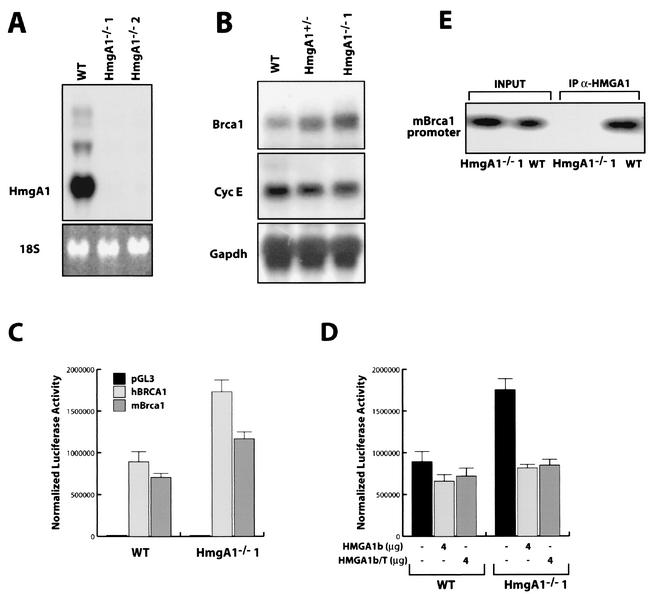
Brca1 expression is higher in Hmga1−/− ES cells than in wild-type ES cells.
Since the mouse Brca1 promoter and the human BRCA1 promoter were similar in their response to the HMGA1 proteins, we used a mouse system for further studies of the interaction of the Hmga1 protein and the Brca1 gene. We investigated the transcription of Brca1 in murine ES cells in which both Hmga1 alleles had been deleted by targeted homologous recombination (Battista et al., unpublished data). Figure 5A shows the absence of Hmga1 mRNA in two different Hmga1−/− ES cells. Northern blot analysis, shown in the Fig. 5B, demonstrated a fivefold increase of Brca1 mRNA expression levels in homozygous Hmga1−/− ES cells when compared to the wild-type ES cells. Western blot analysis results paralleled the data obtained with Northern blot and RT-PCR analyses (data not shown). Two different clones of Hmga1−/− obtained from two different parental Hmga+/− ES cells showed the same difference in Brca1 mRNA levels (data not shown). Interestingly, ES cells lacking only one allele of the Hmga1 gene (i.e., Hmga1+/− ES cells) showed a threefold reduction in Brca1 mRNA when compared to wild-type ES cells (Fig. 5B), suggesting a dose-dependent effect of Hmga1 proteins on Brca1 gene expression. Conversely, when we tested the expression of cyclin E, another gene whose expression is S-phase dependent, no change was observed in the same cell lines (Fig. 5B). The activities of both the human and mouse promoters were twofold higher in the Hmga1−/− ES cells than in the wild-type cells, as measured by luciferase activity (Fig. 5C). Cotransfection of Hmga1−/− ES cells with a vector carrying either the full-length or the truncated HMGA1b cDNA decreased the activity of the human BRCA1 promoter to levels comparable to those seen in the parental ES cells (Fig. 5D). These results clearly demonstrate that for both the human and murine genes, the difference in BRCA1 promoter activity in Hmga1−/− and wild-type ES cells was due to the presence of Hmga1 proteins.
FIG. 5.
Hmga−/− cells express higher levels of BRCA1 mRNA and protein. (A) Hmga1 expression in wild-type (WT) ES cells and in two different Hmga1−/− ES cell clones analyzed by Northern blotting. (B) Northern blot showing the expression of Brca1 (top panel), cyclin E (middle panel), and GAPDH (lower panel) mRNA in WT Hmga1+/− and Hmga1−/−1 ES cell lines. (C) Activity of the human and mouse BRCA1 minimal promoter regions transfected in WT or Hmga1−/−1 ES cells. pGL3 vector was transfected as a control. Error bar, standard deviation. (D) Effect of HMGA1b overexpression on the activity of the human BRCA1 minimal promoter region transfected in WT or Hmga1−/−1 ES cells (Error bar, standard deviation.) Each transfection assay was performed in triplicate and repeated in at least three independent experiments and using two different Hmga1−/− ES cell clones. (E) ChIP assay performed on WT and Hmga1−/− (Hmga1−/−1) ES cells. The purified DNA untreated (input) or immunoprecipitated with an anti-HMGA1 antibody (IP α-HMGA1) was used as a template for the PCRs with primers that amplify the mouse Brca1 promoter region comprised between nt −236 and +44.
ChIP experiments demonstrated that Hmga1 proteins bind the mouse Brca1 promoter in vivo. A specific anti-Hmga1 antibody was able to recover the DNA from wild-type ES cells but not from the Hmga1−/− ES cells (Fig. 5E), indicating that endogenous Hmga1 protein binds the Brca1 promoter in vivo.
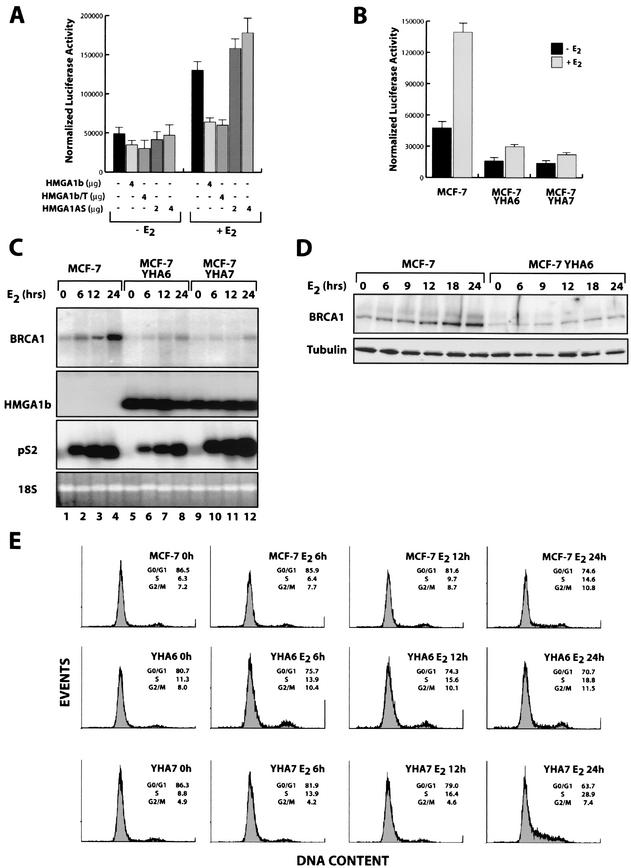
HMGA1b overexpression prevents estrogen-induced BRCA1 expression.
MCF-7 cells are estrogen-receptor positive cells, and it has been reported that BRCA1 expression is enhanced by estrogen treatments (22, 30). We investigated whether HMGA1b overexpression could somehow interfere with the estrogen activity on the BRCA1 promoter. As shown in Fig. 6A, overexpression of HMGA1b protein completely abolished the estrogen-mediated induction of the BRCA1 promoter activity. Interestingly, the elimination of endogenous HMGA1 protein expression (obtained using an expression vector carrying the full length of HMGA1b in antisense orientation) resulted in an increase of BRCA1 promoter activity induced by estrogen treatment (Fig. 6A). Similarly, the activity of the pGL3-202 vector on the MCF-7 YHA6 and MCF-7 YHA7 transfectants was fourfold lower in MCF-7 cells overexpressing the HMGA1b construct than in control cells (Fig. 6B). Moreover, estrogen-induced upregulation of BRCA1 promoter activity was strongly inhibited in both MCF-7 YHA clones (Fig. 6B).
FIG.6.
HMGA1b protein interferes with estrogen-induced upregulation of BRCA1. (A) Effect of HMGA1b, HMGA1b/T, and HMGA1bAS constructs on the activity of the pGL3-BRCA1-202 reporter vector transfected in the MCF-7 cells grown for three days in medium deprived of steroid hormones and stimulated (+E2) or not (−E2) with 10 nM of 17-β-estradiol. (B) Activity of the pGL3-BRCA1-202 vector transfected in the MCF-7, MCF-7 YHA6, and MCF-7 YHA7 cells grown for 3 days in medium deprived of steroid hormones and stimulated (+E2) or not (−E2) with 10 nM of 17-β-estradiol. (C) Northern blot analysis of BRCA1 (upper panel), HMGA1 (second panel), and pS2 (third panel) expression in MCF-7, MCF-7 YHA6, and MCF-7 YHA7 cells grown for 3 days in medium deprived of steroid hormones (lane 0) and stimulated with 10 nM of 17-β-estradiol (E2) for the indicated time. Levels of rRNA were used to monitor RNA amounts in each lane (lower panel). (D) Western blot analysis of BRCA1 (upper panel) expression in MCF-7 and MCF-7 YHA6 cells treated as described for panel C. α-Tubulin expression confirmed the equal amount of loaded proteins (lower panel). (E) Flow cytometry analysis of MCF-7 cells (first row) and its derivative clones, YHA6 (second row) and YHA7 (third row), treated as described in panel C.
Expression of endogenous BRCA1 was analyzed in MCF-7 cells and in the HMGA1b transfectants over a period of 48 h of treatment with estrogen. In MCF-7 control cells, BRCA1 messenger levels started to increase after 6 h of estrogen treatment and progressively increased, peaking at 24 h (Fig. 6C, lanes 1 to 4). In contrast, E2-mediated BRCA1 upregulation was almost completely prevented in the stable HMGA1b transfectants (Fig. 6C, lanes 5 to 12). The expression of pS2 gene, a typical estrogen-responsive gene used as control, was essentially comparable in the MCF-7 and MCF-7 YHA cells (Fig. 6C, third panel).
Results of the Western blot analysis paralleled those of the Northern blot analysis (Fig. 6D). We report the comparison between the MCF-7 and the MCF-7 YHA6 cell lines, which is representative of all the others cell lines examined. As it is clearly visible in Fig. 6D, in MCF-7 cells BRCA1 protein began to accumulate after 6 h of treatment, with progressive accumulation until 24 h after E2 addition (Fig. 6D). By contrast, in the MCF-7 YHA6 cells BRCA1 protein was virtually absent for the first 12 h and then slightly increased (Fig. 6D). The cell cycle distribution of the different cell lines revealed that the MCF-7 cells accumulate in the G1 phase of the cell cycle after 3 days of starvation and enter the S phase after 12 to 18 h of E2 treatment. In contrast, MCF-7 YHA6 and MCF YHA7 transfectants failed to accumulate in G1 and/or entered the S phase 6 to 12 h earlier than the control cells (Fig. 6E). Thus, also in the case of estrogen stimulation the difference in BRCA1 expression could not be ascribed to a defect of the HMGA1b-transfected cells to enter in S phase.
HMGA1 gene overexpression in human breast carcinoma cell lines and tissues: inverse correlation with BRCA1 protein expression.
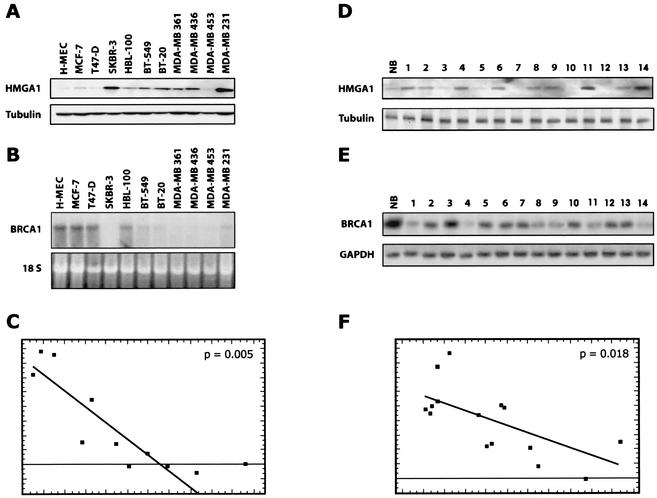
Finally, we analyzed the expression of the HMGA1 and BRCA1 genes in a panel of 10 human mammary carcinoma-derived cell lines (Table 1). HMGA1 expression was increased in all tumor-derived cell lines tested when compared to normal primary cultured H-MEC cells used as a control. The highest level of HMGA1 proteins was found in the highly tumorigenic MDA-MB 231 cells (Fig. 7A). Conversely, BRCA1 mRNA expression level was lower in most of the breast carcinoma cell lines compared to normal H-MEC cells (Fig. 7B). In fact, Northern blot analysis showed that BRCA1 gene expression was reduced by 5- to 10-fold in SK-BR3, BT-20, BT-549, MDA-MB 436, MDA-MB 361, MDA-MB 453, and MDA-MB 231 cell lines in comparison to the MCF-7 and H-MEC cells. The highest levels of BRCA1 mRNA were observed in the MCF-7, T47D, and HBL-100 cell lines, which express the lowest levels of HMGA1 protein among the tumor-derived cell lines. Therefore, an inverse correlation between HMGA1 protein levels and BRCA1-specific mRNA expression was observed in mammary carcinoma cell lines analyzed, with the exception only of the MDA-MB 453 carcinoma cells (Fig. 7A and B). BRCA1 protein levels essentially paralleled the mRNA levels (data not shown), suggesting that the downregulation of BRCA1 expression occurs at the transcription level.
FIG. 7.
Expression of HMGA1 and BRCA1 in primary breast tumors and in tumor-derived cell lines. (A) Expression of HMGA1 proteins in primary cultured epithelial cells (HMEC) and in 10 different mammary carcinoma-derived cell lines. As shown in the lower panel, α-tubulin expression was used to normalize the amount of the loaded proteins. (B) In the upper panel is the Northern blot of the BRCA1 expression in the indicated cell lines. In the lower panel, levels of 18S rRNA were used to monitor RNA amounts in each lane. (D) Western blot analysis of HMGA1 expression in a normal breast tissue (lane NB) and 14 different mammary carcinomas (lanes 1 to 9). α-Tubulin (lower panel) was used to normalize the blot. (E) RT-PCR analysis of BRCA1 expression in a normal breast tissue (lane NB) and 14 different mammary carcinomas. GAPDH expression (lower) was used as internal control of RNA quantity and status. (C and F) Statistical analysis of the HMGA1 protein and BRCA1 mRNA levels in the cell lines (C) and in the primary mammary tumors (F) by linear regression analysis. The expression of HMGA1 protein was determined by Western blot, quantified by densitometric analysis, and normalized for uniform gel loading with the signals obtained with antitubulin antibody. The levels of BRCA1 mRNA were determined by Northern blot (cell lines) or RT-PCR (primary tumors), quantified by densitometric analysis, and normalized. The linear regression plots between HMGA1 proteins ad BRCA1 mRNA levels were calculated by using the GB-STAT program.
Patterns of BRCA1 and HMGA1 gene expression were subsequently examined in 14 surgically removed human breast carcinomas (Fig. 7D shows a typical Western blot for HMGA1). An inverse correlation between HMGA1 protein and BRCA1 mRNA levels was clearly present. HMGA1 protein levels were almost undetectable in normal breast tissue (Fig. 7D, lane NB) but they were highly expressed in most of the tumors analyzed (Fig. 7D, lanes 1 to 14). By contrast, BRCA1 expression was strongly diminished in tumor samples compared to normal breast tissue, as determined by RT-PCR on the RNAs extracted from the same samples (Fig. 7E). The results were analyzed statistically by linear regression analysis. The expression of HMGA1 protein was determined by Western blotting, quantified by densitometric analysis, and normalized for uniform gel loading with the signals obtained with antitubulin antibody. The levels of BRCA1 mRNA were determined by Northern blotting (cell lines) or RT-PCR (primary tumors), quantified by densitometric analysis, and normalized with levels of 18S rRNA and GAPDH mRNA, respectively. Then, the linear regression plots between HMGA1 proteins ad BRCA mRNA levels were calculated by using the GB-STAT program. The correlation coefficients were −0.8945, with P = 0.005, for the cell lines (Fig. 7C) and −0.6202, with P = 0.018, for the mammary tumors (Fig. 7F). Since these values are statistically significant, these data suggest that, also in vivo, the HMGA1 proteins are likely candidates for negative regulators of the BRCA1 expression.
DISCUSSION
Several reports have demonstrated that BRCA1 protein levels are decreased in a subset of sporadic breast carcinomas compared to normal breast tissues (7, 37, 48, 45, 51, 52). The majority of high-grade ductal carcinomas lack BRCA1 protein or have low levels (51), supporting the idea of a potentially central role for BRCA1 in the pathogenesis of a significant percentage of noninherited breast cancers. Epigenetic loss of BRCA1 may occur at the level of transcription or in a subsequent process affecting RNA accumulation (45, 49). Accelerated growth of normal and malignant mammary cells occurs following experimental inhibition of BRCA1 expression with antisense oligonucleotides, suggesting that BRCA1 protein functions as a negative growth regulator in mammary epithelial cell (49).
One possible mechanism for the low levels of BRCA1 expression in breast carcinomas could be the hypermethylation of the BRCA1 promoter. The methylation status of BRCA1 promoter in sporadic breast cancer specimens was extensively investigated by several laboratories (9, 11, 16, 29, 36). However, only the 11% (19 of 178) of breast cancer specimens displayed aberrant methylation of BRCA1, indicating that hypermethylation alone could not explain the 82% of aggressive breast cancer specimens that were reported to have weak or no BRCA1 protein expression (51). Similarly, loss of heterozygosity at the BRCA1 locus proved insufficient as an explanation for the low levels of BRCA1 expression in breast carcinomas, since studies showed that loss of heterozygosity correlated with reduced BRCA1 expression in only a few cases (7, 37, 45, 49).
A possible mechanism responsible for the low BRCA1 protein levels was suggested by our data (Fig. 7) showing an inverse relation between HMGA1 and BRCA1 expression in a panel of 10 mammary carcinoma-derived cell lines and in 15 primary breast cancer tissues. Overexpression of HMGA1 proteins significantly correlated with parameters known to indicate a poor prognosis in patients with colon carcinoma (1, 14). Liu et al. (28) demonstrated a direct correlation between HMGA1 protein levels and the metastatic phenotype of breast human cancer cell lines (28). Moreover, BRCA1 downregulation has been observed primarily in high-grade ductal breast tumors (51), and preliminary data also suggest that HMGA1 overexpression is a common feature in advanced breast cancers (Chiappetta et al., unpublished data).
The data presented here clearly demonstrate that the HMGA1 protein overexpression results in the downregulation of the BRCA1 promoter activity as assessed using different model systems, including primary cultured mammary epithelial cells, two different mammary carcinoma-derived cell lines, and ES cells carrying a targeted deletion of Hmga1 gene.
In both human and mouse model systems BRCA1 expression peaks during S phase and is linked to estrogen and progesterone stimulation (22, 23, 30, 38, 39, 50), suggesting that human and mouse BRCA1 promoters are similarly regulated. The positive regulatory region of BRCA1 promoter described by Thakur and Croce (48) is highly conserved in the mouse Brca1 promoter and located almost in the same position as in the human promoter. This pyrimidine-rich sequence is preceded and followed (both in the human and mouse promoters) by different AT stretches typically bound by the HMGA1 proteins (reviewed in reference 35).
We demonstrated by various techniques that HMGA1b protein inhibits BRCA1 promoter activity in both the human and mouse genes by directly binding to the promoter regions. Interestingly, the promoter region that seems to be necessary for the regulation of BRCA1 transcription in humans is highly conserved in the mouse, suggesting that the mechanisms regulating this promoter are conserved in different species. At this time we cannot completely explain the mechanism by which HMGA1 binding to the BRCA1 promoter downregulates its activity. The possibility that binding of HMGA1 proteins to the DNA somehow impairs the activity of some positive regulatory proteins of the BRCA1 promoter and the possibility that there exists a repressor which is helped by HMGA1 are hypotheses currently under investigation.
It is noteworthy that under physiological conditions, like puberty and pregnancy, both estrogens and BRCA1 expression are significantly increased. Under these conditions, the function of BRCA1 may be to protect the breast from estrogen-induced proliferation or genetic instability (25). In breast cancer the decreased expression of BRCA1 protein could result in the loss of its caretaker role (27) and ultimately in a transformed phenotype. We demonstrated that constitutive expression of HMGA1b protein in MCF-7 cells results in the downregulation of endogenous BRCA1 mRNA and protein and in the prevention of BRCA1 upregulation following estrogen treatment. Flow cytometry analysis coupled to analysis of RNA and proteins revealed that, at least in MCF-7 cells, HMGA1b overexpression was able to disjoint the proliferation induced by estrogen from E2-dependent BRCA1 upregulation. The observation that BRCA1 expression is high during physiological conditions like puberty and pregnancy could be related to the notion that expression of HMGA proteins is very low or completely absent in all normal adult tissues examined so far (12, 53). Perhaps in the absence of HMGA1 proteins, estrogen stimulates both cell proliferation and BRCA1 production in the mammary gland, whereas upregulation of HMGA1 gene expression impairs these two effects of estrogen. The low expression of BRCA1 in highly proliferating cells (like mammary epithelial cells stimulated with estrogen) could result in impaired DNA repair activity and thus could represent an important mechanism acting in mammary gland tumorigenesis. Accordingly, Hartman and Ford suggest that decreased expression of BRCA1 leads to an initial decline in DNA repair activity, resulting in an accumulation of additional mutations that may initiate a set of downstream genetic events, resulting in additional genetic alterations which lead the cells to an invasive cancer phenotype (24). Preliminary results from our laboratory indicate that MCF-7 cells overexpressing HMGA1 are more sensitive to DNA damage agents with respect to the parental MCF-7 cell line (Baldassarre and Fusco, unpublished observation), thus confirming the hypothesis of a central role for BRCA1 expression levels in mammary tumorigenesis.
It has been proposed that wild-type BRCA1 suppresses estrogen-dependent transcriptional pathways related to mammary epithelial cell proliferation, with loss of this function contributing to tumorigenesis (17, 18). Our data seem to confirm this hypothesis, suggesting that HMGA1 overexpression facilitates the estrogen-induced proliferation of MCF-7 cells, at least in part preventing the upregulation of the BRCA1 tumor suppressor gene. Interestingly, Reeves and coworkers reported that MCF-7 cells overexpressing the HMGA1b protein acquire the ability to form both primary and metastatic tumors in nude mice when injected directly into the mammary fat pads, indicating that HMGA1 protein may have a role in the process of mammary carcinogenesis (34).
In conclusion, our data suggest a critical role of the HMGA1 proteins in the negative regulation of BRCA1 gene expression. Since an inverse correlation between HMGA1 and BRCA1 protein expression has been observed in human breast carcinoma cell lines and tissues, it is likely that this mechanism occurs also in vivo and may account, at least in part, for the low amounts of BRCA1 protein observed in advanced breast cancer.
Acknowledgments
We thank R. Baserga and A. Colombatti for critical reading of the manuscript and Barbara J. Rutledge for editing assistance.
This work was supported by grants P01CA76259 and P30CA56036 from the National Cancer Institute to C. M. Croce and by grants from the Associazione Italiana Ricerca sul Cancro (AIRC) to A. Fusco.
REFERENCES
- 1.Abe, N., T. Watanabe, M. Sugiyama, H. Uchimura, G. Chiappetta, A. Fusco, and Y. Atomi. 1999. Determination of high mobility group I(Y) expression level in colorectal neoplasias: a potential diagnostic marker. Cancer Res. 59:1169-1174. [PubMed] [Google Scholar]
- 2.Abe, N., T. Watanabe, T. Masaki, T. Mori, M. Sugiyama, H. Uchimura, Y. Fujioka, G. Chiappetta, A. Fusco, and Y. Atomi. 2000. Pancreatic duct cell carcinomas express high levels of high mobility group I(Y) proteins. Cancer Res. 60:3117-3122. [PubMed] [Google Scholar]
- 3.Baldassarre, G., M. V. Barone, B. Belletti, C. Sandomenico, P. Bruni, S. Spiezia, A. Boccia, M. T. Vento, A. Romano, S. Pepe, A. Fusco, and G. Viglietto. 1999. Key role of the cyclin-dependent kinase inhibitor p27kip1 for embryonal carcinoma cell survival and differentiation. Oncogene 18:6241-6251. [DOI] [PubMed] [Google Scholar]
- 4.Baldassarre, G., M. Fedele, S. Battista, A. Vecchione, A. J. Klein-Szanto, M. Santoro, T. A. Waldmann, N. Azimi, C. M. Croce, and A. Fusco. 2001. Onset of natural killer cell lymphomas in transgenic mice carrying a truncated HMGI-C gene by the chronic stimulation of the IL-2 and IL-15 pathway. Proc. Natl. Acad. Sci. USA 98:7970-7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bandiera, A., D. Bonifacio, G. Manfioletti, F. Mantovani, A. Rustighi, F. Zanconati, A. Fusco, L. Di Bonito, and V. Giancotti. 1998. Expression of HMGI(Y) proteins in squamous intraepithelial and invasive lesions of the uterine cervix. Cancer Res. 58:426-431. [PubMed] [Google Scholar]
- 6.Berlingieri, M. T., G. Manfioletti, M. Santoro, A. Bandiera, R. Visconti, V. Giancotti, and A. Fusco. 1995. Inhibition of HMGI-C protein synthesis suppresses retrovirally induced neoplastic transformation of rat thyroid cells. Mol. Cell. Biol. 15:1545-1553. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Bernard-Gallon, D. J., M. P. De Latour, P. G. Rio, F. M. Penault-Llorca, D. A. Favy, C. Hizel, J. Chassagne, and Y. J. Bignon. 1999. Subcellular localization of BRCA1 protein in sporadic breast carcinoma with or without allelic loss of BRCA1 gene. Int. J. Oncol. 14:653-661. [DOI] [PubMed] [Google Scholar]
- 8.Bennett, L. M., H. A. Brownlee, S. Hagavik, and R. W. Wiseman. 1999. Sequence analysis of the rat Brca1 homolog and its promoter region. Mamm. Genome 10:19-25. [DOI] [PubMed] [Google Scholar]
- 9.Bianco, T., G. Chenevix-Trench, D. C. Walsh, J. E. Cooper, and A. Dobrovic. 2000. Tumour-specific distribution of BRCA1 promoter region methylation supports a pathogenetic role in breast and ovarian cancer. Carcinogenesis 21:147-151. [DOI] [PubMed] [Google Scholar]
- 10.Braunstein, M., A. B. Rose, S. G. Holmes, C. D. Allis, and J. R. Broach. 1993. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 7:592-604. [DOI] [PubMed] [Google Scholar]
- 11.Catteau, A., W. H. Harris, C. F. Xu, and E. Solomon. 1999. Methylation of the BRCA1 promoter region in sporadic breast and ovarian cancer: correlation with disease characteristics. Oncogene 18:1957-1965. [DOI] [PubMed] [Google Scholar]
- 12.Chiappetta, G., V. Avantaggiato, R. Visconti, M. Fedele, S. Battista, F. Trapasso, B. M. Merciai, V. Fidanza, V. Giancotti, M. Santoro, A. Simeone, A. Fusco. 1996. High level expression of the HMGI (Y) gene during embryonic development. Oncogene 13:2439-2446. [PubMed] [Google Scholar]
- 13.Chiappetta, G., G. Tallini, M. C. De Biasio, G. Manfioletti, F. J. Martinez-Tello, F. Pentimalli, F. de Nigris, A. Mastro, G. Botti, M. Fedele, N. Berger, M. Santoro, V. Giancotti, and A. Fusco. 1998. Detection of high mobility group I HMGI(Y) protein in the diagnosis of thyroid tumors: HMGI(Y) expression represents a potential diagnostic indicator of carcinoma. Cancer Res. 58:4193-4198. [PubMed] [Google Scholar]
- 14.Chiappetta, G., G. Manfioletti, F. Pentimalli, N. Abe, M. Di Bonito, M. T. Vento, A. Giuliano, M. Fedele, G. Viglietto, M. Santoro, T. Watanabe, V. Giancotti, and A. Fusco. 2001. High mobility group HMGI(Y) protein expression in human colorectal hyperplastic and neoplastic diseases. Int. J. Cancer. 91:147-151. [DOI] [PubMed] [Google Scholar]
- 15.Chiappetta, G., A. Bandiera, M. T. Berlingieri, R. Visconti, G. Manfioletti, S. Battista, F. J. Martinez-Tello, M. Santoro, V. Giancotti, and A. Fusco. 1995. The expression of the high mobility group HMGI (Y) proteins correlates with the malignant phenotype of human thyroid neoplasias. Oncogene 10:1307-1314. [PubMed] [Google Scholar]
- 16.Dobrovic, A., and D. Simpfendorfer. 1997. Methylation of the BRCA1 gene in sporadic breast cancer. Cancer Res. 57:3347-3350. [PubMed] [Google Scholar]
- 17.Fan, S., J. Wang, R. Yuan, Y. Ma, Q. Meng, M. R. Erdos, R. G. Pestell, F. Yuan, K. J. Auborn, I. D. Goldberg, and E. M. Rosen. 1999. BRCA1 inhibition of estrogen receptor signaling in transfected cells. Science 284:1354-1356. [DOI] [PubMed] [Google Scholar]
- 18.Fan, S., Y. X. Ma, C. Wang, R. Q. Yuan, Q. Meng, J. A. Wang, M. Erdos, I. D. Goldberg, P. Webb, P. J. Kushner, R. G. Pestell, and E. M. Rosen. 2001. Role of direct interaction in BRCA1 inhibition of estrogen receptor activity. Oncogene 20:77-87. [DOI] [PubMed] [Google Scholar]
- 19.Fedele, M., A. Bandiera, G. Chiappetta, S. Battista, G. Viglietto, G. Manfioletti, A. Casamassimi, M. Santoro, V. Giancotti, and A. Fusco. 1996. Human colorectal carcinomas express high levels of high mobility group HMGI(Y) proteins. Cancer Res. 56:1896-1901. [PubMed] [Google Scholar]
- 20.Fedele, M., G. M. Pierantoni, M. T. Berlingieri, S. Battista, G. Baldassarre, N. Munshi, M. Dentice, D. Thanos, M. Santoro, G. Viglietto, and A. Fusco. 2001. Overexpression of proteins HMGA1 induces cell cycle deregulation and apoptosis in normal rat thyroid cells. Cancer Res. 61:4583-4590. [PubMed] [Google Scholar]
- 21.Futreal, P. A., Q. Liu, D. Shattuck-Eidens, C. Cochran, K. Harshman, S. Tavtigian, L. M. Bennett, A. Haugen-Strano, J. Swensen, Y. Miki, et al. 1994. BRCA1 mutations in primary breast and ovarian carcinomas. Science 266:120-122. [DOI] [PubMed] [Google Scholar]
- 22.Gudas, J. M., H. Nguyen, T. Li., and K. H. Cowan. 1995. Hormone-dependent regulation of BRCA1 in human breast cancer cells. Cancer Res. 55:4561-4565. [PubMed] [Google Scholar]
- 23.Gudas, J. M., T. Li, H. Nguyen, D. Jensen, F. J. Rauscher, and K. H. Cowan. 1996. Cell cycle regulation of BRCA1 messenger RNA in human breast epithelial cells. Cell Growth Differ. 7:717-723. [PubMed] [Google Scholar]
- 24.Hartman, A. R., and J. M. Ford. 2002. BRCA1 induces DNA damage recognition factors and enhances nucleotide excision repair. Nat. Gen. 32:180-184. [DOI] [PubMed]
- 25.Hilakivi-Clarke, L. 2000. Estrogens, BRCA1, and breast cancer. Cancer Res. 60:4993-5001. [PubMed] [Google Scholar]
- 26.Jarvis, E. M., J. A. Kirk, and C. L. Clarke. 1998. Loss of nuclear BRCA1 expression in breast cancers is associated with a highly proliferative tumor phenotype. Cancer Genet. Cytogenet. 101:109-115. [DOI] [PubMed] [Google Scholar]
- 27.Kinzler, K., and B. Vogelstein. 1997. Gatekeepers and caretakers. Nature 386:761-763. [DOI] [PubMed] [Google Scholar]
- 28.Liu, W. M., F. K. Guerra-Vladusic, S. Kurakata, R. Lupu, and T. Kohwi-Shigematsu. 1999. HMG-I(Y) recognizes base-unpairing regions of matrix attachment sequences and its increased expression is directly linked to metastatic breast cancer phenotype. Cancer Res. 59:5695-5703. [PubMed] [Google Scholar]
- 29.Magdinier, F., S. Ribieras, G. M. Lenoir, L. Frappart, and R. Dante. 1998. Down-regulation of BRCA1 in human sporadic breast cancer; analysis of DNA methylation patterns of the putative promoter region. Oncogene 17:3169-3176. [DOI] [PubMed] [Google Scholar]
- 30.Marquis, S. T., J. V. Rajan, A. Wynshaw-Boris, J. Xu, G. Y. Yin, K. J. Abel, B. L. Weber, and L. A. Chodosh. 1995. The developmental pattern of Brca1 expression implies a role in differentiation of the breast and other tissues. Nat. Genet. 11:17-26. [DOI] [PubMed] [Google Scholar]
- 31.Miki, Y., J. Swensen, D. Shattuck-Eidens, P. A. Futreal, K. Harshman, S. Tavtigian, Q. Liu, C. Cochran, L. M. Bennett, W. Ding, et al. 1994. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266:66-71. [DOI] [PubMed] [Google Scholar]
- 32.Orlando, V. 2000. Mapping chromosomal proteins in vivo by formaldehyde-crosslinked-chromatin immunoprecipitation. Trends Biochem. Sci. 25:99-104. [DOI] [PubMed] [Google Scholar]
- 33.Ram, T. G., R. Reeves, and H. L. Hosick. 1993. Elevated high mobility group-I(Y) gene expression is associated with progressive transformation of mouse mammary epithelial cells. Cancer Res. 53:2655-2660. [PubMed] [Google Scholar]
- 34.Reeves, R., D. D. Edberg, and Y. Li. 2001. Architectural transcription factor HMGI(Y) promotes tumor progression and mesenchymal transition of human epithelial cells. Mol. Cell. Biol. 21:575-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reeves, R., and L. Beckerbauer. 2001. HMGI/Y proteins: flexible regulators of transcription and chromatin structure. Biochim. Biophys. Acta 1519:13-29. [DOI] [PubMed] [Google Scholar]
- 36.Rice, J. C., H. Ozcelik, P. Maxeiner, I. Andrulis, and B. W. Futscher. 2000. Methylation of the BRCA1 promoter is associated with decreased BRCA1 mRNA levels in clinical breast cancer specimens. Carcinogenesis 21:1761-1765. [DOI] [PubMed] [Google Scholar]
- 37.Rio, P. G., J. C. Maurizis, M. Peffault de Latour, Y. J. Bignon, and D. J. Bernard-Gallon. 1999. Quantification of BRCA1 protein in sporadic breast carcinoma with or without loss of heterozygosity of the BRCA1 gene. Int. J. Cancer 80:823-826. [DOI] [PubMed] [Google Scholar]
- 38.Romagnolo, D., L. A. Annab, T. E. Thompson, J. I. Risinger, L. A. Terry, J. C. Barrett, and C. A. Afshari. 1998. Estrogen upregulation of BRCA1 expression with no effect on localization. Mol. Carcinog. 22:102-109. [DOI] [PubMed] [Google Scholar]
- 39.Ruffner, H., and I. M. Verma. 1997. BRCA1 is a cell cycle-regulated nuclear phosphoprotein. Proc. Natl. Acad. Sci. USA 94:7138-7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russell, P. A., P. D. Pharoah, K. De Foy, S. J. Ramus, I. Symmonds, A. Wilson, I. Scott, B. A. Ponder, and S. A. Gayther. 2000. Frequent loss of BRCA1 mRNA and protein expression in sporadic ovarian cancers. Int. J. Cancer 87:317-321. [DOI] [PubMed] [Google Scholar]
- 41.Scala, S., G. Portella, M. Fedele, G. Chiappetta, and A. Fusco. 2000. Adenovirus-mediated suppression of HMGI(Y) protein synthesis as potential therapy of human malignant neoplasias. Proc. Natl. Acad. Sci. USA 97:4256-4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schafer, J. M., E. S. Lee, R. M. O'Regan, K. Yao, and V. C. Jordan. 2000. Rapid development of tamoxifen-stimulated mutant p53 breast tumors (T47D) in athymic mice. Clin. Cancer Res. 6:4373-4380. [PubMed] [Google Scholar]
- 43.Scully, R., and D. L. Livingston. 2000. In search of the tumor suppressor function of BRCA1 and BRCA2. Nature 408:429-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soriano, P., C. Montgomery, R. Geske, and A. Bradley. 1991. Targeted disruption of the c-src proto-oncogene leads to osteoporosis in mice. Cell 64:693-702. [DOI] [PubMed] [Google Scholar]
- 45.Sourvinos, G., and D. A. Spandidos. 1998. Decreased BRCA1 expression may arrest the cell cycle through activation of p53 checkpoint in human sporadic breast tumors. Biochem. Biophys. Res. Comm. 245:75-80. [DOI] [PubMed] [Google Scholar]
- 46.Tamimi, Y., H. G. van der Poel, M. M. Denyn, R. Umbas, H. F. Karthaus, F. M. Debruyne, and J. A. Schalken. 1993. Increased expression of high mobility group protein I(Y) in high grade prostatic cancer determined by in situ hybridization. Cancer Res. 3:5512-5516. [PubMed] [Google Scholar]
- 47.Thakur, S., H. B. Zhang, Y. Peng, H. Le, B. Carroll, T. Ward, J. Yao, L. M. Farid, F. J. Couch, R. B. Wilson, and B. L. Weber. 1997. Localization of BRCA1 and a splice variant identifies the nuclear localization signal. Mol. Cell. Biol. 17:444-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thakur, S., and C. M. Croce. 1999. Positive regulation of the BRCA1 promoter. J. Biol. Chem. 274:8837-8843. [DOI] [PubMed] [Google Scholar]
- 49.Thompson, M. E., R. A. Jensen, P. S. Obermiller, D. L. Page, and J. T. Holt. 1995. Decreased expression of BRCA1 accelerates growth and is often present during sporadic breast cancer progression. Nat. Genet. 9:444-450. [DOI] [PubMed] [Google Scholar]
- 50.Vaughn, J. P., P. L. Davis, M. D. Jarboe, G. Huper, A. C. Evans, R. W. Wiseman, A. Berchuck, J. D. Iglehart P. A. Futreal, and J. R. Marks. 1996. BRCA1 expression is induced before DNA synthesis in both normal and tumor-derived breast cells. Cell Growth Differ. 7:711-715. [PubMed] [Google Scholar]
- 51.Wilson, C. A., L. Ramos, M. R. Villaseñor, K. H. Anders, M. F. Press, K. Clarke, B. Karlan, J. J. Chen, R. Scully, D. Livingston, R. H. Zuch, M. H. Kanter, S. Cohen, F. J. Calzone, and D. J. Slamon. 1999. Localization of human BRCA1 and its loss in high-grade, non-inherited breast carcinomas. Nat. Gen. 21:236-240. [DOI] [PubMed] [Google Scholar]
- 52.Yoshikawa, K., K. Honda, T. Inamoto, H. Shinohara, A. Yamauchi, K. Suga, T. Okuyama, T. Shimada, H. Kodama, S. Noguchi, A. F. Gazdar, Y. Yamaoka, and R. Takahashi. 1999. Reduction of BRCA1 protein expression in Japanese sporadic breast carcinomas and its frequent loss inBRCA1-associated cases. Clin. Cancer Res. 5:1249-1261. [PubMed] [Google Scholar]
- 53.Zhou, X., K. F. Benson, H. R. Ashar, and K. Chada. 1995. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature 376:771-774. [DOI] [PubMed] [Google Scholar]