Abstract
Toll-like receptor 4 (TLR4) mediates the host response to lipopolysaccharide (LPS) by promoting the activation of pro- and anti-inflammatory cytokine genes. To activate each gene, numerous signal transduction pathways are required. The adaptor proteins MyD88 and TIRAP contribute to the activation of several and possibly all pathways via direct interactions with TLR4's Toll/interleukin-1 receptor (IL-1R) (TIR) domain. However, additional adaptors that are required for the activation of specific subsets of pathways may exist, which could contribute to the differential regulation of target genes. Furthermore, it remains unknown whether direct interactions that have been reported between TIR domains and other proteins are required for TLR4 signaling. To address these issues, we systematically mutated the TLR4 TIR domain in the context of a CD4/TLR4 fusion protein. Several exposed residues defining at least two structural surfaces were required in macrophages for activation of the proinflammatory IL-12 p40 and anti-inflammatory IL-10 promoters, as well as promoters dependent on individual transcription factors. Interestingly, the same residues were required by all promoters tested, suggesting that the signaling pathways diverge downstream of the adaptors. The mutant phenotypes provide a framework for future studies of TLR4 signaling, as the interaction supported by each critical surface residue will need to be defined.
The innate immune system provides a first line of defense against invading foreign microbes. The contribution of macrophages to innate immunity requires the recognition of microbial products by germ line-encoded pattern recognition receptors (35). One well-known example of such a microbial product is the lipopolysaccharide (LPS) found on the cell walls of gram-negative bacteria. When exposed to LPS, macrophages produce numerous regulators of the innate immune response, including pro- and anti-inflammatory cytokines (38, 57). To better understand the innate immune response and its connection to adaptive immunity, the LPS-stimulated signaling pathways that regulate cytokine gene expression must be elucidated.
In macrophages, LPS stimulates numerous signaling pathways, including pathways that activate IκB kinase (2), phosphatidylinositol 3-kinase (21, 49), protein kinase C (33), two Src family kinases, lyn and Hck (9, 14, 55), and three mitogen-activated protein kinases (MAPKs), ERK1/2, p38, and JNK (19). The protein that is largely responsible for transmitting the LPS signal across the plasma membrane is Toll-like receptor 4 (TLR4) (2, 6, 36). The central role of TLR4 in LPS signal transduction is evident from the severe hyporesponsiveness of TLR4 mutant mice (1, 7, 24, 46, 47).
The cytoplasmic domain of TLR4 is commonly referred to as a Toll/interleukin-1 receptor (IL-1R) (TIR) domain on the basis of its extensive homology to the corresponding domains of all members of the Toll, TLR, and IL-1R families (43). The importance of the TIR domain for TLR4 signaling was demonstrated by the constitutive activation of endogenous cytokine genes by a protein containing the TIR and transmembrane domains of TLR4 fused to the extracellular domain of CD4 (CD4/TLR4) (36, 37, 40).
Although the precise signaling pathways required for the induction of a given cytokine gene are not known, multiple pathways usually need to be activated. The proinflammatory IL-12 p40 and anti-inflammatory IL-10 genes, which are most relevant to the present study, can be used as examples. Induction of the murine IL-12 p40 gene, whose regulation has been characterized only partially, requires NF-κB, C/EBP, and AP-1 proteins, as well as the inducible remodeling of a positioned nucleosome spanning the promoter (39, 44, 60, 62). Induction of NF-κB requires dissociation from IκB proteins and often the posttranslational modification of NF-κB subunits (17). Induction of AP-1 activity generally requires phosphorylation of c-Jun by JNK and transcriptional induction of the c-fos gene (52). Induction of C/EBP in macrophages is poorly understood but may require p38 MAPK activity and possibly other events (4, 45). The pathways leading to remodeling of the nucleosome spanning the p40 promoter are not known. However, remodeling appears to be independent of NF-κB, C/EBP, and AP-1 yet may require p38 MAPK (48, 59). Interestingly, LPS induction of the anti-inflammatory IL-10 gene appears to be independent of NF-κB, and functionally relevant binding sites for NF-κB, C/EBP, and AP-1 have not been identified in the IL-10 promoter (5, 8, 30, 41). Instead, IL-10 promoter activity in macrophages is strongly dependent on a binding site for SP1, which may be a direct or indirect target of LPS-stimulated pathways (10, 34).
Elucidation of the complete mechanisms by which the TLR4 TIR domain activates the many pathways required for induction of pro- and anti-inflammatory cytokine genes is an enormous challenge. The only well-documented role of the TIR domain is to support homotypic interactions (1, 37). In particular, dimerization or multimerization of the TIR domain may be required for TLR4 signaling (28, 40). The TLR4 TIR domain can also interact with two TIR domain-containing adaptor proteins, MyD88 and TIRAP (MAL) (16, 22, 37, 40). Disruption of the MyD88 gene resulted in a severe defect in TLR4 signaling (25, 26). Disruption of TIRAP function using a cell-permeable TIRAP peptide revealed that it is also critical for TLR4 signaling (22). The precise relationship between MyD88 and TIRAP remains unknown.
In addition to the direct interactions with MyD88 and TIRAP, TLR4 assembles into complexes containing other important signaling proteins, including IL-1R-associated kinases (IRAKs), tumor necrosis factor receptor-associated factor 6, receptor-interacting protein 2 (11), and protein kinase R (22). However, these interactions appear to be indirect, and at least a subset are mediated by the MyD88 or TIRAP adaptors. Although no other proteins have been reported to interact directly with the mammalian TLR4 TIR domain, the TIR domain of the Drosophila Toll protein can interact directly with Pelle, the Drosophila IRAK homologue (53, 56), and dMyd88, the Drosophila Myd88 homologue (23). Furthermore, there has been speculation that the Drosophila Tube protein may be a Toll adaptor, although a direct interaction has not been reported (3). Therefore, it remains unknown whether oligomerization and the binding of MyD88 and TIRAP are sufficient for TLR4 signaling or whether the TLR4 TIR domain interacts directly with other proteins that may be critical for the induction of some or all downstream pathways. The observation that different TLR proteins stimulate different sets of target genes provides further support for the hypothesis that each TIR domain may interact with unique sets of proteins (13).
Although further characterization of MyD88, TIRAP, and other known proteins will yield valuable information about their functions, it will be difficult to determine when the complete set of TLR4 adaptors has been identified and whether there are adaptors that are dedicated to specific TLR proteins or signaling pathways. To begin to address these issues, we constructed a large panel of TLR4 substitution mutants to identify critical surface residues and to determine whether the residues required for the activation of various promoters are identical or different. The results provide valuable information about the point of divergence of signaling pathways and a framework for future studies of TLR4 signaling.
MATERIALS AND METHODS
Plasmids.
The murine IL-12 p40 chloramphenicol acetyltransferase (CAT) reporter containing sequences from positions −378 to +55 has been described previously (44). A CAT reporter containing three C/EBP sites from the mouse IL-12 p40 promoter (C/EBP IL-12 CAT) was constructed by inserting the following primer and its complement between the BglII and KpnI sites of pCAT T/I (44): CGTGTTGCAATTTGATATCGGGTGTTGCAATTCGCAAGTCTGTGTTGCAAT. A CAT reporter containing three NF-κB sites from the IL-12 p40 promoter (NF-κB IL-12 CAT) was constructed similarly using the following primer: CTAAAATTCCCCGCCTAAAATTCCCCGCCTAAAATTCCCCGCCA. The pAP-1 reporter was from Stratagene. The NF-κB ELAM LUC reporter was a gift from Paul Godowski (Genentech) and Robert Modlin (University of California, Los Angeles). Murine IL-10 promoter-luciferase (LUC) reporters were described previously (10).
pSRα, containing the CD4/TLR4 cDNA, was a gift from Ruslan Medzhitov and Charles Janeway (Yale University). The CD4/TLR4 cDNA was amplified by PCR and cloned between the HindIII and ClaI sites of pFLAG-CMV-1 (Sigma). In the process, the murine CD4 leader (amino acids 1 to 26) was replaced with a preprotrypsin leader and FLAG tag. Mutagenesis of the TIR domain was performed by the Quikchange method (Stratagene). Deletion mutants were created by introducing the TGA stop codon. Plasmids were verified by restriction mapping and sequencing.
Cell lines and reagents.
The RAW 264.7 murine macrophage line and 293T human embryonic kidney line were maintained in Dulbecco modified Eagle medium (Gibco) supplemented with 10% fetal calf serum (Omega Scientific). Superfect (Qiagen) was used to transfect RAW 264.7 cells. The p38 MAPK inhibitor SB203580 was from Calbiochem.
RAW 264.7 cell lines pcDNA3 and IκB-DA were generated by stably transfecting cells with an empty pcDNA3 vector or a plasmid encoding an IκB-estrogen receptor (ER) fusion. Clonal lines expressing IκB-ER were generated by drug selection and limiting dilution. Cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum, 1 mg of G418 (GibcoBRL) per ml, and 1 mM sodium pyruvate (Omega Scientific).
Flow cytometry.
293T cells were transfected with 8 μg of the expression plasmids by the CaPO4 method (27). Cells were stained on ice with phycoerythrin-conjugated anti-mCD4 monoclonal antibody (Pharmingen catalog no. 09005B). A total of 10,000 cells were counted using a FACSCalibur (Becton Dickinson) flow cytometer and CELLQuest software. Cells whose fluorescence signal in the FL2 channel was above that of mock-transfected cells were gated.
Reporter assays.
For CAT and LUC assays, half a million RAW 264.7 cells per well were seeded on six-well plates. The next day, cells were transfected with Superfect (Qiagen). Three microliters of Superfect was used per microgram of DNA; cells were incubated with Superfect/DNA mix for 3 h. Cells were harvested 24 h after transfection and used in reporter assays (44).
Sequence alignment and homology modeling.
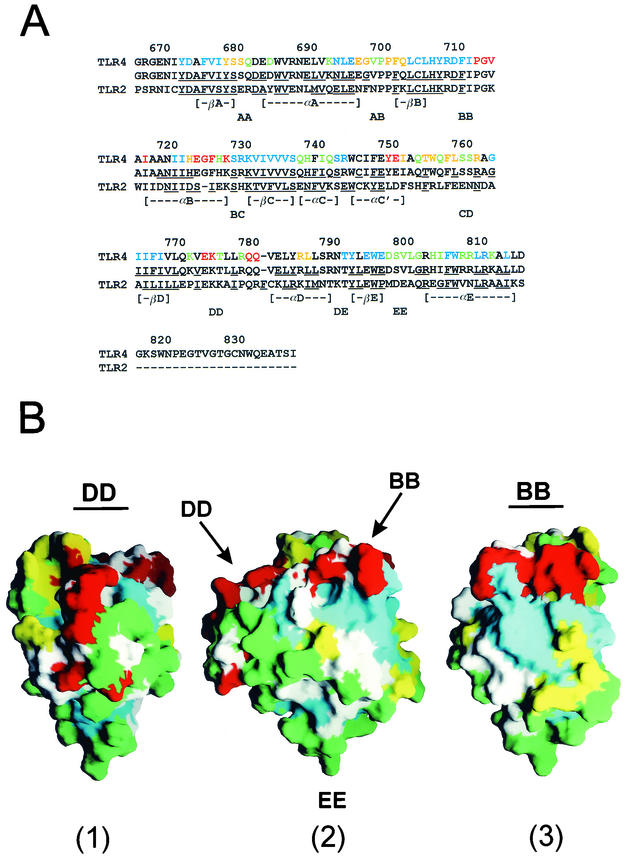
Protein sequence alignment (using CLUSTALW algorithm) and secondary structure predictions were performed on the NPS@ server (http://npsa-pbil.ibcp.fr). Homology modeling was based on a known crystal structure of TLR2 (61) and was performed using the Geno-3D server (http://geno3d-pbil.ibcp.fr). Additional minimization and modeling were performed with the InsightII software package (Accelrys, Inc., San Diego, Calif.) on the SGI Octane workstation. The software program GRASP was used to construct the molecular surfaces presented in Fig. 7 (42). The program MOLMOL (29) was used to calculate solvent-accessible surfaces (1.4-Å probe radius).
FIG.7.
Structural model of the TLR4 TIR domain. (A) Amino acid alignment of TLR2 and TLR4. In the second and third lines, the TLR4 and TLR2 (49) residues that exhibit less than 25% exposed surface area are underlined. In the top line, the residues that were not mutated are shown in black type. Residues that were mutated at the N and C termini are also shown in black. The phenotypes of the remaining substitution mutants are shown using the same colors as in Fig. 6 (see “Molecular modeling of CD4/TLR4 mutants” for description). (B) Three views of the GRASP model of the TLR4 cytoplasmic domain. For orientation purposes, the BB, DD, and EE loops are marked. Residues that were not mutated are shown in white or gray.
RESULTS
The chimeric receptor CD4/TLR4 activates the murine IL-12 p40 promoter.
The goal of this study was to identify residues within the TIR domain of human TLR4 that are essential for the activation of a variety of signal transduction pathways. The strategy was to transiently express a large panel of TLR4 proteins containing alanine substitution mutations in a physiologically relevant cell line and to monitor the effect of each mutation on signaling pathways by cotransfection of appropriate reporter plasmids. The cell line chosen for this analysis was the murine macrophage line RAW 264.7 because it responds well to LPS and therefore is likely to contain most or all of the relevant signal transduction molecules. Because this line expresses endogenous TLR4, it was necessary to use a strategy that allows signaling from the transiently expressed wild-type and mutant proteins to be distinguished from that of endogenous TLR4. The CD4/TLR4 chimeric receptor described by Medzhitov et al. (36) was appropriate for this purpose. When transiently expressed in macrophage cell lines, this chimeric receptor is constitutively active and stimulates endogenous pro-inflammatory cytokine genes, suggesting that it is competent for activation of all of the relevant pathways (36).
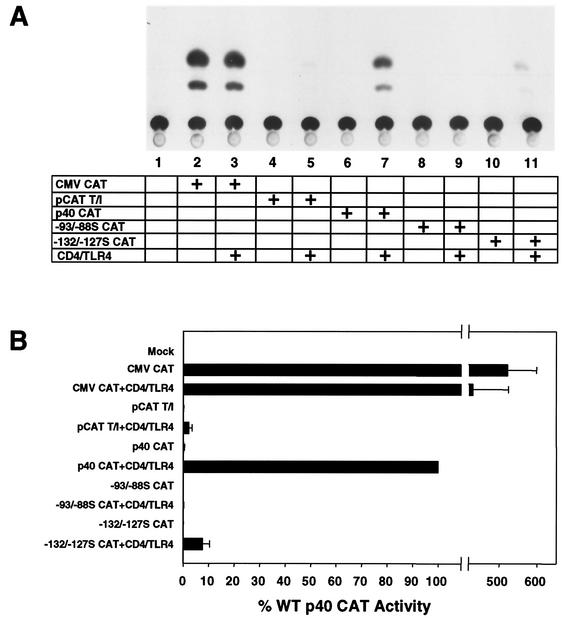
Although a large number of signal transduction pathways are activated by LPS and presumably by CD4/TLR4, we primarily focused on pathways that may contribute to activation of the murine IL-12 p40 promoter. To confirm that signaling via CD4/TLR4 can activate this promoter, RAW 264.7 cells were cotransfected with the CD4/TLR4 expression plasmid and a reporter plasmid, p40 CAT, which contains murine IL-12 p40 promoter sequences from positions −378 to +55 (relative to the transcription start site) fused to a CAT reporter gene (44). This promoter was strongly activated by the chimeric protein (Fig. 1A, lanes 6 and 7). In contrast, the cytomegalovirus (CMV) promoter and a minimal core promoter containing a TATA box and Inr element were only weakly affected by CD4/TLR4 (Fig. 1A, lanes 2 to 5). Activation of the p40 promoter was dependent on binding sites for C/EBP and NF-κB proteins, as a mutation in either site (from positions −93 to −88S [−93/−88S] and −132 to −127S [−132/−127S], respectively) greatly diminished activity (Fig. 1A, lanes 8 to 11). These results, which are reminiscent of our findings following LPS stimulation of RAW 264.7 cells (44), are presented graphically in Fig. 1B.
FIG. 1.
CD4/TLR4 activates the murine IL-12 p40 promoter. (A) RAW 264.7 cells were mock transfected (lane 1) or transfected (+) with the CAT reporter plasmids and CD4/TLR4 expression plasmid as indicated (lanes 2 to 11). Data are from one representative experiment. (B) The means and standard errors of the means (error bars) from four independent experiments (after phosphorimager quantitation) are shown. WT, wild type.
CD4/TLR4 activates NF-κB, C/EBP, and AP-1 signaling pathways.
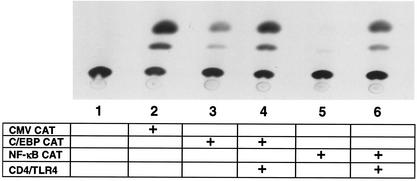
The results presented above suggest that CD4/TLR4 efficiently activates the NF-κB and C/EBP proteins that contribute to p40 induction. To monitor the activation of these proteins individually, CAT reporter plasmids were tested. These plasmids contain three tandem recognition sites for either NF-κB or C/EBP upstream of a minimal core promoter, with the recognition sequences derived from the murine p40 promoter. Strong activation of both promoters by the CD4/TLR4 protein was detected (Fig. 2). A recent study demonstrated that efficient activation of the murine p40 promoter also requires a functional binding site for AP-1 proteins (62). In RAW 264.7 cells, CD4/TLR4 activated transcription from a LUC reporter plasmid containing several AP-1 binding sites (see below). The activation of NF-κB and AP-1 signaling pathways by CD4/TLR4 is consistent with the original analysis of this fusion protein (37).
FIG. 2.
CD4/TLR4 activates NF-κB and C/EBP proteins. RAW 264.7 cells were mock transfected (lane 1) or transfected (+) with the CAT reporter plasmids and CD4/TLR4 expression plasmid as indicated (lanes 2 to 6). Data are shown from one representative experiment.
CD4/TLR4 activates the IL-10 promoter.
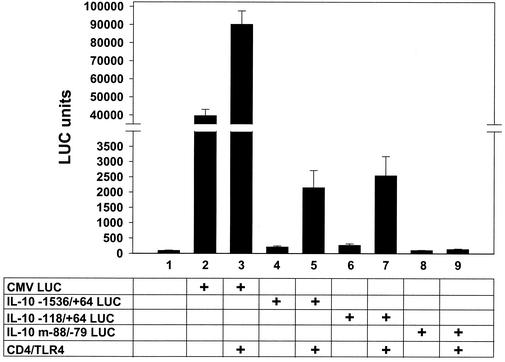
LPS and microbial antigens activate IL-10 gene transcription in human monocytes (10, 18, 58). Induction of the murine IL-10 promoter requires a binding site at positions −88 to −79 for transcription factor Sp1, which may be a direct or indirect target of LPS-activated signal transduction pathways (10, 34). To determine whether TLR4 signaling is sufficient to activate the IL-10 promoter, RAW 264.7 cells were cotransfected with the CD4/TLR4 expression plasmid and LUC reporter plasmids containing either a 1.6-kb fragment of the murine promoter (positions −1536 to +64) or a 182-bp minimal IL-10 promoter containing the Sp1 site (positions −118 to +64). CD4/TLR4 strongly activated transcription from both reporter plasmids (Fig. 3). Promoter activity was abolished by a substitution mutation in the Sp1 binding site that is essential for LPS induction (Fig. 3, m−88/−79) (10).
FIG. 3.
CD4/TLR4 activates the murine IL-10 promoter. RAW 264.7 cells were mock transfected (column 1) or transfected (+) with the LUC reporter plasmids and CD4/TLR4 expression plasmid as indicated (columns 2 to 9). Means and standard errors of the means (error bars) from four independent experiments are shown.
Different signaling pathways activate transcription from the various reporter plasmids.
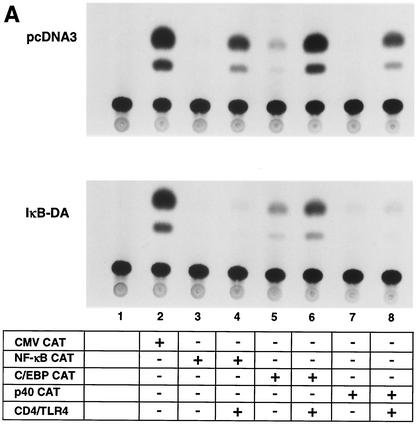
Because a major goal of this analysis was to compare the TLR4 residues required for activation of different signal transduction pathways, it was important to confirm that the promoters within the various reporter plasmids are indeed monitoring different pathways. To determine the degree to which each promoter depends on signaling through IκB, we generated RAW 264.7 cells expressing an IκB-ER fusion protein (see Materials and Methods), which acts as a dominant active repressor of NF-κB activity (32). The IκB component of this fusion protein lacks the IκB kinase phosphoacceptor sites and, therefore, is not targeted for proteasome degradation upon cell activation. Although the initial reason for using an ER fusion protein was to suppress NF-κB activity only in the presence of tamoxifen, suppression was observed in the absence of the drug (M. E. Haberland and G. Cheng, unpublished data).
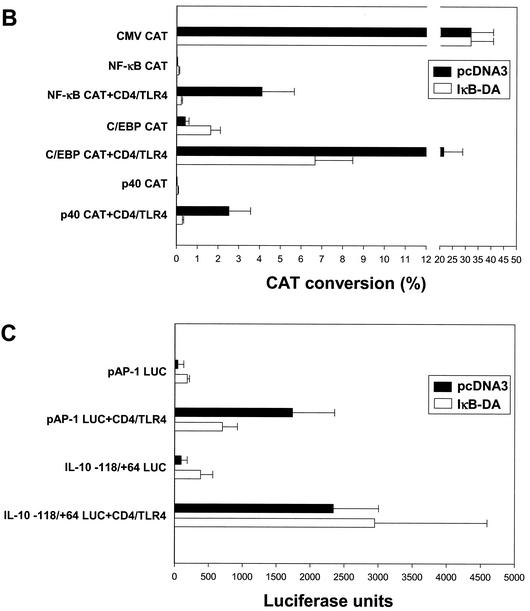
The suppression of NF-κB activity in one IκB-ER-expressing line (IκB-DA) is apparent in Fig. 4A. Although CD4/TLR4 strongly activated transcription from the NF-κB CAT reporter plasmid in a RAW 264.7 line containing a stably integrated pcDNA3 vector, activation was not observed in the IκB-DA line (Fig. 4A, lanes 3 and 4). Activation of the p40 CAT reporter gene was also strongly diminished (lanes 7 and 8); the residual activation of this reporter gene probably results from the activation of C/EBP and AP-1, consistent with the residual promoter activity detected when the NF-κB binding site is disrupted in the p40 promoter (Fig. 1A, lane 11). With the C/EBP CAT reporter gene, activation by CD4/TLR4 was retained in the IκB-DA line, although the CAT conversion was reduced from 20 to 6.5% (Fig. 4A, lanes 5 and 6; Fig. 4B). With a LUC reporter plasmid containing seven AP-1 binding sites, LUC activity following CD4/TLR4 activation was reduced from 1,700 U in the pcDNA3 line to 600 U in the IκB-DA line (Fig. 4C). Interestingly, with both the C/EBP and AP-1 reporters, the basal activities appeared to increase in the IκB-DA line, after normalizing to the activity of a CMV promoter-reporter (Fig. 4B and C). At face value, these data suggest that the IκB signaling pathway makes a significant contribution to C/EBP and AP-1 activation. However, since these pathways have not been connected in previous studies, a more likely explanation is that the stable overexpression of IκB alters the overall physiology of the cells, leading to indirect effects on many signaling pathways. For the purposes of this study, the significant activation retained in the IκB-DA line confirms that the C/EBP and AP-1 reporters can be activated by pathways that are independent of IκB.
FIG. 4.
IκB dependence of reporter plasmids. (A) RAW 264.7 cell lines containing a stably integrated pcDNA3 vector or an IκB-ER expression plasmid (IκB-DA line) were transfected (+) with reporter plasmids and the CD4/TLR4 expression plasmid as indicated. (B) The means and standard errors of the means (error bars) from four independent experiments similar to those shown in panel A are shown, following normalization to CMV-CAT activity. The activities in the pcDNA3 and IkB-DA lines are shown. (C) The pcDNA3 and IκB-DA cell lines were transfected with the CD4/TLR4 expression plasmid and LUC reporter plasmids containing multiple AP-1 sites or the IL-10 promoter. Means and standard errors of the means (error bars) from four independent experiments after normalization to CMV-CAT activity are shown.
Finally, activation of an IL-10 promoter-reporter gene by CD4/TLR4 was monitored in the IκB-DA line. Consistent with previous evidence that IL-10 expression is independent of the IκB/NF-κB pathway (8, 10, 34), IL-10 promoter activity was unaffected (Fig. 4C).
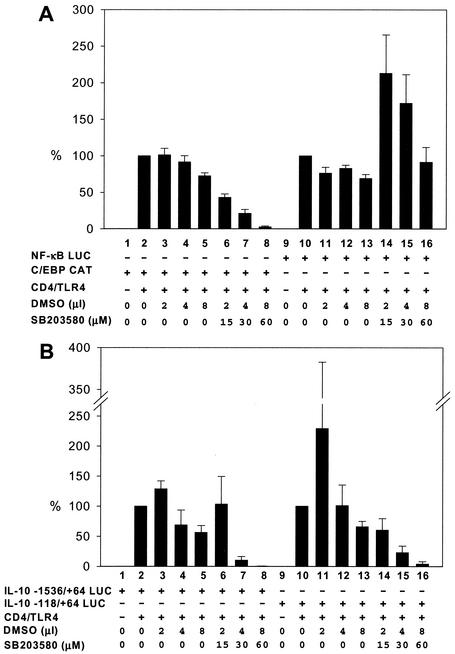
Previous studies suggested that p38 MAPK contributes to LPS induction of C/EBP proteins in macrophages (4, 15). Consistent with these data, activation of the C/EBP CAT reporter gene by CD4/TLR4 was strongly diminished by a p38 inhibitor, SB203580 (12) (Fig. 5A, lanes 6 to 8). In contrast, comparable amounts of the dimethyl sulfoxide solvent used to dissolve the inhibitor had minimal effects (Fig. 5A, lanes 3 to 5). Low concentrations of inhibitor slightly elevated activation of an NF-κB-LUC reporter gene by CD4/TLR4, with activity returning to normal levels in the presence of higher concentrations of inhibitor (Fig. 5A, lanes 10 to 16). The NF-κB-LUC reporter, which served as a p38-independent negative control in this experiment, contains the ELAM-1 promoter, whose activity depends almost exclusively on three NF-κB binding sites (50). Finally, the SB203580 inhibitor was found to diminish activity of the IL-10 promoter (Fig. 5B), suggesting that p38 plays a role in the activation of Sp1 or another protein that contributes to IL-10 promoter activity. This finding is consistent with previous evidence that p38 is involved in transcriptional induction of the human IL-10 gene by LPS (34).
FIG. 5.
p38 MAPK dependence of reporter plasmids. RAW 264.7 cells were transfected (+) with reporter plasmids and the CD4/TLR4 expression plasmid as indicated. The SB203580 p38 MAPK inhibitor or the dimethyl sulfoxide (DMSO) solvent were added as indicated. Activities of the NF-κB and C/EBP promoters (A) and of the IL-10 promoters (B) relative to that of untreated controls (100%) are shown. Means and standard errors of the means (error bars) from four independent experiments are shown.
To summarize, five reporter plasmids that are likely to serve as readouts for signal transduction pathways activated by CD4/TLR4 have been characterized. When analyzing the panel of CD4/TLR4 substitution mutants described below, the activity of the NF-κB reporters will be reduced when mutants that disrupt the IκB signaling pathway are tested. The activity of the AP-1 reporter will presumably be reduced when mutants that disrupt the JNK pathway and perhaps pathways contributing to the induction of Fos family proteins are tested. The activity of the C/EBP reporter will be reduced when mutants that disrupt the p38 pathway and possibly other pathways that may be required for optimal C/EBP activation are tested. Activity of the p40 CAT reporter should be reduced when mutants that disrupt any of the pathways involved in NF-κB, AP-1, or C/EBP activation are tested. Finally, like C/EBP reporter activity, the activity of the IL-10 promoter-reporter should be reduced when mutants that disrupt the p38 pathway are tested. IL-10 promoter activity may also be reduced when mutants that disrupt other pathways that contribute to induction of this anti-inflammatory cytokine gene are tested.
Generation and expression of CD4/TLR4 mutants.
Fifty-seven mutants with alanine substitutions in the TLR4 TIR domain were generated in the context of the CD4/TLR4 fusion protein. For some of the mutants, the amino acids altered were selected before TLR1 and TLR2 TIR domain crystal structures were available (61). After the crystal structures became available, it became apparent that many of the altered amino acids are internal and likely to contribute to the proper folding and stabilization of the TIR domain. Therefore, using a TLR4 model derived from the TLR1 and TLR2 structures as a guide (see below), a number of additional mutants that alter residues at the surface of the protein were generated. The cDNAs for all of the mutant proteins were inserted into the pFLAG-CMV-1 vector, which drives transcription of the cDNA from a CMV promoter-enhancer. This vector encodes a FLAG epitope tag in frame with the N terminus of the expressed protein, such that the protein expressed on the cell surface will contain an extracellular FLAG tag (see Materials and Methods).
In addition to the substitution mutants, three deletion mutants with part of the TLR4 TIR domain deleted were generated in the context of the CD4/TLR4 protein. These three C-terminal deletions, Y794STOP, R809STOP, and W821STOP, were generated by introducing a stop codon into the coding sequence.
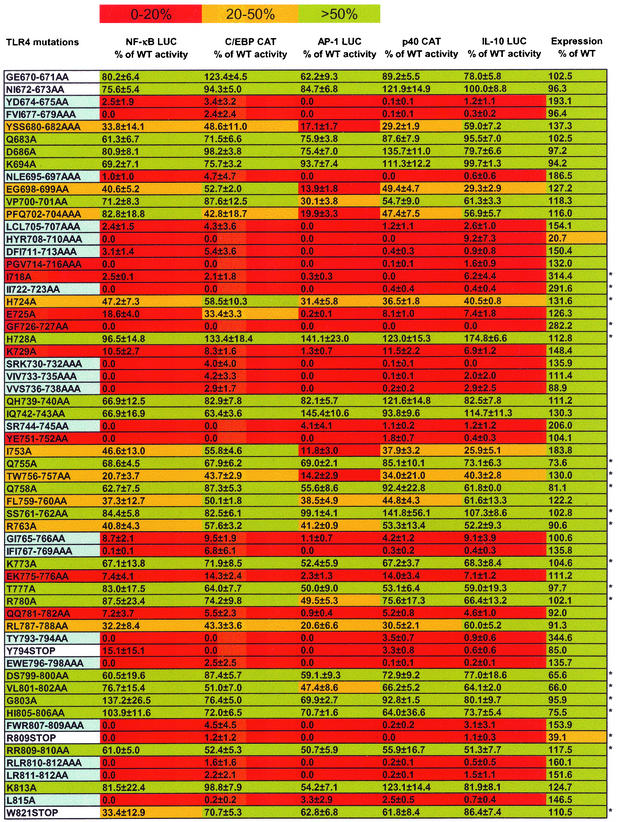
The mutants analyzed are listed in the leftmost column of Fig. 6. The amino acid numbering is based on the numbering of the wild-type human TLR4 sequence (36) (GenBank accession number U93091). To compare their cell surface expression levels to that of wild-type CD4/TLR4, the expression plasmids were transfected into 293T cells. After 48 h, the cells were harvested and analyzed by flow cytometry using a monoclonal antibody to murine CD4. The geometric mean fluorescence observed with each mutant protein relative to the fluorescence for the wild-type protein (100%) is shown in the rightmost column in Fig. 6. All of the mutants, with the exception of mutants HYR708-711AAA and R809STOP, were expressed at approximately wild-type levels on the cell surface. Thus, most mutations did not appear to affect the intracellular localization of the CD4/TLR4 protein. Although expression levels could not be determined in RAW 264.7 cells because of low transfection efficiencies, the relative expression levels of the wild-type and mutant proteins are likely to be comparable to those determined in 293T cells. Attempts to use Western blots to confirm that the expressed proteins were intact were unsuccessful, due to insolubility and aberrant migration of the transmembrane protein when analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, even when conditions that are often compatible with transmembrane proteins were used (data not shown).
FIG. 6.
Phenotypes of the CD4/TLR4 mutants. Residues in the mutant designations are numbered according to the human TLR4 sequence. The second through sixth columns show, for each mutant, the percentage of wild-type (WT) CD4/TLR4 activation of five different reporter plasmids. The rightmost column shows the surface expression of each mutant expressed in 293T cells, as determined by flow cytometry. In columns 2 to 7, the colors depict the percent activity of each mutant in each assay. In column 1, red, yellow, and green indicate the mean reporter activity from columns 2 to 6. Blue depicts the subset of mutants in which at least one residue exhibits 25% surface exposure, if that mutant exhibits an average of 0 to 20% activity in the reporter assays. White indicates the three deletion and two substitution mutants that alter residues that are not included in the homology model in Fig. 7. The data in columns 2 to 6 are the means and standard errors of the means from two (marked with an asterisk) or four experiments.
Molecular phenotypes of CD4/TLR4 mutants.
Expression plasmids for the 60 mutant CD4/TLR4 proteins were transfected into RAW 264.7 cells together with each of the five reporter plasmids described above. The activity of each reporter plasmid when stimulated by each mutant is summarized in Fig. 6. Reporter activities are presented as a percentage of the activity observed with wild-type CD4/TLR4. In each reporter assay, basal activity (i.e., no CD4/TLR4 activator) was subtracted from the activated signal. If the final value was negative, it was set at zero.
The results obtained with all five of the reporter plasmids were remarkably similar (Fig. 6). Most significantly, mutations that reduced the activity of one reporter below 20% of the wild-type value almost always reduced the activity of all other reporters to a comparable extent. These results strongly suggest that the same set of TLR4-mediated interactions is required for the activation of all five promoters studied, including the proinflammatory IL-12 p40 promoter and the anti-inflammatory IL-10 promoter. In other words, divergence of the various TLR4 signaling pathways is likely to occur downstream of the adaptors that interact directly with the TLR4 TIR domain.
Molecular modeling of CD4/TLR4 mutants.
In addition to the key conclusion described above, the results of this analysis pinpoint structural motifs that contribute to TLR4 signaling. For this analysis, a structural model of the TLR4 TIR was needed. Because a crystal structure of TLR4 has not been published yet, the published structures of the homologous TIR domains from human TLR2 and TLR1 (61) were used to prepare a model for the TIR domain of human TLR4. The TLR4 sequence starting at position Y674 and ending at position D817 was used for modeling. The structural model was prepared using the NPS@ and Geno3D servers and the InsightII package (Accelrys, Inc.) and was subsequently visualized using GRASP software (see Materials and Methods). The TLR4 model folded similarly when the TLR1, TLR2, and TLR2 P712H mutant structures (61) were used as the templates, implying that the model is reasonably accurate. Additional support for the validity of the model was provided by the fact that a similar structure was produced with modeling tools built into the InsightII software package (data not shown).
The alignment of the TLR2 and TLR4 primary sequences is shown in Fig. 7A. The top line shows the phenotypes of the TLR4 mutants, using a color code similar to that used for the surface representation model shown in Fig. 7B. The residues that were not mutated are shown in black type. A few residues that were mutated at the N and C termini are also shown in black type because these residues were not included in the TLR4 model or in the crystal structures of TLR1 and TLR2. The residues that exhibit less than 25% exposed surface area in the TLR4 model and TLR2 crystal structure are underlined in the second and third lines of Fig. 7A. Three views of a GRASP model of the TLR4 cytoplasmic domain are shown in Fig. 7B. In Fig. 7B, the residues that were not mutated are shown in white or gray.
In both Fig. 7A and B, residues shown in green are the residues that, when altered, have no effect on promoter activity or reduce promoter activity by an average of less than twofold. Residues shown in yellow are those resides that, when altered, reduce promoter activity by a value between two- and fivefold. Amino acids shown in red are the amino acids that fulfill two conditions: (i) when altered, these amino acids reduce promoter activity by an average value of greater than fivefold, and (ii) they exhibit greater than 25% exposed surface area. Amino acids shown in blue are the amino acids that reduce promoter activity by an average value of greater than fivefold but exhibit less than 25% exposed surface area, suggesting a relatively high probability that the structural integrity of the domain was compromised. If any residue within a double or triple mutant exhibits less than 25% exposed surface area and if the mutant reduced promoter activity greater than fivefold, all amino acids altered in that mutant are shown in blue.
DISCUSSION
Common residues required for multiple signaling pathways.
The results of this analysis suggest that several and possibly all signaling pathways activated by TLR4 diverge downstream of the TIR domain. Of greatest significance is the finding that the residues required for activation of the proinflammatory IL-12 p40 and anti-inflammatory IL-10 promoters were similar. Although different signaling pathways are likely to be required for the activation of these two promoters, we could find no evidence that they require different TLR4 interaction surfaces. The similar results obtained with reporter plasmids regulated by NF-κB, C/EBP, and AP-1 proteins provide further evidence against the existence of pathway-specific interaction surfaces. Although these results suggest that the same set of adaptor proteins may be required for the activation of all downstream pathways, they leave open the possibility that, within a single cell, two different adaptors may interact with the same TLR4 surface on different TLR4 molecules (or sequentially on the same TLR4 molecule). In particular, MyD88 and TIRAP may interact with the same surface yet may stimulate different downstream pathways (16, 22).
The precise pathways required for the activation of each reporter plasmid used for this analysis are difficult to determine. For example, for the reporter plasmids regulated by three NF-κB recognition sites, it is not clear whether reporter activation required only IKK activation or whether it also required the posttranslational modification of NF-κB subunits. The direct activation of specific kinases will ultimately need to be monitored, using either kinase assays or Western blots with phosphorylation-specific antibodies. Unfortunately, these assays could not be used for the current analysis because of the low transfection efficiencies of macrophage cell lines. For this reason, it was also not possible to monitor nucleosome remodeling at the IL-12 p40 promoter; the restriction enzyme accessibility assay used to monitor remodeling requires gene activation in a high percentage of cells. To monitor the TLR4 residues required for remodeling and the activation of specific signaling pathways, it may be possible to use retroviral vectors to express the mutant proteins.
Residues and structural surfaces involved in TLR4 signaling.
In addition to providing strong evidence that multiple signaling pathways require the same TIR domain interactions, the results of this analysis allow a detailed examination of the interaction surfaces on the TLR4 TIR domain model. The TLR4 model, like the TLR1 and TLR2 TIR domain structures, contains a hydrophobic core, which includes 5 β-strands, surrounded by 5 α-helices (61). A crystal structure of the TLR4 TIR domain will be needed to determine the validity of this model.
The models shown in Fig. 7B reveal at least two exposed surfaces that are critical for signaling. These two surfaces include the BB and DD loops. The BB loop can be visualized most easily in view 3. The critical residues shown in red include three that were altered simultaneously within the BB loop itself (P714, G715, and V716) and five within the αB helix that were altered in three separate mutants (I718, E725, G726, F727, and K729). These residues form a contiguous surface that has the potential to contribute to a protein-protein interaction.
The large patch shown in blue in view 3 in Fig. 7B includes five other residues from the BB loop (Y709, R710, D711, F712, and I713) and three residues from the αA helix (E697) and βB strand (L705 and C706). Although mutations in these eight residues eliminated TLR4 signaling, six of the eight (E697, L705, C706, Y709, D711, and F712) are shown in blue because they correspond to residues that exhibit less than 25% surface exposure. The remaining two (R710 and I713) exhibit greater than 25% surface exposure but are shown in blue because they were mutated along with unexposed residues. One of these exposed residues, R710, is a residue that contributes to a critical ion pair interaction (with E697) in TLR1 and TLR2 (61), which greatly increases the probability that these residues play an important structural role. Thus, although the low surface exposure and ion pair contributions of seven of the eight residues in this blue patch limit our ability to determine whether they are required for a protein-protein interaction, there is a reasonable chance that the critical interaction surface extends into this region.
The patch of yellow below the blue patch in view 3 (Fig. 7B) corresponds primarily to the AB loop (E698, P702, and Q704) and indicates moderate importance for signaling. It is not clear whether this region supports a protein-protein interaction that modestly enhances but is not essential for TLR4 signaling or whether the function of this region is solely structural.
It is important to note that residues within the BB loop were previously shown to be critical for TLR and Drosophila Toll signaling. These residues include those that correspond to R710, D711, P714, and G715 in human TLR4 (51, 54, 61). In fact, the widely studied Lpsd mutation in murine TLR4 (46), which changes P712 to histidine, corresponds to P714 in human TLR4. This mutation disrupts the interaction between TLR4 and MyD88 (61), providing strong evidence that the region shown in red in view 3 in Fig. 7B corresponds to the MyD88 interaction interface. Although the previous analyses of the P714 mutation suggested that the BB loop is responsible for the MyD88 interaction, it was not previously known that nearby surface residues within the αB helix are also critical for TLR4 signaling. It will be interesting to determine whether these residues, E725 and K729, are critical for the MyD88 interaction or for an interaction with the related TIRAP protein (16, 22).
The second patch of essential surface residues is best visualized in view 1 in Fig. 7B. This patch contains four residues from the DD loop (E775, K776, Q781, and Q782) and two from the αC′ helix (Y751 and E752). This region may support an interaction with another signaling molecule or may be required for TLR4 oligomerization. It is interesting that two critical residues, E775 and K776, are located on the underside of the protruding region (see view 2), raising the possibility that these residues contribute to an interaction that is distinct from the interaction supported by the other critical residues in this region.
In view 1 in Fig. 7B, a second patch of yellow is apparent in the upper left corner. This region corresponds to the CD loop. The data suggest that this loop may support a protein-protein interaction that contributes to but is not essential for TLR4 signaling. Alternatively, a subset of residues within this loop may make moderately important contributions to the TIR domain structure. Because all of the surface residues in yellow exhibit less than 25% exposed surface area or were mutated simultaneously with residues that exhibit less than 25% exposed surface area, we favor the latter hypothesis.
Although mutations were introduced into a large percentage of the TLR4 surface residues, the analysis was not complete. However, it is unlikely that the unaltered surfaces of sufficient size to support a protein-protein interaction. It is important to note, however, that the alanine-scanning strategy used for this analysis has the potential to miss important interaction surfaces. Loss of one or even two side chains that contribute to an interaction may not reduce binding energy to a sufficient extent to disrupt signaling.
One additional limitation is that many of the mutations made before the crystal structures became available alter two or three amino acids simultaneously. If any one amino acid exhibits less than 25% solvent exposure, we interpreted the results cautiously and highlighted all amino acids in blue. Overall, six amino acids that are greater than 25% exposed in the TLR4 model (R710, I713, R731, R745, T793, and E796) were shown in blue because they were mutated along with unexposed amino acids. Five other amino acids that are greater than 25% exposed in the corresponding positions of the TLR2 crystal structure (D711, I723, S744, G765, and I766) were shown in blue because they were mutated along with unexposed amino acids. These 11 exposed amino acids should be mutated individually to determine whether they are critical for signaling.
Deleting 19 amino acids from the C terminus of TLR4 (mutation W821STOP) had no effect on signaling, while deleting 31 (R809STOP) or 46 (Y794STOP) amino acids completely eliminated activity. The C terminus of TLR2 contains an α-helix after the EE loop (61), and our structural model suggests that the corresponding region of TLR4 also has an α-helical structure. Mutant R809STOP is not expressed properly, and signaling is completely lost. This indicates that the C-terminal α-helix may be essential for structural integrity. This view is supported by the TLR2 crystal structure, which indicates multiple intramolecular bonds between the C-terminal α-helix and other structural elements. In accordance with these results, an earlier study reported that a C-terminal deletion of only 30 amino acids from a related human type I IL-1R causes a null phenotype (20). Also, deletion of 42 amino acids from the C terminus of the mouse type I IL-1R destroys function (31).
In conclusion, we have performed an extensive mutant analysis of the human TLR4 TIR domain, which revealed at least two important surface patches. Careful in vitro studies are needed to determine whether these surfaces correspond to binding sites for adaptor proteins or are involved in receptor oligomerization. Such studies would be of paramount importance for understanding the molecular mechanisms of LPS signaling in macrophages and possibly other immune cells.
.
Acknowledgments
We thank Ruslan Medzhitov and Charles Janeway for the CD4/TLR4 expression plasmid; Robert Modlin and Paul Godowski for the NF-κB ELAM LUC plasmid; and Shomyseh Sanjabi, Michelle Bradley, and Tianyi Wang for critical reading of the manuscript.
G.C. and M.E.H. were supported in part by NIH grant GM57559. S.T.S. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Akira, S. 2000. Toll-like receptors: lessons from knockout mice. Biochem. Soc. Trans. 28:551-556. [DOI] [PubMed] [Google Scholar]
- 2.Akira, S., K. Takeda, and T. Kaisho. 2001. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat. Immunol. 2:675-680. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, K. V. 2000. Toll signaling pathways in the innate immune response. Curr. Opin. Immunol. 12:13-19. [DOI] [PubMed] [Google Scholar]
- 4.Baldassare, J. J., Y. Bi, and C. J. Bellone. 1999. The role of p38 mitogen-activated protein kinase in IL-1 beta transcription. J. Immunol. 162:5367-5373. [PubMed] [Google Scholar]
- 5.Benkhart, E. M., M. Siedlar, A. Wedel, T. Werner, and H. W. Ziegler-Heitbrock. 2000. Role of Stat3 in lipopolysaccharide-induced IL-10 gene expression. J. Immunol. 165:1612-1617. [DOI] [PubMed] [Google Scholar]
- 6.Beutler, B. 2000. Tlr4: central component of the sole mammalian LPS sensor. Curr. Opin. Immunol. 12:20-26. [DOI] [PubMed] [Google Scholar]
- 7.Bihl, F., L. Larivière, S. T. Qureshi, L. Flaherty, and D. Malo. 2001. LPS-hyporesponsiveness of mnd mice is associated with a mutation in Toll-like receptor 4. Genes Immun. 2:56-59. [DOI] [PubMed] [Google Scholar]
- 8.Bondeson, J., K. A. Browne, F. M. Brennan, B. M. Foxwell, and M. Feldmann. 1999. Selective regulation of cytokine induction by adenoviral gene transfer of IκBα into human macrophages: lipopolysaccharide-induced, but not zymosan-induced, proinflammatory cytokines are inhibited, but IL-10 is nuclear factor-κB independent. J. Immunol. 162:2939-2945. [PubMed] [Google Scholar]
- 9.Boulet, I., S. Ralph, E. Stanley, P. Lock, A. R. Dunn, S. P. Green, and W. A. Phillips. 1992. Lipopolysaccharide- and interferon-gamma-induced expression of hck and lyn tyrosine kinases in murine bone marrow-derived macrophages. Oncogene 7:703-710. [PubMed] [Google Scholar]
- 10.Brightbill, H. D., S. E. Plevy, R. L. Modlin, and S. T. Smale. 2000. A prominent role for Sp1 during lipopolysaccharide-mediated induction of the IL-10 promoter in macrophages. J. Immunol. 164:1940-1951. [DOI] [PubMed] [Google Scholar]
- 11.Chin, A. I., P. W. Dempsey, K. Bruhn, J. F. Miller, Y. Xu, and G. Cheng. 2002. Involvement of receptor-interacting protein 2 in innate and adaptive immune responses. Nature 416:190-194. [DOI] [PubMed] [Google Scholar]
- 12.Cuenda, A., J. Rouse, Y. N. Doza, R. Meier, P. Cohen, T. F. Gallagher, P. R. Young, and J. C. Lee. 1995. SB 203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stresses and interleukin-1. FEBS Lett. 364:229-233. [DOI] [PubMed] [Google Scholar]
- 13.Doyle, S. E., S. A. Vaidya, R. O'Connell, H. Dadgostar, P. W. Dempsey, T.-T. Wu, G. Rao, R. Sun, M. E. Haberland, R. L. Modlin, and G. Cheng. 2002. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity 17:251-263. [DOI] [PubMed] [Google Scholar]
- 14.English, B. K., J. N. Ihle, A. Myracle, and T. Yi. 1993. Hck tyrosine kinase activity modulates tumor necrosis factor production by murine macrophages. J. Exp. Med. 178:1017-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng, G. J., H. S. Goodridge, M. M. Harnett, X. Q. Wei, A. V. Nikolaev, A. P. Higson, and F. Y. Liew. 1999. Extracellular signal-related kinase (ERK) and p38 mitogen-activated protein (MAP) kinases differentially regulate the lipopolysaccharide-mediated induction of inducible nitric oxide synthase and IL-12 in macrophages: Leishmania phosphoglycans subvert macrophage IL-12 production by targeting ERK MAP kinase. J. Immunol. 163:6403-6412. [PubMed] [Google Scholar]
- 16.Fitzgerald, K. A., E. M. Palsson-McDermott, A. G. Bowie, C. A. Jefferies, A. S. Mansell, G. Brady, E. Brint, A. Dunne, P. Gray, M. T. Harte, D. McMurray, D. E. Smith, J. E. Sims, T. A. Bird, and L. A. O'Neill. 2001. Mal (MyD88-adapter-like) is required for Toll-like receptor-4 signal transduction. Nature 413:78-83. [DOI] [PubMed] [Google Scholar]
- 17.Ghosh, S., and M. Karin. 2002. Missing pieces in the NF-κB puzzle. Cell 109(Suppl.):S81-S96. [DOI] [PubMed]
- 18.Giambartolomei, G. H., V. A. Dennis, B. L. Lasater, and M. T. Philipp. 1999. Induction of pro- and anti-inflammatory cytokines by Borrelia burgdorferi lipoproteins in monocytes is mediated by CD14. Infect. Immun. 67:140-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guha, M., and N. Mackman. 2001. LPS induction of gene expression in human monocytes. Cell Signal. 13:85-94. [DOI] [PubMed] [Google Scholar]
- 20.Heguy, A., C. T. Baldari, G. Macchia, J. L. Telford, and M. Melli. 1992. Amino acids conserved in interleukin-1 receptors (IL-1Rs) and the Drosophila toll protein are essential for IL-1R signal transduction. J. Biol. Chem. 267:2605-2609. [PubMed] [Google Scholar]
- 21.Herrera-Velit, P., and N. E. Reiner. 1996. Bacterial lipopolysaccharide induces the association and coordinate activation of p53/56lyn and phosphatidylinositol 3-kinase in human monocytes. J. Immunol. 156:1157-1165. [PubMed] [Google Scholar]
- 22.Horng, T., G. M. Barton, and R. Medzhitov. 2001. TIRAP: an adapter molecule in the Toll signaling pathway. Nat. Immunol. 2:835-841. [DOI] [PubMed] [Google Scholar]
- 23.Horng, T., and R. Medzhitov. 2001. Drosophila MyD88 is an adapter in the Toll signaling pathway. Proc. Natl. Acad. Sci. USA 98:12654-12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoshino, K., O. Takeuchi, T. Kawai, H. Sanjo, T. Ogawa, Y. Takeda, K. Takeda, and S. Akira. 1999. Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J. Immunol. 162:3749-3752. [PubMed] [Google Scholar]
- 25.Kaisho, T., O. Takeuchi, T. Kawai, K. Hoshino, and S. Akira. 2001. Endotoxin-induced maturation of MyD88-deficient dendritic cells. J. Immunol. 166:5688-5694. [DOI] [PubMed] [Google Scholar]
- 26.Kawai, T., O. Adachi, T. Ogawa, K. Takeda, and S. Akira. 1999. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity 11:115-122. [DOI] [PubMed] [Google Scholar]
- 27.Kingston, R. E. 1997. Introduction of DNA into mammalian cells, p. 9.0.1-9.0.5. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 1. John Wiley & Sons, Inc., New York, N.Y.
- 28.Kopp, E. B., and R. Medzhitov. 1999. The Toll-receptor family and control of innate immunity. Curr. Opin. Immunol. 11:13-18. [DOI] [PubMed] [Google Scholar]
- 29.Koradi, R., M. Billeter, and K. Wuthrich. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14:29-32, 51-55. [DOI] [PubMed] [Google Scholar]
- 30.Kube, D., C. Platzer, A. von-Knethen, H. Straub, H. Bohlen, M. Hafner, and H. Tesch. 1995. Isolation of the human interleukin 10 promoter. Characterization of the promoter activity in Burkitt's lymphoma cell lines. Cytokine 7:1-7. [DOI] [PubMed] [Google Scholar]
- 31.Kuno, K., S. Okamoto, K. Hirose, S. Murakami, and K. Matsushima. 1993. Structure and function of the intracellular portion of the mouse interleukin 1 receptor (type I). Determining the essential region for transducing signals to activate the interleukin 8 gene. J. Biol. Chem. 268:13510-13518. [PubMed] [Google Scholar]
- 32.Lee, H. H., H. Dadgostar, Q. Cheng, J. Shu, and G. Cheng. 1999. NF-κB-mediated up-regulation of Bcl-x and Bfl-1/A1 is required for CD40 survival signaling in B lymphocytes. Proc. Natl. Acad. Sci. USA 96:9136-9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, M. K., P. Herrera-Velit, R. W. Brownsey, and N. E. Reiner. 1994. CD14-dependent activation of protein kinase C and mitogen-activated protein kinases (p42 and p44) in human monocytes treated with bacterial lipopolysaccharide. J. Immunol. 153:2642-2652. [PubMed] [Google Scholar]
- 34.Ma, W., W. Lim, K. Gee, S. Aucoin, D. Nandan, M. Kozlowski, F. Diaz-Mitoma, and A. Kumar. 2001. The p38 mitogen-activated kinase pathway regulates the human interleukin-10 promoter via the activation of Sp1 transcription factor in lipopolysaccharide-stimulated human macrophages. J. Biol. Chem. 276:13664-13674. [DOI] [PubMed] [Google Scholar]
- 35.Medzhitov, R., and C. A. Janeway, Jr. 1997. Innate immunity: the virtues of a nonclonal system of recognition. Cell 91:295-298. [DOI] [PubMed] [Google Scholar]
- 36.Medzhitov, R., P. Preston-Hurlburt, and C. A. Janeway, Jr. 1997. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 388:394-397. [DOI] [PubMed] [Google Scholar]
- 37.Medzhitov, R., P. Preston-Hurlburt, E. Kopp, A. Stadlen, C. Chen, S. Ghosh, and C. A. Janeway, Jr. 1998. MyD88 is an adaptor protein in the hToll/IL-1 receptor family signaling pathways. Mol. Cell 2:253-258. [DOI] [PubMed] [Google Scholar]
- 38.Moore, K. W., A. O'Garra, R. de Waal Malefyt, P. Vieira, and T. R. Mosmann. 1993. Interleukin-10. Annu. Rev. Immunol. 11:165-190. [DOI] [PubMed] [Google Scholar]
- 39.Murphy, T. L., M. G. Cleveland, P. Kulesza, J. Magram, and K. M. Murphy. 1995. Regulation of interleukin 12 p40 expression through an NF-κB half-site. Mol. Cell. Biol. 15:5258-5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muzio, M., G. Natoli, S. Saccani, M. Levrero, and A. Mantovani. 1998. The human toll signaling pathway: divergence of nuclear factor κB and JNK/SAPK activation upstream of tumor necrosis factor receptor-associated factor 6 (TRAF6). J. Exp. Med. 187:2097-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nemeth, Z. H., G. Hasko, and E. S. Vizi. 1998. Pyrrolidine dithiocarbamate augments IL-10, inhibits TNF-α, MIP-1α, IL-12, and nitric oxide production and protects from the lethal effect of endotoxin. Shock 10:49-53. [DOI] [PubMed] [Google Scholar]
- 42.Nicholls, A., K. A. Sharp, and B. Honig. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11:281-296. [DOI] [PubMed] [Google Scholar]
- 43.O'Neill, L. 2000. The Toll/interleukin-1 receptor domain: a molecular switch for inflammation and host defence. Biochem. Soc. Trans. 28:557-563. [DOI] [PubMed] [Google Scholar]
- 44.Plevy, S. E., J. H. Gemberling, S. Hsu, A. J. Dorner, and S. T. Smale. 1997. Multiple control elements mediate activation of the murine and human interleukin 12 p40 promoters: evidence of functional synergy between C/EBP and Rel proteins. Mol. Cell. Biol. 17:4572-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poli, V. 1998. The role of C/EBP isoforms in the control of inflammatory and native immunity functions. J. Biol. Chem. 273:29279-29282. [DOI] [PubMed] [Google Scholar]
- 46.Poltorak, A., X. He, I. Smirnova, M. Y. Liu, C. V. Huffel, X. Du, D. Birdwell, E. Alejos, M. Silva, C. Galanos, M. Freudenberg, P. Ricciardi-Castagnoli, B. Layton, and B. Beutler. 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282:2085-2088. [DOI] [PubMed] [Google Scholar]
- 47.Qureshi, S. T., L. Larivière, G. Leveque, S. Clermont, K. J. Moore, P. Gros, and D. Malo. 1999. Endotoxin-tolerant mice have mutations in Toll-like receptor 4 (Tlr4). J. Exp. Med. 189:615-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saccani, S., S. Pantano, and G. Natoli. 2002. p38-dependent marking of inflammatory genes for increased NF-κB recruitment. Nat. Immunol. 3:69-75. [DOI] [PubMed] [Google Scholar]
- 49.Salh, B., R. Wagey, A. Marotta, J. S. Tao, and S. Pelech. 1998. Activation of phosphatidylinositol 3-kinase, protein kinase B, and p70 S6 kinases in lipopolysaccharide-stimulated Raw 264.7 cells: differential effects of rapamycin, Ly294002, and wortmannin on nitric oxide production. J. Immunol. 161:6947-6954. [PubMed] [Google Scholar]
- 50.Schindler, U., and V. R. Baichwal. 1994. Three NF-κB binding sites in the human E-selectin gene required for maximal tumor necrosis factor alpha-induced expression. Mol. Cell. Biol. 14:5820-5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schneider, D. S., K. L. Hudson, T. Y. Lin, and K. V. Anderson. 1991. Dominant and recessive mutations define functional domains of Toll, a transmembrane protein required for dorsal-ventral polarity in the Drosophila embryo. Genes Dev. 5:797-807. [DOI] [PubMed] [Google Scholar]
- 52.Shaulian, E., and M. Karin. 2002. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4:E131-E136. [DOI] [PubMed] [Google Scholar]
- 53.Shen, B., and J. L. Manley. 1998. Phosphorylation modulates direct interactions between the Toll receptor, Pelle kinase and Tube. Development 125:4719-4728. [DOI] [PubMed] [Google Scholar]
- 54.Slack, J. L., K. Schooley, T. P. Bonnert, J. L. Mitcham, E. E. Qwarnstrom, J. E. Sims, and S. K. Dower. 2000. Identification of two major sites in the type I interleukin-1 receptor cytoplasmic region responsible for coupling to pro-inflammatory signaling pathways. J. Biol. Chem. 275:4670-4678. [DOI] [PubMed] [Google Scholar]
- 55.Stefanova, I., M. L. Corcoran, E. M. Horak, L. M. Wahl, J. B. Bolen, and I. D. Horak. 1993. Lipopolysaccharide induces activation of CD14-associated protein tyrosine kinase p53/56lyn. J. Biol. Chem. 268:20725-20728. [PubMed] [Google Scholar]
- 56.Towb, P., R. L. Galindo, and S. A. Wasserman. 1998. Recruitment of Tube and Pelle to signaling sites at the surface of the Drosophila embryo. Development 125:2443-2450. [DOI] [PubMed] [Google Scholar]
- 57.Trinchieri, G. 1995. Interleukin-12: a proinflammatory cytokine with immunoregulatory functions that bridge innate resistance and antigen-specific adaptive immunity. Annu. Rev. Immunol. 13:251-276. [DOI] [PubMed] [Google Scholar]
- 58.van Furth, A. M., E. M. Verhard-Seijmonsbergen, J. A. M. Langermans, J. T. van Dissel, and R. van Furth. 1999. Anti-CD14 monoclonal antibodies inhibit the production of tumor necrosis factor alpha and interleukin-10 by human monocytes stimulated with killed and live Haemophilus influenzae or Streptococcus pneumoniae organisms. Infect. Immun. 67:3714-3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weinmann, A. S., D. M. Mitchell, S. Sanjabi, M. N. Bradley, A. Hoffmann, H. C. Liou, and S. T. Smale. 2001. Nucleosome remodeling at the IL-12 p40 promoter is a TLR-dependent, Rel-independent event. Nat. Immunol. 2:51-57. [DOI] [PubMed] [Google Scholar]
- 60.Weinmann, A. S., S. E. Plevy, and S. T. Smale. 1999. Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity 11:665-675. [DOI] [PubMed] [Google Scholar]
- 61.Xu, Y., X. Tao, B. Shen, T. Horng, R. Medzhitov, J. L. Manley, and L. Tong. 2000. Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature 408:111-115. [DOI] [PubMed] [Google Scholar]
- 62.Zhu, C., K. Gagnidze, J. H. Gemberling, and S. E. Plevy. 2001. Characterization of an activation protein-1-binding site in the murine interleukin-12 p40 promoter. Demonstration of novel functional elements by a reductionist approach. J. Biol. Chem. 276:18519-18528. [DOI] [PubMed] [Google Scholar]