Abstract
Topoisomerase I (Top I)-DNA covalent complexes represent a unique type of DNA lesion whose repair and processing remain unclear. In this study, we show that Top I-DNA covalent complexes transiently arrest RNA transcription in normal nontransformed cells. Arrest of RNA transcription is coupled to activation of proteasomal degradation of Top I and the large subunit of RNA polymerase II. Recovery of transcription occurs gradually and depends on both proteasomal degradation of Top I and functional transcription-coupled repair (TCR). These results suggest that arrest of the RNA polymerase elongation complex by the Top I-DNA covalent complex triggers a 26S proteasome-mediated signaling pathway(s) leading to degradation of both Top I and the large subunit of RNA polymerase II. We propose that proteasomal degradation of Top I and RNA polymerase II precedes repair of the exposed single-strand breaks by TCR.
Topoisomerase I (Top I)-DNA covalent complexes, more commonly referred to as TopI cleavable or cleavage complexes, represent a unique type of DNA lesion (12, 21, 23). The antitumor drug camptothecin (CPT) is the first agent demonstrated to induce Top I-DNA covalent complexes, which is believed to be exclusively responsible for the antitumor activity of CPT (5). In addition to Top I-directed antitumor drugs, many DNA lesions (e.g., UV adducts, 1-β-d-arabinofuranosylcytosine-substituted DNA, benzo[a]pyrene-DNA adducts, and 8-oxo-guanine-modified DNA) are now known to trap Top I-DNA covalent complexes to form secondary DNA lesions (16, 22, 32). Despite the increasing importance of Top I-DNA covalent complexes, their repair mechanism(s) remains largely unclear.
Top I-DNA covalent complexes are reversible (21, 23). They represent the covalent intermediates of the cleavage-religation reaction of Top I (12). In the complex, Top I is covalently linked to the 3′-phosphoryl end of the transient single-strand DNA break via a tyrosyl phosphate bond (reviewed in reference 17). CPT, the prototypic Top I poison, is known to block the religation reaction of Top I, resulting in accumulation of Top I-DNA covalent complexes (12, 23). Because of the reversible nature of Top I-DNA covalent complexes, lethality occurs only upon their irreversible conversion to DNA strand breaks (17). Studies using CPT have demonstrated that moving DNA replication forks can process Top I-DNA covalent complexes into potentially lethal DNA lesions, which explains the S-phase-specific cytotoxicity of CPT (6). In addition to lethality, the collision between the DNA replication fork and the Top I-DNA covalent complex is responsible for G2 cell cycle arrest, activation of NF-κB and ATR (for “AT mutated and Rad3 related”), stabilization of p53, increased accumulation of c-fos and c-jun mRNAs, and phosphorylation of Chk1 and RPA (reviewed in reference 16). The induction of these DNA damage responses by CPT is consistent with the notion that Top I-DNA covalent complexes are converted into DNA damage by their collisions with the replication forks. Indeed, studies with a cell-free simian virus 40 DNA replication system have suggested that collisions between the replication forks and Top I-DNA covalent complexes result in irreversible arrest of the fork, the formation of double-strand DNA breaks, and the conversion of reversible Top I-DNA complexes into Top I-linked DNA breaks (6, 13, 29).
Repair of Top I-DNA covalent complexes is conceptually challenging because of the reversibility of the complexes and the bulkiness of the protein-DNA adducts. Recently, CPT has been demonstrated to specifically induce degradation of Top I via a ubiquitin-26S proteasome pathway (8, 9). It has been suggested that degradation of Top I in the Top I-DNA covalent complex represents a potential repair mechanism for Top I-DNA covalent complexes (9). In the present study, we show that Top I-DNA covalent complexes arrest transcription and trigger transcription-dependent degradation of both Top I and the large subunit of RNA polymerase II (RNA Pol II0). Transcription recovery is dependent on both degradation of Top I and functional transcription-coupled repair (TCR). These results are consistent with a model in which arrest of the elongating RNA polymerase complexes by Top I-DNA covalent complexes triggers 26S proteasome-mediated degradation of Top I and subsequent repair of the exposed single-strand breaks.
MATERIALS AND METHODS
Cells.
Monkey kidney fibroblast BSC cells and Chinese hamster lung V79 cells were obtained from the American Type Culture Collection (Manassas, Va.). The human breast cancer cell line ZR-75-1 was kindly provided by K.-V. Chin (The Cancer Institute of New Jersey). The human lymphoblast cell line RPMI 8402 and its CPT-resistant variant CPT-K5 were obtained from Toshiwo Andoh (Soka University, Tokyo, Japan). The human prostate cancer cell line DU145 and its CPT-resistant variant DU145/RC and the human ovarian cancer cell line 2774 and its CPT-resistant variant 2774/RC were kindly provided by Beppino Giovanella (Stehlin Foundation for Cancer Research, Houston, Tex.). The murine leukemia cell line P388 and its Top I-deficient variant P388/CPT45 were kindly provided by M. R. Mattern (Glaxo-SmithKline Pharmaceuticals, King of Prussia, Pa.). The lymphoblast cell line GM01953C and the Cockayne's syndrome group B (CSB) lymphoblast cell line GM01712B were obtained from the NIGMS Human Genetic Mutant Cell Repository, Coriell Institute for Medical Research, Camden, N.J. Both cell lines were transformed with Epstein-Barr virus.
All cells were cultured in RPMI 1640 medium except for V79 and BSC cells, which were grown in Dulbecco's modified Eagle medium. All media were supplemented with 10% fetal bovine serum, l-glutamine (2 mM), penicillin (100 U/ml), and streptomycin (100 μg/ml). All cells were cultured in a 37°C incubator with 5% CO2.
Immunoblotting of Top I.
Cells (106/sample) were treated with CPT (25 μM, 1% dimethyl sulfoxide [DMSO]) for various periods at 37°C. Cells were then lysed either directly (for detection of Top I covalent complexes by the band depletion assay) or incubated in CPT-free fresh medium for another 30 min prior to lysis (for reversal of Top I covalent complexes). Lysis was carried out with 0.2 N NaOH containing 2 mM EDTA as described previously (8, 9). Cell lysates were then neutralized with 1/10 volume of a solution containing 10% NP-40, 1 M Tris (pH 7.4), 0.1 M MgCl2, 0.1 M CaCl2, 10 mM dithiothreitol, 1 mM EGTA, and a 100-μg/ml concentration each of leupeptin, pepstatin, and aprotinin, followed by the addition of another 1/10 volume of 2 N HCl. Neutralized cell lysates were incubated with Staphylococcus aureus nuclease S7 (60 U/reaction) for 20 min on ice. Reactions were terminated by the addition of SDS-PAGE sample buffer. Immunoblotting analysis of cell lysates was carried out with Top I antiserum from scleroderma patients as described previously by the enhanced chemiluminescence (ECL) Western procedure (8, 9).
Immunoblotting of RNA Pol II0.
Cells (106/sample) were treated with CPT (25 μM, 1% DMSO) for various periods at 37°C. After treatment, cells were lysed directly with 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. Cell lysates were then sonicated and analyzed with a 6% SDS-polyacrylamide gel. After transfer of proteins to nitrocellulose membranes, RNA Pol IIa and Pol II0 were detected by immunoblotting with ARNA-3 antibodies (Research Diagnostics Inc., Flanders, N.J.) directed against a non-C-terminal-domain, unphosphorylated epitope (amino acids 806 to 820) by the ECL Western procedure (Pierce).
Uridine incorporation.
Cells (5 × 105/sample) were grown in 35- by 10-mm tissue culture plates and treated with 25 μM CPT for various periods, followed by 15 min of pulse-labeling with 1 μCi of [5,6-3H]uridine (47 Ci/mmol; ICN)/ml. Following uridine labeling, cells were lysed with a solution containing 4 M isothioguanidine, 0.5% Sarkosyl, 2 mM sodium citrate, and 0.1 M β-mercaptoethanol. Samples were directly spotted onto DE81 papers. Filters were washed three times (30 min each) with 0.3 M ammonium formate followed by ethanol wash (10 min). Filters were then dried and counted by using a Beckman liquid scintillation counter.
Analysis of TopI protein levels in normal and tumor tissues from mice treated with topotecan.
MDA-MB-435 human breast cancer cells (1.6 × 106 cells/100 μl) were inoculated into the flanks of female Ncrnu/nu mice. On the 12th day after inoculation, mice were given an intraperitoneal bolus injection of topotecan in 1% DMSO (either 200 or 20 mg/kg of body weight). Control mice were injected with 1% DMSO only. Animals were sacrificed after 0, 4, and 8 h of drug administration. Different normal tissues, peripheral blood cells, and the xenografted tumor tissue were taken and snap-frozen by placing them in liquid nitrogen. For detecting TopI levels, frozen tissues were weighed, cut into small pieces, and placed in test tubes containing SDS sample buffer. Tissue samples were then sonicated with a Tissue-Tearor (Biospec Products, Inc.). Sonicated samples were immediately boiled for 10 min at 100°C and centrifuged at 13,000 × g for 10 min. Cleared supernatants containing SDS-solubilized protein extracts of different tissues were then subjected to SDS-PAGE analysis. Immunoblotting analysis of Top I was carried out with Top I antiserum from scleroderma patients by following the Western procedure described above (9).
RESULTS
Top I-DNA covalent complexes are degraded via 26S proteasome.
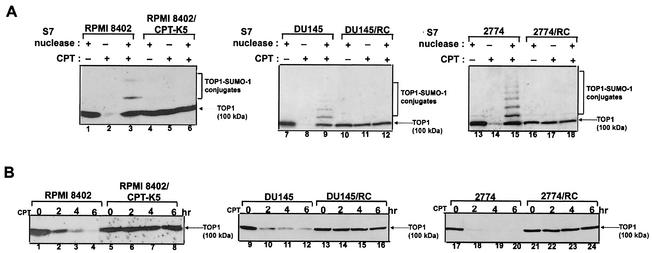
Previous studies have demonstrated that CPT induces 26S proteasome-mediated degradation of Top I (8, 9). In order to demonstrate that degradation of Top I is due to the formation of Top I-DNA covalent complexes, three CPT-resistant cell lines (the human lymphoblast line RPMI 8402/CPT-K5, the prostate cancer line DU145/RC, and the ovarian cancer line 2774/RC cells) were employed in the present study. As shown in Fig. 1A (compare lanes 1, 7, and 13 with lanes 2, 8, and 14, respectively), CPT induced the formation of Top I-DNA covalent complexes in their respective parental wild-type cells (RPMI 8402, DU145, and 2774 cells), as evidenced by depletion of the 100-kDa Top I band. More importantly, S7 nuclease treatment of the cell lysates released Top I from Top I-DNA covalent complexes, as evidenced by the reappearance of the 100-kDa Top I band (Fig. 1A, lanes 3, 9, and 15). The appearance of the SUMO-1-Top I conjugates was again indicative of the formation of Top I-DNA covalent complexes in these wild-type cells (Fig. 1A, lanes 3, 9, and 15) (20). By contrast, no Top I-DNA covalent complexes were detectable (no depletion of the 100-kDa Top I band) in the three CPT-resistant cells (Fig. 1A, compare lanes 4, 10, and 16 with lanes 5, 11, and 17, respectively). The absence of SUMO-1-Top I conjugates in these resistant cells (Fig. 1A, lanes 6, 12, and 18) was expected, since these mutant cell lines are defective in the formation of Top I-DNA covalent complexes. As shown in Fig. 1B, CPT induced time-dependent degradation of Top I in CPT-sensitive RPMI 8402, DU145, and 2774 cells. By contrast, CPT was completely ineffective in inducing Top I degradation in the corresponding CPT-resistant variant cells (RPMI/CPT-K5, DU145/RC, and 2774/RC cells in Fig. 1B). The correlation between Top I degradation and the formation of Top I-DNA covalent complexes is consistent with the notion that Top I-DNA covalent complexes are degraded by the 26S proteasome.
FIG. 1.
Degradation of Top I is dependent on the formation of Top I-DNA covalent complexes. (A) CPT-resistant cells are defective in the formation of Top I-DNA covalent complexes. Three pairs of CPT-resistant cell lines (RPMI 8402 and RPMI 8402/CPTK5, DU145 and DU145/RC, and 2774 and 2774/RC) were treated with CPT (25 μM) for 15 min. Cells were then immediately lysed by the alkaline lysis procedure. Half of the neutralized lysates were treated with Staphylococcus nuclease S7, and the other half were directly mixed with SDS sample buffer for loading onto a 6% SDS-polyacrylamide gel. Immunoblotting was performed with human Top I antibodies derived from scleroderma patients. (B). CPT induces degradation of Top I in wild-type but not CPT-resistant cells. The cell lines used for panel A were treated with CPT (25 μM) for 0, 2, 4, and 6 h. After CPT treatment, cells were incubated in CPT-free medium for another 30 min to reverse Top I covalent complexes prior to lysis by the alkaline lysis procedure. Neutralized lysates were further treated with Staphylococcus nuclease S7 to release trapped Top I from Top I-DNA covalent complexes. Immunoblotting was performed as described for panel A.
Transcription inhibitors block degradation of TopI-DNA covalent complexes.
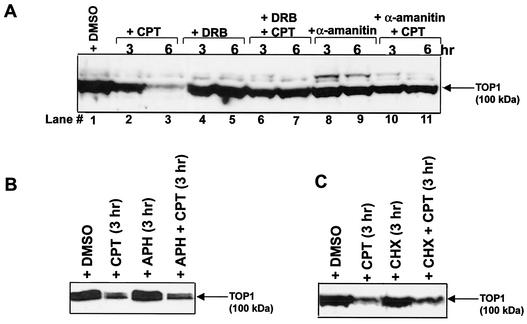
At present, it is unclear what the trigger for the degradation of Top I-DNA covalent complexes is. However, Top I-DNA covalent complexes are known to arrest both replication forks and elongating RNA polymerase complexes (6, 13, 36, 38). To test whether DNA replication or RNA transcription may be involved in Top I degradation, various inhibitors were employed. As shown in Fig. 2A, the Top I level was reduced by 90% in 6 h (lane 3) during continuous treatment with CPT in V79 cells. The transcription inhibitors 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) (150 μM) (Fig. 2A, lanes 6 and 7) and α-amanitin (5 μg/ml) (Fig. 2A, lanes 10 and 11) were found to significantly block CPT-induced degradation of Top I in V79 cells. DRB and α-amanitin did not affect the amounts of Top I-DNA covalent complexes in CPT-treated V79 cells (data not shown).
FIG. 2.
Transcription inhibitors block CPT-induced degradation of Top I. (A) The transcription inhibitors DRB and α-amanitin block CPT-induced degradation of Top I in V79 cells. V79 cells were pretreated with either 150 μM DRB (for 1 h) or 5 μg of α-amanitin per ml (for 16 h) followed by cotreatment with CPT (25 μM) for 3 or 6 h as indicated. Cells were then replenished with fresh CPT-free medium containing either DRB or α-amanitin and incubated at 37°C for another 30 min to reverse Top I covalent complexes prior to lysis by the alkaline lysis procedure. Immunoblotting with anti-human Top I scleroderma antibodies was then performed. Lane 1 contains a 1% DMSO solvent control. (B) The replication inhibitor aphidicolin does not affect CPT-induced degradation of Top I. V79 cells were pretreated with aphidicolin (30 μg/ml in 1% DMSO) for 1 h followed by treatment with CPT (25 μM) in the presence or absence of aphidicolin (APH; 30 μg/ml) for 3 h. Cells were then replenished with aphidicolin- and CPT-free fresh medium and incubated at 37°C for another 30 min to reverse Top I covalent complexes. Cells were lysed by the alkaline lysis procedure. Cell lysates were treated with S7 nuclease and immunoblotted with anti-human Top I antibodies as for panel A. (C) The protein synthesis inhibitor CHX does not affect CPT-induced degradation of Top I. CHX (10 μg/ml) was used to assess the role of protein synthesis in Top I degradation. The procedure was identical to that for panel B except that CHX was used instead of aphidicolin.
The effect of DNA replication on degradation of Top I-DNA covalent complexes was also examined. Aphidicolin (30 μg/ml) was shown to have no effect on CPT-induced Top I degradation (Fig. 2B). The effect of protein synthesis on Top I degradation was evaluated by using the protein synthesis inhibitor cycloheximide (CHX) (10 μg/ml). As shown in Fig. 2C, CHX had little effect on CPT-induced degradation of Top I. The lack of a significant effect of protein synthesis on Top I degradation suggests that the requirement for RNA transcription is not due to the need for new protein synthesis.
Recovery of transcription during prolonged CPT treatment.
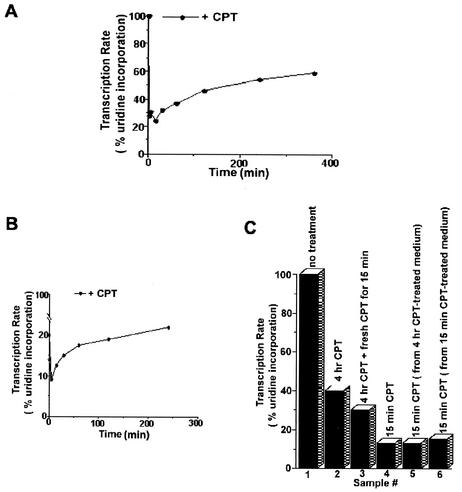
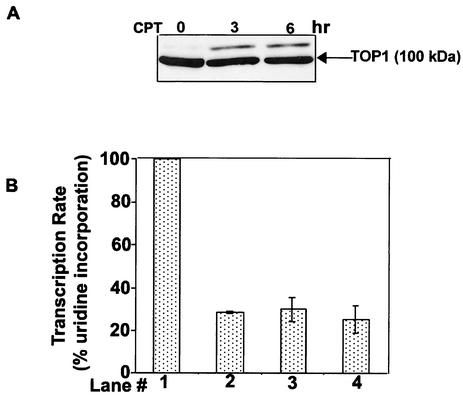
CPT causes transcription arrest due to blockage of the RNA polymerase elongation complexes by Top I cleavable complexes (3, 13, 36, 38). As expected, RNA transcription was rapidly arrested upon addition of CPT (25 μM) to V79 cells (Fig. 3A). Within 10 min, transcription was reduced to 20%, as monitored by [3H]uridine incorporation (Fig. 3A). However, recovery of transcription as monitored by pulse-labeling with [3H]uridine was evident within 15 min and continued gradually in the next 4 h in the presence of CPT (Fig. 3A). After 4 h of CPT treatment, transcription recovered up to 48% of the untreated control. This transcription recovery phenomenon is not unique to V79 cells, as we observed similar recovery in monkey kidney fibroblast BSC cells treated with CPT (Fig. 3B).
FIG. 3.
CPT induces transient transcription inhibition. (A) Transcription inhibition and recovery in V79 cells treated with CPT. V79 cells were treated with CPT (25 μM) in six-well tissue culture plates. Transcription rates were measured by 15 min of pulse-labeling with [3H]uridine at different times following drug treatment. The transcription rate of untreated cells was taken as 100%. (B) Transcription recovery in BSC cells treated with CPT. BSC cells were treated with 50 μM CPT in 24-well tissue culture plates. Transcription rates were measured at different times following drug treatment as for panel A. (C). Recovery of transcription is not due to CPT inactivation. Rates of transcription were measured by 30 min of pulse-labeling with [3H]uridine. Sample 1, control (no drug treatment); sample 2, BSC cells treated with CPT (50 μM) for 4 h; sample 3, fresh CPT (50 μM) was added to BSC cells that had been treated with CPT (50 μM) for 4 h (sample 2). Cells were incubated for another 15 min prior to lysis. Sample 4, BSC cells treated with CPT (50 μM) for 15 min; sample 5, incubated culture medium (50 μM CPT for 4 h) from sample 2 was transferred to fresh untreated cells for 15 min; sample 6, the incubated culture medium (50 μM CPT for 15 min) from sample 4 was transferred to fresh untreated cells for 15 min.
Transcription recovery following CPT treatment could be due to a number of possibilities. First, we tested whether CPT was inactivated in the culture medium of treated cells. In this experiment, 4 h of continuous treatment with CPT caused the transcription rate to recover from 14% (15 min of CPT treatment) (Fig. 3C, sample 4) to 40% (4 h of CPT treatment) (Fig. 3C, sample 2). When CPT-containing medium from treated cells (4 h) (Fig. 3C, sample 2) were transferred to untreated fresh BSC cells for 15 min, transcription was again reduced to 14% (sample 5), suggesting that CPT remained active in the medium during 4 h of incubation. We then tested whether transcription in cells following 4 h of incubation with CPT became resistant to further CPT treatment. As shown in Fig. 3C (sample 3), when an additional aliquot of CPT (a combined final concentration of 100 μM) was added to cells which had already had CPT (50 μM) in the medium for 4 h, the transcription rate (measured after another 15 min of treatment) was reduced only slightly, from 40 to 30% of that of the untreated control. Considering the increased CPT concentration (from 50 to 100 μM), the BSC cells following 4 h of treatment with CPT could be considered to be in a tolerant state which is much less sensitive to further CPT-mediated transcription inhibition. Without tolerance, the rate should have been reduced to less than 14%. These experiments suggested that transcription recovery in cells treated with CPT was due not to inactivation of CPT but to development of a tolerant state in CPT-treated cells.
Recovery of transcription depends on the degradation of Top I-DNA covalent complexes and functional TCR.
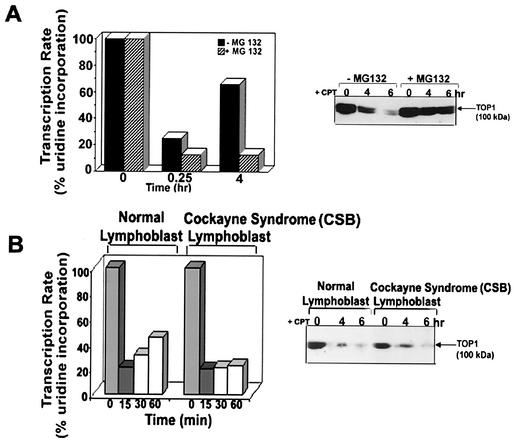
Transcription recovery during prolonged CPT treatment could be due to the 26S proteasome-mediated degradation of Top I. Previous studies have shown that the proteasome inhibitors MG132 and lactacystin can effectively block Top I down-regulation in V79 cells (9). To test this possibility, the proteasome inhibitor MG132 was used to inhibit the 26S proteasome in V79 cells. As shown in Fig. 4A, MG132 (1 μM) blocked transcription recovery during continuous CPT treatment in V79 cells. MG132 by itself had no effect on transcription (Fig. 4A). As expected, MG132 (1 μM) was shown to effectively block CPT-induced degradation of Top I in V79 cells (Fig. 4A, right panel). This result suggests that degradation of Top I by 26S proteasome is necessary for transcription recovery.
FIG. 4.
Transcription recovery requires Top I degradation and functional TCR. (A) Transcription recovery is blocked by the 26S proteasome inhibitor MG132. (Left) V79 cells were pretreated with MG132 for 1 h, followed by CPT treatment for various periods. The transcription rate was measured (by 15 min of pulse-labeling with [3H]uridine) at different times following CPT treatment. (Right) CPT-induced degradation of Top I is blocked by MG132. V79 cells were pretreated with MG132 (1 μM) for 1 h, followed by treatment with CPT (25 μM) for 4 and 6 h. Cells were then incubated in the absence of CPT but in the presence of MG132 for 30 min at 37°C to reverse the cleavable complexes. Cells were lysed by the alkaline lysis procedure. Cell lysates were neutralized, treated with S7 nuclease, and analyzed by immunoblotting. Immunodetection of Top I was done by the ECL Western procedure as described for Fig. 1A. (B) Transcription recovery in CSB cells treated with CPT. (Left) Wild-type lymphoblast GM01953C and CSB lymphoblast GM01712B cells were treated with CPT (25 μM). Transcription rates were measured at different times by 15 min of pulse-labeling with [3H]uridine following CPT treatment. (Right) Degradation of Top I-DNA covalent complexes in wild-type and CSA cells. Normal GM01953C cells and CSB GM01712B cells were treated with 25 μM CPT for various times. Reversal of cleavable complexes and immunodetection of Top I were carried out as described for Fig. 1A.
Previous studies have suggested that TCR is involved in repair of Top I-DNA covalent complexes (19, 30). We therefore tested whether TCR is necessary for transcription recovery in CPT-treated cells. As shown in Fig. 4B, transcription recovery occurred in normal lymphoblasts but not in CSB lymphoblasts treated with CPT. Importantly, Top I was degraded in both normal and CSB lymphoblasts (Fig. 4B, right panel). These results suggest that transcription recovery in CPT-treated cells depends on not only proteasomal degradation of Top I but also functional TCR. We have also found that RNA Pol II was degraded in both wild-type and CSB cells upon treatment with CPT (data not shown). Consequently, degradation of RNA Pol II is not causally linked to TCR repair of Top I-mediated DNA lesion, consistent with a recent report (35).
Most tumor cells are defective in Top I degradation and unable to recover from transcription arrest.
Many tumor cells are hypersensitive to CPT, and such cells are defective in CPT-induced Top I degradation (8). To test whether transcription recovery is also defective in these tumor cells, the rate of transcription was measured in the breast cancer cell line ZR-75-1, which is known to be defective in CPT-induced degradation of Top I (Fig. 5A) (8). As shown in Fig. 5B, no significant recovery of CPT-arrested RNA transcription was observed even after 4 h. In addition to ZR-75-1 cells, a number of other tumor cell lines, including human breast cancer cells (BT474, SKBR3, and BT20) and human colorectal cancer cells (HCT116), were also examined for both Top I degradation and transcription recovery. Except BT474 cells, all other tumor cells were found to be defective in both Top I degradation and transcription recovery (data not shown). BT474 cells, which are known to be highly resistant to CPT (7, 8), are proficient in both Top I degradation and transcription recovery.
FIG. 5.
Transcription recovery does not occur in breast cancer ZR-75-1 cells. (A) CPT-induced Top I degradation is defective in the human breast cancer cell line ZR-75-1. ZR-75-1 cells (106/sample) were treated with CPT (25 μM) for the indicated times. Cells were then replenished with CPT-free fresh medium and incubated at 37°C for another 30 min to reverse the Top I cleavable complexes. Upon removal of the media from plates, cells were immediately lysed by the alkaline lysis procedure, followed by neutralization and S7 nuclease treatment. Samples were analyzed by SDS-6% PAGE and immunoblotted with Top I antiserum from scleroderma patients by the ECL Western procedure. (B) Transcription recovery does not occur in human breast cancer ZR-75-1 cells. ZR-75-1 cells were treated with CPT (25 μM) in six-well tissue culture plates. The transcription rate was measured by 15 min of pulse-labeling with [3H]uridine after 15 min (lane 2), 2 h (lane 3), and 4 h (lane 4) of CPT treatment.
Top I-DNA covalent complexes trigger phosphorylation and proteasomal degradation of RNA Pol II0.
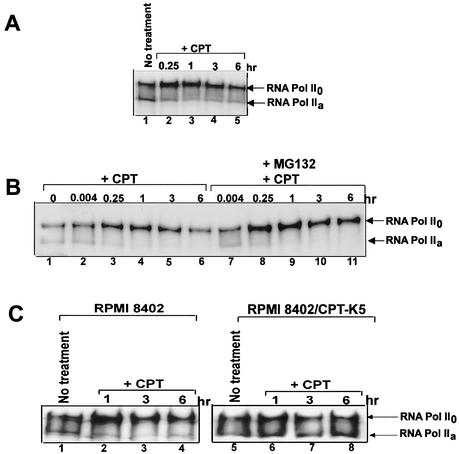
Recent studies have suggested that arrest of the elongating RNA Pol II by DNA lesion (e.g., UV-induced DNA adducts) results in ubiquitin-26S proteasome-dependent degradation of phosphorylated RNA Pol II0 (4, 26). We have thus tested whether Top I-DNA covalent complexes, which are known to arrest RNA polymerase elongating complexes, can trigger proteasomal degradation of RNA Pol II0. Within 15 min of CPT treatment in V79 cells, RNA Pol IIa was converted to its phosphorylated form (RNA Pol II0) (Fig. 6A, compare lanes 1 and 2). Prolonged CPT treatment in V79 cells resulted in gradual degradation of RNA Pol II0 (Fig. 6A, compare lanes 2 to 5).
FIG. 6.
CPT induces proteasomal degradation of RNA Pol II0. (A) CPT induces degradation of RNA Pol II0 in V79 cells. V79 (106) cells were treated with CPT (25 μM) for the indicated times. Cells were immediately lysed with 2× SDS-PAGE sample buffer. Immunoblotting was carried out with ARNA-3 antibodies against RNA Pol II0. (B) CPT induces proteasome-dependent degradation of RNA Pol II0 in human breast cancer ZR-75-1 cells. ZR-75-1 cells were pretreated with the 26S proteasome inhibitor MG132 (1 μg/ml) for 1 h followed by cotreatment with CPT (25 μM) for different periods up to 6 h. Cells were immediately lysed with 2× SDS-PAGE sample buffer. Immunoblotting was carried out with ARNA-3 antibodies to detect RNA Pol II0 as described for panel A. (C). Degradation of RNA Pol II0 depends on the formation of Top I-DNA covalent complexes. Cells of the RPMI 8402 line and its CPT-resistant variant CPT-K5 were treated with CPT (25 μM) for the indicated times. Cells were then immediately lysed with 2× SDS-PAGE sample buffer. Immunoblotting was carried out with ARNA-3 to detect RNA Pol II0.
Accumulation followed by degradation of RNA Pol II0 was also observed in ZR-75-1 cells (Fig. 6B). In this case, we showed that degradation but not accumulation of RNA Pol II0 was inhibited by the proteasome inhibitor MG132 (Fig. 6B). To ascertain that the CPT-induced accumulation and degradation of RNA Pol II0 are indeed due to its arrest by Top I-DNA covalent complexes, RPMI 8402/CPT-K5 cells, which are defective in CPT-induced formation of Top I-DNA covalent complexes (1), were used. As shown in Fig. 6C, both phosphorylation and degradation of RNA Pol II0 were observed in wild-type RPMI 8402 cells but not in CPT-resistant RPMI 8402/CPT-K5 cells, suggesting that Top I-DNA covalent complexes are responsible for both phosphorylation and degradation of RNA Pol II0.
Topotecan induces degradation of Top I in normal but not tumor tissues: studies with nude mice carrying xenografted human breast cancer cells.
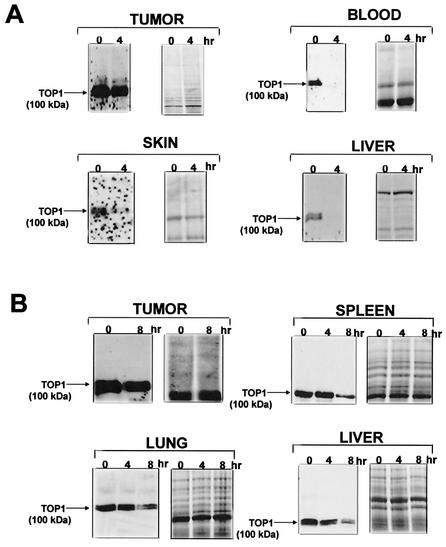
Previous studies demonstrated that transformed and tumor cells were less efficient in CPT-induced degradation of Top I than normal nontransformed cells (8). To test whether Top I in normal cells is more efficiently degraded in an animal model, different doses of topotecan (a CPT derivative in clinical use) were injected into nude mice carrying human breast cancer MDA-MB-435 cells. The tumor tissue as well as various normal tissues (liver, skin, spleen, lung, and peripheral blood cells) were then examined for their Top I protein levels. As shown in Fig. 7A, Top I completely disappeared within 4 h of topotecan treatment (200 mg/kg of body weight) in liver, skin, and peripheral blood cells. On the other hand, the Top I level was reduced very slightly (5% reduction) in the xenografted tumor tissue after 4 h of topotecan treatment. The lack of Top I degradation in tumor tissue was not due to reduced topotecan distribution, since tumor and liver tissues had about the same topotecan concentration (24.44 versus 22.42 μg/g of tissue, measured by a high-pressure liquid chromatography method) (27). A lower concentration of topotecan (20 mg/kg) was also used to evaluate the rate of Top I degradation in various tissues. As shown in Fig. 7B, Top I levels remained unchanged in extracts of normal spleen, lung, and liver after 4 h of topotecan (20 mg/kg) treatment. However, the Top I level was reduced to about 10% in extracts of normal spleen, lung, and liver after 8 h of topotecan treatment. Again, the Top I level remained unchanged in xenografted tumor tissue even after 8 h of topotecan treatment. Together, these results suggest that CPT-induced down-regulation of Top I occurs efficiently in normal cells but is impaired in tumor cells.
FIG. 7.
Topotecan induces degradation of Top I in normal but not tumor tissues. (A) Analysis of Top I levels in normal and tumor tissues following administration of topotecan (200 mg/kg of body weight). Nude mice carrying xenografted breast cancer MDA-MB-435 cells were treated with topotecan (200 mg/kg) for 4 h. SDS extracts of tumor, skin, liver, and peripheral blood were subjected to SDS-6% PAGE. Western blotting was performed with human Top I antibodies derived from scleroderma patients (left panels). The same samples were also loaded on another gel, which was then stained with Coomassie blue to ascertain equal protein loading (right panels). (B) Analysis of Top I levels in normal and tumor tissues following administration of topotecan (20 mg/kg). The experiment was performed as for panel A, except that a lower concentration of topotecan (20 mg/kg) was used and the sampling time was increased to 8 h.
DISCUSSION
Studies using CPT have suggested that Top I-DNA covalent complexes arrest elongating RNA polymerase complexes without a significant effect on transcription initiation in mammalian cells (3, 6, 15, 18, 36-38). Arrest of RNA transcription by Top I-DNA covalent complexes has also been studied in vitro by using purified enzymes. These studies have led to the proposal of a transcription collision model in which collisions between elongating RNA polymerases and Top I-DNA covalent complexes on the template strands results in transcription arrest and the conversion of reversible Top I-DNA covalent complexes into Top I-linked DNA strand breaks (36).
In the present study, we show that Top I-DNA covalent complexes undergo transcription-dependent degradation via a 26S proteasome pathway. We also show that the dependence on RNA transcription is unrelated to synthesis of new proteins, since the protein synthesis inhibitor CHX has no effect on Top I degradation. The transcription collision model can readily explain the observed transcription dependence. According to this model, the collision between the RNA polymerase complex and the Top I-DNA covalent complex triggers 26S proteasome-mediated degradation of Top I. The lack of an effect of aphidicolin on the overall degradation of Top I can be explained by two possibilities: one is that collisions between Top I-DNA covalent complexes with replication forks do not trigger proteasome-mediated degradation of Top I, and the second is that only a small number of Top I-DNA covalent complexes undergo collisions with the replication forks. The majority of the Top I-DNA covalent complexes are known to be located within the transcribed regions and can therefore undergo collisions with the transcription elongation complexes.
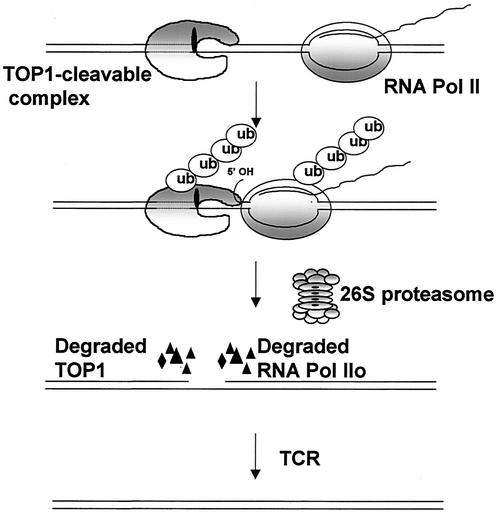
We also show that Top I-DNA covalent complexes induce phosphorylation and 26S proteasome-mediated degradation of RNA Pol II0. Phosphorylation and proteasomal degradation of the large subunit of RNA polymerase have been observed following DNA damage (2, 4, 26). It has been suggested that blockage of the RNA polymerase elongation complex by DNA adducts results in phosphorylation and subsequent proteasomal degradation of RNA Pol II0 (4, 26). Our results are thus consistent with the notion that Top I-DNA covalent complexes arrest the RNA polymerase elongation complexes and thereby trigger 26S proteasome-mediated degradation of both Top I and RNA Pol II0 (see Fig. 8 for a model).
FIG. 8.
Transcription collision model for proteasomal degradation of Top I-DNA covalent complexes. In this model, the collision between the Top I-DNA covalent complex and the elongating RNA polymerase complex triggers proteasomal degradation of both Top I and RNA Pol II0. Degradation of Top I-DNA covalent complexes results in exposure of single-strand breaks. Degradation of RNA Pol II0 may signal repair (4, 26). Repair of the exposed single-strand breaks presumably occurs via TCR. ub, ubiquitin.
Transcription recovery studies have demonstrated that both Top I degradation and functional TCR are necessary for transcription recovery. Top I degradation was shown to occur normally in CSB cells, suggesting that TCR may occur downstream from Top I degradation. The simplest explanation for these results is shown in Fig. 8. In this model, collision between the Top I-DNA covalent complex and the elongating RNA polymerase complex triggers Top I degradation. Top I degradation results in exposure of single-strand breaks. Repair of the exposed single-strand breaks may then occur through TCR (30). The tyrosyl-DNA phosphodiesterase Tdp1, which removes tyrosine from the 3′ end of DNA, has been implicated in the repair of Top I-DNA covalent complexes (24, 33, 34). In our model, Tdp1 could potentially be involved in hydrolysis of the tyrosyl phosphate bond after proteasomal degradation of Top I but prior to TCR.
Top I has been firmly established to be a new molecular target for anticancer drugs (17). However, the molecular basis for the antitumor specificity of Top I-directed anticancer drugs is still unclear. Previous studies have suggested that S-phase-specific cytotoxicity may be important for the antitumor activity of CPT (16). The elevated expression of Top I in many tumor tissues has also been suggested to contribute to antitumor specificity (14). The present study may suggest yet another explanation for the antitumor specificity of Top I-directed anticancer drugs (31). Due to the blockage of the RNA polymerase elongation complexes by Top I-DNA covalent complexes, Top I-directed anticancer drugs are expected to inhibit gene expression and cause severe toxicity. However, the development of a tolerant state due to Top I degradation in normal cells, as demonstrated here, is expected to allow RNA transcription to resume in normal tissues. Consequently, the severe toxic side effect due to transcription inhibition in normal cells may not occur. It is interesting that patients receiving topotecan therapy also exhibit reduced Top I levels in normal peripheral blood cells but not in leukemic cells (11, 28). Thus, Top I degradation in normal tissues can be viewed as a mechanism to evade the toxic effect of Top I-directed anticancer drugs.
Studies with a panel of tumor and normal cells have shown that defective Top I degradation is correlated with increased sensitivity to Top I-directed anticancer drugs (8). Inhibition of 26S proteasome has also been shown to result in increased sensitivity to Top I-directed drugs (8). It appears that Top I degradation impacts cell survival. The present study suggests that Top I degradation precedes TCR repair. Consequently, defective Top I degradation is expected to block repair of Top I-mediated DNA damage and hence increase drug sensitivity. It seems possible that defective Top I degradation in tumor cells may also contribute to the antitumor specificity of Top I-directed anticancer drugs.
The molecular basis for the Top I degradation defect in tumors is unclear. The fact that Top II degradation occurs efficiently in tumor cells that are defective in Top I degradation suggests that the defect is Top I specific and is not due to a general defect in the 26S proteasome activity (19). Recent studies have suggested that TOPORS, a ring finger Top I-interacting protein, is a potential ubiquitin E3 for Top I (10, 25). It remains to be determined whether Top I-specific E2/E3 is defective during tumorigenesis.
Acknowledgments
This work was supported by NIH grants CA 39662 (to L.F.L.), CA 77433 (to L.F.L.), and GM 59170 (to E.H.R.).
We are grateful to Tao-Shih Hsieh for critical reading of the manuscript.
REFERENCES
- 1.Andoh, T., K. Ishii, Y. Suzuki, Y. Ikegami, Y. Kusunoki, Y. Takemoto, and K. Okada. 1987. Characterization of a mammalian mutant with a camptothecin-resistant DNA topoisomerase I. Proc. Natl. Acad. Sci. USA 84:5565-5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baskaran, R., G. G. Chiang, T. Mysliwiec, G. D. Kruh, and J. Y. Wang. 1997. Tyrosine phosphorylation of RNA polymerase II carboxyl-terminal domain by the Abl-related gene product. J. Biol. Chem. 272:18905-18909. [DOI] [PubMed] [Google Scholar]
- 3.Bendixen, C., B. Thomsen, J. Alsner, and O. Westergaard. 1990. Camptothecin-stabilized topoisomerase I-DNA adducts cause premature termination of transcription. Biochemistry 29:5613-5619. [DOI] [PubMed] [Google Scholar]
- 4.Bregman, D. B., R. Halaban, A. J. van Gool, K. A. Henning, E. C. Friedberg, and S. L. Warren. 1996. UV-induced ubiquitination of RNA polymerase II: a novel modification deficient in Cockayne syndrome cells. Proc. Natl. Acad. Sci. USA 93:11586-11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, A. Y., and L. F. Liu. 1994. DNA topoisomerases: essential enzymes and lethal targets. Annu. Rev. Pharmacol. Toxicol. 34:191-218. [DOI] [PubMed] [Google Scholar]
- 6.D'Arpa, P., C. Beardmore, and L. F. Liu. 1990. Involvement of nucleic acid synthesis in cell killing mechanisms of topoisomerase poisons. Cancer Res. 50:6919-6924. [PubMed] [Google Scholar]
- 7.Davis, P. L., W. L. Shaiu, G. L. Scott, J. D. Iglehart, T. S. Hsieh, and J. R. Marks. 1998. Complex response of breast epithelial cell lines to topoisomerase inhibitors. Anticancer Res. 18:2919-2932. [PubMed] [Google Scholar]
- 8.Desai, S. D., T. K. Li, A. Rodriguez-Bauman, E. H. Rubin, and L. F. Liu. 2001. Ubiquitin/26S proteasome-mediated degradation of topoisomerase I as a resistance mechanism to camptothecin in tumor cells. Cancer Res. 61:5926-5932. [PubMed] [Google Scholar]
- 9.Desai, S. D., L. F. Liu, D. Vazquez-Abad, and P. D'Arpa. 1997. Ubiquitin-dependent destruction of topoisomerase I is stimulated by the antitumor drug camptothecin. J. Biol. Chem. 272:24159-24164. [DOI] [PubMed] [Google Scholar]
- 10.Haluska, P., Jr., A. Saleem, Z. Rasheed, F. Ahmed, E. W. Su, L. F. Liu, and E. H. Rubin. 1999. Interaction between human topoisomerase I and a novel RING finger/arginine-serine protein. Nucleic Acids Res. 27:2538-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hochster, H., L. Liebes, J. Speyer, J. Sorich, B. Taubes, R. Oratz, J. Wernz, A. Chachoua, R. H. Blum, and A. Zeleniuch-Jacquotte. 1997. Effect of prolonged topotecan infusion on topoisomerase 1 levels: a phase I and pharmacodynamic study. Clin. Cancer Res. 3:1245-1252. [PubMed] [Google Scholar]
- 12.Hsiang, Y. H., R. Hertzberg, S. Hecht, and L. F. Liu. 1985. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J. Biol. Chem. 260:14873-14878. [PubMed] [Google Scholar]
- 13.Hsiang, Y. H., M. G. Lihou, and L. F. Liu. 1989. Arrest of replication forks by drug-stabilized topoisomerase I-DNA cleavable complexes as a mechanism of cell killing by camptothecin. Cancer Res. 49:5077-5082. [PubMed] [Google Scholar]
- 14.Larsen, A. K., and C. Gobert. 1999. DNA topoisomerase I in oncology: Dr Jekyll or Mr Hyde? Pathol. Oncol. Res. 5:171-178. [DOI] [PubMed] [Google Scholar]
- 15.Li, L. H., T. J. Fraser, E. J. Olin, and B. K. Bhuyan. 1972. Action of camptothecin on mammalian cells in culture. Cancer Res. 32:2643-2650. [PubMed] [Google Scholar]
- 16.Li, T. K., and L. F. Liu. 2001. Tumor cell death induced by topoisomerase-targeting drugs. Annu. Rev. Pharmacol. Toxicol. 41:53-77. [DOI] [PubMed] [Google Scholar]
- 17.Liu, L. F. 1989. DNA topoisomerase poisons as antitumor drugs. Annu. Rev. Biochem. 58:351-375. [DOI] [PubMed] [Google Scholar]
- 18.Ljungman, M., and P. C. Hanawalt. 1996. The anti-cancer drug camptothecin inhibits elongation but stimulates initiation of RNA polymerase II transcription. Carcinogenesis 17:31-35. [DOI] [PubMed] [Google Scholar]
- 19.Mao, Y., S. D. Desai, C. Y. Ting, J. Hwang, and L. F. Liu. 2001. 26 S proteasome-mediated degradation of topoisomerase II cleavable complexes. J. Biol. Chem. 276:40652-40658. [DOI] [PubMed] [Google Scholar]
- 20.Mao, Y., M. Sun, S. D. Desai, and L. F. Liu. 2000. SUMO-1 conjugation to topoisomerase I: a possible repair response to topoisomerase-mediated DNA damage. Proc. Natl. Acad. Sci. USA 97:4046-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nitiss, J. L., and J. C. Wang. 1996. Mechanisms of cell killing by drugs that trap covalent complexes between DNA topoisomerases and DNA. Mol. Pharmacol. 50:1095-1102. [PubMed] [Google Scholar]
- 22.Pommier, Y., G. Kohlhagen, P. Pourquier, J. M. Sayer, H. Kroth, and D. M. Jerina. 2000. Benzo[a]pyrene diol epoxide adducts in DNA are potent suppressors of a normal topoisomerase I cleavage site and powerful inducers of other topoisomerase I cleavages. Proc. Natl. Acad. Sci. USA 97:2040-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Porter, S. E., and J. J. Champoux. 1989. The basis for camptothecin enhancement of DNA breakage by eukaryotic topoisomerase I. Nucleic Acids Res. 17:8521-8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pouliot, J. J., C. A. Robertson, and H. A. Nash. 2001. Pathways for repair of topoisomerase I covalent complexes in Saccharomyces cerevisiae. Genes Cells 6:677-687. [DOI] [PubMed] [Google Scholar]
- 25.Rasheed, Z. A., A. Saleem, Y. Ravee, P. P. Pandolfi, and E. H. Rubin. 2002. The topoisomerase I-binding RING protein, topors, is associated with promyelocytic leukemia nuclear bodies. Exp. Cell Res. 277:152-160. [DOI] [PubMed] [Google Scholar]
- 26.Ratner, J. N., B. Balasubramanian, J. Corden, S. L. Warren, and D. B. Bregman. 1998. Ultraviolet radiation-induced ubiquitination and proteasomal degradation of the large subunit of RNA polymerase II. Implications for transcription-coupled DNA repair. J. Biol. Chem. 273:5184-5189. [DOI] [PubMed] [Google Scholar]
- 27.Rosing, H., D. M. van Zomeren, E. Doyle, W. W. ten Bokkel, J. H. Schellens, A. Bult, and J. H. Beijnen. 1999. Quantification of topotecan and its metabolite N-desmethyltopotecan in human plasma, urine and faeces by high-performance liquid chromatographic methods. J. Chromatogr. B 727:191-203. [DOI] [PubMed] [Google Scholar]
- 28.Rubin, E., V. Wood, A. Bharti, D. Trites, C. Lynch, S. Hurwitz, S. Bartel, S. Levy, A. Rosowsky, D. Toppmeyer, and D. Kufe. 1995. A phase I and pharmacokinetic study of a new camptothecin derivative, 9-aminocamptothecin. Clin. Cancer Res. 1:269-276. [PubMed] [Google Scholar]
- 29.Shao, R. G., C. X. Cao, H. Zhang, K. W. Kohn, M. S. Wold, and Y. Pommier. 1999. Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes. EMBO J. 18:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Squires, S., A. J. Ryan, H. L. Strutt, and R. T. Johnson. 1993. Hypersensitivity of Cockayne's syndrome cells to camptothecin is associated with the generation of abnormally high levels of double strand breaks in nascent DNA. Cancer Res. 53:2012-2019. [PubMed] [Google Scholar]
- 31.Subramanian, D., C. S. Furbee, and M. T. Muller. 2001. ICE bioassay. Isolating in vivo complexes of enzyme to DNA. Methods Mol. Biol. 95:137-147. [DOI] [PubMed] [Google Scholar]
- 32.Subramanian, D., B. S. Rosenstein, and M. T. Muller. 1998. Ultraviolet-induced DNA damage stimulates topoisomerase I-DNA complex formation in vivo: possible relationship with DNA repair. Cancer Res. 58:976-984. [PubMed] [Google Scholar]
- 33.Takashima, H., C. F. Boerkoel, J. John, G. M. Saifi, M. A. Salih, D. Armstrong, Y. Mao, F. A. Quiocho, B. B. Roa, M. Nakagawa, D. W. Stockton, and J. R. Lupski. 2002. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat. Genet. 32:267-272. [DOI] [PubMed] [Google Scholar]
- 34.Vance, J. R., and T. E. Wilson. 2002. Yeast Tdp1 and Rad1-Rad10 function as redundant pathways for repairing Top1 replicative damage. Proc. Natl. Acad. Sci. USA 99:13669-13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woudstra, E. C., C. Gilbert, J. Fellows, L. Jansen, J. Brouwer, H. Erdjument-Bromage, P. Tempst, and J. Q. Svejstrup. 2002. A Rad26-Def1 complex coordinates repair and RNA pol II proteolysis in response to DNA damage. Nature 415:929-933. [DOI] [PubMed] [Google Scholar]
- 36.Wu, J., and L. F. Liu. 1997. Processing of topoisomerase I cleavable complexes into DNA damage by transcription. Nucleic Acids Res. 25:4181-4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu, R. S., A. Kumar, and J. R. Warner. 1971. Ribosome formation is blocked by camptothecin, a reversible inhibitor of RNA synthesis. Proc. Natl. Acad. Sci. USA 68:3009-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, H., J. C. Wang, and L. F. Liu. 1988. Involvement of DNA topoisomerase I in transcription of human ribosomal RNA genes. Proc. Natl. Acad. Sci. USA 85:1060-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]