Abstract
Based on evidence that the von Hippel-Lindau (VHL) tumor suppressor protein is associated with polysomes and interacts with translation regulatory factors, we set out to investigate the potential influence of pVHL on protein translation. To this end, renal cell carcinoma (RCC) cells that either lacked pVHL or expressed pVHL through stable transfection were used to prepare RNA from cytosolic (unbound) and polysome-bound fractions. Hybridization of cDNA arrays using RNA from each fraction revealed a subset of transcripts whose abundance in polysomes decreased when pVHL function was restored. The tumor necrosis factor alpha (TNF-α) mRNA was identified as one of the transcripts that preferentially associated with polysomes in pVHL-deficient cells. Additional evidence that the TNF-α mRNA was a target of translational repression by pVHL was obtained from reporter gene assays, which further revealed that pVHL's inhibitory influence on protein synthesis occurred through the TNF-α 3′-untranslated region. Our findings uncover a novel function for the pVHL tumor suppressor protein as regulator of protein translation.
Mutations in the von Hippel-Lindau (VHL) tumor suppressor gene cause VHL disease, a syndrome in which patients can develop renal cell carcinomas (RCC), retinal angiomas, pheochromocytomas, hemangioblastomas of the cerebellum and spine, endolymphatic sac tumors, and pancreatic adenomas (for review, see reference 22). Mutations in the VHL gene are also found in most sporadic clear-cell RCC (22). Since the identification of the VHL gene one decade ago (28), elucidating its function has been the focus of intense investigation. Given that pVHL bears no homology with other known proteins, efforts to discern the tumor suppressive function of pVHL have progressed through the identification of pVHL-associated proteins and the analysis of pVHL's influence on gene expression.
Association of pVHL with elongin C, elongin B, Cullin 2 (Cul2), and Rbx1 results in the formation of a complex (VCB-CUL2) that functions as an E3 ubiquitin ligase. Accordingly, pVHL is believed to play a key role in the polyubiquitination and subsequent degradation of specific cellular proteins (for review, see reference 19). The best-studied substrates of pVHL-mediated proteolysis are the hypoxia-inducible factor (HIF) alpha subunits. In normal cells growing under regular oxygen conditions, HIF-α becomes hydroxylated and is consequently targeted for pVHL-mediated ubiquitination and rapid degradation by the proteasome. However, under low-oxygen conditions, HIF-α accumulates and transcriptionally activates the expression of several genes that are important for angiogenesis and erythropoiesis, such as the vascular endothelial growth factor (VEGF), Glut-1, platelet-derived growth factor-α, and erythropoietin (10, 45). Nevertheless, pVHL also appears to exert functions unrelated to its influence on HIF-α levels, as evidenced by the findings that certain disease-causing VHL alleles retain the ability to ubiquitinate HIF-α, that pVHL and HIF regulate overlapping but not identical sets of genes, and that expression of HIF-1α variants that escape pVHL regulation does not recapitulate the formation of VHL-related tumors and cysts (6, 8, 55). pVHL has also been proposed to participate in the surveillance of protein processing, based on the observations that pVHL-deficient cells are hypersensitive to the toxicity of endoplasmic reticulum (ER) stress agents and that pVHL appears to alter the function of the cytosolic protein-folding complex CCT (13, 16). Similarly, pVHL has been postulated to influence extracellular matrix function by affecting fibronectin deposition, a function that likely relies on pVHL's association with the ER and its potential involvement in both the retrograde transport of misprocessed fibronectin and integrin assembly (25, 38). Indeed, while pVHL appears to reside in both the nucleus and the cytoplasm (6, 17, 32), cytoplasmic pVHL has been found in association with the endoplasmic reticulum and the polysomal fraction (40, 43). Moreover, pVHL was shown to regulate the polysomal association of important RNA-binding proteins such as hnRNP A2 (whose abundance is regulated by pVHL) and hnRNAP L (40).
A different approach to elucidating the tumor suppressor function of the VHL protein has been taken by several research teams seeking to investigate pVHL's influence on the patterns of expressed genes. Examples of pVHL-regulated genes include carbonic anhydrases 9 (CA9) and 12 (CA12), plasminogen activator inhibitor 1, transforming growth factor alpha, VEGF, platelet-derived growth factor, and the glucose transporter Glut-1 (18, 53; reviewed in reference 19). In addition, high-throughput analyses of gene expression using DNA arrays and serial analysis of gene expression have revealed a broad range of genes whose expression is influenced by pVHL (4, 58). A correspondingly complex array of mechanisms has been proposed to explain pVHL's influence on gene expression patterns. pVHL has been reported to influence transcription initiation through its association with transcription factors such as Sp1 (35) and transcription elongation through its inhibitory binding to elongin/SIII-containing complexes, as illustrated for the tyrosine hydroxylase gene (7, 26). mRNA turnover has also been shown to be influenced by pVHL, as exemplified by the VEGF mRNA, whose stability is markedly enhanced in pVHL-deficient cells (11, 18). pVHL also regulates gene expression by directly influencing protein degradation, as mentioned above. Given that pVHL promotes the degradation of proteins such as HIF-α under normoxic conditions, pVHL-deficient cells have higher constitutive levels of HIF-α and therefore exhibit upregulated expression of hypoxia-regulated genes bearing hypoxia-response elements in their promoter regions (34, 37). Finally, a recent report demonstrated that pVHL binds to and enhances the stability of the target protein Jade-1, a homeodomain-containing protein that is abundantly expressed in proximal tubule cells in the kidney (59). The multiple and diverse mechanisms whereby pVHL influences gene expression are believed to be a reflection of the variety of proteins with which pVHL associates. Remarkably, this influence is exerted through transcriptional, posttranscriptional, and posttranslational mechanisms of gene regulation.
In order to identify additional pVHL-interacting proteins that might mediate pVHL's influence on gene expression patterns, we performed a yeast two-hybrid analysis using full-length pVHL as bait. The discovery of two candidate pVHL interaction partners that were translational regulators, along with the finding (40) that a fraction of pVHL was associated with the cell's polysomes (strings of ribosomes engaged in active translation of a given mRNA), prompted us to examine the potential influence of pVHL on the process of protein synthesis. Comparison of stably transfected 786-0 RCC cells that either lacked or expressed the VHL protein indicated that global protein synthesis was not influenced by pVHL. However, hybridization of cDNA arrays using either polysome-bound mRNA or unbound, cytosolic mRNA (not associated with polysomes) revealed a subset of mRNAs that were preferentially associated with polysomes in cells lacking pVHL but not in cells in which pVHL function was restored, suggesting that pVHL caused specific translational repression. One such mRNA, encoding tumor necrosis factor alpha (TNF-α), was further investigated to demonstrate the influence of pVHL on protein translation. Our findings uncover a previously unknown function of pVHL as an inhibitor of protein synthesis.
MATERIALS AND METHODS
Cell culture and transfection.
Matched pairs of the human RCC lines 786-0 and UOK121 (UOK) that either were pVHL-deficient (VHL−) or expressed the wild-type VHL (VHL+) allele through stable transfection were used (11, 17); the parent 786-0 and UOK lines (pVHL deficient) had been transfected with the corresponding empty vectors and selected accordingly (11, 17). All cell lines used were maintained in high-glucose Dulbecco's modified essential medium (DMEM; Gibco-BRL) containing 10% fetal bovine serum (HyClone, Logan, Utah).
The effect of pVHL on global protein translation was studied by incubating 106 786-0 cells with 500 μCi of l-[35S]methionine and l-[35S]cysteine (Easy Tag EXPRESS; NEN/Perkin-Elmer, Boston, Mass.) per 100-mm plate for 5 h. Label incorporation into whole-cell protein lysates was then assessed either by electrophoresis of protein aliquots through sodium dodecyl sulfate-containing 12% polyacrylamide gels or by quantitation of radioactivity after precipitating protein samples in 20% trichloroacetic acid (TCA).
The influence of pVHL on TNF-α translation was assessed by transiently transfecting either plasmid pGL3-LUC (pGL3-control vector) or plasmid pGL3-LUC+TNF-α(3′), constructed by insertion of the entire 801-bp 3′-untranslated region (3′UTR) of human TNF-α immediately after the 3′ end of the luciferase coding region, in the XbaI site of the pGL3-control vector plasmid. Cotransfection of plasmid pSV-β-gal for normalization was carried out as previously described (30). Transfections were performed using SuperFect (Qiagen, Valencia, Calif.). Expression of the corresponding chimeric luciferase mRNAs, as well as control β-galactosidase mRNA, was monitored by Northern blotting (described below) in transfected cells. To investigate changes in protein translation, alterations in the levels of chimeric mRNAs were compared with changes in activity associated with luciferase and galactosidase proteins, assessed using standard methodologies (30).
Polysome analysis.
Five million cells were used per sucrose gradient. Cells were incubated for 3 min with 0.1 mg of cycloheximide/ml at 37°C and then lysed in 1 ml of PEB lysis buffer (0.3 M NaCl, 15 mM MgCl2, 15 mM Tris-Cl [pH 7.6], 1% Triton X-100, 1 mg of heparin/ml) as well as 0.1 mg of cycloheximide/ml to interrupt protein translation and thus prevent the disassembly of polysomes “running off” of a given mRNA. After a 10-min incubation on ice, nuclei were pelleted by centrifugation at 10,000 × g (4°C, 10 min), and the resulting supernatant was carefully layered onto a 10-to-50% linear sucrose gradient. Gradients were centrifuged at 35,000 × g for 3 h at 4°C, and fractions were collected at a rate of 1 ml per min using a system comprising a syringe pump, needle-piercing system (Brandel, Gaithersburg, Md.), UV-6 detector, and fraction collector (ISCO, Lincoln, Nebr.). RNA in each fraction was extracted with 8 M guanidine-HCl and used for Northern blotting. Equal volumes from each fraction were used for Northern blot analysis.
Northern blotting, Western blotting, and enzyme-linked immunosorbent assay (ELISA).
Whole-cell RNA, isolated using STAT-60 (TEL-TEST, INC., Friendswood, Tex.), or RNA from sucrose fractions was subjected to Northern blot analysis as previously described (14, 51). The TNF-α and Glut-1 cDNA fragments, obtained from plasmid pGEM3zTNFα (spanning positions 271 to 1002 of NM_000594) and by PCR amplification (4), respectively, were labeled by random priming using [α-32P]dATP and Klenow enzyme. Expression of mRNAs containing the luciferase coding region and of β-galactosidase mRNA was detected using the oligomers GTGACGAACGTGTACATCGACTGAAATCCCTGGTAATCCG and TTCAGACGGCAAACGACTGTCCTGGCCGTAACCGACCCAG, respectively; an oligonucleotide recognizing the 18S rRNA was used to monitor the amount and integrity of the RNA samples (12). All oligonucleotides were end labeled using [α-32P]dATP and terminal transferase. For Western blot analysis, equal volumes from each fraction were size fractionated using Tris-HCl gels (Bio-Rad Life Science, Hercules, Calif.) and transferred onto polyvinylidene difluoride membranes as previously described (50). Hybridizations were carried out in the presence of monoclonal antibodies recognizing either pVHL (NeoMarkers, Fremont, Calif.) or β-actin (Abcam, Cambridge, United Kingdom). Following incubation with the appropriate secondary antibodies, Western blotting signals were visualized using enhanced chemiluminescence (Amersham).
TNF-α levels secreted by 200,000 cells into the culture medium (DMEM) during a 24-h period were ascertained using an ELISA kit (BioSource International, Camarillo, Calif.) and normalized using a 3(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide thiazolyl blue (MTT) assay (Sigma).
cDNA array analysis.
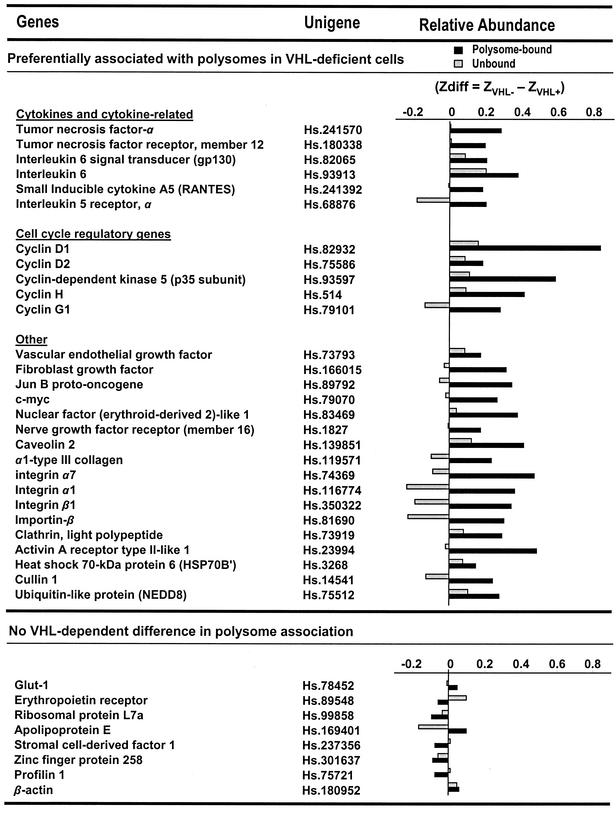
RNA was prepared from polysome-bound fractions (fractions 5 to 11) or from unbound fractions (fractions 1 and 2), reverse transcribed in the presence of [α-33P]dCTP, and used to hybridize cDNA arrays (focused array; 4,608 genes; described on the website http://www.grc.nia.nih.gov/branches/rrb/dna/array.htm), employing previously reported methodologies (9, 49). All of the data were first analyzed using the Array Pro software (Media Cybernetics, Inc., Carlsbad, Calif.) and then normalized by Z score transformation (5). In brief, the log10 of each original spot intensity was adjusted to the mean and divided by the standard deviation of all the spot intensities. Changes in gene expression between different RNA groups were then calculated by subtracting the average of replicate measurements. This value, referred to as the Z difference (Zdiff; average Z in pVHL-deficient cells minus the average Z in pVHL-expressing cells) was tested for significance using a two-tailed Z test (Z ≥ 2.4; P ≤ 0.05). The data reflect three independent experiments. Genes were considered to be preferentially associated with polysomes in pVHL-deficient cells (see Fig. 4, top) if Zdiff values in fractions 5 to 11 (polysome bound) were significantly different between pVHL-expressing and pVHL-deficient cells and the Zdiff values obtained from fractions 1 and 2 (unbound) were not significantly different. The complete cDNA array data are available at http://www.grc.nia.nih.gov/branches/rrb/dna/dnapubs.htm. Several genes illustrating no pVHL dependence in polysome association are listed below in Fig. 4 (bottom). For these genes, Zdiff values were not significantly different by Z test in either unbound or polysome-bound fractions.
FIG. 4.
Collection of genes encoding mRNAs that are specifically bound to polysomes in pVHL-deficient cells. For each gene on the cDNA arrays, the relative presence of the encoded mRNA in polysome-bound and unbound fractions was compared between pVHL-expressing and pVHL-deficient 786-0 cell lines. Cell fractionations and cDNA array analysis were performed three independent times. Differences in Z averages (Zdiff) served to assess the relative abundance of a given transcript in polysomes (polysome bound; black bars) or in unbound fractions (gray bars) of pVHL-deficient and pVHL-expressing cells and were calculated as explained in Materials and Methods and in reference 5. The top group(preferentially associated with polysomes in pVHL-deficient cells) includes genes potentially subject to translational repression by pVHL, since they are encoded by mRNAs that were significantly more abundant in polysome-bound fractions prepared from pVHL-deficient cells than in those prepared from pVHL-expressing cells. *, Zdiff values for the polysome-bound mRNA are significant, using two tests (Z ≥ 2.4; P ≤ 0.05). For these genes, unbound mRNAs were not significantly different between pVHL-expressing and pVHL-deficient populations. The bottom group (no pVHL-dependent difference in polysome association) lists genes encoding mRNAs that exhibited no significant differences in their relative distribution (polysomal and unbound) in cells with different pVHL status.
Yeast two-hybrid analysis and yeast mating.
To identify pVHL-interacting proteins, we performed a yeast two-hybrid screen using the human full-length pVHL as bait (subcloned from the g7 clone [28]), downstream of the LexA DNA-binding domain (pLexA-Vector; pGilda; Clontech, Heidelberg, Germany) and a human kidney cDNA library as target (pB42AD vector; Matchmarker LexA cDNA library; Clontech). Target and bait constructs (library in pB42AD and pGilda-VHL, respectively) were cotransformed into EGY48-p8op-lacZ Saccharomyces cerevisiae and selected for uracil, histidine, and tryptophan prototrophy. Colonies with potential pVHL-target protein interactions were identified by the activation of two independent reporter genes that conferred leucine prototrophy and β-galactosidase activity. Plasmids in positive colonies were recovered in the Escherichia coli strain KC8, sequenced, and analyzed for sequence homology.
For yeast mating confirmation of protein-protein interactions, library targets were selected and transformed into strain EGY48 (MATα), while pGilda-VHL was transformed into strain YM4271 (MATa). Diploid yeast strains bearing the three plasmid vectors p8op-lacZ, pGilda-VHL, and pB42AD were selected for prototrophy for uracil, histidine, and tryptophan.
RESULTS
Candidate pVHL-interacting proteins and the polysome association of pVHL suggest potential influence of pVHL in regulating protein synthesis in RCC cells.
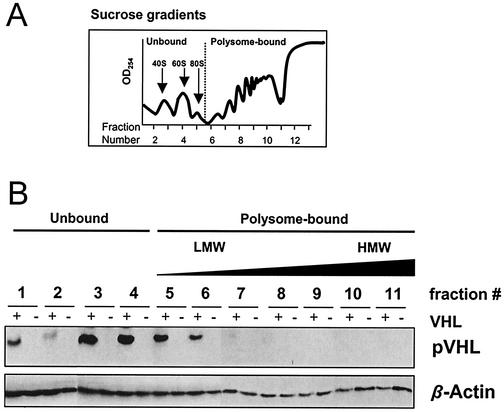
Since pVHL function relies, at least in part, on its interaction with other proteins, we sought to take a systematic approach towards the identification of additional cellular proteins that associate with pVHL. To this end, we set up a yeast two-hybrid screen using full-length pVHL as bait and a kidney cDNA library as target. We identified several independent cDNAs encoding potential interaction partners, among which were 15 genes that were subsequently confirmed by yeast mating (Table 1). In addition to finding a previously described pVHL target, the pVHL-binding protein (VBP-1 [48]), we identified several stress response proteins (Hsp60 and Hsp90), one protein involved in proteolysis (Tat-binding protein-1), and the translational regulators eIF3 and EF-1α among the putative pVHL-interacting proteins (Table 1). The identification of the latter group of proteins, along with the recent discovery that pVHL associates with polysomes (40), led us to postulate that pVHL may influence protein translation. In order to test this hypothesis, we employed two RCC cell lines. One, 786-0 (VHL−), has a frameshift mutation in the VHL gene and fails to express VHL protein; the other, 786-0 (VHL+), is derived from 786-0 by stable transfection of a vector expressing human pVHL (17). Cytoplasmic components from both 786-0 lines were fractionated through continuous sucrose gradients (Fig. 1A) (explained in Materials and Methods), and the presence of pVHL in the various fractions was examined. As shown in Fig. 1B, pVHL was found in both the soluble fraction (unbound to polysomes) and in the polysome-bound fractions. Among the latter fractions, pVHL appeared to localize primarily with low-molecular-weight (LMW) polysomes. To ascertain whether the presence of pVHL in polysomes was associated with gross changes in protein synthesis, we monitored the incorporation of l-[35S]methionine and l-[35S]cysteine into nascent polypeptide chains in 786-0 cells that either lacked or expressed pVHL. As shown, pVHL-deficient and pVHL-expressing cells exhibited very similar patterns of labeled polypeptides in whole-cell lysates (Fig. 2A) and comparable incorporation of the labeled amino acids into TCA-precipitable product (Fig. 2B). Thus, pVHL does not appear to alter overall protein synthesis.
TABLE 1.
pVHL can associate with translation regulatory factors in yeasta
| Candidate pVHL-interacting proteins | GenBank accession no. | Function | No. of clones |
|---|---|---|---|
| Translation initiation factor 3 subunit 4 (eIF3) | NP003746 | Translational regulation | 1 |
| Translation elongation factor 1-alpha (EF-1α) (EF-TU) | AAA52367 | Translational regulation | 4 |
| Glucose-regulated protein (GRP 78) | H56456S1 | Molecular chaperone | 1 |
| Heat shock 60-kDa protein 1 (HSP 60) | P10809 | Molecular chaperone | 1 |
| Heat shock protein 90 (HSP 90) | HHHU86 | Molecular chaperone | 2 |
| VHL-binding protein 1 (VBP-1) | Q15765 | Cytoplasmic export of pVHL | 1 |
| 26S protease regulatory subunit (Tat-binding protein 1) | P17980 | Protein degradation | 1 |
| Gelsolin | NP000168 | Other | 1 |
| Metallothionein 2A | H56373S3 | Other | 1 |
| Filamin (actin-binding protein 280) ABP-280 | NM001456 | Other | 1 |
| RIE 2 sid 2705 | BAA84708 | Other | 1 |
| CGI-111 protein | AAD34106 | Other | 1 |
| Antioxidant enzyme BP 66 | AAF03750 | Other | 1 |
| Gene SA protein | I54401 | Other | 1 |
| X-Pro dipeptidase | L10320 | Other | 2 |
Shown are the results of a yeast two-hybrid survey leading to the identification of potential pVHL-interacting proteins from a human kidney cDNA library. Some of the clones were represented several times (number of clones, right column). The interaction of pVHL with all of the gene products listed was confirmed by yeast mating (explained in Materials and Methods).
FIG. 1.
pVHL fractionates with LMW polysomes. (A) Cytoplasmic extracts from RCC 786-0 cells that either lacked (VHL−) or expressed (VHL+) pVHL were centrifuged through continuous sucrose gradients to obtain fractions of increasing molecular weight. Absorbance at 254 nm served to monitor the presence of ribosomal subunits, ribosomes, and polysomes. Fractions 1 and 2 were completely devoid of ribosomes and ribosome components, fractions 3 and 4 typically included the free small and large ribosomal subunits, respectively, and fractions 5 to 11 contained polysomes of increasing weight. (B) Representative Western blot analysis to monitor pVHL abundance in each of the continuous fractions prepared from sucrose gradients in both pVHL-expressing and pVHL-deficient cells; equal volumes were loaded from each fraction. To monitor the quality of the protein preparations, blots were stripped and rehybridized using an antibody that recognizes β-actin.
FIG. 2.
pVHL does not appear to influence global protein synthesis in 786-0 cells. 786-0 cells that either lacked (VHL−) or expressed (VHL+) pVHL were incubated with l-[35S]methionine and l-[35S]cysteine for 5 h, whereupon whole-cell lysates were prepared and newly synthesized, radiolabeled proteins were assessed. (A) Representative gel depicting radiolabeled proteins following electrophoresis through sodium dodecyl sulfate-containing 12% polyacrylamide gels. (B) Quantitation of radioactivity incorporated into nascent proteins, measured after precipitation in 20% TCA. Data shown are the means and standard errors of the means from three different experiments.
cDNA array-based identification of mRNAs associating with polysomes in a pVHL-dependent fashion.
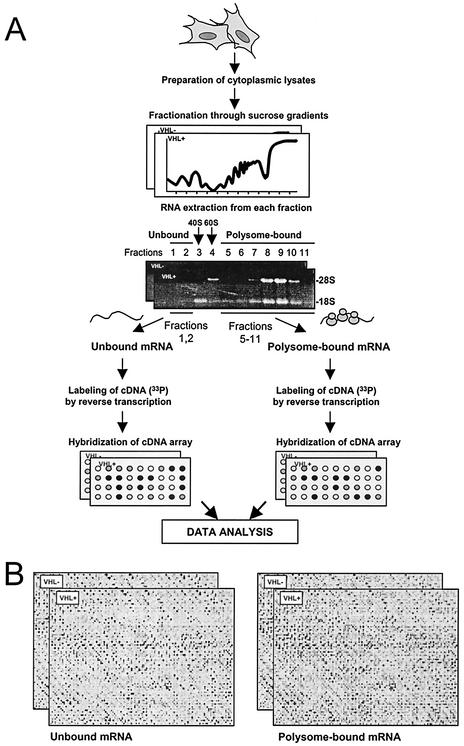
While pVHL did not exert a noticeable influence on global protein synthesis, we sought to examine whether pVHL could regulate the translation of specific mRNAs. Towards this goal, we used an approach based on the analysis of cDNA arrays (21, 60). Cytoplasmic lysates from both pVHL-deficient and pVHL-expressing 786-0 cells were centrifuged through sucrose gradients and then collected in 11 continuous fractions, from lightest (cytosolic components devoid of ribosomes) to heaviest (largest polysomes).
As shown in the schematic of Fig. 3A, fractions 1 and 2 lacked any ribosomes or ribosome subunits, with no rRNA detectable by ethidium bromide staining; pooled mRNA present in these two fractions constitutes the unbound mRNA and represents the subset of mRNAs that are not being translated. Ribosome components typically began to appear in fraction 3, first with the small subunit (40S, ∼fraction 3), then the large subunit (60S, ∼fraction 4), followed by monosomes (single ribosomes, 80S, ∼fraction 5), LMW polysomes (∼fractions 6 and 7), and high-molecular-weight (HMW) polysomes (∼fractions 8 to 11). Fractions 5 to 11 contained both 18S and 28S rRNA; pooled mRNA present in these fractions was termed polysome-bound mRNA and represents the collection of mRNAs engaged in translation. Fractions 3 and 4 were discarded to ensure a clean separation between mRNAs not bound to ribosomes and ribosome-bound mRNAs. Both sets of mRNAs (unbound and polysome bound) from each cell line were used in reverse transcription reactions to prepare radiolabeled probes that were subsequently used to hybridize cDNA arrays (4,608 genes; Human Focused Array, described on the website http://www.grc.nia.nih.gov/branches/rrb/dna/array.htm).
FIG. 3.
Strategy to study pVHL-mediated regulation of mRNA translation in RCC cells. (A) Cytoplasmic extracts from 786-0 cells that either lacked (VHL−) or expressed (VHL+) pVHL were centrifuged through sucrose gradients to obtain fractions of increasing molecular weight. Absorbance at 254 nm served to monitor the presence of ribosomal subunits, ribosomes, and polysomes. Pooled fractions 1 and 2 contained mRNAs that were completely devoid of ribosomes and ribosome components (they constitute the unbound mRNA component); the mRNA in fractions 3 and 4, which typically included the free small and large ribosomal subunits, respectively, were discarded; pooled fractions 5 to 11 contained mRNAs bound to one or several ribosomes (the polysome-bound mRNA component). From each 786-0 cell population, two sets of radiolabeled cDNA were prepared through reverse transcription in the presence of [α-33P]dCTP: one from unbound mRNA and one from polysome-bound mRNA. Probes were used for hybridization of cDNA arrays (described in reference 5). (B) Representative cDNA arrays to illustrate the hybridization signals corresponding to either unbound mRNA (left) or polysome-bound mRNA (right). Cell fractions and cDNA array analyses were performed three times, each as a completely independent experiment.
Pairwise comparisons of unbound (untranslated) versus polysome-bound (translationally active) mRNAs were carried out in pVHL-deficient and pVHL-expressing cells, and representative cDNA array signals are shown in Fig. 3B. For each gene, the signal on membranes corresponding to polysome-bound mRNA was compared between pVHL-expressing and pVHL-deficient cells; similar comparisons were done for unbound mRNA. Differences in the relative distribution of mRNAs when comparing pVHL-deficient and pVHL-expressing cells were quantitated using Z averages (5). mRNAs that were found in greater abundance in polysomes of pVHL-deficient cells, i.e., mRNAs that were potential targets of pVHL-mediated translational repression, were considered significant if they met the criteria explained in Materials and Methods. Listed in Fig. 4 are several genes out of a total of approximately 3.8% of genes on the array (174 individual genes) meeting the significance criteria described below. Briefly, the degree to which each mRNA listed was preferentially associated with polysomes in pVHL-deficient cells (Fig. 4, top) was assessed by comparing Zdiff values: average Z in pVHL-deficient compared with pVHL-expressing cells (ZVHL− − ZVHL+). For the genes listed in this group, Zdiff values for the polysome-bound mRNA were considered significant based on a two-tailed Z test (Z ≥ 2.4; P ≤ 0.05). For the genes listed on the top part of Fig. 4, Zdiff values for the unbound mRNA were not significantly different by Z test between pVHL-expressing and pVHL-deficient cells. Interestingly, among the mRNAs preferentially bound to polysomes in pVHL-deficient cells were several encoding proteins that are elevated in pVHL-deficient cells, such as VEGF and cyclin D1. Expression of VEGF, for example, has been shown to be regulated in a pVHL-dependent fashion through a variety of mechanisms, including transcription and mRNA turnover. The observation that VEGF mRNA is significantly more abundant in polysomes of cells lacking pVHL suggests that VEGF expression may be controlled by pVHL at one additional level, protein translation. Several mRNAs whose relative abundance in the unbound and polysome-bound fractions are not significantly different when comparing pVHL-expressing and pVHL-deficient cells are listed under the category “no VHL-dependent difference in polysome association.” Three sets of unbound and polysome-bound RNA preparations were independently prepared and analyzed using cDNA arrays. The complete list of genes and the relative abundance in unbound and polysome-bound fractions is available at the website http://www.grc.nia.nih.gov/branches/rrb/dna/dnapubs.htm.
As indicated above, the cDNA array screen also revealed that the TNF-α mRNA was preferentially associated with polysomes in pVHL-deficient cells. Given Caldwell et al.'s recent studies describing pVHL's sensitization of RCC cells to the cytotoxic effects of TNF-α (4), we sought to examine the influence of pVHL on TNF-α expression in greater detail and thereby also ascertain the usefulness of this cDNA array-based methodology to study translational regulation in this system.
pVHL-dependent regulation of TNF-α translational efficiency and TNF-α expression.
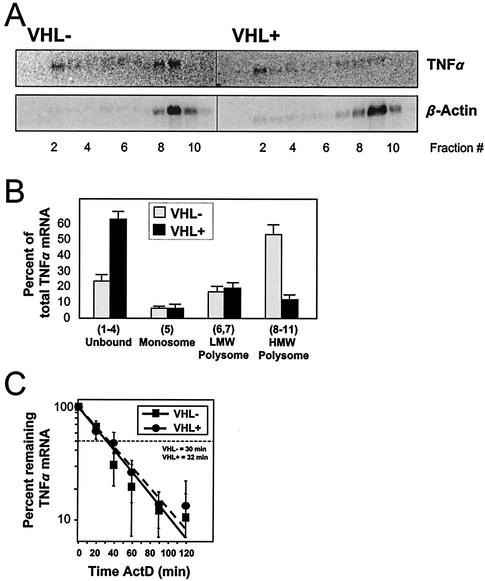
Cytoplasmic lysates prepared from 786-0-deficient (VHL−) and 786-0-expressing (VHL+) cells were fractionated through sucrose gradients, and the distribution of TNF-α mRNA in the fractions was assessed by Northern blotting (Fig. 5A). TNF-α mRNA signals in the various fractions were quantitated and are represented as the percentage of the total (sum of signals in all fractions). As shown in Fig. 5B, pVHL-deficient cells exhibited substantially higher levels of polysome-bound TNF-α mRNA, particularly in the HMW fraction, while unbound TNF-α mRNA was preferentially found in the nonpolysomal fraction in the pVHL-expressing cells. It was of interest to determine if the TNF-α mRNA stability was influenced by the cell's pVHL status, given several reports demonstrating that the expression of TNF-α could be regulated through altered mRNA turnover (29, 33). However, despite marked differences in the subcellular localization of TNF-α mRNA in cells that either expressed or lacked pVHL, the half-life of the TNF-α mRNA, as assessed by actinomycin D (ActD)-based determinations, was essentially unchanged (t1/2, ∼30 min in each cell line) (Fig. 5C). Briefly, for calculation of the TNF-α mRNA half-life, ActD was added to block new mRNA transcription and whole-cell RNA was prepared at various times thereafter (20 min, 40 min, etc.). RNA was then used for Northern blot analysis to monitor the rate of clearance of the TNF-α transcript. Following quantitation of signals from TNF-α mRNA and 18S rRNA (the latter was used to normalize for differences in loading and transfer of samples), the TNF-α mRNA half-life was calculated as the time necessary to achieve a reduction to 50% of the signal measured before adding ActD.
FIG. 5.
TNF-α mRNA is associated with HMW polysomes in pVHL-deficient cells. Cytoplasmic lysates from 786-0 cells that either lacked (VHL−) or expressed (VHL+) pVHL were fractionated through sucrose gradients, and the RNA prepared from each of the 11 fractions was subjected to Northern blot analysis. (A) Representative TNF-α Northern blotting signals; β-actin mRNA signals were included to assess differences in loading and transfer of samples. (B) Quantitation of the relative abundance of TNF-α mRNA in the indicated fractions prepared from sucrose gradients. TNF-α mRNA signals were normalized to β-actin mRNA signals and then shown as the percent abundance of the TNF-α mRNA in a given RNA subset relative to the total cytoplasmic TNF-α mRNA. (C) For mRNA half-life assessments, ActD (2 μg/ml) was added to 786-0 cells and total RNA prepared at the times indicated. mRNA half-lives were calculated after measurement of Northern blotting signals, normalization to 18S rRNA signals, plotting on logarithmic scales, and determination of the time period required to decrease the abundance of the TNF-α transcript to 50% of the initial signal intensity. Data in graphs represent the means ± standard errors of the means from three independent experiments.
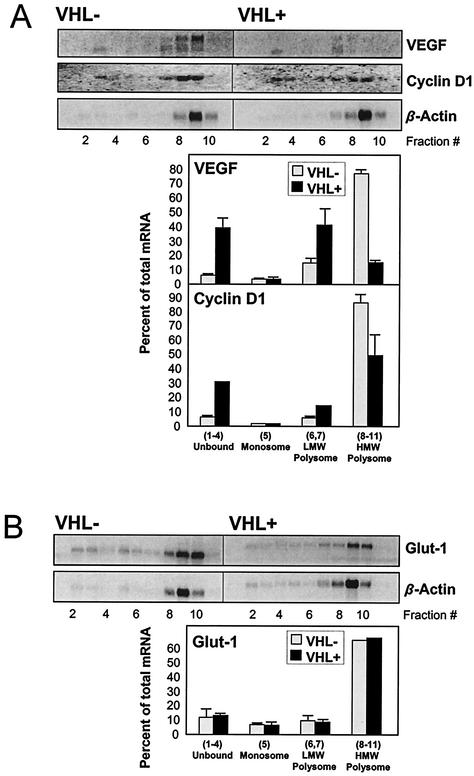
Additional mRNAs were examined to test the validity of this approach. As shown in Fig. 6A, mRNAs encoding VEGF and cyclin D1, which were predicted to preferentially associate with polysomes in pVHL-deficient cells (Fig. 4, top), indeed appeared more abundant in RNA preparations obtained from polysome-bound material in pVHL-deficient cells. Interestingly, a pVHL-dependent difference in cyclin D1 expression was recently reported (3, 58). By contrast, similar Northern blot analysis revealed that Glut-1 mRNA, whose abundance was predicted not to be preferentially associated with polysomes in a pVHL-dependent fashion (Fig. 4, bottom), displayed a similar relative distribution in both cell lines (Fig. 6B), although overall Glut-1 mRNA expression was higher in pVHL-deficient cells, keeping with earlier reports (3, 18). Expression levels of β-actin, another transcript whose relative polysomal association was not pVHL dependent (Fig. 4, bottom), was comparable between pVHL-expressing and pVHL-deficient cells, as anticipated. β-Actin mRNA signals were routinely used to test sample quality as well as evenness of loading of Northern blotting samples (Fig. 5 and 6).
FIG. 6.
Validation of mRNAs displaying either preferential or nonpreferential polysomal association in a pVHL-dependent fashion. (A) Abundance of mRNAs encoding VEGF and cyclin D1, which preferentially associated with polysomes in pVHL-deficient cells, was assessed by Northern blotting and quantitated as described in the legend for Fig. 5. (B) Abundance of Glut-1 mRNA, which is not preferentially associated with polysomes in pVHL-deficient cells, was assessed as described in the legend for Fig. 5. β-Actin mRNA signals served to both validate the results in Fig. 5 and monitor the evenness in loading and transfer of samples.
Previous reports have shown that TNF-α expression can be regulated by controlling the translation of its mRNA, and these studies have linked this regulatory mechanism to a region within the TNF-α 3′UTR that was a target of translational repression (15, 39). These reports, taken together with our observation that reintroduction of pVHL correlated with a substantial reduction in the translationally active fraction of the TNF-α mRNA, strongly suggested that pVHL might inhibit TNF-α translation through the TNF-α 3′UTR. We tested this hypothesis by monitoring mRNA expression and protein production from a chimeric reporter construct that encompassed the 3′UTR of TNF-α directly downstream of the luciferase coding sequence. Initial efforts to analyze nascent TNF-α by using a pulse incubation in the presence of 35S-labeled amino acids were unfruitful due to the very low expression levels of TNF-α in these cells. The alternative approach employed here involved the transient transfection of cells with plasmids expressing either the luciferase mRNA (LUC) or a chimeric luciferase-TNF-α(3′UTR) mRNA [LUC+TNFα(3′)] (Fig. 7A); comparison of mRNA levels with luciferase activity levels in each cell line allowed the assessment of pVHL-dependent changes in translation rates. In addition, in order to control for potential cell line-specific effects, we also included a second, independently developed pair of pVHL-deficient and pVHL-expressing RCC cell lines. The parent UOK cell line lacked pVHL function (VHL−); pVHL expression was restored by stable transfection of UOK cells with a human pVHL cDNA (VHL+) (11). As shown, all mRNAs transcribed from the transfected plasmids, including those expressed from control transfected plasmids encoding β-galactosidase, were found to be expressed with the same abundance in cells with different pVHL status (Fig. 7B). Importantly, when synthesis of the encoded proteins was measured (by measuring luciferase and β-galactosidase activities), all luciferase activities were found to be comparable, except in populations transfected with plasmid that expressed LUC+TNF-α (3′) mRNA, where pVHL-expressing cells produced significantly less luciferase, approximately 50% of that measured in pVHL-deficient cells (Fig. 7C). This finding indicated that addition of the TNF-α 3′UTR to a heterologous transcript (luciferase, in this case) caused it to be translated less efficiently in cells expressing functional pVHL. Such observations were made in both the 786-0 and the UOK cells, strongly suggesting that the effect on TNF-α synthesis was truly dependent on the cell's pVHL status.
FIG. 7.
The 3′UTR of TNF-α mRNA confers translational repression in a pVHL-dependent manner. (A) pGL3-luciferase (pGL3-LUC) and pGL3-luciferase containing the 3′UTR of TNF-α [pGL3-LUC+TNF-α(3′)] were transiently transfected along with control plasmid pSV-β-gal into 786-0 cells that either expressed or lacked pVHL function (VHL+ and VHL−, respectively). (B) Representative Northern blot analysis used to ascertain the expression levels of LUC+TNF-α(3′) transcript (and LUC in pGL3-LUC transfections [data not shown]) and the β-galactosidase mRNA in transfected 786-0 and UOK populations; determination of 18S signals served to monitor the quality of RNA samples. Graphs represent the relative abundance of mRNAs containing the luciferase coding region [either LUC mRNA or LUC+TNF-α(3′) mRNA] in pVHL-deficient cells (100%) compared with pVHL-expressing cells. In each transfection group [pGL3-LUC and pGL3-LUC+TNF-α(3′)], luciferase mRNA values were normalized to β-galactosidase mRNA values. (C) Luciferase activity in 786-0 and UOK cells transfected with either pGL3-LUC or pGL3-LUC+TNF-α(3′); measurements of luciferase activity were normalized to measurements of β-galactosidase activity and plotted as percent luciferase activity/galactosidase activity in pVHL-expressing cells (VHL+) relative to that measured in pVHL-deficient cells (VHL−) (100%). Data represent the means and standard errors of the means from three independent experiments.
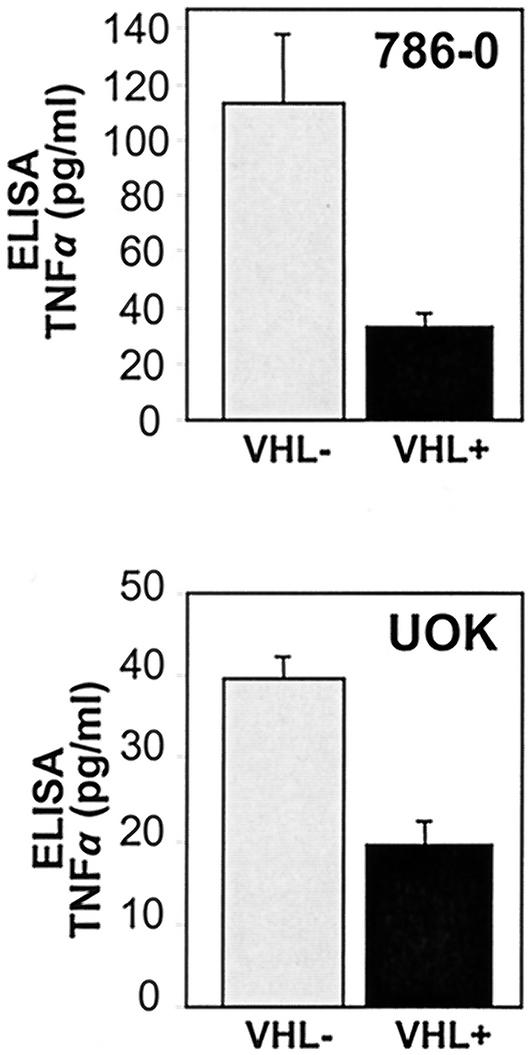
Finally, restoration of pVHL function in both RCC lines, an intervention that was shown to inhibit TNF-α mRNA association with polysomes (Fig. 5) and repress translation of transcripts carrying the TNF-α 3′UTR (Fig. 7), significantly reduced the amount of TNF-α synthesized, as determined by ELISA (Fig. 8). The specific distribution profiles of TNF-α mRNA in the UOK cells (data not shown) were also consistent with the levels of secreted TNF-α protein. For comparison, cultured macrophages and lymphocytes (106/ml) can produce 3 to 5 ng of TNF-α/ml after lipopolysaccharide and CD3 plus CD28 stimulation, respectively. In conclusion, the TNF-α mRNA shifts to higher-molecular-weight polysomes in pVHL-deficient cells, and TNF-α protein is synthesized and secreted. Together, our findings demonstrate that pVHL functions in selectively excluding the TNF-α mRNA from translationally active polysomes and specifically reduces TNF-α expression in RCC cells.
FIG. 8.
Determination of TNF-α synthesis by ELISA. Abundance of TNF-α production by 786-0 and UOK cultures was quantified in conditioned media using an ELISA, as explained in Materials and Methods. Data represent the means and standard errors of the means from three independent experiments.
DISCUSSION
The present study provides evidence in support of a role for pVHL in the translational repression of TNF-α biosynthesis. The finding that cytoplasmic pVHL associates with polysomes and translation factors (40) (Table 1; Fig. 1) prompted us to hypothesize that pVHL might be involved, directly or indirectly, in regulating protein synthesis. To test this possibility, we used RCC cell lines that either lack pVHL function (VHL−) or express pVHL through stable transfection (VHL+). Cytoplasmic lysates prepared from 786-0 RCC cells were further fractionated into polysome-bound mRNA (engaged in translation) and mRNA not bound to ribosomes (hence not being translated), and each mRNA collection was reverse transcribed and used in hybridization of cDNA arrays. Comparison of the resulting data revealed a group of mRNAs that preferentially associated with polysomes in cells lacking pVHL; for these genes, restoration of pVHL expression greatly reduced their association with ribosomes and thus presumably their translation.
Such pVHL-dependent translational repression was further demonstrated for TNF-α, a gene product that was found to be markedly cytotoxic for RCC cells expressing VHL protein (4). In addition, TNF-α expression is of particular interest in the context of the cancer-fraught VHL syndrome, given TNF-α's close links to angiogenesis and cancer, as discussed below. Our studies show that the pVHL-dependent reduction in the association of TNF-α mRNA with the translationally active, polysome-bound fraction was linked to decreased TNF-α protein synthesis. In addition, we have provided evidence that the TNF-α 3′UTR, previously shown to participate in the translational regulation of TNF-α expression (20, 27, 41, 52), was responsible for conferring translational repression upon a chimeric transcript. According to our unpublished results, neither the levels of several translational regulatory proteins examined (i.e., TIAR, TIA-1, EF-1α) nor their association with in vitro-synthesized TNF-α 3′UTR transcripts appeared to be substantially influenced by the cell's pVHL status (results not shown). Experiments are under way to investigate whether TIAR and TIA-1 (attractive candidate proteins reportedly involved in translational regulation [15]) bind to the TNF-α 3′UTR transcripts in vivo and whether this association is direct or results from their being part of larger, multiprotein complexes (23). It is unlikely that pVHL influences TNF-α translation by directly binding the TNF-α mRNA, given the absence of discernible RNA-binding domains in the VHL protein. Instead, in its capacity as a key component of an E3 (VCB-CUL2) ligase, pVHL may assist in the targeting for ubiquitin-mediated degradation of other (as-yet-unidentified) translational regulatory proteins. Formal testing of this possibility awaits the identification of additional protein targets of VCB-CUL2-mediated ubiquitination. Alternatively, pVHL could associate with other cytosolic, ER-associated, or polysomal proteins and thereby modulate their function. These possibilities also remain to be examined directly.
What might be the significance of the heightened TNF-α expression in pVHL-deficient RCC cells? Despite the antitumor effects of local, high-dose TNF-α administration (1, 44), a growing body of evidence paradoxically supports a tumor-promoting role for TNF-α. It is expressed by a wide range of tumors, including RCC (1, 36, 57), and its presence is generally associated with poor prognosis. Local chronic production of TNF-α in the tumor appears to contribute to chronic inflammatory conditions that favor cancer development (2). In this regard, TNF-α has been shown to induce the expression of angiogenic factors (VEGF, bFGF), matrix metalloproteinases, and chemokines, all of which can remodel the tumor stroma and promote cancer spread (31). TNF-α has also been shown to be a transforming agent in vitro and can function as a growth factor for cultured cancer cells (24, 54). Furthermore, loss of pVHL greatly diminished the cytotoxic effects of TNF-α on cultured RCC cells (4), suggesting that tumor suppression by pVHL may rely, at least in part, on its ability to enhance cell death by TNF-α. The precise effects of TNF-α in RCC cells arising from pVHL mutations, as well as TNF-α's influence on other VHL disease-related tumors remain to be examined directly. Chemotherapeutic approaches that inhibit TNF-α expression and/or function (such as TNF antagonists, thalidomide, nonsteroidal antiinflammatory drugs, etc.) are showing great promise for cancer treatment (42, 47, 56). Although the usefulness of these therapies in the prevention or treatment of VHL disease-related lesions remains to be investigated, there is evidence for their efficacy in the treatment of RCC (46).
Acknowledgments
We are grateful to J. Gnarra for providing the UOK121 cells and O. Iliopoulos and W. G. Kaelin for providing the 786-0 cells. We thank K. Becker, P. Ghosh, and I. Espinoza-Delgado for helpful discussions and the DNA Array Unit (NIA-IRP) for providing cDNA arrays for analysis.
S.G. was sponsored by a German Academic Exchange Service (DAAD) grant; J.B. and J.D. were sponsored by the German Research Council (DFG 356-3/3-4 and SFB 519-C2).
REFERENCES
- 1.Balkwill, F. 2002. Tumor necrosis factor or tumor promoting factor? Cytokine Growth Factor Rev. 13:135-141. [DOI] [PubMed] [Google Scholar]
- 2.Balkwill, F., and A. Mantovani. 2001. Inflammation and cancer: back to Virchow? Lancet 357:539-545. [DOI] [PubMed] [Google Scholar]
- 3.Bindra, R. S., J. R. Vasselli, R. Stearman, W. M. Linehan, and R. D. Klausner. 2002. VHL-mediated hypoxia regulation of cyclin D1 in renal carcinoma cells. Cancer Res. 62:3014-3019. [PubMed] [Google Scholar]
- 4.Caldwell, M. C., C. Hough, S. Fürer, W. M. Linehan, P. J. Morin, and M. Gorospe. 2002. Serial analysis of gene expression in renal carcinoma cells reveals VHL-dependent sensitivity to TNF-α cytotoxicity. Oncogene 21:929-936. [DOI] [PubMed] [Google Scholar]
- 5.Cheadle, C., M. P. Vawter, W. J. Freed, and K. G. Becker. Analysis of microarray data using Z score transformation. J. Mol. Diagn., in press. [DOI] [PMC free article] [PubMed]
- 6.Corless, C. L., A. Kibel, O. Iliopoulos, and W. G. Kaelin. 1997. Immunostaining of the von Hippel-Lindau gene product (pVHL) in normal and neoplastic human tissues. Hum. Pathol. 28:459-464. [DOI] [PubMed] [Google Scholar]
- 7.Duan, D. R., A. Pause, W. Burgess, T. Aso, D. Y. T. Chen, K. P. Garrett, R. C. Conaway, J. W. Conaway, W. M. Linehan, and R. D. Klausner. 1995. Inhibition of transcriptional elongation by the VHL tumor suppressor protein. Science 269:1402-1406. [DOI] [PubMed] [Google Scholar]
- 8.Elson, D. A., G. Thurston, L. E. Huang, D. G. Ginzinger, D. M. McDonald, R. S. Johnson, and J. M. Arbeit. 2001. Induction of hypervascularity without leakage or inflammation in transgenic mice overexpressing hypoxia-inducible factor-1alpha. Genes Dev. 15:2520-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan, J., X. Yang, W. Wang, W. H. Wood III, K. G. Becker, and M. Gorospe. 2002. Global analysis of stress-regulated mRNA turnover by using cDNA arrays. Proc. Natl. Acad. Sci. USA 99:10611-10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forsythe, J. A., B. H. Jiang, N. V. Iyer, F. Agani, S. W. Leung, R. D. Koos, and G. L. Semenza. 1996. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol. Cell. Biol. 16:4604-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gnarra, J. R., S. Zhou, M. J. Merrill, J. R. Wagner, A. Krumm, E. Papavassiliou, E. H. Oldfield, R. D. Klausner, and W. M. Linehan. 1996. Post-transcriptional regulation of vascular endothelial growth factor mRNA by the product of the VHL tumor suppressor gene. Proc. Natl. Acad. Sci. USA 93:10589-10594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorospe, M., Y. Liu, Q. Xu, F. J. Chrest, and N. J. Holbrook. 1996. Inhibition of G1 cyclin-dependent kinase activity during growth arrest of human breast carcinoma cells by prostaglandin A2. Mol. Cell. Biol. 16:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorospe, M., J. M. Egan, B. Zbar, M. Lerman, L. Geil, I. Kuzmin, and N. J. Holbrook. 1999. Protective function of von Hippel-Lindau protein against impaired protein processing in renal carcinoma cells. Mol. Cell. Biol. 19:1289-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorospe, M., and N. J. Holbrook. 1996. Role of p21 in prostaglandin A2-mediated cellular arrest and death. Cancer Res. 56:475-479. [PubMed] [Google Scholar]
- 15.Gueydan, C., L. Droogmans, P. Chalon, G. Huez, D. Caput, and V. Kruys. 1999. Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor-α mRNA. J. Biol. Chem. 274:2322-2326. [DOI] [PubMed] [Google Scholar]
- 16.Hansen, W. J., M. Ohh, J. Moslehi, K. Kondo, W. G. Kaelin, and W. J. Welch. 2002. Diverse effects of mutations in exon II of the von Hippel-Lindau (VHL) tumor suppressor gene on the interaction of pVHL with the cytosolic chaperonin and pVHL-dependent ubiquitin ligase activity. Mol. Cell. Biol. 22:1947-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iliopoulos, O., A. Kibel, S. Gray, and W. G. Kaelin, Jr. 1995. Tumour suppression by the human von Hippel-Lindau gene product. Nat. Med. 1:822-826. [DOI] [PubMed] [Google Scholar]
- 18.Iliopoulos, O., A. P. Levy, C. Jiang, W. C. Kaelin, Jr., and M. A. Goldberg. 1996. Negative regulation of hypoxia-inducible genes by the von Hippel-Lindau protein. Proc. Natl. Acad. Sci. USA 93:10595-10599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivan, M., and W. G. Kaelin. 2001. The von Hippel-Lindau tumor suppressor protein. Curr. Opin. Genet. Dev. 11:27-34. [DOI] [PubMed] [Google Scholar]
- 20.Iyer, S., D. Kontoyiannis, D. Chevrier, J. Woo, N. Mori, M. Cornejo, G. Kollias, and R. Buelow. 2000. Inhibition of tumor necrosis factor mRNA translation by a rationally designed immunomodulatory peptide. J. Biol. Chem. 275:17051-17057. [DOI] [PubMed] [Google Scholar]
- 21.Johannes, G., M. S. Carter, M. B. Eisen, P. O. Brown, and P. Sarnow. 1999. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc. Natl. Acad. Sci. USA 96:13118-13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaelin, W. G. 2002. Molecular basis of the VHL hereditary cancer syndrome. Nat. Rev. Cancer 2:673-682. [DOI] [PubMed] [Google Scholar]
- 23.Kedersha, N. L., M. Gupta, W. Li, I. Miller, and P. Anderson. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J. Cell Biol. 147:1431-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Komori, A., J. Yatsunami, M. Suganuma, S. Okabe, S. Abe, A. Sakai, K. Sasaki, and H. Fujiki. 1993. Tumor necrosis factor acts as a tumor promoter in BALB/3T3 cell transformation. Cancer Res. 53:1982-1985. [PubMed] [Google Scholar]
- 25.Koochekpour, S., M. Jeffers, P. H. Wang, C. Gong, G. A. Taylor, L. M. Roessler, R. Stearman, J. R. Vasselli, W. G. Stetler-Stevenson, W. G. Kaelin, Jr., W. M. Linehan, R. D. Klausner, J. R. Gnarra, and G. F. Vande Woude. 1999. The von Hippel-Lindau tumor suppressor gene inhibits hepatocyte growth factor/scatter factor-induced invasion and branching morphogenesis in renal carcinoma cells. Mol. Cell. Biol. 19:5902-5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kroll, S. L., W. R. Paulding, P. O. Schnell, M. C. Barton, J. W. Conaway, R. C. Conaway, and M. F. Czyzyk-Krzeska. 1999. von Hippel-Lindau protein induces hypoxia-regulated arrest of tyrosine hydroxylase transcript elongation in pheochromocytoma cells. J. Biol. Chem. 274:30109-30114. [DOI] [PubMed] [Google Scholar]
- 27.Kruys, V., and G. Huez. 1994. Translational control of cytokine expression by 3′ UA-rich sequences. Biochimie 76:862-866. [DOI] [PubMed] [Google Scholar]
- 28.Latif, F., K. Tory, J. Gnarra, M. Yao, F. M. Duh, M. L. Orcutt, T. Stackhouse, I. Kuzmin, W. Modi, L. Geil, et al. 1993. Identification of the von Hippel-Lindau disease tumor suppressor gene. Science 260:1317-1320. [DOI] [PubMed] [Google Scholar]
- 29.Lieberman, A. P., P. M. Pitha, and M. L. Shin. 1990. Protein kinase regulates tumor necrosis factor mRNA stability in virus-stimulated astrocytes. J. Exp. Med. 172:989-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, S., W. Wang, G. M. Wilson, X. Yang, G. Brewer, N. J. Holbrook, and M. Gorospe. 2000. Down-regulation of cyclin D1 expression by prostaglandin A2 is mediated by enhanced cyclin D1 mRNA turnover. Mol. Cell. Biol. 20:7903-7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Locksley, R. M., N. Killeen, and M. J. Lenardo. 2001. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell 104:487-501. [DOI] [PubMed] [Google Scholar]
- 32.Los, M., G. H. Jansen, W. G. Kaelin, C. J. Lips, G. H. Blijham, and E. E. Voest. 1996. Expression pattern of the von Hippel-Lindau protein in human tissues. Lab. Investig. 75:231-238. [PubMed] [Google Scholar]
- 33.Mahtani, K. R., M. Brook, J. L. Dean, G. Sully, J. Saklatvala, and A. R. Clark. 2001. Mitogen-activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor alpha mRNA stability. Mol. Cell. Biol. 21:6461-6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maxwell, P. H., M. S. Wiesener, G.-W. Chang, S. C. Clifford, E. C. Vaux, M. E. Cockman, C. C. Wykoff, C. W. Pugh, E. R. Maher, and P. J. Ratcliffe. 1999. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399:271-275. [DOI] [PubMed] [Google Scholar]
- 35.Mukhopadhyay, D., B. Knebelmann, H. T. Cohen, S. Ananth, and V. P. Sukhatme. 1997. The von Hippel-Lindau tumor suppressor gene product interacts with Sp1 to repress vascular endothelial growth factor promoter activity. Mol. Cell. Biol. 17:5629-5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakajima, K., M. Sasaki, D. Nojima, B. R. Oh, N. Ishii, K. Miura, and R. Dahiya. 2001. Tumor necrosis factor-α gene mutations and genotype changes in renal cell carcinoma. J. Urol. 165:612-615. [DOI] [PubMed] [Google Scholar]
- 37.Ohh, M., C. W. Park, M. Ivan, M. A. Hoffman, L. E. Huang, V. Chau, and W. G. Kaelin. 2000. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. J. Biol. Chem. 275:25733-25741. [DOI] [PubMed] [Google Scholar]
- 38.Ohh, M., R. L. Yauch, K. M. Lonergan, J. M. Whaley, A. O. Stemmer-Rachamimov, D. N. Louis, B. J. Gavin, N. Kley, W. G. Kaelin, Jr., and O. Iliopoulos. 1998. The von Hippel-Lindau tumor suppressor protein is required for proper assembly of an extracellular fibronectin matrix. Mol. Cell 1:959-968. [DOI] [PubMed] [Google Scholar]
- 39.Piecyk, M., S. Wax, A. R. Beck, N. Kedersha, M. Gupta, B. Maritim, S. Chen, C. Gueydan, V. Kruys, M. Streuli, and P. Anderson. 2000. TIA-1 is a translational silencer that selectively regulates the expression of TNF-α. EMBO J. 19:4154-4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pioli, P. A., and W. F. Rigby. 2001. The von Hippel-Lindau protein interacts with heteronuclear ribonucleoprotein A2 and regulates its expression. J. Biol. Chem. 276:40346-40352. [DOI] [PubMed] [Google Scholar]
- 41.Prichett, W., A. Hand, J. Sheilds, and D. Dunnington. 1995. Mechanism of action of bicyclic imidazoles defines a translational regulatory pathway for tumor necrosis factor-α. J. Inflamm. 45:97-105. [PubMed] [Google Scholar]
- 42.Richardson, P., T. Hideshima, and K. Anderson. 2002. Thalidomide: emerging role in cancer medicine. Annu. Rev. Med. 53:629-657. [DOI] [PubMed] [Google Scholar]
- 43.Schoenfeld, A., E. Davidowitz, and R. A. Burk. 1998. A second major native von Hippel-Lindau gene product, initiated from an internal translation start site, functions as a tumor suppressor. Proc. Natl. Acad. Sci. USA 95:8817-8822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Selby, P., S. Hobbs, C. Viner, E. Jackson, A. Jones, D. Newell, A. H. Calvert, T. McElwain, K. Fearon, J. Humphreys, et al. 1987. Tumour necrosis factor in man: clinical and biological observations. Br. J. Cancer 56:803-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Semenza, G. L., and G. L. Wang. 1992. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol. Cell. Biol. 12:5447-5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stebbing, J., C. Benson, T. Eisen, L. Pyle, K. Smalley, H. Bridle, I. Mak, F. Sapunar, R. Ahern, and M. E. Gore. 2001. The treatment of advanced renal cell cancer with high-dose oral thalidomide. Br. J. Cancer 85:953-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tisdale, M. J. 1997. Biology of cachexia. J. Natl. Cancer Inst. 89:1763-1773. [DOI] [PubMed] [Google Scholar]
- 48.Tsuchiya, H., T. Iseda, and O. Hino. 1996. Identification of a novel protein (VBP-1) binding to the von Hippel-Lindau (VHL) tumor suppressor gene product. Cancer Res. 56:2881-2885. [PubMed] [Google Scholar]
- 49.Vawter, M. P., T. Barrett, C. Cheadle, B. P. Sokolov, W. H. Wood, D. Donovan, M. Webster, W. J. Freed, and K. G. Becker. 2001. Application of cDNA microarrays to examine gene expression differences in schizophrenia. Brain Res. Bull. 55:641-650. [DOI] [PubMed] [Google Scholar]
- 50.Wang, W., S. Lin, C. M. Caldwell, H. Furneaux, and M. Gorospe. 2000. HuR regulates cyclin A and cyclin B1 mRNA stability during the cell division cycle. EMBO J. 19:2340-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, W., H. Furneaux, H. Cheng, M. C. Caldwell, D. Hutter, Y. Liu, N. J. Holbrook, and M. Gorospe. 2000. HuR regulates p21 mRNA stabilization by UV light. Mol. Cell. Biol. 20:760-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Willeaume, V., V. Kruys, T. Mijatovic, and G. Huez. 1995. Tumor necrosis factor-α production induced by viruses and by lipopolysaccharides in macrophages: similarities and differences. J. Inflamm. 46:1-12. [PubMed] [Google Scholar]
- 53.Wizigmann-Voos, S., G. Breier, W. Risau, and K. Plate. 1995. Up-regulation of vascular endothelial growth factor and its receptors in von Hippel-Lindau disease-associated and sporadic hemangioblastomas. Cancer Res. 55:1358-1364. [PubMed] [Google Scholar]
- 54.Wu, S., C. M. Boyer, R. S. Whitaker, A. Berchuck, J. R. Wiener, J. B. Weinberg, and R. C. Bast, Jr. 1993. Tumor necrosis factor-α as an autocrine and paracrine growth factor for ovarian cancer: monokine induction of tumor cell proliferation and tumor necrosis factor-α expression. Cancer Res. 53:1939-1944. [PubMed] [Google Scholar]
- 55.Wykoff, C., C. Pugh, P. Maxwell, A. Harris, and P. Ratcliffe. 2000. Identification of novel hypoxia-dependent and -independent target genes of the von Hippel-Lindau (VHL) tumor suppressor by mRNA differential expression profiling. Oncogene 19:6297-6305. [DOI] [PubMed] [Google Scholar]
- 56.Yoneda, T., R. M. Lyall, M. M. Alsina, P. E. Persons, A. P. Spada, A. Levitzki, A. Zilberstein, and G. R. Mundy. 1991. The antiproliferative effects of tyrosine kinase inhibitors tyrphostins on a human squamous cell carcinoma in vitro and in nude mice. Cancer Res. 51:4430-4435. [PubMed] [Google Scholar]
- 57.Yoshida, N., S. Ikemoto, K. Narita, K. Sugimura, S. Wada, R. Yasumoto, T. Kishimoto, and T. Nakatani. 2002. Interleukin-6, tumour necrosis factor-α and interleukin-1β in patients with renal cell carcinoma. Br. J. Cancer 86:1396-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zatyka, M., N. F. da Silva, S. C. Clifford, M. R. Morris, M. S. Wiesener, K. U. Eckardt, R. S. Houlston, F. M. Richards, F. Latif, and E. R. Maher. 2002. Identification of cyclin D1 and other novel targets for the von Hippel-Lindau tumor suppressor gene by expression array analysis and investigation of cyclin D1 genotype as a modifier in von Hippel-Lindau disease. Cancer Res. 62:3803-3811. [PubMed] [Google Scholar]
- 59.Zhou, M. I., H. Wang, J. J. Ross, I. Kuzmin, C. Xu, and H. T. Cohen. 2002. The von Hippel-Lindau (VHL) tumor suppressor stabilizes novel PHD protein Jade-1. J. Biol. Chem. 277:39887-39898. [DOI] [PubMed] [Google Scholar]
- 60.Zong, Q., M. Schummer, L. Hood, and D. R. Morris. 1999. Messenger RNA translation state: the second dimension of high-throughput expression screening. Proc. Natl. Acad. Sci. USA 96:10632-10636. [DOI] [PMC free article] [PubMed] [Google Scholar]