Abstract
DNA polymerases are defined as such because they use deoxynucleotides instead of ribonucleotides with high specificity. We show here that polymerase mu (pol μ), implicated in the nonhomologous end-joining pathway for repair of DNA double-strand breaks, incorporates both ribonucleotides and deoxynucleotides in a template-directed manner. pol μ has an approximately 1,000-fold-reduced ability to discriminate against ribonucleotides compared to that of the related pol β, although pol μ's substrate specificity is similar to that of pol β in most other respects. Moreover, pol μ more frequently incorporates ribonucleotides when presented with nucleotide concentrations that approximate cellular pools. We therefore addressed the impact of ribonucleotide incorporation on the activities of factors required for double-strand break repair by nonhomologous end joining. We determined that the ligase required for this pathway readily joined strand breaks with terminal ribonucleotides. Most significantly, pol μ frequently introduced ribonucleotides into the repair junctions of an in vitro nonhomologous end-joining reaction, an activity that would be expected to have important consequences in the context of cellular double-strand break repair.
A large number of mammalian DNA polymerases have been identified within the last 3 years, and initial characterization suggests that they possess a surprisingly diverse array of substrate specificities. We investigate here the substrate specificity of one of these recently described polymerases, polymerase mu (pol μ).
pol μ, in addition to pol β, pol λ, pol σ, and terminal deoxynucleotidyltransferase (TdT), is a member of the Pol X family (5). pol μ is most similar to TdT; the two polymerases have similar domain organizations and are ∼40% identical in sequence, but both are less than 25% identical to remaining Pol X family members (e.g., pol β) (7). Moreover, pol μ and TdT are likely both involved in the nonhomologous end-joining (or end-joining) pathway for repair of double-strand breaks (DSBs) in vertebrates. We have recently demonstrated that pol μ and TdT form essentially identical complexes with the end-joining factors Ku and the XRCC4-ligase IV complex (X4-LIV) (14). End joining is a major pathway for repair of DSBs introduced by exogenous sources (e.g., ionizing radiation) in all cell types and is required for resolution of DSB intermediates in V(D)J recombination, the lymphocyte-specific process required for assembly of mature antigen receptor genes (reviewed in reference 9). TdT is expressed only in cells active in V(D)J recombination (reviewed in reference 10) and thus participates in end-joining reactions only in this context. In contrast, the more widely expressed pol μ seems likely to play an important role in general end-joining DSB repair.
TdT's substrate specificity is uniquely suited for its role in promoting diversity in V(D)J recombination. TdT is template independent, adding random nucleotides only to 3′ single-stranded or blunt DNA ends (reviewed in reference 6). Indeed, TdT shows generally reduced specificity in substrate selection, efficiently utilizing even triphosphate esters with nonnucleoside groups (e.g., p-nitrophenylethyl triphosphate) (2). TdT thus can incorporate both ribonucleotides and deoxynucleotides during synthesis with similar efficiency in vitro (4, 12, 20, 21). Ribonucleotide incorporation may be tolerated in vivo due to the combination of TdT's highly restricted expression pattern and the beneficial impact of this enzyme on diversification of the immune repertoire.
In contrast, pol μ is apparently more ubiquitously expressed (1, 7). Moreover, in most respects pol μ's substrate specificity bears more similarity to that of pol β, in that pol μ is also primarily a template-directed (7, 26), gap-filling (as demonstrated below) polymerase. This is consistent with a role for pol μ in promoting accuracy in general end-joining DSB repair. However, we show here that pol μ incorporates both deoxynucleotides and ribonucleotides opposite DNA templates with similar efficiency. Such relaxed sugar selectivity has previously been observed for template-dependent DNA polymerases only when nonphysiological cations (Mn2+) are used or following polymerase mutation (reviewed in reference 11). Furthermore, we show that pol μ is primarily an RNA polymerase using typical cellular nucleotide pools and remains primarily an RNA polymerase in the context of an in vitro end-joining reaction.
MATERIALS AND METHODS
Purified proteins.
The cloning, expression, and purification of human versions of pol μ and TdT (14) and Ku and the X4-LIV complex (16) have been previously described. A predicted catalytic mutant of human pol μ (D330E/D332E) was generated by using QuikChange site-directed mutagenesis (Stratagene) and purified as was wild-type pol μ. Highly purified DNA-dependent protein kinase catalytic subunit (DNA-PKcs) (>95% pure) was recovered from human placenta (D. Ramsden and V. Yeturu, unpublished data). pol β was purified after overexpression in Escherichia coli (gift of S. Wilson, National Institute of Environmental Health Sciences [NIEHS]) as previously described (17).
Oligonucleotide substrates.
RNA (Dharmacon Research) and DNA (University of North Carolina [UNC] Nucleic Acids Core Facility) oligonucleotides were gel purified. All polymerization substrates were based on a 1-nucleotide (nt)-gapped DNA substrate (shown schematically in Fig. 1C, “1 nt gap”) generated by annealing a primer strand (5′-GCTTGAAGACTGGTGAAGACTTGAG-3′) and a downstream strand (5′-TACAGGTCGATTCATGGAGT-3′) to a template strand (5′-CCATGAATCGACCTGTACCTCAAGTCTTCACCAGTCTTCA-3′). Substrates were 5′ phosphorylated by T4 kinase (New England BioLabs) with [γ-32P]ATP on the primer strand and cold ATP on the downstream strand, except for experiments in Fig. 2 and Table 2, where only the downstream strand was phosphorylated with [γ-32P]ATP, and Table 1, where only the downstream strand was phosphorylated with cold ATP. The templating base was varied in experiments for Fig. 1B and Tables 1 and 2 by replacing the C at position 18 (underlined in the sequence above) with A, G, or T. Frameshift synthesis at dinucleotide repeats (26) was prevented in Fig. 1B by changing the base pair directly 3′ to the gap from T/A to C/G (primer/template) for template A and the base pair directly 5′ to the gap from G/C to C/G for template C. Substrates in Fig. 1C were assembled by annealing alternate downstream strands shorter by 1 (2-nt gap) or 2 (3-nt gap) nt relative to the original downstream strand, by not phosphorylating the downstream strand (no 5′-P), by not including the downstream strand (recess), or by replacing the three different DNA strands with RNA or mixed nucleic acid versions as noted. Nick ligation substrates (Fig. 3A) were generated by annealing the standard primer and downstream strands to a template strand with the nucleotide at position 18 of the original template sequence removed, eliminating the 1-nt gap.
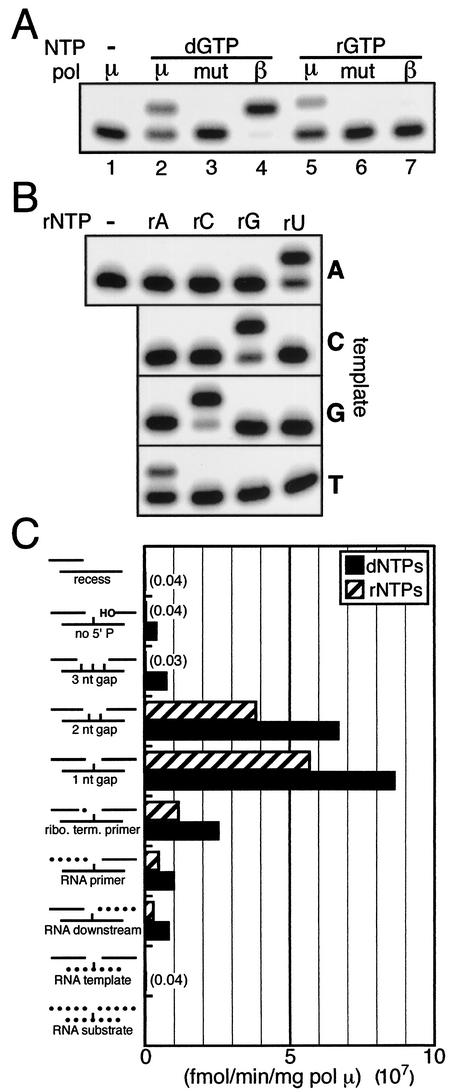
FIG. 1.
Substrate specificity of pol μ. (A) A single-nucleotide-gapped DNA substrate with template C was present (5 nM) in all reaction mixtures (diagrammed in panel C; “1 nt gap”). dGTP or rGTP (100 μM) was included as indicated. Wild-type pol μ (μ; 0.5 nM), pol β (β; 0.5 nM), or mutant pol μ (mut; 50 nM) was added as noted. Reaction time was 1 min. (B) pol μ (5 nM), rNTP substrate (1 mM), and 1-nt-gapped DNA substrate (5 nM) (as in panel A) with either A, C, G, or T as the templating base were included as indicated. Reaction time was 1 min. (C) The specific activity of pol μ (femtomoles of product per minute per milligram of pol μ) was determined with 5 nM nucleic acid substrate and a 25 μM concentration of each of the four dNTPs or rNTPs. Concentrations of pol μ and reaction times were varied such that reactions were in the linear range. Dotted lines indicate RNA strands substituted for DNA in the standard 1-nt-gappedsubstrate. The means of three replicate experiments performed in the linear range are shown. The standard deviation was less than 15% of the mean for all substrates tested.
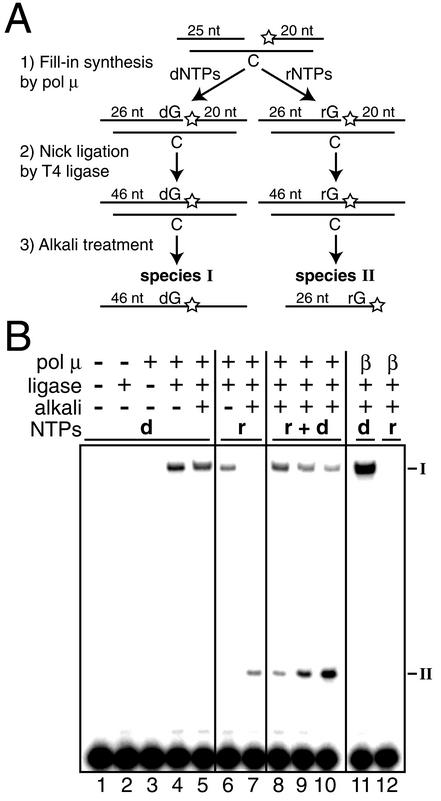
FIG. 2.
pol μ ribonucleotide incorporation with mixed nucleotide pools. (A) Alkali cleavage assay for incorporation of ribonucleotides. A single-nucleotide-gapped DNA substrate with template C was 5′ 32P labeled as indicated (star). Alkali treatment of products containing incorporated ribonucleotides results in product cleavage, as well as transfer of the 32P from the 20-nt downstream strand to the primer strand, generating a novel 26-nt species (species II). (B) DNA substrate (5 nM) as in panel A was present in all reaction mixtures. Where indicated, 0.05 U of T4 ligase (+) and 0.5 nM pol μ (+) or 5 pM pol β (β) were added. Following a 1-min polymerization-ligation reaction, samples were treated with alkali as noted (+). A 25 μM concentration of each dNTP (d) or rNTP (r) was present as noted. Reaction mixtures 8 to 10 contained a mixture of both dNTPs and rNTPs (d + r), with 25 μM each dNTP and 25 μM (lane 8), 125 μM (lane 9), or 500 μM (lane 10) each rNTP. I, species I; II, species II.
TABLE 2.
Ribonucleotide incorporation by pol μ with mixed nucleotide pools
TABLE 1.
Kinetic measurement of templateda nucleotide incorporation by pol X polymerases
| pol | NTP | Km (μM) | kcat (s−1) | kcat/Km (mM−1 s−1) | Sugar selectivityb |
|---|---|---|---|---|---|
| pol μ | dATP | 15.6 ± 0.7 | 0.12 ± 0.01 | 7.9 ± 1.2 | 11 |
| rATP | 114 ± 15 | 0.08 ± 0.02 | 0.70 ± 0.19 | ||
| dCTP | 40.2 ± 9.2 | 0.12 ± 0.03 | 3.13 ± 0.49 | 1.4 | |
| rCTP | 78 ± 25 | 0.16 ± 0.02 | 2.17 ± 0.45 | ||
| dGTP | 11.6 ± 1.5 | 0.54 ± 0.12 | 46.5 ± 5.8 | 4.0 | |
| rGTP | 24.1 ± 6.4 | 0.28 ± 0.05 | 11.7 ± 1.0 | ||
| dTTP | 31.3 ± 5.7 | 0.21 ± 0.01 | 7.0 ± 1.9 | 7.2 | |
| rUTP | 189 ± 24 | 0.18 ± 0.02 | 0.97 ± 0.04 | ||
| TdT | dATP | 113 ± 17 | 0.74 ± 0.18 | 6.6 ± 1.6 | 8.9 |
| rATP | 108 ± 10 | 0.08 ± 0.01 | 0.74 ± 0.15 | ||
| dCTP | 254 ± 66 | 1.11 ± 0.05 | 4.51 ± 0.87 | 2.6 | |
| rCTP | 300 ± 8.4 | 0.53 ± 0.01 | 1.76 ± 0.05 | ||
| pol β | dATP | 2.40 ± 0.35 | 0.71 ± 0.13 | 299 ± 84 | 6,000 |
| rATP | 1,040 ± 430 | 0.05 ± 0.03 | 0.05 ± 0.02 | ||
| dCTP | 0.32 ± 0.01 | 0.81 ± 0.01 | 2,540 ± 95 | 2,000 | |
| rCTP | 62.1 ± 1.6 | 0.08 ± 0.02 | 1.25 ± 0.29 | ||
| rGTP | 606 ± 280 | 0.07 ± 0.03 | 0.11 ± 0.03 | NDc | |
| rUTP | 1,320 ± 30 | 0.08 ± 0.01 | 0.06 ± 0.01 | ND |
Using standard 1-nt-gapped substrates (Fig. 1), with template base varied for each tested NTP as appropriate; detectable pol μ and pol β activity was template dependent, while TdT activity was independent of the template base, as expected. Data are means (± standard deviations) from replicate experiments.
Sugar selectivity = [(kcat/Km)dNTP]/[(kcat/Km)rNTP].
ND, not determined.
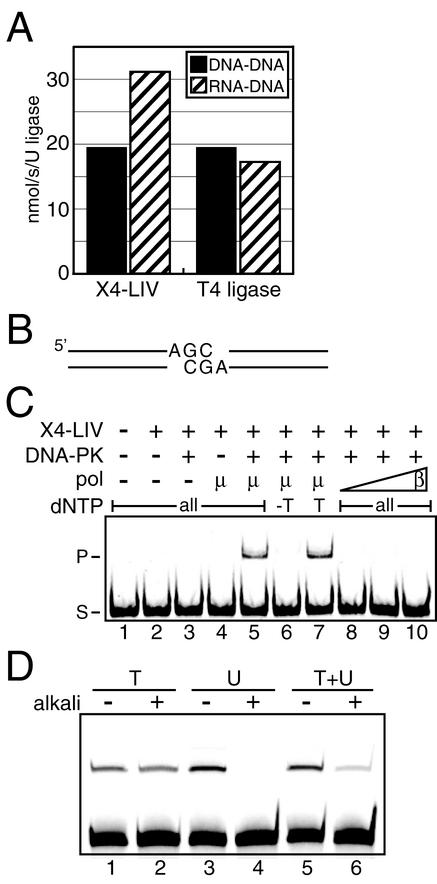
FIG. 3.
pol μ ribonucleotide incorporation during end joining. (A) DNA-DNA, nicked DNA substrate; RNA-DNA, nicked DNA substrate with a single ribonucleotide at the 3′ terminus of the upstream strand. Nick ligation activity is shown as nanomoles of product per second per unit of ligase. The means of three replicate experiments are shown. The standard deviation was less than 10% of the mean for all substrates tested. (B) The 300-bp DNA end-joining substrate, aligned as required to form head-to-head ligation products as generated in panels C and D. (C) A 20 nM concentration of DNA substrate as in panel B was present in all reaction mixtures. Purified X4-LIV (50 nM) and DNA-PK (25 nM Ku plus 25 nM DNA-PKcs) were added as indicated (+). pol μ (μ; 25 nM) or pol β (β; 10, 25, or 50 nM [lanes 8 to 10, respectively]) was added as noted. All, 25 μM each dNTP; −T, 25 μM each dATP, dCTP, and dGTP; T, 25 μM dTTP only; S, substrate; P, ligation product. (D) All reactions as in panel C, lane 7, except that 25 μM dTTP (T), 500 μM rUTP (U), or 25 μM dTTP plus 500 μM rUTP (T+U) was included as noted. Following an in vitro end-joining reaction, samples were treated with alkali as indicated (+).
Polymerase activity assays.
32P-labeled DNA substrate (5 nM) was incubated in 10 μl of standard reaction buffer (25 mM Tris [pH 7.5], 100 mM NaCl, 25 mM KCl, 0.1 mM EDTA, 5 mM MgCl2, 0.05% Triton X-100, 50 μg of bovine serum albumin/ml, 1% glycerol, and 2 mM dithiothreitol [DTT]). Deoxynucleoside triphosphates (dNTPs) or ribonucleoside triphosphates (rNTPs) were added as noted. Reactions were initiated by addition of the indicated amount of polymerase and transfer to 37°C, were stopped by addition of an equal volume of formamide loading dye, and were analyzed by electrophoresis on a denaturing 10% polyacrylamide gel (denaturing 10% polyacrylamide gel electrophoresis).
Experiments in Fig. 2 were supplemented with 0.1 mM ATP and 0.05 U of T4 ligase (Roche). After reactions were stopped with 20 mM EDTA, KOH was added to 0.3 M, and reaction mixtures were incubated for 2 h at 55°C and analyzed by denaturing PAGE as described above. Experiments in Table 2 were performed as for Fig. 2, except that a mix of all eight nucleotides at the concentrations noted in Table 2 was added (25), MgCl2 was added to 10 mM, and the reaction time was 5 min. All detection and quantification of radiolabeled DNA were performed with a PhosphorImager and ImageQuanNT software (Molecular Dynamics).
Kinetic assays were performed in standard reaction buffer with 1-nt-gapped substrates (50 μM) that were identical except for the templating base (dA, dC, dG, or dT) and various concentrations of the complementary [α-32P]dNTP or [α-32P]rNTP. In order to more accurately compare the activities of the three polymerases tested, we used DNA substrates that contained both a 1-nt gap (the preferred substrate of pol μ and pol β) and a terminal 3′ overhang (the preferred substrate of TdT). Importantly, we confirmed that with this substrate and under the conditions used here essentially all activity, with dNTPs or rNTPs, of both pol β and pol μ was at the gap and template dependent, while essentially all activity of TdT was template independent (S. A. Nick McElhinny and D. A. Ramsden, unpublished data). Each experiment tested six different concentrations of the complementary nucleotide, with a minimum range of 0.2 to 6 × Km (except for pol β with rNTPs, where Mg2+ concentrations limited the concentration of rNTPs to a maximum of 2.5 mM). For each nucleotide concentration used, five time points were stopped by 30-fold dilution into 50 mM EDTA-5% Na2HPO4 and applied to DE81 Whatman paper with a Bio-Rad Bio-Dot 96-well apparatus, and unincorporated nucleotide was removed by washing with 5% Na2HPO4. Incorporated radiolabeled nucleotide was detected and quantified with a PhosphorImager and ImageQuanNT software. Km and kcat values were determined by fitting data to an Eadie-Hofstee plot (v versus v/[S]). Standard deviations were determined from replicate experiments.
Ligation assays.
For nick ligation reactions, 5 nM 32P-labeled DNA substrate was incubated at 37°C in 10 μl of standard reaction buffer supplemented with 0.1 mM ATP and various concentrations of ligase such that reactions were in the linear range. One unit of T4 ligase (Roche) was equivalent to ∼350 μg of X4-LIV.
The end-joining substrate described for Fig. 3B was generated by PCR amplification in the presence of [α-33P]dCTP of a 300-bp fragment including the Jκ1 coding region with the primers 5′-GCTCACGCTGTGGACGTTCGGTGGAGGC-3′ and 5′-GGCTACCCTGCTTCTTTGAGC-3′, digestion of this fragment with DraIII (recognition site embedded in the sequence of the first primer) to generate the described 3′ overhang, and purification of the digested fragment with a QIAquick PCR purification kit (Qiagen). Ku and DNA-PKcs were preincubated with 20 nM DNA substrate for 15 min in a buffer containing 25 mM Tris (pH 7.5), 75 mM NaCl, 72.5 mM KCl, 2 mM DTT, 50 μg of bovine serum albumin/ml, 0.025% Triton X-100, 5% glycerol, 0.1 mM EDTA, and 10% (wt/vol) polyethylene glycol (molecular mass, >8,000 kDa). Ligation was initiated by addition of polymerase, X4-LIV, dNTPs and/or rNTPs as noted, MgCl2 to 5 mM, 50 ng of plasmid competitor, and 0.1 mM ATP. Reaction mixtures were incubated at 37°C for 10 min, reactions were stopped, and reaction mixtures were deproteinized prior to analysis by either native 6% PAGE (Fig. 3C) or denaturing 5% PAGE with 25% formamide (Fig. 3D). Alkali treatment was performed as described for Fig. 2 experiments. Radiolabeled DNA was detected and quantified with a PhosphorImager and ImageQuant software.
RESULTS
Substrate specificity of pol μ.
Polymerases are characterized as either DNA or RNA polymerases based on whether they selectively utilize dNTPs or rNTPs as substrate. We show here that purified pol μ uses both dGTP and rGTP for synthesis opposite template C with similar efficiency (Fig. 1A, lanes 2 and 5; also Table 1). Similar results were observed with a variety of conditions (pH 7 to 8) and with different divalent metal cation cofactors, including Mg2+, Co2+, Mn2+, or a combination of Mg2+ and Zn2+ (negligible activity was observed with Zn2+ alone) (Nick McElhinny and Ramsden, unpublished). Experiments shown here were performed with a pH of 7.5 and the most relevant cofactor in cells (Mg2+). pol μ thus possesses both DNA and RNA polymerase activities. RNA polymerase activity is not a contaminant of our pol μ preparation, as a similarly prepared catalytic mutant of pol μ was inactive irrespective of the type of NTP added, even when 100-fold-higher concentrations of polymerase were used (Fig. 1A, lanes 3 and 6). In contrast to pol μ, RNA polymerase activity by the related pol β was negligible under these conditions (Fig. 1A, lane 7). pol μ RNA polymerase activity is also template directed (Fig. 1B), as previously shown for its DNA polymerase activity (7, 26; Nick McElhinny and Ramsden, unpublished).
We next addressed the nucleic acid substrate specificity of pol μ, under conditions designed to stringently assess pol μ's ability to interact with nucleic acid. Activities on 1- and 2-nt-gapped DNA substrates were comparable but were reduced 10-fold on a 3-nt gap and ∼100-fold in the absence of a downstream strand (recessed end) (Fig. 1C). pol μ is therefore primarily a short-gap-filling polymerase. Like pol β (23), pol μ appears to interact with the 5′-phosphate of the downstream strand, as removal of this phosphate results in a 20-fold reduction in activity (Fig. 1C, compare “1 nt gap” to “no 5′ P”).
Since pol μ shows reduced sugar selectivity with respect to NTP substrate, we determined if pol μ showed similarly reduced sugar selectivity with respect to nucleic acid substrate. The specific activity of pol μ was reduced 10- to 15-fold when either the primer or downstream strand of a gapped DNA substrate was replaced with RNA (Fig. 1C, compare “1 nt gap” to “RNA primer” and “RNA downstream”). However, pol μ activity was most dramatically affected by the presence of an RNA template, with activity decreased more than 100-fold (Fig. 1C, compare “1 nt gap” to “RNA template” and “RNA substrate”). pol μ is thus a DNA-directed DNA/RNA polymerase.
We next asked if ribonucleotide incorporation inhibited further synthesis by pol μ (are ribonucleotides chain terminators?). A substrate with a DNA primer terminating in a single 3′ ribonucleotide caused an approximately fourfold reduction in activity relative to an entirely DNA substrate (Fig. 1C, compare “1 nt gap” to “ribo. term. primer”). Ribonucleotide incorporation by pol μ therefore has only a mild chain-terminating effect.
To quantitatively address the sugar selectivity of pol μ, we determined the catalytic efficiency of pol μ on its favored substrate, a 1-nt-gapped DNA duplex, by using steady-state kinetics. For comparison, the catalytic efficiencies of TdT and pol β were determined in parallel, and all three enzymes were assayed using DNA substrates with both a 1-nt gap (the preferred substrate of pol μ and pol β) and a terminal 3′ overhang (the preferred substrate of TdT). We further confirmed that under the conditions used here pol μ was active only at the gap and was template dependent (had no detectable terminal transferase activity) (Nick McElhinny and Ramsden, unpublished). The catalytic efficiencies of pol μ for dNTPs relative to rNTPs (sugar selectivities) favor utilization of dNTPs only 1.4- to 11-fold, depending on the identity of the base (Table 1). Strikingly similar results are observed for A and C NTPs with TdT (Table 1), though TdT activity was template independent, as expected (Nick McElhinny and Ramsden, unpublished). In contrast, the corresponding sugar selectivities for pol β are approximately 500- to 1,000-fold higher (Table 1). The reduced sugar selectivities of pol μ and TdT relative to those of pol β are primarily due to reduced (∼50- to 100-fold) catalytic efficiency with dNTPs (pol μ and TdT are less effective DNA polymerases). However, pol μ and TdT are also more effective RNA polymerases than pol β, with ∼1.5- to 100-fold-greater catalytic efficiency with rNTPs.
pol μ ribonucleotide incorporation with mixed nucleotide pools.
In cells, pol μ will have both dNTPs and rNTPs available, with rNTPs typically 10- to 200-fold more abundant than dNTPs (Table 2) (25). We therefore developed an assay to determine relative levels of ribonucleotide and deoxynucleotide incorporation when pol μ is given a mixed pool of both dNTPs and rNTPs. Concurrent polymerization and ligation reactions were performed on a single-nucleotide-gapped DNA substrate, generating either a chain entirely composed of DNA if pol μ used a dNTP as substrate or a DNA chain interrupted by a single ribonucleotide if pol μ used an rNTP (Fig. 2A). Treatment of such a mixed polynucleotide chain with alkali and heat causes the 2′-hydroxyl of the ribonucleotide to cleave the adjacent phosphodiester bond, producing a strand break with 3′-phosphate and 5′-hydroxyl termini. Alkali treatment of the polymerization-ligation reaction products had no effect if pol μ incorporated a deoxynucleotide (Fig. 2, species I, compare lanes 4 and 5 in panel B) but generated a novel cleaved species (species II) if pol μ incorporated a ribonucleotide (Fig. 2, species II, compare lanes 6 and 7 in panel B). The amount of ribonucleotide incorporation relative to deoxynucleotide incorporation is therefore directly proportional to the amount of species II relative to species I.
We determined that the amount of ribonucleotide incorporation by pol μ increased in parallel with the ratio of rNTP to dNTP concentration, increasing from 26% when the two were present at equal concentrations to 81% ribonucleotide incorporation when rNTPs were present at a 20-fold excess over dNTPs (Fig. 2B, lanes 8 to 10; Table 2). As shown above (Fig. 1A), pol β had negligible activity in the presence of rNTPs (Fig. 2B, compare lanes 7 and 12).
The cellular ratios of rNTP to dNTP concentrations (Table 2) (25), as well as the sugar selectivities of pol μ (Table 1), vary widely according to base identity. We therefore determined the frequency of ribonucleotide incorporation for all four template bases using a mix of the eight common NTPs. The concentrations for each nucleotide in this mix (Table 2) were equal to average concentrations as determined from sampling a wide variety of mammalian cell types (25). Ribonucleotides were incorporated over 70% of the time in all cases (Table 2), indicating that pol μ is primarily an RNA polymerase when nucleotide concentrations approximate cellular pools.
pol μ ribonucleotide incorporation during end joining.
Our recent evidence favors a role for pol μ in repairing gaps prior to the ligation step in end-joining DSB repair (14). However, ribonucleotide incorporation by pol μ could block end joining, as X4-LIV, the ligase required for end joining, is the only mammalian ligase unable to join two RNA strands (19). We show here that X4-LIV is slightly more active (1.60- ± 0.05-fold) on a nicked DNA substrate with a single ribonucleotide substituted at the 3′ terminus of the upstream strand (Fig. 3A), a substrate with more relevance to end joining than two wholly RNA strands. Ribonucleotide incorporation by pol μ is thus compatible with X4-LIV activity.
pol μ and TdT form a functional complex with end-joining factors that could induce a conformational change in pol μ and TdT, preventing their ability to utilize rNTPs. We therefore tested the ability of pol μ to incorporate ribonucleotides in the context of an in vitro end-joining reaction. We used a 300-bp linear DNA substrate with one 3′ overhang (Fig. 3B), constructed such that a head-to-head ligation product is formed only by the combined activity of X4-LIV, DNA-PK (Ku and DNA-PKcs), and a DNA polymerase, in the presence of the appropriate NTP (Fig. 3C, lanes 1 to 7). pol β was unable to substitute for pol μ under these conditions (Fig. 3C, compare lane 5 to lanes 8 to 10), arguing that joining is dependent on the ability of pol μ to form a specific complex with end-joining factors. Substitution of rUTP for dTTP at typical physiological concentrations supported joining with slightly elevated efficiency (Fig. 3D, compare lane 1 to lane 3), indicating that pol μ can incorporate ribonucleotides when complexed with end-joining factors.
We then tested joining in the presence of a mixed pool of dTTP and rUTP, again using physiologically appropriate nucleotide concentrations, and assessed levels of ribonucleotide incorporation by alkali treatment of reaction products. As before, only chains with incorporated ribonucleotides were sensitive to alkali treatment (Fig. 3D, compare lane 2 to lane 4). Significantly, when both dTTP and rUTP were present, more than 75% of reaction products were cleaved upon alkali treatment (Fig. 3D, compare lane 5 to lane 6). A ribonucleotide (rU) was thus incorporated in 75% of all end-joining products, essentially the same ratio of rU/dT incorporation observed in the gap-filling assay (Table 2, template A). RNA polymerase activity of pol μ is thus in no way inhibited in the context of an end-joining reaction.
DISCUSSION
pol μ in vitro is a DNA template-directed, gap-filling polymerase. These characteristics, in addition to the ability of pol μ to direct synthesis from very short aligned regions of complementary sequence (Fig. 3C) (14, 26), are consistent with pol μ's putative role in repairing gaps during end-joining DSB repair (14).
pol μ's template-directed gap-filling activity is similar to that of pol β, while the more closely related TdT is template independent and most active on 3′ overhangs. However, unlike pol β, both pol μ and TdT efficiently use rNTPs for synthesis, and both form complexes with the end-joining factors Ku and X4-LIV. With respect to TdT, it could be argued that ribonucleotide incorporation is simply a consequence of overall reduced nucleotide specificity; this might be tolerated in cells due to TdT's limited expression pattern and the importance of its reduced nucleotide specificity in generating antigen receptor diversity. In contrast, pol μ specifically recognizes nucleotide substrate by incorporating the appropriate template-directed base. pol μ is also widely expressed, and evidence supports a general role in promoting templated DSB repair (14). This suggests that ribonucleotide incorporation may be a property specific to, and thus beneficial for, DNA repair synthesis in the context of cellular end joining.
Are pol μ and TdT primarily RNA polymerases in cells? Our data argue that, at the very least, pol μ and TdT will incorporate ribonucleotides in vivo far more often than other polymerases (e.g., pol β). Furthermore, we present evidence against the possibility that ribonucleotide incorporation by pol μ is reduced in the context of end joining (e.g., by allostery). In principle, ribonucleotide incorporation by these enzymes could be reduced in cells if local nucleotide pools were somehow manipulated to favor use of dNTPs. For example, it has been proposed that dNTPs are directly channeled to the replication machinery in S phase. However, most evidence does not support a channeling mechanism for replication (reviewed in reference 18). The evolution of a channeling mechanism specific to end joining seems even less likely, given that completing an end-joining repair reaction requires far fewer dNTPs than does completing replication of the genome.
Regulation of nucleotide pools in mammalian nuclei during the cell cycle suggests a possible benefit of RNA polymerase activity. Most DNA-directed polymerases are active in S phase, when nuclear dNTP pools are highest. End joining, however, is the predominant repair pathway in G1 (24) and likely noncycling cells as well, when nuclear dNTP pools are lowest (3- to 20-fold lower than the average values listed in Table 2) (3, 25). Since rNTP pools remain high throughout the cell cycle and in noncycling cells (15, 25), pol μ and TdT may have evolved RNA polymerase activity to be more effective under conditions when end joining is most active. At the same time, the mild chain-terminating effect of RNA incorporation, together with the low catalytic efficiency of pol μ and TdT, limits these polymerases to short tracts of synthesis. This will also benefit end joining, as more efficient polymerases (even pol β) are more likely to destroy the alignment of partially complementary overhangs (such as those shown in Fig. 3B) by displacing the downstream strand of the aligned end. Ribonucleotides incorporated in DNA might also be beneficial, as they could “flag” sites of repair, possibly to activate a checkpoint response or target other repair mechanisms (see, e.g., below).
Conversely, ribonucleotide incorporation by pol μ and TdT may have negative consequences. Ribonucleotides incorporated in genomic DNA will be more likely to generate mutations and strand breaks (13) and may be inappropriately targeted by RNA-metabolizing enzymes (e.g., RNA-editing enzymes). Additionally, replicative polymerases cannot use wholly RNA templates (see, e.g., reference 13); thus, DNA replication might be expected to arrest at chromosomal sites of end-joining repair with embedded ribonucleotides. A replication block could be avoided if the ribonucleotide was bypassed by a translesion polymerase, but a more likely possibility is that the inserted ribonucleotide is removed from the genome by an excision repair mechanism argued to specifically remove such misincorporated ribonucleotides (8, 22). A single ribonucleotide embedded in a DNA duplex substrate (exactly as generated in the end-joining reactions described for Fig. 3) is precisely and efficiently removed in eukaryotic cell extracts by the combined action of a type 2 RNase H, which makes a nick 5′ of the inserted ribonucleotide (8, 22), and FEN-1, which cleaves the bond 3′ of the ribonucleotide (22).
Experiments presented here argue that pol μ, and likely TdT as well, will frequently introduce ribonucleotides into sites of end-joining DSB repair in cells. We suggest that this property is advantageous to end joining, as it allows repair synthesis to proceed when dNTP pools are low and limits repair synthesis to short tracts. Possible disadvantages of RNA polymerase activity (e.g., a replication block) may be avoided if ribonucleotides are removed by a cellular repair pathway already in place (8, 22). Future experiments characterizing and manipulating ribonucleotide incorporation by pol μ and TdT in vivo will be required to further explore the biological significance of this activity.
Acknowledgments
We thank S. Wilson (NIEHS) for providing the pol β expression strain, R. Prasad (NIEHS) for advice in expression and purification of pol β, T. Traut (UNC) and K. Weeks (UNC) for helpful discussion, and A. Sancar (UNC) and M. Cordeiro-Stone (UNC) for critical reading of the manuscript.
This work was supported by American Cancer Society grant RSG-03-045-01-GMC to D.A.R. and a National Science Foundation Graduate Research Fellowship to S.A.N.M.
REFERENCES
- 1.Aoufouchi, S., E. Flatter, A. Dahan, A. Faili, B. Bertocci, S. Storck, F. Delbos, L. Cocea, N. Gupta, J. C. Weill, and C. A. Reynaud. 2000. Two novel human and mouse DNA polymerases of the polX family. Nucleic Acids Res. 28:3684-3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arzumanov, A. A., L. S. Victorova, M. V. Jasko, D. S. Yesipov, and A. A. Krayevsky. 2000. Terminal deoxynucleotidyl transferase catalyzes the reaction of DNA phosphorylation. Nucleic Acids Res. 28:1276-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjursell, G., and L. Skoog. 1980. Control of nucleotide pools in mammalian cells. Antibiot. Chemother. 28:78-85. [DOI] [PubMed] [Google Scholar]
- 4.Boule, J. B., F. Rougeon, and C. Papanicolaou. 2001. Terminal deoxynucleotidyl transferase indiscriminately incorporates ribonucleotides and deoxyribonucleotides. J. Biol. Chem. 276:31388-31393. [DOI] [PubMed] [Google Scholar]
- 5.Burgers, P. M., E. V. Koonin, E. Bruford, L. Blanco, K. C. Burtis, M. F. Christman, W. C. Copeland, E. C. Friedberg, F. Hanaoka, D. C. Hinkle, C. W. Lawrence, M. Nakanishi, H. Ohmori, L. Prakash, S. Prakash, C. A. Reynaud, A. Sugino, T. Todo, Z. Wang, J. C. Weill, and R. Woodgate. 2001. Eukaryotic DNA polymerases: proposal for a revised nomenclature. J. Biol. Chem. 276:43487-43490. [DOI] [PubMed] [Google Scholar]
- 6.Chang, L. M., and F. J. Bollum. 1986. Molecular biology of terminal transferase. Crit. Rev. Biochem. 21:27-52. [DOI] [PubMed] [Google Scholar]
- 7.Dominguez, O., J. F. Ruiz, T. Lain de Lera, M. Garcia-Diaz, M. A. Gonzalez, T. Kirchhoff, A. C. Martinez, A. Bernad, and L. Blanco. 2000. DNA polymerase mu (Pol mu), homologous to TdT, could act as a DNA mutator in eukaryotic cells. EMBO J. 19:1731-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eder, P. S., and J. A. Walder. 1991. Ribonuclease H from K562 human erythroleukemia cells. Purification, characterization, and substrate specificity. J. Biol. Chem. 266:6472-6479. [PubMed] [Google Scholar]
- 9.Gellert, M. 2002. V(D)J recombination: rag proteins, repair factors, and regulation. Annu. Rev. Biochem. 71:101-132. [DOI] [PubMed] [Google Scholar]
- 10.Gilfillan, S., C. Benoist, and D. Mathis. 1995. Mice lacking terminal deoxynucleotidyl transferase: adult mice with a fetal antigen receptor repertoire. Immunol. Rev. 148:201-219. [DOI] [PubMed] [Google Scholar]
- 11.Joyce, C. M. 1997. Choosing the right sugar: how polymerases select a nucleotide substrate. Proc. Natl. Acad. Sci. USA 94:1619-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato, K. I., J. M. Goncalves, G. E. Houts, and F. J. Bollum. 1967. Deoxynucleotide-polymerizing enzymes of calf thymus gland. II. Properties of the terminal deoxynucleotidyltransferase. J. Biol. Chem. 242:2780-2789. [PubMed] [Google Scholar]
- 13.Kornberg, A., and T. Baker. 1992. DNA replication, 2nd ed. W. H. Freeman and Company, New York, N.Y.
- 14.Mahajan, K. N., S. A. Nick McElhinny, B. S. Mitchell, and D. A. Ramsden. 2002. Association of DNA polymerase μ (pol μ) with Ku and ligase IV: role for pol μ in end-joining double-strand break repair. Mol. Cell. Biol. 22:5194-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCormick, P. J., L. L. Danhauser, Y. M. Rustum, and J. S. Bertram. 1983. Changes in ribo- and deoxyribonucleoside triphosphate pools within the cell cycle of a synchronized mouse fibroblast cell line. Biochim. Biophys. Acta 756:36-40. [DOI] [PubMed] [Google Scholar]
- 16.Nick McElhinny, S. A., C. M. Snowden, J. McCarville, and D. A. Ramsden. 2000. Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol. Cell. Biol. 20:2996-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prasad, R., A. Kumar, S. G. Widen, J. R. Casas-Finet, and S. H. Wilson. 1993. Identification of residues in the single-stranded DNA-binding site of the 8-kDa domain of rat DNA polymerase beta by UV cross-linking. J. Biol. Chem. 268:22746-22755. [PubMed] [Google Scholar]
- 18.Reichard, P. 1988. Interactions between deoxyribonucleotide and DNA synthesis. Annu. Rev. Biochem. 57:349-374. [DOI] [PubMed] [Google Scholar]
- 19.Robins, P., and T. Lindahl. 1996. DNA ligase IV from HeLa cell nuclei. J. Biol. Chem. 271:24257-24261. [DOI] [PubMed] [Google Scholar]
- 20.Roychoudhury, R. 1972. Enzymic synthesis of polynucleotides. Oligodeoxynucleotides with one 3′-terminal ribonucleotide as primers for polydeoxynucleotide synthesis. J. Biol. Chem. 247:3910-3917. [PubMed] [Google Scholar]
- 21.Roychoudhury, R., and H. Kossel. 1971. Synthetic polynucleotides. Enzymic synthesis of ribonucleotide terminated oligodeoxynucleotides and their use as primers for the enzymic synthesis of polydeoxynucleotides. Eur. J. Biochem. 22:310-320. [DOI] [PubMed] [Google Scholar]
- 22.Rydberg, B., and J. Game. 2002. Excision of misincorporated ribonucleotides in DNA by RNase H (type 2) and FEN-1 in cell-free extracts. Proc. Natl. Acad. Sci. USA 99:16654-16659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singhal, R. K., and S. H. Wilson. 1993. Short gap-filling synthesis by DNA polymerase beta is processive. J. Biol. Chem. 268:15906-15911. [PubMed] [Google Scholar]
- 24.Takata, M., M. S. Sasaki, E. Sonoda, C. Morrison, M. Hashimoto, H. Utsumi, Y. Yamaguchi-Iwai, A. Shinohara, and S. Takeda. 1998. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 17:5497-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Traut, T. W. 1994. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 140:1-22. [DOI] [PubMed] [Google Scholar]
- 26.Zhang, Y., X. Wu, F. Yuan, Z. Xie, and Z. Wang. 2001. Highly frequent frameshift DNA synthesis by human DNA polymerase mu. Mol. Cell. Biol. 21:7995-8006. [DOI] [PMC free article] [PubMed] [Google Scholar]