Abstract
The SAP family transcription factor myocardin functionally synergizes with serum response factor (SRF) and plays an important role in cardiac development. To determine the function of myocardin in the smooth muscle cell (SMC) lineage, we mapped the pattern of myocardin gene expression and examined the molecular mechanisms underlying transcriptional activity of myocardin in SMCs and embryonic stem (ES) cells. The human and murine myocardin genes were expressed in vascular and visceral SMCs at levels equivalent to or exceeding those observed in the heart. During embryonic development, the myocardin gene was expressed abundantly in a precise, developmentally regulated pattern in SMCs. Forced expression of myocardin transactivated multiple SMC-specific transcriptional regulatory elements in non-SMCs. By contrast, myocardin-induced transactivation was not observed in SRF−/− ES cells but could be rescued by forced expression of SRF or the SRF DNA-binding domain. Furthermore, expression of a dominant-negative myocardin mutant protein or small-interfering-RNA-induced myocardin knockdown significantly reduced SM22α promoter activity in SMCs. Most importantly, forced expression of myocardin activated expression of the SM22α, smooth muscle α-actin, and calponin-h1 genes in undifferentiated mouse ES cells. Taken together, these data demonstrate that myocardin plays an important role in the SRF-dependent transcriptional program that regulates SMC development and differentiation.
The diverse functions mediated by smooth muscle cells (SMCs) in organ systems throughout the body are ultimately dependent upon the expression of a unique set of SMC-restricted contractile and cytoskeletal proteins that distinguish this cell lineage from cardiac and skeletal myocytes. A distinguishing feature of the SMC lineage is the capacity of SMCs to reversibly modulate their phenotype and proliferate in response to a variety of stimuli during postnatal development (for a review, see reference 35). In the vasculature, SMCs in the tunica medium of arteries and veins are cell cycle arrested and express a set of lineage-restricted genes, including those for smooth muscle (SM) myosin heavy chain, SM α-actin, SM22α, and calponin, which together define the unique contractile properties of this muscle cell lineage. However, in response to arterial injury, SMCs downregulate expression of contractile genes and concomitantly upregulate a set of genes required for synthetic, migratory, and proliferative functions. This phenotypic modulation has been implicated in the pathogenesis of diseases, including atherosclerosis, restenosis following coronary angioplasty and/or stent implantation, pulmonary hypertension, and asthma (14, 33, 39, 41).
Because it is expressed exclusively and abundantly in SMCs during postnatal development (23, 42), our group and others have utilized the SMC-restricted SM22α promoter as a model system to elucidate the molecular mechanisms that regulate SMC differentiation and modulation of the SMC phenotype (4, 19, 24, 28, 34, 42, 44). The 441-bp mouse SM22α promoter functions in an SMC-specific fashion and restricts gene expression to arterial SMCs in transgenic mice (19, 24, 28). The SM22α promoter contains six nuclear protein binding sites, designated SME 1 to 6, two of which contain consensus CArG motifs (SME-1 and SME-4) that bind the MADS box transcription factor serum response factor (SRF) (19). Mutations that abolish binding of SRF to the SM22α promoter totally abolish promoter activity in transgenic mice (19). Moreover, a multimerized copy of the proximal SM22α CArG motif and 21 nucleotides of 5′-flanking sequence (bp −171 to −136) is necessary and sufficient to restrict gene expression to arterial SMCs in F0 transgenic mouse embryos (44). Consistent with these data, functionally important CArG boxes have been identified in multiple other SMC-restricted genes, including the SM myosin heavy chain (MyHC) promoter and intragenic enhancer, the SM α-actin promoter and intragenic enhancer, the telokin promoter, and the γ-enteric actin promoter (12, 25, 26, 38). Moreover, SRF plays an important role in regulating SMC differentiation and specification from undifferentiated mesenchyme (22).
However, it remains unclear how SRF, a transcription factor that is expressed ubiquitously and which activates expression of growth-responsive genes, including c-fos and egr-1, can differentially activate genes expressed in an SMC lineage-restricted pattern. In this regard, it is noteworthy that SRF may be activated by posttranslational modifications and that alternative SRF isoforms have been identified, some of which function in a dominant-negative fashion (3, 17). SRF binds to the CArG box as a homodimer, via its MADS domain, and recruits transcriptional cofactors in order to integrate combinatorial signaling and modulate target gene expression (for a review, see reference 45). In skeletal muscle, SRF physically associates with MyoD and synergistically activates skeletal muscle-specific transcriptional regulatory elements (40). Similarly, in cardiac myocytes, SRF physically associates with the cardiac lineage-restricted transcription factors Nkx-2.5 and GATA4 and synergistically activates cardiac-specific transcriptional regulatory elements (5). Consistent with this paradigm, UV-crosslinking analyses of nuclear proteins that bind to the SME-4 CArG box in the SM22α promoter with SRF revealed an 80- to 90-kDa band that was expressed in an SMC lineage-restricted fashion (44). These data suggested that one, or more, SMC lineage-restricted SRF cofactors may function in concert with SRF and activate transcription of SMC lineage-restricted genes.
Therefore, it was noteworthy when Olson and colleagues performed a search in silico and identified a novel cardiac-restricted transcription factor, designated myocardin, which binds to and functionally synergizes with SRF in the heart (48). Myocardin shows high-level sequence identity with members of the SAP (SAF-A/B, Acinus, PIAS) family of nuclear proteins, including the recently identified proteins MRTF-A/MAL/MKL1 and MRTF-B (1, 49). Each member of this family contains a conserved SAP domain that binds to A/T-rich genomic sequences or scaffold attachment regions (SARs). Related SAP family members have been shown to play a role in high-order transcriptional regulation, chromatin remodeling, and apoptosis-mediated condensation and fragmentation of chromosomal DNA (1, 10, 20).
Olson and colleagues reported that myocardin is an early marker of the cardiac muscle cell lineage, where it continues to be expressed throughout development (48). In addition, they reported that myocardin is expressed transiently in some SMCs during embryonic development. Interestingly, myocardin transactivates multiple cardiac-restricted promoters but notably only those containing consensus CArG boxes or SRF binding sites (48). Consistent with this finding, the N-terminal domain of myocardin binds directly to the MADS box of SRF (48). In Xenopus laevis, forced expression of a dominant negative myocardin mutant protein resulted in a dramatic reduction in genes encoding cardiac myocyte-restricted contractile protein isoforms and the cardiac-restricted homeobox transcription factor Nkx-2.5 (48).
Given these data and the previously defined role of SRF in the SMC lineage, it was of interest to determine what role, if any, myocardin plays in regulating differentiation of the SMC lineage. In the studies described in this report, we demonstrated that the myocardin gene is expressed in visceral and vascular SMCs at levels equivalent to or exceeding those observed in the heart, the myocardin gene is developmentally regulated in visceral and vascular SMCs during embryonic development, myocardin transactivates multiple SMC-specific transcriptional regulatory elements in an SRF-dependent fashion, SM22α promoter activity in SMCs is myocardin dependent, and, remarkably, forced expression of myocardin in undifferentiated embryonic stem (ES) cells activates transcription and expression of the endogenous SM22α gene as well as expression of the endogenous SM-α-actin and calponin-h1 genes. Taken together, these data demonstrate that myocardin plays a previously unrecognized and important role in the transcriptional program regulating SMC development and differentiation.
MATERIALS AND METHODS
Cloning of full-length human myocardin cDNA.
To clone the human myocardin cDNA, a Blast search of the GenBank database was performed with the deduced amino acid sequence of mouse myocardin (GenBank accession number AF384055). A sequence from chromosome 17 (accession number AC005358) that encoded a protein 74% identical to mouse myocardin was identified. This genomic nucleotide sequence was used to generate sense (5′-ATGAAGCTGAAAAGAGCCCGACTCG-3′) and antisense (5′-CTACCACTGCTGCAAGTGAAGGTCC-3′) oligonucleotide primers, and PCR was performed with human heart cDNA to generate the human myocardin cDNA. The resulting PCR products were cloned into pCRII (Invitrogen), and four clones were sequenced. Subsequently, 5′- and 3′-rapid amplification of cDNA end (RACE) reactions were performed with both murine and human heart RNA and nested primers with RNA ligase-mediated RACE (Ambion) reaction conditions to generate the full-length human and murine myocardin cDNAs.
Plasmids.
To generate pcRII-mMyocardin, the coding region of the mouse myocardin cDNA was generated by reverse transcription (RT)-PCR as described previously (37) and cloned into pcRII (Invitrogen). To generate pcRII-hMyocardin, the coding region of the human myocardin cDNA was generated by RT-PCR with oligonucleotide primers and human heart cDNA as described (37) and subcloned into pcRII (Invitrogen). pmMyocardin1.1-BS contains the mouse myocardin cDNA (bp 1 to 1144) subcloned into ApaI- and SacI-digested pBluescriptIIKS (Stratagene). The pcDNA-SRF and pcDNA-Myocardin expression plasmids were generated by subcloning the coding region of the mouse SRF and myocardin cDNAs, respectively, into pcDNA3 (Invitrogen).
The pcDNA-SRFΔC expression plasmid encodes a mouse SRF deletion mutant lacking the C-terminal SRF transcriptional activation domain subcloned into pcDNA3 (16). The pcDNA-SRF-DB expression plasmid encodes the previously characterized SRF DNA binding domain (amino acids 112 to 265) subcloned into pcDNA3 (13). The −441SM22.luc reporter plasmid contains the mouse SM22α promoter (bp −441 to +41) subcloned into pGL2-Basic (Promega) (42). The −441μCArG.luc reporter plasmid is identical to −441.luciferase except that the italic nucleotides in SME-1 (5′-TCCTGCCCATAGATCTTTTTTCCC-3′) and SME-4 (5′-GCTCCAACTTGGTGTCTTTCCCCGGATATGGAGCCT-3′) were mutated to abolish SRF binding activity (19). p-90.luc contains the 90-bp SM22α promoter subcloned into pGL2-Basic as described previously (42). pSME4×4.luc contains four copies of SME-4 (bp −171 to −136) subcloned immediately 5′ of the 90-bp mouse SM22α promoter in p-90.luc (19).
pCArGx4.luc contains four copies of the SME-4 CArG box alone (bp −150 to −141) subcloned immediately 5′ of the 90-bp SM22α promoter in p-90.luc (44). The p5′SME4×4.luc contains four copies of the SME-4 CArG box and 5′-flanking sequence (bp −171 to −141) subcloned 5′ of the 90-bp SM22α promoter in p-90.luc. p3′SME4×4.luc contains four copies of the SME-4 CArG box and its 3′-flanking sequences (bp −151 to −136) subcloned 5′ of the 90-bp SM22α promoter in p-90.luc (44). The pPIAct-luc and pPIMyo-luc luciferase reporter plasmids contain the SM-α-actin promoter and intragenic enhancer (bp −2560 to +2784) and the SM-MyHC promoter and intragenic enhancer (bp −4216 to +11795) subcloned into the pGL3-Basic plasmid, respectively, and have been described previously (25, 26).
Northern blot and in situ hybridization analyses.
Northern blot analyses were performed with the human 12-lane multiple-tissue Northern (MTN), human muscle MTN, and human cardiovascular MTN blots prepared by Clontech (Palo Alto, Calif.), and RNA was harvested from various tissues of adult C57BL/6 mice as described previously (37). The 32P-labeled human and mouse myocardin cDNA probes were generated by random-primed labeling as described previously (37). In situ hybridization analyses were performed on sections of staged murine embryos as described previously (29). To generate 35S-labeled antisense and control sense riboprobes, the pmMyocardin1.1-BS plasmid was linearized with SacI (antisense) and KpnI (sense) and used as the template in T3 or T7 in vitro transcription reactions in the presence of [35S]dUTP to generate antisense and control sense cRNA probes.
Generation of SRF-deficient ES cells.
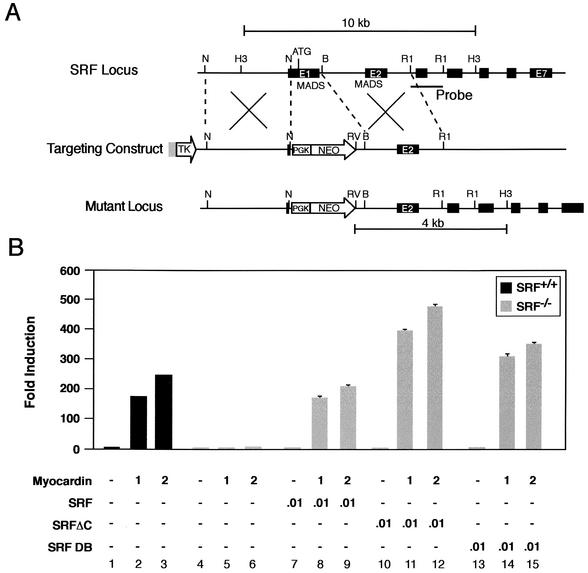
A genomic clone including the 5′ end of the murine SRF gene was isolated from an Sv129 mouse library as described previously (31). The targeting vector was generated with the pPNT plasmid (47), which contains the PGK-neomycin resistance-poly(A) and phosphoglycerate kinase (PGK)-thymidine kinase-poly(A) cassettes for positive and negative selection, respectively. As shown in Fig. 5A, the pPNT-SRF targeting vector contains a 5-kb genomic subfragment that includes approximately 4.9 kb of SRF 5′-flanking sequence and SRF exon 1 sequence terminating at the NotI site 123 bp 5′ of the initiation codon (ATG) and a 3-kb BamHI-EcoRI SRF genomic subfragment that includes intron 1, exon 2, and intron 2 sequences subcloned 3′ of the PGK-neomycin cassette of pPNT (Fig. 5A). In properly targeted ES cell clones, the exon 1 sequence, which includes the initiation codon (Fig. 5A, ATG) and nucleotide sequences encoding approximately one half of the MADS box domain, is replaced by the PGK-neomycin cassette. This gene targeting strategy is similar to that employed by Nordheim and coworkers to generate SRF-deficient mice (2).
FIG. 5.
Myocardin-induced transactivation of embryonic stem cells is SRF dependent. (A) Schematic representation of the SRF gene targeting strategy. (Top) Partial restriction endonuclease map of the murine SRF genomic locus showing NotI (N), HindIII (H3), BamHI (B), and EcoRI (R1) sites. Exons are shown in black. The MADS box domain (MADS) is encoded in exons 1 and 2. The location of the probe used in Southern blot analysis is shown. (Middle) SRF targeting vector containing the neomycin resistance (NEO) and herpes simplex virus thymidine kinase (TK) genes under the control of the PGK promoter. (Bottom) Structure of the targeted mutant SRF allele. (B) Myocardin-induced transactivation of wild type and SRF−/− ES cells. Wild type SRF+/+ (black bars) and homozygous SRF−/− (gray bars) ES cells were transiently cotransfected with the −441.SM22.luc reporter plasmid and the indicated amounts (in micrograms) of myocardin, SRF, SRFΔC, and SRF DB expression plasmids. Data are reported as induction in luciferase activity observed when cotransfected with pcDNA-Myocardin relative to that obtained with pcDNA3. Results are expressed as the mean ± SEM. All transfections were repeated at least three times to ensure reproducibility.
The SRF targeting construct was linearized and electroporated into RW ES cells as described previously (31). Neomycin-resistant transfectants were selected in 250 μg of G418 per ml and 1 μM ganciclovir as described (31). DNA from resistant ES cell clones was digested with HindIII and EcoRV and analyzed by Southern blot analysis with a 1-kb EcoRI genomic subfragment containing SRF gene sequences, as shown in Fig. 5A. SRF−/− ES cells were obtained by subjecting independently derived SRF+/− clones to selection in G418 at a concentration of 1.5 μg/ml as described before (32). Targeted clones were checked for single integration by hybridization with a neo probe after digestion with EcoRV. Northern blot and Western blot analyses were performed as described previously (50) to confirm that the gene-targeting strategy had generated a null allele (data not shown).
Cell culture, cotransfections, and myocardin knockdown experiments.
COS-7 cells were maintained in Dulbecco's modified essential medium with 10% fetal bovine serum as described previously (30). Transient cotransfection of 105 COS-7 cells was performed with Fugene6 (Roche) and 200 ng to 1 μg of the indicated luciferase reporter plasmid, 100 ng to 2 μg of the indicated expression plasmid, and 10 ng of the phRL-TK(-Int) reference plasmid (Promega).
Wild-type mouse ES cells and SRF−/− ES cells were grown and maintained as described before (6). ES cells were plated at 106 cells/well and transfected with Lipofectamine 2000 (Invitrogen) and 0 to 200 ng of the −441SM22.luc reporter plasmid, 0 to 2 μg the of pcDNA3myocardin expression plasmid, 2 to 4 μg of the pcDNA3 control expression plasmid, and 10 ng of phRL-TK(-Int) reference plasmid. Primary rat aortic SMCs were cotransfected with Transit-LT1 (Mirus) and 200 ng of either the −441.luc reporter plasmid or pGL2-Control (Promega) and between 0 and 3 μg of the pcDNA-CΔ585 expression plasmid. Myocardin knockdown experiments in A7r5 SMCs were performed with small interfering RNA (siRNA) as described before (7). Double-stranded siRNA directed against myocardin (5′-AATGCAACTGCAGAAGCAGAA-3′) and myocardin scrambled control siRNA (5′-AATAGATGAGAGACACACAGC-3′) were generated with the Silencer siRNA construction kit (Ambion). A total of 105 A7r5 SMCs were cotransfected with 2 μg of the −441SM22.luc reporter plasmid, 100 ng of phRL-TK(-Int) reference plasmid, and either myocardin siRNA or a control scrambled myocardin siRNA at a final concentration of 12.5 nM or 25 nM, with Transit-TKO (Mirus) and Transit-LT1 (Mirus) according to the manufacturer's instructions. Luciferase activity was measured with the dual luciferase assay system (Promega) and corrected for transfection efficiency as described previously (42). All experiments were repeated at least three times to ensure reproducibility. Data are reported as mean normalized relative light units (RLU) ± standard error of the mean (SEM).
Immunohistochemistry and quantitative RT-PCR.
Wild-type mouse ES cells, SRF−/− ES cells, and ES cells harboring a knock-in of the lacZ gene into the initiation codon of the SM22α gene (50) were grown and maintained as described previously (6). ES cells were plated at 106 cells/well and transfected with Lipofectamine 2000 (Invitrogen) and 4 μg of pcDNA3, 4 μg of pcDNA-Myocardin, 5 ng of pcDNA-SRF, or 4 μg of pcDNA-Myocardin plus 5 ng of pcDNASRF. At 24 to 48 h posttransfection, cells were harvested for quantitative RT-PCR or fixed and stained for β-galactosidase activity determinations as described previously (48). Quantitative RT-PCR was performed with Applied Biosystems SYBR Green PCR Master Mix and MJ Research DNA Engine Opticon 2 real-time detection system according to the manufacturer's instructions. All RT and PCRs were performed in duplicate with and without RT controls.
A total of 26 PCR cycles were run to ensure that saturation had not been reached as monitored by real-time fluorescence. The following primer pairs were utilized: SM-α-actin (5′-GAGAAGCCCAGCCAGTCG-3′ and 5′-CTCTTGCTCTGGGCTTCA-3′), yielding a 240-bp product; myocardin (5′-CTGTGTGGAGTCCTCAGGTCAAACC-3′ and 5′-GATGTGCTGCGGGCTCTTCAG-3′), yielding a 108-bp product; calponin-h1 (5′-CCCAGAAATACGACCATCAGCG-3′ and 5′-CACCCCCTCAATCCACTCTCTCAG-3′), yielding a 57-bp product; SM22α (5′-AGCCAGTGAAGGTGCCTGAGAAC-3′ and 5′-TGCCCAAAGCCATTAGAGTCCTC-3′), yielding a 181-bp product; and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (5′-GTGGCAAAGTGGAGATTGTTGCC-3′ and 5′-GATGATGACCCGTTTGGCTCC-3′), yielding a 291-bp product.
RESULTS
Isolation and structural characterization of human myocardin cDNA and gene.
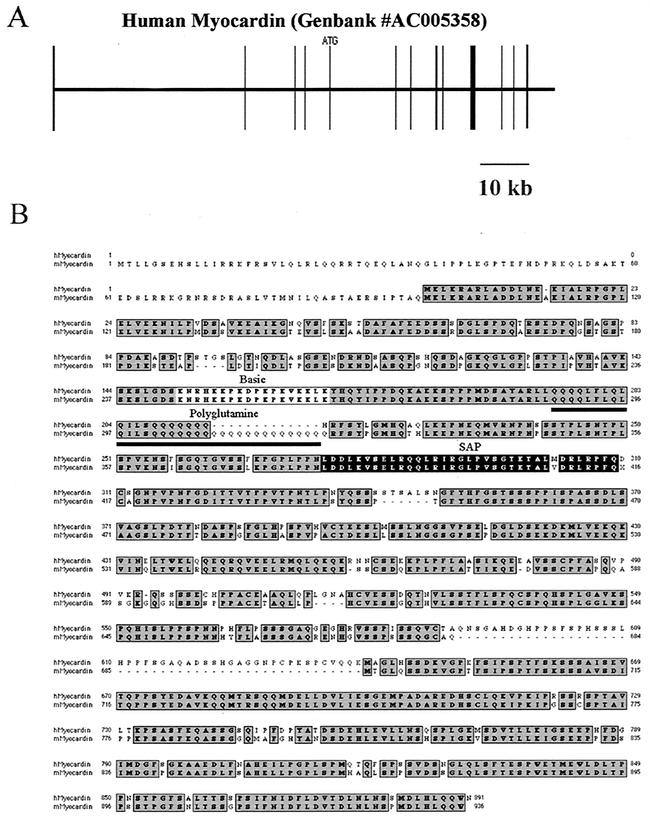
To clone the human myocardin cDNA, a Blast search of the GenBank database was performed with the deduced amino acid sequence of mouse myocardin (48). This search revealed a human nucleotide sequence on chromosome 17 that was 74% identical to the sequence of mouse myocardin (GenBank accession no. AC005358). The human myocardin gene is encoded on 13 exons, spanning approximately 92 kb of genomic DNA on chromosome 17 (Fig. 1A). The human myocardin cDNA contains a 2,674-bp open reading frame encoding a 95.7-kDa protein.
FIG. 1.
Primary structure of the human myocardin gene and alignment of the deduced amino acid sequences of the human and murine myocardin proteins. (A) A schematic map of the human myocardin gene structure. Performing a Blast search of the GenBank database identified the human myocardin gene. The myocardin gene is located on human chromosome 17 and is identified as GenBank accession number AC005358. Exons are shown as vertical boxes. The initiation codon (ATG) in exon 5 is shown. A 10-kb scale is shown below the map. (B) Alignment of the deduced amino acid sequences of the deduced human and murine myocardin proteins. The deduced amino acid sequence of mouse myocardin is shown below the sequence of the human myocardin protein. The conserved amino acid residues are shaded dark gray. The conserved basic domain (light gray box), polyglutamine tract (underline), and SAP box (black box) are shown.
As shown in Fig. 1B, high-grade amino acid sequence identity between mouse and human myocardin was observed across the previously identified basic domain (light gray shading), polyglutamine tract (underlined), and SAP domain (black shading). However, several differences in the coding region of the human and mouse myocardin proteins were observed: the human polyglutamine tract was 13 glutamine residues shorter than the mouse polyglutamine tract, and the C terminus of human myocardin contained a 49-amino-acid insertion that was not present in the mouse protein (Fig. 1B). Of note, one of the four human myocardin clones that was isolated and sequenced contained an Alu insertion (43), in frame, between the polyglutamine region and the SAP domain.
Myocardin is expressed in SMC-containing tissues during postnatal development.
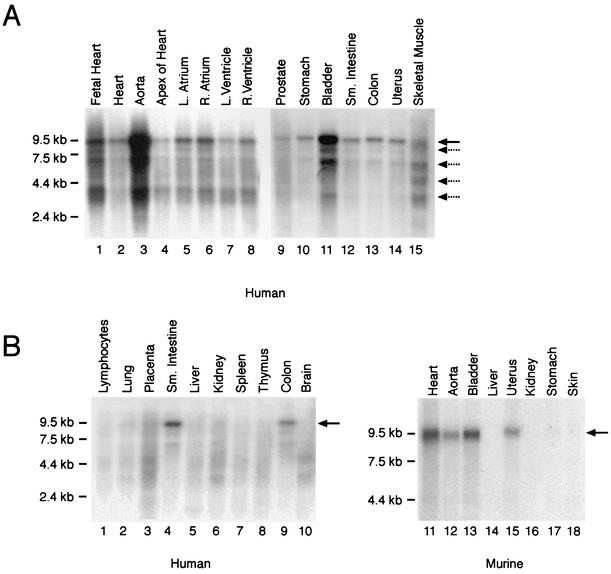
Olson and colleagues reported that the mouse myocardin gene is expressed in a cardiac-restricted fashion during postnatal development and transiently in some embryonic SMCs (48). To determine the pattern of myocardin gene expression in human tissues, the human myocardin cDNA probe was hybridized to Northern blots containing mRNA harvested from a variety of human tissues. As anticipated, in both the adult human heart (all chambers) and the fetal heart, the radiolabeled myocardin cDNA hybridized to multiple mRNA species (Fig. 2A lanes 1, 2, and 4 to 8). However, the most intense myocardin hybridization signal was observed in the lane containing mRNA harvested from the adult human aorta (Fig. 2A, lane 3). Moreover, the human myocardin gene was expressed in multiple other SMC-containing tissues, including the stomach, bladder, small intestine, colon, and uterus (Fig. 2A, lanes 10 to 14).
FIG. 2.
In vivo tissue distribution of myocardin gene expression. (A) Northern blot analyses of muscle cell-containing tissues. Membranes containing 2 μg of poly(A)+ RNA per lane isolated from embryonic and adult human tissues were hybridized to the radiolabeled human myocardin cDNA probe (1 week of exposure). RNA markers are shown to the left of the blot (in kilobases). The human myocardin probe hybridized to a predominant mRNA species of approximately 9.5 kb (black arrow). In addition, four to five additional low-abundance transcripts (dashed arrows) migrating between 8.5 and 3.0 kb were observed. Myocardin mRNA was observed in the heart and smooth muscle cell-containing tissues, including the aorta, stomach, bladder, small intestine, colon, and uterus. (B) Northern blot analysis of mRNA samples isolated from adult human (left panel) and murine (right panel) tissues hybridized to the radiolabeled human (left panel) and murine (right panel) myocardin cDNA probes (1 week of exposure). Myocardin mRNA (black arrow) was observed in human and murine heart and SMC-containing tissues but not in other tissues.
In the heart and SMC-containing tissues, the predominant myocardin transcript was 9.5 kb (black arrow), although four to five less abundant mRNA species were detected migrating between 8.5 and 3.0 kb (dashed arrows). Of note, the relative level of myocardin gene expression varied greatly in these SMC-containing tissues, with the highest levels observed in the aorta and bladder (Fig. 2A, lanes 3 and 11). In addition, upon prolonged autoradiographic exposure, faint but detectable hybridization of the human myocardin probe to mRNA harvested from prostate and skeletal muscle was observed (Fig. 2A, lanes 9 and 15).
In contrast, the predominant 9.5-kb myocardin transcript was not detectable in mRNA harvested from human lymphocytes, liver, kidney, spleen, thymus, and brain and was barely detectable in the lung and placenta, perhaps reflecting the vascular component of these tissues (Fig. 2B, lanes 1 to 3, 5 to 8, and 10). To confirm that the mouse myocardin gene is also expressed in SMC-containing tissues during postnatal development, a radiolabeled mouse myocardin cDNA probe was hybridized to Northern blots containing RNA harvested from adult murine tissues (Fig. 2B, lanes 11 to 18). Consistent with the results observed in humans, a 9.5-kb signal (arrow) was observed in the heart, aorta, bladder, and uterus and, upon prolonged autoradiographic exposure, in the stomach (Fig. 2B, lanes 11 to 13, 15, and 17) but not in the liver, kidney, or skin (lanes 14, 16, and 18). Hybridization of each blot to a radiolabeled GAPDH cDNA probe confirmed that the lanes contained equal amounts of mRNA (data not shown). These data demonstrate that in humans and mice, myocardin is expressed in the heart as well as multiple SMC-containing tissues, including the aorta, bladder, stomach, intestine, colon, and uterus.
Myocardin is expressed in a precise developmentally regulated pattern in visceral and vascular SMCs during murine embryogenesis.
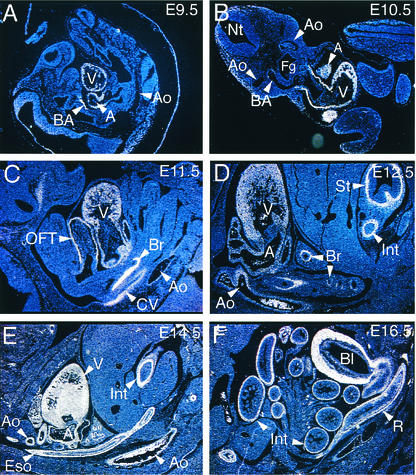
To determine the temporal and spatial pattern of myocardin gene expression during mouse embryonic development, in situ hybridization was performed on staged murine embryos with radiolabeled antisense and control sense mouse myocardin riboprobes. In the embryonic day 9.5 (E9.5) mouse embryo, myocardin mRNA (white signal) was restricted to the common atrial and ventricular chambers of the primitive heart (Fig. 3A). Despite the fact that SM22α and SM-α-actin gene expression is detectable in the dorsal aorta at E9.5 (23), at the level of sensitivity afforded by these in situ hybridization analyses, myocardin gene expression was not detected in the dorsal aorta at E9.5 (Fig. 3A) or E10.5 (Fig. 3B). At E10.5, abundant myocardin gene expression was observed throughout the heart. In addition, a faint hybridization signal was detectable in the third branchial arch artery, where it communicates with the dorsal aorta (Fig. 3B). This finding was confirmed in whole-mount in situ hybridization studies performed with a digoxigenin-labeled myocardin antisense riboprobe (data not shown).
FIG. 3.
Myocardin gene is expressed in a developmentally regulated fashion in vascular and visceral SMCs. In situ hybridization analyses were performed on staged CD-1 mouse embryos with a 35S-labeled myocardin antisense probe. (A) In this sagittal section of an E9.5 embryo, hybridization of the myocardin riboprobe (white signal) to the ventricular chamber (V), common atria (A), and branchial arch artery (BA) was observed. Myocardin mRNA was not detectable in the dorsal aorta (Ao) at E9.5. Magnification, ×5. (B) In this transverse section of an E10.5 embryo, hybridization of the myocardin riboprobe to the ventricular chamber (V), atria (A), and cardiac outflow tract was observed. A faint signal, above background levels, was also observed in the third branchial arch artery (BA) near its communication with the aorta (Ao). Magnification, ×5. (C) In this sagittal section of the thoracic cavity of an E11.5 embryo, hybridization of the myocardin riboprobe to the ventricle (V), atria, cardiac outflow tract (OFT), primitive bronchi (Br) of the lung bud, common cardinal vein (CV), and dorsal aorta (Ao) was observed. Magnification, ×12.5. (D) In this sagittal section of the thoracic cavity and abdomen of an E12.5 embryo, hybridization of the myocardin riboprobe to the ventricle (V), atria (A), bronchi (Br) in the lung bud, aorta (Ao), stomach (St), and intestine (Int) was observed. Magnification, ×12.5. (E) In this sagittal section of the thoracic cavity and abdomen of an E14.5 embryo, hybridization of the myocardin riboprobe to the ventricle (V), atria (A), aorta (Ao), esophagus (Eso), and intestine (Int) was observed. Intense hybridization of the riboprobe to the urogenital ridge was also observed at this stage (not shown). Magnification, ×5. (F) In this sagittal section of the abdomen of an E16.5 embryo, hybridization of the myocardin riboprobe to the small and large intestine (Int), rectum (R), and muscular wall of the bladder (Bl) was observed. Magnification, ×5. No hybridization of the control sense myocardin riboprobe was observed.
At E11.5, myocardin gene expression was observed in the ventricle, atria, cardiac outflow tract, primitive bronchi, and the cardinal veins and at low but detectable levels in the dorsal aorta (Fig. 3C). These data suggest that during angiogenesis, the myocardin gene is differentially expressed throughout the primitive vasculature but that in some arteries (including the dorsal aorta), expression of SMC markers may precede expression of myocardin.
By contrast, prior to or coincident with differentiation of visceral SMCs from surrounding mesenchyme, myocardin gene expression is detectable in presumptive SMCs. At E11.5, myocardin mRNA was observed in primitive bronchi (Fig. 3C). By E12.5, myocardin gene expression was easily detectable in the muscularis mucosa layer of the stomach and intestine (Fig. 3D). In the primitive lung buds, expression of myocardin was also detectable and was restricted to presumptive bronchial SMCs (Fig. 3D, Br). At E12.5, myocardin mRNA was also observed throughout the heart and was now readily observed in the dorsal aorta (Fig. 3D). At E14.5, myocardin expression was observed in the aorta as well as the heart, esophagus, bronchi, stomach (not shown), and intestine (Fig. 3E). In addition, at E12.5 to 14.5, intense hybridization of the myocardin riboprobe to the urogenital ridge, which gives rise to the bladder, was observed (data not shown).
At E16.5, the myocardin gene was expressed throughout the heart and major arteries (not shown) and in presumptive visceral SMCs in the trachea and lungs (not shown), esophagus (not shown), stomach (not shown), small intestine (Fig. 3F), colon and rectum (Fig. 3F), and bladder (Fig. 3F). These data demonstrate that in the mouse embryo, the myocardin gene is restricted to cardiac myocytes as well as vascular and visceral SMCs. Of note, despite the fact that myocardin mRNA was detected upon prolonged autoradiographic exposure on Northern blots containing mRNA harvested from human skeletal muscle (Fig. 2A, lane 15), in these in situ hybridization analyses, myocardin mRNA was not observed in the myotomal component of the somites or embryonic SMCs.
Myocardin transactivates the SMC-restricted SM22α promoter in non-SMCs.
The 441-bp CArG box-dependent SM22α promoter is both necessary and sufficient to restrict transgene expression to arterial SMCs in transgenic mice (19, 24, 28, 44). To confirm that myocardin transactivates the mouse SM22α promoter in non-SMCs, COS-7 cells were transiently cotransfected with the pcDNA-Myocardin expression plasmid and the −441SM22.luc reporter plasmid (Fig. 4B, −441.luc) containing the firefly luciferase reporter gene placed under the transcriptional control of the 441-bp mouse SM22α promoter. Consistent with the findings of Olson and coworkers (48), cotransfection of the pcDNA-Myocardin expression plasmid resulted in an 836-fold increase in luciferase activity (Fig. 4A). In contrast, forced expression of myocardin failed to transactivate the SM22α promoter harboring mutations in SME-1 and SME-4 that abolish binding of SRF (Fig. 4A, −441μCArG.luc).
FIG. 4.
Myocardin-induced transactivation of multiple SMC-specific transcriptional regulatory elements in COS-7 and undifferentiated mouse ES cells. (A) cis-acting mechanisms regulating myocardin-induced transactivation of the SM22α promoter in COS-7 cells. The nucleotide sequence of SME-4 (bp −171 to −136) and SME-4 deletion mutants CArG (bp −150 to −141), 5′CArG (bp −171 to −141), and 3′CArG (bp −151 to −136) are shown. COS-7 cells were cotransfected with the pcDNA-Myocardin expression plasmid, the phRL-TK(-Int) reference plasmid, and the indicated reporter plasmid. Luciferase activities were measured 48 h posttransfection. Myocardin-induced transcriptional activation of luciferase reporter plasmids placed under the transcriptional control of the 441-bp mouse SM22α promoter (−441.luc), the SM22α promoter containing mutations that abolish SRF binding (−441μCArG.luc), the 90-bp SM22α promoter linked to four copies of SME-4 (SME4×4.luc), the 90-bp SM22α promoter linked to four copies of the CArG oligonucleotide (CarGx4.luc), the 90-bp SM22α promoter linked to four copies of the 5′CArG oligonucleotide (5′CarGx4.luc), and the 90-bp SM22α promoter linked to four copies of the 3′CArG oligonucleotide (3′CarGx4.luc). Luciferase activity is reported as induction of luciferase activity observed when each reporter plasmid was cotransfected with pcDNA-Myocardin relative to the activity observed with pcDNA3. Results are expressed as the mean ± SEM. (B) Myocardin-induced transactivation of multiple SMC-specific transcriptional regulatory elements in mouse ES cells. Undifferentiated mouse ES cells were cotransfected with the pcDNA-Myocardin expression plasmid, the phRL-TK(-Int) reference plasmid, and the indicated reporter plasmid. Myocardin-induced transcriptional activation of luciferase reporter plasmids placed under the transcriptional control of the 441-bp mouse SM22α promoter (−441.luc), the SM-α-actin promoter and intragenic enhancer (pPIAct-luc), and the SM-MyHC promoter and intragenic enhancer (pPIMyo-luc) is reported as the induction in luciferase activity observed when cotransfected with pcDNA-Myocardin relative to the activity observed with pcDNA3. Results are expressed as the mean ± SEM. All transfections were repeated at least three times to ensure reproducibility.
To determine whether the SME-4 CArG box nucleotide sequence alone may be transactivated by myocardin or whether transactivation of the CArG-box containing SME-4 nuclear protein binding site requires additional flanking nucleotides, COS-7 cells were cotransfected with the pcDNA-Myocardin expression plasmid and luciferase reporter plasmids under the transcriptional control of the 90-bp SM22α promoter linked to either four copies of SME-4 (bp −171 to −136), the SME-4 CArG box alone (bp −150 to −141), the SME-4 CArG box and 21 nucleotides of 5′-flanking sequence (bp −171 to −141), or the SME-4 CArG box and five nucleotides of 3′-flanking sequence (bp −151 to −136) (Fig. 4A, top panel). Consistent with analysis of the SM22α promoter in transgenic mice (44), high-level luciferase activity was observed in COS-7 cells cotransfected with luciferase reporter plasmids containing either four copies of SME-4 (Fig. 4A, SME4×4.luc) or four copies of the SME-4 CArG box and 21 bp of its 5′-flanking sequence (Fig. 4A, 5′CArGx4.luc). In contrast, myocardin-induced transactivation was not observed with luciferase reporter plasmids containing four copies of the SME-4 CArG box alone (Fig. 4A, CArGx4.luc) or four copies of the SME-4 CArG box and its additional 3′-flanking nucleotides (Fig. 4A, 3′CArGx4.luc). These data demonstrate that myocardin-induced transactivation of a multimerized SME-4 nuclear protein binding site requires additional nucleotides flanking its embedded CArG box.
Myocardin transactivates multiple SMC-specific transcriptional regulatory elements in ES cells.
To determine whether forced expression of myocardin transactivates other SMC-restricted transcriptional regulatory elements that have been validated in transgenic mice, undifferentiated mouse ES cells were transiently cotransfected with an expression plasmid encoding myocardin and luciferase reporter plasmids under the transcriptional control of the mouse SM22α promoter, the SM-α-actin promoter-enhancer (25), and the SM-MyHC promoter-enhancer (26) (Fig. 4B). Remarkably, in mouse ES cells, forced expression of myocardin resulted in a 65-fold induction in SM22α promoter activity, a 370-fold induction in the activity of the SM-α-actin promoter-enhancer, and a 12-fold induction in the activity of the SM-MyHC promoter-enhancer, which is a relatively late marker of the SMC lineage (35). Together, these data strongly suggest that myocardin activates the SRF-dependent transcriptional regulatory program that controls SMC differentiation.
Myocardin-induced activation of the SM22α promoter is dependent upon expression of SRF in embryonic stem cells.
Mice harboring a null mutation in SRF do not survive past gastrulation, precluding assessment of the function of SRF in the SMC lineage (2). To determine whether SRF is required for myocardin-induced transactivation of the SMC-restricted SM22α promoter, we tested whether forced expression of myocardin transactivates the SM22α promoter in SRF-deficient ES cells. Heterozygous SRF+/− and homozygous deficient SRF−/− ES cells were generated with the gene targeting strategy outlined in Fig. 5A. Consistent with the results observed in COS-7 cells, a 160- to 240-fold induction in luciferase activity was observed in wild-type ES cells (black bars) cotransfected with the pcDNA-Myocardin expression plasmid and the −441SM22.luc reporter plasmid (Fig. 5B, lanes 1 to 3). In contrast, basal levels of luciferase activity were observed when SRF−/− ES cells (gray bars) were cotransfected with −441SM22.luc and pcDNA-Myocardin (Fig. 5B, lanes 4 to 6).
To prove that the loss of myocardin-induced transactivation of SRF−/− ES cells resulted from the absence of SRF and not from another event that may have altered these cells during the selection process, the capacity of the pcDNA-SRF expression plasmid to rescue myocardin-induced transactivation of SRF−/− ES cells was assessed. As anticipated, expression of SRF in the SRF−/− ES cells restored the observed myocardin-induced transactivation of the SM22α promoter (Fig. 5B, lanes 7 to 9).
Next, to determine whether myocardin-induced transactivation of the SM22α promoter in ES cells requires the SRF transcriptional activation domain, SRF−/− ES cells were cotransfected with −441SM22.luc, pcDNA-Myocardin, and the pcDNA-SRFΔC expression plasmid, encoding a mutated SRF protein lacking its transcriptional activation domain (16). Remarkably, a 400- to 500-fold induction in myocardin-induced transcriptional activity of the SM22α promoter was observed (Fig. 5B, lanes 10 to 12). Finally, to determine whether the SRF DNA-binding domain (which includes the MADS box) is necessary and sufficient to rescue myocardin-induced transactivation of the SM22α promoter in ES cells, the capacity of the pcDNA-SRF-DB plasmid encoding the mouse SRF DNA binding domain to rescue myocardin-induced transactivation of the SM22α promoter in SRF−/− ES cells was assessed. Once again, a 300- to 400-fold induction in luciferase activity was observed (Fig. 5B, lanes 13 to 15). Taken together, these data prove that myocardin-induced transactivation of the SMC-restricted SM22α promoter requires SRF. Moreover, forced expression of the SRF DNA-binding domain alone is necessary and sufficient to rescue myocardin-induced transactivation of the SM22α promoter in SRF−/− ES cells.
Dominant-negative myocardin represses activity of the SM22α promoter in SMCs.
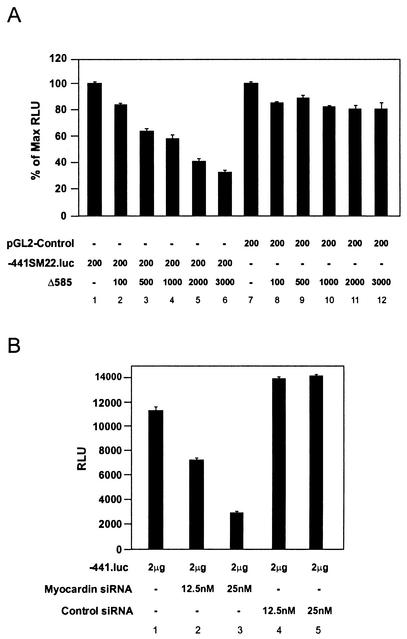
The data described above support the hypothesis that myocardin activates the SM22α promoter in SMCs. To determine whether transcriptional activity of the SM22α promoter in SMCs is in fact dependent upon myocardin, primary rat aortic SMCs were transiently cotransfected with the −441SM22.luc reporter plasmid (Fig. 6A, −441.luc) and increasing amounts of the pcDNA-Δ585 expression plasmid (Fig. 6A, Δ585), encoding a dominant negative myocardin mutant protein (48). Consistent with our previous reports (42), an approximately 100-fold induction in luciferase reporter activity was observed in SMCs transfected with the −441SM22.luc reporter plasmid (Fig. 6A, lane 1) above the levels observed with the promoterless pGL2-Basic plasmid. However, cotransfection with increasing amounts of the pcDNA-Δ585 expression plasmid resulted in a stepwise reduction in activity of the 441-bp SM22α promoter to levels approximately 30% of that of the control (Fig. 6A, lanes 2 to 6). To confirm that this finding was not the result of generalized transcriptional squelching, the pcDNA-Δ585 expression plasmid was cotransfected into primary rat aortic SMCs with the pGL2-Control luciferase plasmid, containing the simian virus 40 promoter and enhancer, and luciferase activities exceeding 80% of the control value were observed (Fig. 6A, lanes 7 to 12). These data demonstrate that SM22α promoter activity in primary rat aortic SMCs is myocardin dependent.
FIG. 6.
SM22α promoter activity is myocardin dependent in SMCs. (A) Dominant-negative myocardin suppresses SM22α promoter activity in primary rat aortic SMCs. Primary rat aortic SMCs were transiently cotransfected with either the pGL2-Control plasmid (lanes 7 to 12) or the −441SM22.luc (lanes 1 to 6) reporter plasmids and the indicated amount (in nanograms) of the pcDNA-Δ585 expression plasmid, encoding a dominant-negative myocardin mutant protein. Luciferase activity is expressed as the percentage of luciferase activity observed when SMCs were cotransfected with the pcDNA-Δ585 expression plasmid relative to that observed when transfected with −441SM22.luc alone (lanes 1 to 6) or pGL2-Control alone (lanes 7 to 12). Data are expressed as mean ± SEM. (B) siRNA-mediated knockdown of myocardin reduces activity of the SM22α promoter in A7r5 SMCs. A7r5 SMCs were placed in medium containing the indicated concentration of myocardin siRNA (lanes 2 and 3) or control siRNA (lanes 4 and 5) and transfected with 2 μg of the −441SM22.luc (−441.luc) reporter plasmid. Cells were harvested 48 h posttransfection, and luciferase activities were calculated. Data are expressed as normalized relative light units (RLU) ± SEM. Transfections were repeated at least three times to ensure reproducibility.
Myocardin siRNA inhibits activity of the SM22α promoter in A7r5 SMCs.
To confirm that the activity of the SM22α promoter is myocardin dependent in vascular SMCs, a complementary series of gene knockdown experiments were performed with siRNA (7). In these experiments, A7r5 SMCs were cotransfected with increasing concentrations of myocardin siRNA or control scrambled myocardin siRNA and the −441SM22.luciferease reporter plasmid. Luciferase activity was reduced by 40% in A7r5 cells exposed to 12.5 nM myocardin siRNA and by 75% in A7r5 cells exposed to 25 nM concentrations of myocardin siRNA (Fig. 6B, lanes 1 to 3). In contrast, exposure of A7r5 SMCs to 12.5 nM and 25 nM concentrations of control scrambled myocardin siRNA did not significantly decrease the activity of the 441-bp SM22α promoter in SMCs (Fig. 6B, lanes 4 and 5). Together with the results of the experiments described above, these data demonstrate that the activity of the SMC-restricted SM22α promoter is dependent upon myocardin in SMCs.
Forced expression of myocardin activates endogenous SMC gene expression in mouse embryonic stem cells.
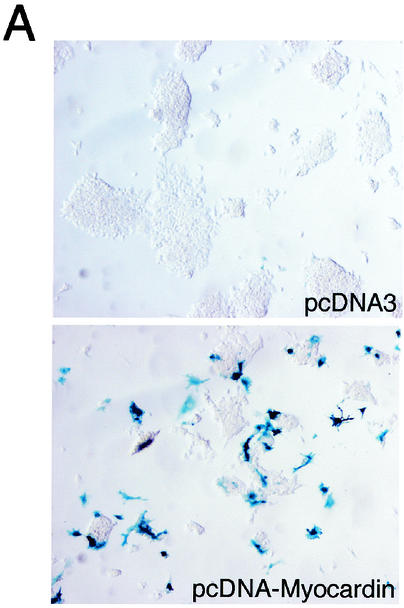
To determine whether forced expression of myocardin activates transcription of the endogenous SM22α gene in ES cells, mouse ES cells in which the bacterial lacZ gene has been knocked into the initiation codon of the endogenous SM22α gene (50) were transfected with the control plasmid pcDNA3 and the pcDNA-Myocardin expression plasmid. At 48 h posttransfection, the undifferentiated ES cells were fixed and stained for β-galactosidase activity determination. As anticipated, β-galactosidase activity (blue staining) was not observed in undifferentiated ES cells, which do not express SM22α (Fig. 7A, upper panel). In contrast, multiple dark-blue-stained ES cells were observed following transfection with pcDNA-Myocardin, demonstrating that forced expression of myocardin activates transcription of the endogenous SM22α gene in mouse ES cells (Fig. 7A, bottom panel). Of note, the frequency of staining (approximately 30% of cells) corresponded to the transfection efficiency in these cells.
FIG. 7.
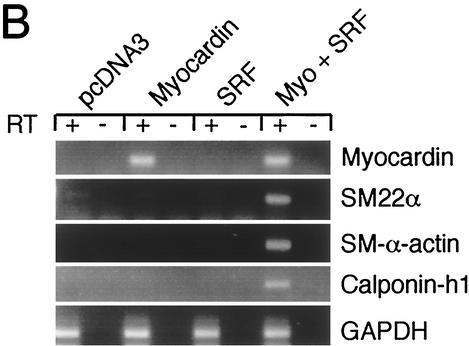
Forced expression of myocardin induces expression of endogenous SMC genes in undifferentiated mouse ES cells. (A) Forced expression of myocardin in SM22α+/lacZ ES cells induces transcription of the endogenous SM22α gene. SM22α+/lacZ ES cells were transfected with the pcDNA3 control plasmid (upper panel) or the pcDNA-Myocardin expression plasmid (lower panel). At 48 h posttransfection, cells were fixed and stained for β-galactosidase activity. β-Galactosidase activity (blue staining) was not observed in the cells transfected with pcDNA3. In contrast, β-galactosidase activity (intense blue staining) was observed in the SM22α+/lacZ ES cells transfected with pcDNA-Myocardin. (B) Myocardin-induced SMC gene expression in undifferentiated ES cells is SRF dependent. SRF−/− ES cells were transiently transfected with the control plasmid pcDNA3 or expression plasmids encoding myocardin, SRF, or myocardin and SRF. At 48 h posttransfection, the cells were harvested and RNA was prepared. Quantitative real-time PCR was performed with Applied Biosystems SYBR Green PCR Master Mix and MJ Research DNA Engine Opticon 2 real-time detection system. All RT-PCRs were performed in duplicate with (+) and without (−) RT controls. Primer pairs were designed to quantitatively amplify the mouse myocardin, SM22α, SM-α-actin, calponin-h1,
To determine whether forced expression of myocardin activates expression of genes encoding other SMC lineage-restricted markers and to test whether the induction of SMC genes is both SRF and myocardin dependent, SRF−/− ES cells were transiently transfected with either the pcDNA3 control plasmid or expression plasmids encoding myocardin, SRF, or myocardin plus SRF. At 24 h posttransfection, the ES cells were harvested, and the expression of SMC genes was measured by quantitative real-time RT-PCR (Fig. 7B). As anticipated, in SRF−/− ES cells transfected with the pcDNA-Myocardin expression plasmid with or without pcDNA-SRF, myocardin mRNA was observed (Fig. 7B, row 1). More importantly, expression of the SMC lineage-restricted SM22α, SM-α-actin and calponin-h1 genes was only observed when SRF−/− ES cells were transfected with expression plasmids encoding both SRF and myocardin (Fig. 7B, rows 2 to 4). Neither plasmid alone activated expression of these gene products in SRF-deficient ES cells. As a positive control, GAPDH was amplified in reactions mixtures containing RT (Fig. 7B, row 5). Together, these data demonstrate that forced expression of myocardin induces expression of multiple SMC lineage-restricted genes in undifferentiated mouse ES cells. Moreover, these data demonstrate that myocardin-induced expression of multiple endogenous SMC genes is SRF dependent in undifferentiated mouse ES cells.
DISCUSSION
Previous reports by our group and others suggested strongly that the MADS box transcription factor SRF plays an essential role in regulating SMC differentiation and modulation of SMC phenotype (for a review, see reference 36). However, it remains unclear how SRF, a transcription factor that is expressed ubiquitously, can differentially activate and/or repress transcription of sets of SMC lineage-restricted genes. We hypothesized that SRF regulates smooth muscle cell gene transcription by physically associating with and synergistically activating transcription with one or more SMC lineage-restricted transcription factors (36).
In the studies described in this report, the gene encoding the human myocardin homologue was cloned, and its pattern of expression was characterized. Surprisingly, myocardin gene expression was observed not only in the heart but also in embryonic and adult SMCs. Consistent with analyses of the SMC-restricted SM22α promoter in transgenic mice (4, 44), myocardin-induced transactivation of the SM22α promoter was CArG box dependent. In addition, we demonstrated that myocardin-induced transactivation was SRF dependent. Moreover, forced expression of a dominant negative myocardin mutant protein or myocardin knockdown by siRNA significantly repressed activity of the SM22α promoter in SMCs. Most importantly, we observed that forced expression of myocardin in undifferentiated mouse ES cells activated transcription of the endogenous SM22α gene as well as expression of multiple other SMC-restricted genes. Taken together, these data provide fundamental new insights into how SRF and myocardin regulate the expression of SMC-restricted genes.
Myocardin, a member of the SAP domain family of nuclear proteins (1), functions as an SRF cofactor and is involved in differentiation of cardiac myocytes (48). Some SAP domain family members, including MRTF-A/MAL/MKL1, topoisomerase II, histone H1, lamin B1, HMG-I/Y, and nucleolin, are expressed ubiquitously, while others, such as myocardin, MRTF-B, SATB1, and p114, are expressed in a cell lineage-restricted fashion (20, 49). SAP family members have been shown to regulate nuclear architecture and chromatin organization by binding to specialized AT-rich regions of genomic DNA designated as scaffold-associated regions, some of which are located within transcriptional enhancers (8, 15, 20, 21, 27, 46).
Interestingly, the SAF-Box shows significant amino acid sequence homology to helixes 1 and 2 of the homeodomains. However, most homeodomain proteins contain a third helix that is required for DNA-binding activity (5, 11). Moreover, myocardin does not contain the conserved C-terminal RGG domain present in SAP family members, including SAF-A and SAF-B, that mediates physical association of these family members with heteronuclear RNA and regulates packaging and processing of heteronuclear RNA (18). Further studies are necessary to determine whether, like SAF-A, SAF-B, Acinus and PIAS, myocardin serves as a molecular linker capable of effecting the collapse of chromatin and nuclear fragmentation in response to apoptotic stimuli (9, 10).
Olson and coworkers reported that myocardin is expressed as early as E7.5 in the cardiac crescent and is expressed throughout the heart during embryonic and postnatal development (48). In addition, they observed transient expression of myocardin in a subset of SMCs during embryonic development. However, this earlier report did not assess whether myocardin gene expression in the embryo precedes or occurs coincident with induction of SMC differentiation markers. Moreover, this study did not address whether the myocardin gene is expressed in visceral and vascular SMCs during postnatal development.
In the mouse embryo, early SMC marker genes, including SM22α and SM-α-actin, are observed in the dorsal aorta at E9.5 (35). In fact, β-galactosidase activity is detected in the cells surrounding the dorsal aorta of mice harboring a knock-in of the bacterial lacZ gene under the control of the SM22α promoter at E8.0 (50). However, at the level of sensitivity afforded by in situ hybridization analyses, the myocardin gene was not expressed in the dorsal aorta of the mouse embryo at E9.5 and was barely detectable at E11.5. In contrast, myocardin mRNA was observed in the branchial arch arteries at E9.5. These data reveal that myocardin gene expression is differentially regulated in subsets of vascular SMCs during embryonic development. Moreover, the finding that at the level of sensitivity afforded by in situ hybridization, the SM22α was expressed in the dorsal aorta at E9.5 but the myocardin gene was not is consistent with a model in which alternative mechanisms or transcription factors may regulate the activity of the SM22α gene in some vascular SMCs. In this regard, it is noteworthy that Olson and colleagues have recently identified two myocardin-related factors, MRTF-A and MRTF-B, that may serve partially redundant functions with myocardin (49).
By contrast, expression of myocardin in visceral SMCs occurs coincident with or precedes expression of genes encoding SMC markers. High levels of myocardin mRNA were observed in the embryonic foregut, hindgut, esophagus, stomach, intestine, colon, bronchi, and, most impressively, bladder. Moreover, myocardin mRNA was observed in visceral SMC-containing human tissues during postnatal development. Taken together, these data suggest strongly that myocardin may be an essential SRF cofactor in tissue-restricted SMC subsets, most notably visceral SMCs, but that myocardin may not be an essential cofactor in some SMCs. In this regard, it is possible that one or more related SAP domain factors expressed in SMCs may be functionally redundant with myocardin. These data are consistent with previous experimental findings from our laboratory suggesting that distinct transcriptional programs may regulate and distinguish tissue-restricted subsets of SMCs (36).
The data presented herein reveal the importance of myocardin in regulating SMC gene expression and differentiation of the SMC lineage. In addition to activating transcription of the SM22α promoter, forced expression of myocardin transactivated the SM-α-actin promoter-enhancer and the SM-MyHC promoter-enhancer. Notably, SM-MyHC is considered the most definitive marker of the SMC lineage, as it is not expressed in other muscle cell lineages. Surprisingly and remarkably, we also observed that forced expression of myocardin in undifferentiated ES cells induced expression of multiple endogenous genes encoding SMC markers, including SM22α, SM-α-actin, and calponin-h1. This was unanticipated, as the inductive cues and cell-cell interactions required for specification of muscle cell lineages from undifferentiated ES cells were not provided under the ES cell culture conditions employed in these experiments (6). Whether forced expression of myocardin leads to a definitive SMC phenotype without these cues remains to be determined. However, these data strongly suggest that myocardin lies upstream in a transcriptional cascade that promotes SMC differentiation.
The demonstration that myocardin failed to transactivate the SM22α promoter in SRF-deficient ES cells is the most direct evidence to date that SMC-specific transcription is SRF dependent. Furthermore, the finding that myocardin-induced transactivation of the SM22α promoter in SRF−/− ES cells can be rescued by expression of the SRF DNA-binding domain alone is consistent with a molecular model in which SRF binds directly to the SM22α promoter and also physically associates with myocardin, presumably through the N terminus of the protein (48), thereby activating transcription of SM22α. The finding that the transcriptional activation domain of SRF was not required in this model is unanticipated and suggests that SRF may regulate SMC-specific transcription primarily by binding to and thereby linking the lineage-restricted transcription factor myocardin to specific cis-acting regulatory elements rather than through direct transcriptional activation of these regulatory elements. This model can be expanded to incorporate the requirement that myocardin-induced activation of SMC-specific CArG elements is dependent upon specific nucleotides within as well as flanking CArG boxes. As reported previously, substitution of the c-fos serum response element for the CArG boxes embedded within SME-1 and SME-4 in the SM22α promoter totally abolishes the arterial SMC-restricted activity of this element in transgenic mice (44).
While these experiments provide new insights into the transcriptional mechanisms controlling SMC differentiation, they also raise several important questions. SM22α is transiently expressed in the embryonic heart through E15, and thereafter the gene is not expressed during embryonic or postnatal development in the heart (23, 42). However, both myocardin and SRF are expressed in the heart during late embryonic and postnatal development. Therefore, molecular mechanisms must have evolved to suppress myocardin-induced activation of the SM22α promoter in the adult heart. If true, additional muscle-restricted trans-acting repressive factors that modulate the activity of myocardin may exist. Alternatively, the capacity of myocardin and/or SRF to activate the SM22α promoter may be modulated by other cell lineage-restricted signaling pathways. Conversely, in SMCs that do not express myocardin, it remains to be determined how the SM22α promoter (as well as other SMC-restricted promoters) is activated. In this regard, the recently identified muscle-restricted SAP factor MRTF-B and the more ubiquitously expressed factor MRTF-A are potentially intriguing candidates (48). Future studies addressing these and other questions should provide fundamental insights into the transcriptional programs that regulate SMC development and modulation of the SMC phenotype.
Acknowledgments
This work was supported in part by NIH grant HL56915 to M.S.P. and an allocation from the Commonwealth of Pennsylvania. M.S.P. is an Established Investigator of the American Heart Association.
We thank Lisa Gottschalk for expert preparation of the figures and Angelica Boyce for secretarial assistance.
REFERENCES
- 1.Aravind, L., and E. V. Koonin. 2000. SAP—a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 25:112-114. [DOI] [PubMed] [Google Scholar]
- 2.Arsenian, S., B. Weinhold, M. Oelgeschlager, U. Ruther, and A. Nordheim. 1998. Serum response factor is essential for mesoderm formation during mouse embryogenesis. EMBO J. 17:6289-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belaguli, N. S., W. Zhou, T.-H. T. Trinh, M. W. Majesky, and R. J. Schwartz. 1999. Dominant negative murine serum response factor: alternative splicing within the activation domain inhibits transactivation of serum response factor binding targets. Mol. Cell. Biol. 19:4582-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, P. S., L. Li, J. McAnally, and E. N. Olson. 2001. Muscle specificity encoded by specific serum response factor-binding sites. J. Biol. Chem. 276:17206-17212. [DOI] [PubMed] [Google Scholar]
- 5.Chen, C. Y., and R. J. Schwartz. 1996. Recruitment of the tinman homolog Nkx-2.5 by serum response factor activates cardiac alpha-actin gene transcription. Mol. Cell. Biol. 16:6372-6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doetschman, T. C., H. Eistetter, M. Katz, W. Schmidt, and R. Kemler. 1985. The in vitro development of blastocyst-derived embryonic stem cell lines: formation of visceral yolk sac, blood islands and myocardium. J. Embryol. Exp. Morphol. 87:27-45. [PubMed] [Google Scholar]
- 7.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 8.Garrard, W. T. 1990. Chromosomal loop organization in eukaryotic genomes, p. 163-176. In D. J. M. Lilley (ed.), Nucleic acids and molecular biology. Spinger-Verlag, Heidelberg, Germany.
- 9.Gohring, F., and F. O. Fackelmayer. 1997. The scaffold/matrix attachment region binding protein hnRNP-U (SAF-1) is directly bound to chromosomal DNA in vivo: a chemical cross-linking study. Biochemistry 36:8267-8283. [DOI] [PubMed] [Google Scholar]
- 10.Gohring, F., B. L. Schwab, P. Nicotera, M. Leist, and F. O. Fackelmayer. 1997. The novel SAR-binding domain of scaffold attachment factor a (SAF-A) is a target in apoptotic nuclear breakdown. EMBO J. 16:7361-7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grueneberg, D. A., S. Natesan, C. Alexandre, and M. Z. Gilman. 1992. Human and Drosophila homeodomain proteins that enhance the DNA-binding activity of serum response factor. Science 257:1089-1095. [DOI] [PubMed] [Google Scholar]
- 12.Herring, B. P., and A. F. Smith. 1997. Telokin expression in A10 smooth muscle cells requires serum response factor. Am. J. Physiol. 272:C1394-C1404. [DOI] [PubMed] [Google Scholar]
- 13.Hill, C. S., J. Wynne, and R. Treisman. 1994. Serum-regulated transcription by serum response factor (SRF): a novel role for the DNA binding domain. EMBO J. 13:5421-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.James, A. L., P. D. Pare, and J. C. Hogg. 1989. The mechanics of airway narrowing in asthma. Am. Rev. Respir. Dis. 139:242-246. [DOI] [PubMed] [Google Scholar]
- 15.Jenuwein, T., W. C. Forrester, L. A. Fernandez-Herrero, G. Laible, M. Dull, and R. Grosschedl. 1997. Extension of chromatin accessibility by nuclear matrix attachment regions. Nature 385:269-272. [DOI] [PubMed] [Google Scholar]
- 16.Johansen, F. E., and R. Prywes. 1993. Identification of transcriptional activation and inhibitory domains in serum response factor (SRF) by with GAL4-SRF constructs. Mol. Cell. Biol. 13:4640-4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kemp, P. R., and J. C. Metcalfe. 2000. Four isoforms of serum response factor that activate or inhibit smooth muscle-specific promoter activity. Biochem. J. 345:445-451. [PMC free article] [PubMed] [Google Scholar]
- 18.Kiledjian, M., and G. Dreyfuss. 1992. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 11:2655-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, S., H. S. Ip, M. M. Lu, C. Clendenin, and M. S. Parmacek. 1997. A serum response factor-dependent transcriptional regulatory program identifies distinct smooth muscle cell sublineages. Mol. Cell. Biol. 17:2266-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kipp, M., F. Gohring, T. Ostendorp, C. M. van Drunen, R. van Driel, M. Przybylski, and F. O. Fackelmayer. 2000. SAF-Box, a conserved protein domain that specifically recognizes scaffold attachment region DNA. Mol. Cell. Biol. 20:7480-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laemmli, U. K., E. Kas, L. Poljak, and Y. Adachi. 1992. Scaffold associated regions: cis-acting determinants of chromatin structural loops and functional domains. Curr. Opin. Genet. Dev. 2:275-285. [DOI] [PubMed] [Google Scholar]
- 22.Landerholm, T. E., X.-R. Dong, N. S. Belaguli, R. J. Schwartz, and M. W. Majesky. 1999. A role for serum response factor in coronary smooth muscle differentiation from proepicardial cells. Development 126:1053-2062. [DOI] [PubMed] [Google Scholar]
- 23.Li, L., J. M. Miano, P. Cserjesi, and E. N. Olson. 1996. SM22α, a marker of adult smooth muscle, is expressed in multiple myogenic lineages during embryogenesis. Circ. Res. 78:188-195. [DOI] [PubMed] [Google Scholar]
- 24.Li, L., J. M. Miano, B. Mercer, and E. N. Olson. 1996. Expression of the SM22α promoter in transgenic mice provides evidence for distinct transcriptional regulatory programs in vascular and visceral smooth muscle cells. J. Cell Biol. 132:849-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mack, C. P., and G. K. Owens. 1999. Regulation of smooth muscle α-actin expression in vivo is dependent on CArG elements within the 5′ and first intron promoter regions. Circ. Res. 84:852-861. [DOI] [PubMed] [Google Scholar]
- 26.Madsen, C. S., C. P. Regan, J. E. Hungerford, S. L. White, I. Manabe, and G. K. Owens. 1998. Smooth muscle-specific expression of the smooth muscle myosin heavy chain gene in transgenic mice requires 5′-flanking and first intronic DNA sequence. Circ. Res. 82:908-917. [DOI] [PubMed] [Google Scholar]
- 27.Mirkovitch, J., M. E. Mirault, and U. K. Laemmli. 1984. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell 39:223-232. [DOI] [PubMed] [Google Scholar]
- 28.Moessler, H., M. Mericskay, Z. Li, S. Nagl, D. Paulin, and J. V. Small. 1996. The SM22 promoter directs tissue-specific expression in arterial but not in venous or visceral smooth muscle cells in transgenic mice. Development 122:2415-2425. [DOI] [PubMed] [Google Scholar]
- 29.Morrisey, E. E., H. S. Ip, M. M. Lu, and M. S. Parmacek. 1996. GATA-6: a zinc finger transcription factor that is expressed in multiple cell lineages derived from lateral mesoderm. Dev. Biol. 177:309-322. [DOI] [PubMed] [Google Scholar]
- 30.Morrisey, E. E., H. S. Ip, Z. Tang, and M. S. Parmacek. 1997. GATA-4 activates transcription via two novel domains that are conserved within the GATA-4/5/6 subfamily. J. Biol. Chem. 272:8515-8524. [DOI] [PubMed] [Google Scholar]
- 31.Morrisey, E. E., Z. Tang, K. Sigrist, M. M. Lu, F. Jiang, H. S. Ip, and M. S. Parmacek. 1998. GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes Dev. 12:3579-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mortensen, R. M., D. A. Conner, S. Chao, A. A. T. Geisterfer-Lowrance, and J. G. Seidman. 1992. Production of homozygous mutant ES cells with a single targeting construct. Mol. Cell. Biol. 12:2391-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newby, A. C., and A. B. Zaltsman. 1999. Fibrous cap formation or destruction — the critical importance of vascular smooth muscle cell proliferation, migration and matrix formation. Cardiovasc. Res. 41:345-360. [PubMed] [Google Scholar]
- 34.Osbourn, J. K., P. L. Weissberg, and C. M. Shanahan. 1995. A regulatory element downstream of the rat SM22α gene transcription start point enhances reporter gene expression in vascular smooth muscle cells. Gene 154:249-253. [DOI] [PubMed] [Google Scholar]
- 35.Owens, G. K. 1998. Molecular control of vascular smooth muscle cell differentiation. Acta Physiol. Scand. 164:623-635. [DOI] [PubMed] [Google Scholar]
- 36.Parmacek, M. S. 2001. Transcriptional programs regulating vascular smooth muscle cell development and differentiation. Curr. Top. Dev. Biol. 51:69-89. [DOI] [PubMed] [Google Scholar]
- 37.Parmacek, M. S., and J. M. Leiden. 1989. Structure and expression of the murine slow/cardiac troponin C gene. J. Biol. Chem. 264:13217-13225. [PubMed] [Google Scholar]
- 38.Qian, J., A. Kumar, J. C. Szuscik, and J. L. Lessard. 1996. Tissue and developmental specific expression of murine smooth muscle γ-actin fusion genes in transgenic mice. Dev. Dynam. 207:135-144. [DOI] [PubMed] [Google Scholar]
- 39.Ross, R. 1999. Mechanisms of disease: atherosclerosis — an inflammatory disease. N. Engl. J. Med. 340:115-126. [DOI] [PubMed] [Google Scholar]
- 40.Sartorelli, V., K. A. Webster, and L. Kedes. 1990. Muscle-specific expression of the cardiac α-actin gene requires MyoD1, CArG-box binding factor, and Sp1. Genes Dev. 4:1811-1822. [DOI] [PubMed] [Google Scholar]
- 41.Schwartz, S. M., and C. E. Murry. 1998. Proliferation and the monoclonal origins of atherosclerotic lesions. Annu. Rev. Med. 49:437-460. [DOI] [PubMed] [Google Scholar]
- 42.Solway, J., J. Seltzer, F. F. Samaha, S. Kim, L. E. Alger, Q. Niu, E. E. Morrisey, H. S. Ip, and M. S. Parmacek. 1995. Structure and expression of a smooth muscle cell-specific gene SM22α. J. Biol. Chem. 270:13460-13469. [DOI] [PubMed] [Google Scholar]
- 43.Stoneking, M., J. J. Fontius, S. L. Clifford, H. Soodyall, S. S. Arcot, N. Saha, T. Jenkins, M. A. Tahir, P. L. Deininger, and M. A. Batzer. 1997. Alu insertion polymorphisms and human evolution: evidence for a larger population size in Africa. Genome Res. 7:1061-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strobeck, M., S. Kim, J. C. Zhang, C. Clendenin, K. L. Du, and M. S. Parmacek. 2001. Binding of serum response factor to CArG box sequences is necessary but not sufficient to restrict gene expression to arterial smooth muscle cells. J. Biol. Chem. 276:16418-16424. [DOI] [PubMed] [Google Scholar]
- 45.Treisman, R. 1995. DNA-binding proteins. Inside the MADS box. Nature 376:468-469. [DOI] [PubMed] [Google Scholar]
- 46.Tsutsui, K., S. Okada, S. Watarai, S. Seki, T. Yasuda, and T. Shohmori. 1993. Identification and characterization of a nuclear scaffold protein that binds the matrix attachment region DNA. J. Biol. Chem. 268:12886-12894. [PubMed] [Google Scholar]
- 47.Tybulewicz, V. L. J., C. E. Crawford, P. K. Jackson, R. T. Bronson, and R. C. Mulligan. 1991. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell 65:1153-1163. [DOI] [PubMed] [Google Scholar]
- 48.Wang, D., P. S. Chang, Z. Wang, L. Sutherland, J. A. Richardson, E. Small, P. A. Krieg, and E. N. Olson. 2001. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell 105:851-862. [DOI] [PubMed] [Google Scholar]
- 49.Wang, D., S. Li, D. Hockemeyer, L. Sutherland, Z. Wang, G. Schratt, J. A. Richardson, A. Nordheim, and E. N. Olson. 2002. Potentiation of serum response factor activity by a family of myocardin-related transcription factors. Proc. Natl. Acad. Sci. USA 99:14855-14860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, J. C., S. Kim, B. P. Helmke, W. W. Yu, K. L. Du, M. M. Lu, M. Strobeck, Q. Yu, and M. S. Parmacek. 2001. Analysis of SM22α-deficient mice reveals unanticipated insights into smooth muscle cell differentiation and function. Mol. Cell. Biol. 21:1336-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]