Abstract
The nuclear lamina is an important determinant of nuclear architecture. Mutations in A-type but not B-type lamins cause a range of human genetic disorders, including muscular dystrophy. Dominant mutations in nuclear lamin proteins have been shown to disrupt a preformed lamina structure in Xenopus egg extracts. Here, a series of deletion mutations in lamins A and B1 were evaluated for their ability to disrupt lamina structure in Chinese hamster ovary cells. Deletions of either the lamin A “head” domain or the C-terminal CaaX domain formed intranuclear aggregates and resulted in the disruption of endogenous lamins A/C but not lamins B1/B2. By contrast, “head-less” lamin B1 localized to the nuclear rim with no detectable effect on endogenous lamins, whereas lamin B1 CaaX domain deletions formed intranuclear aggregates, disrupting endogenous lamins A/C but not lamins B1/B2. Filter binding assays revealed that a head/CaaX domain lamin B1 mutant interacted much more strongly with lamins A/C than with lamins B1/B2. Regulated induction of this mutant in stable cell lines resulted in the rapid elimination of all detectable lamin A protein, whereas lamin C was trapped in a soluble form within the intranuclear aggregates. In contrast to results in Xenopus egg extracts, dominant negative lamin B1 (but not lamin A) mutants trapped replication proteins involved in both the initiation and elongation phases of replication but did not effect cellular growth rates or the assembly of active replication centers. We conclude that elimination of the CaaX domain in lamin B1 and elimination of either the CaaX or head domain in lamin A constitute dominant mutations that can disrupt A-type but not B-type lamins, highlighting important differences in the way that A- and B-type lamins are integrated into the lamina.
INTRODUCTION
The nuclear lamina is a filamentous meshwork of intermediate filament (IF) proteins that lines the nucleoplasmic face of the inner nuclear membrane (Gerace and Blobel, 1980; Aebi et al., 1986; McKeon et al., 1986; Goldberg and Allen, 1992; Zhang et al., 1996). Lamin proteins fall into two subgroups, the A-type and B-type lamins. B-type lamins are expressed in all tissues (Lehner et al., 1987; Wolin et al., 1987; Vorburger et al., 1989b), whereas expression of A-type lamins is restricted to differentiated tissues and is dispensable for both cell proliferation and mouse development (Lehner et al., 1987; Rober et al., 1989; Sullivan et al., 1999). A-type lamins are clearly important, however, because loss of A-type lamins has been implicated in several diseases, including muscular dystrophy and disorders of adipose tissue and insulin action (Morris and Manilal, 1999; Sullivan et al., 1999; Flier, 2000; Shackleton et al., 2000). Both types of lamin contain a sequence motif CaaX (C, cysteine; a, aliphatic amino acid, X, any amino acid) at the carboxyl terminus that serves as a site for modification by isoprenylation (Wolda and Glomset, 1988; Vorburger et al., 1989a; Beck et al., 1990; Firmbach-Kraft and Stick, 1993) and methylation (Chelsky et al., 1987). In addition, a splicing variant of the A-type lamins, termed lamin C (Fisher et al., 1986), lacks codons for the final 82 amino acids, including the CaaX motif. A- and B-type lamins differ in that the CaaX motif in lamin A is removed by proteolytic cleavage after nuclear import (Vorburger et al., 1989a; Weber et al., 1989; Beck et al., 1990). As with all IF proteins, the lamins have a central α-helical domain flanked by “head” and “tail” domains. Only limited structural detail concerning the way in which lamin filaments are assembled is available because lamins form paracrystals in preference to 10–13-nm filaments in vitro (Aebi et al., 1986; Moir et al., 1991; Heitlinger et al., 1992). Under the appropriate conditions, lamins do form head-to-tail assemblies in vitro for which sequences at both the N terminus and C terminus of the central α-helical rod domain appear to be important (Moir et al., 1991; Heitlinger et al., 1992).
In vivo lamins form a flattened orthagonol array of filaments at the nucleoplasmic face of the nuclear membrane (Aebi et al., 1986; Zhang et al., 1996). This filament network is disassembled and then reassembled during mitosis, this process being regulated by phosphorylation/dephosphorylation of sequences flanking the central rod domain (Heald and McKeon, 1990; Peter et al., 1990; Ward and Kirschner, 1990). Mutational analysis has also demonstrated that both CaaX modification (Holtz et al., 1989; Krohne et al., 1989; Kitten and Nigg, 1991; Hennekes and Nigg, 1994) and certain residues within the central rod domain (Holtz et al., 1989) are essential for lamina filament assembly in vivo.
More recently, the effects of deletion mutants of Xenopus lamin B1 (Ellis et al., 1997) or human lamin A (Spann et al., 1997) on nuclei assembled in Xenopus egg extracts have been investigated. Both investigations concluded that deletion of the N-terminal head domain of lamins leads to the creation of dominant negative proteins capable of preventing lamina assembly and of disrupting a preformed lamina. Both investigations also concluded that normal lamina assembly is a requirement for DNA replication. In the case of the human lamin A head deletion mutant, the localization of replication fork proteins PCNA and RFC was altered such that they were found within intranucleoplasmic lamin aggregates. Because the localization of prereplication complex proteins XMcm3 and XORC2 and initiation protein DNA polymerase α was not disrupted, these investigators proposed that a properly assembled nuclear lamina is required for the elongation phase of replication but not for the assembly of prereplication complexes (Spann et al., 1997; Moir et al., 2000). However, in the case of the Xenopus lamin B1 mutants it was demonstrated that, once sites of DNA replication have been established, disruption of the lamina does not inhibit the elongation phase of replication (Ellis et al., 1997). Because efficient replication in Xenopus egg extracts can take place in the complete absence of a nucleus (Walter et al., 1998), it remains to be determined what role, if any, the nuclear lamina plays in DNA replication.
To examine the effects of potential dominant negative mutants on lamin organization in mammalian cells, we constructed a number of head and CaaX deletion mutants in both lamin A and B1, and expressed them in Chinese hamster ovary (CHO) cells. CaaX deletion mutants of lamin B1 and lamin A resulted in the formation of intranuclear aggregates. Head deletions of lamin A but not lamin B1 also resulted in the formation of intranuclear aggregates. Endogenous A-type lamins but not B-type lamins relocated to the aggregates. In addition, significant quantities of the replication proteins PCNA, Mcm2, and Mcm7 were trapped in aggregates formed from lamin B1 mutant proteins but not lamin A mutant proteins. Deletion of coil 1a and part of coil 1b from lamin B1 CaaX-less mutants led to the formation of nuclear aggregates that failed to trap either A-type lamins or replication proteins. None of the mutant proteins affected either cell proliferation or DNA replication. These results highlight fundamental differences in the organization and behavior of A-type and B-type lamins. The implications of these results for our understanding of lamin filament assembly and the role of lamins in DNA replication are discussed.
MATERIALS AND METHODS
Plasmid Constructions
Green Fluorescent Protein (GFP) Fusions.
Construction of the expression vectors for GFP fusion proteins, ptetGFP and ptetGFPins, has been described (Izumi and Gilbert, 2000). Xenopus lamin B1 was amplified by polymerase chain reaction (PCR) and subcloned into pGEM-T (Promega, Madison, WI) as described in Ellis et al. (1997). To make ptetGFP-WTLMB1, the ptetGFP vector was cut with EcoRI and SacII, the EcoRI site was filled with Klenow, and an MscI/SacII fragment containing the entire coding region for Xenopus lamin B1 was cloned into ptetGFP. Δ2+ was sublconed into the SalI-NotI sites of pGEX-4T-3 as described in Ellis et al. (1997). The SalI/NotI fragment containing Δ2+ was subcloned into the Sal-NotI sites of ptetGFP and ptetGFPins to construct ptetGFPΔ2+ and ptetGFPinsΔ2+, respectively. To construct pEGFPΔ1, a BamHI-EcoRI fragment containing the Xenopus lamin B1 cDNA was subcloned into the BglII-EcoRI sites of pEGFP-C1 (Clonetech, Palo Alto, CA). To construct ptetGFPΔ3, the 0.8-kb MluI fragment of ptetGFPΔ2+ was substituted by the 0.9-kb MluI fragment of ptetGFP-WTLMB1.
To make the CaaX-less lamin B1 and Δ3 constructs, a fragment containing the corresponding coding sequences was amplified by using the primer 5′-AGG ATA TGC TAG CTA AGG-3′ and the mutagenic primer 5′-TAT TAG GGC CCT CAG TTT TTA TTT CCA GAC TTC TG-3′. The amplified cDNA was verified by sequencing, digested with NheI and Bsp120I, and cloned into the Bsp120I and NheI sites of tetGFP-WTLMB1 and ptetGFP-Δ3.
Human lamin A cDNA was kindly provided by H. Worman. The N-terminal part of the cDNA was amplified by using the primers 5′-ATT ACT CGA GAG ACC CCG TCC CAG CGG CG-5′ and 5′-ATT AGA ATT CGA TGT AGA CCG CCA AGC GAT, digested with XhoI and EcoRI, and then subcloned into the XhoI-EcoRI site of pBluescript KSII(+) (Stratagene, La Jolla, CA) to construct phLA-N. Next, the two oligonucleotides 5′-TCG AGA CCT GCA GGA GCT CAA TGA TCG CTT GGC GGT CTA CG-3′ and 5′-AAT TCG TAG ACC GCC AAG CGA TCA TTG AGC TCC TGC AGG TC-3′ were annealed and subcloned into the XhoI-EcoRI site of pBluescript KSII(+) to construct phLA-N33. The C-terminal part of the cDNA was amplified by using the primers 5′-ATT AGA ATT CCA TGG GCA ATT GGC AGA TCA-3′ and 5′-ATT AGG ATC CTT ACA TGA TGC TGC AGT TCT-3′, digested with EcoRI and BamHI, then subcloned into the phLA-N and phLA-N33 to construct phLA-N/C and phLA-N33/C, respectively. The CaaX-less 3′-terminal part of cDNA was amplified by using the primers 5′-ATT AGA ATT CCA TGG GCA ATT GGC AGA TCA-3′ and 5′-ATT AGG ATC CTT AGT TCT GGG GGC TCT GGG TTC G-3′, digested with EcoRI and BamHI, and then subcloned into the phLA-N and phLA-N33 to construct phLA-N/CX and phLA-N33/CX, respectively. The AccI-NcoI fragment of human lamin A cDNA was purified and subcloned into the phLA-N/C, phLA-N33/C, phLA-N/CX, and phLA-N33/CX to construct phLA-WT, phLA-head, phLA-CaaX, and phLA-haed/CaaX, respectively. The XhoI-BamHI fragments of phLA-WT, phLA-head, phLA-CaaX, and phLA-head/CaaX were subcloned into the XhoI-BamHI sites of pEGFP-C1 construct expression vectors pEGFPhLA-WT, pEGFPhLA-head, pEGFP hLA-CaaX, and pEGFPhLA-head/CaaX.
HA-tagged Fusions.
To make ptetHAins, two oligonucleotides with the sequence of 5′-CCG GTC GCC ACC ATG GTG TAC CCA TAC GAC GTC CCA GAC TAC GCT G-3′ and 5′-GGC CCA GCG TAG TCT GGG ACG TCG TAT GGG TAC ACC ATG GTG GCG A-3′ were annealed and cloned into the AgeI-Bsp120I sites of ptetGFPins. ptetHAins was digested with BamHI, filled with Klenow, redigested with MluI, and the resulting 3.7-kb ptetHA-BamHI/MluI fragment was purified. ptetGFP-WTLB1 was digested with EcoRI, filled with Klenow, and then digested with MluI. The resulting 1.2-kb EcoRI/MluI fragment was purified and ligated to the ptetHA-BamHI/MluI fragment to construct ptetHA-LMB1-A. ptetGFP-Δ2+ was digested with EcoRI, filled with Klenow, and then digested with MluI, and the resulting 1.1-kb EcoRI/MluI fragment was subcloned into the ptetHA-B/M fragment to construct ptetHA-LMB1-B. The 0.3-kb MluI fragment of ptetGFP-Δ2+ was subcloned in to the MluI site of ptetHA-LMB1-B to construct ptetHA-Δ2+. The 0.8-kb MluI fragment of ptetGFP-WTLB1 was subcloned in to the MluI site of ptetHA-LMB1-N to construct ptetHA-WTLMB1. The 0.8-kb MluI fragment of ptetGFP-WTLB1 was subcloned in to the MluI site of ptetHA-WTLMB1 to construct ptetHA-Δ3.
Cell Culture and Transfection
CHOC 400 cells, a derivative of CHO in which the dihydrofolate reductase gene has been amplified ∼500-fold (Hamlin et al., 1994), were cultured in DMEM supplemented with 5% fetal calf serum and nonessential amino acids. For transient expression, 3 × 105 cells in 35-mm dishes were grown for 24 h, and transfected with 1.0 μg of lamin expression vector and 0.1 μg of pTRE (Clontech, Palo Alto, CA) by using Lipofectoamine reagent (Life Technologies, Gaithersburg, MD) for 5 h. Typical transfection efficiencies were 10–30%.
For stable transfections, ptetGFPins-Δ2+ and ptetGFPins-Δ3 was linearized with ApaLI and transfected into Bsr26, which is a CHOC 400 derivative stably transformed with the tetracycline (tet) transactivator tTA (Izumi and Gilbert, 2000). Transfected cells were selected with 0.7 mg/ml G418 (Life Technologies) for 2 wk in the presence of 2 μg/ml tet. Individual colonies were screened for clonal populations that induced the GFP-tagged lamin mutant in 100% of the cells in the population, both by microscopic observation and by flow cytometry (Izumi and Gilbert, 2000). Stable transformant cell lines were maintained in the presence of G418 and tet and induced in the absence of tet.
Indirect Immunofluorescence Microscopy
Unless otherwise indicated, cells grown on coverslips were washed twice in phosphate-buffered saline (PBS) and fixed in ice-cold 95% ethanol:5% acetic acid at −20°C for 5 min. In experiments where cells were first extracted before fixation, cells were washed three times in ice-cold CSK buffer [10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), pH 6.8, 10 mM KCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EDTA, 0.025 mM phenylmethylsufonyl fluoride (PMSF), 10 μg/ml aprotinin], and then incubated in CSK buffer containing 0.5%Triton X-100 for 5 min at 4°C. Cells were then rinsed three times in ice-cold CSK-buffer and fixed with ethanol:acetic acid (19:1) at −20°C for 10 min. In some experiments, cells were treated with DNaseI and extracted with ammonium sulfate before fixation as described in Dyer et al. (1997), with similar results. Endogenous lamin A/C and B2 proteins were detected by using mouse monoclonal antibodies Jol2 and LN43 (Dyer et al., 1997) and Texas Red-conjugated donkey anti-mouse IgG (Jackson ImmunoResearch, West Grove, PA). Endogenous lamin B1 was detected with a goat polyclonal antibody (sc-6216; Santa Cruz Biotechnology, Santa Cruz, CA) and Alexa Fluor-conjugated anti-goat (A-11058; Molecular Probes, Eugene, OR) or Texas Red anti-goat (Jackson ImmunoResearch) antibodies. PCNA (Seikagaku Corp., Japan), Mcm7 (sc-9966; Santa Cruz Biotechnology), and nucleolin (sc-8031; Santa Cruz Biotechnology) were all detected with mouse monoclonal antibodies. Mcm2 was detected with a rabbit anti-Mcm2 antibody (gift of I. Todorov, Cythera, Inc., San Diego, CA; Dimitrova et al., 1999). To double-stain for both protein localization and bromodeoxyuridine (BrdU) incorporation, cells were labeled with 30 μg/ml BrdU for 30 min, fixed as described above, incubated for 30 min at room temperature in 1.5 N HCl, and then washed with PBS and incubated with the following two antibodies simultaneously. BrdU incorporation was detected by using a sheep anti-BrdU primary antibody (BioDesign, New York, NY) and secondary Texas Red-conjugated rabbit anti-sheep antibody (Jackson ImmunoResearch). Lamin A/C was detected by using Jol2 primary monoclonal antibody and fluorescein isothiocyanate-conjugated anti-mouse antibody. Incubations with antibodies were carried out in a humidified chamber for 1 h at room temperature. All washes after antibody incubations were done with PBS at room temperature. Photographs were taken on Kodak TRI-X400 film with a Nikon Labophot-2 microscope with a 100× 1.4 NA oil-immersion Nikon PlanApo objective. Slides were scanned with a Nikon Coolscan device and assembled by using Adobe Photoshop and Claris Draw software.
Nuclear Matrix Extraction, Immunoblotting, and Blot Overlay Assays
Cells (5 × 106) were cultured in the absence of tet for 24 h, washed twice with PBS, and scraped into 2 ml of PBS. Cell pellets were washed once with CSK and extracted with 500 μl of CSK containing 0.5% Triton X-100 at 4°C for 5 min. Nuclei and insoluble components were pelleted by centrifugation and the supernatants were saved for Western analysis (fraction 1). Pelleted material was washed twice with digestion buffer (10 mM PIPES, pH 6.8, 300 mM sucrose, 50 mM NaCl, 3 mM MgCl2, 1 mM EGTA, 0.5% Triton, 0.025 mM PMSF, 10 μg/ml aprotinin), resuspended in 500 μl of digestion buffer containing 500 U of RNase-free DNaseI (Boehringer Mannheim, Indianapolis, IN), and incubated at 30°C for 45 min. Nuclei and insoluble components were again pelleted by centrifugation and the supernatants were saved for Western analysis (fraction 2). Finally, insoluble components were extracted twice with 500 μl of extraction buffer (10 mM PIPES, 250 mM ammonium sulfate, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 0.025 mM PMSF, 10 μg/ml aprotinin) at 4°C for 5 min. After centrifugation, supernatants from both extractions were pooled (fraction 3) and pellets were taken as the nuclear matrix fraction (fraction 4).
Extracts from 100,000 cells were subjected to SDS-PAGE and transferred to polyvinylidene difluoride membrane. GFP was detected with a rabbit polyclonal antibody (Clontech) followed by peroxidase conjugated-goat anti-rabbit polyclonal antibody (Vector Laboratories, Burlingame, CA). Lamins A and C were detected with a mouse monoclonal antibody (Berkeley Antibody, Richmond, CA) followed by peroxidase conjugated-goat anti-mouse IgM polyclonal antibody (Kappel). Lamin B2 was detected with mouse monoclonal antibody (Progen, Heidelberg, Germany) followed by peroxidase conjugated-goat anti-mouse polyclonal antibody (Vector Laboratories). PCNA and Mcm7 were detected with mouse monoclonal antibodies described above followed by peroxidase conjugated-goat anti-mouse polyclonal antibody (Vector Laboratories). Lamin B1 was detected with the goat polyclonal antibody described above followed by peroxidase conjugated anti-goat antibody (Vector Laboratories). All antigens were detected by enhanced chemiluminescence.
Blot overlay assays were performed essentially as described previously (Smythe et al., 2000). Nuclear matrix fractions were prepared from HeLa by using the method described above. The fractions were resolved on SDS-PAGE, transferred to nitrocellulose, and either blotted with type specific anti-lamin antibodies or overlayed with glutathione-S-transferase (GST)-tagged-Δ1 or GST-Δ2+ and probed with anti-GST antibodies.
RESULTS
Effect of Mutant Lamin Expression on the Endogenous Lamina
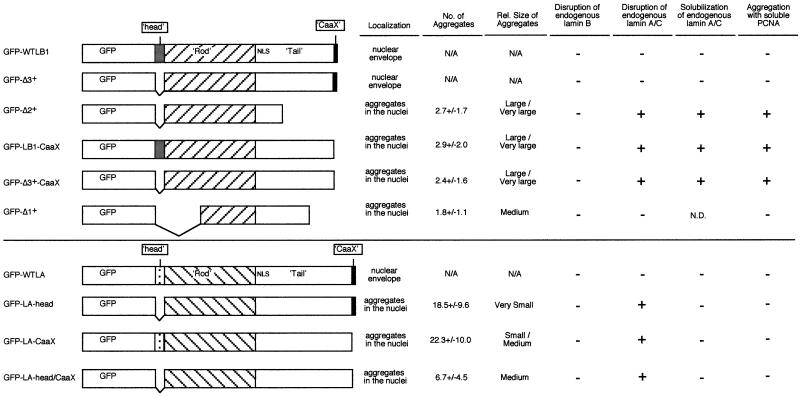
Figure 1 summarizes the lamin deletion mutants evaluated in this report and provides a brief description of the phenotypes (described in detail below) observed upon transient transfection of these constructs into CHO fibroblasts. Most of these constructions remove either the head domain (first 33 amino acids), the CaaX domain (last 4 amino acids), or both. In one case (Δ2+), 159 amino acids of the C-terminal tail domain have been removed along with the head and CaaX domains. A second mutant (Δ1+) has part of the rod domain deleted along with 66 amino acids of the tail domain (Ellis, 1997). The effects of these latter two lamin B1 mutants (Ellis et al., 1997), as well as the head-less human lamin A mutant (Spann et al., 1997) on the assembly of Xenopus sperm nuclei in Xenopus egg extracts have been described. We constructed our mutations from human lamin A and Xenopus lamin B1 to compare our results in mammalian cells with these previous studies. The effects of these mutants on lamina structure were found to be identical in Xenopus tissue culture cells (XLK-2) as well as in several other mammalian cell lines (HeLa, HDF, SW13, and HEK293; our unpublished results). All proteins were constructed as N-terminal fusions with GFP. Three of these proteins (wt, Δ2+, and Δ3+) were also constructed as fusions with the 11 amino acid hemigglutinin (HA) tag to verify that the rather large (26-kDa) GFP adduct did not effect lamin localization. Transient transfection results were identical with HA- and GFP-tagged proteins. All constructs were expressed from a tetracycline-regulatable promoter, in anticipation of constructing stable inducible cell lines, and were introduced by cotransfection with a tTA-expressing plasmid. Aliquots of transfected cells were removed at 24, 48, and 72 h thereafter. To evaluate the effects of these mutations on the integrity of the nuclear lamina, cells were fixed and stained with monoclonal antibodies specific to either mammalian lamin B2 (LN43) or lamin A/C (antibody Jol2, which recognizes both lamin splice variants A and C [Dyer et al., 1999]) and visualized with Texas Red-conjugated secondary antibody. Identical results for lamin A/C were obtained with two additional monoclonal antibodies against human lamin A/C and one additional polyclonal antibody against Chinese hamster lamin A/C. GFP-tagged lamin mutants were directly visualized by fluorescence microscopy.
Figure 1.
Schematic diagram of lamin expression constructs. Xenopus lamin B1 and human lamin A cDNAs were used to construct mutants. All constructs were tagged with GFP. Three of these proteins (wt, Δ2+, and Δ3+) were also constructed as fusions with the 11 amino acid HA tag, giving identical results. A summary of the results obtained by transfection of each mutant construct is shown to the right. The number of aggregates per cell was counted directly and shown is the mean ± SEM for two experiments in which 100 cells each were counted. N/A, not applicable. N.D., not determined.
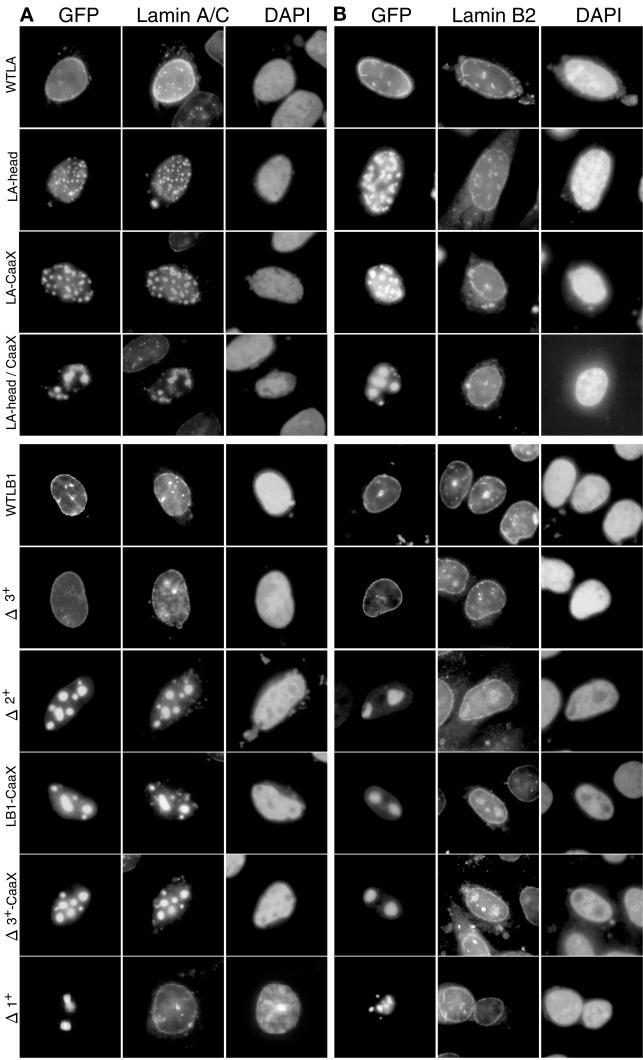
Figure 2 illustrates exemplary results of transient transfections with lamin A deletion mutants. Ectopically expressed wild-type lamin A localized to the nuclear rim, as expected, while the head-less lamin A protein aggregated in the interior of nucleus and endogenous lamin A/C was eliminated from the nuclear rim (Figure 2A). Mutants lacking either the CaaX domain or both the head and CaaX domains displayed the same phenotype as the head domain deletion, although the double deletion formed consistently larger nuclear aggregates (Figure 2, A and B). Hence, both the CaaX and the head domain of lamin A were necessary to direct ectopic lamin A protein to the nuclear envelope. None of the mutants disturbed localization of endogenous lamin B2. We conclude that ectopically expressed lamin A head domain deletion mutants disrupt endogenous lamin A/C but do not disrupt endogenous lamin B2 in mammalian cells.
Figure 2.
Effect of lamin mutants on the distribution of endogenous CHO lamin proteins. CHO cells were transfected with expression vectors for GFP-tagged lamin mutants (indicated on left of each row). After 24 h, cells were fixed and stained with anti-lamin A/C (Jol 2) (A) or anti-lamin B2 (LN43) (B) monoclonal antibody and 4, 6-diamino-2-phenylindole (DAPI) as described in MATERIALS AND METHODS. Intranuclear aggregates of lamin proteins exclude DNA, and hence do not stain with DAPI. Because anti-lamin A/C recognizes both endogenous CHO lamin A/C and transfected human lamin A, it is not certain that displaced CHO lamin A/C accumulates in the human lamin A aggregates. Also, when stained with lamin A/C antibody, transfected cells appear more brightly staining because the antibody recognizes both endogenous and ectopic lamins in the nuclear lamina.
Figure 2 also shows results of transfections with lamin B1 deletion mutants. As expected, wild-type lamin B1 localized exclusively to the nuclear rim. Surprisingly, in contrast to the head-less lamin A mutant that localized to intranuclear aggregates, localization of a head-less mutant lamin B1 protein (Δ3+) was indistinguishable from wild-type. Deletion mutant Δ2+, which has been shown to disrupt a preformed lamina in Xenopus egg extracts (Ellis et al., 1997), formed intranuclear aggregates that were able to disrupt endogenous lamin A/C localization (Figure 2A) but not endogenous lamin B2 (Figure 2B) in CHO cells. Detectable lamin A/C protein was eliminated at the nuclear rim and was relocated to the Δ2+ intranuclear aggregates. Δ2+ is missing both the N-terminal head domain that is also absent in Δ3+ and part of the C-terminal tail domain, including the CaaX domain. Hence, we deleted the CaaX domain from Δ3+ as well as from the full-length lamin B1. Neither of these mutants was directed to the nuclear periphery and instead aggregated in the nuclear interior, causing a relocalization of endogenous lamin A/C to intranuclear aggregates, similar to Δ2+ (Figure 3A). Hence, the head domain of lamin B1, unlike lamin A, is dispensable for localization to the nuclear periphery, whereas the CaaX domain of both lamins A and B1 is required for peripheral localization. Deletion mutant Δ1+, which is missing a segment of the rod domain, formed intranuclear aggregates but was not able to disrupt lamin A/C, suggesting that an intact rod domain may be required for the interaction of lamin A/C with lamin B1. None of these mutants affected endogenous lamin B2 localization (Figure 2B).
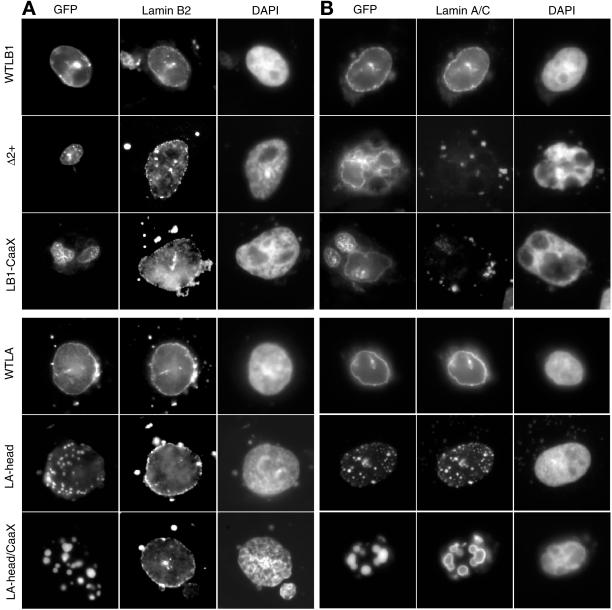
Figure 3.
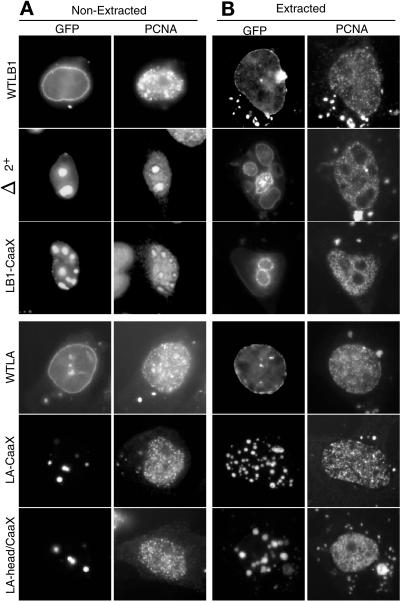
Effect of lamin mutants on the solubility of endogenous CHO lamin proteins. CHO cells transfected with GFP-lamins were extracted with Triton X-100 as described in MATERIALS AND METHODS before fixation and antibody staining as in Figure 2. Similar results were obtained when cells were extracted with high salt or treated with DNaseI before fixation. Δ3+, Δ3+-CaaX, and LA-CaaX gave results identical to WTLB1, LB1-CaaX, and LA-head, respectively. The halo-like staining of anti-lamin A/C in cells transfected with LA-CaaX and LA-head/CaaX was seen with or without (Figure 2) prior extraction of cells and is believed to be due to the inability of the anti-lamin A/C antibody to access proteins within the tightly packed GFP-lamin A aggregates.
To investigate the solubility of ectopic and endogenous lamins in transfected cells, we extracted cells with Triton X-100 before fixation (Figure 3). Wild-type GFP-lamin A and B1 proteins, as well as Δ3+ (our unpublished results), were maintained in the nuclear periphery after extraction. In addition, all lamin A deletion mutants that aggregated in the nuclear interior were resistant to extraction (e.g., Figure 3, LA-CaaX and LA-head/CaaX). In contrast, lamin B1 aggregates were partially solubilized and/or dispersed, often leaving a “halo”-like appearance, and endogenous lamin A/C was rendered completely soluble by all disruptive lamin B1 mutants (e.g., Figure 3, Δ2+ and LB1-CaaX). It was not possible for us to assess whether endogenous lamin A/C remained soluble in cells transfected with GFP-lamin A mutants because our antibodies recognize epitopes in both the endogenous (hamster) and ectopic (human) lamin A. Endogenous lamin B2 remained intact under all conditions. We conclude that dominant negative lamin B1 mutants form larger (Figure 1), more loosely associated aggregates that trap lamin A/C in a soluble form, whereas lamin A mutants form smaller insoluble aggregates (Figure 1). Importantly, none of the mutant lamin proteins affected the solubility of endogenous lamin B2.
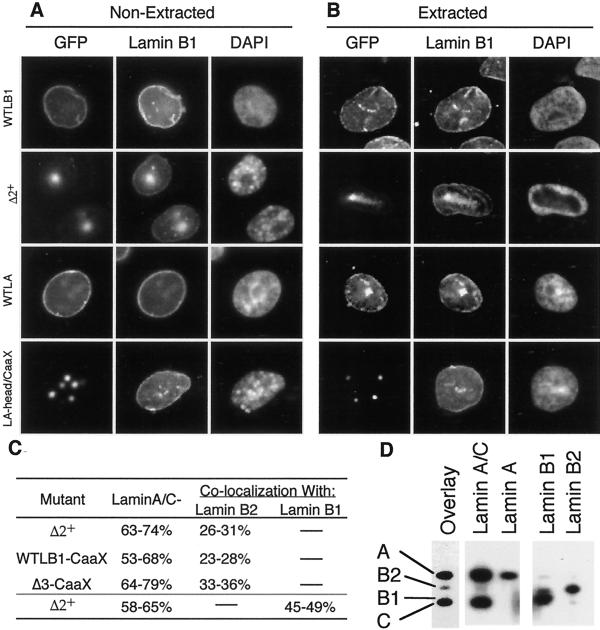
In addition to lamin B2, CHO cells contain lamin B1. Lamins B1 and B2 are the products of distinct genes and differ significantly in sequences particularly within the tail domain (Quinlan et al., 1995). To determine whether there are any differences in the interaction of our lamin mutants with the two B-type lamins, cells transfected with either wild-type or disruptive mutant lamins A and B1 were stained with an antibody specific for endogenous lamin B1 protein. Results (Figure 4, A and B) revealed that, whether or not cells were extracted with Triton-X-100 before fixation, a significant amount of endogenous lamin B1 signal remained at the nuclear periphery in the presence of mutant lamin proteins. Hence, the effect of these mutant lamin proteins on endogenous lamin B1 is similar to their effect on lamin B2.
Figure 4.
Interactions between transfected mutant lamins and endogenous lamins. (A and B) CHO cells were transfected with GFP-lamins as in Figure 2. After 48 h, cells were either fixed immediately (A) or first extracted with Triton X-100 before fixation (B). Cells were then stained with antibody against CHO lamin B1 as described in MATERIALS AND METHODS. (C) Percentage of cells transfected (GFP+ cells) with the indicated mutant lamin B1 construct that either had lost detectable lamin A/C signal at the nuclear rim (Lamin A/C−) or that displayed colocalization of endogenous B-type lamins with the mutant GFP-lamins (Lamin B colocalization) was scored. Shown are the percentages obtained in two independent experiments. (D) A nuclear matrix fraction was prepared from HeLa cells and multiple aliquots of this fraction were resolved on 10% SDS-PAGE gels. Samples were transferred to nitrocellulose, the nitrocellulose was cut into separate lanes, and individual lanes were blotted with type specific anti-lamin antibodies (indicated over each lane) to reveal the positions of the lamins (indicated to the left of the overlay lane), or overlayed with GST-Δ2+ (overlay). Parallel gels, overlayed with GST-Δ1+, did not reveal any interaction of Δ1+ with any of the endogenous lamins.
Although none of our lamin mutants eliminated the peripheral localization of lamins B1 or B2, high expression levels of CaaX-less lamin B1 mutants did result in a low but detectable level of colocalization of endogenous B-type lamins with the mutant lamin B1 (but, not mutant lamin A) aggregates (Figure 2). This colocalization was specific to disruptive lamin B1 mutants, and was not seen with disruptive lamin A mutants or with Δ1+. The amount of B-type lamins trapped in these aggregates was much less than the amount of lamin A-type lamins (although somewhat stronger for lamin B1 than for lamin B2), and was observed in less than half of transfected cells (Figure 4C). To confirm that this degree of colocalization reflected the relative ability of lamin B1 mutants to interact with A-type versus B-type lamins, HeLa cell matrix preparations were separated by SDS-PAGE, transferred to a membrane, and overlayed with GST-tagged Δ2+ or Δ1+ (Figure 4D). Consistent with the immunofluorescence experiments, Δ2+ interacted strongly with both lamins A and C but very weakly with B-type lamins, whereas Δ1+ did not show any detectable interaction with any lamin proteins (our unpublished results). We conclude that lamin B1 deletion mutants interact more strongly with A-type lamins than with B-type lamins and that this interaction requires an intact coil domain (absent in Δ1+).
Effect of Mutant Lamin Expression on PCNA Localization and DNA Replication
Several laboratories have demonstrated that nuclear lamin proteins are essential for DNA replication in Xenopus egg extracts. Without lamin proteins, nuclear membranes assemble around Xenopus sperm chromatin but do not initiate replication. This result has been observed when lamina-less nuclei are assembled either by immunodepleting extracts of lamin proteins (Newport et al., 1990; Meier et al., 1991; Jenkins et al., 1993) or by supplementing extracts with either Δ2+ (Ellis et al., 1997) or head-less lamin A protein (Spann et al., 1997). In one case (Spann et al., 1997) it was shown that the replication fork protein PCNA is relocated from replication centers to the intranuclear lamin aggregates. Hence, we investigated whether these same deletion mutants sequester PCNA from replication centers in transfected mammalian cells.
Figure 5 shows the results of experiments in which CHO cells were transfected with the lamin constructs shown in Figure 1 and then immunostained for PCNA 48 and 72 h thereafter. All CaaX-less lamin B1 mutants that were capable of disrupting endogenous lamin A/C also trapped PCNA into intranuclear aggregates (Figure 5A). Δ1+ was the only CaaX-less lamin B1 mutant that did not aggregate with PCNA (our unpublished results), suggesting that the segment of the rod domain deleted in Δ1+ is necessary for interaction with both lamin A/C (Figure 2) and PCNA. In contrast, PCNA did not colocalize with any of the human lamin A mutants. Furthermore, when cells were first extracted with Triton X-100 before fixation (Figure 5B), all of the detectable PCNA trapped within intranuclear aggregates was solubilized, whereas PCNA associated with replication centers remained intact. Hence, neither the disruption of lamin A/C nor the trapping of soluble PCNA into intranuclear aggregates affected the ability of PCNA to be recruited to replication centers.
Figure 5.
Effect of lamin mutants on the distribution of CHO PCNA. CHO cells were transfected with GFP-lamins as in Figure 2. After 48 h, cells were either fixed immediately (A) or first extracted with Triton X-100 before fixation (B). Cells were then stained with antibody against PCNA as described in MATERIALS AND METHODS. Δ3+, Δ3+-CaaX, and LA-head gave results identical to WTLB1, LB1-CaaX, and LA-CaaX, respectively.
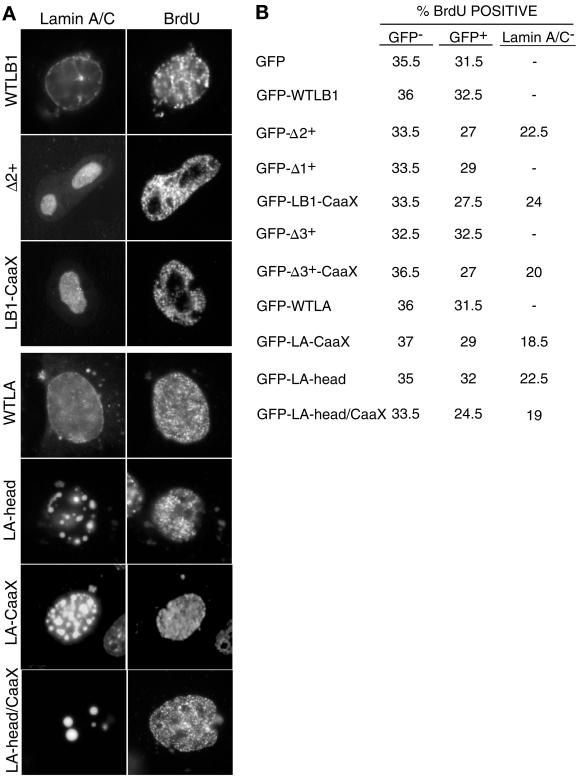
To determine whether lamin A/C-disrupted nuclei were able to form active replication centers, transfected CHO cells were pulse labeled with BrdU, fixed, and stained both for lamin A/C and for incorporation of BrdU into punctate sites of DNA replication. Results (Figure 6A) revealed that even cells that completely lacked detectable lamin A/C staining at the nuclear rim efficiently incorporated BrdU. Furthermore, the expression of these mutant GFP-lamin proteins had no effect on the percentage of cells labeled with BrdU compared with cells transfected with the GFP tag alone (Figure 6B). There was a slight reduction in the percentage of BrdU-positive cells among those transfected cells that no longer displayed any detectable lamin A/C at the nuclear periphery. This reduction was clearly not due to the sequestration of PCNA because the same effect was observed when endogenous lamin A/C was disrupted with lamin A mutants, which do not sequester PCNA (Figure 6B). Hence, this modest reduction in S-phase cells is most likely due to altered cell-cycle dynamics, possibly resulting from the high levels of ectopic GFP lamin expression in some cells after transient transfection. Furthermore, we found no evidence for differences in the intensity of BrdU labeling in cells transfected with wild-type versus mutant lamin proteins or in cells in which PCNA was sequestered into mutant lamin B1 aggregates. We conclude that, although dominant negative lamin B1 mutants can trap PCNA within intranuclear aggregates, sufficient PCNA remained available to support DNA replication.
Figure 6.
Effect of lamin mutants on DNA replication. (A) CHOC400 cells were transiently transfected with expression vectors for lamin mutants. 48 h after transfection, cells were labeled for 30 min with BrdU and stained with anti-BrdU and anti-lamin A/C antibodies as described in MATERIALS AND METHODS. (B) From the transfection shown in A, the percentage of BrdU-positive cells was scored from three distinct populations of cells: untransfected cells (GFP−), transfected cells with incomplete lamina disruption (GFP+), or transfected cells lacking detectable lamin A/C staining at the nuclear rim. Although only patterns of DNA synthesis indicative of early S-phase are shown, patterns of BrdU labeling corresponding to mid- and late-S-phase were also observed at normal frequencies (Dimitrova and Gilbert, 1999). Shown are the average results from two independent experiments.
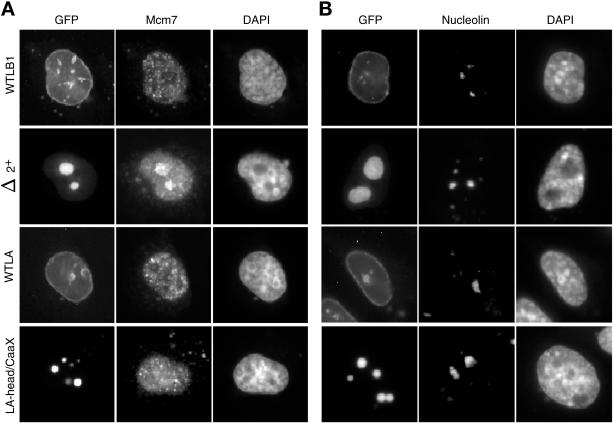
To examine the specificity of the interaction between mutant lamin proteins and replication proteins, we examined the distribution of Mcm proteins within cells transfected with lamin B1 and lamin A mutants. A previous report (Spann et al., 1997) found that lamin A head domain deletions could sequester PCNA but not Mcm3 in Xenopus egg extracts. Because Mcm proteins are part of prereplication complexes (Dimitrova et al., 1999), these investigators concluded that lamin disruption effects the activity of proteins involved in the elongation of replication forks but not initiation proteins. As shown in Figure 7A, our results revealed that both Mcm7 and Mcm2 (our unpublished results) were associated with lamin B1 Δ2+ aggregates. The association is not nonspecific because nucleolin was not trapped in these aggregates (Figure 7B) and aggregates of lamin A mutants did not trap either PCNA (Figure 5) or Mcm (Figure 7A) proteins. In a recent study we have also shown that the nuclear matrix protein lamina-associated polypeptide 2α (LAP2α) is recruited to Δ2+ aggregates, whereas NuMa is not (Dechat et al., 2000). Hence, sequestration of nuclear proteins by lamin B1 mutants is not restricted to enzymes that are exclusively involved in the elongation phase of replication.
Figure 7.
Effect of lamin mutants on the distribution of CHO Mcm7 and Nucleolin. CHO cells were transfected with GFP-lamins as in Figure 2. After 48 h, cells were fixed and stained with antibody against Mcm7 (A) or nucleolin (B) as described in MATERIALS AND METHODS. Results with an antibody against Mcm2 were indistinguishable to those seen for Mcm7 and PCNA.
Construction of Stable Tetracycline-regulatable Cell Lines Expressing Lamin Mutants
Transient transfection experiments deliver mutant proteins to only a fraction (20–30%) of the cells in a population. This limitation precluded the use of biochemical fractionation to evaluate the effect of mutant lamin expression on the solubility of endogenous lamin proteins because the majority of cells in the population would not express the mutant lamins. For these reasons, we constructed stable cell lines expressing GFP-tagged Δ2+ and Δ3+ mutants. Due to the potential toxicity of mutant lamin proteins, it was important that they be expressed from an inducible promoter. Methodology to achieve stable and homogeneous tet-regulatable gene expression in CHO cells has been described (Izumi and Gilbert, 2000), including the construction of a CHO cell line (Bsr26) that constitutively expresses the fusion protein (tTA) consisting of the tet repressor linked to the potent transcriptional activating domain from the herpes simplex virus transactivator VP16. Bsr26 cells were transfected with plasmids encoding GFP-Δ2+ or GFP-Δ3+ placed under the control of the tet-responsive promoter (Ptet). Cells were selected for the linked neomycin marker in the continuous presence of tet (to maintain tTA in an inactive state). After the expansion of colonies, tet was removed from aliquots of these clones and the expression of GFP-tagged lamin mutant was evaluated both by fluorescence microscopy and flow cytometry. Only cell lines in which GFP-Δ2+ or GFP-Δ3+ was induced in nearly 100% of the cells were chosen for further analysis.
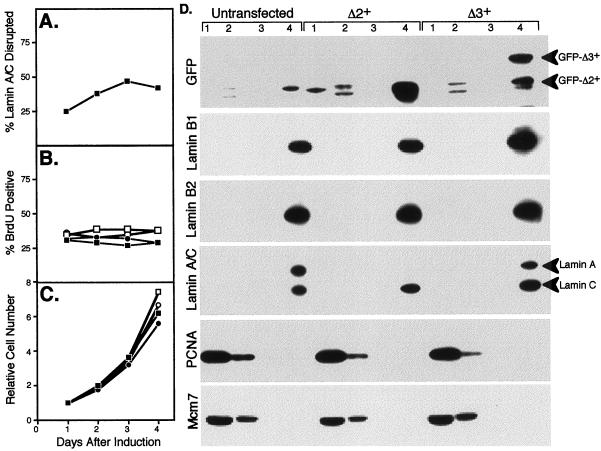
Results of these experiments (Figure 8) showed that GFP-Δ2+ rapidly accumulated into several small intranuclear aggregates that recruited endogenous lamin A/C. Within 24 h, 25% of cells had lost detectable lamin A/C signal in the nuclear rim (Figure 8A), which had become trapped in the intranuclear Δ2+ aggregates, as for transient transfections. By 2–4 d after induction, GFP-Δ2+ had accumulated in progressively larger aggregates, at which point up to 50% of cells had lost detectable lamin A/C signal in the nuclear rim (Figure 8A). Despite the lack of perinuclear lamin A/C and the presence of large intranuclear Δ2+ aggregates, there was no change in the percentage of cells in S-phase (Figure 8B) and cells were still able to grow at nearly normal rates (Figure 8C). Eventually (>4 d), levels of GFP-Δ2+ accumulated until nuclei became fragmented and GFP-Δ2+ protein could be observed leaking out into the cytoplasm. In contrast, GFP-Δ3+ localized to the nuclear rim with no effect on the localization of endogenous lamins. As with GFP-Δ2+, cells with induced GFP-Δ3+ exhibited no change in the percentage of cells in S-phase (Figure 8B) and continued to grow at normal rates over the course of at least 4 d (Figure 8C). Levels of GFP-Δ3+ accumulated until, at 4 d, ∼10% of cells exhibited levels of expression so high that GFP-Δ3+ was observed to form intranuclear tracks (also observed with a small percentage of cells after transient transfection with WTLB1 or Δ3+). These cells no longer incorporated BrdU.
Figure 8.
Tet-regulatable induction of Δ2+ leads to a loss of detectable lamin A but does not effect the solubility of lamins B or C. CHO cell lines in which either GFP-Δ2+ or GFP-Δ3+ was under the control of a tet-repressible promoter were each divided into four equal cultures and grown in medium lacking tet to induce the respective GFP-lamin protein. In all cases >80% of cells in each population induced GFP-lamin protein within 5–6 h. As a control, parallel cultures were maintained in the presence of tet. Cultures are designated as follows: GFP-Δ2+ −tet (▪); GFP-Δ2+ +tet (□); GFP-Δ3+ −tet (●); GFP-Δ3+ +tet (○). At daily intervals, the percentage of GFP-Δ2+-expressing cells that lacked dectectable lamin A/C staining at the nuclear periphery was scored as in Figure 6 (A); the percentage of BrdU-positive cells was scored as in Figure 6 (B); the total number of cells was plotted relative to that counted on day 1 after removal of tet (C). Data shown are the average of two experiments. (D) After induction for 24 h, cells were subjected to the nuclear matrix extraction protocol described in MATERIALS AND METHODS. Equal cell number equivalents were loaded on each lane of an immunoblot and probed with antibodies to the indicated proteins. Fraction 1, triton soluble protein; fraction 2, protein solubilized by DnaseI; fraction 3, protein solubilized by ammonium sulfate; and fraction 4, nuclear matrix fraction. Cell cultures not transfected with either expression vector were analyzed as a control (untransfected). Results for uninduced cells (grown in the presence of tet) were identical to untransfected cells. The anti-GFP antibody detected cross-reacting background bands in lanes 2 and 4 of all cell preparations, including untransfected cells.
The relatively homogeneous expression level of these cell lines allowed us to monitor the solubility of endogenous lamin proteins by immunoblotting after extraction. GFP-Δ2+ and GFP-Δ3+ cell lines were induced for 24 h and immunofluorescence experiments revealed results indistinguishable from those presented (Figures 2–5) for transient transfections. These same cells were then subjected to a nuclear matrix extraction procedure that allowed us to analyze each fraction by immunoblotting (Figure 6D) with antibodies against GFP, lamin B1, lamin B2, lamin A/C, PCNA, and Mcm7. Most (∼90%) GFP-Δ2+ and all GFP-Δ3+ was detected in the nuclear matrix fraction. Endogenous lamin B2 also remained in the nuclear matrix fraction in both cell lines. Interestingly, results with the lamin A/C antibody revealed different behavior for lamins A and C, which could be distinguished by their differing molecular weight. Whereas lamin C remained insoluble, lamin A was completely undetectable in all fractions from cells expressing GFP-Δ2+. This result suggests that GFP-Δ2+ expression results in lamin A degradation because all cellular fractions were equally represented in this experiment. Hence, the remaining lamin A/C signal detected by immunofluorescence in GFP-Δ2+ expressing cells must be entirely due to lamin C. Control immunoblots with uninduced cells (our unpublished results) were indistinguishable from those of untransfected cells (Figure 6D), demonstrating that the lack of lamin A signal in GFP-Δ2+-expressing cells was specifically due to the presence of GFP-Δ2+. By contrast, expression of GFP-Δ3+ had no effect on the solubility of lamin A or C. The results for PCNA and Mcm7 were indistinguishable between cell lines or expression conditions despite the differential ability of Δ2+ but not Δ3+ to trap PCNA and Mcm7 into intranuclear aggregates. Approximately 67% of PCNA and 80% of Mcm7 was found in the soluble fraction, whereas the remainder was found in the DnaseI-soluble chromatin fraction under all conditions. We conclude that deletion of the N-terminal head domain of lamin B1 (Δ3+) does not affect localization or solubility of lamin proteins in somatic cells. However, removal of the CaaX domain (Δ2+) causes the formation of intranuclear aggregates, the elimination of lamin A, disruption of lamin C, and sequestration of some of the soluble pool of PCNA and Mcm7 but, does not directly affect B-type lamins, the assembly of replication centers or cell proliferation.
DISCUSSION
We have investigated the influence of head domain and CaaX mutations on lamin filament organization and DNA replication in CHO cells. We report that a number of mutant proteins will exert a dominant negative influence over A-type lamins by causing the formation of intranuclear aggregates that act as a sink for those lamins. Differences in the behavior of lamin A mutant proteins and lamin B1 mutant proteins were observed. For lamin B1, deletion of the CaaX was essential for the formation of a dominant negative lamin mutant because a head domain deletion of lamin B1 (GFP-Δ3+) did not act as a dominant negative mutant but was stably incorporated into the nuclear envelope. Double mutants incorporating both head domain deletions and C-terminal deletions (GFP-Δ2+ and GFP-Δ3+-CaaX) were effective as dominant negative mutants but their effects were almost identical to the CaaX only deletion (GFP-LB1-CaaX). Elimination of coil 1a and most of coil 1b from C-terminal deletion mutants created a mutant protein (GFP-Δ1+) which formed aggregates but which had no dominant negative effects on endogenous lamins. Deletion of either the head (GFP-LA-head) or CaaX (GFP-LA-CaaX) domain of lamin A, or both (GFP-LA-head/CaaX), all led to the creation of dominant negative mutant proteins. Interestingly, all of the dominant negative lamin B1 mutants, but none of the dominant negative lamin A mutants, trapped a significant proportion of the replication proteins PCNA Mcm2 and Mcm7. However, this sequestration of replication proteins had no effect on the assembly or maintenance of active replication centers and did not affect cell growth and proliferation.
Deletion of the CaaX Creates Dominant Negative Lamin Mutants
Our results demonstrate that deletion of the CaaX in either lamin A or lamin B1 leads to the creation of dominant negative mutants. The formation of nuclear aggregates by CaaX-less lamin A has been reported previously although the effects on endogenous lamins were not investigated (Holtz et al., 1989). In a second study, lamin B2 CaaX-less mutant proteins were reported not to form nuclear aggregates but instead were distributed diffusely throughout the nucleoplasm (Nigg et al., 1992). Again the effects on endogenous lamins were not investigated. The slight discrepancy in the behavior of our mutant lamin B1 protein and the mutant lamin B2 protein used by Kitten and Nigg (Nigg et al., 1992) could reflect genuine differences between lamin B1 and lamin B2. However, in the study by Kitten and Nigg (Nigg et al., 1992) stably transfected cell lines were used, in which the levels of expression of CaaX mutated or deleted proteins were up to fourfold less than the level of endogenous lamins. In our assays, transient transfection of GFP-fusion proteins and inducible expression in stably transfected cell lines both led to an overexpression of protein ranging from two- to fourfold. Therefore, it is also possible that the formation of aggregates depends upon the level of expression of the mutant protein rather than differences between lamin B1 and lamin B2.
Two classes of endogenous lamins lack a CaaX. Lamin C is an alternatively spliced variant of lamin A that lacks the final 82 amino acids of lamin A with a sequence of eight residues at its C terminus being unique to lamin C (Fisher et al., 1986). Mature lamin A is processed at the nuclear envelope by proteolytic cleavage of an 18 amino acid peptide, including the modified C-terminal cysteine residue (Vorburger et al., 1989a; Weber et al., 1989; Beck et al., 1990). Why then are mature lamin A and lamin C not normally assembled into intranuclear aggregates? In fact, in proliferating human fibroblasts and mouse 3T3 cells nuclear aggregates containing lamins A and C are observed in cells that are in early G1 phase of the cell cycle but the aggregates later disappear (Goldman et al., 1992; Bridger et al., 1993). Moreover, microinjection of fluorescently labeled lamin C into proliferating 3T3 cells leads to the formation of intranuclear aggregates that persist for ∼3 h (Pugh et al., 1997). Finally, prelamin A assembles as intranuclear aggregates when isoprenylation is inhibited in cultured cells by the addition of lovastatin (Lutz et al., 1992). Consistent with this observation, when human prelamin A is added to cell-free extracts of Xenopus eggs that support nuclear assembly, intranuclear aggregates form initially but again the lamin eventually relocates to the nuclear envelope (Dyer et al., 1999). Thus, the formation of intranuclear aggregates may be explained by a general requirement for prenylation and methylation of the C-terminal cysteine residue to target lamins to the inner nuclear membrane (Holtz et al., 1989; Krohne et al., 1989; Kitten and Nigg, 1991; Hennekes and Nigg, 1994). In the absence of this modification (i.e., in prelamin A, mature lamin A and lamin C, or in CaaX-less mutants) lamins initially form intranuclear aggregates. When relatively low levels of these “unmodified” lamins are present they are eventually guided to the nuclear envelope (Pugh et al., 1997; Dyer et al., 1999). However, in situations of overexpression the aggregates may become stable and act as a sink for unprenylated lamins. Two nuclear membrane proteins bind to B-type lamins in domains that are not deleted in any of the lamin B1 dominant negative mutants reported here, namely, LBR (Mical and Monteiro, 1998) and LAP2β (Furukawa et al., 1998). Although it has been concluded previously that the LBR binding domain is not sufficient for nuclear envelope localization of lamin B (Mical and Monteiro, 1998), LAP2β does appear to have an important role in lamin filament assembly (Yang et al., 1997). Our results reinforce previous conclusions that prenylation of B-type lamins is the dominant factor in guiding these lamins to their nuclear envelope location.
Do Head Domain Mutants Have Dominant Negative Effects?
A surprising finding of our study was that head-less lamin B1 does not act as a dominant negative mutant. Two previous reports had suggested that deletion of the first 33 amino acids of both lamin B1 (Ellis et al., 1997) and lamin A (Spann et al., 1997) was important for the dominant negative effects of these proteins observed in vitro. Moreover, the phosphorylation status of a cdc2 phosphorylation site at Ser16 in the head domain of lamins largely determines their state of assembly (Heald and McKeon, 1990; Peter et al., 1990; Ward and Kirschner, 1990). Similarly, type II cytoplasmic intermediate filament fragility arises through point mutations in the head domain (Chan et al., 1993; Rugg et al., 1993; Chipev et al., 1994). Headless lamin A does act as a dominant negative mutant (see above; Spann et al., 1997). However, because ectopically expressed wild-type GFP-lamin A undergoes normal C-terminal processing (Broers et al., 1999), it seems likely that head-less lamin A should also become processed to a mature form in which the terminal 18 residues are eliminated. Under these circumstances, it could be argued that the head-less lamin A mutant protein would be effectively CaaX-less. On the other hand, endogenous lamin A would be similarly processed, yet GFP-WT lamin A becomes stably associated with the nuclear envelope. Thus, we conclude that although head-less lamin B1 does not display dominant negative effects, head-less lamin A does. This further suggests that the assembly properties of lamins A and B1 are different as we discuss below.
Dominant Negative Mutants Reveal Differences in the Assembly Properties of A-type and B-type Lamins
A surprising feature of this study was that every dominant negative mutant created affected the distribution of A-type but not B-type lamins. This finding contrasts with previous investigations in which one of the mutant proteins used here (Δ2+) as well as headless lamin A were both reported to disrupt lamin B3 in cell-free extracts of Xenopus eggs. Nevertheless, our findings suggest distinct differences in the organization of these two classes of lamins within lamina filaments. Recently, we demonstrated that in cell-free nuclear assembly extracts, the association of exogenous lamin A with the nuclear envelope was dependent upon the presence of the endogenous lamin B3 (Dyer et al., 1999). One explanation for our previous findings was that integration of A-type lamins into the lamina was by building these lamins into existing B-type lamina filaments. The data presented here provide further compelling evidence for a distinct difference in the ways in which each lamin subtype is built into lamin filaments. One explanation for this difference is related to the way in which the different lamins are anchored to the nuclear envelope. B-type lamins are permanently isoprenylated and carboxy methylated (Chelsky et al., 1987; Wolda and Glomset, 1988; Vorburger et al., 1989a; Beck et al., 1990; Firmbach-Kraft and Stick, 1993). However, prenylation itself, although necessary, is not sufficient for anchorage at the nuclear envelope (Firmbach-Kraft and Stick, 1993), and a prenyl receptor is required for this association. In addition, the integral membrane protein LAP 2β also binds specifically to B-type lamins (Foisner and Gerace, 1993) through a region in coil 1b of the rod domain (Furukawa and Kondo, 1998). Moreover, peptides corresponding to the lamin binding domain of LAP 2β prevent complete lamina assembly when injected into mitotic cells (Yang et al., 1997). Therefore, it seems likely that B-type lamins are anchored to the nuclear envelope by interactions with integral membrane proteins at two separate points along the axis of the lamin (one at the tail and one toward the N terminus of the rod). This may force the lamin to polymerize as a flattened two-dimensional array that is then rigidly associated with the interphase nuclear envelope. In contrast, A-type lamins are not anchored through their tail domains and although there are integral membrane proteins that bind to A-type lamins (LAPs 1A, 1B, and emerin) (Chipev et al., 1994; Fairley, 1999; Senior and Gerace, 1988) these proteins also bind to B-type lamins. Therefore, specific associations between A-type lamins and integral membrane proteins may either not occur or may be less stable than associations between integral membrane proteins and B-type lamins. We have suggested previously that dominant negative lamin mutants exert their effects by trapping lamins that are in a dynamic equilibrium between a filamentous and soluble nucleoplasmic state (Schmidt et al., 1994; Ellis et al., 1997). Presumably in somatic cells A-type lamins are more mobile than B-type lamins for the reasons stated above. Indeed, when nuclei are sequentially extracted to prepare nuclear matrices, a significant fraction of A-type lamins are soluble, whereas B-type lamins are completely insoluble (Venables, Quinlan, and Hutchison, unpublished data). Thus, a combination of the greater affinity of A-type lamins for the mutant proteins and the greater mobility/solubility of A-type lamins means that these proteins are readily sequestered from the lamina to nucleoplasmic lamin aggregates, whereas B-type lamins are not.
Is the Lamina Required for DNA Synthesis?
In this study DNA replication was not influenced by the presence of dominant negative mutant lamin proteins, even when overexpressed. However, mutant lamin B1 proteins containing coils 1a and 1b did trap significant amounts of PCNA, Mcm2, and Mcm7. PCNA exists in two pools in S-phase cells one soluble and one that is associated with replication centers (Bravo and Macdonald-Bravo, 1987; Dimitrova and Gilbert, 2000). Sequestration of PCNA from the soluble pool to lamin aggregates was clearly incomplete because replication centers containing PCNA still formed (Figure 4) and the same fraction of total PCNA was associated with insoluble replication factories (Figure 6D). The interaction of lamin B1 with PCNA is consistent with the observation of Moir et al. (1994) that B-type lamins associate with late replication centers. Moir et al. (1994) suggested that the appearance of lamin B1 at late replication centers resulted from a dynamic redistribution of this lamin from the lamina to replication centers. Our results indicate that this explanation is unlikely because we find that both lamins B1 and B2 are at all times tightly associated with the lamina. Moreover, PCNA does not interact with either A-type or B-type lamins in yeast two-hybrid assays (Venables, Hutchison, Warbrick, and Quinlan, unpublished data). Because the mutant proteins that we describe failed to disrupt B-type lamins we were unable to investigate whether an intact lamin B structure is required for DNA synthesis, as it appears to be in Xenopus egg extracts. However, the fact that both PCNA and the prereplication complex Mcm proteins were localized to mutant lamin aggregates demonstrates that, in mammalian cells, the association of nuclear proteins with dominant negative lamin mutants is not restricted to proteins exclusively present at the replication fork as has been concluded from studies in Xenopus egg extracts (Moir et al., 2000). Hence, our results point to some potentially important differences in lamina structure and function in somatic mammalian cells versus Xenopus egg extracts. First, disruption of the embryonic Xenopus lamin B3 can be achieved with head domain deletions of either human lamin A (Spann et al., 1997) or Xenopus lamin B3 (Ellis et al., 1997), whereas mammalian lamin B structures were not disrupted with even the highest levels of mutant protein expression. Second, Spann et al. (1997) reported that a human lamin A head domain deletion mutant sequestered proteins involved in the elongation phase of DNA replication into intranuclear aggregates within nuclei assembled in Xenopus egg extracts, whereas we found no evidence for this with the same lamin A mutants expressed in mammalian cells. Finally, in mammalian cells the range of proteins that are found in association with mutant lamin aggregates is not restricted to proteins involved in the synthesis of DNA at replication forks. Although we cannot formally exclude a direct involvement of lamins at replication centers (as has been suggested by Spann et al., 1997) our data casts serious doubt on this hypothesis. Future work should determine whether the introduction of a combination of LAP2β peptides such as those described by Yang et al. (1997) and dominant negative lamin mutants disrupts B-type lamins in S-phase cells.
ACKNOWLEDGMENTS
We thank H. Worman for providing human lamin A cDNA, I. Todorov for providing Mcm2 antibody, A. McNairn for critical reading of the manuscript, and J. Chen for technical assistance. This work was supported by a grant from the “Biodesign Research Program” of the Institute of Physical and Chemical Research (RIKEN), by a grant from the Wellcome Trust to C.J.H, and by National Institutes of Health Grant GM-57233-01 to D.M.G.
REFERENCES
- Aebi U, Cohn J, Buhle L, Gerace L. The nuclear lamina is a meshwork of intermediate-type filaments. Nature. 1986;323:560–564. doi: 10.1038/323560a0. [DOI] [PubMed] [Google Scholar]
- Beck LA, Hosick TJ, Sinensky M. Isoprenylation is required for the processing of the lamin A precursor. J Cell Biol. 1990;110:1489–1499. doi: 10.1083/jcb.110.5.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo R, Macdonald-Bravo H. Existence of two populations of cyclin/proliferating cell nuclear antigen during the cell cycle: association with DNA replication sites. J Cell Biol. 1987;105:1549–1554. doi: 10.1083/jcb.105.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridger JM, Kill IR, O'Farrell M, Hutchison CJ. Internal lamin structures within G1 nuclei of human dermal fibroblasts. J Cell Sci. 1993;104:297–306. doi: 10.1242/jcs.104.2.297. [DOI] [PubMed] [Google Scholar]
- Broers JL, Machiels BM, van Eys GJ, Kuijpers HJ, Manders EM, van Driel R, Ramaekers FC. Dynamics of the nuclear lamina as monitored by GFP-tagged A-type lamins. J Cell Sci. 1999;112:3463–3475. doi: 10.1242/jcs.112.20.3463. [DOI] [PubMed] [Google Scholar]
- Chan YM, Yu QC, Fine JD, Fuchs E. The genetic basis of Weber-Cockayne epidermolysis bullosa simplex. Proc Natl Acad Sci USA. 1993;90:7414–7418. doi: 10.1073/pnas.90.15.7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelsky D, Olson JF, Koshland DE., Jr Cell cycle-dependent methyl esterification of lamin B. J Biol Chem. 1987;262:4303–4309. [PubMed] [Google Scholar]
- Chipev CC, Yang JM, DiGiovanna JJ, Steinert PM, Marekov L, Compton JG, Bale SJ. Preferential sites in keratin 10 that are mutated in epidermolytic hyperkeratosis. Am J Hum Genet. 1994;54:179–190. [PMC free article] [PubMed] [Google Scholar]
- Dechat T, Korbei B, Vaughan OA, Vlceck S, Hutchison CJ, Foisner R. Intranuclear lamina-associated polypeptide 2α binds A-type lamins. J Cell Sci. 2000;113:3473–3484. doi: 10.1242/jcs.113.19.3473. [DOI] [PubMed] [Google Scholar]
- Dimitrova DS, Gilbert DM. The spatial position and replication timing of chromosomal domains are both established in early G1-phase. Mol Cell. 1999;4:983–993. doi: 10.1016/s1097-2765(00)80227-0. [DOI] [PubMed] [Google Scholar]
- Dimitrova DS, Gilbert DM. Stability and nuclear distribution of mammalian replication protein A heterotrimeric complex. Exp Cell Res. 2000;254:321–327. doi: 10.1006/excr.1999.4770. [DOI] [PubMed] [Google Scholar]
- Dimitrova DS, Todorov IT, Melendy T, Gilbert DM. Mcm2, but not RPA, is a component of the mammalian early G1-phase pre-replication complex. J Cell Biol. 1999;146:709–722. doi: 10.1083/jcb.146.4.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer JA, Kill IR, Pugh G, Quinlan RA, Lane EB, Hutchison CJ. Cell cycle changes in A-type lamin associations detected in human dermal fibroblasts using monoclonal antibodies. Chromosome Res. 1997;5:383–394. doi: 10.1023/a:1018496309156. [DOI] [PubMed] [Google Scholar]
- Dyer JA, Lane BE, Hutchison CJ. Investigations of the pathway of incorporation and function of lamin A in the nuclear lamina [In Process Citation] Microsc Res Tech. 1999;45:1–12. doi: 10.1002/(SICI)1097-0029(19990401)45:1<1::AID-JEMT1>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Ellis DJ. An investigation of lamin filament assembly using Xenopus egg extracts. Ph.D. Thesis. University of Dundee, Scotland, U.K.; 1997. [Google Scholar]
- Ellis DJ, Jenkins H, Whitfield WG, Hutchison CJ. GST-lamin fusion proteins act as dominant negative mutants in Xenopus egg extract and reveal the function of the lamina in DNA replication. J Cell Sci. 1997;110:2507–2518. doi: 10.1242/jcs.110.20.2507. [DOI] [PubMed] [Google Scholar]
- Fairley EA, Kendrick-Jones J, Ellis JA. The emery-driefuss muscular dystrophy phenotype arises from aberrant targeting and binding of emerin at the inner nuclear membrane. J Cell Sci. 1999;112:2571–2582. doi: 10.1242/jcs.112.15.2571. [DOI] [PubMed] [Google Scholar]
- Firmbach-Kraft I, Stick R. The role of CaaX-dependent modifications in membrane association of Xenopus nuclear lamin B3 during meiosis and the fate of B3 in transfected mitotic cells. J Cell Biol. 1993;123:1661–1670. doi: 10.1083/jcb.123.6.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher DZ, Chaudhary N, Blobel G. cDNA sequencing of nuclear lamins A and C reveals primary and secondary structural homology to intermediate filament proteins. Proc Natl Acad Sci USA. 1986;83:6450–6454. doi: 10.1073/pnas.83.17.6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flier JS. Pushing the envelope on lipodystrophy [news; comment] Nat Genet. 2000;24:103–104. doi: 10.1038/72734. [DOI] [PubMed] [Google Scholar]
- Foisner R, Gerace L. Integral membrane proteins of the nuclear envelope interact with lamins and chromosomes, and binding is modulated by mitotic phosphorylation. Cell. 1993;73:1267–1279. doi: 10.1016/0092-8674(93)90355-t. [DOI] [PubMed] [Google Scholar]
- Furukawa K, Fritze CE, Gerace L. The major nuclear envelope targeting domain of LAP2 coincides with its lamin binding region but is distinct from its chromatin interaction domain. J Biol Chem. 1998;273:4213–4219. doi: 10.1074/jbc.273.7.4213. [DOI] [PubMed] [Google Scholar]
- Furukawa K, Kondo T. Identification of the lamina-associated-polypeptide-2-binding domain of B-type lamin. Eur J Biochem. 1998;251:729–733. doi: 10.1046/j.1432-1327.1998.2510729.x. [DOI] [PubMed] [Google Scholar]
- Gerace L, Blobel G. The nuclear envelope lamina is reversibly depolymerized during mitosis. Cell. 1980;19:277–287. doi: 10.1016/0092-8674(80)90409-2. [DOI] [PubMed] [Google Scholar]
- Goldberg MW, Allen TD. High resolution scanning electron microscopy of the nuclear envelope: demonstration of a new, regular, fibrous lattice attached to the baskets of the nucleoplasmic face of the nuclear pores. J Cell Biol. 1992;119:1429–1440. doi: 10.1083/jcb.119.6.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman AE, Moir RD, Montag LM, Stewart M, Goldman RD. Pathway of incorporation of microinjected lamin A into the nuclear envelope. J Cell Biol. 1992;119:725–735. doi: 10.1083/jcb.119.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlin JL, Mosca PJ, Levenson VV. Defining origins of replication in mammalian cells. Biochim Biophys Acta. 1994;1198:85–111. doi: 10.1016/0304-419x(94)90008-6. [DOI] [PubMed] [Google Scholar]
- Heald R, McKeon F. Mutations of phosphorylation sites in lamin A that prevent nuclear lamina disassembly in mitosis. Cell. 1990;61:579–589. doi: 10.1016/0092-8674(90)90470-y. [DOI] [PubMed] [Google Scholar]
- Heitlinger E, Peter M, Lustig A, Villiger W, Nigg EA, Aebi U. The role of the head and tail domain in lamin structure and assembly: analysis of bacterially expressed chicken lamin A and truncated B2 lamins. J Struct Biol. 1992;108:74–89. doi: 10.1016/1047-8477(92)90009-y. [DOI] [PubMed] [Google Scholar]
- Hennekes H, Nigg EA. The role of isoprenylation in membrane attachment of nuclear lamins. A single point mutation prevents proteolytic cleavage of the lamin A precursor and confers membrane binding properties. J Cell Sci. 1994;107:1019–1029. doi: 10.1242/jcs.107.4.1019. [DOI] [PubMed] [Google Scholar]
- Holtz D, Tanaka RA, Hartwig J, McKeon F. The CaaX motif of lamin A functions in conjunction with the nuclear localization signal to target assembly to the nuclear envelope. Cell. 1989;59:969–977. doi: 10.1016/0092-8674(89)90753-8. [DOI] [PubMed] [Google Scholar]
- Izumi M, Gilbert DM. Homogeneous tetracycline-regulatable gene expression in mammalian fibroblasts. J Cell Biochem. 2000;76:280–289. doi: 10.1002/(sici)1097-4644(20000201)76:2<280::aid-jcb11>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Jenkins H, Holman T, Lyon C, Lane B, Stick R, Hutchison C. Nuclei that lack a lamina accumulate karyophilic proteins and assemble a nuclear matrix. J Cell Sci. 1993;106:275–285. doi: 10.1242/jcs.106.1.275. [DOI] [PubMed] [Google Scholar]
- Kitten GT, Nigg EA. The CaaX motif is required for isoprenylation, carboxyl methylation, and nuclear membrane association of lamin B2. J Cell Biol. 1991;113:13–23. doi: 10.1083/jcb.113.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krohne G, Waizenegger I, Hoger TH. The conserved carboxy-terminal cysteine of nuclear lamins is essential for lamin association with the nuclear envelope. J Cell Biol. 1989;109:2003–2011. doi: 10.1083/jcb.109.5.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner CF, Stick R, Eppenberger HM, Nigg EA. Differential expression of nuclear lamin proteins during chicken development. J Cell Biol. 1987;105:577–587. doi: 10.1083/jcb.105.1.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz RJ, Trujillo MA, Denham KS, Wenger L, Sinensky M. Nucleoplasmic localization of prelamin A: implications for prenylation-dependent lamin A assembly into the nuclear lamina [published erratum appears in Proc. Natl. Acad. Sci. USA (1992) 89, 5699] Proc Natl Acad Sci USA. 1992;89:3000–3004. doi: 10.1073/pnas.89.7.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeon FD, Kirschner MW, Caput D. Homologies in both primary and secondary structure between nuclear envelope and intermediate filament proteins. Nature. 1986;319:463–468. doi: 10.1038/319463a0. [DOI] [PubMed] [Google Scholar]
- Meier J, Campbell KH, Ford CC, Stick R, Hutchison CJ. The role of lamin LIII in nuclear assembly and DNA replication, in cell-free extracts of Xenopus eggs. J Cell Sci. 1991;98:271–279. doi: 10.1242/jcs.98.3.271. [DOI] [PubMed] [Google Scholar]
- Mical TI, Monteiro MJ. The role of sequences unique to nuclear intermediate filaments in the targeting and assembly of human lamin B: evidence for lack of interaction of lamin B with its putative receptor. J Cell Sci. 1998;111:3471–3485. doi: 10.1242/jcs.111.23.3471. [DOI] [PubMed] [Google Scholar]
- Moir R, Donaldson A, Stewart M. Expression in E. coli of human lamins A and C: influence of head and tail domains on assembly properties and paracrystal formation. J Cell Sci. 1991;99:363–372. doi: 10.1242/jcs.99.2.363. [DOI] [PubMed] [Google Scholar]
- Moir RD, Montag LM, Goldman RD. Dynamic properties of nuclear lamins: lamin B is associated with sites of DNA replication. J Cell Biol. 1994;125:1201–1212. doi: 10.1083/jcb.125.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir RD, Spann TP, Herrmann H, Goldman RD. Disruption of nuclear lamin organization blocks the elongation phase of DNA replication. J Cell Biol. 2000;149:1179–1192. doi: 10.1083/jcb.149.6.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GE, Manilal S. Heart to heart: from nuclear proteins to Emery-Dreifuss muscular dystrophy. Hum Mol Genet. 1999;8:1847–1851. doi: 10.1093/hmg/8.10.1847. [DOI] [PubMed] [Google Scholar]
- Newport JW, Wilson KL, Dunphy WG. A lamin-independent pathway for nuclear envelope assembly. J Cell Biol. 1990;111:2247–2259. doi: 10.1083/jcb.111.6.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA, Kitten GT, Vorburger K. Targeting lamin proteins to the nuclear envelope: the role of CaaX box modifications. Biochem Soc Trans. 1992;20:500–504. doi: 10.1042/bst0200500. [DOI] [PubMed] [Google Scholar]
- Peter M, Nakagawa J, Doree M, Labbe JC, Nigg EA. In vitro disassembly of the nuclear lamina and M phase-specific phosphorylation of lamins by cdc2 kinase. Cell. 1990;61:591–602. doi: 10.1016/0092-8674(90)90471-p. [DOI] [PubMed] [Google Scholar]
- Pugh GE, Coates PJ, Lane EB, Raymond Y, Quinlan RA. Distinct nuclear assembly pathways for lamins A and C lead to their increase during quiescence in Swiss 3T3 cells. J Cell Sci. 1997;110:2483–2493. doi: 10.1242/jcs.110.19.2483. [DOI] [PubMed] [Google Scholar]
- Quinlan R, Hutchison C, Lane B. Intermediate filament proteins. Protein Profile. 1995;2:795–952. [PubMed] [Google Scholar]
- Rober RA, Weber K, Osborn M. Differential timing of nuclear lamin A/C expression in the various organs of the mouse embryo and the young animal: a developmental study. Development. 1989;105:365–378. doi: 10.1242/dev.105.2.365. [DOI] [PubMed] [Google Scholar]
- Rugg EL, Morley SM, Smith FJ, Boxer M, Tidman MJ, Navsaria H, Leigh IM, Lane EB. Missing links: Weber-Cockayne keratin mutations implicate the L12 linker domain in effective cytoskeleton function. Nat Genet. 1993;5:294–300. doi: 10.1038/ng1193-294. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Tschodrich-Rotter M, Peters R, Krohne G. Properties of fluorescently labeled Xenopus lamin A in vivo. Eur J Cell Biol. 1994;65:70–81. [PubMed] [Google Scholar]
- Senior A, Gerace L. Integral membrane proteins specific to the inner nuclear membrane and associated with the nuclear lamina. J Cell Biol. 1988;107:2029–2036. doi: 10.1083/jcb.107.6.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleton S, Lloyd DJ, Jackson SN, Evans R, Niermeijer MF, Singh BM, Schmidt H, Brabant G, Kumar S, Durrington PN, Gregory S, O'Rahilly S, Trembath RC. LMNA, encoding lamin A/C, is mutated in partial lipodystrophy [see comments] Nat Genet. 2000;24:153–156. doi: 10.1038/72807. [DOI] [PubMed] [Google Scholar]
- Smythe C, Jenkins HE, Hutchison CJ. Incorporation of the nuclear pore basket protein nup153 into nuclear pore structures is dependent upon lamina assembly: evidence from cell-free extracts of Xenopus eggs [In Process Citation] EMBO J. 2000;19:3918–3931. doi: 10.1093/emboj/19.15.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann TP, Moir RD, Goldman AE, Stick R, Goldman RD. Disruption of nuclear lamin organization alters the distribution of replication factors and inhibits DNA synthesis. J Cell Biol. 1997;136:1201–1212. doi: 10.1083/jcb.136.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorburger K, Kitten GT, Nigg EA. Modification of nuclear lamin proteins by a mevalonic acid derivative occurs in reticulocyte lysates and requires the cysteine residue of the C-terminal CXXM motif. EMBO J. 1989a;8:4007–4013. doi: 10.1002/j.1460-2075.1989.tb08583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorburger K, Lehner CF, Kitten GT, Eppenberger HM, Nigg EA. A second higher vertebrate B-type lamin. cDNA sequence determination and in vitro processing of chicken lamin B2. J Mol Biol. 1989b;208:405–415. doi: 10.1016/0022-2836(89)90505-6. [DOI] [PubMed] [Google Scholar]
- Walter J, Sun L, Newport J. Regulated chromosomal DNA replication in the absence of a nucleus. Mol Cell. 1998;1:519–529. doi: 10.1016/s1097-2765(00)80052-0. [DOI] [PubMed] [Google Scholar]
- Ward GE, Kirschner MW. Identification of cell cycle-regulated phosphorylation sites on nuclear lamin C. Cell. 1990;61:561–577. doi: 10.1016/0092-8674(90)90469-u. [DOI] [PubMed] [Google Scholar]
- Weber K, Plessmann U, Traub P. Maturation of nuclear lamin A involves a specific carboxy-terminal trimming, which removes the polyisoprenylation site from the precursor; implications for the structure of the nuclear lamina. FEBS Lett. 1989;257:411–414. doi: 10.1016/0014-5793(89)81584-4. [DOI] [PubMed] [Google Scholar]
- Wolda SL, Glomset JA. Evidence for modification of lamin B by a product of mevalonic acid. J Biol Chem. 1988;263:5997–6000. [PubMed] [Google Scholar]
- Wolin SL, Krohne G, Kirschner MW. A new lamin in Xenopus somatic tissues displays strong homology to human lamin A. EMBO J. 1987;6:3809–3818. doi: 10.1002/j.1460-2075.1987.tb02717.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Guan T, Gerace L. Lamin-binding fragment of LAP2 inhibits increase in nuclear volume during the cell cycle and progression into S phase. J Cell Biol. 1997;139:1077–1087. doi: 10.1083/jcb.139.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Jenkins H, Goldberg M, Allen T, Hutchison C. Nuclear lamina and nuclear matrix organization in sperm pronuclei assembled in Xenopus egg extract. J Cell Sci. 1996;109:2275–2286. doi: 10.1242/jcs.109.9.2275. [DOI] [PubMed] [Google Scholar]