Abstract
The cell wall is the major limiting factor for plant growth. Wall extension is thought to result from the loosening of its structure. However, it is not known how this is coordinated with wall synthesis. We have identified two novel allelic cellulose-deficient dwarf mutants, kobito1-1 and kobito1-2 (kob1-1 and kob1-2). The cellulose deficiency was confirmed by the direct observation of microfibrils in most recent wall layers of elongating root cells. In contrast to the wild type, which showed transversely oriented parallel microfibrils, kob1 microfibrils were randomized and occluded by a layer of pectic material. No such changes were observed in another dwarf mutant, pom1, suggesting that the cellulose defect in kob1 is not an indirect result of the reduced cell elongation. Interestingly, in the meristematic zone of kob1 roots, microfibrils appeared unaltered compared with the wild type, suggesting a role for KOB1 preferentially in rapidly elongating cells. KOB1 was cloned and encodes a novel, highly conserved, plant-specific protein that is plasma membrane bound, as shown with a green fluorescent protein–KOB1 fusion protein. KOB1 mRNA was present in all organs investigated, and its overexpression did not cause visible phenotypic changes. KOB1 may be part of the cellulose synthesis machinery in elongating cells, or it may play a role in the coordination between cell elongation and cellulose synthesis.
INTRODUCTION
A major question in plant biology is how cells expand despite the presence of the cell wall. Cell walls are sufficiently rigid to resist the high osmotic pressure of the protoplast and sufficiently plastic to allow cells to grow. The measurement of turgor pressure in expanding cells has shown that, in general, cells grow as a result of the loosening of the wall rather than an increase in turgor (Cosgrove, 1993). In vitro studies have shown that expansins are involved in cell wall plasticity (McQueen-Mason et al., 1992). These small cell wall proteins are suspected to promote the slippage between cellulose and xyloglucan chains by breaking the hydrogen bonds that hold these polymers together (McQueen-Mason et al., 1992).
Other cell wall enzymes also may contribute to growth by hydrolyzing, grafting, or modifying xyloglucans or pectins. These wall modifications may lead in turn to changes in the pore size or viscosity of the wall matrix. In most models of growth based on experiments with isolated cell walls, cell wall synthesis is ignored and considered not to play a critical role. This view is further supported by observations that chemical inhibition of cellulose synthesis does not necessarily lead to growth inhibition (Brummell and Hall, 1985). However, the strong growth defects observed in mutants with relatively mild defects in the synthesis of cellulose (Arioli et al., 1998; Nicol et al., 1998; Fagard et al., 2000) suggest that cellulose synthesis actively contributes to the control of cell elongation.
In addition to its role in cell elongation, cellulose synthesis also is crucial for cell plate formation in dividing cells and secondary wall formation after cessation of growth. This is demonstrated clearly by the abnormal cell plate formation in dividing cells in the presence of 2,6-dichlorobenzonitrile (DCB), an inhibitor of cellulose synthesis (Wells et al., 1994) and the collapsing of xylem cells observed in the irregular xylem (irx) mutants as a result of a specific defect in cellulose synthesis within the secondary cell wall (Turner and Somerville, 1997). Together, these results indicate that cellulose synthesis plays a central role in three stages of the life of a cell: division, expansion, and differentiation.
The synthesis of cellulose remains a poorly understood process. The site of synthesis is a plasma membrane–bound hexameric protein complex referred to as “terminal complex” or “rosette” (Mueller and Brown, 1980). The composition of the complex is not known, although it is thought to contain multiple cellulose synthase catalytic subunits (CESAs) (Kimura et al., 1999). CESAs are encoded by a multigene family in plants and are represented by 10 isoforms in the Arabidopsis genome (Richmond, 2000).
Another protein essential for normal cellulose synthesis is KOR, a membrane-bound endo-β-1,4-glucanase (Nicol et al., 1998; Lane et al., 2001; Sato et al., 2001). The exact role of this protein is not known. kor mutants show reduced cellulose synthesis in growing cells as well as in secondary walls (S.R. Turner, personal communication), and strong alleles show cytokinesis defects (Zuo et al., 2000), suggesting that KOR is involved in cellulose synthesis during cell division, cell elongation, and secondary cell wall formation.
By contrast, genetic evidence shows that CESA isoforms are specialized in terms of function. Indeed, strong mutant alleles of rsw1 and prc1 mutated in CESA1 and CESA6, respectively, show cell expansion defects but are not affected in cytokinesis (Fagard et al., 2000; Gillmor et al., 2002). This finding may indicate that CESA1 and CESA6 are functionally redundant with other CESAs for cell plate formation or functionally specialized for elongating cells. Finally, irregular xylem mutants, irx1, irx3, and irx5, which show cellulose defects specifically in secondary cell wall, are mutated in CESA8, CESA7, and CESA4, respectively (Taylor et al., 1999, 2000).
The functional specialization of CESAs for cellulose synthesis may simply reflect the different constraints that are imposed on the cellulose synthesis machinery during cytokinesis, cell elongation, or differentiation. This may relate to differences in carbon metabolism, the size of the microfibrils, the orientation of the microfibrils, or the stability or intracellular localization of the cellulose synthase complex. Against this background, the identification of novel mutants affected specifically in the synthesis of cellulose during particular cellular stages may be highly informative.
Here, we describe a new locus, KOBITO1 (KOB1), that was identified by mutations that cause a defect in the synthesis of cellulose during the elongation phase of the cell. By contrast, in dividing cells and in secondary thickenings of xylem vessels of the mutants, microfibrils appeared unaffected. KOB1 encodes a novel plasma membrane–anchored protein. Possible roles for KOB1 in the synthesis of cellulose during cell elongation are discussed.
RESULTS
Kob1 Phenotype
A screen for mutants showing reduced growth anisotropy in the dark-grown hypocotyl led to the identification of kob1-1 and kob1-2. The mutants were isolated from a T-DNA–mutagenized population in the accession Wassilewskija (Ws) and an ethyl methanesulfonate–mutagenized population in Columbia (Col0), respectively. Both mutations are recessive, monogenic, and nuclear, and complementation tests showed that they are allelic (data not shown).
Hypocotyls of the mutant grown in the dark for 7 days were five times shorter than those of the wild type (Figure 1E) and showed a reduced apical hook compared with wild-type seedlings. When grown in the light, kob1 hypocotyls and roots were shorter than those of the wild type (Figure 1F). Mature greenhouse-grown plants homozygous for kob1 showed a strong dwarf phenotype: inflorescence stems were short with small internodes, and floral organs presented a miniature morphology (Figures 1A to 1D).
Figure 1.
Phenotype of kob1.
(A) to (D) Adult plants grown for 6 weeks in a greenhouse.
(A) From left to right: Col0, Ws, kob1-2 (Col0 background), and kob1-1 (Ws background).
(B) Detail of kob1-2.
(C) Detail of kob1-1.
(D) Siliques of kob1-1. Mutant kob1 plants are severely dwarfed and sterile.
(E) Seven-day-old dark-grown wild-type (left) and kob1-1 (right) seedlings (two seedlings each).
(F) Seven-day-old light-grown wild-type (left) and kob1-1 (right) seedlings (two seedlings each).
Mutant plants were sterile. Root hairs showed normal length and morphology, indicating that kob1 mutations do not affect tip-growing cells (data not shown). This was confirmed by the absence of a segregation bias for kob1 alleles (data not shown), which suggests that pollen tube growth also was unaffected in the mutant. The phenotype of kob1-2 was stronger than that of kob1-1.
The Kob1 phenotype is not a result of a brassinosteroid, auxin, or gibberellin deficiency, because none of these hormones added to the growth medium rescued the dwarf phenotype (data not shown). In addition, the Kob1 phenotype is not attributable to an overproduction of ethylene, because various concentrations of an inhibitor of ethylene perception (aminoethoxyvinylglycine) did not rescue the growth defect (data not shown).
Cellular Defects in kob1
To investigate the cellular effects of kob1 mutations, we studied transverse sections halfway along the fully elongated hypocotyls of 7-day-old dark-grown seedlings. In this organ, cortical and epidermal cells do not undergo significant cell divisions during postembryonic development (Gendreau et al., 1997). The overall anatomy of the mutant hypocotyl was unaltered, but it showed an increase in girth as a result of a dramatically increased diameter of all cell types (Figures 2A and 2B).
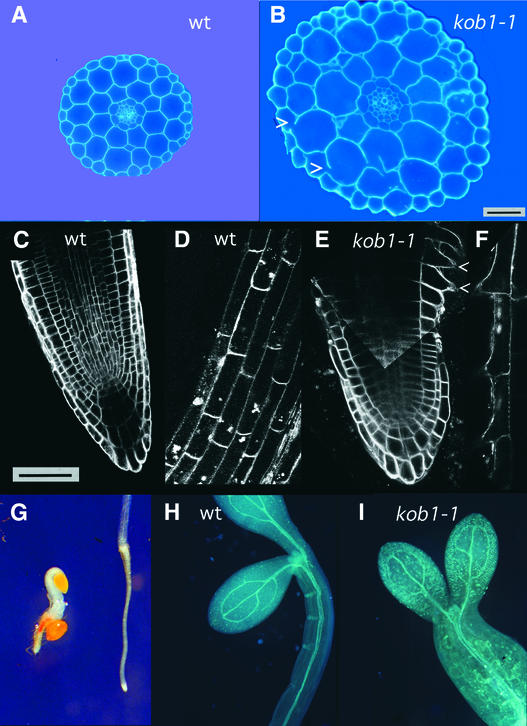
Figure 2.
Cellular Phenotype of kob1-1 Hypocotyls and Roots.
(A) and (B) Transverse sections halfway along hypocotyls of 5-day-old dark-grown wild-type (A) and kob1-1 (B) seedlings after Calcofluor staining. Mutant hypocotyls show unaltered anatomy, but all cells show increased radial expansion compared with the wild type. Mutant cell walls are interrupted frequently (arrowheads in [B]). wt, wild type. Bar in (B) = 50 μm for (A) and (B).
(C) to (F) Confocal optical median sections, stained in vivo with FM1-43, of 7-day-old light-grown wild-type ([C] and [D]) and kob1-1 ([E] and [F]) roots. Root meristem ([C] and [E]), elongation zone (D), and mature part (F) are shown. Note the dramatically reduced elongation zone in kob1-1, as shown by the premature appearance of root hairs (arrowheads). Bar in (C) = 50 μm for (C) to (F).
(G) Phloroglucinol staining of 4-day-old dark-grown kob1-1 (left) and wild-type (right) seedlings showing red staining of ectopic lignin in the mutant root.
(H) and (I) Sirofluor staining of 4-day-old dark-grown wild-type (H) and kob1-1 (I) seedlings showing light blue staining of ectopic callose in the mutant.
The cellular phenotype also was investigated in roots using longitudinal optical root sections obtained by confocal microscopy (Figures 2C to 2F). The advantage of studying this organ is that all stages of cell development can be examined simultaneously. The root meristem is located at the root apex and includes a set of continuously dividing cells that produce the basic cell types and define their spatial organization. The elongation zone is delineated distally by the end of the mitotic zone, which in general coincides with the proximal end of the lateral root cap (Beemster and Baskin, 1998), and proximally by the differentiation zone, which is characterized by the appearance of root hairs.
In mutant roots, the organization in cell files and division planes appeared unaltered (Figure 2E), suggesting that the mutation does not affect cytokinesis. The absence of a cytokinesis phenotype also was shown in mutant embryos using confocal microscopy. The homozygous plants being sterile, we studied 12 embryos from the progeny of a heterozygous plant; no differences compared with the wild type were observed in any of them (data not shown). By contrast, cell elongation was affected dramatically in kob1-1 roots.
After cessation of mitosis, cells had barely elongated before entering the differentiation zone, as indicated by the premature appearance of root hairs. However, in contrast to hypocotyl cells, which showed a greatly increased diameter (Figures 2A and 2B), mature roots cells were significantly shorter but not wider than those of the wild type (Figures 2D and 2F). Mature root cortex cells of kob1-1 were at least two times shorter (wild type, 79.59 ± 11.19 μm [n = 30]; kob1-1, 37.10 ± 6.24 μm [n = 30]) but not wider (wild type, 28.71 ± 3.83 μm; kob1-1, 23.96 ± 3.67 μm) than those of the wild type.
Cell Wall Defects in kob1
As shown in Figures 2A and 2B, incomplete cell walls were visible in epidermal, cortical, and endodermal cell layers on transverse sections through dark-grown hypocotyls. Incomplete walls also have been found in wild-type seedlings treated with the cellulose synthesis inhibitors DCB and isoxaben and in prc1 seedlings mutated for the cellulose synthase isoform CESA6 (Fagard et al., 2000). Incomplete cell walls may reflect abortive cytokinesis or may arise during cell elongation. Because no or very few cell divisions take place during postembryonic hypocotyl elongation (Gendreau et al., 1997) and no cell wall defects were observed in mature embryos, the defects most likely are not a result of a cytokinesis defect.
Changes in Noncellulosic Polysaccharides
Cell wall material isolated from wild-type and kob1-1 seedlings was hydrolyzed with 2 N trifluoroacetic acid (TFA), and the neutral sugar and uronic acid contents were determined. kob1-1 fractions showed minor changes in the composition of noncellulosic polymers compared with the wild type (Figure 3A). A relative increase in galacturonic acid suggested an increased pectin content enriched for homogalacturonan relative to rhamnogalacturonan, as suggested by the unaltered rhamnose content.
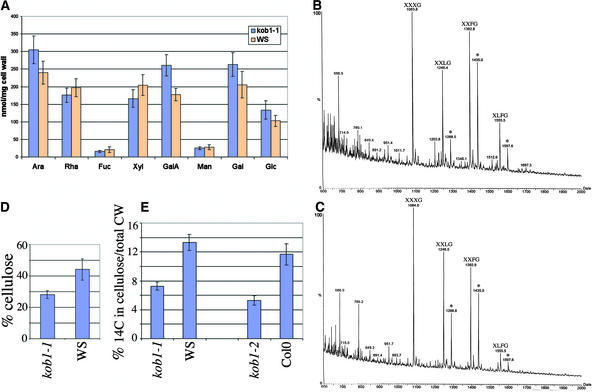
Figure 3.
Cell Wall Alterations in Dark-Grown kob1-1.
(A) Neutral sugar and galacturonic acid contents of TFA-hydrolyzable cell wall fractions of wild-type (Ws) and kob1-1 seedlings. Mutant cell walls show increased pectin content, as shown by the higher galacturonic acid levels compared with the wild-type control.
(B) and (C) Matrix-assisted laser desorption ionization–time of flight mass spectra of the fragments produced from wild-type (B) and kob1-1 (C) cell walls. No significant differences in the composition of xyloglucan fragments released by endo-β-1,4-glucanase treatment was observed in the mutant compared with the wild type. XXXG, XXLG, XXFG, and XLFG refer to the xyloglucan fragments according to the nomenclature of Fry et al. (1993). The acetylated versions of the fragments XXLG, XXFG, and XLFG (from left to right) are indicated by asterisks.
(D) and (E) Cellulose content (D) and incorporation in the cellulosic fraction of 14C-Glc (percentage of total cell wall [CW] material) (E). Cell walls of kob1-1 show reduced relative cellulose content compared with the wild type, and both kob1 alleles incorporate less 14C-Glc into the cellulosic fraction.
Error bars indicate sd values; n = 3 ([A] and [D]) and n = 5 (E).
We also investigated the xyloglucan structure in kob1-1. To this end, xyloglucan fragments were released by an endo-1,4-β-glucanase treatment of cell wall material prepared from dark-grown wild-type and kob1-1 seedlings. This enzyme cleaves the xyloglucan backbone behind nonsubstituted Glc residues and releases heptasaccharide XXXG to decasaccharide XLFG fragments, according to the nomenclature reported by Fry et al. (1993).
Figures 3B and 3C show the matrix-assisted laser desorption ionization–time of flight mass spectra of the fragments produced from wild-type and kob1-1 cell walls. In both spectra, main ions were assigned to (M+Na)+ adducts of XXXG, XXLG, XXFG, and XLFG on the basis of their molecular masses and literature data (Zablackis et al., 1995). Furthermore, ions having 42 additional mass units were assigned to fragments having acetyl groups on Gal residues, as observed in other plant species (York et al., 1984), which confirms that Arabidopsis xyloglucan is acetylated in muro (Pauly et al., 2001). Comparison of the spectra in Figures 3B and 3C did not reveal qualitative differences between wild-type and kob1-1 xyloglucan subunits.
Ectopic Callose and Lignin
Dark-grown wild-type and mutant seedlings were stained for lignin using phloroglucinol. Whole mounts showed a significant accumulation of ectopic lignin, especially in the mature part of the root (Figure 2G). Sirofluor staining also showed callose accumulation in kob1 hypocotyls (Figures 2H and 2I). The accumulation of ectopic lignin and callose is consistent with a defect in the synthesis of cellulose, because it also has been shown in wild-type seedlings treated with cellulose synthesis inhibitors such as DCB or isoxaben (Desprez et al., 2002). Callose accumulation also has been observed in cellulose-deficient cyt1 embryos (Lukowitz et al., 2001).
Cellulose Content and Cellulose Synthase Activity
After hydrolysis of the cell wall material with TFA, the insoluble cellulosic fraction was separated and hydrolyzed after dissolution in 72% H2SO4. The sugar contained in both TFA-insoluble and TFA-soluble fractions was quantified by gas chromatography. The percentages of cellulose expressed as the ratio between Glc contained in the TFA-insoluble fraction and total sugars are presented in Figure 3D and demonstrate a 33% reduction in the relative amount of cellulose in kob1-1 compared with the wild type.
We also compared the relative in vivo cellulose synthesis activity in mutant and wild-type cells. To this end, cell walls were prepared from dark-grown seedlings cultured for 4 days in the presence of 14C-Glc, and the incorporation of the label in the acetic acid:nitric acid–insoluble fraction was measured (Figure 3E). This acid-insoluble fraction is 97% crystalline cellulose (Peng et al., 2000), and comparison of the radioactivity in this fraction with that in the total cell wall provides a good measure of the amount of crystalline cellulose synthesized. Both mutant alleles showed a highly reproducible reduction in the ratio between acid-insoluble and acid-soluble material, indicating a relative reduction in the synthesis of crystalline cellulose.
Cellulose Microfibril Orientation
To visualize the microfibrils directly, wild-type and mutant roots were studied using field emission scanning electron microscopy (FESEM), as described by Sugimoto et al. (2000). The technique consists of removing one or more longitudinal cryosections to expose the internal surface in the fixed root and then extracting sufficient cytoplasm to reveal the most recently deposited cell wall layers. Figures 4A and 4B show an overview of the cryosections made from wild-type and mutant roots. In kob1, root hairs appeared very close to the meristematic zone, whereas in wild-type siblings, the distance was much greater. We also observed normally elongated root hairs in kob1-1.
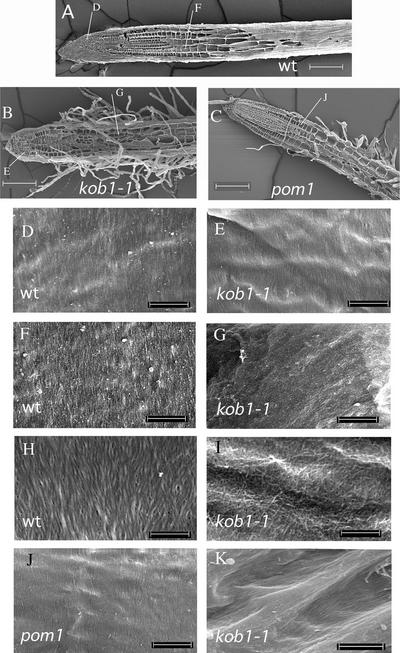
Figure 4.
Microfibril Organization Is Altered in Elongating Cells of kob1-1.
FESEM of cryosectioned roots of the wild type (Ws) ([A], [D], [F], and [H]), kob1-1 ([B], [E], [G], [I], and [K]), and pom1 ([C] and [J]) at low magnification (×100) ([A] to [C]) and ×25,000 magnification of hypochlorite-treated samples ([D] to [G] and [J]) and ×50,000 magnification of hypochlorite-pectolyase–treated samples ([H] and [I]). Microfibrils are parallel and oriented transversely to the elongation axis in cells in the division zone (D) and the elongation zone (F) in the wild type. In kob1-1, microfibrils are indistinguishable from those of the wild type in the division zone (E); however, in the elongation zone (G), only amorphous material can be seen. This is primarily pectic material, because pectolyase treatment of the same cells reveals microfibrils in kob1-1 (I). These remaining microfibrils are disordered, as opposed to the ordered microfibrils seen in the wild type treated in the same way (H). This microfibril defect is unlikely an indirect effect of reduced cell elongation in kob1-1, because in pom1, a mutant with a comparable cell elongation defect in the root (C), microfibrils are indistinguishable from those of the wild type (J). In contrast to the findings in elongating cells, in secondary thickenings in xylem cells of kob1-1 roots, microfibrils show a normal parallel orientation (K). Bars = 100 μm in (A) to (C), 1 μm in (D) to (G) and (J), and 500 nm in (H) and (I).
In the meristematic zone, cellulose microfibrils were oriented transversely to the elongation axis, and no difference was observed between the wild type and kob1-1 (Figures 4D and 4E). In addition, the secondary wall thickening on the inner side of the xylem cell wall presented transversely orientated cellulose microfibrils in both kob1-1 (Figure 4K) and the wild type (data not shown).
In the elongation zone of wild-type roots, microfibrils also showed a transverse orientation with respect to the elongation axis (Figure 4F). By contrast, no transverse microfibrils were observed in the elongation zone of kob1-1 (Figure 4G). Instead, an amorphous mass was visible. Given the increased pectin content in kob1-1 cell walls, we investigated whether this corresponded to pectic material. Indeed, treatment of the sections with pectolyase revealed a sparse network of randomly oriented microfibrils in the mutant (Figure 4I) compared with the dense microfibril arrays in the wild type (Figure 4H).
These observations are consistent with the idea that in kob1-1, cellulose synthesis is inhibited in elongating cells and the continuous supply of pectic polysaccharides in these cells would lead to the occlusion of the remaining microfibrils. It remains conceivable that the kob1 mutations affect cell elongation and that this results only indirectly in the observed changes in cellulose deposition. That this is unlikely was shown by the analysis of microfibril orientation in pompom1 (pom1), another root elongation mutant (Figure 4C) (Hauser et al., 1995). In this mutant, the elongation zone was reduced dramatically, as shown by the appearance of root hairs close to the cell division zone. In contrast to kob1, transversely oriented microfibrils of pom1 also were visible in elongating cells (Figure 4J).
KOB1 Encodes a Novel Protein Specific for Plants
kob1-1 was isolated from a T-DNA–mutagenized population. The kanamycin resistance marker cosegregated with the mutant phenotype and was located at a maximum distance of 0.5 centimorgan from the kob1-1 mutation. T-DNA flanking sequences were isolated and used as a probe to isolate full-length cDNA and genomic clones. The T-DNA was inserted in the first intron of a predicted gene located on chromosome 3 (Figure 5).
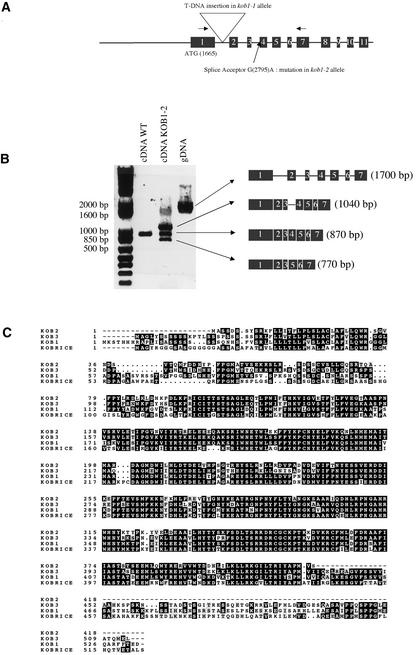
Figure 5.
Cloning of KOB1.
(A) Overview of the intron-exon organization of KOB1 and positions of mutations in kob1-1 and kob1-2.
(B) PCR with the primers indicated by arrows in (A) on cDNA from wild-type seedlings (lane 1) and heterozygous kob1-2 seedlings (lane 2) and on genomic DNA (gDNA) as a control (lane 3). Note the aberrant splicing of the third intron in kob1-2 seedlings leading to two distinct products, one still containing the third intron and a second lacking the fourth exon. WT, wild type.
(C) Sequence alignment of KOB1, KOB2, KOB3, and KOBRICE, a homolog in rice.
To confirm that the mutation in this gene is responsible for the mutant phenotype, the kob1-2 allele was sequenced. kob1-2 contains a G2795A transition that abolishes the splice acceptor site of the third intron. The effect of the kob1-2 mutation on the maturation of KOB1 mRNA was analyzed by reverse transcriptase–mediated (RT)–PCR using primers straddling the first three introns. A fragment of the expected size for a correctly spliced mRNA was observed in wild-type extracts.
Interestingly, in kob1-2 heterozygous plants, three fragments were observed. Sequencing of these fragments showed that they represent three mRNA species: the largest fragment corresponds to a mRNA unspliced for the second intron; the middle fragment was a correctly spliced mRNA presumably derived from the wild-type allele; and the smallest fragment was a mRNA in which the splice acceptor site of the third intron was used instead, leading to a mRNA lacking the fourth exon (Figure 5B). For both incorrectly spliced mRNAs, conceptual translation produces a protein truncated at a premature stop codon (data not shown).
Low levels of KOB1 mRNA were detected in all organs investigated, as shown by RNA gel blot analysis using specific probes as well as RT-PCR (data not shown). In addition, KOB1 mRNA levels were constitutive throughout seedling development both in the dark and in the light (data not shown). Comparison of the KOB1 cDNA and genomic sequences showed that the gene contains 11 exons. The cDNA carries an open reading frame of 1.6 kb encoding a predicted protein of 543 amino acids. The sequence did not match any protein of known function or any functional motifs in the public databases. The amino acid sequence shows four predicted N-glycosylation sites and a predicted transmembrane anchor at the N terminus. The protein is predicted to be a type II membrane protein with the N terminus exposed to the cytosol.
Two additional KOB1 isoforms are encoded by genes in the Arabidopsis genome located on chromosomes 2 and 3, showing 70 and 66% amino acid sequence identity, respectively, with KOB1 (Figure 5C). Other sequences matching KOB1 were found in various plant species, including the dicotyledons cotton, tomato, and poplar and the monocotyledons rice and maize. KOB1 showed 65% identity with the protein predicted from the full-length rice sequence (Figure 5C).
Interestingly, KOB1 ESTs also were detected in a library from 6-day-old elongating cotton fibers and in developing cambium cells of poplar (T. Teeri, personal communication). No database matches were found with sequences from nonplant species, including the completely sequenced genomes of various bacteria, yeast, Caenorhabditis elegans, and Drosophila melanogaster. In conclusion, KOB1 encodes a novel, highly conserved, plant-specific protein carrying a putative N-terminal membrane anchor.
KOB1 Is Localized in the Plasma Membrane but Not in All Cell Types
To investigate the subcellular localization of KOB1, a construct expressing a translational fusion between green fluorescent protein (GFP) and the N terminus of KOB1 under the control of the constitutive 35S promoter of Cauliflower mosaic virus was introduced into plants. This construct is functional, as shown by the complementation of the mutant phenotype when expressed in a kob1-1 background (data not shown). Moreover, transformants overexpressing the GFP-KOB1 fusion protein showed no changes in growth compared with the untransformed control (data not shown).
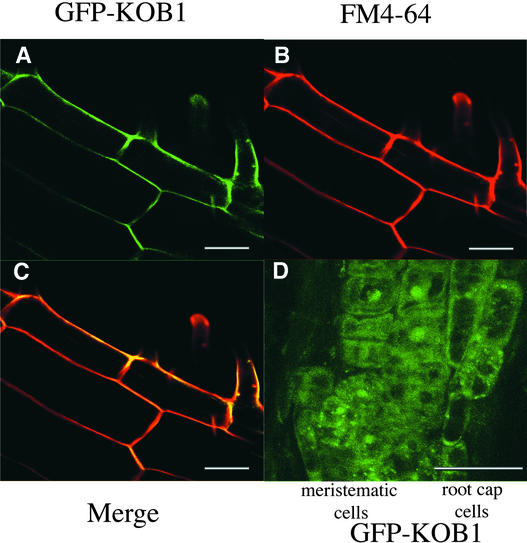
The GFP-KOB1 localization was investigated by confocal analysis in the different cell types in roots of kob1-1 seedlings complemented by the construct. The GFP signal was observed at the cell surface of elongated epidermal or cortical cells (Figure 6A). The plasma membrane localization of GFP-KOB1 in these cells was demonstrated by the colocalization with the vital dye FM4-64, which stains specifically the phospholipids of the plasma membrane (Figures 6B and 6C). These results are consistent with the prediction from the amino acid sequence, which suggested that KOB1 is a type II membrane protein.
Figure 6.
Plasma Membrane Localization of GFP-KOB1 in Elongated Cells.
Fluorescence of GFP-KOB1 ([A] and [D]) and the plasma membrane dye FM4-64 (B) in the root of the GFP-KOB1 transformant. Bars = 50 μm.
(A) to (C) Elongated cells. (C) shows a merged image.
(D) Cell division zone of the root. GFP-KOB1 is present in the plasma membrane in elongated cells. By contrast, within the cell division zone, GFP fluorescence is present in intracellular structures and not in the plasma membrane.
Interestingly, the plasma membrane localization of GFP-KOB1 was not observed in all cell types. Indeed, in cells within the meristem and the root cap, GFP fluorescence accumulated in the cytoplasm and in distinct punctate patches, which may correspond to an unidentified intracellular compartment (Figure 6D).
DISCUSSION
Mutations in KOB1 Cause the Cellulose-Deficient Dwarf Phenotype
Two lines of evidence show that the observed growth defect and the cellulose deficiency are the results of mutations in KOB1. First, both independent alleles are cellulose-deficient dwarfs and carry mutations in the KOB1 gene; second, the mutant phenotype of kob1-1 was complemented by the expression of a KOB1 cDNA or a GFP-KOB1 fusion protein under the control of the duplicated 35S promoter of Cauliflower mosaic virus (data not shown). Both mutants may correspond to complete loss-of-function alleles, because they express aberrant transcripts that encode severely truncated fragments of KOB1. The possibility is not excluded, however, that low levels of correctly spliced wild-type transcripts persist in the mutants. In addition, it is not known whether the related KOB2 and KOB3 genes are partially redundant with KOB1, because RT-PCR experiments showed that all three KOB genes are at least expressed in light-grown seedlings (data not shown).
To understand the function of KOB1, the causal relationship between the growth phenotype and the cellulose deficiency needs to be established. The following observations suggest that the cellulose deficiency is a primary defect of the kob1 mutations and not an indirect result of the cell elongation defect.
First, the dwarf phenotype with a radially expanded hypocotyl is very similar to that of other cellulose-deficient mutants (Arioli et al., 1998; Nicol et al., 1998; Fagard et al., 2000) and wild-type seedlings treated with the cellulose synthesis inhibitors DCB or isoxaben (Desprez et al., 2002). Second, we observed a highly reproducible cellulose deficiency in both kob1 alleles using two different chemical techniques. The close similarity between kob1 mutants and other cellulose-deficient mutants became even clearer when Fourier transform infrared microspectroscopy of dark-grown hypocotyls was used to classify >80 cell wall mutants.
We observed that both kob1 alleles clustered together inside a larger group containing exclusively cellulose-deficient mutants (rsw1, kor/rsw2, pom1, prc1) and wild-type plants treated with DCB or isoxaben. Other dwarfs, not deficient for cellulose, formed separate clusters (G. Mouille, unpublished results). In addition, the accumulation of ectopic lignin and callose as well as the presence of gapped walls in transverse hypocotyl sections also are characteristic of cellulose-deficient mutants.
Third, in cells within the cell elongation zone of kob1 roots, microfibrils were oriented randomly and occluded completely by pectic material at the cytoplasmic side of the wall. A reduced cellulose/pectin ratio in kob1 was confirmed by a chemical analysis of mutant and wild-type cell walls. Increased pectin content has been observed in other cellulose-deficient mutants (His et al., 2001; Sato et al., 2001) and may be an indirect result of cellulose deficiency. Randomly oriented microfibrils also have been observed in elongating root cells of the cellulose-deficient mutant rsw1 at restrictive temperatures and in the wild type treated with 1 μM DCB, indicating that a severe reduction in the synthesis of cellulose also leads to a randomization of the remaining microfibrils (Sugimoto et al., 2001).
Why this randomization occurs is not understood, but it may indicate that a critical density of nascent microfibrils is required to coordinate the orientation of cellulose deposition. Alternatively, the disorganized microfibrils in the mutant may correspond to the deeper cell wall layers that are covered by the pectin-rich and cellulose-depleted superficial layers in the mutant. Indeed, microfibrils were shown to lose their characteristic transverse alignment in deeper layers of the cell walls of expanded cells (Sugimoto et al., 2000).
Other observations also make it unlikely that the cellulose defect is an indirect result of the cell expansion defect in kob1. pom1 roots showed a reduction of the cell elongation zone comparable to that of kob1 but still contained normal transverse microfibrils in elongating cells. The same effect was observed for prc1 (G. Refrégier, unpublished data), which is mutated in CESA6. In addition, no evidence exists to date that reduced cell expansion can lead to changes in the ratio between cellulose and other wall polysaccharides. For instance, despite dramatic differences in the growth rate of dark-grown and light-grown hypocotyls and a much higher level of cell wall synthesis in light-grown seedlings, the ratio between cellulose and other cell wall polysaccharides remained constant (G. Refrégier, unpublished data). Together, these observations strongly suggest that the cellulose defect is a direct result of mutations in KOB1 and not an indirect result of the growth defect.
What Causes the Growth Defect in Cellulose-Deficient Mutants?
All mutants with cellulose defects in the primary wall that have been described to date are dwarfs. In general, reduced organ growth in these mutants is interpreted as a result of the loss of growth anisotropy, which is caused by the absence of the skeleton of ordered microfibrils that constrain growth in one preferential direction. This may explain at least part of the phenotype of those organs of rsw1 (Williamson et al., 2001), prc1 (Fagard et al., 2000), kor (Nicol et al., 1998; Lane et al., 2001), and kob1, which show dramatically increased radial expansion. However, mature root cells in kob1 were less elongated than those of the wild type, with no detectable increase in width. The same was observed for root cells of lit (Hauser et al., 1995), an allele of kor, suggesting that at least in these cells the reduced cell elongation is not exclusively the result of the loss of growth anisotropy.
Therefore, the alternative possibility should be considered: that the inhibition of cellulose synthesis may lead to an active inhibition of cell elongation, suggesting that at least in these cases growth is linked to cellulose synthesis through feedback control mechanisms. Why different organs show differences in radial expansion behavior in the mutants remains to be determined (for a more detailed discussion, see Williamson et al., 2001).
Potential Function of KOB1
The amino acid sequence did not provide insights into the function of KOB1. The plasma membrane localization of the functional GFP-KOB1 fusion protein in elongated cells is compatible with a role in the synthesis of cellulose suggested by the loss-of-function phenotype. Interestingly, the fusion protein expressed from a constitutive promoter showed an intracellular localization within the root cap and the cell division zone of the root. It remains conceivable that this is the result of the ectopic expression of KOB1 in cells not equipped for its targeting to the plasma membrane. Nevertheless, the ability to target KOB1 to the plasma membrane is regulated developmentally.
The mutant phenotype also corroborated a role for KOB1 in elongating cells as opposed to meristematic cells. Indeed, cell division planes were unaltered in mutant root meristems and embryos, indicating that KOB1 is not essential for cytokinesis. Also, microfibrils in the meristematic zone of the mutant were indistinguishable from those of the wild type, suggesting that the absence of KOB1 becomes critical only in cells that have ceased dividing and that enter a rapid elongation phase. Together, these observations suggest that KOB1 has a specific role in cellulose synthesis in postmitotic cells and that its activity requires the presence of the protein in the plasma membrane.
It is not clear from our data whether KOB1 also has a role in cellulose deposition in secondary walls. The observation that microfibrils appeared normal in the secondary wall thickenings of xylem cells suggests that normal cellulose deposition may resume in the mutant once growth has ceased. Preliminary FESEM data suggest that this also is the case for microfibrils in secondary walls of other cell types in kob1 roots (G. Refrégier, unpublished data).
What could be the role of KOB1 in cellulose synthesis? From its plasma membrane localization, KOB1 may be part of the cellulose synthesis machinery or it may play a regulatory role. A functional specialization of the cellulose synthesis machinery in postmitotic cells may reflect specific constraints that are imposed on cellulose synthesis in rapidly growing cells. In addition, KOB may play a role in the coordination between cellulose synthesis and cell expansion. The molecular analysis of KOB1 will provide new insights in the role of this protein in any of these processes.
METHODS
In Vitro Growth Conditions
Arabidopsis thaliana seedlings were grown on medium as described by Estelle and Somerville (1987) without Suc at 25°C. Seeds were cold treated for 48 h to synchronize germination. For dark growth, seeds were exposed to fluorescent white light (200 μmol·m−2·s−1) for 4 h to induce germination, and the plates were wrapped in three layers of aluminum foil. The age of the seedlings was counted from the beginning of the light treatment.
Microscopy and Image Analysis
For cross-sections, seedlings were fixed in PBS buffer containing 4% paraformaldehyde and 0.2% glutaraldehyde and were embedded in historesin (Technovit 7100; Hereaus Kulzer, Wehrheim, Germany) according to the manufacturer's instructions. Sections 3 μm thick were cut using a Jung RM2055 microtome (Leica Instruments, Nussloch, Germany). The cross-sections were stained with a 0.005% aqueous solution of Calcofluor (fluorescent brightener 28; Sigma) for 2 min and visualized under UV light using a Nikon microphot FXA microscope (Nikon, Champigny-sur-Marne, France).
Cells in living roots were stained with 0.1 μg/mL FM1-43 (Molecular Probes, Leiden, The Netherlands). The samples were washed twice after staining before observation with a TCSNT confocal microscope (Leica, Heidelberg, Germany) equipped with an argon/krypton laser (Omnichrome, Chino, CA) Length and width of root cells were measured using the image analysis software Optimas version 5 (Bioscan, Suresne, France).
Field emission scanning electron microscopy (FESEM) was performed as described by Sugimoto et al. (2000) using a Philips XL30 FESEM device (FEI, Eindhoven, The Netherlands).
Preparation of Cell Wall Material and Sugar Composition
Homozyous kob1-1 seeds were selected from the progeny and then heated at 70°C for 15 min in 70% ethanol to inactivate enzymes. The tissues then were ground in a homogenizer, and the homogenate was washed two times with 70% ethanol at 70°C and one time with water. The remaining pellet corresponding to the cell walls was freeze-dried. Cell wall material was hydrolyzed with 2 N trifluoroacetic acid (TFA), and sugar compositions were determined by gas chromatography analysis of trimethylsilyl methyl ester methyl glycoside derivatives according to York et al. (1985) using inositol as an internal standard.
For the analysis of cellulose content, cell walls were hydrolyzed for 2 h at 110°C with 800 μL of 2 M TFA containing 20 μg of inositol; after centrifugation, the supernatant was collected, neutralized, and lyophilized. The TFA-insoluble cellulosic material was washed with water and then dissolved in 30 μL of 72% H2SO4 at room temperature for 1 h. After dilution to 3% H2SO4 with water and the addition of 20 μg of inositol, the solution was heated at 110°C for 2 h.
The sample then was neutralized with BaCO3 and centrifuged, and the supernatant was lyophilized. Sugar in both cellulosic fractions and the TFA-soluble fraction was quantified by gas chromatography using inositol as an internal standard. The percentage of cellulose was expressed as the ratio between the amount of Glc in the cellulosic fractions and the amount of total sugar in the TFA-soluble and TFA-insoluble fractions.
For cellulose incorporation assays, heterozygous kob1 seeds were sown and germinated in the dark as described above, and homozygous seedlings were selected from the progeny and grown overnight in liquid medium. After liquid preincubation, they were washed three times with 15 mL of Glc-free growth medium before being resuspended in 1 mL of growth medium with 1.0 μCi/mL 14C-Glc (DuPont–New England Nuclear, Boston, MA). The seedlings then were incubated for 1 h in the dark at 15°C in glass tubes.
After treatment, seedlings were washed three times with 6 mL of Glc-free growth medium. The seedlings were extracted with 5 mL of boiling absolute ethanol for 20 min. This step was repeated three times. Next, seedlings were resuspended in 3 mL of chloroform: methanol (1:1), extracted for 20 min at 45°C, and finally resuspended in 3 mL of acetone for 15 min at room temperature with gentle shaking.
The remaining material was resuspended in 500 μL of an acetic acid:nitric acid:water solution (8:1:2) as described by Updegraff (1969) for 1 h in a boiling-water bath. Acid-soluble and acid-insoluble materials were separated by vacuum filtration through glass microfiber filters (2.5 cm; Whatman, Maidstone, UK). Filters were washed with 1.5 mL of water. The acid solution and water wash constituted the “acid-soluble fraction.” Filters were washed with 30 mL of water and 20 mL of ethanol. This “acid-insoluble fraction” consisted essentially of crystalline cellulose. The amount of label in both fractions was determined by scintillation counting in a Kontron Betamatic (Kontron Instruments, Montigny le Bretonneux, France) using Ultima-Flo AP (Packard, Groningen, The Netherlands) as the scintillation liquid. The percentage incorporation was expressed as follows: label in the cellulosic faction/acid-soluble fraction + cellulosic fraction × 100.
For the analysis of xyloglucan structure, xyloglucan fragments were generated by treating 500 μg of cell wall material with 5 units of endo-1,4-β-glucanase (EC 3.2.1.4) (catalog number E-CELTR; Megazyme International, Bray, Ireland) in 500 μL of buffer for 18 h. Matrix-assisted laser desorption ionization–time of flight mass spectra of the resulting xyloglucan-solubilized fragments were recorded on a Micromass mass spectrometer (Manchester, UK). Mass spectra were made in the reflectron mode using 2,5-dihydroxybenzoic acid as a matrix.
RNA Gel Blot Analysis
RNA was extracted as described previously (Brusslan and Tobin, 1992). RNA gel blot analysis on Hybond N membranes and labeling of the probes by random priming in the presence of 50 μCi of 32P-dCTP were performed according to the manufacturer's instructions (Amersham, Buckinghamshire, UK). The membrane was hybridized overnight at 42°C in 10 mL of SDS-rich buffer as described by Church and Gilbert (1984) with 50% formamide and 100 μg/mL salmon sperm DNA after overnight prehybridization. Washes were performed at 65°C in 2 × SSC and 0.1% SDS for 15 min and in 1 × SSC and 0.1% SDS for 15 min (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate). The KOB1 probe was prepared with a 300-bp fragment covering the C-terminal region that is specific for KOB1.
Upon request, all novel materials described in the article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Accession Numbers
The accession numbers for the sequences mentioned in this article are AC012562.5 for KOB1 and AP002481.1 for KOBRICE.
Acknowledgments
Estelle Aletti and Jocelyne Kronenberger are thanked for their skilled technical assistance, Olivier Grandjean for help with the confocal microscopy, and Guislaine Refrégier and Soizic Rochange for their help with the FESEM. Catherine Bellini and Thierry Desnos are thanked for the isolation of kob1-2. This work was financed in part by GENOPLANTE Grant 19990025 to H.H. and P.L., European Economic Community Framework 5 Grant GEMINI, a grant from the French Ministery of Science and Technology, and a grant from Centre de Coopération Internationale en Recherche Agronomique pour le Développement to S.P.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.002873.
References
- Arioli, T., et al. (1998). Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279, 717–720. [DOI] [PubMed] [Google Scholar]
- Beemster, G., and Baskin, T. (1998). Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiol. 116, 1515–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummell, D.A., and Hall, J.L. (1985). The role of cell wall synthesis in sustained auxin-induced growth. Physiol. Plant. 63, 406–412. [Google Scholar]
- Brusslan, J.A., and Tobin, E.M. (1992). Light-independent developmental regulation of cab gene expression in Arabidopsis thaliana seedlings. Proc. Natl. Acad. Sci. USA 89, 7791–7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church, G.M., and Gilbert, W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove, D.J. (1993). Water uptake by growing cells: An assessment of the controlling roles of wall relaxation, solute uptake, and hydraulic conductance. Int. J. Plant Sci. 154, 10–21. [DOI] [PubMed] [Google Scholar]
- Desprez, T., Vernhettes, S., Fagard, M., Refrégier, G., Desnos, T., Aletti, E., Py, N., Pelletier, S., and Höfte, H. (2002). Resistance against herbicide isoxaben and cellulose deficiency caused by distinct mutations in same cellulose synthase isoform CESA6. Plant Physiol. 128, 482–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estelle, M.A., and Somerville, C.R. (1987). Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Mol. Gen. Genet. 206, 200–206. [Google Scholar]
- Fagard, M., Desnos, T., Desprez, T., Goubet, F., Refrégier, G., Mouille, G., McCann, M., Rayon, C., Vernhettes, S., and Höfte, H. (2000). PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell 12, 2409–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry, S.C., York, W.S., and Albersheim, P. (1993). An unambiguous nomenclature for xyloglucan-derived oligosaccharide. Physiol. Plant. 89, 1–3. [Google Scholar]
- Gendreau, E., Traas, J., Desnos, T., Grandjean, O., Caboche, M., and Höfte, H. (1997). Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 114, 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillmor, C.S., Poindexter, P., Lorieau, J., Palcic, M.M., and Somerville, C. (2002). α-Glucosidase I is required for cellulose biosynthesis and morphogenesis in Arabidopsis. J. Cell Biol. 156, 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser, M.T., Morikami, A., and Benfey, P.N. (1995). Conditional root expansion mutants of Arabidopsis. Development 121, 1237–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- His, I., Driouich, A., Nicol, F., Jauneau, A., and Höfte, H. (2001). Altered pectin composition in primary cell walls of korrigan, a dwarf mutant of Arabidopsis deficient in a membrane-bound endo-1,4-beta-glucanase. Planta 212, 348–358. [DOI] [PubMed] [Google Scholar]
- Kimura, S., Laosinchai, W., Itoh, T., Cui, X., Linder, C.R., and Brown, R.M., Jr. (1999). Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant Vigna angularis. Plant Cell 11, 2075–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane, D.R., et al. (2001). Temperature-sensitive alleles of rsw2 link the korrigan endo-1,4-beta- glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiol. 126, 278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz, W., Nickle, T.C., Meinke, D.W., Last, R.L., Conklin, P.L., and Somerville, C.R. (2001). Arabidopsis cyt1 mutants are deficient in a mannose-1-phosphate guanylyltransferase and point to a requirement of N-linked glycosylation for cellulose biosynthesis. Proc. Natl. Acad. Sci. USA 98, 2262–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQueen-Mason, S., Durachko, D.M., and Cosgrove, D.J. (1992). Two endogenous proteins that induce cell wall extension in plants. Plant Cell 4, 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, S.C., and Brown, R.M., Jr. (1980). Evidence for an intramembrane component associated with a cellulose microfibril-synthesizing complex in higher plants. J. Cell Biol. 84, 315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol, F., His, I., Jauneau, A., Vernhettes, S., Canut, H., and Höfte, H. (1998). A plasma membrane-bound putative endo-1,4-beta-d-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 17, 5563–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly, M., Eberhard, S., Albersheim, P., Darvill, A., and York, W.S. (2001). Effects of the mur1 mutation on xyloglucans produced by suspension-cultured Arabidopsis thaliana cells. Planta 214, 67–74. [DOI] [PubMed] [Google Scholar]
- Peng, L., Hocart, C.H., Redmond, J.W., and Williamson, R.E. (2000). Fractionation of carbohydrates in Arabidopsis root cell walls shows that three radial swelling loci are specifically involved in cellulose production. Planta 211, 406–414. [DOI] [PubMed] [Google Scholar]
- Richmond, T. (2000). Higher plant cellulose synthases. Genome Biol. 1, 30001.1–30001.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, S., Kato, T., Kakegawa, K., Ishii, T., Liu, Y.G., Awano, T., Takabe, K., Nishiyama, Y., Kuga, S., Nakamura, Y., Tabata, S., and Shibata, D. (2001). Role of the putative membrane-bound endo-1,4-beta-glucanase KORRIGAN in cell elongation and cellulose synthesis in Arabidopsis thaliana. Plant Cell Physiol. 42, 251–263. [DOI] [PubMed] [Google Scholar]
- Sugimoto, K., Williamson, R.E., and Wasteneys, G.O. (2000). New techniques enable comparative analysis of microtubule orientation, wall texture, and growth rate in intact roots of Arabidopsis. Plant Physiol. 124, 1493–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto, K., Williamson, R.E., and Wasteneys, G.O. (2001). Wall architecture in the cellulose-deficient rsw1 mutant of Arabidopsis thaliana: Microfibrils but not microtubules lose their transverse alignment before microfibrils become unrecognizable in the mitotic and elongation zones of roots. Protoplasma 215, 172–183. [DOI] [PubMed] [Google Scholar]
- Taylor, N.G., Laurie, S., and Turner, S.R. (2000). Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 12, 2529–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, N.G., Scheible, W.R., Cutler, S., Somerville, C.R., and Turner, S.R. (1999). The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11, 769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, S.R., and Somerville, C.R. (1997). Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9, 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegraff, D.M. (1969). Semi-determination of cellulose in biological materials. Anal. Biochem. 32, 420–424. [DOI] [PubMed] [Google Scholar]
- Wells, B., McCann, M.C., Shedletzky, E., Delmer, D., and Roberts, K. (1994). Structural features of cell walls from tomato cells adapted to grow on the herbicide 2,6-dichlorobenzonitrile. J Microsc. 173, 155–164. [Google Scholar]
- Williamson, R., Burn, J., Birch, R., Baskin, T., Arioli, T., Betzner, A., and Cork, A. (2001). Morphology of rsw1, a cellulose-deficient mutant of Arabidopsis thaliana. Protoplasma 215, 116–127. [DOI] [PubMed] [Google Scholar]
- York, W., Darvill, A., and Albersheim, P. (1984). Inhibition of 2,4-dichlorophenoxyacetic acid-stimulated elongation of pea stem segments by a xyloglucan oligosaccharide. Plant Physiol. 75, 295–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York, W., Darvill, A., McNeil, M., and Albersheim, P. (1985). Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol. 118, 3–40. [Google Scholar]
- Zablackis, E., Huang, J., Muller, B., Darvill, A.G., and Albersheim, P. (1995). Characterization of the cell-wall polysaccharides of Arabidopsis thaliana leaves. Plant Physiol. 107, 1129–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, J., Niu, Q.W., Nishizawa, N., Wu, Y., Kost, B., and Chua, N.H. (2000). KORRIGAN, an Arabidopsis endo-1,4-β-glucanase, localizes to the cell plate by polarized targeting and is essential for cytokinesis. Plant Cell 12, 1137–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]