Abstract
The B motif is a signature of type-B response regulators (ARRs) involved in His-to-Asp phosphorelay signal transduction systems in Arabidopsis. Homologous motifs occur widely in the GARP family of plant transcription factors. To gain general insight into the structure and function of B motifs (or GARP motifs), we characterized the B motif derived from a representative ARR, ARR10, which led to a number of intriguing findings. First, the B motif of ARR10 (named ARR10-B and extending from Thr-179 to Ser-242) possesses a nuclear localization signal, as indicated by the intracellular localization of a green fluorescent protein–ARR10-B fusion protein in onion epidermal cells. Second, the purified ARR10-B molecule binds specifically in vitro to DNA with the core sequence AGATT. This was demonstrated by several in vitro approaches, including PCR-assisted DNA binding site selection, gel retardation assays, and surface plasmon resonance analysis. Finally, the three-dimensional structure of ARR10-B in solution was determined by NMR spectroscopy, showing that it contains a helix-turn-helix structure. Furthermore, the mode of interaction between ARR10-B and the target DNA was assessed extensively by NMR spectroscopy. Together, these results lead us to propose that the mechanism of DNA recognition by ARR10-B is essentially the same as that of homeodomains. We conclude that the B motif is a multifunctional domain responsible for both nuclear localization and DNA binding and suggest that these insights could be applicable generally to the large GARP family of plant transcription factors.
INTRODUCTION
His-to-Asp (His→Asp) phosphorelays (or two-component regulatory systems) are a conserved signal transduction mechanism involved in a wide variety of cellular responses to environmental stimuli (Parkinson and Kofoid, 1992). To date, numerous instances of such His→Asp phosphorelay systems have been reported, not only for many prokaryotic species (Mizuno, 1997) but also for certain eukaryotic species (Wurgler-Murphy and Saito, 1997). Results of recent molecular studies of the higher plant Arabidopsis revealed that plant signal transduction often involves His→Asp phosphorelays, particularly in response to hormones (e.g., ethylene and cytokinin) (for reviews, see Chang and Stewart, 1998; D'Agostino and Kieber, 1999; Schaller, 2000; Urao et al., 2000).
In general, a given His→Asp phosphorelay system consists of two or more conserved signal transducers: a sensor exhibiting His kinase activity, a His-containing phosphotransfer intermediate, and a response regulator containing a phospho-accepting Asp in its receiver domain (Mizuno, 1998). An inspection of the entire genome sequence of Arabidopsis revealed 11 His kinases (Arabidopsis Genome Initiative, 2000), including ethylene receptors (ETR1, ETR2, ERS1, ERS2, and EIN4) (Chang et al., 1993; Hua and Meyerowitz, 1998) and cytokinin sensors (AHK2, AHK3, and AHK4/CRE1) (Inoue et al., 2001; Suzuki et al., 2001; Yamada et al., 2001).
These His kinase hormone receptors most likely modulate the function of downstream components, such as response regulators, through His→Asp phosphorelay signaling. In fact, Arabidopsis has at least 22 members of the response regulator family (collectively named ARRs). These can be classified into two subtypes: type A (11 members) and type B (11 members) (D'Agostino and Kieber, 1999; Imamura et al., 1999; Kiba et al., 1999), the latter containing putative transcription factors (Sakai et al., 2000; Lohrmann et al., 2001). In this context, a multistep AHK→AHP (Arabidopsis His-containing phosphotransfer factor)→ARR phosphorelay pathway(s) was considered to be involved in a signaling network in response to cytokinin (Suzuki et al., 2002).
Two recent reports on Arabidopsis provide genetic and biological evidence in support of this notion by showing that a type-B ARR functions as a transcriptional activator for a type-A ARR, presumably in AHK-mediated cytokinin signaling (Hwang and Sheen, 2001; Sakai et al., 2001). In this study, we focus our attention on the type-B ARR family, with special reference to the structure and function of these DNA binding transcription factors.
The completion of the Arabidopsis genome sequence allows comparative and statistical analyses of plant transcription factors. Interestingly, Arabidopsis transcription factors that belong to families common to all eukaryotes do not necessarily share significant sequence similarity with those of the other kingdoms beyond the conserved DNA binding domains (Riechmann et al., 2000). One of the largest such families of Arabidopsis transcription factors is classified as the Myb-(R1)R2R3 family, consisting of 5 Myb-R1R2R3 members and 125 Myb-R2R3 members (Stracke et al., 2001). Each member of this family of transcription factors has a putative DNA binding domain similar to that found originally in the mammalian c-Myb oncoprotein.
In addition to these Myb-(R1)R2R3 proteins, Arabidopsis contains other transcription factors that are characterized by a more divergent Myb motif, which is present either as a single copy or as a repeat. Often, these are referred to collectively as the Myb-related family, which consists of ∼50 members. In this context, the Arabidopsis type-B ARR proteins, which are involved in His→Asp phosphorelay signaling, are particularly interesting in that each has a conserved signature motif that somewhat resembles the classic Myb repeat (Imamura et al., 1999). This motif consists of ∼60 amino acids and was referred to originally as the B motif. According to Riechmann's classification (2000), the B motif appears to be a representative of the plant single Myb-related domains, which belong to the GARP subfamily.
Other members of the GARP family are maize GOLDEN2 (G2) (Hall et al., 1998), Arabidopsis ARRs (Imamura et al., 1999), Chlamydomonas reinhardtii PSR1 (Wykoff et al., 1999), and Arabidopsis PHR1 (Rubio et al., 2001). A GARP motif also was found in the recently identified KANADI gene product, which regulates organ polarity in Arabidopsis (Kerstetter et al., 2001). Little is known about the structure and function of plant Myb-related domains, including the B motif (or GARP motif).
In the hope of better understanding the features of these widespread plant Myb-related motifs, we have characterized a representative B motif (from ARR10) at the molecular level, showing that it functions in sequence-specific DNA recognition. Furthermore, the three-dimensional structure of this B motif was determined in solution by NMR spectroscopy, revealing three α-helices and a flexible arm at the N terminus. The NMR studies were extended to the complex formed by the B motif and its target DNA. Our results suggest that the B motif recognizes the major groove of DNA with residues located in a helix-turn-helix (HTH) variant motif and contacts the adjacent minor groove with the N-terminal flexible arm.
RESULTS
Structure and Function of the Type-B Family of ARRs
Recently, it was suggested that some members (e.g., ARR1 and ARR2) of the type-B ARR family function as DNA binding signal transduction factors (Sakai et al., 2000; Lohrmann et al., 2001). These type-B family members have a common structural design, as represented by ARR10 (Figure 1A). Each of them has a highly conserved phospho-accepting receiver domain of ∼120 amino acids containing a phosphorylated Asp residue at the center. In each protein, the common region is followed by a large C-terminal extension.
Figure 1.
The Type-B Response Regulator 10 (ARR10).
(A) The structural design of ARR10, a member of the type-B ARR family, is shown schematically compared with those of three other plant proteins (LHY, CCA1, and Myb-like) and the classic mouse c-Myb oncoprotein. The B motif is similar in length and primary sequence to the Myb repeat of c-Myb and the Myb-related domains of the plant proteins. Two truncated polypeptides were used in this study: ARR10-B, corresponding to the B motif of ARR10; and ARR10-RB, containing both the phospho-accepting receiver domain and the B motif.
(B) Amino acid sequences of the B motifs of the type-B ARRs and the corresponding motifs of other GARP proteins. These sequences are compared with those of the plant Myb-related domains and mouse Myb repeats of the proteins listed in (A), and the conserved amino acids are highlighted.
Although these extensions are highly variable in length and amino acid sequence (Imamura et al., 1999), they all have a short stretch of conserved amino acids, named the B motif, as aligned in Figure 1B. In other words, this B motif of ∼60 amino acids is a signature of the type-B family of ARRs. It also is related to plant GARP motifs, as emphasized in the Introduction (Riechmann et al., 2000), and its inferred consensus sequence shows similarity to the so-called plant Myb-related domains that are found in many plant proteins (Sakai et al., 1998). These include the well-characterized CCA1 (circadian clock–associated protein) and LHY (late elongated hypocotyl), both of which were classified as single Myb-related transcription factors of Arabidopsis (Schaffer et al., 1998; Wang and Tobin, 1998) (Figure 1A).
The authentic mammalian Myb repeat is a sequence of ∼60 amino acids and is repeated three times in the classic mouse c-Myb oncoprotein (R1R2R3; Figure 1). These repeats together can recognize DNA in a sequence-specific manner on the basis of its fine three-dimensional structure, characteristics of which have been well examined at the molecular level (Ogata et al., 1994). In short, the amino acid sequences of the B motifs are distantly related to that of the authentic mammalian Myb repeat (Figure 1B), but little is known about the structure and function of these intriguing B motifs and Myb-related domains in plants.
Based on these considerations, in this study, we addressed the crucial questions regarding the B motif in ARR10. Does the single B motif bind to DNA in a sequence-specific manner? Is the plant B motif related to the mammalian Myb repeat at the level of molecular structure? The answers to these critical questions should provide insight into the structure and function of plant Myb-related domains generally and, in particular, to type-B ARR family members that are involved in His→Asp phosphorelay signal transduction.
Isolation and Purification of the B Motif of ARR10
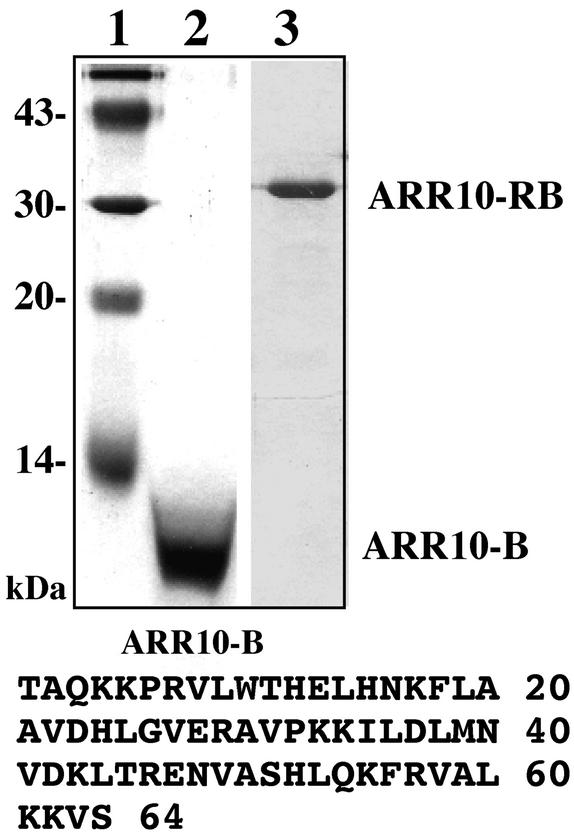
The 64–amino acid B motif (Thr-179 to Ser-242; Figure 2) was expressed and purified in Escherichia coli (see Methods). A large amount of this polypeptide (named ARR10-B) was purified to near homogeneity in a soluble form (Figure 2, lane 2). The purified protein has an apparent molecular mass (8500 D) that is approximately in agreement with the calculated molecular mass (7367.6 D), and its determined N-terminal amino acid sequence was identical to that predicted from its cDNA sequence.
Figure 2.
Isolation of ARR10-Derived Polypeptides.
The polypeptides used in this study were analyzed by SDS-PAGE followed by staining with Coomassie blue. Lane 1, molecular mass marker; lane 2, ARR10-B; lane 3, ARR10-RB. The sequence of ARR10-B's 64 amino acids is shown at bottom.
Similarly, a longer ARR10 derivative (named ARR10-RB) also was expressed in and purified from E. coli (Figure 2, lane 3). ARR10-RB corresponds to the N-terminal portion of ARR10 (extending from Met-1 to Ser-263) and contains both the receiver domain and the B motif (Figure 1). Note also that the purified ARR10-RB polypeptide contains a His tag (His × 6) at its C terminus. ARR10-B and ARR10-RB were used for functional and structural analyses, as described below.
The B Motif Amino Acid Sequence Contains a Nuclear Localization Signal
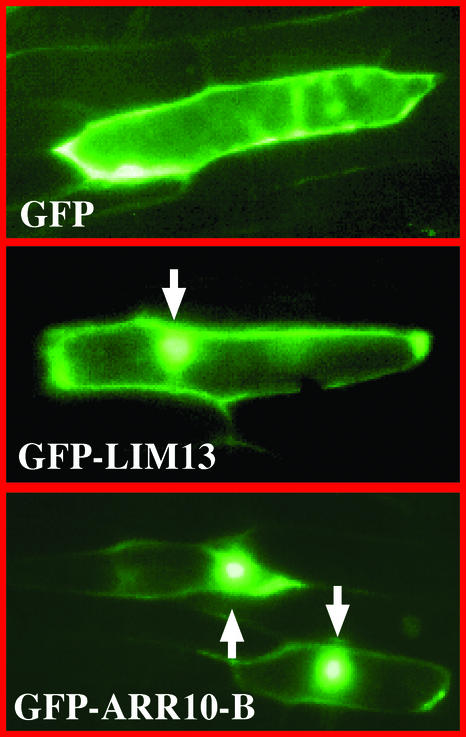
To gain an insight into the function of the B motif, the 64–amino acid region corresponding to the B motif sequence of ARR10 (Thr-179 to Ser-242) was fused to green fluorescent protein (GFP). This was done using a conventional GFP fusion recombinant plasmid, as described previously (Makino et al., 2000). The gene encoding GFP-ARR10-B was introduced into onion skin epidermal cells using a particle-mediated DNA delivery procedure. The onion epidermal cells expressing the GFP fusion proteins were examined with a fluorescence microscope (Figure 3). For comparison, GFP alone was used as a control for cytoplasmic localization and GFP-LIM13 was used as a control for nuclear localization.
Figure 3.
Nuclear Localization of the GFP Fusion Derivative of ARR10-B.
The recombinant genes encoding GFP alone, GFP-LIM13, and GFP-ARR10-B were constructed to be under the control of the plant 35S promoter. These DNA constructs were bombarded individually into onion skin epidermal cells. The expression and localization of each product was observed after 24 h of incubation by fluorescence microscopy. Arrows indicate fluorescent nuclei.
In the case of GFP alone, fluorescence was detected mainly in the cytoplasmic space, whereas in the case of GFP-LIM13, fluorescence was detected as a dense glittering spot in the cells (Figure 3). This spot corresponded well with the position of the nucleus, as visualized by 4′,6-diamidino-2-phenylindole staining (data not shown). LIM13 is a meiosis-associated gene from Lilium longiflorum, the product of which was demonstrated previously to be a nucleus-localizing protein (Ogata et al., 1999). Based on these data, it was found that GFP–ARR10-B is capable of localizing predominantly, if not exclusively, to the nuclei (Figure 3). This finding suggests that the B motif amino acid sequence characterized in this study includes a nuclear localization signal (NLS) and is responsible for the recently discovered nuclear localization of the intact ARR10 protein (Imamura et al., 2001).
Identification of a DNA Sequence Recognized Specifically by ARR10
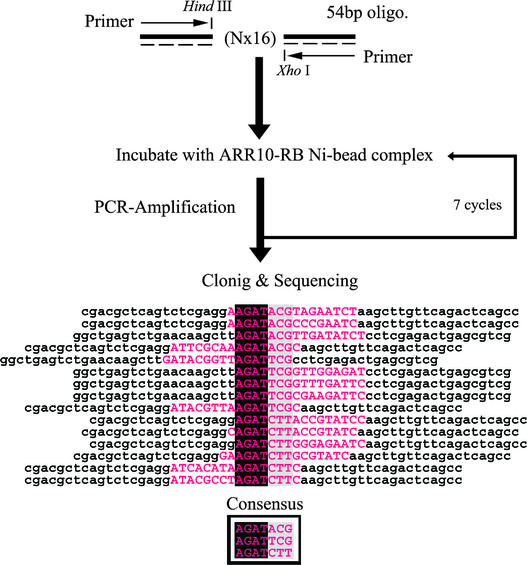
As mentioned above, ARR10 is assumed to be a DNA binding transcription factor. In the hope of identifying a specific nucleotide sequence recognized by ARR10, the well-established random oligonucleotide selection method, combined with PCR amplification, was applied to purified ARR10-RB polypeptides immobilized on Ni beads (see Methods). This strategy, using random nucleotide sequences (N × 16) and adapted for ARR10-RB, is shown schematically in Figure 4.
Figure 4.
PCR-Assisted Binding Site Selection.
The scheme illustrates the experimental approach for selecting DNA sequences with high affinity for ARR10-RB. A mixture of 54-base oligonucleotides, in which the central 16 bases were of random sequence, was generated by incorporating each of the four nucleotides at equimolar concentration. Each round of selection consisted of incubation of the 54-bp DNA with ARR10-RB Ni bead complexes, with subsequent purification and amplification of bound DNA for use in the next round of selection. The nucleotide sequences of DNA molecules cloned after the seventh round of selection were aligned to infer the consensus sequence. Note that the flanking invariant nucleotides (lowercase letters) are not included in the selected and aligned sequences (uppercase letters in magenta). Four more selected clones also have been sequenced. They were found to contain tandem AGAT or GATTCG, which also are well explained by the proposed consensus sequences. However, they were not included in the alignment for clarity in the figure.
Using this strategy, we were able to isolate a number of clones, each likely to contain a putative ARR10-RB binding sequence. The determined nucleotide sequences were aligned to infer the most common sequence (Figure 4), allowing us to identify the core sequence AGAT. This conserved sequence seems to be followed by a preferred set of extra sequences (ACG, TCG, or CTT). Thus, we tentatively concluded that ARR10-RB has the ability to bind to DNA in a sequence-specific manner and that its recognition sequence contains the core sequence AGAT, followed by A/TCG or CTT.
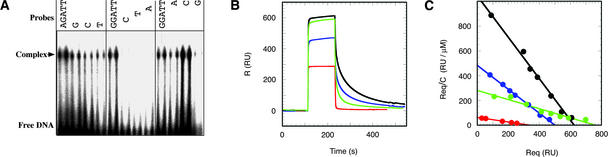
To verify this conclusion, an in vitro DNA binding gel-shift assay was performed with a set of synthetic 34-bp oligonucleotides, each containing the core sequence (AGAT) or its variant (GGAT), as shown in Figure 5A. The presumed minimum DNA binding domain of 64 amino acids (i.e., ARR10-B) was used for the gel-shift assays. The results are shown in Figure 5A. The purified ARR10-B displayed an ability to associate efficiently with AGATT and to associate slightly less efficiently with GGATT, CGATT, and TGATT, whereas GcATT, GGtTT, GGAaT, and GGATg were unfavorable.
Figure 5.
DNA Binding Properties of ARR10-B.
(A) Purified ARR10-B was subjected to DNA binding gel-shift assay using synthetic 32P-labeled DNA 34-mers containing the core AGAT sequence or its variant GGAT.
(B) Surface plasmon resonance sensorgrams for the binding of ARR10-B to four different immobilized DNA 12-mers at a protein concentration of 10 μM: DNA 1 (d-CACTGATTCAGG/d-CCTGAATCAGTG; red), DNA 2 (d-GCAAGATTCGGC/d-GCCGAATCTTGC; black), DNA 3 (d-GCAAGATCAGGC/d-GCCTGATCTTGC; green), and DNA 4 (d-GCAAGATCTTGC/d-GCAAGATCTTGC; blue). The amounts of immobilized DNAs 1, 2, 3, and 4 were 658, 512, 1136, and 476 resonance units (RU), respectively. The control signals, which were generated by flowing ARR10-B through a blank channel without DNA, have been subtracted.
(C) Scatchard plots (Req/C versus Req) of signal intensities at the steady states. Kd values were determined to be 5.4, 0.6, 2.7, and 1.0 μM for DNA 1 (red), DNA 2 (black), DNA 3 (green), and DNA 4 (blue), respectively, by fitting the experimental data with the function Req/C = Rmax/Kd − Req/Kd, where Rmax is the maximum response.
The apparent enhancement in binding affinity observed for GGATC, compared with GGATT, could be attributed to the creation of another GAT sequence in the complementary strand as a result of the replacement of T at the fifth position with C. Collectively, the purified B motif of ARR10 appeared to associate preferentially with the a/gGATc/t sequence in vitro. These results are essentially consistent with the recognition sequences proposed for ARR10-RB (Figure 4) and show that the isolated single B motif is capable of binding to DNA in a sequence-specific manner.
To quantitate the DNA binding ability of ARR10-B, the binding affinity of the protein to synthetic oligonucleotides was measured using a surface plasmon resonance (SPR) biosensor on a BIAcore system (Figure 5B). For this purpose, we designed and synthesized four 12-bp oligonucleotides: DNA 1 with TGATT, DNA 2 with AGATT, DNA 3 with AGATC, and DNA 4 with two AGATC sequences in both strands of the DNA duplex. DNAs 1 and 2 contain the core sequences TGATT and AGATT and display the lowest and highest binding ability, respectively, in the gel-shift assay among the oligonucleotides with GATT as the core sequence (Figure 5A).
DNAs 3 and 4, with the core sequence AGATC, were designed to determine the preferred base after the AGAT sequence. Judging from the equilibrium Kd, determined by a Scatchard plot of signal intensities at the steady states (Figure 5C), ARR10-B displays nine times higher affinity to DNA 2 (0.6 μM) than to DNA 1 (5.4 μM). These results indicate that this B motif binds to the AGATT sequence more tightly than to TGATT, which is in good agreement with the results of the gel-shift assay (Figure 5A).
The lower affinity observed for DNA 3 (Kd = 2.7 μM), compared with that for DNA 2, suggests that ARR10-B preferentially binds to AGATT over AGATC. The existence of two recognition sequences in both strands of the DNA duplex increases the affinity, as indicated by the fact that DNA 4 (Kd = 1.0 μM) has approximately three times higher affinity than DNA 3. However, the binding affinity of ARR10-B to DNA 4 still is lower than that to DNA 2. Taking the results of the DNA binding gel-shift assay and the SPR study together, we conclude that the optimal recognition sequence of ARR10-B is AGATT.
Molecular Structure and Thermal Stability of ARR10-B in Solution
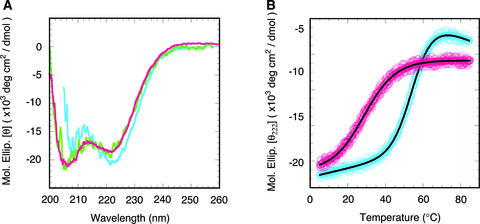
Because our results indicated that ARR10-B most likely is a DNA binding motif and is widespread in plants, we next analyzed its molecular structure in solution by means of circular dichroism (CD) and NMR spectroscopy. The CD spectra of ARR10-B at two different protein concentrations, 27 and 7 μM, are nearly superimposable (Figure 6A), indicating no aggregation tendency of the polypeptide in this concentration range. The free form of ARR10-B showed typical α-helical characteristics, such as minima at 208 and 222 nm, in the CD spectra at 10°C. This polypeptide lost its tertiary and secondary structures with increasing temperature but recovered the native structure with decreasing temperature. These results indicate that the thermal denaturation process of ARR10-B is reversible.
Figure 6.
Secondary Structure and Thermal Stability of ARR10-B.
(A) CD spectra of 7.4 μM (green) and 27 μM (magenta) free ARR10-B in 10 mM sodium phosphate buffer, pH 7.0, and of 40 μM ARR10-B bound with DNA 2 (blue) in the same buffer containing 160 mM NaCl. The spectrum of the DNA-bound form of ARR10-B was obtained by subtracting the spectrum of DNA 2 from that of the mixture of ARR10-B and DNA 2.
(B) Thermal denaturation curves of the free and DNA-bound forms of ARR10-B. The curve of the free form (magenta circles) was measured for the solution containing 27 μM polypeptide. The curve of the DNA-bound form (blue circles) was obtained by subtracting the curve of DNA 2 from that of the mixture of ARR10-B and DNA 2 at a protein concentration of 40 μM. The solid lines are the best fits to the experimental data based on a two-state model.
We analyzed the thermal denaturation curve using a two-state model between the native and unfolded forms (Figure 6B) and obtained thermodynamic parameters for the transition of Tm = 29°C, ΔHm = 22 kcal·mol−1, and ΔSm = 73 cal·mol−1·K−1 where Tm is the midpoint of a temperature transition, and ΔHm and ΔSm are, respectively, the enthalpy and entropy differences between denatured and native states. Although we analyzed the same curve using a three-state model, the amount of any intermediate in the unfolding pathway was negligible. The same results were obtained for the transition at a protein concentration of 7 μM, suggesting that the thermal denaturation process does not involve concentration-dependent phenomena such as protein–protein interactions or aggregation of the unfolded polypeptides.
The effects of DNA on the molecular structure and thermal stability of ARR10-B were examined by CD spectroscopy. The CD spectrum and the thermal denaturation curve for the DNA-bound ARR10-B were derived by subtracting the data for DNA alone from those for the mixture of ARR10-B and DNA. The 12-bp oligonucleotide with the highest affinity, DNA 2, was used for these experiments. The DNA-bound form of ARR10-B also displayed typical α-helical characteristics in the CD spectrum and slightly higher helix content compared with its free form at the same temperature (Figure 6A).
More remarkably, the bound form had significantly higher thermal stability than the free form (Figure 6B). The denaturation curve could be fit using a two-state model, as in the case of the free form. The Tm, ΔHm, and ΔSm values for the transition were estimated to be 55°C, 39 kcal·mol−1, and 120 cal·mol−1·K−1, respectively. These results indicate that binding to DNA greatly stabilizes the molecular structure of ARR10-B.
Three-Dimensional Structure of ARR10-B in Solution
As a first step in investigating the molecular interactions between ARR10-B and DNA at atomic-level resolution, we determined the three-dimensional structure of ARR10-B in solution by NMR spectroscopy. The 1H-15N HSQC spectra at higher temperatures (>30°C) showed only a few peaks for ARR10-B among the 75 expected (64 amino acid residues plus side chains) because of denaturation of the polypeptide. Decreasing the temperature to <20°C dramatically improved the spectrum, which displayed a reasonable number of well-dispersed, intense signals, indicating a well-defined protein structure.
The NMR observation that ARR10-B undergoes thermal denaturation in a reversible manner is in agreement with the results obtained by CD analysis. Increased salt concentration also improved protein stability, such that good-quality spectra were obtained for 1 mM ARR10-B in 50 mM sodium phosphate buffer containing 500 mM NaCl, pH 6.9, at 15°C. The NMR studies did not detect any dimerization or higher aggregates of ARR10-B under this condition.
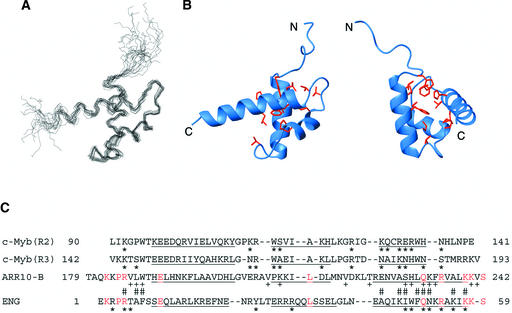
The three-dimensional structure of ARR10-B (Thr-179 to Ser-242) was determined by a hybrid distance geometry–dynamic simulated annealing approach (Nilges et al., 1988) based on 1040 experimental restraints derived from NMR spectroscopy. The superposed backbone N, Cα, and C′ coordinates of the final 15 structures (Figure 7A) were well aligned, except for residues 179 to 187 at the N terminus and residues 240 to 242 at the C terminus. For these residues (188 to 239), the root-mean-square deviation for backbone heavy atoms was 0.72 ± 0.15 Å; for all heavy atoms, it was 1.21 ± 0.16 Å.
Figure 7.
Three-Dimensional Solution Structure of ARR10-B.
(A) The best-fit superposition of the backbone (N, Cα, and C′) atoms of the final 15 structures of ARR10-B. The structures were superimposed against the energy-minimized average structure using the backbone coordinates of residues 188 to 239.
(B) Two different views of a ribbon diagram of the energy-minimized average structure of ARR10-B. Side-chain heavy atoms of Trp-188, Leu-192, Phe-196, Val-205, Pro-210, Ile-213, Met-217, Val-219, Leu-222, His-230, and Leu-231 are shown as ball-and-stick models. Hydrophobic interactions between these residues stabilize the three-helix protein structure. The left view is in the same orientation as (A), whereas the right view was obtained by ∼90° clockwise rotation about the vertical axis.
(C) Structure-based alignment of the amino acid sequences of ARR10-B, the engrailed homeodomain (ENG), and the second and third repeats of c-Myb. The residues in the α-helices of each structure are underlined. Identical residues between ARR10-B and ENG are highlighted in magenta. Asterisks indicate the residues of c-Myb and ENG that interact with their target DNAs. Residues of ARR10-B with + display large chemical shift changes upon binding to DNA 2, and those with # directly contact DNA 2 (see text).
Ribbon diagrams (Figure 7B) representing the backbone conformation of the restrained, energy-minimized mean structure of ARR10-B show that its structure consists of three α-helices (α1, residues 190 to 202; α2, 210 to 217, α3, 224 to 239). The helices α1 and α2 are connected by a type-I β-turn (Val-205 to Ala-208), whereas α2 and α3 are connected by a type-II β-turn (Val-219 to Lys-222). The three-helix structure is stabilized by a hydrophobic core formed by residues Trp-188, Leu-192, Phe-196, Val-205, Pro-210, Ile-213, Met-217, Val-219, Leu-222, His-230, and Leu-231, as shown in Figure 7B.
The helices α2 and α3, along with their connecting type-II β-turn, form a HTH variant motif, a fundamental DNA-recognition unit observed in many DNA binding proteins, such as c-Myb repeats and homeodomains. In addition, ARR10-B has a positively charged surface around the HTH motif, especially around the presumed DNA-recognition helix α3, as observed in the other DNA binding proteins (data not shown).
A specific DNA complex with the minimal DNA binding domains of c-Myb has revealed that two DNA-recognition helices from the two tandem repeats are closely packed in the major groove of DNA in a cooperative manner (Ogata et al., 1994). On the other hand, a single engrailed homeodomain (ENG) molecule has been shown to form a stable complex with DNA, in which the third helix binds in the major groove of DNA and provides the sequence-specific interactions (Kissinger et al., 1990). In the same complex, the N-terminal basic residue–rich region (a so-called flexible arm) of ENG binds in the minor groove on the opposite side of the DNA molecule, and side chains of the basic residues interact nonspecifically with the DNA's phosphate groups (Kissinger et al., 1990).
It should be noted that the DNA-recognition helix of ENG is longer than that in each of the c-Myb repeats. Comparing the structure of ARR10-B with those of other DNA binding proteins with HTH variant motifs, we observed more similarity to homeodomains composed of one Myb-related domain, such as ENG, than to the classic Myb-like proteins with multiple repeats. This conclusion is based on (1) the length of α3-helix, and (2) the existence of the basic residue–rich N-terminal region in front of the α1-helix (Figure 7C). Although the primary sequences of ARR10-B and ENG are only 15% identical, most of the shared amino acid residues are located in the α3-helix and the N-terminal region.
Specific Residues in ARR10-B Interact with DNA
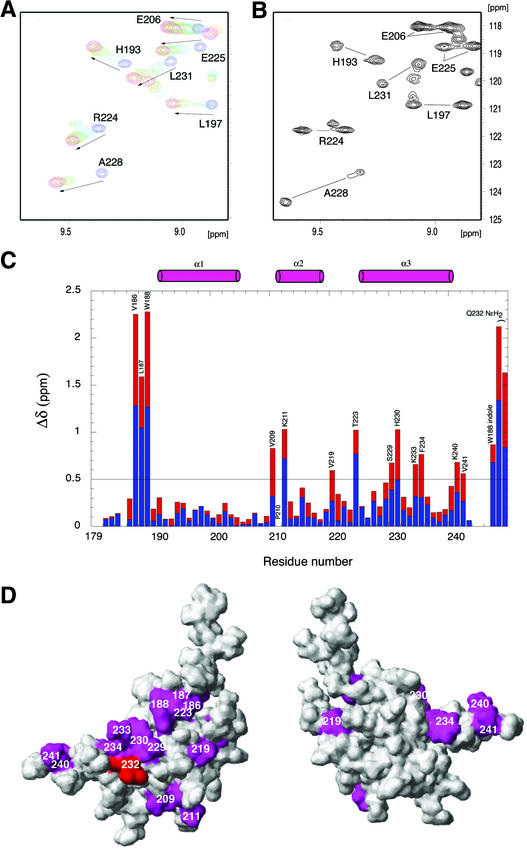
To identify specific residues in ARR10-B that interact with DNA, we recorded ARR10-B amide 15N and 1H chemical shifts as a function of the concentration of the 12-bp DNA 2 (Kd = 0.6 μM). It was anticipated that residues proximal to DNA would exhibit large changes in their chemical shifts as a result of DNA binding, because the chemical shift of a nucleus depends on its magnetic environment. For comparison, the 12-bp DNA 1 (Kd = 5.4 μM) also was included in such studies. The results of titration experiments indicate that DNA 1 binds to ARR10-B in a fast exchange manner on the NMR time scale (Figure 8A), whereas DNA 2 binds in a slow exchange manner (Figure 8B). This is clear NMR evidence that ARR10-B binds more tightly to DNA 2 than to DNA 1, supporting the results of binding analyses by SPR.
Figure 8.
NMR Chemical Shift Perturbation of ARR10-B upon Binding to DNA.
(A) Overlay of a selected region of the 1H-15N HSQC spectra obtained at 15°C of ARR10-B in the absence and presence of DNA 1, providing evidence that the binding of ARR10-B to DNA 1 is in fast exchange on the NMR time scale. The arrows show the direction of shifts. ARR10-B/DNA 1 = 1:0 (blue), 1:0.5 (aqua), 1:1 (green), 1:1.5 (orange), and 1:2.5 (magenta).
(B) Selected region of the single 1H-15N HSQC spectrum obtained at 15°C of ARR10-B in the presence of 0.3 equivalents of DNA 2, which shows two sets of ARR10-B amide signals from the free and DNA-bound forms in slow exchange.
(C) Changes in the NMR chemical shifts of ARR10-B induced by complex formation with DNA 2 and calculated with the function Δδ = |ΔδHN (blue)| + |0.17Δδ15N (red)|. The positions of the α-helices are indicated schematically as cylinders at the top of the figure.
(D) Two views of the ARR10-B surface colored magenta to indicate residues whose backbone amide chemical shifts were most sensitive (chemical shift perturbations Δδ > 0.5 ppm) to binding with DNA 2. Note that the side chain of Glu-54, shown in red, was perturbed. The left view is in the same orientation as Figure 7A, whereas the right view was obtained by ∼180° clockwise rotation about the vertical axis.
As shown in Figure 8C, 14 residues (Val-186, Leu-187, Trp-188 and its side chain, Val-209, Lys-211, Val-219, Thr-223, Ser-229, His-230, Lys-233, Phe-234, Lys-240, Val-241, and the Gln-232 side chain) display significant chemical shift perturbations upon complex formation with DNA 2. Binding to DNA 1 induces similar chemical shift changes for the same residues. It should be mentioned, however, that the amide signals of Val-186, Leu-187, and Trp-188 were not observed for the ARR10-B/DNA 1 complex because of the extreme broadening of the signal, an observation we ascribe to chemical exchange between the free and bound forms on the chemical shift time scale.
The residues whose chemical shifts are most sensitive (chemical shift perturbations Δδ = |ΔδHN| + |0.17Δδ15N| > 0.5 ppm) to complex formation with DNA 2 are mapped on the free-form structure of ARR10-B (Figure 8D). These residues, with the exception of Val-186, Leu-187, and Trp-188, are localized on one side of the molecule around the HTH motif from α2 and α3 and form the major DNA binding surface. The location of this DNA binding surface is in good agreement with the contact sites of ENG in the crystal structure of its complex with DNA (Kissinger et al., 1990). Note that the residues Val-186, Leu-187, and Trp-188, which show the largest chemical shift perturbations, as well as their preceding basic residue–rich region (Lys-182–Lys-183–Pro-184–Arg-185), closely resemble the flexible arm of ENG (Lys-2–Arg-3–Pro-4–Arg-5–Thr-6–Ala-7–Phe-8), which binds in the minor groove of its target DNA (Figure 7C).
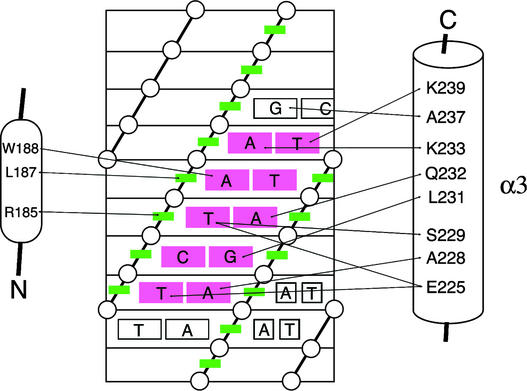
To obtain direct information regarding the mode of DNA recognition by ARR10-B, we analyzed three-dimensional 1H-15N NOESY-HSQC (13C filtered in F1) and three-dimensional 1H-13C NOESY-HSQC (13C selected in F1 and filtered in F3) spectra of the 1:1 15N/13C-labeled ARR10-B/DNA 2 complex. We were able to unambiguously assign 20 intermolecular nuclear overhauser effects (NOEs) between the polypeptide and DNA, as illustrated schematically in Figure 9. The majority of the identified intermolecular NOEs were observed between residues in the α3-helix of ARR10-B and bases around the AGATT core sequence of DNA 2.
Figure 9.
Scheme of the ARR10-B–DNA Interactions.
The contacts were derived from intermolecular NOEs between ARR10 and DNA 2. The DNA is represented as a cylindrical projection with phosphates indicated by circles and sugar rings indicated by green boxes. The critical AGATT base pairs are shown in red.
It should be mentioned that the side chain of Arg-185, in the N-terminal flexible arm, shows intermolecular NOEs with the minor groove side of the sugar ring of the thymine at bp 6. Hence, we conclude that a single ARR10-B molecule forms a complex with its target DNA, in which residues of the α3-helix located in the HTH motif recognize the major groove, and the N-terminal flexible arm contacts the minor groove of the DNA.
DISCUSSION
The B motif, originally identified as a signature of the type-B ARRs involved in His→Asp phosphorelay systems in Arabidopsis (Imamura et al., 1999), is a representative of the GARP motifs commonly observed in GARP family plant transcription factors, including maize G2 and C. reinhardtii PSR1 (Riechmann et al., 2000). G2 controls the differentiation of a photosynthetic cell type of the maize leaf (Hall et al., 1998), whereas PSR1 is a regulator of phosphorus metabolism (Wykoff et al., 1999). Arabidopsis PHR1, a protein homologous with PSR1, is involved in phosphate starvation signaling (Rubio et al., 2001). It has been reported that the recently identified KANADI gene product, which possesses a GARP motif, regulates organ polarity in Arabidopsis (Kerstetter et al., 2001). In any case, proteins in the GARP family appear to be involved in plant-specific processes. Although their amino acid sequences are related distantly to those of the authentic mammalian Myb repeat, little is known about the structure and function of the B motifs (or GARP motifs).
In this study, we performed extensive biochemical and structural studies on ARR10-B, the B motif (Thr-179 to Ser-242) of ARR10, a representative of the type-B family of ARRs. We found that GFP-ARR10-B is capable of localizing predominantly, if not exclusively, to the nucleus, suggesting that the B motif possesses an NLS in its amino acid sequence. Although there is no consensus sequence, NLS generally are short sequences that contain a high proportion of positively charged amino acids.
Inspection of the primary sequences of various B motifs revealed that a short stretch in front of the first α-helix, corresponding to residues 182 to 185 (KKPR) in the case of ARR10-B (Figures 1B and 7C), has the highest density of positively charged amino acids. This stretch also fulfills a prerequisite that the NLS be exposed on the surface of the protein to function. Hence, we propose that the short, positively charged stretch located in the flexible region preceding the α-helix of type-B ARRs may serve as an NLS, at least in part.
The B motif has been presumed to be a functional unit for DNA binding because its primary sequence weakly resembles the actual DNA binding domains of the classic mouse c-Myb. The results presented in this study clearly demonstrate that the B motif binds to DNA in a sequence-specific manner and that the optimal target sequence is AGATT. The same sequence has been proposed as the best target for two other type-B ARRs, ARR1 and ARR2, based on studies using their B motifs fused with glutathione S-transferase (Sakai et al., 2000).
It is quite reasonable that ARR10, ARR1, and ARR2 preferentially recognize the same sequence because of the high degree of conservation between the primary sequences of their B motifs. Although nuclear localization and DNA binding are functionally separable, NLS are observed in the DNA binding domains of other classes of transcription factors, including POU homeodomains (Sock et al., 1996) and HMG box (Poulat et al., 1995), HLH (Tapscott et al., 1988), and bZip proteins (Waeber and Habener, 1991). These observations indicate that the B motif is a multifunctional domain of type-B ARRs.
The three-dimensional solution structure of ARR10-B, determined by NMR and CD analyses in this study, is similar to the Myb repeat and homeodomain and consists of a three-helix structure containing an HTH variant motif from the second to third helices. A careful comparison of these structures revealed that ARR10-B more closely resembles the homeodomain than the Myb repeat, based on the length of the third helix and the existence of a basic residue–rich N-terminal flexible arm in front of the first helix.
This idea is further strengthened by NMR studies of the protein–DNA interactions of the ARR10-B/DNA complex. As in homeodomains, a single ARR10-B molecule recognizes the major groove of DNA with residues located on the solvent-exposed surface of the third helix and contacts the adjacent minor groove with the N-terminal flexible arm (Figures 8 and 9). Complex formation between ARR10-B and DNA significantly increases the thermal stability of the polypeptide: Tm = 29°C for the free form and 55°C for the bound form. Also, a slight increase in helix content is observed that is attributable, at least in part, to elongation of the third helix. Detailed structural analysis of the ARR10-B/DNA 2 complex is in progress, which could aid in fully understanding the structural basis for DNA recognition by the B motif of ARR10.
Our observation that ARR10-B binds the target DNA as a monomer is in sharp contrast to the report that PHR1, another member of the GARP family, has been shown to bind as a dimer to an imperfect palindromic DNA sequence, GNATATNC (Rubio et al., 2001). PHR1 has a coiled-coil (CC) domain that is potentially involved in dimerization as well as the GARP motif responsible for the sequence-specific DNA recognition. By contrast, ARR10 does not have such a CC domain in its entire sequence. Hence, the intact ARR10 could bind DNA as a monomer.
These findings prompted us to examine whether other GARP proteins contain such a dimerization CC domain; for this, we used COILS (www.ch.embnet.org/software/COILS_form.html). The results revealed that the GARP family could be divided into two subgroups: subgroup I includes G2, ARRs, and KANADI without a CC domain; subgroup II includes PHR1 and PSR1 with a CC domain. Based on this classification, we predict that the GARP proteins belonging to subgroup I bind DNA as a monomer, whereas those of subgroup II bind DNA as a dimer. The mechanism of DNA recognition by the former class of proteins could be the same as that of ARR10-B determined in this study.
Interruption of dimer formation in PHR1 leads to a loss of DNA binding affinity to the target sequence, GNATATNC (Rubio et al., 2001), suggesting that this sequence is not optimum for the single GARP motif of PHR1. Rather, cooperative DNA binding by two GARP motifs of the dimeric PHR1 may establish a high affinity to the imperfect palindromic sequence. Our modeling study shows that two recognition α3-helices of the GARP motifs of PHR1 could enter the major groove of DNA in opposite directions in a manner similar to Max, a dimeric basic/HTH/Leu zipper transcription factor (Ferré-D'Amaré et al., 1993). In this model, each GARP motif of the dimeric PHR1 recognizes half of the target sequence, GNAT, just as ARR10-B binds the first four bases of its target, AGATT.
In summary, in this study, we clarified the functional and structural features of the DNA binding B motif of Arabidopsis response regulators, providing a general and molecular basis for DNA recognition by the GARP family of transcription factors, which are widespread in higher plants.
METHODS
Plants and Related Materials
The Columbia ecotype of Arabidopsis thaliana was used. Plants were grown with 16-h-light/8-h-dark fluorescent illumination at 22°C on soil or on agar plates containing Murashige and Skoog (1962) salts and 2% Suc.
Escherichia coli and Related Materials
E. coli K-12 strain DZ225 (F-, ΔenvZ, lacU169, araD139, rpsL, relA, flgB, thiA), carrying the plasmid-borne multicopy arcB gene, was used to prepare cytoplasmic membranes (Nagasawa et al., 1993). E. coli cells were grown in Luria broth, and the urea-treated cytoplasmic membranes were purified as described previously (Tokishita et al., 1990). The cytoplasmic membranes were used to phosphorylate the Arabidopsis AHP phosphotransmitter proteins in vitro, as described previously (Makino et al., 2000). E. coli strain BL21(DE3) was used as a host for preparations of Arabidopsis ARR10-RB and ARR10-B polypeptides.
PCR Amplification
Using appropriate pairs of primers, PCR was performed to prepare DNA segments. An Arabidopsis cDNA library in the λgt11 expression vector was a gift from K. Shinozaki (RIKEN, Genomic Sciences Center, Yokohama, Japan). PCR conditions usually were 94°C for 1 min, 55°C for 2 min, and 72°C for 3 min for 25 cycles in a Thermal Cycler 480 (Takara Shuzo, Kyoto, Japan). A PCR kit was used according to the supplier's instructions.
Plasmid Construction
For overproduction of the ARR10-RB and ARR10-B polypeptides, plasmids pET-ARR10-RB and pET-ARR10-B were constructed as follows. Each coding sequence was amplified from the cDNA clone using an appropriate pair of PCR primers. These amplified DNA segments were cloned into an E. coli expression vector, pET22b(+) (Novagen, Madison, WI). In the case of pET-ARR10-RB, but not pET-ARR10-B, the coding sequence was designed to include a C-terminally extended His tag.
Purification of Polypeptides
E. coli BL21(DE3) cells carrying appropriate plasmids, as described above, were grown in M9-Glc medium containing 0.2% casamino acids and 15% Suc. A cleared cell lysate was obtained using an Amino French pressure cell (FA073; Aminco, SLM Instruments, Urbana, IL) followed by centrifugation at 100,000g for 3 h (Tokishita et al., 1990). In the case of ARR10-RB, the resulting soluble protein samples were applied to a Ni column with the rapid affinity purification pET His-Tag system (Novagen). Other details were as recommended by the supplier.
The B motif (ARR10-B) was purified as follows. E. coli cells transformed with pET-ARR10-B were grown to the late logarithmic growth phase in the presence of 2 mM isopropyl-β-d-thiogalactopyranoside. To 10 g of cells was added 30 mL of sodium phosphate buffer, pH 7.1, containing 5 mM MgSO4, 2 mM 2-mercaptoethanol, and 0.1 mM phenylmethylsulfonyl fluoride. The suspension was passed through an Amino French pressure cell (FA073) twice at 10,000 p.s.i. and centrifuged at 100,000g for 3 h at 4°C to isolate the clear supernatant fraction. The volume of the suspension was brought up to 150 mL by the addition of 100 mL of a column buffer containing 50 mM Tris-HCl, pH 8.0, 0.5 mM EDTA, and 2 mM 2-mercaptoethanol. Solid ammonium sulfate was added to 75% saturation at 4°C, and precipitates were discarded.
Precipitates formed at 100% saturation were recovered by centrifugation and then dissolved in 10 mL of the column buffer. After dialysis against the same buffer, the sample was loaded onto the Mono-S column of a fast protein liquid chromatography system (Pharmacia-LKB). Proteins were eluted with a 200-mL linear gradient from 20 to 600 mM NaCl. The peak fractions containing the B motif polypeptide were collected and used.
Cellular Localization Assays with Green Fluorescent Protein Fusion Proteins
The green fluorescent protein (GFP) expression vector p221-EGFP-C1 is a recombinant derivative of pBI221/pEGFP-C1 (a gift from K. Hiratsuka, Nara Institute of Science and Technology) (Chiu et al., 1996). The coding sequences of ARR10-B and LIM13 were inserted into p221-EGFP-C1 to be fused in frame to the C-terminal end of the GFP coding sequence (Ogata et al., 1999). DNA fragments carrying each of the resulting fusion genes (0.4 μg each) were introduced into onion skin epidermal cells using a particle gun (GIE-III; Tanaka Co., Sapporo, Japan) (Makino et al., 2000). The conditions of bombardment were as follows: vacuum of 640 mm Hg, helium pressure of 3.8 kg·cm−2, and target distance of 3 cm. After bombardment, tissues were incubated for 12 h at 25°C in the dark. Histochemical observation was made with an Olympus fluorescence microscope (BH2-RFCA; Tokyo, Japan) using a 380- to 490-nm excitation filter.
PCR-Assisted DNA Binding Site Selection from Random Oligonucleotides
To identify the target binding sequence of ARR10-RB, we synthesized a mixture of 54-base oligonucleotides in which the middle 16 bases consisted of random sequences generated by incorporating each of the four nucleotides at equimolar concentration. These oligonucleotides (2.5 μmol) were converted to double-stranded DNA using the DNA polymerase Klenow fragment, extending from a primer that hybridizes to the 18 bases at their 3′ ends (5′-CGACGCTCAGTCTCGAGG-3′). The double-stranded DNA then was incubated with ARR10-RB (10 μg) in a buffer containing 20 mM Tris-HCl, pH 8.0, and 80 mM KCl. ARR10-RB was recovered by precipitation with Ni beads, and the precipitate was washed extensively with the binding buffer.
The precipitate, presumably containing ARR10-RB binding oligonucleotides, was subjected to PCR amplification with an appropriate pair of primers (5′-GGCTGAGTCTGAACAAGCTT-3′ and 5′-CGACGCTCAGTCTCGAGG-3′). Resulting DNA fragments of appropriate size were purified by electrophoresis through a polyacrylamide gel (8%) and then used for a second cycle of precipitation with ARR10-RB, as described above. After seven repeated cycles of such amplification, the recovered DNA fragments were cloned into pUC19 in E. coli, and their DNA sequences were determined. As shown in Figure 4, these procedures are essentially the same as those reported previously (Chittenden et al., 1991; Sun and Baltimore, 1991).
DNA Binding Gel-Shift Assay
Purified ARR10-B was incubated with appropriate 32P-labeled 34-bp DNA fragments, and the resulting samples were analyzed on a 6% polyacrylamide gel (30:0.5, monomer:bis). The running buffer comprised 40 mM Tris-acetate, pH 7.4, 5 mM Na-acetate, and 1 mM EDTA. DNA fragments in the gels were detected by autoradiography.
Surface Plasmon Resonance Measurements
DNA binding affinity of ARR10-B was measured by surface plasmon resonance at 20°C on a BIAcore biosensor system (BIAcore, Uppsala, Sweden). Four 12-bp oligonucleotides (with their different core sites underlined) were examined: double-stranded DNA (d)-CACTGATTCAGG/d-CCTGAATCAGTG (DNA 1), d-GCAAGATTCGGC/d-GCCGAATCTTGC (DNA 2), d-GCAAGATCAGGC/d-GCCTGATCTTGC (DNA 3), and d-GCAAGATCTTGC/d-GCAAGATCTTGC (DNA 4). The oligonucleotides were biotinylated at their 5′ ends, dissolved in running buffer (10 mM potassium phosphate, pH 7.0, containing 150 mM NaCl and 1 mM EDTA) at a DNA concentration of 1 μg/mL, and immobilized on a streptavidin preimmobilized biosensor chip (BIAcore).
Immobilization was performed at a flow rate of 5 μL/min and an exposure time of 20 min, resulting in the capture of 658, 512, 1136, and 476 resonance units of DNAs 1, 2, 3, and 4, respectively. The flow cell was primed with running buffer before sample injection. For binding analyses, ARR10-B, dissolved in the running buffer at different concentrations from 0.1 to 20 μM, was injected at a flow rate of 20 μL/min for 2 min. The apparent rate constant for dissociation was measured in the free running buffer flowing at the same rate for 3 min, after the end of the association phase. After each binding experiment, the sensor chip was regenerated with 0.05% Triton containing 3 M NaCl.
Circular Dichroism Measurements
Circular dichroism spectra of ARR10-B were recorded on a J-720WI spectropolarimeter (JASCO, Tokyo, Japan) in the far-UV range (200 to 260 nm) using a 0.1-cm thermostatted cell. The spectra for the free form were obtained for 7 and 27 μM protein in 10 mM sodium phosphate buffer, pH 7.0, whereas those for the DNA-bound form were measured for 40 μM protein in the same buffer containing 1.2 equivalents of DNA 2 and 160 mM NaCl. Thermal transition curves were obtained by monitoring molecular ellipticity at 222 nm, [θ222], as a function of temperature with a 0.1-cm path length cell and were analyzed by fitting to a two-state model (Yadav and Ahmad, 2000). The temperature was increased or decreased at a rate of 20°C/h.
NMR Spectroscopy
Recombinant, uniformly 15N- and 15N/13C-labeled ARR10-B was expressed in E. coli grown in M9 minimal medium and purified as described above. To determine the best solution conditions for NMR spectroscopy, temperature, pH, salt concentration, and protein concentration for 15N-labeled ARR10-B were varied systematically. The results were judged based on the appearance of the 1H-15N HSQC. Pilot experiments suggested that relatively high concentrations of salt and low temperatures were desirable. NMR samples used for structure determination of ARR10-B contained ∼1 mM 15N or 15N/13C-labeled protein and 500 mM NaCl in 50 mM sodium phosphate buffer, pH 6.9.
All NMR spectra were acquired at 15°C on a Bruker DMX 750 spectrometer equipped with pulse-field gradients (Billerica, MA). 1H, 13C, and 15N sequential resonance assignments were obtained using two-dimensional (2D) double resonance and three-dimensional (3D) double and triple resonance through bond-correlation experiments (Clore and Gronenborn, 1991, 1998; Bax and Grzesiek, 1993): 2D 1H-15N HSQC, 2D 1H-13C CT-HSQC optimized for observation of either aliphatic or aromatic signals; 3D 15N-separated HOHAHA-HSQC, 3D HNHA, 3D HNHB, 3D HNCO, 3D CBCA(CO)NH, 3D HBHA(CBCACO)NH, 3D HNCACB, 3D C(CO)NH, 3D HCABGCO, and 3D HCCH-TOCSY.
Stereospecific assignments of α- and β-protons and the methyl side chains of Val and Leu residues were achieved by a combination of quantitative J measurements and NOESY data. 3J couplings were measured using quantitative 2D and 3D J correlation spectroscopy (Clore and Gronenborn, 1998). Interproton distance restraints were derived from multidimensional nuclear overhauser effect (NOE) spectra (Clore and Gronenborn, 1991, 1998; Bax and Grzesiek, 1993): a 3D 15N-separated NOESY-HSQC spectrum with a mixing time of 100 ms, a 3D 13C/15N-separated NOESY-HSQC spectrum with a mixing time of 100 ms, and a four-dimensional 13C/13C-separated HMQC-NOESY-HMQC spectrum with a mixing time of 100 ms.
All spectra were processed using NMRPipe software (Delaglio et al., 1995) and analyzed using Capp/Pipp/Stapp software (Garrett et al., 1991). 1H, 13C, and 15N chemical shifts were referenced, respectively, to mono-deuterated water (4.87 ppm at 15°C), indirectly to sodium 3-(trimethylsilyl)-propionate (13C) (Bax and Subramanian, 1986), and to liquid ammonia (15N) (Live et al., 1984).
A series of 2D 1H-15N HSQC spectra were recorded, as a function of DNA concentration, for ARR10-B (1.0 mM) in 10 mM sodium phosphate buffer, pH 6.9, containing 150 mM NaCl. Two oligonucleotide sequences, d-CACTGATTCAGG/d-CCTGAATCAGTG (DNA 1) and d-GCAAGATTCGGC/d-GCCGAATCTTGC (DNA 2), were examined. The HSQC signals of ARR10-B bound to DNA 2 were assigned by analyzing 3D triple resonance through bond-correlation experiments performed on the solution containing 1.0 mM 15N/13C-labeled ARR10-B and 1.2 mM DNA 2 using the same procedure used for the signal assignments of the free form (see above). Assignments of the HSQC signals of ARR10-B bound to DNA 1 were made by tracing the peaks during titration and were confirmed by analyzing 3D triple-resonance experiments performed on the solution containing 1.0 mM 15N/13C-labeled ARR10-B and 2.5 mM DNA 1.
To assign intermolecular NOEs between the DNA and the protein, the following isotope-filtered experiments were performed on the 1:1 15N/13C-labeled ARR10-B/DNA 2 complex: (1) a 3D 15N-separated, 13C-filtered NOESY-HSQC experiment with a 100-ms mixing time, and (2) a 3D 13C-separated, 13C-filtered NOESY-HSQC experiment with a 100-ms mixing time. The proton signals of DNA 2 were assigned using a combination of 2D 13C-filtered (in F1 and F2) COSY and NOESY spectra (Ikura and Bax, 1992), the latter obtained with a mixing time of 100 ms.
Structure Calculations
NOE-derived interproton distance restraints were classified into four ranges: 1.8 to 2.7 Å, 1.8 to 3.3 Å, 1.8 to 4.3 Å, and 1.8 to 5.0 Å, corresponding to strong, medium, weak, and very weak NOEs. An additional 1.5 Å was added to the upper bound for NOEs involving methyl protons and methylene protons that were not assigned stereospecifically (Wüthrich et al., 1983). Torsion angle restraints on φ and ψ were derived from 3JHNHα coupling constants (Vuister and Bax, 1993), short-range NOEs, and a database analysis of backbone (13Cα, 13Cβ, 13C′, 1Hα, 15N) chemical shifts using the program TALOS (Cornilescu et al., 1999). The side-chain χ1 angle restraints were derived from HOHAHA connectivities and short-range NOEs and ROEs combined with 3JHαHβ and 3JNHβ coupling constants.
Structures of ARR10-B were calculated using the hybrid distance geometry–dynamic simulated annealing method (Nilges et al., 1988), as contained in X-PLOR 3.1 (Brünger, 1993). For structure calculations, we used 893 interproton distance restraints [comprising 402 intraresidue, 246 sequential (|i − j| = 1), 169 medium-range (1 < |i − j| < 5), and 76 long-range (|i − j| ≥ 5) restraints] obtained from heteronuclear 3D and four-dimensional NOE spectra. In addition to the NOE-derived distance restraints, 147 dihedral angle restraints (49 φ, 51 ψ, 37 χ1, and 10 χ2) were included in the structure calculation. A final set of 15 lowest energy structures was selected from 100 calculations. None of them had NOE or dihedral angle violations of >0.5 Å or 5°, respectively.
The structural statistics calculated for the final 15 structures are summarized in Table 1. The average coordinates of the ensembles of the final 15 structures were subjected to 500 cycles of Powell restrained-energy minimization to improve stereochemistry and nonbonded contacts. The structural statistics for the restrained-energy minimized average structure also are summarized in Table 1. Figures were generated using MOLMOL (Koradi et al., 1996).
Table 1.
Structural Statistics for ARR10-B
| Parameter | {SA} | <SA>r |
|---|---|---|
| X-PLOR energies (kcal·mol−1)a | ||
| Etotal | 77.6 ± 3.2 | 75.8 |
| Ebond | 3.39 ± 0.27 | 3.46 |
| Eangle | 58.1 ± 1.3 | 58.3 |
| Eimproper | 6.49 ±0.34 | 5.97 |
| Evdw | 3.46 ± 0.99 | 4.04 |
| Enoe | 5.76 ± 0.84 | 3.70 |
| Ecdih | 0.46 ± 0.15 | 0.34 |
| Root-mean-square deviation from idealized geometry |
||
| Bond (Å) | 0.0018 ± 0.00007 | 0.0018 |
| Angle (°) | 0.44 ± 0.005 | 0.44 |
| Improper (°) | 0.28 ± 0.01 | 0.27 |
| Root-mean-square deviation from experimental restraintsb |
||
| Distance (Å) | 0.011 ± 0.0008 | 0.0091 |
| Dihedral angle (°) | 0.22 ± 0.04 | 0.19 |
| Ramachandran plot analysis (%)c | ||
| Residues in most favored regions | 92.8 (83.7) | |
| Residues in additional allowed regions |
6.4 (13.0) | |
| Residues in generously allowed regions |
0.8 (3.0) | |
| Residues in disallowed regions | 0.0 (0.3) | |
| Root-mean-square deviation from mean structure (Å)d |
||
| Backbone (N, Cα, C′) atoms | 0.72 ± 0.158 | |
| All nonhydrogen atoms | 1.21 ± 0.164 |
The notation of the NMR structures is as follows: {SA} is the ensemble of the final 15 simulated annealing structures; <SA>r is the restrained regularized mean structure obtained by averaging the backbone coordinates of the individual SA structures best fitted to each other (residues 188 to 239), followed by restrained regularization of the mean structure.
a These values were estimated using X-PLOR 3.1. The final values of the force constants used for the calculations are as follows: 1000 kcal·mol−1·Å−2 for bond lengths, 500 kcal·mol−1·rad−2 for bond angles and improper torsions, 4 kcal·mol−1·Å−4 for the quartic van der Waals term with van der Waals radii set to 0.75 times the values used in the CHARMM empirical energy function, 50 kcal·mol−1·Å−2 for NOE-derived and hydrogen bonding distance restraints with the ceiling value of 1000 kcal·mol-1, and 200 kcal·mol-1·rad-2 for dihedral angle restraints.
b None of the structures exhibited interproton distance violations > 0.5 Å or dihedral angle violations > 5°. The distance restraints comprise 402 intraresidue, 246 sequential (|i−j|= 1), 169 medium-range (1 < |i−j| < 5) and 76 long-range (|i−j| ≧ 5) NOEs. The dihedral angle restraints involve 49 φ, 51 ψ, 37 χ1, and 10 χ2.
c The values were calculated for residues 188 to 239 using PROCHECK (Laskowski et al., 1996). The corresponding values calculated for all residues (179 to 242), are shown in parentheses.
d The atomic root-mean-square deviation values were calculated for residues 188 to 239.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Accession Numbers
The coordinates of the final structures and the structural constraints used for the calculations have been deposited in the RCSB Protein Data Bank (accession code 1IRZ). The chemical shift values of the 1H, 13C, and 15N resonances have been deposited in the BioMagResBank (http://www.brmb.wisc.edu) database (accession number 5174).
Acknowledgments
Thanks are due to A. Oka and T. Aoyama (Kyoto University) for their gifts (oligonucleotides used in Figure 5A). This work was supported by a grant from the Bio-Oriented Technology Research Advancement Institution of Japan to T.Y., by a grant for the Rice Genome Project (PR-2201) from the Ministry of Agriculture, Forestry, and Fisheries of Japan to E.K., and by a grant-in-aid for scientific research on a priority area (12142201 and COE) from the Ministry of Education, Science, Sport, and Culture of Japan to T.M.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.002733.
References
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Bax, A., and Grzesiek, S. (1993). Methodological advances in protein NMR. Acc. Chem. Res. 26, 131–138. [Google Scholar]
- Bax, A., and Subramanian, S. (1986). Sensitivity enhanced two-dimensional heteronuclear shift correlation NMR spectroscopy. J. Magn. Reson. 67, 565–569. [Google Scholar]
- Brünger, A.T. (1993). X-PLOR Manual Version 3.1. (New Haven, CT: Yale University).
- Chang, C., Kwok, S.F., Bleeker, A.B., and Meyerowitz, E.M. (1993). Arabidopsis ethylene-response gene ETR1: Similarity of product to two-component regulators. Science 262, 539–544. [DOI] [PubMed] [Google Scholar]
- Chang, C., and Stewart, R.C. (1998). The two-conponent system: Regulation of diverse signaling pathways in prokaryotes and eukaryotes. Plant Physiol. 117, 723–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chittenden, T., Livingston, D.M., and Kaelin, W.G., Jr. (1991). The T/E1A-binding domain of the retinoblastoma product can interact selectively with a sequence-specific DNA-binding protein. Cell 65, 1073–1082. [DOI] [PubMed] [Google Scholar]
- Chiu, W.-I., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H., and Sheen, J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6, 325–330. [DOI] [PubMed] [Google Scholar]
- Clore, G.M., and Gronenborn, A.M. (1991). Structures of larger proteins in solution: Three- and four-dimensional heteronuclear NMR spectroscopy. Science 252, 1390–1399. [DOI] [PubMed] [Google Scholar]
- Clore, G.M., and Gronenborn, A.M. (1998). Determining the structures of larger proteins and protein complexes by NMR. Trends Biotechnol. 16, 22–34. [DOI] [PubMed] [Google Scholar]
- Cornilescu, G., Delaglio, F., and Bax, A. (1999). Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 113, 289–302. [DOI] [PubMed] [Google Scholar]
- D'Agostino, I.B., and Kieber, J.J. (1999). Phosphorelay signal transduction: The emerging family of plant response regulators. Trends Biol. Sci. 24, 452–456. [DOI] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., and Bax, A. (1995). NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Ferré-D'Amaré, A.R., Prendgergast, G.C., Ziff, E.B., and Burley, S.K. (1993). Recognition by Max of its cognate DNA through a dimeric b/HLH/Z domain. Nature 363, 38–45. [DOI] [PubMed] [Google Scholar]
- Garrett, D.S., Powers, R., Gronenborn, A.M., and Clore, G.M. (1991). A common sense approach to peak picking in two-, three- and four-dimensional spectra using automatic computer analysis of contour diagrams. J. Magn. Reson. 95, 214–220. [DOI] [PubMed] [Google Scholar]
- Hall, L.N., Rossini, L., Cribb, L., and Langdale, J.A. (1998). GOLDEN2: A novel transcriptional regulator of cellular differentiation in the maize leaf. Plant Cell 10, 925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, J., and Meyerowitz, E.M. (1998). Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94, 261–271. [DOI] [PubMed] [Google Scholar]
- Hwang, I., and Sheen, J. (2001). Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413, 383–389. [DOI] [PubMed] [Google Scholar]
- Ikura, M., and Bax, A. (1992). Isotope-filtered 2D NMR of a protein-peptide complex: Study of skeletal muscle myosin light chain kinase fragment bound to calmodulin. J. Am. Chem. Soc. 114, 2433–2440. [Google Scholar]
- Imamura, A., Hanaki, N., Nakamura, A., Suzuki, T., Taniguchi, M., Kiba, T., Ueguchi, C., Sugiyama, T., and Mizuno, T. (1999). Compilation and characterization of Arabidopsis response regulators implicated in His-Asp phosphorelay signal transduction. Plant Cell Physiol. 40, 733–742. [DOI] [PubMed] [Google Scholar]
- Imamura, A., Yoshino, Y., and Mizuno, T. (2001). Cellular localization of the signaling components of Arabidopsis His-to-Asp phosphorelay. Biosci. Biotechnol. Biochem. 65, 2113–2117. [DOI] [PubMed] [Google Scholar]
- Inoue, T., Higuchi, M., Hashimoto, Y., Seki, M., Kobayashi, M., Kato, T., Tabata, S., Shinozaki, K., and Kakimoto, T. (2001). Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409, 1060–1063. [DOI] [PubMed] [Google Scholar]
- Kerstetter, R.A., Bollman, K., Taylor, A., Bomblies, K., and Poethig, R.S. (2001). KANADI regulates organ polarity in Arabidopsis. Nature 411, 706–709. [DOI] [PubMed] [Google Scholar]
- Kiba, T., Taniguchi, M., Imamura, A., Ueguchi, C., Mizuno, T., and Sugiyama, T. (1999). Differential expression of genes for response regulators in response to cytokinins and nitrate in Arabidopsis thaliana. Plant Cell Physiol. 40, 767–771. [DOI] [PubMed] [Google Scholar]
- Kissinger, C.R., Liu, B., Martin-Blanco, E., Kornberg, T.B., and Pabo, C.O. (1990). Crystal structure of an engrailed homeodomain-DNA complex at 2.8 Å resolution: A framework for understanding homeodomain-DNA interactions. Cell 63, 579–590. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Wüthrich, K. (1996). MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., Rullmannn, J.A., MacArthur, M.W., Kaptein, R., and Thornton, J.M. (1996). AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486. [DOI] [PubMed] [Google Scholar]
- Live, D.H., Davis, D.G., Agosta, W.C., and Cowburn, D. (1984). Long-range hydrogen bond mediated effects in peptides: 15N NMR study of gramicidin S in water and organic solvents. J. Am. Chem. Soc. 106, 1934–1941. [Google Scholar]
- Lohrmann, J., Sweere, U., Zabaleta, E., Baurle, I., Kietel, C., Kozma-Bognar, L., Brennicke, A., Schafer, E., Kudla, J., and Harter, A. (2001). The response regulator ARR2: A pollen-specific transcription factor involved in the expression of nuclear genes for components of mitochondrial complex I in Arabidopsis. Mol. Genet. Genomics 265, 2–13. [DOI] [PubMed] [Google Scholar]
- Makino, S., Kiba, T., Imamura, A., Hanaki, N., Nakamura, A., Suzuki, T., Taniguchi, M., Ueguchi, C., Sugiyama, T., and Mizuno, T. (2000). Genes encoding pseudo-response regulators: Insight into His-to-Asp phosphorelay and circadian rhythm in Arabidopsis thaliana. Plant Cell Physiol. 41, 791–803. [DOI] [PubMed] [Google Scholar]
- Mizuno, T. (1997). Compilation of all genes encoding two-component phosphotransfer signal transducers in the genome of Escherichia coli. DNA Res. 4, 161–168. [DOI] [PubMed] [Google Scholar]
- Mizuno, T. (1998). His-Asp phosphotransfer signal transduction. J. Biochem. 123, 555–563. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Nagasawa, S., Tokishita, S., Aiba, H., and Mizuno, T. (1993). A novel sensor-regulator protein that belongs to the homologous family of signal transduction proteins involved in adaptive responses in Escherichia coli. Mol. Microbiol. 6, 799–807. [DOI] [PubMed] [Google Scholar]
- Nilges, M., Gronenborn, A.M., Brunger, A.T., and Clore, G.M. (1988). Determination of three-dimensional structures of proteins by simulated annealing with interproton distance restraints: Application to crambin, potato carboxypeptidase inhibitor and barley serine proteinase inhibitor 2. Protein Eng. 2, 27–38. [DOI] [PubMed] [Google Scholar]
- Ogata, K., Morikawa, S., Nakamura, H., Sekikawa, A., Inoue, T., Kanai, H., Sarai, A., Ishii, S., and Nishimura, Y. (1994). Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell 79, 639–648. [DOI] [PubMed] [Google Scholar]
- Ogata, S., Takase, H., Hiratsuka, K., and Hotta, Y. (1999). Mutational analysis of the signal for a nuclear localization of proteins which accumulate specifically during meiosis in lily microsporocytes. Plant Cell Rep. 19, 101–105. [DOI] [PubMed] [Google Scholar]
- Parkinson, J.S., and Kofoid, E.C. (1992). Communication modules in bacterial signalling proteins. Annu. Rev. Genet. 26, 71–112. [DOI] [PubMed] [Google Scholar]
- Poulat, F., Girard, F., Chevron, M.P., Goze, C., Rebillard, X., Calas, B., Lamb, N., and Berta, P. (1995). Nuclear localization of the testis determining gene product SRY. J. Cell Biol. 128, 737–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann, J.L., et al. (2000). Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. [DOI] [PubMed] [Google Scholar]
- Rubio, V., Linhares, F., Solano, R., Martin, A.C., Iglesias, J., Leyva, A., and Paz-Ares, J. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15, 2122–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai, H., Aoyama, T., Bono, H., and Oka, A. (1998). Two-component response regulators from Arabidopsis thaliana contain a putative DNA-binding motif. Plant Cell Physiol. 39, 1232–1239. [DOI] [PubMed] [Google Scholar]
- Sakai, H., Aoyama, T., and Oka, A. (2000). Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J. 24, 703–711. [DOI] [PubMed] [Google Scholar]
- Sakai, H., Honma, T., Aoyama, T., Sato, S., Kato, T., Tabata, S., and Oka, A. (2001). ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294, 1519–1521. [DOI] [PubMed] [Google Scholar]
- Schaffer, R., Ramsay, N., Samach, A., Corden, S., Putterill, J., Carre, I.A., and Coupland, G. (1998). The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93, 1219–1229. [DOI] [PubMed] [Google Scholar]
- Schaller, G.E. (2000). Histidine kinases and the role of two-component systems in plants. Adv. Bot. Plant Pathol. 32, 109–148. [Google Scholar]
- Sock, E., Enderich, J., Rosenfeld, M.G., and Wegner, M. (1996). Identification of the nuclear localization signal of the POU domain protein Tst-1/Oct6. J. Biol. Chem. 271, 17512–17518. [DOI] [PubMed] [Google Scholar]
- Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. [DOI] [PubMed] [Google Scholar]
- Sun, X.-H., and Baltimore, D. (1991). An inhibitory domain of E12 transcription factor prevents DNA binding in E12 homodimers but not in E12 heterodimers. Cell 64, 459–470. [DOI] [PubMed] [Google Scholar]
- Suzuki, T., Ishikawa, K., Yamashino, T., and Mizuno, T. (2002). An Arabidopsis histidine-containing phosphotransfer (HPt) factor implicated in phosphorelay signal transduction: Overexpression of AHP2 in plants results in hypersensitiveness to cytokinin. Plant Cell Physiol. 43, 123–129. [DOI] [PubMed] [Google Scholar]
- Suzuki, T., Miwa, K., Ishikawa, K., Yamada, H., Aiba, H., and Mizuno, T. (2001). The Arabidopsis sensor His-kinase, AHK4, can respond to cytokinins. Plant Cell Physiol. 42, 107–113. [DOI] [PubMed] [Google Scholar]
- Tapscott, S.J., Davis, R.L., Thayer, M.J., Cheng, P.F., Weintraub, H., and Lassar, A.B. (1988). MyoD1: A nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science 242, 405–411. [DOI] [PubMed] [Google Scholar]
- Tokishita, S., Yamada, H., Aiba, H., and Mizuno, T. (1990). Transmembrane signal transduction and osmoregulation in Escherichia coli. II. The osmotic sensor, EnvZ, located in the isolated cytoplasmic membrane displays its phosphorelation and dephosphorelation abilities as to the activator protein, OmpR. J. Biochem. 108, 488–493. [DOI] [PubMed] [Google Scholar]
- Urao, T., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2000). Two-component systems in plant signal transduction. Trends Plant Sci. 5, 67–74. [DOI] [PubMed] [Google Scholar]
- Vuister, G.W., and Bax, A. (1993). Quantitative J correlation: A new approach for measuring homonuclear three-bond J(HNHα) coupling constants in 15N-enriched proteins. J. Am. Chem. Soc. 115, 7772–7777. [Google Scholar]
- Waeber, G., and Habener, J.F. (1991). Nuclear translocation and DNA recognition signals colocalized within the bZIP domain of cyclic adenosine 3′,5′-monophosphate response element-binding protein CREB. Mol. Endocrinol. 5, 1431–1438. [DOI] [PubMed] [Google Scholar]
- Wang, Z.-Y., and Tobin, E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93, 1207–1217. [DOI] [PubMed] [Google Scholar]
- Wurgler-Murphy, S.M., and Saito, H. (1997). Two-component signal transducers and MAPK cascades. Trends Biol. Sci. 22, 172–176. [DOI] [PubMed] [Google Scholar]
- Wüthrich, K., Billetter, M., and Braun, W. (1983). Pseudo-structures for 20 common amino acids for use on studies of protein conformations by measurements of intramolecular proton-proton distance constraints with nuclear magnetic resonance. J. Mol. Biol. 169, 949–961. [DOI] [PubMed] [Google Scholar]
- Wykoff, D.D., Grossman, A.R., Weeks, D.P., Usuda, H., and Shimogawar, K. (1999). Psr1, a nuclear localized protein that regulates phosphorus metabolism in Chlamydomonas. Proc. Natl. Acad. Sci. USA 96, 15336–15341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav, S., and Ahmad, F. (2000). A new method for the determination of stability parameters of proteins from their heat-induced denaturation curves. Anal. Biochem. 283, 207–213. [DOI] [PubMed] [Google Scholar]
- Yamada, H., Suzuki, T., Terada, K., Takei, K., Ishikawa, K., Miwa, K., and Mizuno, T. (2001). The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol. 42, 1017–1023. [DOI] [PubMed] [Google Scholar]