Abstract
S-Adenosyl-l-methionine:phosphoethanolamine N-methyltransferase (PEAMT; EC 2.1.1.103) catalyzes the key step in choline (Cho) biosynthesis, the N-methylation of phosphoethanolamine. Cho is a vital precursor of the membrane phospholipid phosphatidylcholine, which accounts for 40 to 60% of lipids in nonplastid plant membranes. Certain plants use Cho to produce the osmoprotectant glycine betaine, which confers resistance to salinity, drought, and other stresses. An Arabidopsis mutant, t365, in which the PEAMT gene is silenced, was identified using a new sense/antisense RNA expression system. t365 mutant plants displayed multiple morphological phenotypes, including pale-green leaves, early senescence, and temperature-sensitive male sterility. Moreover, t365 mutant plants produced much less Cho and were hypersensitive to salinity. These results demonstrate that Cho biosynthesis not only plays an important role in plant growth and development but also contributes to tolerance to environmental stresses. The temperature-sensitive male sterility caused by PEAMT silencing may have a potential application in agriculture for engineering temperature-sensitive male sterility in important crop plants.
INTRODUCTION
Choline (Cho) is a key metabolite in plants because it is needed to synthesize the major membrane lipid phosphatidylcholine (PC), which accounts for 40 to 60% of lipids in nonplastid plant membranes (Moore, 1990; Bolognese and McGraw, 2000). Studies have shown that the synthesis of PC is affected by the plant growth regulator indole-3-acetic acid (Price-Jones and Harwood, 1983), suggesting that PC has a fundamental function in plant growth and development. Evidence also has indicated that freezing tolerance is correlated with changes in the amount and degree of polyunsaturation of PC (Sikorska and Kacperska-Palacz, 1980; Kinney et al., 1987). In addition, it has been shown that salinity stress induces an increase in the turnover of PC in Arabidopsis suspension-cultured cells (Pical et al., 1999), suggesting that PC also may play an important role in plant responses to salt and other environmental stresses.
In certain plants, such as spinach, Cho is oxidized to glycine betaine (GlyBet) by chloroplast enzymes (Rhodes and Hanson, 1993; Sakamoto and Murata, 2000). GlyBet is a strong osmoprotectant that confers tolerance to salinity, drought, and other stresses (Gorham, 1995). The enzymes that oxidize Cho to GlyBet also have been found in many bacteria (Rhodes and Hanson, 1993; Sakamoto and Murata, 2000). However, many plants, such as tobacco and Arabidopsis, do not have the enzymes for Cho oxidation (McNeil et al., 2000, 2001). To enhance their tolerance to environmental stresses, plant or bacterial genes for Cho oxidation have been used to engineer GlyBet synthesis in these species (Hayashi et al., 1997; Alia et al., 1998, 1999; Holmstrom et al., 2000; Sakamoto et al., 2000; Sakamoto and Murata, 2001).
Although the engineered plants have shown increased stress tolerance, the improvements in stress tolerance are relatively small because the levels of GlyBet obtained by this engineering strategy are low (Nuccio et al., 1999; Huang et al., 2000). It has been shown that low levels of GlyBet in the engineered plants are attributable to an inadequate capacity for Cho biosynthesis (Nuccio et al., 1998, 1999; Huang et al., 2000; McNeil et al., 2001). Therefore, researchers' attention has focused on the enzymes involved in the biosynthesis of Cho and the regulatory mechanism of Cho biosynthesis in plants.
Cho biosynthesis has been investigated in diverse plants. Evidence from these investigations indicated that Cho can be produced via three parallel, interconnected pathways involving sequential methylations of an ethanolamine moiety at the free base, phospho-base (P-base), and phosphatidyl-base (Ptd-base) levels (Rhodes and Hanson, 1993; McNeil et al., 2000). In plants, both the P-base and Ptd-base pathways exist in leaves, whereas the free base route has been found only in endosperm (Prud'homme and Moore, 1992a, 1992b). It has been demonstrated in tobacco that the first methylation in leaves occurs only at the P-base level (McNeil et al., 2000). The second and third methylations occur mainly at the P-base level (83 to 92% and 65 to 85%, respectively), with the remainder occurring at the Ptd-base level (McNeil et al., 2000). Evidence from leaves or cultured cells of species representing five families all are consistent with this result (Rhodes and Hanson, 1993; McNeil et al., 2000), although there are variations among these species.
The three methylation steps at the P-base level from phosphoethanolamine to phosphocholine all are catalyzed by the cytosolic enzyme S-adenosyl-l-methionine:phosphoethanolamine N-methyltransferase (PEAMT; EC 2.1.1.103), and the first methylation step exerts major control over flux through the entire pathway (Datko and Mudd, 1988a, 1988b; Nuccio et al., 2000; McNeil et al., 2001). The activity of PEAMT is feedback inhibited when phosphocholine or Cho is supplied in the medium (Mudd and Datko, 1989b, 1989c) and is induced in response to salt stress and light (Mudd and Datko, 1989a; Summers and Weretilnyk, 1993; Weretilnyk et al., 1995). Therefore, PEAMT is the key regulatory enzyme in Cho biosynthesis in plants.
Despite the importance of PEAMT as the regulatory enzyme in the biosynthesis of Cho moieties, it remains unclear how PEAMT affects plant growth and development and the plant response to environmental stresses. Considering the fundamental functions of Cho biosynthesis, a null mutation is likely lethal. No such mutants have been isolated to date.
In this study, we report the characterization of an Arabidopsis transgenic mutant, t365, which was identified using a new sense/antisense RNA expression (SARE) system developed to determine gene functions on a genome scale, and cloning of the gene responsible for the t365 mutant phenotype. The T365 gene encodes a PEAMT, which catalyzes all three methylation steps required to convert phosphoethanolamine to phosphocholine. Expression of the T365 transgene causes silencing of endogenous PEAMT and results in temperature-sensitive male sterility, decrease in Cho production, and salt hypersensitivity in Arabidopsis plants.
RESULTS
A SARE System
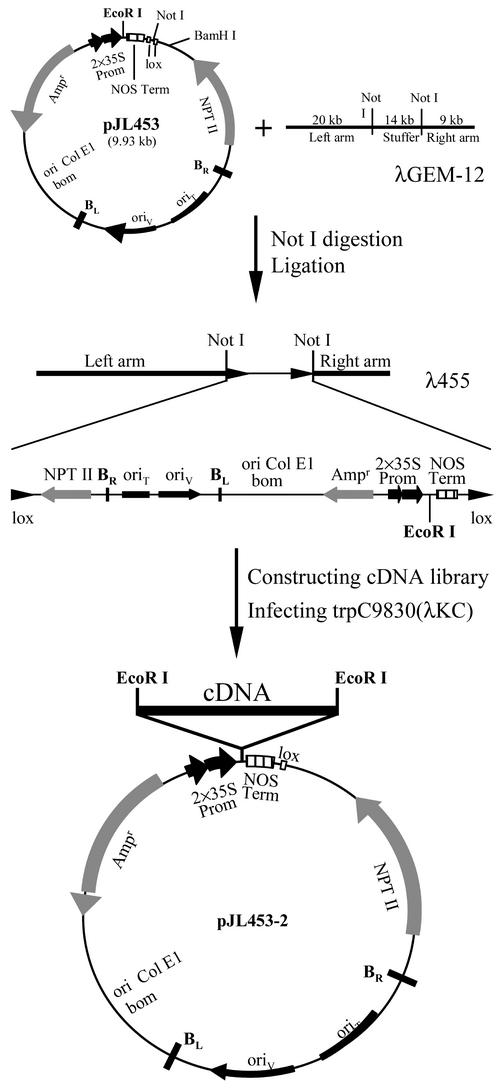
To introduce sense or antisense RNA into Arabidopsis plants, we first designed a plant expression vector, λ455, by replacing the central stuffer of λGEM-12 with the plasmid pJL453 (Figure 1). λ455 contains the colE1 replicon, a conditional mini-RK2 replicon, and the gene encoding β-lactamase, which allow replication and selection in Escherichia coli and Agrobacterium tumefaciens. The vector also carries the T-DNA border sequences for the transfer and integration of cDNA into a plant genome, the double 35S promoter of Cauliflower mosaic virus (2×35S) and the nopaline synthase (NOS) terminator for in planta expression of the cDNA, and the neomycin phosphotransferase gene for the selection of transgenic plants. The plasmid portion of λ455 is flanked by two lox direct repeats, which are used for site-specific recombination under catalysis of the cre protein (Sternberg et al., 1986; Elledge et al., 1991).
Figure 1.
The SARE System.
A plant expression vector, λ455, was constructed by replacing the central stuffer of λGEM-12 with the plasmid pJL453 (see Methods). A cDNA library was constructed in λ455 using the unique EcoRI site between the 2×35S promoter and the NOS terminator. The cDNA library was converted subsequently to a binary plasmid cDNA library using cre-lox site-specific recombination (Elledge et al., 1991). Sequences for replication and selection in E. coli and Agrobacterium are indicated. The T-DNA border sequences for the transfer and integration of cDNA into a plant genome also are indicated. BL, left border; BR, right border; NPT, neomycin phosphotransferase.
After infecting E. coli strain trpC9830(λKC) (Li et al., 1995) cells that produce cre protein, a precise excision occurred in λ455 between the lox sites, resulting in a binary plasmid. Through this recombination system, a cDNA library constructed in λ455 could be converted into a library of binary plasmids, which then could be transformed into plants by Agrobacterium. After transformation, the cDNA inserts introduced into transgenic plants were isolated by PCR using primers designed to anneal to the sequences of the 35S promoter and the NOS terminator. The genes that were disrupted by the T-DNA insertion could be identified through plasmid rescue or inverse PCR.
Transgenic Plants Generated by the SARE System
To generate a large number of transgenic lines containing different sense or antisense cDNA inserts, a cDNA library was constructed in λ455 using mRNA from Arabidopsis ecotype Columbia (Col-0). After infecting E. coli strain trpC9830(λKC) cells, a binary plasmid cDNA library was collected and transformed into Agrobacterium strain GV3101(pMP90RK) (Koncz and Schell, 1986). After transformation, the Agrobacterium cells were screened on Luria-Bertani agar plates supplemented with 50 μg/mL rifamycin, 20 μg/mL gentamicin, 50 μg/mL kanamycin, and 50 μg/mL carbenicillin.
For this experiment, ∼2 × 105 independent transformants were obtained. Transformants were allowed to grow on plates for 3 days and then were pooled and cultured for 2 h at 28°C. Arabidopsis Col-0 wild-type plants were transformed with these agrobacteria via vacuum infiltration (Bechtold et al., 1993). T2 seeds from the first 600 primary transgenic lines were used to evaluate the system. The phenotype of each line was observed under normal growth conditions.
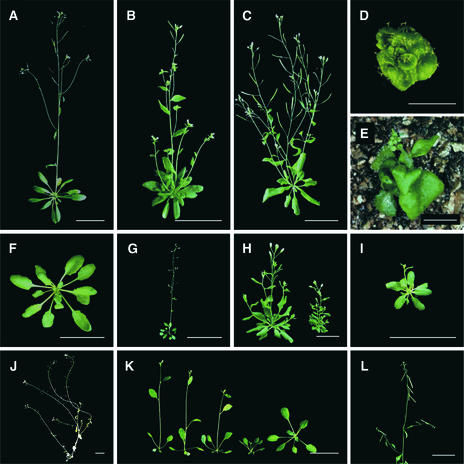
Among these 600 lines, 19 were found to exhibit apparent morphological abnormalities, including decreased plant size, altered leaf shape, bushy inflorescence, and reduced fertility (Figures 2A to 2J; Table 1, t189, t104, t77, t101, t113, t30, t307, t44, and t365). Mutants with early flowering time, short flower pedicels, and propendent siliques also were identified (Figures 2K and 2L; Table 1, t59 and t414). Genetic analyses of the T3 seeds from the 19 mutant lines showed that they were all capable of transmitting the mutant phenotypes to their progeny (data not shown), indicating that the phenotypes are heritable.
Figure 2.
Morphology of the Transgenic Mutants Generated by SARE.
Transgenic plants showing morphological abnormalities were grown under continuous light at 23°C and photographed at the number of days after germination indicated.
(A) Wild-type plant at day 40.
(B) Transgenic line t189 plant at day 35.
(C) Transgenic line t104 plant at day 40.
(D) Transgenic line t77 plant at day 40.
(E) Transgenic line t101 plant at day 40.
(F) Transgenic line t113 plant at day 25.
(G) Transgenic line t30 plant at day 40.
(H) Transgenic line t307 plant at day 40 (left, heterozygous; right, homozygous).
(I) Transgenic line t44 plant at day35.
(J) Transgenic line t365 plant at day 45.
(K) Transgenic line t59 plant at day 25.
(L) Transgenic line t414 plant at day 40.
Bars in (A) to (C) and (F) to (L) = 2 cm; bars in (D) and (E) = 0.5 cm.
Table 1.
Phenotypes of the Transgenic Mutants Generated by SARE
| Mutant | Phenotypes |
|---|---|
| t25 | Small plant, curly leaves, single short inflorescence stem, small siliques |
| t30 | Small dark-green round leaves with smooth edges |
| t38 | Small plant, small epinasty leaves |
| t44 | Small slightly dark-green leaves, short petioles, smooth leaf edges |
| t59 | Early flowering |
| t62 | Small plant |
| t67 | Small plant, curly and slightly glassy leaves, decreased fertility |
| t69 | Laterally curling leaves, limp inflorescence stem |
| t77 | Tiny plant, dark-green and thickened leaves, late flowering and senescence |
| t97 | Small plant |
| t101 | Small plant, dark-green leaves, male sterility |
| t104 | Curly leaves, numerous axillary inflorescences and lateral branches |
| t113 | Glabrous and light-green plant |
| t130 | Small plant, curly leaves |
| t189 | Multiple rosettes, dark-green plant, increased number of trichomes, short petioles |
| t247 | Small plant, dark-green leaves, single inflorescence stem |
| t307 | Small rosette, elongated internodes, numerous axillary inflorescences |
| t365 | Pale-green plant, early senescence, male sterility |
| t414 | Short flower pedicels, propendent siliques |
Morphology of t365 Mutant Plants
To validate the SARE system in identifying gene functions, one of the transgenic mutant lines, t365, was chosen for in-depth analysis. When grown under continuous light at 22°C, t365 rosette leaves were pale green in juvenile stages (Figures 3A and 3B) and senesced early in the late reproductive stage (Figures 3C and 3D). Compared with the wild type (Figure 3E), the t356 mutant plants had shorter siliques and produced fewer seeds (Figure 3F). Microscopic examination of wild-type and t365 flowers revealed that t365 mutant plants produced less pollen than the wild type (Figures 3G and 3H), suggesting that the reduced fertility may be caused by abnormal male fertility in t365 mutant plants.
Figure 3.

Morphology of t365 Mutant Plants.
Plants were grown under continuous light at 22°C and photographed at the number of days after germination indicated.
(A) and (B) Wild-type and t365 mutant plants at day 35, showing pale-green plants of t365 (B).
(C) and (D) Wild-type and t365 mutant plant rosette leaves at day 40, showing early senescence of t365 mutant plants (D).
(E) and (F) Wild-type and t365 mutant inflorescences, showing poor development of t365 siliques (F).
(G) and (H) Magnification of flowers, showing decreased fertility of t365 mutant plants (H).
Bars in (A) to (F) = 2 cm; bars in (G) and (H) = 0.25 mm.
To test this possibility, we conducted cross-pollination experiments. Cross-pollination of t365 mutant plants with wild-type pollen resulted in normal seed set. In reciprocal crosses, using pollen from t365 mutant plants to pollinate wild-type stigmas, fewer seeds were produced (data not shown). Thus, the reduced seed set observed in t365 mutant plants was caused by a decrease in male fertility.
Genetic Analysis of the t365 Mutant
The abnormal morphology of t365 was observed in the T1 generation, suggesting that the t365 mutant phenotype is inherited as a dominant trait. To confirm this hypothesis, t365 T2 transgenic plants were grown on Murashige and Skoog (1962) (MS) medium plates containing kanamycin to test for antibiotic resistance and were grown in soil to examine the segregation of the morphological phenotype. The segregation of 112 kanamycin-resistant to 36 kanamycin-sensitive plants fits the expectation for a single-locus insertion of the transgene (χ2 = 0.036, P > 0.80). However, the segregation of 70 t356-looking to 47 wild-type-looking plants did not fit any simple Mendelian ratio.
To determine which plants harbored the T-DNA insert, PCR was used to amplify the DNA samples from the individual T2 plants. A PCR product was detected in all of the 70 t356-looking plants and in 18 of the 47 wild-type-looking plants. This result was confirmed by DNA gel blot analysis using the 35S promoter DNA as a probe (data not shown). The segregation ratio of 88 to 29 fits the expectation for a single-locus insertion of the transgene (χ2 = 0.0028, P > 0.90). Seeds of the 18 wild-type-looking plants with the T-DNA insertion and the 29 wild-type-looking plants without the T-DNA insertion were sown on soil. Those plants with the T-DNA insertion segregated for the wild-type and mutant phenotypes, similar to T2 transgenic plants. However, all of the progeny of those plants without the T-DNA insertion displayed a wild-type phenotype. These results suggest that the phenotype of t365 probably is caused by expression of the cDNA insert.
Cloning of the T365 Gene
The cDNA fragment integrated into the t365 genome was isolated by PCR. DNA sequencing indicated that the cDNA fragment was 745 bp and inserted in a sense orientation. Using this cDNA fragment as a probe, we screened an Arabidopsis cDNA library (Col-0). Among the 21 cDNA clones obtained, the longest cDNA was 1457 bp with a 22-bp poly(A) tail. To determine whether this cDNA was full length, we performed rapid amplification of cDNA ends–PCR to identify the 5′ sequence of T365 cDNA. The longest cDNA identified was 1904 bp, in which an in-frame TGA stop codon was found 66 nucleotides upstream of the first ATG, suggesting that we had identified the full-length cDNA.
T365 Encodes a PEAMT in Arabidopsis
Comparison between the cDNA and the genomic DNA sequences indicated that the T365 gene contains 12 exons and encodes a polypeptide of 492 amino acid residues with a calculated molecular mass of 56.1 kD (data not shown). A BLASTN (Altschul et al., 1997) search in the GenBank database revealed an identical Arabidopsis cDNA that encodes PEAMT (Bolognese and McGraw, 2000) and a highly homologous gene encoding PEAMT in spinach (Nuccio et al., 2000). Alignment of the predicted T365 amino acid sequence with the spinach PEAMT protein revealed that they share 87% amino acid identity (data not shown), confirming that T365 encodes a PEAMT in Arabidopsis (Bolognese and McGraw, 2000).
The t365 Mutant Phenotype Results from Silencing of the Endogenous PEAMT
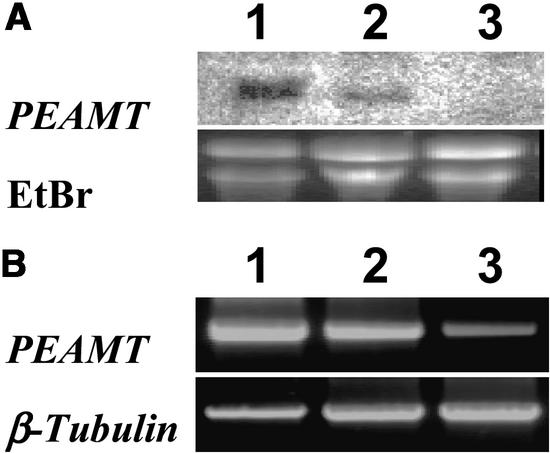
One explanation for the mutant phenotype of t365 is that the endogenous PEAMT gene is suppressed as a result of the expression of the T365 transgene. To investigate this possibility, the steady state level of the PEAMT transcripts was examined by RNA gel blot analysis using the 1457-bp cDNA as a probe (Figure 4A). The abundance of the endogenous PEAMT transcripts in t365 wild-type-looking plants (lane 2) was much lower than that in wild-type plants (lane 1), whereas the transcripts were not detected in t365 mutant plants (lane 3).
Figure 4.
Analysis of PEAMT Transcript Accumulation in t365 Transgenic Plants.
(A) RNA gel blot analysis of total RNA prepared from wild-type Col-0 plants (lane 1) and t365 transgenic plants displaying wild-type phenotypes (lane 2) and mutant phenotypes (lane 3). EtBr, ethidium bromide.
(B) Reverse transcriptase–PCR analysis of PEAMT expression in wild-type Col-0 plants (lane 1) and t365 transgenic plants displaying wild-type phenotypes (lane 2) and mutant phenotypes (lane 3).
To determine further if there were PEAMT transcripts in t365 mutant plants, we used reverse transcriptase–PCR to detect the transcripts. Although the amount was significantly lower than that in wild-type plants (Figure 4B, lane 1) and t365 wild-type-looking plants (Figure 4B, lane 2), the transcripts did exist in t365 mutant plants (Figure 4B, lane 3). Therefore, it is likely that the amount of the PEAMT transcripts in the wild-type-looking plants was sufficient to support normal plant growth and development, whereas the residual level of PEAMT transcripts in t365 mutant plants only supported plant survival.
To confirm that the t365 mutant phenotype was caused by suppression of the endogenous PEAMT by the T365 transgene, the 745-bp cDNA fragment isolated from t365 transgenic plants was cloned into vector pJL453-2 (Figure 1) in both the sense and antisense orientations and retransformed into Arabidopsis. Of 13 and 11 independent sense and antisense transgenic lines, 6 and 4 lines showed morphology similar to that of t365, as shown in Figure 5A. The same effect on the endogenous PEAMT transcripts was observed in these transgenic lines as in t365 (Figure 5B).
Figure 5.
Transgenic Mutant Plants Obtained by Retransformation of the 745-bp T365 Transgene.
(A) Morphology of transgenic plants expressing the 745-bp T365 fragment in both the sense and antisense orientations.
(B) Analysis of PEAMT transcript accumulation in transgenic plants. RNA gel blot analysis of total RNA prepared from wild-type Col-0 plants (lane 2), transgenic plants containing the T365 transgene in the sense orientation that display wild-type phenotypes (lane 3), and transgenic plants containing the T365 transgene in the sense (lane 4) and antisense (lane 1) orientations that display similar phenotypes to the t365 mutant plants. EtBr, ethidium bromide.
Besides the original t365 mutant, independent antisense and sense-suppressed lines were used for further temperature-sensitive male sterility and saline sensitivity studies (see below). All of the phenotypes of the original t365 mutant also were observed in the independent antisense and sense-suppressed lines. These results demonstrate that the t365 mutant phenotypes are caused by silencing of the endogenous PEAMT rather than by disruption of a gene function caused by the T-DNA insertion.
Silencing of PEAMT Leads to a Decrease in Cho Content in t365 Mutant Plants
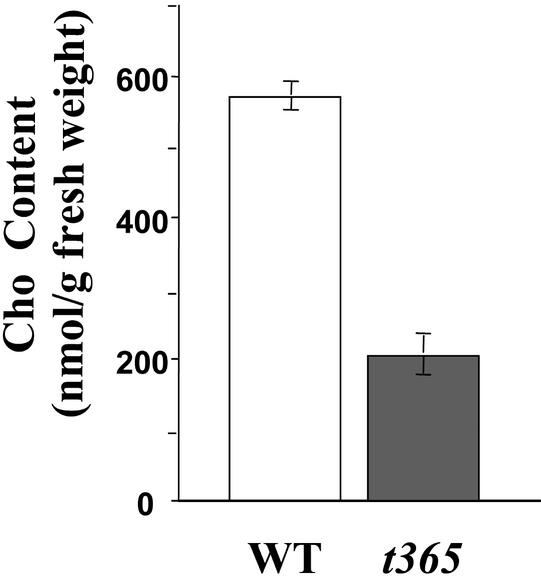
Because PEAMT is the key enzyme in the plant Cho synthesis pathway, we measured the Cho contents in wild-type and t365 mutant plants. As shown in Figure 6, Cho content of t365 mutant plants was ∼64% lower than that of the wild type, indicating that silencing of PEAMT in the t365 mutant did affect Cho biosynthesis, which in turn caused the t365 mutant phenotypes.
Figure 6.
Comparison of Cho Contents between t365 and Wild-Type Plants.
Cho content was determined in 0.5 g of fresh rosette leaves from 4-week-old plants. The Cho contents are shown as means ± se (n = 3). WT, wild type.
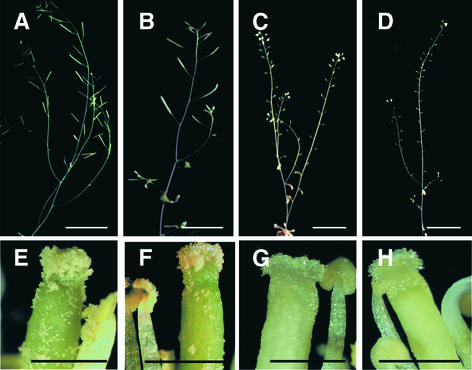
t365 Mutant Plants Show Temperature-Sensitive Male Sterility
In plants, PEAMT catalyzes the committing methylation step, the methylation of phosphoethanolamine to phosphomonomethylethanolamine, and the subsequent two methylation steps at the P-base level for de novo PC biosynthesis (Nuccio et al., 2000; McNeil et al., 2001). Deficiency in the two terminal phospholipid N-methyltransferase activities required for de novo biosynthesis of PC in Saccharomyces cerevisiae opi3 mutant strains leads to a temperature-sensitive growth phenotype (McGraw and Henry, 1989).
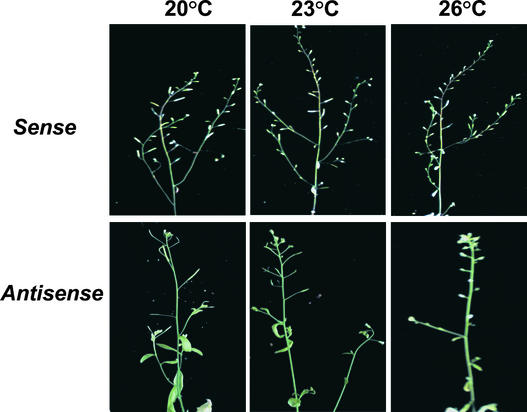
To investigate the influence of temperature on the phenotype of t365 mutant and independent sense/antisense transgenic plants, we grew them under continuous light at 20, 23, and 26°C, respectively. Although temperature had no effect on the pale-green color of t365 leaves, it had a dramatic effect on the fertility of t365 mutant plants. When grown at 20°C, t365 mutant plants produced a lot of seeds (Figure 7B), although fewer than wild-type plants (Figure 7A). By contrast, t365 mutant plants grown at both 23 and 26°C showed markedly decreased fertility. No seeds were produced in the abnormal siliques (Figures 7C and 7D). Compared with wild-type plants (Figure 7E), t365 mutant plants grown at temperatures of 23°C or greater had diminished pollen production (Figures 7G and 7H), which confirmed that the decreased fertility under these growth conditions was caused by male sterility. The similar temperature-sensitive male sterility phenotypes of sense/antisense transgenic plants also were observed, as shown in Figure 8.
Figure 7.
Effect of Temperature on the Male Fertility of t365 Mutant Plants.
(A) and (E) Wild-type plant and flower grown at 23°C. Wild-type plants grown at 20 or 26°C also display normal fertility.
(B) and (F) t365 mutant plant and flower grown at 20°C.
(C) and (G) t365 mutant plant and flower grown at 23°C.
(D) and (H) t365 mutant plant and flower grown at 26°C.
Bars in (A) to (D) = 2 cm; bars in (E) to (H) = 0.25 mm.
Figure 8.
Temperature Effect on the Male Fertility of the Sense and Antisense Transgenic Plants.
t365 Mutant Plants Are Hypersensitive to Salt Stress
After conversion from phosphoethanolamine through the three methylation steps, phosphocholine is either incorporated into PC or metabolized to Cho (Summers and Weretilnyk, 1993; McNeil et al., 2000). Some plants such as spinach have chloroplast enzymes that catalyze the two-step oxidation of Cho to GlyBet, which has strong osmoprotectant properties and confers tolerance to salinity and other stresses (Rhodes and Hanson, 1993; Gorham, 1995; Sakamoto and Murata, 2000). Arabidopsis lacks the genes for Cho oxidation (McNeil et al., 2000, 2001). However, evidence has shown that changes in the amount of PC are correlated with salt stress (Kinney et al., 1987; Pical et al., 1999), suggesting that PC may be involved in the response to high salinity in plant cells.
To determine if the silencing of PEAMT in t365 mutant plants has an effect on their response to salinity, we performed a series of salt stress experiments with different concentrations of NaCl. Figure 9A shows the phenotype of soil-grown t365 mutant plants after treatment with 200 mM NaCl. Compared with wild-type plants (Figure 9A, left), t365 mutant plants showed a hypersensitivity phenotype (Figure 9A, right). The sensitivity also was tested on plates containing 100 mM NaCl, as described previously (Liu and Zhu, 1998). On regular MS nutrient medium, t365 mutant seedlings were indistinguishable from wild-type seedlings (Figure 9C). However, under high salt stress, the growth of t365 mutant plants (Figure 9B, right) was inhibited to a greater extent than that of wild-type plants (Figure 9B, left).
Figure 9.
Morphology of Wild-Type and t365 Mutant Plants Exposed to High-Salt Stress.
(A) Two-week-old soil-grown wild-type and t365 mutant plants treated with 200 mM NaCl, showing salt hypersensitivity of the t365 mutant plants (at right). The photograph was taken 10 days after treatment.
(B) to (D) Salt sensitivity determination on plates. Seedlings were grown on half-strength MS agar plates for 3 days and then transferred to vertical half-strength MS agar plates, half-strength MS agar plates supplemented with 100 mM NaCl, and half-strength MS agar plates supplemented with 100 mM NaCl plus 5 mM Cho. The photographs were taken 10 days after transfer. The growth of the t365 mutant (at right in [B]) was inhibited to a greater extent than that of the wild type (at left in [B]), whereas the two plants were indistinguishable when grown on regular half-strength MS agar medium (C). The salt inhibition to t365 mutant plants was greatly ameliorated by adding Cho to the medium (at right in [D]).
To confirm that the salt hypersensitivity was caused by the decrease in Cho content as a result of the silencing of PEAMT in t365 mutant plants, Cho was supplied to the medium containing 100 mM NaCl. The salt inhibition to t365 mutant plants was ameliorated significantly (Figure 9D, right). These results demonstrate that the silencing of PEAMT in t365 mutant plants results in hypersensitivity to high salinity.
DISCUSSION
Here, we have described a system based on SARE in transgenic plants for the identification of gene functions on a genome scale. Using this system, we identified 19 transgenic mutants with apparent abnormal morphological phenotypes from the first 600 transgenic lines. We systematically characterized one of these mutants, t365, and cloned the gene responsible for the t365 mutant phenotypes. The T365 gene encodes a PEAMT that catalyzes the three methylation steps required to convert phosphoethanolamine to phosphocholine in de novo Cho or PC biosynthesis. Expression of a T365 transgene in t365 mutant plants causes silencing of the endogenous PEAMT, which leads to abnormal morphological phenotypes, temperature-sensitive male sterility, reduction in Cho biosynthesis, and salt hypersensitivity.
In plants, PC is a dominant constituent of membrane phospholipids and is necessary for a wide array of structural and biochemical functions (Moore, 1990; Bolognese and McGraw, 2000). Silencing of PEAMT in t365 mutant plants leads to a temperature-sensitive phenotype, which is consistent with the data for yeast temperature-sensitive opi3 mutant strains. The opi3 mutations cause a deficiency in the two terminal phospholipid N-methyltransferase activities required for the de novo synthesis of PC.
Under certain growth conditions, opi3 mutants produce membranes virtually devoid of PC (McGraw and Henry, 1989). The mutant loses its viability at the stationary phase of the cell cycle in the absence of dimethylethanolamine or Cho and is temperature sensitive for growth at 37°C, especially in media containing monomethylethanolamine (McGraw and Henry, 1989). These growth defects are correlated with the presence of specific phospholipids in the membrane. Therefore, the temperature-sensitive phenotype observed in t365 mutant plants may result from a lack of PC, the presence of specific phospholipids in membranes, or both. Further investigation is needed to clarify this issue.
Temperature mainly affects the fertility of t365 mutant plants. When grown at low temperatures, such as 20°C, t365 mutant plants are fertile and can produce some seeds. However, at temperatures greater than 23°C, t365 mutant plants showed dramatically decreased fertility, and no seeds were produced in the abnormal siliques. Cross-pollination experiments indicated that the sterility observed in t365 mutant plants is attributable to failure to produce functional pollen. The ability to induce male sterility in t365 mutant plants by growth at high temperatures suggests that silencing of PEAMT may provide an efficient biotechnological approach to engineer temperature-sensitive male sterility in agriculturally important plants.
Evidence has shown that salinity and hyperosmotic stress induce dramatic increases in the levels of phosphatidylinositol 4,5-bisphosphate and diacylglycerol pyrophosphate and also affect the turnover of PC (Sikorska and Kacperska-Palacz, 1980; Pical et al., 1999), suggesting that PC may play an important role in salt stress response. The hypersensitivity to salinity of t365 mutant plants, which results from the silencing of PEAMT, is consistent with this suggestion. However, it remains unclear whether the hypersensitivity to salinity of t365 mutant plants is caused by decreased salt tolerance, reduced plant vigor, or both. Although the t365 mutants display relatively normal root growth in the absence of salt stress, they do show abnormal growth phenotypes such as pale-green leaves and decreased fertility. Thus, PEAMT is more likely a gene essential for both normal growth and salt tolerance.
In plants, it has been shown that PEAMT is the key enzyme of Cho biosynthesis. Overexpression of PEAMT in transgenic tobacco increases the levels of phosphocholine by 5-fold and free Cho by 50-fold without affecting PC content or plant growth (McNeil et al., 2001). The silencing of PEAMT in t365 mutant plants causes a reduction of ∼64% in Cho content, which leads to temperature-sensitive male sterility and salt hypersensitivity, further supporting the hypothesis that PEAMT controls the metabolic flux to Cho in plants.
Plants have evolved various protective mechanisms to acclimate themselves to unfavorable environments for continued survival and growth. Cho biosynthesis is one such mechanism. However, to date, the precise function of the Cho biosynthesis pathway in stress tolerance has not been well studied as a result of the lack of mutants of the Cho biosynthesis enzymes. In t365 mutant plants, expression of one of the Cho biosynthesis enzymes, PEAMT, is suppressed, allowing us to determine the roles of the Cho biosynthesis pathway in plant stress tolerance.
Successful isolation of the t365 mutant and cloning of the T365 gene demonstrated the applicability of the SARE system. This system has several advantages. First, the cre-lox site-specific recombination system used to convert the λ cDNA library to the plasmid library was simple and efficient (Elledge et al., 1991). Second, a 2×35S promoter was used to drive a strong sense or antisense expression of the cDNA inserts in plants. Third, the cDNA fragments introduced into transgenic plants can be recovered easily by PCR using primers designed to anneal to the 35S promoter and NOS terminator sequences. The cDNA fragments of interest and the sense/antisense orientations can be determined easily by sequencing of the PCR products. Finally, the colE1 origin of replication and the β-lactamase gene are included within the T-DNA; therefore, the genomic DNA fragment flanking the T-DNA insertion can be isolated by plasmid rescue. This is useful for cloning of genes disrupted by the T-DNA insertion.
The SARE system is based on the expression of sense or antisense RNA in transgenic plants, which has been demonstrated to suppress the expression of an endogenous gene or genes with sequence homology (van der Krol et al., 1988; Elmayan et al., 1998; Furner et al., 1998; Vaucheret et al., 1998). This type of gene suppression may affect every member of a gene family and is especially useful in studying the function of multiple-gene families (Jorgensen, 1990; Li et al., 1995). In addition, sense or antisense suppression usually is observed in a limited number (2 to 50%) of primary transformants, and the level of suppression varies among transformants (Hofgen et al., 1994; van Blokland et al., 1994). This variance may provide a possible means to examine the function of essential genes, because total obliteration of the gene function would lead to a dominant lethal phenotype (Martienssen, 1998). Thus, the SARE system is especially useful in identifying essential genes whose disruption will lead to lethality and multiple-copy genes whose functional redundancy makes it difficult to detect the mutant phenotype in well-established genetic model species such as Arabidopsis and rice. The SARE system also may provide a feasible genome-scaled technology for the study of gene function in polyploid species such as rapeseed and wheat. Therefore, the SARE system can complement conventional and insertional mutagenesis to provide a comprehensive strategy for determining plant gene functions.
METHODS
Plant Growth
Arabidopsis thaliana plants were grown on vermiculite saturated with 0.3 × B5 medium under continuous illumination (80 to 120 μE·m−2·s−1) at 23°C as described previously (Mou et al., 2000) or under the conditions specified. For temperature treatment, plants were germinated and grown in three Versatile Environmental Test Chambers (Sanyo, Tokyo, Japan) under continuous illumination (100 μE·m−2·s−1) at 20, 23, and 26°C, respectively. For salt stress treatment, 2-week-old soil-grown plants were watered with 50, 100, 200, and 400 mM NaCl. The phenotype was examined 10 days after NaCl treatment. To determine salt sensitivity on plates, seedlings were grown on Murashige and Skoog (1962) nutrient (GIBCO) agar plates for 3 days and then transferred to vertical Murashige and Skoog (1962) agar plates supplemented with 100 mM NaCl. The phenotype was examined 10 days after transfer to 100 mM NaCl.
Construction of λ455
The plasmids used to construct pJL453 included pSE936 (Elledge et al., 1991), pBluescript II KS+/− (Stratagene), pBI101 (Clontech, Palo Alto, CA), pUC118 (Boehringer Mannheim), and pSS (Becker, 1990). The two lox sites in the BglII (blunt ended)-SalI (blunt ended) fragment from pSE936 were introduced into the EcoRV site of pBluescript II KS+/− to form pJL420. The nopaline synthase terminator was removed from pBI101 by digestion with EcoRI plus SacI and cloned into the EcoRI-SacI sites of pJL420 to yield pJL422. The EcoRI site in pJL422 was destroyed by treatment with the Klenow fragment of DNA polymerase I (Promega) to make pJL423. The SacI-SalI (blunt ended) fragment of this plasmid was inserted between the SacI and SmaI sites of pUC118 to generate pJL452. The BamHI-EcoRI fragment of pJL452 was used to replace the BamHI-EcoRI fragment of pJL429, which was derived from pSS by the destruction of the NotI site using the DNA polymerase I Klenow fragment, to make pJL453. pJL453 was finally linearized with NotI and ligated into gel-purified NotI-cleaved λGEM-12 (Promega) arms to generate the plant cDNA expression vector λ455.
Construction of an Arabidopsis cDNA Library in λ455
Arabidopsis mRNA was isolated from the aerial parts of 2- to 6-week-old Columbia (Col-0) wild-type plants using a QuickPrep Micro mRNA Purification Kit (Pharmacia), and the cDNA was synthesized using the TimeSaver cDNA Synthesis Kit (Pharmacia). The first strand of cDNA was primed with random hexamers provided with the kit to produce cDNAs without poly(A) sequence. The EcoRI/NotI adaptor was ligated to each end of the cDNA, and the adapted cDNA was purified on the spin column as described in the protocol provided. λ455 DNA was prepared using the Wizard Lamda Preps DNA Purification System (Promega). A total of 4 μg of λ455 DNA was digested completely in TA buffer (Epicentre) with 30 units of EcoRI in a reaction volume of 50 μL and then dephosphorylated by adding 4 units of HK (heat-killable) phosphatase (Epicentre) and incubating at 30°C for 1 h. The HK phosphatase was heat inactivated at 65°C for 15 min. The λ455 vector DNA then was precipitated using 5 μL of 3 M sodium acetate and 110 μL of cold (−20°C) ethanol, incubated at −20°C for 30 min, pelleted at 16,900g for 15 min, and washed once with 1 mL of 70% (v/v) ethanol. The pellet was resuspended in 8 μL of TE buffer (10 mM Tris-HCl, pH 8.0; 1 mM EDTA). Fifteen microliters of cDNA column eluate was ligated to 2 μg of EcoRI-digested, phosphatase-treated λ455 vector DNA in a volume of 25 μL at 15°C overnight and packaged using the Ready-To-Go Lamda Packaging Reaction System (Pharmacia).
Automatic Conversion of the Phage cDNA Library into a Binary Plasmid Library and Generation of Transgenic Plants
The λ phage competent cells (Escherichia coli strain XL1-Blue) were prepared as described (Sambrook et al., 1989). The cDNA library λ phages were incubated with 1 mL of a freshly prepared culture of E. coli strain trpC9830(λKC) (Li et al., 1995) cells in 10 mM MgCl2 and 1 mL of Luria-Bertani (LB) medium at 37°C for 30 min without shaking. Cells then were plated on 150-mm plates with ampicillin at 50 μg/mL and with kanamycin at 50 μg/mL and incubated overnight at 37°C. Ampicillin- and kanamycin-resistant cells were scraped from the plates, resuspended in 250 mL of LB medium with ampicillin and kanamycin, and incubated with shaking at 37°C for 1 h.
The plasmid DNA was extracted and purified with polyethylene glycol as described (Sambrook et al., 1989). This DNA was introduced into Agrobacterium tumefaciens strain GV3101(pMP90RK) (Koncz and Schell, 1986) by electroporation, and transformants were screened on LB agar plates supplemented with 50 μg/mL rifamycin, 20 μg/mL gentamicin, 50 μg/mL kanamycin, and 50 μg/mL carbenicillin. Transformants were allowed to grow on plates for 3 days and then were pooled and cultured for 2 h at 28°C.
Arabidopsis Col-0 plants were transformed with these agrobacteria via vacuum infiltration (Bechtold et al., 1993). Plants prepared for transformation were grown on vermiculite saturated with 0.3 × B5 medium under continuous illumination (80 to 120 μE·m−2·s−1) at 23°C. Transformed plants (T0) were allowed to self, and T1 seeds were harvested. Transgenic plants were selected by screening T1 seeds with 50 μg/mL kanamycin. The kanamycin-resistant plants then were transferred to soil, and the T2 seeds from each T1 plant were harvested individually.
Mutant Screening and Genetic Analysis
T2 transgenic plants grown in soil under normal growth conditions were examined for morphological abnormalities, including plant size, hypocotyl length, leaf shape, trichomes, floral structure, flowering time, and fertility. Fine structures were observed with a stereomicroscope (model SZX-12; Olympus, Tokyo, Japan). For each mutant line, segregation of mutant phenotype and kanamycin resistance was performed in the T2 and T3 generations, and T-DNA copy number was determined by DNA gel blot analysis using 35S promoter DNA as a probe. T4 seeds from the homozygous T-DNA plants with mutant phenotypes were used for further analysis
Isolation of the T365 Gene
The cDNA fragment integrated into t365 transgenic plants was isolated by PCR with primers 35S-F2 (5′-ACCACGTCTTCAAAGCAAGTG-3′) and NOS-R2 (5′-TATGATAATCATCGCAAGACCG-3′) under the following conditions: 94°C for 3 min, 45 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 1 min, and a final extension at 72°C for 10 min. The PCR products were cloned into pBluescript II SK+/− (Stratagene) and sequenced with the DYEnamic Direct dGTP Sequencing Kit (Amersham) in a 373A DNA sequencer (Applied Biosystems, Foster City, CA). An Arabidopsis cDNA library was screened using the t365 insert cDNA as a probe.
The 5′ sequence of the T365 cDNA was obtained by rapid amplification of cDNA ends–PCR (RACE-PCR) with a 5′-RACE Kit (Boehringer Mannheim). The three specific primers used in the RACE reactions were SP1 (5′-GTTTGTGAGCACTGGTGGAC-3′), SP2 (5′-TGGCCAAAGACACGCTCATAG-3′), and SP3 (5′-GACATAAGC-TCCAATGCACTTG-3′). The PCR products were cloned into pBluescript II SK+/− and sequenced as described above.
Confirmation That the T365 Transgene Leads to the t365 Mutant Phenotype
To determine whether the 745-bp T365 transgene cosegregated with the t365 mutant phenotypes, DNA samples from 76 t365 transgenic mutant plants were amplified with the T-DNA vector primer 35S-F2 and the T365 cDNA fragment–specific primer T365-R (5′-GATGAGAACTTTACCTCCCG-3′). The PCR products were separated on an agarose gel. Meanwhile, the 745-bp EcoRI cDNA fragment (isolated from t365 transgenic plant DNA) was cloned into EcoRI-digested, calf intestinal alkaline phosphatase (Gibco BRL)–treated pJL453-2 in both the sense and antisense orientations to make pJL710 and pJL711. The plasmids were introduced into Agrobacterium strain GV3101(pMP90RK) by electroporation, and the agrobacteria were used to transform Col-0 wild-type plants via vacuum infiltration (Bechtold et al., 1993). The phenotypes of the transgenic plants were scored in T1 transformants and T2 progeny.
RNA Gel Blot and Reverse Transcriptase–PCR Analyses
Total RNA from 4-week-old wild-type and phenotypically normal or male-sterile t365 transgenic plants was prepared by a guanidine thiocyanate extraction method (Hu et al., 2000). RNA (15 μg/lane) was separated on an agarose gel containing 10% formaldehyde and blotted onto a Hybond-N Plus membrane (Amersham). RNA gel blot analysis was performed as described previously (Hu et al., 2000) using PEAMT cDNA as the probe.
For reverse transcriptase–PCR analysis, 2 μg of total RNAs was used in a 20-μL reverse transcription reaction as described (Li et al., 1998). Two microliters of reaction products was used in a 50-μL PCR with the S-adenosyl-l-methionine:phosphoethanolamine N-methyltransferase (PEAMT) gene-specific primers T365RT-F (5′-ATGTGTGCTGATGTTACATCC-3′) and T365RT-R (5′-TGCATAAACTGATCA-GTACGG-3′) under the following conditions: 94°C for 3 min, 40 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 1.5 min, and a final extension at 72°C for 10 min. As an internal control, primers for the Arabidopsis β-tubulin gene (5′-CGTGGATCACAGCAATACAGAGCC-3′ and 5′-CCTCCTGCACTTCCACTTCGTCTTC-3′) were included in the PCR as described (Li et al., 1998).
Determination of Choline
Fresh tissue (0.5 g) from rosette leaves of 4-week-old plants was pulverized in liquid N2 for choline (Cho) determination. Cho was extracted and assayed according to the method described by Nuccio et al. (1998). Cho oxidase, horseradish peroxidase, and aminoantipyrine were purchased from Sigma.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Accession Numbers
The GenBank accession numbers for the sequences mentioned in this article are AB019230 (genomic DNA), AF197940 (a cDNA that encodes PEAMT in Arabidopsis), and AF237633 (a highly homologous gene encoding PEAMT in spinach).
Acknowledgments
We thank Xinnian Dong (Duke University) and Wendy E. Durrant (Duke University) for comments on the manuscript. The gift of Agrobacterium strain GV3101(pMP90RK) from Csaba Koncz and Jeff Schell (Max-Plank-Institut für Zuchtungsforschung, Koln, Germany) is much appreciated. This research was supported by grants from the State Transgenic Plant Program and the State Hi-Tech Program (863), by the National Natural Science Foundation of China Grant 39889003, and by a National Distinguished Young Scholar Award to J.L.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.001701.
References
- Alia, Hayashi, H., Sakamoto, A., and Murata, N. (1998). Enhancement of the tolerance of Arabidopsis to high temperatures by genetic engineering of the synthesis of glycinebetaine. Plant J. 16, 155–161. [DOI] [PubMed] [Google Scholar]
- Alia, Kondo, Y., Sakamoto, A., Nonaka, H., Hayashi, H., Saradhi, P.P., Chen, T.H., and Murata, N. (1999). Enhanced tolerance to light stress of transgenic Arabidopsis plants that express the codA gene for a bacterial choline oxidase. Plant Mol. Biol. 40, 279–288. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of Arabidopsis thaliana plants. C. R. Acad. Sci. Paris 316, 1194–1199. [Google Scholar]
- Becker, I. (1990). Construction of Chimeric Phosphoenolpyruvate Carboxylase Gene and Its Expression in Potato. PhD dissertation (Aachen, Germany: Rheinisch Westfälische Technische Hochschule).
- Bolognese, C.P., and McGraw, P. (2000). The isolation and characterization in yeast of a gene for Arabidopsis S-adenosylmethionine:phospho-ethanolamine N-methyltransferase. Plant Physiol. 124, 1800–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datko, A.H., and Mudd, S.H. (1988. a). Enzymes of phosphatidyl-choline synthesis in Lemna, soybean and carrot. Plant Physiol. 88, 1338–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datko, A.H., and Mudd, S.H. (1988. b). Phosphatidylcholine synthesis. Plant Physiol. 88, 854–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge, S.J., Mulligan, J.T., Ramer, S.W., Spottswood, M., and Davis, R.W. (1991). Lambda YES: A multifunctional cDNA expression vector for the isolation of genes by complementation of yeast and Escherichia coli mutations. Proc. Natl. Acad. Sci. USA 88, 1731–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmayan, T., Balzergue, S., Beon, F., Bourdon, V., Daubremet, J., Guenet, Y., Mourrain, P., Palauqui, J.C., Vernhettes, S., Vialle, T., Wostrikoff, K., and Vaucheret, H. (1998). Arabidopsis mutants impaired in cosuppression. Plant Cell 10, 1747–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furner, I.J., Sheikh, M.A., and Collett, C.E. (1998). Gene silencing and homology-dependent gene silencing in Arabidopsis: Genetic modifiers and DNA methylation. Genetics 149, 651–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorham, J. (1995). Betaines in higher plants: Biosynthesis and role in stress metabolism. In Amino Acids and Their Derivatives in Higher Plants, R.M. Wallsgrove, ed (Cambridge, UK: Cambridge University Press), pp. 171–203.
- Hayashi, H., Alia, Mustardy, L., Deshnium, P., Ida, M., and Murata, N. (1997). Transformation of Arabidopsis thaliana with the codA gene for choline oxidase: Accumulation of glycinebetaine and enhanced tolerance to salt and cold stress. Plant J. 12, 133–142. [DOI] [PubMed] [Google Scholar]
- Hofgen, R., Axelsen, K.B., Kannangara, C.G., Schuttke, I., Pohlenz, H.D., Willmitzer, L., Grimm, B., and von Wettstein, D. (1994). A visible marker for antisense mRNA expression in plants: Inhibition of chlorophyll synthesis with a glutamate-1-semialdehyde aminotransferase antisense gene. Proc. Natl. Acad. Sci. USA 91, 1726–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmstrom, K.O., Somersalo, S., Mandal, A., Palva, T.E., and Welin, B. (2000). Improved tolerance to salinity and low temperature in transgenic tobacco producing glycine betaine. J. Exp. Bot. 51, 177–185. [DOI] [PubMed] [Google Scholar]
- Hu, Y., Bao, F., and Li, J. (2000). The promotive effect of brassinolide on cell division involves a distinct cycD3-induction pathway in Arabidopsis. Plant J. 24, 693–710. [DOI] [PubMed] [Google Scholar]
- Huang, J., Hirji, R., Adam, L., Rozwadowski, K.L., Hammerlindl, J.K., Keller, W.A., and Selvaraj, G. (2000). Genetic engineering of glycinebetaine production toward enhancing stress tolerance in plants: Metabolic limitations. Plant Physiol. 122, 747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, R. (1990). Altered gene expression in plants due to trans interactions between homologous genes. Trends Biotechnol. 8, 340–344. [DOI] [PubMed] [Google Scholar]
- Kinney, A.J., Clarkson, D.T., and Loughman, B.C. (1987). The regulation of phosphatidylcholine biosynthesis in rye (Secale cereale) roots: Stimulation of the nucleotide pathway by low temperature. Biochem. J. 242, 755–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz, C., and Schell, J. (1986). The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204, 383–396. [Google Scholar]
- Li, H., Wu, G., Ware, D., Davis, K.R., and Yang, Z. (1998). Arabidopsis Rho-related GTPases: Differential gene expression in pollen and polar localization in fission yeast. Plant Physiol. 118, 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Zhao, J., Rose, A.B., Schmidt, R., and Last, R.L. (1995). Arabidopsis phosphoribosylanthranilate isomerase: Molecular genetic analysis of triplicate tryptophan pathway genes. Plant Cell 7, 447–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., and Zhu, J.K. (1998). A calcium sensor homolog required for plant salt tolerance. Science 280, 1943–1945. [DOI] [PubMed] [Google Scholar]
- Martienssen, R.A. (1998). Functional genomics: Probing plant gene function and expression with transposons. Proc. Natl. Acad. Sci. USA 95, 2021–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw, P., and Henry, S.A. (1989). Mutations in the Saccharomyces cerevisiae opi3 gene: Effects on phospholipid methylation, growth and cross-pathway regulation of inositol synthesis. Genetics 122, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil, S.D., Nuccio, M.L., Rhodes, D., Shachar-Hill, Y., and Hanson, A.D. (2000). Radiotracer and computer modeling evidence that phospho-base methylation is the main route of choline synthesis in tobacco. Plant Physiol. 123, 371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil, S.D., Nuccio, M.L., Ziemak, M.J., and Hanson, A.D. (2001). Enhanced synthesis of choline and glycine betaine in transgenic tobacco plants that overexpress phosphoethanolamine N-methyltransferase. Proc. Natl. Acad. Sci. USA 98, 10001–10005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, T.S. (1990). Biosynthesis of phosphatidylinositol. Inositol Metab. Plant 9, 107–112. [Google Scholar]
- Mou, Z., He, Y., Dai, Y., Liu, X., and Li, J. (2000). Deficiency in fatty acid synthase leads to premature cell death and dramatic alterations in plant morphology. Plant Cell 12, 405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd, S.H., and Datko, A.H. (1989. a). Synthesis of EA and its regulation in Lemna paucicostata. Plant Physiol. 91, 587–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd, S.H., and Datko, A.H. (1989. b). Synthesis of methylated EA moieties: Regulation by Cho in Lemna. Plant Physiol. 90, 296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd, S.H., and Datko, A.H. (1989. c). Synthesis of methylated EA moieties: Regulation by Cho in soybean and carrot. Plant Physiol. 90, 306–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Nuccio, M.L., Rhodes, D., McNeil, S.D., and Hanson, A.D. (1999). Metabolic engineering of plants for osmotic stress resistance. Curr. Opin. Plant Biol. 2, 128–134. [DOI] [PubMed] [Google Scholar]
- Nuccio, M.L., Russell, B.L., Nolte, K.D., Rathinasabapathi, B., Gage, D.A., and Hanson, A.D. (1998). The endogenous choline supply limits glycine betaine synthesis in transgenic tobacco expressing choline monooxygenase. Plant J. 16, 487–496. [DOI] [PubMed] [Google Scholar]
- Nuccio, M.L., Ziemak, M.J., Henry, S.A., Weretilnyk, E.A., and Hanson, A.D. (2000). cDNA cloning of phosphoethanolamine N-methyltransferase from spinach by complementation in Schizosaccharomyces pombe and characterization of the recombinant enzyme. J. Biol. Chem. 275, 14095–14101. [DOI] [PubMed] [Google Scholar]
- Pical, C., Westergren, T., Dove, S.K., Larsson, C., and Sommarin, M. (1999). Salinity and hyperosmotic stress induce rapid increases in phosphatidylinositol 4,5-bisphosphate, diacylglycerol pyrophosphate, and phosphatidylcholine in Arabidopsis thaliana cells. J. Biol. Chem. 274, 38232–38240. [DOI] [PubMed] [Google Scholar]
- Price-Jones, M.J., and Harwood, J.L. (1983). Hormonal regulation of phosphatidylcholine synthesis in plants: The inhibition of cytidylyltransferase activity by indol-3-ylacetic acid. Biochem. J. 216, 627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud'homme, M.-P., and Moore, T.S. (1992. a). Phosphatidylcholine synthesis in castor bean endosperm: Free bases as intermediates. Plant Physiol. 100, 1527–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud'homme, M.-P., and Moore, T.S. (1992. b). Phosphatidylcholine synthesis in castor bean endosperm: The occurrence of an S-adenosyl-L-methionine:ethanolamine N-methyltransferase. Plant Physiol. 100, 1536–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes, D., and Hanson, A.D. (1993). Quaternary ammonium and tertiary sulfonium compounds in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 44, 357–384. [Google Scholar]
- Sakamoto, A., and Murata, N. (2000). Genetic engineering of glycinebetaine synthesis in plants: Current status and implications for enhancement of stress tolerance. J. Exp. Bot. 51, 81–88. [PubMed] [Google Scholar]
- Sakamoto, A., and Murata, N. (2001). The use of bacterial choline oxidase, a glycinebetaine-synthesizing enzyme, to create stress-resistant transgenic plants. Plant Physiol. 125, 180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, A., Valverde, R., Alia, Chen, T.H., and Murata, N. (2000). Transformation of Arabidopsis with the codA gene for choline oxidase enhances freezing tolerance of plants. Plant J. 22, 449–453. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sikorska, E., and Kacperska-Palacz, A. (1980). Frost-induced phospholipid changes in cold-acclimated and nonacclimated rape leaves. Physiol. Plant. 48, 201–206. [Google Scholar]
- Sternberg, N., Sauer, B., Hoess, R., and Abremski, K. (1986). Bacteriophage P1 cre gene and its regulatory region: Evidence for multiple promoters and for regulation by DNA methylation. J. Mol. Biol. 187, 197–212. [DOI] [PubMed] [Google Scholar]
- Summers, P., and Weretilnyk, E.A. (1993). Choline synthesis in spinach in relation to salt stress. Plant Physiol. 103, 1269–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Blokland, R., van der Geest, N., Mol, J.N.M., and Kooter, J.M. (1994). Transgene-mediated suppression of chalcone synthase expression in Petunia hybrida results from an increase in RNA turnover. Plant J. 6, 861–877. [Google Scholar]
- van der Krol, A.R., Mol, J.N., and Stuitje, A.R. (1988). Antisense genes in plants: An overview. Gene 72, 45–50. [DOI] [PubMed] [Google Scholar]
- Vaucheret, H., Beclin, C., Elmayan, T., Feuerbach, F., Godon, C., Morel, J.B., Mourrain, P., Palauqui, J.C., and Vernhettes, S. (1998). Transgene-induced gene silencing in plants. Plant J. 16, 651–659. [DOI] [PubMed] [Google Scholar]
- Weretilnyk, E.A., Smith, D.D., Wilch, G.A., and Summers, P.S. (1995). Enzymes of Cho synthesis in spinach: Response of P-base N-methyltransferase activities to light and salinity. Plant Physiol. 109, 1085–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]