Abstract
Survivin, a member of the inhibitor of apoptosis protein (IAP) family, is detected in most common human cancers but not in adjacent normal cells. Previous studies suggest that survivin associates with the mitotic spindle and directly inhibits caspase activity. To further investigate the function of survivin, we used a survivin antisense (AS) oligonucleotide to downregulate survivin expression in normal and cancer cells. We found that inhibition of survivin expression increased apoptosis and polyploidy while decreasing colony formation in soft agar. Immunohistochemistry showed that cells without survivin can initiate the cleavage furrow and contractile ring, but cannot complete cytokinesis, thus resulting in multinucleated cells. These findings indicate that survivin plays important roles in a late stage of cytokinesis, as well as in apoptosis.
Keywords: survivin, apoptosis, antisense, oligonucleotides, cytokinesis
Introduction
Inhibitors of apoptosis proteins (IAPs) were originally identified as baculovirus genes that complement loss of the caspase inhibitor, p35 [1]. All IAPs contain at least one copy of a 70 amino acid motif called the baculovirus IAP repeat (BIR) domain [2]. BIR-containing proteins have been identified in yeast, Caenorhabditis elegans, Drosophila, mice, and humans, representing a structurally and functionally diverse family [3–9]. Whereas yeast and C. elegans BIR-containing proteins (bir1 and BIR-1, respectively) are involved in cell division [3,4], other IAPs (DIAP, XIAP, c-IAP1 and cIAP2) regulate apoptosis by directly binding and inhibiting certain caspase-family cell death proteases [10–12].
Survivin, a human IAP, has only one BIR domain and a coiled-coil domain [8,13]. Based on sequence comparisons, survivin has been placed in a subfamily along with yeast and C. elegans BIR-family proteins, and with the human BRUCE protein (Apollon), which apparently do not regulate apoptosis [3,4]. Survivin associates with the mitotic spindle and can partially complement the cytokinesis defect caused by BIR-1 deficiency in C. elegans, suggesting that survivin may play a role in cytokinesis [4,13]. However, survivin can suppress apoptosis induced by various apoptotic stimuli, indicating survivin is a typical IAP [8,14]. Consistent with the possibility of a role in cell cycle, survivin reportedly is expressed primarily in the G2/M phase of the cell cycle. Survivin mRNA is undetectable in G1 phase and increases by 40-fold in G2/M phase in HeLa cells [13]. Survivin expression has been detected in most lung, breast, colorectal, prostate and pancreas tumor samples and transformed cell lines examined [8,14–17]. In embryonic and fetal tissue, survivin expression is prominent and developmentally regulated [18]. In normal adult tissues, however, survivin expression was shown to be limited to the thymus [8]. In patients with cancer, elevated expression of survivin in tumors is associated with poor prognosis and increased cancer recurrence [16,17]. Because it remains unclear how survivin contributes to tumor progression, we sought to investigate the consequences of downregulating survivin in normal and cancer cells using antisense oligonucleotides.
Materials and Methods
Cell Lines
Tumor cell lines (HCT15, HeLa, H460, HCT116) and immortalized human mammary gland cells (MCF-10F) were obtained from American Type Culture Collection (Rockville, MD). Normal human umbilical vascular endothelial cells (HUVEC) and Normal human fibroblasts (NHF) were obtained from Clonetics (Walkersville, MD). Cells were cultured according to the recommendations of the suppliers.
Antisense Oligonucleotide Design and Transfection
Forty 2′-O-methoxyethyl chimeric phosphorothioate oligonucleotides designed to hybridize to human survivin mRNA were synthesized and screened for inhibition of survivin gene expression using a quantitative TaqMan RT-PCR assay [19]. The most potent one was further modified by replacing the phosphorothioate linkages with natural phosphodiester linkages in the portion of the molecule containing 2′-O-methoxyethyl modifications resulting in ISIS 28599 [20]. The sequence of the survivin antisense (AS) oligonucleotide (oligo) and the mismatch control (MS) oligonucleotide are as follows: survivin AS, 5′-TGTGC-TATTCTGTGAATT-3′; MS oligonucleotide, 5′-TAAGC-TGTTCTATGTGTT-3′. The underlined portion corresponds to 2′-O-methoxyethyl modified nucleotides. Subconfluent cells were transfected with AS or MS oligos using Lipofectin with a mixture of Lipofectin reagent (Gibco, MD) and oligos in Opti-MEM media (Gibco) at a ratio of 3 µl Lipofectin/ml media per 100 nM oligo or 9 µl Lipofectin/ml media per 300 nM oligo. After culturing at 37°C for 4 hours, cells were incubated with normal complete media.
RNA Preparation and Quantitative PCR Analysis
Survivin mRNA was quantitated with TaqMan® Real Time QPCR (Quantitative Polymerase Chain Reaction) methodology using the ABI Prism 7700 (Perkin-Elmer Applied Biosystems) as described previously [21]. Human survivin sequences used: forward primer 515F, 5′-GCACCACTTCCAGGGTTTATTC-3′; reverse primer 590R, 5′-TCTCCTTTCCTAAGACATTGCTAAGG-3′; TaqMan® probe 539T, 5′-[FAM] TGGTGCCACCAGCCTTCCTGTG [TAM-RA].
Antibodies
High-titer antisera specific for survivin were generated in New Zealand white rabbits using a multiple boosting technique and recombinant GST-survivin as immunogen, as described [14,22]. Antibody reactivity exclusively with survivin was confirmed by immunoblot analysis of in vitro translated survivin versus other IAP/BIR-family members (cIAPI, cIAP2, NAIP, XIAP, BRUCE) and other various control proteins. Analysis of lysates from 293T cells transfected with Myc-survivin versus Myc-tagged cIAPI, cIAP2, NAIP, XIAP or BRUCE (BIR domain region) as well as other control proteins, revealed the antibody reacted solely with the expected ∼15-kDa survivin protein. Specificity of antisera was further determined by comparisons of immune and pre-immune serum and by pre-adsorption of anti-survivin antisera with excess immunogen (GST-survivin) at 5 µg per 50 µl antiserum.
Immunoblot Assay
Cells (1 x 105) were collected for both control and transfected cells 48 hours after transfections. Samples were lysed in Laemmli sample buffer (Sigma, MO) and protein extracts were separated by 15% SDS-PAGE and transferred to nitrocellulose membranes (Schleicher and Schuell, NH), which were blocked with 10% nonfat dry milk in Tris-buffered saline (TBS, Sigma) and incubated with rabbit anti-survivin IgG in TBS with 0.05% Tween-20 (TBST). Membranes were washed with TBST and incubated with goat anti-rabbit IgG conjugated to horse radish peroxidase. After washing, survivin protein was detected using an ECL Plus method (Amersham, NJ).
Flow Cytometric Analysis
Cells were prepared for flow cytometry by pooling attached and floating cells recovered from cultures and pelleting by centrifugation at 800g for 5 minutes. The cells were resuspended in 0.5 ml ice-cold staining solution (5 µg/ml propidium iodide (PI), 40 U/ml RNase, 0.5% Triton X-100, in PBS). After 1 hour at 4°C in the dark the DNA content was analyzed using a Becton Dickinson ExCalibur Flow Cytometer (San Jose, CA).
Immunostaining and Confocal Analysis
Immunolocalization of survivin was accomplished by confocal microscopy. Subconfluent transfected cells were fixed with 10% phosphate-buffered formalin on eight-chamber slides (Nalge Nunc, Naperville, IL). After washing with PBS, cells were incubated with rabbit anti-survivin IgG diluted 1:1000 in PBS for 1 hour at 37°C. The cells were washed three times with PBS, and incubated with Alexa 546 conjugated goat anti-rabbit IgG (Molecular Probes, diluted 1:1000) for 30 minutes at 37°C. Cells were further washed three times with PBS. Microtubules were stained using an FITC conjugated antitubulin IgG (Sigma) diluted 1:75 in PBS for 1 hour at 37°C. Cells were further washed three times with PBS. Microfilaments were stained using Bodipy-FL phallacidin (Molecular Probes, Eugene, OR) as previously described [23]. The DNA was counterstained with Hoechst (15 µg/ml) for 10 seconds. Cells were mounted with Citifluor and imaged with a BioRad MRC-1000 confocal attached to an inverted Nikon microscope fitted with epi-fluorescence optics and a 60x (n.a.=1.4) objective.
Colony Formation Assay
Cells (104) were plated in complete culture medium containing 0.3% agarose on top of 0.6% agarose in the same medium. After 16 days, 0.5 mg/ml of p-iodonitrotetrazo violet (Sigma) were added to stain colonies for 24 hours. Colonies were photographed using a Sony 3CCD color video camera and quantified using the image analysis program Image-Pro plus (Media Cybernetics, Silver Spring, MD).
Results
Antisense Oligonucleotides Inhibit Survivin Expression in Tumor and Normal Cells
To examine whether an antisense approach could be used to inhibit survivin expression, normal (HUVEC, MCF-10F, NHF) and tumor (HeLa, HCT116, HCT15, H460) cells were transfected with an AS survivin oligo or an MS control oligo. Because the transfection efficiency and survivin expression levels in each cell line are different, a variety of concentrations of oligo for each cell line were tested to optimize downregulation of survivin protein levels (data not shown). The optimal concentration for each cell line was determined to be 100 nM for HUVEC and 300 nM for all other cell lines.
The effect of AS oligo on survivin mRNA and protein levels in representative tumor and normal cell lines (HeLa and HUVEC, respectively) are shown in Figure 1. At 24 hours posttransfection, the levels of survivin mRNA in the 300 nM AS oligo-transfected HeLa and 100 nM AS oligo-transfected HUVEC cells, respectively, were reduced by ≈90% compared with the cells treated with the MS oligo (Figure 1a). At 48 hours posttransfection, survivin protein levels in HUVEC cells transfected with 100 nM AS were decreased ≈90% compared with MS controls, whereas survivin protein levels in HeLa cells transfected with 100 and 300 nM AS were reduced ≈40% and ≈90%, respectively (Figure 1b).
Figure 1.
Reduction in survivin expression in antisense oligo-transfected HeLa and HUVEC cells, (a) Twenty-four hours after MS or AS oligo-transfection of HeLa (300 nM) and HUVEC cells (100 nM), total mRNA was extracted from both transfected and nontransfected control cells. The expression level of survivin mRNA was quantified using QPCR measurement. (b) The expression of survivin protein was evaluated by immunoblot assay using total protein extracted from 1 x 105 cells 48 hours after transfection. Data below each band represents their relative densitometric measurement.
Inhibition of Survivin Expression Leads to Increased Apoptosis and Cytokinesis Defects
Previously, an expression plasmid producing AS survivin transcripts was shown to induce apoptosis in HeLa cells [24]. We examined cellular DNA content by flow cytometry to determine whether a reduction in survivin expression obtained using AS oligo would also induce apoptosis. We observed an increase in hypoploidy (DNA<2N) consistent with apoptosis as well as polyploidy (DNA>4N) consistent with failed cytokinesis in both HeLa and HUVEC cells 48 hours following transfection with AS oligo, but not in the cells treated with the MS control oligo (Figure 2a). Similar observations were made in three other tumor cell lines (HCT116, HCT15, H460), normal immortalized cells (MCF-10F), and in normal human fibroblasts (NHF) (Figure 2, b and c).
Figure 2.
Flow cytometric analysis of DNA content in various cell lines 48 hours following transfection with various concentrations of AS or MS oligo. (a) Histograms of MS or AS oligo-transfected HeLa (300 nM) and HUVEC cells (100 nM). (b) Percent apoptotic cells in AS oligo-transfected cells divided by percent apoptosis cells in MS oligo-transfected cells, (c) Percent polyploidy cells in AS oligo-transfected cells divided by percent apoptosis cells in MS oligo-transfected cells. Data represent mean±SD (n=3; apop=apoptotic cells).
To further evaluate the apparent cytokinesis blockade induced by survivin AS oligo, cells were stained for microtubules using a fluorescein conjugated IgG specific for α-tubulin and DNA using Hoechst dye following oligo transfection. Large, multinucleated cells (white arrows), as well as apoptotic cells (red arrows) were observed in both HUVEC (Figure 3B) and HeLa cells (Figure 3D) at 48 hours following AS oligo transfection. In contrast, similar multinucleated cells (white arrows) were not seen in the MS controls (Figure 3, A and C). Interestingly, we observed many multinucleated cells in AS-transfected cells apparently undergoing apoptosis evidenced by DNA condensation using Hoechst stain (red arrows). We also observed that multinucleated cells in metaphase AS oligo-transfected cells had normal, albeit in some cases, multiple mitotic spindle apparatus (Figure 3, F and H) unlike cells in the MS oligo-transfected cells which had a single mitotic apparatus (Figure 3, E and G). This suggests that survivin is probably not required during the formation of the mitotic spindle apparatus.
Figure 3.
Immunohistochemistry in HeLa and HUVEC cells. Microtubules were stained with an FITC conjugated antibody (green) and DNA was stained with Hoechst dye (blue). White arrows indicate multinucleated cells; red arrows indicate apoptotic cells (A–H). (A) MS oligo-transfected HUVEC cells. (B) AS oligo-transfected HUVEC cells. (C) MS oligo-transfected HeLa cells. (D) AS oligo-transfected HeLa cells. (E) A metaphase MS oligo-transfected HUVEC cell. (F) A metaphase AS oligo-transfected Huvec cell. (G) A metaphase MS oligo-transfected HeLa cell. (H) A metaphase AS oligo-transfected HeLa cell. Localization of survlvin, microtubules, microfilaments and DNA in late telophase MS and AS oligo-transfected HeLa cells were determined by triple labeled for survivin (red), DNA (blue, Hoechst) and either microfilaments (a and c, green) or microtubules (e and g green). (a) MS oligo-transfected cell. (b) Same as (a), but survlvin stain only. (c) AS oligo-transfected cell. (d) Same as (c), but survivin stain only. (e) MS oligo-transfected cell. (f) Same as (e), but survivin stain only. (g) AS oligo-transfected cell. (h) Same as (g), but survivin stain only. Scale bars = 15 µm. First scale bar (A–D), second scale bar (E–H), third scale bar (a–h).
Survivin is Required for a Late Stage of Cytokinesis
Yeast BIR deletion mutants are unable to undergo the metaphase to anaphase transition because of failure to extend their mitotic spindle [3]. However, C. elegans embryos lacking bir-1 can initiate the cleavage furrow but they cannot complete the closure to separate the two daughter cells [4]. To further investigate how survivin affects cytokinesis in mammalian cells, we examined the localization of survivin in MS and AS oligo-transfected cells throughout the cell cycle by immunohistochemistry. Two rabbit anti-survivin IgG antibodies from two different sources were used, giving the same results.
We observed that survivin was located at the midbody during telophase in cells transfected with the MS oligo (Figure 3, a, b, e and f). In contrast, survivin was not found in the midbody in AS oligo-transfected cells (Figure 3, c, d, g and h). This is consistent with the effective downregulation of survivin protein by AS oligo in these cells (Figure 1). Staining of actin microfilaments (Figure 3, a and c) and microtubules (Figure 3, e and g) were performed to further investigate the progress of cytokinesis in MS and AS oligo-transfected cells. We observed that both survivin-expressing and survivin-deficient cells could elongate their mitotic spindle (Figure 3, e and g), form a cleavage furrow and a contractile ring (Figure 3, a and c). However, survivin-deficient cells failed to complete cytokinesis, resulting in multinucleated cells (white arrows), whereas survivin-containing cells divided as usual. Subsequently, the cleavage furrow regressed in AS-treated cells lacking survivin resulting in multinucleated cells (Figure 3, g and h).
Inhibition of Survivin Expression Reduces Anchorage-Independent Human Tumor Cell Growth
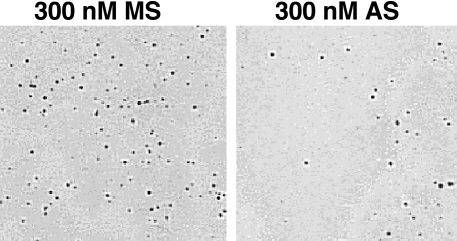
Because downregulation of survivin leads to cytokinesis defects and increased apoptosis, we explored further the consequences of AS-mediated reduction in survivin levels on tumor cell growth. Soft-agar colony formation of HeLa cells was used to assess anchorage-independent cell growth following AS oligo transfection. Anchorage-independent growth of HeLa cells was inhibited by 51±3% following transfection with 300 nM AS oligo, compared with 300 nM MS oligo (Figure 4). In addition to the reduction in colony number we also observed that colony size was smaller following AS oligo treatment.
Figure 4.
Down-regulation of survivin reduces HeLa anchorage-independent cell growth. P-iodonitrotetrazo violet staining of MS and AS oligo-transfected HeLa cell colonies on soft agar. There was 51 ±3% reduction in colony number in AS oligo-transfected cells as compared with MS oligo-transfected cells.
Discussion
Most IAPs had been shown to function as apoptosis regulators by directly binding and inhibiting caspases [10–12]. Previous studies suggest that survivin has the ability to block apoptosis induced by a variety of death signals such as Bax, etoposide and Fas [14]. In this study, we show that a survivin AS oligo can downregulate survivin expression, causing an increase in apoptosis. This results therefore lend further support to the notion that survivin is a suppressor of apoptosis.
Structurally, yeast and C. elegans BIRs and human survivin are members of the same subfamily of BIR/IAP family proteins [3]. However, yeast and C. elegans BIR proteins do not regulate apoptosis, instead they control cytokinesis [3,4]. Here we show that reduced expression of survivin also leads to accumulation of multinucleated cells, suggesting that survivin also plays an important role in cell division in mammalian cells. This is consistent with the results that survivin can partially complement the defects caused by BIR-1 deficiency in C. elegans. However, it is unclear how survivin affects cytokinesis. It has been reported that survivin expressed in bacteria interacts with polymerized tubulin through its C-terminus in vitro [13]. Yeast BIR protein deletion mutants were unable to undergo the metaphase to anaphase transition because of an inability to extend their mitotic spindle [3], whereas C. elegans embryos lacking bir-1 can initiate cleavage furrow but fail to complete cytokinesis [4]. With triple staining (DNA, survivin, microtubules/actin microfilament), we observed that survivin is normally located predominantly at the midbody during telophase. Moreover, AS oligo-transfected cells with reduced survivin elongated their mitotic spindle, formed a cleavage furrow and a contractile ring, but were unable to complete cytokinesis. Therefore, the survivin-deficient phenotype produced using AS oligo is similar to that seen in C. elegans embryos in which bir-1 expression is inhibited using RNA interference (RNAi) [4]. These observations suggest that functionally, human survivin resembles more closely the C. elegans bir-1 protein than the yeast BIR proteins, serving an important function during a late stage of cytokinesis.
Taken together, these and previously published data indicate that survivin has dual functions: a) cytokinesis and b) apoptosis regulation. Our results clearly show that the cytokinesis effect of survivin is not limited to cancer cells. Because cytokinesis is universal to all dividing cells, it is not surprising that we detected survivin in normal dividing cells in vitro. The observation that multinucleated cells resulting from inhibition of survivin expression eventually undergo apoptosis suggests that survivin may also link apoptosis to genetic stability. Reduction of colony formation in soft agar suggests that targeting survivin may have anti-tumor efficacy, however, it should be cautioned that our results are based on in vitro cultured cell lines. Further investigations of survivin AS oligos in vivo are warranted to evaluate survivin as a therapeutic target for cancer treatment.
Acknowledgements
We thank Dario Altieri for providing rabbit anti-survivin antibody, Regina Reilly, Ed Han, Xuesong Liu, Haichao Zhang, Robert Simmer, Don Halbert and Perry Nisen for helpful discussions.
Abbreviations
- IAP
inhibitors of apoptosis proteins
- BIR
baculovirus IAP repeat
- AS
antisense
- MS
mismatch control
- Oligo
oligonucleotides
Footnotes
The work in Reed's lab was supported partially by NIH grant AG-15402.
These authors contributed equally to the study.
References
- 1.Crook NE, Clem RJ, Miller LK. An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J Virol. 1993;67:2168–2174. doi: 10.1128/jvi.67.4.2168-2174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bimbaum MJ, Clem RJ, Miller LK. An apoptosis-inhibiting gene from a nuclear polyhedrosis virus encoding a polypeptide with Cys/His sequence motifs. J Virol. 1994;68:2521–2528. doi: 10.1128/jvi.68.4.2521-2528.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uren AG, Beilharz T, O'Connell MJ, Bugg SJ, van Driel R, Vaux DL, Lithgow T. Role for yeast inhibitor of apoptosis (IAP)-like proteins in cell division. Proc Natl Acad Sci USA. 1999;96:10170–10175. doi: 10.1073/pnas.96.18.10170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fraser AG, James C, Evan GI, Hengartner MO. Caenorhabditis elegans inhibitor of apoptosis protein (IAP) homologue BIR-1 plays a conserved role in cytokinesis. Curr Biol. 1999;9:292–301. doi: 10.1016/s0960-9822(99)80137-7. [DOI] [PubMed] [Google Scholar]
- 5.Hay BA, Wassarman DA, Rubin GM. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell. 1995;83:1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- 6.Hauser HP, Bardroff M, Pyrowolakis G, Jentsch S. A giant ubiquitin-conjugating enzyme related to IAP apoptosis inhibitors. J Cell Biol. 1998;141:1415–1422. doi: 10.1083/jcb.141.6.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobayashi K, Hatano M, Otaki M, Ogasawara T, Tokuhisa T. Expression of a murine homologue of the inhibitor of apoptosis protein is related to cell proliferation. Proc Natl Acad Sci USA. 1999;96:1457–1462. doi: 10.1073/pnas.96.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nature Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 9.Chen Z, Naito M, Hori S, Mashima T, Yamori T, Tsuruo T. A human IAP-family gene, Apollon, expressed in human brain cancer cells. Biochem Biophys Res Commun. 1999;264:847–854. doi: 10.1006/bbrc.1999.1585. [DOI] [PubMed] [Google Scholar]
- 10.Roy N, Deveraux QL, Takahashi R, Salvesen GS, Reed JC. The c-IAP-1 and c-IAP-2 proteins are direct inhibitors of specific caspases. EMBO J. 1997;16:6914–6925. doi: 10.1093/emboj/16.23.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deveraux QL, Takahashi R, Salvesen GS, Reed JC. X-linked IAP is a direct inhibitor of cell-death proteases. Nature. 1997;17:300–304. doi: 10.1038/40901. [DOI] [PubMed] [Google Scholar]
- 12.Deveraux QL, Leo E, Stennicke HR, Welsh K, Salvesen GS, Reed JC. Cleavage of human inhibitor of apoptosis protein XIAP results in fragments with distinct specificities for caspases. EMBO J. 1999;18:5242–5251. doi: 10.1093/emboj/18.19.5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li F, Ambrosini G, Chu EY, Plescia J, Tognin S, Marchisio PC, Altieri DC. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396:580–584. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- 14.Tamm I, Wang Y, Sausville E, Scudiero DA, Vigna N, Oltersdorf T, Reed JC. IAP-family protein survivin inhibits caspase activity and apoptosis induced by Fas (CD95), Bax, and anticancer drugs. Cancer Res. 1998;58:5315–5320. [PubMed] [Google Scholar]
- 15.Lu CD, Altieri DC, Tanigawa N. Expression of a novel antiapoptosis gene, survivin, correlated with tumor cell apoptosis and p53 accumulation in gastric carcinomas. Cancer Res. 1998;58:1808–1812. [PubMed] [Google Scholar]
- 16.Kawasaki H, Altieri DC, Lu CD, Toyoda M, Tenjo T, Tanigawa N. Inhibition of apoptosis by survivin predicts shorter survival rates in colorectal cancer. Cancer Res. 1998;58:5071–5074. [PubMed] [Google Scholar]
- 17.Adida C, Berrebi D, Peuchmaur M, Reyes-Mugica M, Altieri DC. Anti-apoptosis gene, survivin, and prognosis of neuroblastoma. Lancet. 1998;351:882–883. doi: 10.1016/S0140-6736(05)70294-4. [DOI] [PubMed] [Google Scholar]
- 18.Adida C, Crotty PL, McGrath J, Berrebi D, Diebold J, Altieri DC. Developmentally regulated expression of the novel cancer anti-apoptosis gene survivin in human and mouse differentiation. Am J Pathol. 1998;152:43–49. [PMC free article] [PubMed] [Google Scholar]
- 19.McKay RA, Miraglia LJ, Cummins LL, Owens SR, Sasmor H, Dean NM. Characterization of a potent and specific class of antisense oligonucleotide inhibitor of human protein kinase C-alpha expression. J Biol Chem. 1999;274:1715–1722. doi: 10.1074/jbc.274.3.1715. [DOI] [PubMed] [Google Scholar]
- 20.Baker BF, Lot SS, Condon TP, Cheng-Flournoy S, Lesnik EA, Sasmor HM, Bennett CF. 2′-O-(2-methoxy)ethyl-modified anti-intercellular adhesion molecule 1 (ICAM-1) oligonucleotides selectively increase the ICAM-1 mRNA level and inhibit formation of the ICAM-1 translation initiation complex in human umbilical vein endothelial cells. J Biol Chem. 1997;272:11994–2000. doi: 10.1074/jbc.272.18.11994. [DOI] [PubMed] [Google Scholar]
- 21.Heid C, Junko S, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 22.Krajewski S, Krajewska M, Shabaik A, Wang HG, Irie S, Fong L, Reed JC. Immunohistochemical analysis of in vivo patterns of Bcl-X expression. Cancer Res. 1994;54:5501–5507. [PubMed] [Google Scholar]
- 23.Tahir SK, Trogadis JE, Stevens JK, Zimmerman AM. Cytoskeletal organization following cannabinoid treatment in undifferentiated and differentiated PC12 cells. Biochem Cell Biol. 1992;70:1159–1173. doi: 10.1139/o92-162. [DOI] [PubMed] [Google Scholar]
- 24.Ambrosini G, Adida C, Sirugo G, Altieri DC. Induction of apoptosis and inhibition of cell proliferation by survivin gene targeting. J Biol Chem. 1998;273:11177–11182. doi: 10.1074/jbc.273.18.11177. [DOI] [PubMed] [Google Scholar]