Abstract
The plant genes required for the growth and reproduction of plant pathogens are largely unknown. In an effort to identify these genes, we isolated Arabidopsis mutants that do not support the normal growth of the powdery mildew pathogen Erysiphe cichoracearum. Here, we report on the cloning and characterization of one of these genes, PMR6. PMR6 encodes a pectate lyase–like protein with a novel C-terminal domain. Consistent with its predicted gene function, mutations in PMR6 alter the composition of the plant cell wall, as shown by Fourier transform infrared spectroscopy. pmr6-mediated resistance requires neither salicylic acid nor the ability to perceive jasmonic acid or ethylene, indicating that the resistance mechanism does not require the activation of well-described defense pathways. Thus, pmr6 resistance represents a novel form of disease resistance based on the loss of a gene required during a compatible interaction rather than the activation of known host defense pathways.
INTRODUCTION
When a plant interacts with a pathogen, there are two possible outcomes: either the pathogen grows and reproduces normally on the plant, in which case the plant is said to be susceptible, or the pathogen fails to grow and reproduce normally, in which case the plant is said to be resistant. For obvious reasons, a tremendous amount of research has focused on identifying the plant genes involved in mediating resistance to a diverse array of pathogens.
From this intense scrutiny, a picture of the pathways leading from perception of the pathogen by resistance genes to the activation of host defenses is emerging. In Arabidopsis, two pathways through which defense genes can be activated have been described. One pathway requires salicylic acid (SA) as a signaling intermediary, and the other requires the simultaneous perception of ethylene and jasmonic acid (JA). Numerous mutations that affect signaling through these pathways have been identified, and some of the corresponding genes have been cloned.
In contrast to our knowledge of the plant genes involved in resistance, little is known about the plant genes required for susceptibility. The best examples of susceptibility factors are plant metabolites that serve as chemoattractants or cues for the induction of bacterial genes involved in symbiosis or pathogenesis (Stachel et al., 1985; Stachel and Zambryski, 1986; Denarié et al., 1992).
In an effort to identify plant genes required for susceptibility, we began studying the interaction between Arabidopsis and the powdery mildew Erysiphe cichoracearum UCSC1 (Adam and Somerville, 1996) because the underlying complexity of plant–powdery mildew interactions suggests that many plant genes are involved (Schulze-Lefert and Vogel, 2000). Three events must occur for a powdery mildew pathogen to grow and reproduce on its host. First, the pathogen must invade an epidermal cell and establish a feeding structure, the haustorium, through which the fungus derives all of its nutrition. Second, the fungus must suppress or evade host defenses that, if fully activated, would arrest or kill the pathogen (Frye and Innes, 1998; Reuber et al., 1998). Third, because the powdery mildew pathogen cannot survive on dead host tissue, it must ensure that the underlying host cells remain viable until the pathogen's life cycle is complete.
Given the intimacy and complexity of this interaction, we reasoned that many plant factors would be required for powdery mildew growth. Assuming that these factors are not redundant, mutations in the corresponding genes, if not lethal, would be expected to reduce pathogen growth. Such mutations can be considered a novel form of disease resistance based not on resistance genes or the activation of host defenses but on the loss of the host's ability to support the pathogen.
Previously, we screened 26,000 M2 ethyl methanesulfonate mutants for individuals that were resistant to powdery mildew. In an effort to avoid mutants in which host defenses were activated constitutively, we excluded mutants that constitutively expressed PR1 or formed spontaneous lesions. This left 32 confirmed powdery mildew–resistant mutants. Twelve of these mutants formed lesions after inoculation and were set aside. Crosses between the remaining 20 mutants defined four recessive loci, pmr1 to pmr4 (Vogel and Somerville, 2000).
Neither the SA-dependent nor the JA/ethylene-dependent defense pathway was activated constitutively in pmr1 to pmr4, indicating that the resistance was not attributable to the constitutive activation of known defense pathways. Also notable was the absence of cell death at infection sites, indicating that resistance was not mediated by cell death. Thus, the resistance observed in these mutants is qualitatively different from resistance gene–mediated resistance, which typically involves a hypersensitive response. pmr-mediated resistance also is qualitatively different from previously described disease-resistant mutants that either constitutively express host defenses (e.g., cpr, lsd, cim, acd, and mlo) (Jørgensen, 1992; Greenberg and Ausubel, 1993; Bowling et al., 1994; Dietrich et al., 1994) or form lesions after pathogen challenge (e.g., edr1) (Frye and Innes, 1998).
It is important to note that for the two previously described powdery mildew–resistant mutants, mlo and edr1, resistance seems to be mediated by an enhancement of host defenses rather than by the loss of susceptibility factors. The barley mlo mutant is a lesion-mimic mutant in which resistance is correlated with a greatly enhanced papilla (a callose-rich barrier deposited at the site of attack) response (Wolter et al., 1993). Resistance in the Arabidopsis edr1 mutant is correlated with enhanced expression of defense genes and massive cell death beneath infection sites (Frye and Innes, 1998). Thus, it seems likely that both mutants are resistant as a result of enhanced host defense rather than a loss of susceptibility factors. In fact, because of their phenotypes, neither mlo-like nor edr1-like mutants would have been retained in the screen for pmr mutants.
In addition to powdery mildew resistance, pmr1, pmr3, and pmr4 mutants display obvious phenotypic defects: pmr1 has a defect in pollen tube growth, pmr3 is a conditional dwarf, and pmr4 has epinastic leaves. Thus, PMR1, PMR3, and PMR4 play roles in normal plant growth and development. This is to be expected for susceptibility factors because it is unlikely that the plant evolved genes with the sole function of servicing pathogens. Rather, the fungus has evolved to use the plant's normal machinery to create an environment favorable to disease. Here, we describe the cloning and characterization of PMR6.
RESULTS
Mutant Isolation and Fungal Growth
During the initial screen in which pmr1 to pmr4 were identified, we also identified six additional recessive mutants at two other loci, pmr5 (one allele) and pmr6 (five alleles). pmr6 plants are highly resistant to powdery mildew (Figure 1). To quantify that resistance and determine if pmr6 resistance was correlated with a block at a defined stage of fungal development, we measured fungal growth at various times after inoculation (Vogel and Somerville, 2000).
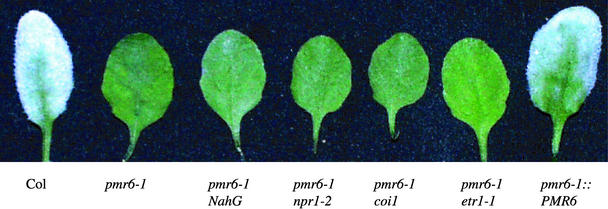
Figure 1.
Phenotypes of pmr6 Plants and Double Mutants.
Plants were inoculated with powdery mildew 8 days before being photographed. Note the abundant fungal growth on Col and the lack of macroscopic symptoms on pmr6-1, irrespective of blocks in defense signaling. Also note that pmr6-1::PMR6 plants are fully susceptible.
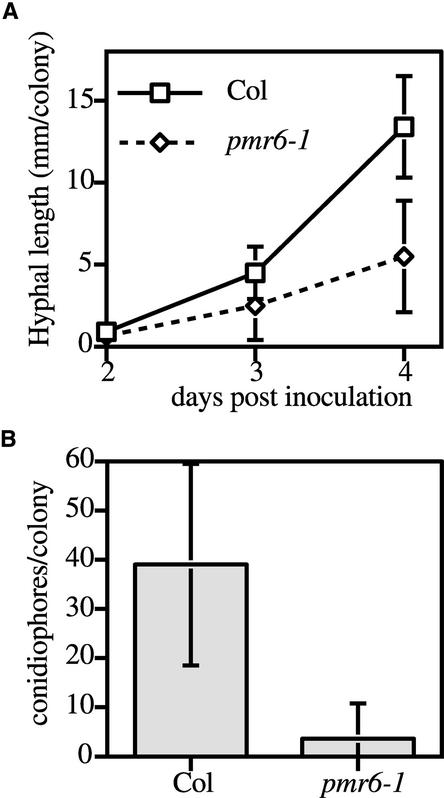
In general, fungal spores germinated and began to grow in a wild-type manner on pmr6-1 plants. However, by 2 days after inoculation (DAI) and beyond, colonies on pmr6-1 plants were significantly smaller (P < 0.05 by t test) than those on the wild type (Figure 2A). Eventually, most of the colonies on pmr6-1 plants consisted of shriveled hyphae, which were attached loosely to the leaf surface (data not shown). The production of asexual conidiospores also was reduced significantly (P < 0.0005 by t test) on pmr6-1 plants (Figure 2B). Thus, pmr6 resistance was not correlated with a block at a defined stage in fungal development.
Figure 2.
Quantification of E. cichoracearum Growth.
(A) Hyphal length per colony.
(B) Conidiophores per colony at 6 DAI.
Means ± sd based on 15 colonies are plotted. The entire experiment was repeated once with similar results.
Host Defenses
A crucial question regarding the resistance mechanism operating in pmr6 is what role, if any, do known host defense pathways play. If PMR6 is a susceptibility factor, then pmr6 resistance should be largely independent of defense pathways. In Arabidopsis, two pathways through which defense genes can be activated are well characterized (Glazebrook, 2001). To assess the contribution of these pathways to pmr6 resistance, we examined the expression of defense genes in response to powdery mildew infection and determined the effect of mutations or transgenes that block signaling through these pathways.
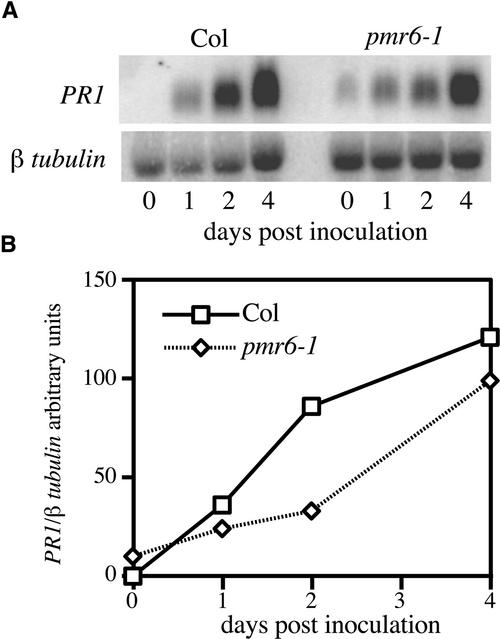
To determine if pmr6 resistance was correlated with enhanced activation of the SA pathway, we measured the expression of the SA-responsive defense gene PR1 at various times after inoculation. After inoculation, PR1 mRNA levels in the mutant were similar to those in the wild type (Figure 3), indicating that resistance is not mediated by hyperactivation of the SA pathway. Uninfected pmr6 plants (all alleles) expressed a very low, and variable, level of PR1, indicating that the SA pathway is usually slightly activated in this mutant.
Figure 3.
Time Course of PR1 mRNA Levels.
(A) RNA gel blot probed with PR1 and β-tubulin.
(B) PR1 intensity from (A) was normalized to β-tubulin intensity and plotted versus DAI. Similar trends were observed in two other experiments using slightly different time points.
To more directly assess the contribution of the SA pathway to resistance, we crossed the NahG transgene (Lawton et al., 1995) into the pmr6-1 background. NahG encodes a salicylate hydroxylase that blocks the SA pathway by degrading SA. Importantly, pmr6-1 NahG plants were resistant to E. cichoracearum (Figure 1), even though pmr6-1 NahG plants no longer accumulated detectable levels of PR1 mRNA (data not shown).
To further exclude a requirement for the SA pathway, we created a pmr6-1 npr1-2 double mutant. The npr1-2 mutant was identified by its inability to express PR1 in response to SA treatment (Cao et al., 1994). NPR1 has since been shown to play a key role in mediating many defense responses. Consistent with the pmr6-1 NahG results, pmr6-1 npr1-2 plants were resistant to powdery mildew (Figure 1), even though pmr6-1 npr1-2 plants no longer accumulated detectable levels of PR1 mRNA (data not shown).
Thus, pmr6 resistance does not require NPR1. Importantly, because the poorly understood phenomenon of induced systemic resistance requires NPR1 (Pieterse et al., 1998), it seems unlikely that pmr6 resistance is mediated by the constitutive activation of this pathway.
As in the experiments with PR1, the plant defensin PDF1.2 was used as a marker for the activation of the JA/ethylene pathway (Penninckx et al., 1996). Unlike the results with PR1, no induction of PDF1.2 was observed in either wild-type or pmr6-1 plants after inoculation with E. cichoracearum (data not shown). Thus, it is not surprising that the coi1 mutation (Xie et al., 1998), which blocks JA perception, and the etr1-1 mutation (Chang et al., 1993), which blocks ethylene perception, had no effect on pmr6-1 resistance (Figure 1).
Cell Death
A common outcome of many incompatible interactions mediated by resistance genes is the hypersensitive necrosis response in which one or a few cells at the infection site die. To determine if resistance in pmr6 is mediated by cell death, we stained infected leaves with trypan blue to highlight dead cells at 1 and 5 DAI. In both wild-type and pmr6-1 plants, no dead cells were observed underneath fungal colonies at 1 DAI. At 5 DAI, dead cells were rarely observed underneath colonies on both pmr6-1 and wild-type plants. Importantly, on pmr6-1 plants, many obviously dead and dying colonies were found on top of living cells. Thus, we conclude that pmr6 resistance is not mediated by host cell death.
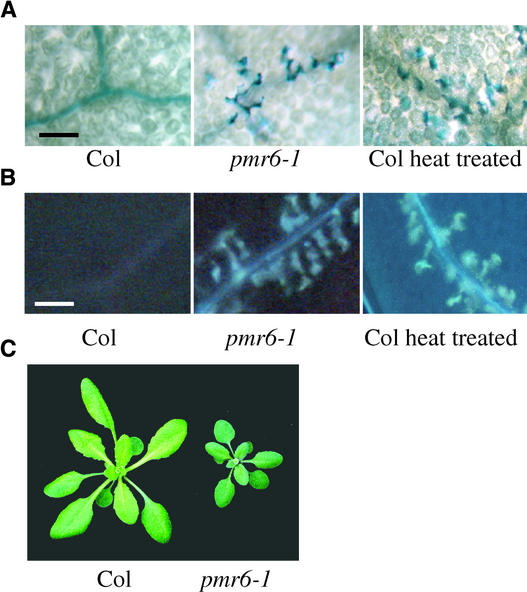
We did note that some leaves on pmr6 plants, all alleles, had a small number of individual dead mesophyll cells. These dead cells typically clustered along veins toward the outer edge of the leaf on a subset of the oldest leaves (Figure 4A). Also, these same cells were autofluorescent, suggesting that phenolic compounds were cross-linked into the cell wall (Figure 4B). It is important to note that these microlesions were seen in only a subset of the oldest leaves, but all leaves were resistant to powdery mildew. Also, there was no obvious increase in microlesions after inoculation. Therefore, resistance is not correlated with microlesions.
Figure 4.
Pleiotropic Effects of the pmr6 Mutation.
(A) Uninfected leaves stained with trypan blue. Dead cells stain dark blue. Individual mesophyll cells along the veins are stained in both pmr6-1 leaves and heat-treated Col leaves. Heat treatment was conducted by transferring plants to 37°C for 20 h. Leaves were stained 2 days after heat treatment. Veins appear as dark lines.
(B) Accumulation of autofluorescent compounds, as indicated by bright blue spots, follow the same pattern as dead cells in both pmr6-1 and heat-treated Col leaves. Veins appear as blue lines.
(C) Mature rosettes were photographed as they began to bolt. Note that pmr6-1 is smaller and greener than Col.
Plants were grown under a 14-h photoperiod for ∼3 weeks before being photographed. Bars in (A) and (B) = 100 μm.
We regard the microlesions as a pleiotropic effect of the pmr6 mutations not associated with resistance. These microlesions are most likely the stimulus for the low constitutive expression of PR1 observed in pmr6 plants. Interestingly, this vein-associated lesion phenotype can be phenocopied by heat treating wild-type plants (Figures 4A and 4B). Heat-treated wild-type plants were fully susceptible to E. cichoracearum (data not shown). Regardless of the mechanism of cell death, the fact that uninfected wild-type plants can be induced to form similar lesions without pathogen challenge and remain susceptible to subsequent powdery mildew attack suggests that the lesions are not strictly indicative of an activation of host defenses and are not associated with pmr6-mediated resistance.
Other Pathogens
If PMR6 is a susceptibility factor, we predict that it would be required by most powdery mildews, but not by unrelated pathogens. This assumes that unrelated pathogens use different methods to gain access to the nutrients guarded by the plant. Therefore, we challenged pmr6-1 plants with diverse pathogens, including the bacterium Pseudomonas syringae pv tomato and the oomycete Peronospora parasitica.
pmr6-1, like the wild type, was fully susceptible to Pseudomonas DC3000 pLAFR3 (Whalen et al., 1991), as measured by both disease symptoms and bacterial growth. After inoculation with bacterial suspensions, typical water-soaked lesions appeared on both pmr6-1 and wild-type plants (data not shown). To determine if the symptoms were indicative of bacterial growth, plants were syringe-infiltrated with a bacterial suspension, and bacterial growth was monitored by dilution plating of ground-up leaf discs. The growth curve for pmr6-1 was indistinguishable from that for the wild type (data not shown).
Similar to the results with Pseudomonas, pmr6-1 was fully susceptible to the biotrophic pathogen Peronospora Emco5 (Dangl et al., 1992). Average sporangiophore production per seedling was 64 ± 16 for wild-type Columbia (Col) and 64 ± 13 for pmr6-1. Values are means ± sd based on 20 seedlings. The entire experiment was repeated once with similar results.
We also challenged pmr6-1 plants with the closely related powdery mildew Erysiphe orontii MGH1 (Plotnikova et al., 1998). Nine of nine pmr6-1 plants from five independent experiments had no visible growth of E. orontii at 7 DAI. The wild-type controls showed extensive fungal growth. Thus, PMR6 is required for a compatible interaction with isolates from two powdery mildew species. This result is consistent with the idea that PMR6 is a susceptibility factor because closely related powdery mildews likely require a largely overlapping set of host genes for compatibility. That pmr6-1 is resistant to isolates from two different powdery mildew species also underscores the fact that pmr6-mediated resistance is fundamentally different from resistance gene–mediated race-specific resistance.
Cloning of PMR6
To gain a fuller understanding of pmr6-mediated resistance, we cloned the corresponding gene using a T-DNA–tagging approach. T-DNA mutants were identified by screening 40,000 T-DNA lines for powdery mildew resistance. Two mutants, pmr6-3 and pmr6-4, that were allelic to pmr6-1 were identified. pmr6-3 was backcrossed to the wild type to determine if the T-DNA insert cosegregated with powdery mildew resistance. A total of 111 F2 progeny were tested for powdery mildew resistance and then sprayed with Finale (the T-DNA confers resistance to Finale) to determine the presence of the T-DNA insert.
All 33 powdery mildew–resistant individuals in the F2 generation also were resistant to Finale. However, only nine individuals were susceptible to Finale, indicating the presence of more than one T-DNA insert. To convincingly demonstrate cosegregation, it was necessary to identify lines with single T-DNA inserts. To this end, 24 F2 plants that were powdery mildew susceptible and Finale resistant were allowed to set seed. The resultant F3 populations were tested for powdery mildew resistance and then sprayed with Finale.
The segregation results for one line that appeared to contain a single T-DNA insert are as follows: 91 plants were powdery mildew and Finale resistant; 204 plants were powdery mildew susceptible and Finale resistant; and 94 plants were powdery mildew susceptible and Finale sensitive. No plants were powdery mildew resistant and Finale sensitive. These results are consistent with a 1:2:1 segregation ratio (χ2 = 0.97, P = 0.61) that would be expected if there was a single T-DNA insert in PMR6. Thus, powdery mildew resistance cosegregated with the T-DNA insert.
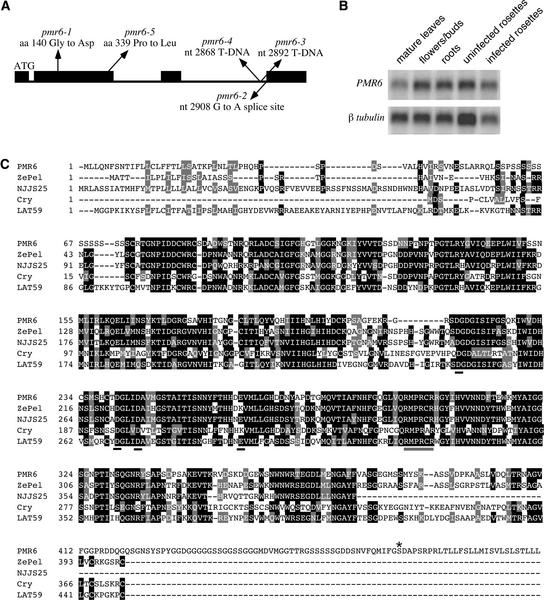
The gene disrupted in pmr6-3 was identified by cloning DNA flanking the T-DNA insert using a PCR-based approach. To confirm that we had cloned the correct gene, the PMR6 genes from three ethyl methanesulfonate alleles and an additional T-DNA allele were sequenced to identify mutations (Figure 5A). Finally, to corroborate that we had identified the correct gene, we complemented the mutation by introducing a 6.9-kb DNA fragment containing a wild-type version of PMR6 into pmr6-1 plants.
Figure 5.
Structure, Expression, and Alignment of PMR6.
(A) Scheme of PMR6. Exons are indicated by black boxes. The location and nature of the mutations in the five pmr6 alleles are indicated. The A in the start codon, ATG, is the first numbered nucleotide. aa, amino acid; nt, nucleotide.
(B) Expression of PMR6 in different organs. A RNA gel blot using 1 μg of poly(A)+ RNA from the indicated organs was probed with a 385-bp gene-specific probe from the C terminus of PMR6. The same blot was probed with β-tubulin as a loading control.
(C) Alignment of PMR6 with three plant-produced pectate lyases, ZePel, NJJS25, Cry j I, and one pectate lyase–like gene, LAT59. Identical amino acids are indicated by black shading, and similar amino acids are indicated by gray shading. Residues thought to be involved in binding Ca2+ or the active site are underlined with black or gray bars, respectively. The amino acid to which the GPI anchor is predicted to be attached is marked with an asterisk. Note that the last 27 amino acids are predicted to be cleaved off during the creation of the GPI anchor.
Forty of 40 T1 plants were susceptible to powdery mildew, whereas 200 of 200 control plants (transformed with a vector not containing PMR6) were resistant to powdery mildew (Figure 1). A comparison of the cDNA sequence with the genomic sequence revealed that PMR6 contains three introns (Figure 5A). PMR6 mRNA is expressed approximately equally in all organs tested: mature leaves, flowers/buds, rosettes, and roots (Figure 5B). PMR6 mRNA levels were not affected significantly by powdery mildew infection at 3 DAI (Figure 5B).
PMR6 contains a pectate lyase–like domain fused to a novel 84–amino acid C-terminal domain (Figure 5C). Pectate lyases cleave α-1,4-glycosidic linkages in pectate (demethylated pectin) by β-elimination and require Ca2+ for activity (Barras et al., 1994). PMR6 shares a proposed active site and three Asp residues thought to be required for Ca2+ binding with ZePel, a pectate lyase from Zinnia (Domingo et al., 1998). A Lys residue found in ZePel that is thought to be involved in Ca2+ binding is changed to Glu in PMR6. However, this residue also is Glu in NJJS25, a pectate lyase from strawberry, indicating that Lys at this residue is not absolutely required for enzymatic activity (Medina-Escobar et al., 1997).
The high degree of similarity between PMR6 and proteins with demonstrated pectate lyase activity suggests a similar function for PMR6. However, this notion must be tempered by the fact that researchers were unable to demonstrate pectate lyase activity for two pectate lyase–like proteins from tomato, Lat56 and Lat59 (Dircks et al., 1996) (Figure 5C). Thus, these proteins either do not have lyase activity or have specificity for an untested pectic substrate. Given the structural diversity of pectins and the large number of pectate lyase–like genes, we favor the latter possibility. Alternatively, these proteins may simply bind pectate, as was demonstrated for a pectate lyase–like protein from Pseudomonas (Charkowski et al., 1998).
Unfortunately, PMR6 has proven difficult to express in Escherichia coli, Pichia pastoris, and a baculoviral system (data not shown), so its enzymatic activity remains untested. Although pathogen-produced pectate lyases are well-characterized virulence factors of various soft rot pathogens, plant-produced pectate lyases have not been shown to play a role in plant–pathogen interactions. Because powdery mildews do not macerate tissue, the role PMR6 plays during a compatible interaction is not directly analogous to the role played by pathogen-produced pectate lyases.
The novel C-terminal domain corresponding to the last exon of PMR6 is required for PMR6 function because the transition mutation in pmr6-2 eliminates the C-terminal domain only by altering a splice site to introduce a frameshift mutation in the mature mRNA. Because of a predicted N-terminal endoplasmic reticulum transport sequence and a predicted C-terminal glycosyl-phosphatidylinositol (GPI) modification (Ferguson and Williams, 1988), PMR6 is predicted to be attached to the exterior surface of the plasma membrane by a GPI anchor.
This feature is not represented in any of the other 26 predicted pectate lyase–like genes in Arabidopsis with significant similarity to PMR6. The presence of a GPI anchor also raises the possibility that PMR6 may need to be released from the plasma membrane to gain access to pectin in the cell wall. The unique features of PMR6 suggest that it serves a specialized function that cannot be compensated for by other members of the gene family.
Cell Wall Analysis
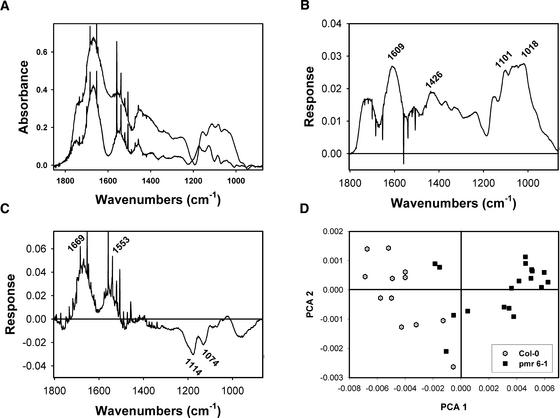
To determine if mutations in PMR6 altered cell wall composition, Fourier transform infrared (FTIR) spectroscopy using a synchrotron light source (Raab and Martin, 2001) was used to survey the composition of pmr6-1 and wild-type cell walls. FTIR spectroscopy has been established as a powerful tool for the analysis of plant cell walls (Chen et al., 1998). Visual inspection of the absorbance spectra from pmr6-1 revealed greater absorbance in the 1743 cm−1 shoulder region attributed to pectin, although the shoulder is noticeably narrowed in the mutants relative to Col. Thus, pmr6-1 cell walls appeared to be enriched for pectin (Figure 6A). This result is not surprising given the fact that PMR6 may be a pectin-degrading enzyme.
Figure 6.
Comparison of pmr6-1 and Col Cell Walls by FTIR Spectroscopy.
(A) Averaged midinfrared absorption spectra from the abaxial leaf surface of Col (lower curve; n = 12) and pmr6-1 (upper curve; n = 18). Note both the increased absorbance and narrower bandshape for pmr6-1 in the region responsive to pectin, at 1720 to 1743 cm−1 (Fillipov, 1972). Two cellulose peaks (Cael et al., 1975) seen in Col (1130 and 1087 cm−1) shift down in energy in pmr6-1 (1119 and 1082 cm−1). Likewise, the xyloglucan peak at 1174 cm−1 (Kacurakova et al., 1998) in Col decreases in energy to 1158 cm−1 in pmr6-1. These changes in cellulose and xyloglucan peak energy suggest an altered hydrogen bonding environment along cellulose microfibrils. The shoulder at ∼1426 cm−1 seen in pmr6-1 arises from free carboxyl stretching of pectins (Wellner et al., 1998). The spikes present in both spectra are the result of water vapor above the samples.
(B) and (C) First and second principal components from the covariance-matrix separation of the full infrared data sets summarized in (A). Wavenumbers of features deviating from the mean centered spectra are shown.
(B) The first principal component suggests that pmr6-1 cell walls are enriched in pectins with a lower degree of esterification than the wild type, as shown by the features at 1609 cm−1 (Fillipov, 1972; Coimbra et al., 1998), 1426 cm−1 (Wellner et al., 1998), 1101 cm−1 (Coimbra et al., 1998), and 1018 cm−1 (Coimbra et al., 1998; Wellner et al., 1998). The loading feature at 1426 cm−1 has two equally plausible assignments: (1) a carboxylate stretch of pectate, or (2) cellulose with altered intermolecular hydrogen bonding (Cael et al., 1975; Kataoka and Kondo, 1998).
(C) The second principal component suggests that epidermis is slightly enriched for protein (1669 and 1553 cm−1) and pectin (1114 and 1074 cm−1) (Wellner et al., 1998).
(D) Biplot showing the separation of Col and pmr6-1 midinfrared spectra generated by the covariance-matrix approach for principal components analysis (PCA) (Kemsley, 1998). Col and pmr6-1 samples form two distinct populations.
Absorbance peaks attributed to both cellulose and xyloglucan shift down in energy and broaden in the spectra from pmr6-1 cell walls, indicating that pmr6-1 cellulose forms a different hydrogen bond network than does wild-type cellulose (Vinogradov and Linell, 1971) or that the rotational environment of the -CH2OH group of cellulose has changed (Figure 6A) (Cael et al., 1975; Kataoka and Kondo, 1998).
Principal component analysis was used to identify features that differ between pmr6-1 and the wild type but that are not obvious in the raw spectra. The first three principal components explained 85, 5, and 3.6% of the variation in the full data set. The signature peaks of the first principal component suggested that pmr6-1 cell walls are enriched for pectins with a lower degree of esterification and an alteration in the H bonding environment of cellulose microfibrils (Figure 6B).
The second principal component, which by itself cannot separate Col from pmr6-1, demonstrated weak signals associated with protein and pectin (Figure 6C). The third principal component did not correspond to any known compounds (data not shown). Col and pmr6 samples formed two distinct populations in a biplot of the first two principal components (Figure 6D).
Pleiotropic Effects
pmr6 mutations are pleiotropic, indicating that PMR6 plays a unique role in normal plant growth and development. One obvious feature of pmr6 plants is that they are smaller than wild-type plants (Figure 4C). To quantify this reduction in stature, the largest diameter of 19 3-week-old rosettes was measured for Col (70.4 ± 3.2 mm) and pmr6-1 (53.9 ± 2.8 mm). The area of representative epidermal cells then was measured. For Col, 69 cells from four leaves were measured (4608 ± 1499 μm2/cell), and for pmr6-1, 87 cells from four leaves were measured (2352 ± 854 μm2/cell). Values shown are means ± sd.
Thus, pmr6-1 rosette diameter was 0.77 times the diameter of wild-type rosettes, and the average area of a pmr6-1 epidermal cell was 0.51 times the area of wild-type epidermal cells. By comparing the square of rosette diameter ratio (0.772 = 0.59) with the ratio of the epidermal cell area (0.51), we conclude that the reduction in the size of pmr6 rosettes was attributable largely to a decrease in cell expansion.
pmr6 leaves were shorter, rounder, and cupped slightly upward compared with wild-type leaves, which curled down (Figures 1 and 4C). The cupping phenotype indicated that the abaxial epidermal surface expanded more than the adaxial surface in pmr6 mutants, whereas the opposite was true in wild-type plants. Along with susceptibility, normal stature and leaf curling were restored by introducing the PMR6 gene into pmr6-1.
DISCUSSION
Because mutations in the PMR6 gene confer strong powdery mildew resistance, PMR6 is required for powdery mildew susceptibility. Two hypothesis can explain this resistance. Either PMR6 is required to promote fungal growth, or the loss of PMR6 somehow results in the activation of host defenses that retard fungal infection. Our characterization focused on differentiating between these possibilities.
Because high levels of PR1 or PDF1.2 mRNA were not observed in uninfected pmr6 plants, the resistance was not mediated by the constitutive activation of the SA-dependent or the JA/ethylene-dependent defense pathway. Likewise, because the levels of PR1 and PDF1.2 mRNA were not greater than those in the wild type after infection, pmr6 resistance is not caused by hyperactivation of the SA or JA/ethylene pathway. This sets pmr6 apart from a previously described powdery mildew–resistant mutant, edr1, in which resistance was correlated with enhanced PR1 expression (Frye and Innes, 1998).
To further exclude the involvement of previously described defense pathways, we constructed double mutants between pmr6-1 and npr1-2, coi1, or etr1-1 and introduced the NahG transgene into pmr6-1 plants. The resultant lines all were still resistant to powdery mildew, indicating that pmr6-mediated resistance is not dependent on signaling through the SA or JA/ethylene pathway. Thus, PMR6 either facilitates powdery mildew growth or somehow activates an undescribed defense pathway.
However, if pmr6 mutations activate a novel defense pathway, it must have a narrow spectrum, because pmr6 plants remain susceptible to Pseudomonas and Peronospora. This is in contrast to the broad-spectrum resistance observed in previously identified disease-resistant mutants (Dietrich et al., 1994; Bowling et al., 1997; Frye and Innes, 1998) and indicates that pmr6 resistance is not mediated by the activation of a broad-spectrum defense pathway such as systemic acquired resistance.
As the next step in trying to assign pmr6 a role in susceptibility or defense, we cloned the corresponding gene. PMR6 shows strong similarity to pectate lyase, suggesting a pectin-degrading activity for PMR6. Importantly, PMR6 does not resemble genes shown previously to be involved in host defense. Consistent with the putative pectin-degrading activity, pmr6 cell walls contain more pectin than wild-type cell walls (Figure 6). Although we do not know the exact role PMR6 plays in the cell, it clearly affects cell wall composition.
The alterations in pmr6 cell wall composition may have made pmr6 plants poor hosts for Erysiphe spp. One possibility is that in a wild-type plant, PMR6 degrades pectin in the extrahaustorial matrix, the space between the fungal haustorial cell wall and the plant extrahaustorial membrane, which envelopes the haustorium. Thus, the loss of PMR6 function in the mutant may lead to pectin accumulation and increased hydrogen bonding in the extrahaustorial matrix, resulting in decreased nutrient availability to the pathogen. This idea is consistent with the slow growth of powdery mildew observed on the mutant.
Another possibility is that fungal growth is reduced as a result of decreased penetration efficiency, possibly as a result of increased pectin content. However, no dramatic reduction in the frequency of colony initiation was observed during the measurement of fungal growth. Thus, at least the first penetration occurs at approximately the same frequency on pmr6-1 as on the wild type.
Mutations in PMR6 result in altered leaf morphology and decreased size. The reduction in size is explained largely by a decrease in cell expansion and is consistent with the putative role pectins play in regulating the complex process of cell wall loosening. The defect in cell expansion may be responsible for the microlesions observed in pmr6, because differential expansion may rip apart individual cells.
The similar levels of PMR6 mRNA observed in all tissues examined fit well with the overall reduction in the size of pmr6 plants. The pleiotropic effects of pmr6 mutations are consistent with a role as a susceptibility factor, because it is highly unlikely that a plant would possess genes that function solely to service pathogens. Rather, the pathogen probably evolved the ability to use plant genes to commandeer host functions for its own needs.
As a result of our analysis, we are left with a gene that, when mutated, leads to powdery mildew resistance independent of a resistance gene, cell death, and the activation of host defenses controlled by SA, JA, or ethylene. In addition, pmr6 plants are fully susceptible to Pseudomonas and Peronospora, indicating that the resistance is not caused by the activation of a novel broad-spectrum resistance mechanism. Thus, pmr6 resistance can be considered a special form of disease resistance, possibly based on the loss of a host susceptibility gene required by the pathogen for growth and development. In addition to increasing our understanding of compatible plant–pathogen interactions, pmr6 resistance potentially can be engineered into crop plants to create durable resistance in the field.
METHODS
Growth Conditions, Inoculations, RNA Gel Blot Analysis, and Microscopy
Except as noted, all experiments were performed as described previously (Vogel and Somerville, 2000). Bacterial inoculations were conducted as described by Vogel and Somerville (2000). To observe symptoms, Arabidopsis thaliana plants were inoculated with bacterial suspensions containing 106, 107, and 108 colony-forming units/mL. At least 12 leaves from four plants were scored for each concentration, and all experiments were conducted at least two times. For bacterial growth measurements, plants were syringe-infiltrated with a bacterial suspension containing 106 colony-forming units/mL in 10 mM MgCl2. Bacterial growth was monitored by dilution plating of ground-up leaf discs at 0, 1, and 2 days after inoculation.
pmr6 Alleles
Because no obvious differences in the strength of the five pmr6 alleles were noted after two backcrosses, we chose to use a representative allele, pmr6-1, for most experiments. Unusual results were verified by examining all alleles as noted. Mutant pmr6-1 has been deposited in the ABRC (Columbus, OH).
Construction of Double Mutants
To create the pmr6-1 etr1-1 double mutant, pmr6-1 was crossed to etr1-1. To identify plants containing the dominant etr1-1 mutation, F2 seeds were plated on Murashige and Skoog (1962) medium supplemented with 10 μM 1-aminocyclopropane-1-carboxylic acid (ACC) (ethylene-resistant plants are tall in the presence of ACC). Several ethylene-resistant plants were transferred to soil, grown for ∼2 weeks, and inoculated with powdery mildew to identify plants homozygous for pmr6-1.
To identify plants homozygous for both etr1-1 and pmr6-1, the ethylene-resistant and powdery mildew–resistant F2 plants were allowed to set seed. The resultant F3 seeds were plated on Murashige and Skoog (1962) medium supplemented with ACC to identify lines homozygous for etr1-1. One homozygous line was chosen as the double mutant.
To introduce the NahG transgene into pmr6-1 plants, pmr6-1 was crossed to a Columbia (Col) plant containing the NahG transgene (a gift from Iain Wilson, CSIRO Plant Industry, Canberra, Australia). F2 plants were inoculated with powdery mildew to identify pmr6-1 homozygous plants. These plants then were allowed to set seed. F3 seeds were plated onto Murashige and Skoog (1962) medium containing 50 μg/mL kanamycin (the NahG-containing construct confers resistance to kanamycin). A line homozygous for kanamycin resistance was chosen as the pmr6-1 NahG double mutant/transgene.
To create the pmr6-1 npr1-2 double mutant, pmr6-1 was crossed to npr1-2. F2 plants were inoculated with powdery mildew to identify plants homozygous for pmr6-1. Several of these plants were allowed to set seed. To identify plants homozygous for npr1-2, a RNA gel blot prepared with total RNA from F3 plants inoculated with powdery mildew (3 days after inoculation) was probed with PR1. A line that showed no detectable PR1 expression was chosen as the pmr6-1 npr1-2 double mutant.
The male-sterile phenotype of homozygous coi1 plants was used to follow the coi1 mutation during the construction of the pmr6-1 coi1 double mutant. A male-sterile coi1 plant was crossed to pmr6-1. F2 plants then were inoculated with powdery mildew and allowed to grow to determine fertility. Powdery mildew–resistant, male-sterile plants were homozygous for both pmr6-1 and coi1. However, because coi1 must be maintained in a heterozygous condition, several plants that were powdery mildew resistant and fertile were allowed to set F3 seed. The resultant F3 plants were grown to determine if the population segregated for male sterility and by inference coi1.
Cloning and Complementation
DNA flanking the T-DNA insertion in pmr6-3 was recovered using the Universal Genome Walker kit (Clontech, Palo Alto, CA) according to the manufacturer's instructions. The flanking DNA was sequenced, and a BLAST search revealed matches with a sequenced BAC.
The PMR6 gene, containing 2.2 kb of upstream DNA and 1.3 kb of downstream DNA, was amplified from wild-type genomic DNA using oligonucleotide primers. The resultant 6.9-kb PCR product was cloned into the shuttle vector pGEM-T (Promega). The fragment then was excised from pGEM-T and cloned into the plant transformation vector pCAMBIA3300 (CAMBIA, Canberra, Australia). The resultant plasmid was transformed into pmr6-1 plants using Agrobacterium tumefaciens–mediated transformation. Transformants were selected on soil using Finale (AgrEvo, Berlin, Germany) herbicide (0.13% active ingredient). T1 transformants were challenged with powdery mildew.
To determine the sequence of the PMR6 mRNA, oligonucleotide primers were designed to amplify a nearly full-length reverse transcriptase–mediated (RT)–PCR product. Multiple RT-PCR products then were sequenced to determine the cDNA sequence. After sequencing of the RT-PCR products, a database search using PMR6 identified an apparently full-length cDNA clone. This clone was end-sequenced using M13fwd and M13rev primers to determine the 5′ and 3′ ends of the mature PMR6 mRNA. The complete cDNA sequence was submitted to GenBank.
Alignment
ClustalX (http://www-igbmc.u-strasbg.fr/BioInfo/ClustalX/) was used to create the alignment of PMR6, ZePel, NJJS25, Cry j I, and LAT59.
Fourier Transform Infrared Analysis
Leaf discs from 12 Col plants and 18 pmr6-1 plants were cleared with a solution of chloroform and methanol (1:1) (Fry, 1988) in borosilicate scintillation vials with four changes of solvent until no pigments could be detected in the supernatant. The cleared discs then were air-dried overnight.
Collimated synchrotron infrared light from the Advanced Light Source Beamline 1.4.3 (Martin and McKinney, 1998) served as an external input to a Nicolet Instruments Magna 760 Fourier transform infrared spectrometer (Madison, WI), providing a nearly diffraction-limited spot size (10 μm × 10 μm) at the sample plane. The modulated light was passed to a Nic-Plan infrared microscope for reflectance spectromicroscopy through an ×15 Schwarzchild objective. The microscope-spectrometer combination was controlled by OMNIC software from Thermo Nicolet.
Each single-beam reflectance spectrum was ratioed to a reflection spectrum from a vapor-deposited gold surface. For each leaf disc, 512 scans were coadded for Fourier transform processing and absorbance spectra calculation. Spectra were collected over the infrared from 4000 to 650 cm−1 at a resolution of 2 cm−1 with a liquid N2-cooled mercury-cadmium-tellurium detector. Water vapor and CO2 in the ambient air were subtracted from each raw spectrum using a library spectrum at equivalent resolution. Each spectrum was manually baseline corrected in OMNIC over the full collection range (4000 to 2000 cm−1; 1850 to 650 cm−1). The baseline-corrected spectra were saved in JCAMP.DX format for further processing.
Exploratory data analysis used the principal components analysis–covariance matrix approach in the Win-DAS software package (Kemsley, 1998). All infrared spectra were area-normalized before principal components analysis processing.
Rosette Size Determination
Epidermal cell size was measured by clearing leaves in 95% ethanol, equilibrating the leaves in water, and examining them microscopically using Nomarski optics. Images were recorded using a digital camera fitted to the microscope. Individual epidermal cells were outlined, and the area was calculated using NIH Image software (http://rsb.info.nih.gov/nih-image/).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Accession Numbers
The GenBank accession numbers for the sequences mentioned in this article are AF534079 (PMR6 complete cDNA sequence), AL049655 (BAC clone), AV526313 (cDNA clone), At3g54920 (locus designation for PMR6), CAA70735 (ZePel), AAB71208 (NJJS25), BAA07020 (Cry j I), and S27098 (LAT59).
Acknowledgments
We thank Iain Wilson for NahG transgenic plants, CAMBIA for pCAMBIA vectors, the Kasusa DNA Research Institute for cDNA clones, and John Turner for coi1 seeds. This work was supported by National Institutes of Health fellowship F32 GN19499-01, by the U.S. Department of Energy Biological Energy Research Program, and by Syngenta. The Advanced Light Source is supported by the U.S. Department of Energy (Grant DE-AC03-76SF00098). This is publication 1496 from the Carnegie Institution of Washington.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.003509.
References
- Adam, L., and Somerville, S.C. (1996). Genetic characterization of five powdery mildew disease resistance loci in Arabidopsis thaliana. Plant J. 9, 341–356. [DOI] [PubMed] [Google Scholar]
- Barras, F., Van Gijsegem, F., and Chatterjee, A.K. (1994). Extracellular enzymes and pathogenesis of soft-rot Erwinia. Annu. Rev. Phytopathol. 32, 201–234. [Google Scholar]
- Bowling, S.A., Clarke, J.D., Liu, Y., Klessig, D.F., and Dong, X. (1997). The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9, 1573–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling, S.A., Guo, A., Cao, H., Gordon, A.S., Klessig, D.F., and Dong, X. (1994). A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6, 1845–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cael, J.J., Gardner, K.H., Koenig, J.L., and Blackwell, J. (1975). Infrared and Raman spectroscopy of carbohydrates. V. Normal coordinate analysis of cellulose I. J. Chem. Phys. 62, 1145–1153. [Google Scholar]
- Cao, H., Bowling, S.A., Gordon, A.S., and Dong, X.N. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C., Kwok, S.F., Bleecker, A.B., and Meyerowitz, E.M. (1993). Arabidopsis ethylene-response gene Etr1: Similarity of product to 2-component regulators. Science 262, 539–544. [DOI] [PubMed] [Google Scholar]
- Charkowski, A.O., Alfano, J.R., Preston, G., Yuan, J., He, S.Y., and Collmer, A. (1998). The Pseudomonas syringae pv. tomato HrpW protein has domains similar to harpins and pectate lyases and can elicit the plant hypersensitive response and bind to pectate. J. Bacteriol. 180, 5211–5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L., Carpita, N.C., Reiter, W.-D., Wilson, R.H., Jeffries, C., and McCann, M.C. (1998). A rapid method to screen for cell-wall mutants using discriminant analysis of Fourier transform infrared spectra. Plant J. 16, 385–392. [DOI] [PubMed] [Google Scholar]
- Coimbra, M.A., Barros, A., Barros, M., Rutledge, D.N., and Delgadillo, I. (1998). Multivariate analysis of uronic acid and neutral sugars in whole pectic samples by FT-IR spectroscopy. Carbohydr. Polym. 37, 241–248. [Google Scholar]
- Dangl, J.L., Holub, E.B., Debener, T., Lehnackers, H., Ritter, C., and Crute, I.R. (1992). Genetic definition of loci involved in Arabidopsis-pathogen interactions. In Methods in Arabidopsis Research, C. Koncz, N.H. Chua, and J. Schell, eds (Singapore: World Scientific), pp. 393–418.
- Denarié, J., Debelle, F., and Rosenberg, C. (1992). Signaling and host range variation in nodulation. Annu. Rev. Microbiol. 46, 497–531. [DOI] [PubMed] [Google Scholar]
- Dietrich, R.A., Delaney, T.P., Uknes, S.J., Ward, E.R., Ryals, J.A., and Dangl, J.L. (1994). Arabidopsis mutants simulating disease resistance response. Cell 77, 565–577. [DOI] [PubMed] [Google Scholar]
- Dircks, L.K., Vancanneyt, G.U.Y., and McCormick, S. (1996). Biochemical characterization and baculovirus expression of the pectate lyase-like LAT56 and LAT59 pollen proteins of tomato. Plant Physiol. Biochem. 34, 509–520. [Google Scholar]
- Domingo, C., Roberts, K., Stacey, N.J., Connerton, I.A.N., Ruiz Teran, F., and McCann, M.C. (1998). A pectate lyase from Zinnia elegans is auxin inducible. Plant J. 13, 17–28. [DOI] [PubMed] [Google Scholar]
- Ferguson, M.A.J., and Williams, A.F. (1988). Cell-surface anchoring of proteins via glycosyl-phosphatidylinositol structures. Annu. Rev. Biochem. 57, 285–320. [DOI] [PubMed] [Google Scholar]
- Fillipov, M.P. (1972). IR spectra of pectin films. J. Appl. Spectrosc. 17, 1052–1054. [Google Scholar]
- Fry, S.C. (1988). The Growing Plant Cell Wall. (Harlow, UK: Longman).
- Frye, C.A., and Innes, R.W. (1998). An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell 10, 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J. (2001). Genes controlling expression of defense responses in Arabidopsis: 2001 status. Curr. Opin. Plant Biol. 4, 301–308. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T., and Ausubel, F.M. (1993). Arabidopsis mutants compromised for the control of cellular damage during pathogenesis and aging. Plant J. 4, 327–341. [DOI] [PubMed] [Google Scholar]
- Jørgensen, J. (1992). Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica 63, 141–152. [Google Scholar]
- Kacurakova, M., Belton, P.S., Wilson, R.H., Hirsch, J., and Ebringerova, A. (1998). Hydration properties of xylan-type structures: An FTIR study of xylooligosaccharides. J. Sci. Food Agric. 77, 38–44. [Google Scholar]
- Kataoka, Y., and Kondo, T. (1998). FT-IR microscopic analysis of changing cellulose crystalline structure during wood cell wall formation. Macromolecules 31, 760–764. [Google Scholar]
- Kemsley, E.K. (1998). Discriminant Analysis and Class Modeling of Spectroscopic Data. (New York: John Wiley & Sons).
- Lawton, K., Weymann, K., Friedrich, L., Vernooij, B., Uknes, S., and Ryals, J. (1995). Systemic acquired resistance in Arabidopsis requires salicylic acid but not ethylene. Mol. Plant-Microbe Interact. 8, 863–870. [DOI] [PubMed] [Google Scholar]
- Martin, M.C., and McKinney, W.R. (1998). The first synchrotron infrared beamlines at the Advanced Light Source: Microspectroscopy and fast timing. Proc. Mater. Res. Soc. 524, 11–24. [Google Scholar]
- Medina-Escobar, N., Cardenas, J., Moyano, E., Caballero, J.L., and Munoz Blanco, J. (1997). Cloning, molecular characterization and expression pattern of a strawberry ripening-specific cDNA with sequence homology to pectate lyase from higher plants. Plant Mol. Biol. 34, 867–877. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Penninckx, I., Eggermont, K., Terras, F.R.G., Thomma, B., Desamblanx, G.W., Buchala, A., Metraux, J.P., Manners, J.M., and Broekaert, W.F. (1996). Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid–independent pathway. Plant Cell 8, 2309–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse, C.M.J., van Wees, S.C.M., van Pelt, J.A., Knoester, M., Laan, R., Gerrits, N., Weisbeek, P.J., and van Loon, L.C. (1998). A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10, 1571–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotnikova, J.M., Reuber, T.L., and Ausubel, F.M. (1998). Powdery mildew pathogenesis of Arabidopsis thaliana. Mycologia 90, 1009–1016. [Google Scholar]
- Raab, T.K., and Martin, M.C. (2001). Visualizing rhizosphere chemistry of legumes with midinfrared synchrotron radiation. Planta 213, 881–887. [DOI] [PubMed] [Google Scholar]
- Reuber, T.L., Plotnikova, J.M., Dewdney, J., Rogers, E.E., Wood, W., and Ausubel, F.M. (1998). Correlation of defense gene induction defects with powdery mildew susceptibility in Arabidopsis enhanced disease susceptibility mutants. Plant J. 16, 473–485. [DOI] [PubMed] [Google Scholar]
- Schulze-Lefert, P., and Vogel, J. (2000). Closing the ranks to attack by powdery mildew. Trends Plant Sci. 5, 343–348. [DOI] [PubMed] [Google Scholar]
- Stachel, S.E., Messens, E., Van Montagu, M., and Zambryski, P. (1985). Identification of the signal molecules produced by wounded plant cells that activate T-DNA transfer in Agrobacterium tumefaciens. Nature 318, 624–629. [Google Scholar]
- Stachel, S.E., and Zambryski, P.C. (1986). VirA and VirG control the plant-induced activation of the T-DNA transfer process of A. tumefaciens. Cell 46, 325–333. [DOI] [PubMed] [Google Scholar]
- Vinogradov, S., and Linell, R.H. (1971). Hydrogen Bonding. (New York: Van Nostrand Reinhold).
- Vogel, J., and Somerville, S. (2000). Isolation and characterization of powdery mildew-resistant Arabidopsis mutants. Proc. Natl. Acad. Sci. USA 97, 1897–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellner, N., Kacurakova, M., Malovikova, A., Wilson, R.H., and Belton, P.S. (1998). FT-IR study of pectate and pectinate gels formed by divalent cations. Carbohydr. Res. 308, 123–131. [Google Scholar]
- Whalen, M.C., Innes, R.W., Bent, A.F., and Staskawicz, B.J. (1991). Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 3, 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolter, M., Hollricher, K., Salamini, F., and Schulze-Lefert, P. (1993). The mlo resistance alleles to powdery mildew infection in barley trigger a developmentally controlled defence mimic phenotype. Mol. Gen. Genet. 239, 122–128. [DOI] [PubMed] [Google Scholar]
- Xie, D.X., Feys, B.F., James, S., NietoRostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. [DOI] [PubMed] [Google Scholar]