Abstract
The PMK1 mitogen-activated protein kinase gene regulates appressorium formation and infectious hyphae growth in the rice blast fungus. To further characterize this mitogen-activated protein kinase pathway, we constructed a subtraction library enriched for genes regulated by PMK1. Two genes identified in this library, GAS1 and GAS2, encode small proteins that are homologous with gEgh16 of the powdery mildew fungus. Both were expressed specifically during appressorium formation in the wild-type strains, but neither was expressed in the pmk1 mutant. Mutants deleted in GAS1 and GAS2 had no defect in vegetative growth, conidiation, or appressoria formation, but they were reduced in appressorial penetration and lesion development. Interestingly, deletion of both GAS1 and GAS2 did not have an additive effect on appressorial penetration and lesion formation. The GAS1–green fluorescent protein and GAS2–green fluorescent protein fusion proteins were expressed only in appressoria and localized in the cytoplasm. These two genes may belong to a class of proteins specific for filamentous fungi and function as novel virulence factors in fungal pathogens.
INTRODUCTION
The ascomycete Magnaporthe grisea is pathogenic to economically important crops such as barley, wheat, rice, and millet. Rice blast, which is caused by this heterothallic haploid fungus, is one of the most severe fungal diseases of rice throughout the world (Valent, 1990). Genetic studies of this pathogen during the past decade have made the Magnaporthe–rice pathosystem an excellent model for investigating fungus–plant interactions.
Magnaporthe infects rice plants with specialized infection structures called appressoria. Enormous turgor pressure generated in appressoria by the accumulation of high concentrations of glycerol is the force used to penetrate the underlying plant surface (de Jong et al., 1997). Mutants blocked at appressorium formation or appressorial turgor generation fail to infect healthy rice plants (Valent et al., 1991). After penetration, infectious hyphae grow in and between plant cells and eventually result in lesion formation. The blast fungus attacks all aboveground parts of the rice plant, and seedlings can be killed during epidemics.
Appressorium formation and penetration processes have been studied extensively in Magnaporthe (Dean, 1997; Hamer and Talbot, 1998). One mitogen-activated protein (MAP) kinase gene, PMK1, was found to be essential for appressorium formation and infectious hyphae growth in Magnaporthe (Xu and Hamer, 1996). Although PMK1 is nonessential for vegetative growth and conidiation in culture, pmk1 mutants fail to form appressoria and infect rice leaves. Recently, PMK1 homologs were characterized in several phytopathogenic fungi (reviewed by Xu, 2000). In all four appressorium-forming fungal pathogens examined to date (Magnaporthe, Colletotrichum lagenarium, Cochliobolus heterostrophus, and Pyrenophora teres), the PMK1 homologs are essential for appressorium formation (Xu and Hamer, 1996; Lev et al., 1999; Takano et al., 2000; Ruiz-Roldan et al., 2001).
Like pmk1 mutants in Magnaporthe, gene replacement mutants of PTK1 in P. teres and CMK1 in C. lagenarium are nonpathogenic and fail to colonize healthy or wounded host tissues (Takano et al., 2000; Ruiz-Roldan et al., 2001). In C. heterostrophus, a PMK1 homolog is necessary for appressorium formation and lesion formation but dispensable for colonizing plant tissues (Lev et al., 1999). The PMK1 homologs BMP1 and FMK1 also are essential for fungal pathogenicity in the necrotrophic pathogen Botrytis cinerea (Zheng et al., 2000) and the vascular wilt pathogen Fusarium oxysporum f. sp. lycopersici (Di Pietro et al., 2001). The B. cinerea bmp1 mutants fail to penetrate and kill plant cells and are nonpathogenic on a variety of plants tested (Zheng et al., 2000). GMK1 from Gaeumannomyces graminis fully complemented the defects of the Magnaporthe pmk1 mutant in appressorial development and invasive growth, indicating that the function of PMK1 MAP kinase may be conserved in this root pathogen (Dufresne and Osbourn, 2001). Even in the biotrophic, nonappressorium-forming pathogen Claviceps purpurea, the PMK1 homolog is essential for colonizing rye ovarian tissues (Mey et al., 2002). These data suggest that the PMK1 pathway is conserved in many, if not all, fungi for appressorium formation and other plant infection processes.
Although PMK1 homologs have been identified in several fungi, it is not clear how PMK1 is activated and what kinds of genes are regulated by this MAP kinase pathway. In this study, we constructed a subtraction library enriched for genes regulated by PMK1 during appressorium formation. Two genes identified in this library, GAS1 and GAS2, also were isolated by screening of an appressorium cDNA library by differential hybridization. Both of them were expressed specifically during appressorium formation and localized in the cytoplasm of appressoria. Gene deletion mutants of GAS1 and GAS2 had no defect in growth, conidiation, germination, or appressorium formation but were reduced in penetration and virulence on rice and barley seedlings.
RESULTS
Identification of the gEgh16 Homologs GAS1 and GAS2
To identify genes regulated by PMK1 during appressorium formation, we constructed a subtraction library using Guy11 cDNA as the tester and nn78 cDNA as the driver. Sequence analysis with the first 96 clones of this library revealed that two of them, MBC4 and MBE5, are homologous with the Erysiphe graminis EST clone gEgh16, a protein expressed during the early infection stage (Justesen et al., 1996). Interestingly, MBC4 and MBE5 were found to correspond to MAS1 and MAS3, respectively. MAS1 and MAS3 are the most redundant and the third most redundant MAS (Magnaporthe appressoria specific) sequences identified in an independent differential hybridization analysis of an appressorium cDNA library constructed from strain 70-15.
Among 621 cDNA clones that exhibited upregulated expression during appressorium formation, 77 and 53 clones are represented by MAS1 and MAS3 sequences, respectively. The 0.35- and 0.4-kb inserts amplified from clones MBE5 and MBC4, respectively, were used as probes to screen a Guy11 cosmid genomic library. Two corresponding open reading frames were identified after sequencing of cosmid clones that hybridized to these two probes; they were named GAS1 and GAS2 (gEgh16 homologs expressed in appressorium stage).
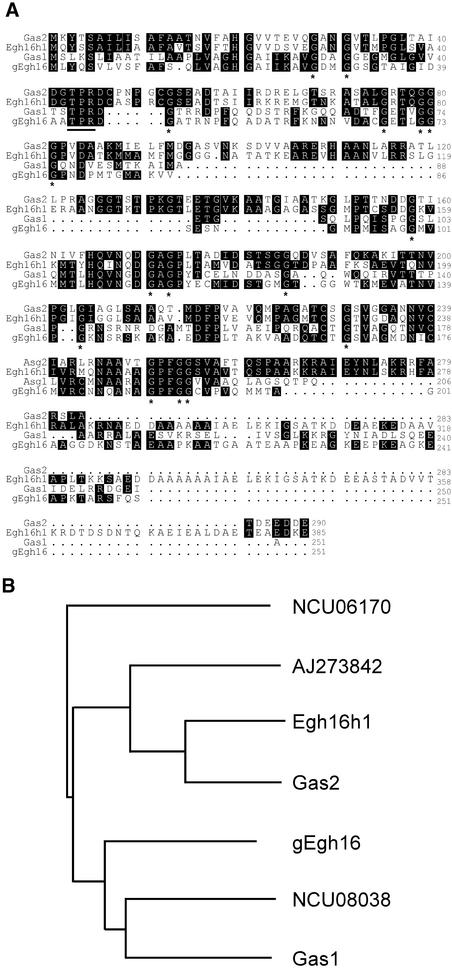
The GAS1 gene contains one 63-bp intron and encodes a 251–amino acid protein. The GAS2 gene has no intron and encodes a 290–amino acid protein. The amino acid sequences of GAS1 and GAS2 share 42% identity, and both are homologous with gEgh16 and its homolog Egh16H1. The highest homology is between GAS2 and Egh16H1 (67% identity). The Gas1 and Gas2 proteins are rich in Gly (12.7 and 12.6%, respectively) and Ala (12.0 and 15.8%, respectively) residues. Many Gly and Ala residues are conserved among GAS1, GAS2, and their homologs (Figure 1).
Figure 1.
Amino Acid Sequences and Relatedness of GAS1 and GAS2.
(A) Alignment of the predicted amino acid sequences of GAS1 and GAS2 with their homologs from E. graminis (gEgh16 and Egh16h). Identical residues are shown in black boxes. Gly residues conserved in all of these proteins are marked with asterisks underneath. The putative protein kinase C phosphorylation site is underlined.
(B) Phylogenetic relationship among Gas1, Gas2, and their homologs. The amino acid sequences of these fungal proteins were analyzed using the Clustal X algorithm to create the dendrogram. NCU08038 and NCU06170 are two hypothetical proteins from N. crassa. AJ273824 is an EST from the infected insect cuticle library of M. anisopliae.
Putative homologs of GAS1 and GAS2 also were found in the Neurospora crassa genome sequenced at the Whitehead Research Institute (www-genome.wi.mit.edu/annotation/fungi/neurospora/), in the Aspergillus fumigatus genome (tigrblast.tigr.org/ufmg), and in the ESTs of the entomopathogenic fungus Metarhizium anisopliae from the cDNA library of infected insect cuticles. However, none of these Gas1/Gas2 homologs has been characterized functionally, and no other close homologs of Gas1/Gas2 with an identified function were found in BLAST searches. Interestingly, there is no homolog of GAS1 or GAS2 in Saccharomyces cerevisiae, Schizosaccharomyces pombe, Candida albicans, or other sequenced eukaryote genomes, indicating that these two genes are specific for filamentous fungi.
Both GAS1 and GAS2 Are Expressed Specifically at the Appressorium Formation Stage
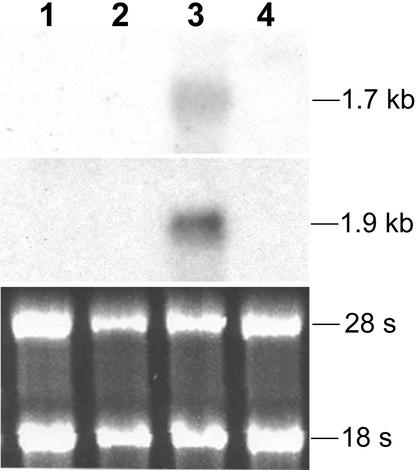
RNA gel blots of RNAs isolated from vegetative hyphae and appressoria formed on wax paper for 36 h were hybridized with GAS1 and GAS2. Bands of 1.7 and 1.9 kb were detected with the GAS1 and GAS2 probes, respectively, in RNA isolated from Guy11 appressoria (Figure 2). No expression of GAS1 and GAS2 was detected in RNA isolated from Guy11 vegetative hyphae grown in 5 × YEG (see Methods). In the pmk1 deletion mutant nn78, no detectable signal was observed for GAS1 or GAS2 in conidia germinated on wax paper for 36 h or in vegetative hyphae growing in 5 × YEG, suggesting that the transcription of GAS1 and GAS2 is regulated by PMK1.
Figure 2.
Expression Patterns of GAS1 and GAS2.
The top gel shows an RNA gel blot hybridized with GAS1. The middle gel shows the same blot stripped and rehybridized with GAS2. The bottom gel shows the RNA gel stained with ethidium bromide. Approximately 10 μg of total RNA was loaded in each well. Lanes 1 and 2 contained RNAs isolated from vegetative hyphae of Guy11 (wild type) and nn78 (pmk1), respectively. Lanes 3 and 4 contained RNAs isolated from Guy11 and nn78 conidia germinated on wax paper for 36 h. Both GAS1 and GAS2 were detectable in Guy11 only under the appressorium formation condition.
To further confirm that GAS1 and GAS2 are expressed specifically during appressorium formation, RNAs from ungerminated conidia, conidia germinated in liquid 5 × YEG for 4 h, and rice leaves infected with Guy11 for 60 h and 5 days were isolated. No detectable hybridization signal was observed in RNA gel blots of these RNAs hybridized with GAS1 and GAS2 (data not shown). Most likely, GAS1 and GAS2 are expressed only during appressorium formation in Magnaporthe.
Approximately 1.2- and 1.4-kb upstream sequences of GAS1 and GAS2 were sequenced and analyzed with several programs, including TRES (www.bioportal.bic.nus.edu.sg/tres), Expasy (www.expasy.org), and SoftBerry (www.softberry.com). Several pyrimidine-rich repeat sequences (CT box) were located downstream from the putative TATA boxes at −257 in GAS1 and −230 in GAS2. The promoter region of GAS1 also has a putative CAAT box (−276) upstream from the start codon. Even though both GAS1 and GAS2 are expressed specifically in appressoria, no other known sequence element was found to be common in their promoter areas.
gas1 Mutants Are Defective in Penetration
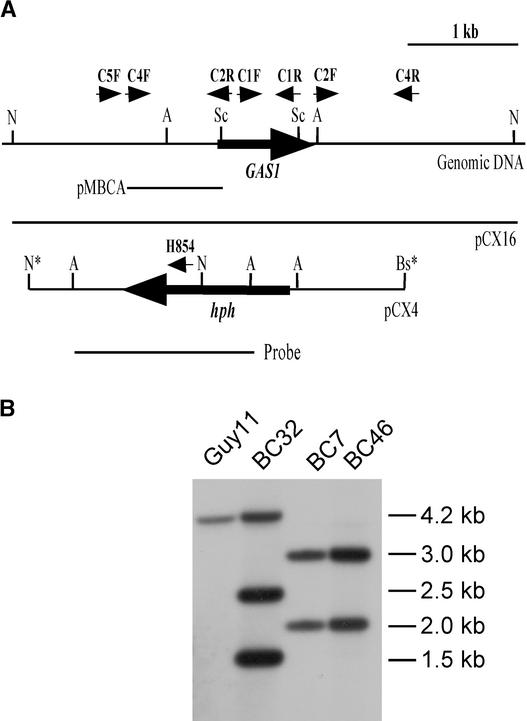
The gene replacement vector pCX4 was linearized with BstXI and transformed into Guy11. Seventy hygromycin-resistant transformants were isolated and screened by PCR with primers C5F and H854. A 1.4-kb fragment was amplified in two putative gas1 deletion transformants, BC7 and BC46 (data not shown). DNA gel blot analysis indicated that BC7 and BC46 had the 2- and 3-kb NcoI bands but lost the wild-type 4.2-kb NcoI band when hybridized with a 1.6-kb AflII fragment from pCX4 as the probe (Figure 3). One of the randomly selected transformants, BC32, contained the wild-type 4.2-kb band and two additional bands, of 1.5 and 2.5 kb, resulting from ectopically integrated pCX4.
Figure 3.
GAS1 Gene Replacement Vector and Transformants.
(A) Physical map of the GAS1 genomic region and gene replacement vector pCX4. Large arrows indicate orientations of the GAS1 and hph genes. Asterisks mark the restriction enzymes derived from the cloning vectors. The positions and orientations of primers C5F, C4F, C2R, C1F, C1R, C2F, C4R, and H854 are labeled with small arrows. A, AflII; Bs, BstXI; N, NcoI; Sc, SacI.
(B) DNA gel blot hybridized with the 1.6-kb AflII fragment (probe) from pCX4. Lanes from left to right represent genomic DNAs from wild-type strain Guy11, ectopically integrated transformant BC32, and gas1 deletion mutants BC7 and BC46. All DNA samples were digested with NcoI. The estimated size of each band is labeled at right (in kb).
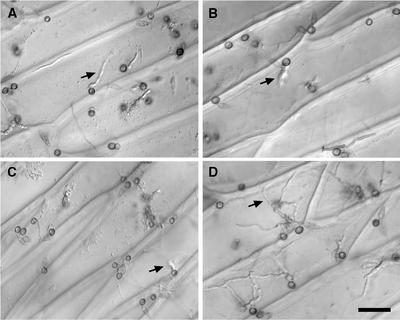
The gas1 deletion mutants BC7 and BC46 had no obvious defects in vegetative growth, conidiation, or sexual reproduction, and they formed typical grayish wild-type colonies. Conidia produced by these mutants were normal in germination and appressorium formation (Table 1). Appressoria formed by gas1 mutants were melanized and regular in shape (Figure 4). However, the percentage of appressoria that penetrated onion epidermal cells was reduced in the gas1 mutants. Although >70% of Guy11 and BC32 appressoria penetrated and developed infectious hyphae after 48 h of incubation, only ∼25% of appressoria formed by gas1 mutants penetrated under the same conditions (Table 1, Figure 4). Thus, the majority of appressoria formed by gas1 mutants were defective in appressorial penetration.
Table 1.
Appressorial Penetration and Lesion Development in gas1 and gas2 Mutants
| Variable | Guy11 (Wild Type) | BE71 (Ectopic) | BC46 (gas1) | BE55 (gas2) | DS21 (gas1 gas2) |
|---|---|---|---|---|---|
| Appressorium formationa (%) | 93.8 ± 4.1 | 90.2 ± 6.1 | 90.5 ± 3.6 | 87.7 ± 6.4 | 91.2 ± 2.7 |
| Appressorium penetration (%) | 77.3 ± 3.8 | 72.2 ± 5.4 | 25.7 ± 10.8 | 20.1 ± 11.6 | 19.9 ± 16.8 |
| Lesion formationb (lesions/5-cm leaf tip) | 46.7 ± 5.7 | 47.9 ± 4.9 | 13.0 ± 10.2 | 10.8 ± 7.0 | 9.7 ± 3.0 |
Appressorium formation and penetration were assayed on onion epidermis. The percentage of germ tubes differentiated into appressoria and the percentage of appressoria that developed infectious hyphae were determined at 48 h after inoculation.
Lesion formation was examined on infected rice leaves 7 days after inoculation. Means and sd values were calculated from at least three independent experiments.
Figure 4.
Appressorial Penetration Assays on Onion Epidermal Cells.
Conidia from gas1 mutant BC46 (A) and gas2 mutant BE55 (B) germinated and formed melanized appressoria, but only a few of them penetrated onion epidermal cells and produced infectious hyphae. The gas1 gas2 double mutant DS21 (C) also was reduced in appressorial penetration. Most appressoria formed by the wild-type strain Guy11 (D) penetrated and produced infectious hyphae. Photographs were taken 48 h after inoculation. Arrows indicate infectious hyphae. Bar in (D) = 15 μm for (A) to (D).
gas1 Mutants Are Reduced in Virulence
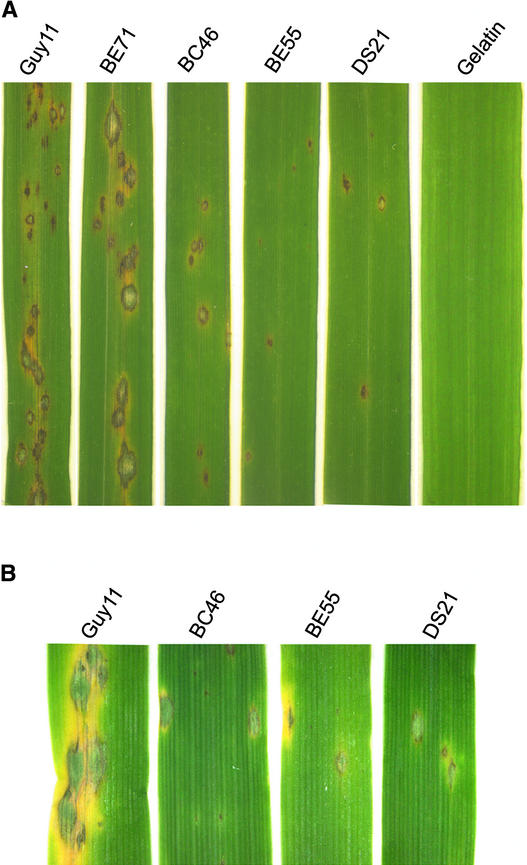
Rice leaves of CO39 seedlings sprayed with conidia of gas1 mutants developed blast lesions 7 days after inoculation (Figure 5A). However, the number of lesions caused by the gas1 mutant BC46 was much lower than that caused by Guy11 under the same conditions (Table 1). On average, Guy11 caused ∼45 lesions per 5-cm leaf tip, whereas leaves inoculated with BC46 conidia had ∼13 lesions per 5-cm leaf tip (Table 1), an ∼70% reduction compared with Guy11. On barley leaves inoculated with BC46, the number of lesions formed also was reduced (Figure 5B). In addition, lesions formed by BC46 on rice or barley leaves usually were smaller and less spreading than the lesions formed by Guy11 (Figure 5A). A similar reduction in lesion formation and lesion size also was observed in the gas1 deletion mutant BC7 (data not shown).
Figure 5.
Infection Assays on Rice and Barley Leaves.
(A) Seedlings from rice cv CO39 were spray inoculated with wild-type Guy11, ectopic integration transformant BE71, gas1 mutant BC46, gas2 mutant BE55, or gas1 gas2 double mutant DS21. For controls, leaves were sprayed with 0.25% gelatin solution.
(B) From left to right are barley leaves inoculated with Guy11, gas1, gas2, and the gas1 gas2 double mutant. Typical leaves are shown 7 days after inoculation.
For cosegregation assays, we isolated and characterized 24 progeny from a cross between BC46 and 2539. Although all 12 hygromycin-resistant progeny were reduced in appressorial penetration and lesion formation on rice leaves, the 12 hygromycin-sensitive progeny all were phenotypically similar to Guy11. These data indicated that the hygromycin resistance cosegregated with the reduced virulence of gas1 mutants.
We also transformed pCX16 containing the full-length GAS1 gene (Figure 3) into gas1 mutant BC46 by cotransformation with pAC905. Among 24 zeocin-resistant transformants screened by PCR with primers C1F and C1R, four with the GAS1 gene integrated ectopically were identified and confirmed further by DNA gel blot analysis (data not shown). These four complemented GAS1 transformants were as virulent as Guy11. Thus, reintroduction of the wild-type GAS1 allele restored the appressorial penetration and lesion development defects in gas1 mutants.
gas2 Mutants Have Phenotypes Similar to Those of gas1 Mutants
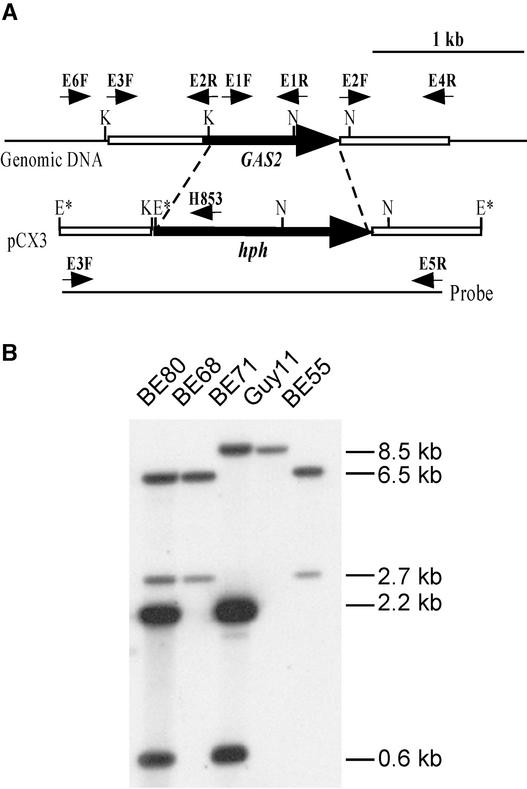
Three putative gas2 gene replacement mutants, BE55, BE68, and BE80, were identified by screening with the PCR primers E6F and H853 (data not shown). When genomic DNAs were digested with EcoRI and hybridized with the 2.6-kb fragment amplified from pCX3 as the probe, the wild-type strain Guy11 had an 8.5-kb band (Figure 6). The gas2 gene replacement mutants BE55, BE68, and BE80 all had the 2.7- and 6.5-kb bands but not the wild-type 8.5-kb band (Figure 6). A randomly selected transformant, BE71, contained both the wild-type 8.5-kb EcoRI fragment and the 0.6- and 2.2-kb EcoRI fragments resulting from ectopically integrated pCX3. Because BE80 had hybridization bands resulting from both homologous recombination and ectopic integration (Figure 6), only BE68 and BE55 were used in further analyses.
Figure 6.
Gene Replacement of GAS2.
(A) Physical map of GAS2 and the gene replacement vector pCX3. Large arrows indicate orientations of the GAS2 and hph genes. The EcoRI sites derived from the cloning vectors are marked with asterisks. The positions and orientations of primers E6F, E3F, E2R, E2F, E4R, E5R, and H853 are labeled with small arrows. E, EcoRI; K, KpnI; N, NcoI.
(B) DNA gel blot hybridized with the 2.6-kb fragment (probe) amplified with primers E3F and E5R. Genomic DNAs isolated from Guy11 and four transformants (BE80, BE68, BE71, and BE55) were digested with EcoRI. The gas2 deletion mutants BE80, BE68, and BE55 had the 2.7- and 6.5-kb hybridization bands but not the wild-type 8.5-kb band.
The gas2 mutants also were normal in vegetative growth and sexual and asexual reproduction, but they were reduced in appressorial penetration and lesion formation (Table 1, Figure 4). On average, the gas2 mutants usually produced ∼10 lesions on the 5-cm leaf tip, an ∼77% reduction compared with Guy11 under the same conditions (Table 1). Morphologically, gas1 and gas2 mutants were not distinguishable. Twenty-six progeny were isolated from a cross between BE55 and 2539. Reduction in lesion formation was observed in all 10 hygromycin-resistant progeny but not in the 16 hygromycin-sensitive progeny (data not shown). Thus, the phenotypes observed in gas2 mutants were associated directly with the deletion of GAS2. We also generated gas1 deletion mutants in strain 70-15 and found that they had phenotypes similar to those of gas1 mutants in Guy11 (data not shown), indicating that the function of GAS2 is not strain specific.
gas1 gas2 Double Mutants Have Phenotypes Similar to Those of gas1 or gas2 Mutants
One MAT1-1 gas2 progeny (MS12) derived from the cross BE55 × 2539 was crossed with the gas1 mutant BC46 (MAT1-2). Among 24 progeny isolated from this cross, four gas1 gas2 double mutants, DS14, DS18, DS21, and DS32, were identified by PCR screening with primers C5F/H854 and E6F/H853 and confirmed further by DNA gel blot analysis (data not shown). All four of these double mutants had normal hyphal growth, colony morphology, and conidiation. Conidia germination and appressoria morphology also were normal (Table 1).
To our surprise, the reduction in the appressoria penetration and lesion formation of gas1 gas2 double mutants was similar to that of the gas2 or gas1 mutants (Table 1). We repeated the infection and appressorial penetration assays several times and observed no significant difference between gas2 and gas1 gas2 double mutants. It is likely that the deletion of both GAS1 and GAS2 did not have any additive effect in gas1 gas2 double mutants.
Both GAS1 and GAS2 Localize to Cytoplasm in the Appressoria
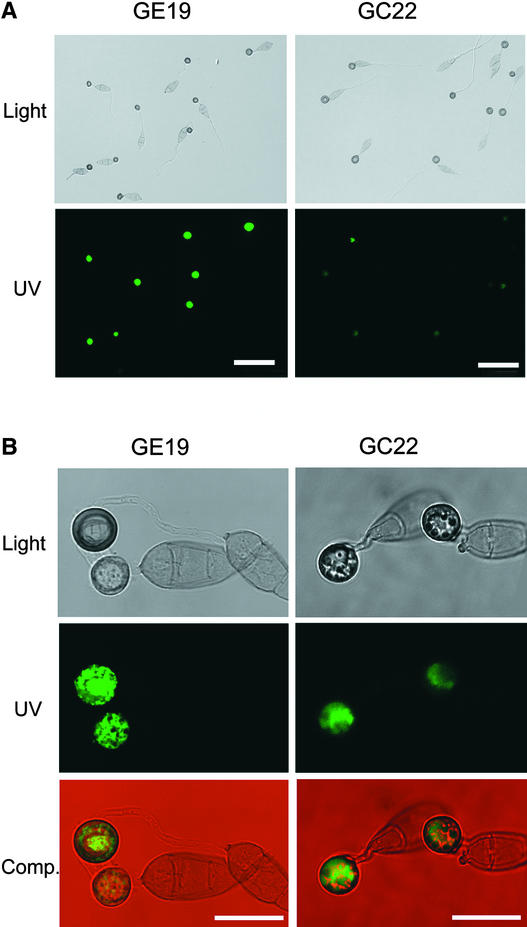
To determine the localization pattern of Gas1 and Gas2 proteins, the GAS1–green fluorescent protein (GFP) fusion construct pCX13 and the GAS2-GFP fusion construct pCX14 were transformed into Guy11. Two pCX13 transformants (GC22 and GC24) and two pCX14 transformants (GE6 and GE19) were identified by PCR and confirmed by DNA gel blot analysis (data not shown). In transformants expressing GAS1-GFP and GAS2-GFP, no obvious change in growth or virulence was observed. In both pCX13 and pCX14 transformants, green fluorescence was observed in the majority of appressoria after 24 h of incubation (Figure 7A). However, no detectable signals were observed in the germ tubes, conidia, or vegetative hyphae (Figure 7A).
Figure 7.
Expression and Localization of Gas1 and Gas2.
(A) GFP tagging of GAS1 and GAS2. Appressoria formed by transformants expressing GAS1-GFP (GC22) and GAS2-GFP (GE19) were normally melanized when examined using Nomarski microscopy (top) at 24 h. Most of these appressoria had green fluorescence, but the intensity of fluorescence varied among different appressoria when examined using epifluorescence microscopy (bottom). Bars = 50 μm.
(B) Confocal microscopy examination of the Gas1-GFP and Gas2-GFP fusion proteins. GFP fusion proteins were localized in the cytoplasm of appressoria formed by GE19 (left) and GC22 (right) at 24 h. Micrographs of the same field taken under bright-field illumination (Light) and UV light (UV) with a confocal laser fluorescence microscope were used to generate the composite images (Comp.) with the Thumbsplus 4 program (Cerious Software, Charlotte, NC). Bars = 15 μm.
We also examined conidiophores, young conidia, perithecia, and asci produced by GC22 and GE19 and did not observe any GFP signals. These data indicate that the Gas1-GFP and Gas2-GFP fusion proteins were expressed specifically in appressoria. The GFP fusion constructs pCX13 and pCX14 also were transformed into the gas1 mutant BC46 and the gas2 mutant BE55, respectively. MCX13 and MCX14 were two resulting transformants that were confirmed by DNA gel blot analysis to contain a single-copy integration of transforming vector pCX13 or pCX14. Both MCX13 and MCX14 were as efficient as Guy11 in appressorial penetration and lesion formation (data not shown), indicating that the Gas1-GFP and Gas2-GFP fusion proteins function normally in Magnaporthe.
When examined by epifluorescence microscopy, the majority of appressoria expressing Gas1-GFP and Gas2-GFP fusion proteins had green fluorescence in the cytoplasm but not in the cell wall. We further confirmed these observations by examining the localization of GFP fusion proteins under confocal microscopy. Appressoria formed by both GE19 and GC22 had uneven fluorescent peripheries and strong green fluorescence in the cytoplasm (Figure 7B), indicating that Gas1-GFP and Gas2-GFP fusion proteins were localized in the cytoplasm but not in the cell wall or cytoplasm membrane.
These GFP fusion proteins were not distributed evenly in the cytoplasm, however, and there were certain areas in the appressorium cytoplasm, possibly the lipid or glycogen bodies and nucleus, that had no detectable green fluorescence. Close examination indicated that the cellular localizations of Gas1 and Gas2 were slightly different. Although Gas2-GFP fusion proteins were distributed somewhat evenly throughout the cytoplasm, Gas1 proteins were localized preferentially in the vacuole in appressoria (Figure 7B).
Interestingly, not all of the appressoria produced by transformants GE19 or GC22 displayed green fluorescence (Figure 7A). We then transformed pCX35 into GC22 and isolated transformants that contained GAS1-GFP and GAS2–yellow fluorescent protein (YFP) fusion constructs. In DT2, one of the transformants expressing both GAS1-GFP and GAS2-YFP, growth and virulence were normal (data not shown). However, even in DT2, there was always a small portion of appressoria that displayed no detectable fluorescence under epifluorescence microscopy (Figure 8). It is possible that the fluorescent fusion proteins were not expressed or were expressed in these appressoria at a level lower than the detection limit of the epifluorescence microscopy.
Figure 8.
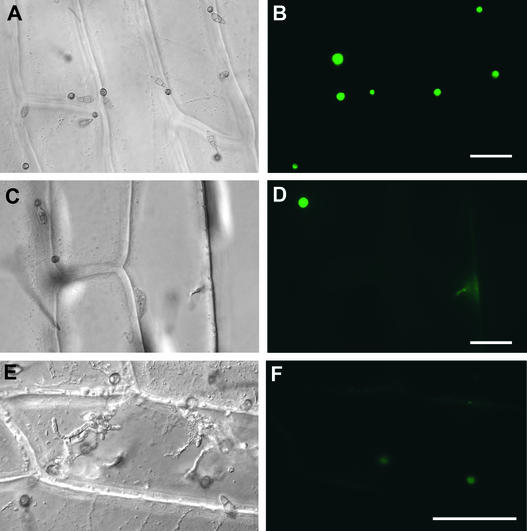
Expression of GAS1 and GAS2 in Infectious Hyphae.
Conidia from DT2, a strain expressing both Gas1-GFP and Gas2-YFP fusion proteins, were inoculated onto onion epidermal cells. At left are photographs taken using bright-field microscopy. At right are the same fields photographed using epifluorescence microscopy. Fluorescence in appressoria ([A] and [B]), penetration pegs and primary infectious hyphae ([C] and [D]), and well-developed infectious hyphae ([E] and [F]) were examined at 24, 48, and 60 h, respectively. After penetration, only the penetration pegs and primary infectious hyphae, but not the well-developed infectious hyphae, had faint fluorescence. Many appressoria that failed to penetrate remained fluorescent, but those that penetrated were no longer fluorescent. Bars = 50 μm.
We also transformed pCX27, a GAS2-GFP fusion construct with the 3′ untranslated region sequence of GAS2, into Guy11. In MCX27 and other transformants expressing this GAS2-GFP-TGAS2 construct, we also observed variation of the GFP signal among appressoria, and some appressoria had no detectable GFP signal at all (data not shown). No obvious difference in GFP signals was observed between appressoria formed by MCX27 and GE19 (Figure 7), indicating that the 3′ untranslated region of GAS2 has no significant effect on the accumulation and localization of Gas2-GFP fusion proteins.
Expression Patterns of GAS1 and GAS2 Are Different during Appressorium Formation
For appressoria produced by GC22, only very faint green fluorescence was detectable after incubation for 9 h (Table 2). Green fluorescence became stronger in the majority of appressoria when examined at 12 and 24 h. At 48 h, the green fluorescence in most appressoria was less intense, and only a small percentage of appressoria had green fluorescence after 4 days (Table 2), suggesting that the Gas1-GFP fusion protein accumulated in appressoria at the earlier stages and then was degraded later (after 24 h). The Gas2-GFP fusion protein, however, was expressed earlier than Gas1-GFP and could be detected as early as 4 h (Table 2) in appressoria formed by GE19.
Table 2.
Expression of GFP Fusion Proteins in Transformants of Magnaporthe
| Appressoria with Detectable GFP Signal (%)
|
||
|---|---|---|
| Time (h) | GE19 (Gas1-GFP) | GC22 (Gas2-GFP) |
| 2 | NDa | ND |
| 4 | Faintb | ND |
| 6 | 42.6 ± 2.6 | ND |
| 9 | 64.4 ± 13.2 | Faint |
| 12 | 82.3 ± 5.5 | 85.4 ± 12.3 |
| 24 | 90.8 ± 0.8 | 80.5 ± 1.9 |
| 36 | 81.1 ± 3.8 | 82.7 ± 1.3 |
| 48 | 84.2 ± 7.6 | 73.1 ± 8.5 |
| 96 | 45.9 ± 2.7 | 16.4 ± 1.0 |
ND, no detectable fluorescence.
Faint fluorescence was observed in some appressoria, but the signal was too weak to count reliably.
At 6 h, >40% of appressoria formed by GE19 had strong green fluorescence. When examined from 12 to 48 h, the majority of appressoria (>80%) had strong green fluorescence. The Gas2-GFP fusion protein appeared to be more stable than Gas1-GFP, because ∼45% of GE19 appressoria remained fluorescent after 4 days of incubation. Interestingly, appressoria formed by transformants expressing GAS2-GFP usually had stronger fluorescence than those formed by transformants expressing GAS1-GFP (Figure 7) at any time points examined (data not shown), indicating that the expression level of GAS2-GFP was higher than that of GAS1-GFP in appressoria.
To determine whether GAS1 and GAS2 can complement each other, we introduced pCX14 into BC46 and pCX13 into BE55. The resulting transformants had phenotypes similar to those of the original gas1 and gas2 mutants BC46 and BE55. No cross-complementation effect on appressorial penetration or lesion development was observed in gas1 and gas2 mutants transformed with pCX14 and pCX13, respectively. It is likely that GAS1 and GAS2 play some overlapping but different functions at different stages of appressorium formation.
GAS1 and GAS2 Are Not Expressed in Infectious Hyphae
To determine whether GAS1 and GAS2 were expressed in the infectious hyphae, conidia from DT2 were used to infect onion epidermal cells and barley leaves. Although appressoria formed by DT2 on onion epidermal cells had strong green fluorescence, there was no detectable fluorescence in well-developed infectious hyphae (Figure 8). Similar results were obtained with infectious hyphae formed by DT2 in barley epidermal cells (data not shown). These data indicated that GAS1 and GAS2 were not expressed in infectious hyphae.
In some but not all primary infectious hyphae or pegs that penetrated into onion epidermal cells, weak fluorescence was observed (Figure 8), suggesting that the fluorescent fusion proteins accumulated in appressoria may be transported into the primary infectious hyphae or that the fusion constructs were expressed transiently at this penetration stage. Interestingly, many appressoria that failed to penetrate remained fluorescent, but none of the appressoria that produced infectious hyphae had any detectable fluorescence (Figure 8).
It is likely that Gas1 and Gas2 proteins are functional only in appressoria and possibly are involved in penetration peg and primary infectious hyphae development. After penetration, these fusion proteins are either degraded rapidly or transported into penetration pegs. Data from these assays are consistent with our earlier observation that GAS1 and GAS2 were not detectable by RNA gel blot analysis or reverse transcriptase–mediated PCR with RNAs isolated from rice leaves collected at 60 h or 5 days after inoculation.
DISCUSSION
To further characterize the PMK1 MAP kinase pathway in Magnaporthe, two genes identified in a subtraction library enriched for genes regulated by PMK1 were characterized functionally in this study. Both GAS1 and GAS2 are highly expressed during appressorium formation and rich in Ala and Gly. Interestingly, there is a putative protein kinase C phosphorylation site (Figure 1) that is well conserved among GAS1, GAS2, and their homologs from other fungi. In Magnaporthe, pharmacological studies had indicated that protein kinase C may be involved in appressorium formation (Eckhard et al., 1998).
GAS1 or GAS2 or both are dispensable for mycelial growth, conidiation, and sexual reproduction, but they are important for appressorial penetration and lesion development. In Magnaporthe, several genes, such as MPG1, PTH11, and ABC1, are dispensable for vegetative growth and sexual or asexual reproduction but play important roles in appressorium formation and plant infection. Mutants disrupted in MPG1, a hydrophobin gene, were reduced by 74 and 80% in appressorium formation and lesion development, respectively (Talbot et al., 1993). Both abc1 and pth11 deletion mutants are reduced substantially in virulence, but they still form appressoria and cause rare lesions on infected rice leaves (DeZwaan et al., 1999; Urban et al., 1999).
Because the expression pattern and cellular localization of the Gas1-GFP and Gas2-GFP fusion proteins are slightly different, it is reasonable to assume that GAS1 and GAS2 may have different functions during appressorial penetration and infectious hyphal growth. Surprisingly, the gas1 gas2 double mutants have phenotypes similar to those of gas2 or gas1 mutants. No additive effect of gas1 and gas2 deletions was observed in the gas1 gas2 double mutants. One possible explanation for this finding is that GAS1 and GAS2 function sequentially in the penetration and lesion development processes. Mutants deleted in either the GAS1 or GAS2 gene will block the infection process to a level similar to that of the gas1 gas2 double mutants.
The other possibility is that GAS1 and GAS2 may interact with each other and form a complex that is required for efficient penetration and lesion development. However, the Magnaporthe genome may contain additional GAS1 or GAS2 homolog(s) that can compensate partially for the deletion of GAS1 or GAS2. In E. graminis, two isoforms of Egh16H1 (A and B) that differ in only a few amino acid residues have been isolated as genes expressed in early infection stages. The exact biological function of gEgh16 and Egh16H1 isoforms in E. graminis is not clear.
Both GAS1 and GAS2 are expressed specifically in appressoria and play important roles in appressorium penetration. A few fungal genes are known to be expressed specifically in appressoria and are critical for plant infection. One of them is PLS1 in Magnaporthe, which encodes a transmembrane protein expressed only in appressoria, as determined by GFP-tagging assays (Clergeot et al., 2001). The pls1 deletion mutants fail to form penetration pegs and thus are nonpathogenic on rice (Clergeot et al., 2001).
Two other Magnaporthe genes were reported to be expressed specifically in appressoria but were not characterized functionally (Lee and Dean, 1993). In Colletotrichum gloeosporioides, CAP20 was isolated as a gene expressed specifically during appressorium formation (Hwang et al., 1995). The cap20 gene disruption mutants were reduced in virulence on avocado and tomato fruits. Like GAS1 and GAS2 in Magnaporthe, CAP20 encodes a protein that is specific for filamentous fungi but with no functionally characterized homolog in GenBank.
Both Gas1-GFP and Gas2-GFP fusion proteins localized to the cytoplasm in appressoria. When analyzed with various programs available on the World Wide Web for extracellular or subcellular localization, no consistent predictions were obtained for either Gas1 or Gas2. Interestingly, putative signal peptides at the N termini of Gas1 and Gas2 were identified by SignalP version 2.0 (Nielsen et al., 1997). However, no signal peptide sequence was detected in either Gas1 or Gas2 when analyzed with PSORT II (psort.nibb.ac.jp).
We were unable to observe green fluorescence in the surrounding areas of either fluorescent or nonfluorescent appressoria formed by GE19, GC22, or DT2 (Figures 7 and 8). After removing appressoria by rubbing with gloved fingers, no green fluorescence was observed in the appressorial mucilage left behind (data not shown). Most likely, the Gas1 and Gas2 fusion proteins were not secreted from these appressoria. We were unable to detect green fluorescence in infectious hyphae or the transcript of GAS1 or GAS2 in infected rice leaves, indicating that these two genes were not expressed in infectious hyphae.
However, it remains possible that the fluorescent fusion proteins may be expressed and secreted into plant cells at a level lower than the detection limit. Plant cells penetrated by Magnaporthe usually accumulate autofluorescent materials as part of the defense response, making it impossible to detect small amounts of secreted fluorescent fusion proteins. It may be necessary to isolate extracellular proteins secreted by Magnaporthe and analyze them by protein gel blot analysis.
In the GFP-tagging experiments with GAS1 and GAS2, there were always some normally shaped appressoria without any detectable green fluorescence. This is similar to what has been reported in transformants expressing the Pls1-GFP fusion protein, which had strong fluorescence only in 80% of appressoria (Clergeot et al., 2001). We also observed that the strength of fluorescent signals varied among appressoria formed by transformants expressing Gas1-GFP, Gas2-GFP, or both. In Magnaporthe, appressorium formation and maturation involve the deposition of additional cell wall layers, mobilization and use of carbohydrate stores, generation of turgor, cytoskeleton reorganization, and other steps (Bourett and Howard, 1990).
The expression of GAS1 and GAS2 may be regulated developmentally in these processes. The variation in GFP signal among appressoria may reflect different developmental stages of the appressoria we observed. MST12 is one of the transcription factors regulated by PMK1 to control appressorial penetration and infectious hyphae growth (Park et al., 2002). Our preliminary data indicated that the expression of Gas2-GFP is normal in appressoria formed by the mst12 mutant MK23 transformed with pCX14. Thus, the expression of GAS2 is not regulated by MST12. GAS1 and GAS2 may be controlled by other transcription factors specific for appressorium formation and maturation and infectious hyphal growth. Although their homologs exist in the saprophytic fungus N. crassa, GAS1 and GAS2 may belong to a class of genes that are specific for filamentous fungi and that play important roles during the early infection stage in fungal pathogens. It will be interesting to isolate and characterize transcription factors that may regulate their expression during appressorium formation.
METHODS
Strains and Culture Conditions
Wild-type Magnaporthe grisea strains and various transformants generated in this study (Table 3) were cultured at 25°C on oatmeal agar plates or V8 juice agar under fluorescent light to induce conidiation (Xu and Hamer, 1996; Choi and Dean, 1997). Mycelia collected from 2-day-old 5 × YEG (5 mg/ml yeast extract and 10 mg/ml glucose; Zheng et al., 2000) cultures shaken at 150 rpm at room temperature were used for the isolation of fungal DNA and protoplasts. Genetic crosses and isolation of progeny were performed as described (Xu and Hamer, 1996). The mating type of each progeny was determined by mating tests and PCR with the MAT1-1–specific primers MATA1 (5′-AGCCTCATCAACGGCAA-3′) and MATA5 (5′-GGCACGAACATGCGATG-3′) and the MAT1-2–specific primers MATB15 (5′-CTCAATCTCCGTAGTAG-3′) and MATB16 (5′-ACAGCAGTATAGCCTAC-3′).
Table 3.
Wild-Type Strains and Mutants of Magnaporthe Used in This Study
| Strain | Brief Description | Reference |
|---|---|---|
| Guy11 | Wild-type MAT1-2 | Leung et al., 1988 |
| nn78 | Δpmk1 of Guy11 | Xu and Hamer, 1996 |
| 2359 | Wild-type MAT1-1 | Leung et al., 1988 |
| 70-15 | Wild-type MAT1-2 | Chao and Ellingboe, 1991 |
| BC32 | Ectopic transformant of Guy11 transformed with pCX4 | This study |
| BE71 | Ectopic transformant of Guy11 transformed with pCX3 | This study |
| BC7 | Δgas1 of Guy11 | This study |
| BC46 | Δgas1 of Guy11 | This study |
| BE55 | Δgas2 of Guy11 | This study |
| BE68 | Δgas2 of Guy11 | This study |
| DS21 | Δgas1 Δgas2 double mutant of Guy11 | This study |
| GC22 | Guy11 transformed with pCX14 (GAS1-GFP) | This study |
| GC24 | Guy11 transformed with pCX14 (GAS1-GFP) | This study |
| GE6 | Guy11 transformed with pCX13 (GAS2-GFP) | This study |
| GE19 | Guy11 transformed with pCX13 (GAS2-GFP) | This study |
| DT2 | GC22 transformed with pCX35 (GAS2-YFP) | This study |
| MCX13 | BC46 transformed with pCX13 (GAS1-GFP) | This study |
| MCX14 | BE55 transformed with pCX14 (GAS2-GFP) | This study |
| MCX27 | Guy11 transformed with pCX27 (GAS2-GFP + 3′ untranslated region) | This study |
Molecular Manipulations with DNA and RNA
Fungal DNAs were extracted using the cetyl-trimethyl-ammonium bromide protocol (Xu and Hamer, 1996). Plasmid DNAs were isolated with Qiagen plasmid preparation kits (Valencia, CA) and sequenced with the ABI Prism BigDye Terminator cycle sequencing kit (PE Applied Biosystems, Foster City, CA). Standard molecular biology procedures were followed for RNA and DNA gel blot analyses and enzymatic manipulations with DNAs and RNAs (Sambrook et al., 1989). Homolog searches of DNA/protein sequence databases were performed with the BLAST programs (Altschul et al., 1997). Amino acid sequence comparisons and alignments were made with the BESTFIT, PILEUP, T-COFFEE, and BOXSHADE programs in the GCG Wisconsin software package (Accelrys, San Diego, CA).
Construction of a Subtraction Library Enriched for Genes Regulated by PMK1
RNAs were prepared with the Trizol reagent (Invitrogen, Carlsbad, CA) from the wild-type strain Guy11 and pmk1 mutant nn78 conidia germinated on wax paper (Reynolds, Richmond, VA) for 18 h. Under these conditions, the wild-type strains form appressoria, but pmk1 mutants form only subapical swollen bodies (Xu and Hamer, 1996). A subtraction library was constructed with the PCR-Select kit (Clontech, Palo Alto, CA) using the cDNA synthesized from Guy11 RNA as the tester and the cDNA synthesized from nn78 as the driver. Subtractive PCR products were cloned in the pGEM-T Easy vector (Promega, Madison, WI). After transformation into Escherichia coli DH5α, 550 white colonies were picked and preserved individually as the subtraction library.
Construction and Differential Screening of an Appressorium-Stage cDNA Library
RNA isolated using the guanidine isothiocyanate method (Sambrook et al., 1989) from conidia of strain 70-15 germinated on cellophane membranes for 6 h was used to synthesize cDNA with a cDNA synthesis kit (Stratagene, La Jolla, CA). After digestion with EcoRI and XhoI, the resulting cDNA was ligated to pBluescript II SK+ (Stratagene) and transformed into E. coli DH10B. A total of 18,432 individual recombinant clones were transferred by a Q-Bot robotic work station (Genetix, New Milton, UK) to 384-well microplates and preserved. High-density filters of this library were hybridized differentially with cDNA probes prepared from total RNAs extracted from vegetative hyphae grown in liquid complete medium and from the appressorial stage. The cDNA clones that hybridized exclusively to the appressorium cDNA probe were selected for sequencing and RNA gel blot analysis.
Construction of the GAS1 and GAS2 Gene Replacement Vectors
The genomic sequences of GAS1 and GAS2 were obtained by sequencing the corresponding cosmid clones isolated from a Guy11 pMOcosX cosmid genomic library (Xu and Hamer, 1996) by primer walking. A 0.8-kb upstream sequence of GAS1 was amplified with PCR primers C4F (5′-AAGGTCCTCAGAGCAGCTTG-3′) and C2R (5′-ATGCTCTCAACATCGTTCTG-3′) and cloned into the pGEM-T Easy vector as pMBCA. The 1.4-kb SalI fragment containing the bacterial hygromycin phosphotransferase gene (hph) was released from plasmid pCB1003 (Carroll et al., 1994) and cloned into the SalI site on pMBCA. The resulting construct was linearized with SacI and ligated with a downstream fragment of GAS1 amplified with primers C2F (5′-TTAAGCGCCTTTCACCTCAA-3′) and C4R (5′-TAGAGCTCT-GTAGGAGCCTCAAGTC-3′) to generate the gene replacement vector pCX4.
A similar approach was used to construct the GAS2 gene replacement vector pCX3. The 0.6-kb upstream sequence amplified with primers E3F (5′-CAATGTGCAAGAGAGTCAGCA-3′) and E2R (5′-AGTGCCATCGATGGCGGTCA-3′) was cloned into pGEM-T Easy as pMBEA. The hph cassette released from pCB1003 and the downstream flanking sequence of GAS2 amplified with primers E2F (5′-GAGGATGACGAGTAGAGTGT-3′) and E4R (5′-GAGTGTTGAGAG-TTGAGACA-3′) were cloned between the KpnI and BamHI sites and the BamHI and BstXI sites on pMBEA, respectively, to generate pCX3.
Isolation of gas1 and gas2 Deletion Mutants
The gene replacement vectors pCX4 and pCX3 were linearized with BstXI and transformed into Guy11 protoplasts as described (Sweigard et al., 1992). Monoconidial cultures of transformants resistant to 150 μg/mL hygromycin B (Calbiochem, La Jolla, CA) were screened by PCR with primers C5F (5′-CTTCACCCGACCTCTCCTAAC-3′) and H854 (5′-AACAATGTCCTGACGGACAA-3′) or E6F (5′-TGGACGTACAAGCATGACTA-3′) and H853 (5′-GACAGACGT-CGCGGTGAGTT-3′). Because the C5F primer sequence was not present in pCX4 (Figure 3), only transformants that had undergone homologous recombination between pCX4 and endogenous GAS1 would have the 1.4-kb PCR product. Similarly, primers E6F and H853 will amplify a 1.1-kb product only in gas2 gene replacement transformants with homologous recombination between pCX3 and GAS2.
All putative gene replacement mutants identified in PCR screens were confirmed further by DNA gel blot analyses. For complementation assays, a 4.2-kb NcoI fragment containing the full-length GAS1 gene (Figure 3) was cloned into pGEM-5Zf (Promega) and cotransformed into the gas1 mutant BC46 with the bleomycin-resistant vector pAC905 (Zheng et al., 2000). Transformants resistant to 200 μg/mL zeocin (Invitrogen) were isolated and screened by PCR with primers C1F and C1R, which are located in the GAS1 region deleted in pCX4 (Figure 3).
Assaying Germination, Appressorium Formation, and Penetration
Conidia were harvested from 10-day-old oatmeal agar cultures, filtered once through Miracloth (Calbiochem), and resuspended to 5 × 104 conidia/mL in sterile distilled water. Drops of conidial suspensions (50 μL) were placed on plastic microscope cover slips (Fisher Scientific, Pittsburgh, PA) and incubated in a moist chamber at room temperature. Conidial germination and appressorium formation were examined after incubating for 0.5, 2, 6, 12, and 24 h. Appressorial penetration on onion epidermal cells was assayed as described previously (Xu et al., 1997). Appressorium formation and infectious hyphal growth were examined after incubation at room temperature for 24 to 72 h.
Plant Infection Assays
Conidia were collected from 10-day-old oatmeal agar cultures and resuspended to 2 × 104 conidia/mL in a 0.25% gelatin solution. Two-week-old seedlings of rice (Oryza sativa cv CO39) and 8-day-old seedlings of barley (Hordeum vulgare cv Golden Promise) were used for infection assays. Plant incubation and inoculation were performed as described (Valent et al., 1991; Xu et al., 1997). Lesion formation was examined 7 days after inoculation. The mean number of lesions formed on 5-cm leaf tips was determined as described previously (Talbot et al., 1993, 1996).
Construction and Expression of Gas1–Green Fluorescent Protein and Gas2–Green Fluorescent Protein Fusion Proteins
The GAS1 coding region and its 1.2-kb upstream sequence were amplified with primers CU3F (5′-TCAAGCTTCCAGTGCTGGTCT-AGTTC-3′) and CU2R (5′-ATGGATCCGCTTTAGCGATCTCG-3′) using Vent DNA polymerase (New England Biolabs, Beverly, MA) and cloned between the HindIII and BamHI sites on pEGFP (Clontech) as pCX13. The stop codon TAA of GAS1 was changed to AAA in the primer CU2R, and the resulting GAS1–green fluorescent protein (GFP) fusion construct was under the control of the native GAS1 promoter. A similar procedure was used to generate the GAS2-GFP fusion construct. The GAS2 open reading frame and its 1.4-kb upstream region were amplified with primers EU2F (5′-ATGGGC-CCTTGCAAATAGAGCTAG-3′) and EU2R (5′-ATGGATCCTTCTCGTCATCCTCCTC-3′) and cloned into pEGFP as pCX14.
We also cloned the PCR product amplified by EU2F and EU2R into pEYFP (Clontech) as pCX26. The GAS2–yellow fluorescent protein (YFP) fusion construct was released from pCX26 with HindIII and NotI digestion and cloned into pCB1004 (Carroll et al., 1994) as pCX35. An 825-bp region downstream from the GAS2 open reading frame was amplified with primers BE5F (5′-TCGCGGCCGCAGGATGACGAGTAGAGTGT-3′) and BE5R (5′-ACGGGCCCGAGTGTT-GAGAGTTGAGACA-3′) and cloned between the NotI and ApaI sites on pCX14 as pCX27. All of the fusion constructs were confirmed by sequencing to be correct and in frame.
pCX13 and pCX14 were introduced individually into Guy11 by cotransformation with pAC905. Zeocin-resistant transformants were screened by PCR with primers GP1F (5′-CATCCTGGTCGAGCT-GGA-3′) and GP1R (5′-CTTGTACAGCTCGTCCATG-3′) and confirmed by DNA gel blot analysis. The expression of GFP was examined on both glass cover slips (Fisher Scientific) and onion epidermis with a Nikon Eclipse E800 epifluorescence microscope (Tokyo, Japan). The Chroma Endow GFP filter on this microscope is suitable to detect both GFP and YFP but not to distinguish them. The subcellular GFP localization also was examined with a Bio-Rad MRC 1024 UV/Vis System confocal laser scanning microscopy (Hercules, CA) on infected barley leaves and glass cover slips. Transformants expressing both GAS1-GFP and GAS2-YFP fusion constructs were generated by transforming pCX35 into GC22, a Guy11 transformant carrying GAS1-GFP.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Accession Numbers
The GenBank accession numbers for GAS1 and GAS2 are AF363065 and AF264035, respectively.
Acknowledgments
We thank Larry Dunkle, Stephen Goodwin, and Charles Woloshuk at Purdue University for critical reading of the manuscript. We also thank Richard Howard at Dupont for useful suggestions on the GFP-tagging experiments and Mark Farman at the University of Kentucky for communicating unpublished data. This work was supported by U.S. Department of Agriculture National Research Initiative Grant 2001-35319-09924 to J.-R.X. and by grants from the National Science Foundation to R.A.D.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.003426.
References
- Altschul, S.F., Madden, T.L., Shaffer, A.A., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourett, T.M., and Howard, R.J. (1990). In vitro development of penetration structures in the rice blast fungus Magnaporthe grisea. Can. J. Bot. 68, 329–342. [Google Scholar]
- Carroll, A.N., Sweigard, J.A., and Valent, B. (1994). Improved vectors for selecting resistance to hygromycin. Fungal Genet. Newsl. 41, 22. [Google Scholar]
- Chao, C.C.T., and Ellingboe, A.H. (1991). Selection for mating competence in Magnaporthe grisea pathogenic to rice. Can. J. Bot. 69, 130–134. [Google Scholar]
- Choi, W., and Dean, R.A. (1997). The adenylate cyclase gene MAC1 of Magnaporthe grisea controls appressorium formation and other aspects of growth and development. Plant Cell 9, 1973–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clergeot, P.H., Gourgues, M., Cots, J., Laurans, F., Latorse, M.P., Pepin, R., Tharreau, D., Notteghem, J.L., and Lebrun, M.H. (2001). PLS1, a gene encoding a tetraspanin-like protein, is required for penetration of rice leaf by the fungal pathogen Magnaporthe grisea. Proc. Natl. Acad. Sci. USA 98, 6963–6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, R.A. (1997). Signal pathways and appressorium morphogenesis. Annu. Rev. Phytopathol. 35, 211–234. [DOI] [PubMed] [Google Scholar]
- de Jong, J.C., McCormack, B.J., Smirnoff, N., and Talbot, N.J. (1997). Glycerol generates turgor in rice blast. Nature 389, 244–245. [Google Scholar]
- DeZwaan, T.M., Carroll, A.M., Valent, B., and Sweigard, J.A. (1999). Magnaporthe grisea pth11p is a novel plasma membrane protein that mediates appressorium differentiation in response to inductive substrate cues. Plant Cell 11, 2013–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro, A., Garcia-Maceira, F.I., Meglecz, E., and Roncero, M.I.G. (2001). A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol. Microbiol. 39, 1140–1152. [PubMed] [Google Scholar]
- Dufresne, M., and Osbourn, A.E. (2001). Definition of tissue-specific and general requirements for plant infection in a phytopathogenic fungus. Mol. Plant-Microbe Interact. 14, 300–307. [DOI] [PubMed] [Google Scholar]
- Eckhard, T., Frank, E., Olov, S., and Heidrun, A. (1998). Inhibitors of appressorium formation in Magnaporthe grisea: A new approach to control rice blast disease. Pestic. Sci. 54, 314–316. [Google Scholar]
- Hamer, J.E., and Talbot, N.J. (1998). Infection-related development in the rice blast fungus Magnaporthe grisea. Curr. Opin. Microbiol. 1, 693–697. [DOI] [PubMed] [Google Scholar]
- Hwang, C.S., Flaishman, M.A., and Kolattukudy, P.E. (1995). Cloning of a gene expressed during appressorium formation by Colletotrichum gloeosporioides and a marked decrease in virulence by disruption of this gene. Plant Cell 7, 183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justesen, A., Somerville, S., Christiansen, S., and Giese, H. (1996). Isolation and characterization of two novel genes expressed in germinating conidia of the obligate biotroph Erysiphe graminis f. sp. hordei. Gene 170, 131–135. [DOI] [PubMed] [Google Scholar]
- Lee, Y.H., and Dean, R.A. (1993). State-specific gene expression during appressorium formation of Magnaporthe grisea. Exp. Mycol. 17, 215–222. [Google Scholar]
- Leung, H., Borromeo, E.S., Bernardo, M.A., and Notteghem, J.L. (1988). Genetic analysis of virulence in the rice blast fungus Magnaporthe grisea. Phytopathology 78, 1227–1233. [Google Scholar]
- Lev, S., Sharon, A., Hadar, R., Ma, H., and Horwitz, B.A. (1999). A mitogen-activated protein kinase of the corn leaf pathogen Cochliobolus heterostrophus is involved in conidiation, appressorium formation, and pathogenicity: Diverse roles for mitogen-activated protein kinase homologs in foliar pathogens. Proc. Natl. Acad. Sci. USA 96, 13542–13547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mey, G., Oesey, B., Lebrun, M.H., and Tudzynski, P. (2002). The biotrophic, nonappressorium-forming grass pathogen Claviceps purpurea needs a Fus3/Kss1 homologous MAP kinase for colonization of rye ovarian tissue. Mol. Plant-Microbe Interact. 15, 303–312. [DOI] [PubMed] [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. (1997). Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10, 1–6. [DOI] [PubMed] [Google Scholar]
- Park, G., Xue, C., Zheng, L., Lam, S., and Xu, J.R. (2002). MST12 regulates infectious growth but not appressorium formation in the rice blast fungus Magnaporthe grisea. Mol. Plant-Microbe Interact. 15, 183–192. [DOI] [PubMed] [Google Scholar]
- Ruiz-Roldan, M.C., Maier, F.J., and Schafer, W. (2001). PTK1, a mitogen-activated-protein kinase gene, is required for conidiation, appressorium formation, and pathogenicity of Pyrenophora teres on barley. Mol. Plant-Microbe Interact. 14, 116–125. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sweigard, J.A., Chumley, F.G., and Valent, B. (1992). Cloning and analysis of CUT1, a cutinase gene from Magnaporthe grisea. J. Gen. Microbiol. 232, 174–182. [DOI] [PubMed] [Google Scholar]
- Takano, Y., Kikuchi, T., Kubo, Y., Hamer, J.E., Mise, K., and Furusawa, I. (2000). The Colletotrichum lagenarium Map kinase gene CMK1 regulates diverse aspects of fungal pathogenesis. Mol. Plant-Microbe Interact. 13, 374–383. [DOI] [PubMed] [Google Scholar]
- Talbot, N.J., Ebbole, D.J., and Hamer, J.E. (1993). Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell 5, 1575–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot, N.J., Kershaw, M.J., Wakley, G.E., de Vries, O.M.H., Wessels, J.G.H., and Hamer, J.E. (1996). MPG1 encodes a fungal hydrophobin involved in surface interactions during infection-related development of Magnaporthe grisea. Plant Cell 8, 985–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban, M., Bhargava, T., and Hamer, J.E. (1999). An ATP-driven efflux pump is a novel pathogenicity factor in rice blast disease. EMBO J. 18, 512–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valent, B. (1990). Rice blast as a model system for plant pathology. Phytopathology 80, 33–36. [Google Scholar]
- Valent, B., Farral, L., and Chumley, F.G. (1991). Magnaporthe grisea genes for pathogenicity and virulence identified through a series of backcrosses. Genetics 127, 87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J.R. (2000). MAP kinases in fungal pathogens. Fungal Genet. Biol. 31, 137–152. [DOI] [PubMed] [Google Scholar]
- Xu, J.R., and Hamer, J.E. (1996). MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev. 10, 2696–2706. [DOI] [PubMed] [Google Scholar]
- Xu, J.R., Urban, M., Sweigard, J.A., and Hamer, J.E. (1997). The CPKA gene of Magnaporthe grisea is essential for appressorium penetration. Mol. Plant-Microbe Interact. 10, 187–194. [Google Scholar]
- Zheng, L., Campbell, M., Murray, J., Lam, S., and Xu, J.R. (2000). The BMP1 gene is essential for pathogenicity in the gray mold fungus Botrytis cinerea. Mol. Plant-Microbe Interact. 13, 724–732. [DOI] [PubMed] [Google Scholar]