Abstract
ANTHOCYANIN1 (AN1) of petunia is a transcription factor of the basic helix-loop-helix (bHLH) family that is required for the synthesis of anthocyanin pigments. Here, we show that AN1 controls additional aspects of cell differentiation: the acidification of vacuoles in petal cells, and the size and morphology of cells in the seed coat epidermis. We identified an1 alleles, formerly known as ph6, that sustain anthocyanin synthesis but not vacuolar acidification and seed coat morphogenesis. These alleles express truncated proteins lacking the C-terminal half of AN1, including the bHLH domain, at an ∼30-fold higher level than wild-type AN1. An allelic series in which one, two, or three amino acids were inserted into the bHLH domain indicated that this domain is required for both anthocyanin synthesis and vacuolar acidification. These findings show that AN1 controls more aspects of epidermal cell differentiation than previously thought through partially separable domains.
INTRODUCTION
The synthesis of anthocyanin pigments in floral tissue provides an excellent marker to study the differentiation of cells. Most of the structural genes that encode the ∼15 enzymes required for anthocyanin synthesis and modification have been isolated (Holton and Cornish, 1995; Winkel-Shirley, 2001). By mutational analyses in Antirrhinum, Arabidopsis, maize, and petunia, several regulatory genes have been identified that are required for the tissue-specific transcription of the structural anthocyanin genes (for reviews, see Mol et al., 1998; Winkel-Shirley, 2001).
ANTHOCYANIN1 (AN1) of petunia encodes a basic helix-loop-helix (bHLH) protein that activates the transcription of the structural anthocyanin gene DIHYDROFLAVONOL REDUCTASE (DFR) and a putative regulatory gene (MYB27) whose function is unknown (Spelt et al., 2000). The JAF13 gene encodes a bHLH protein that is homologous with DELILA (DEL) from snapdragon and R from maize (Quattrocchio et al., 1998) but functionally and evolutionary distinct from AN1 (Spelt et al., 2000). The expression of AN1 is regulated by AN2 and AN4 (Spelt et al., 2000).
AN2 is expressed in the petal limb only and encodes a MYB domain protein (Quattrocchio et al., 1999) that is functionally interchangeable with C1 from maize (Quattrocchio et al., 1998) and that may be orthologous with either TRANSPARENT TESTA2 (TT2) (Nesi et al., 2001) or PRODUCTION OF ANTHOCYANIN PIGMENT1 (PAP1) and PAP2 (Borevitz et al., 2000) from Arabidopsis. AN4 encodes (or controls the expression of) a paralogous MYB protein that is expressed in anthers (C. Spelt, A. Kroon, and R. Koes, unpublished data). The activity of one or more of these transcription factors appears to be regulated post-transcriptionally by a cytosolic WD40 repeat protein encoded by AN11 in petunia and by TRANSPARENT TESTA GLABRA (TTG) in Arabidopsis (de Vetten et al., 1997; Walker et al., 1999).
In Arabidopsis, two other transcription factors, the homeodomain protein ANTHOCYANINLESS2 (ANL2) and the zinc finger protein TT1, have been implicated in the accumulation of proanthocyanidin polymers in the seed coat (Kubo et al., 1999; Sagasser et al., 2002). However, it is unclear whether ANL2 and TT1 coregulate the anthocyanin-specific genes together with the MYB, bHLH, and WD40 proteins or control a distinct set of structural anthocyanin genes.
Although the regulatory genes mentioned above were identified initially by their role in anthocyanin synthesis, an increasing amount of evidence indicates that they control additional aspects of cell differentiation. Expression of a C1-R fusion protein in maize suspension cells causes the activation or repression of hundreds of genes (Bruce et al., 2000), many more than are required for anthocyanin synthesis. In Arabidopsis, ttg1 mutations cause the loss of trichomes on leaves and stems, whereas in roots, they cause the formation of ectopic root hairs (Galway et al., 1994).
Ectopic expression of R in Arabidopsis has the opposite effect: the formation of extra trichomes in stems and leaves (Lloyd et al., 1992, 1994) and a reduction of root hair formation (Galway et al., 1994). Mutation of ANL2 also affects root architecture, causing the formation of extra cells between the cortex and the epidermis (Kubo et al., 1999). Interestingly, in petunia, an1 or an11 mutations do not have a clear effect on the formation of trichomes or root hairs, but they affect seemingly unrelated cellular processes: the acidification of the vacuole and the morphogenesis of the seed coat epidermis (this work).
The vacuole of plant cells plays a central role in pH homeostasis, osmoregulation, ion transport, and the sequestration of (toxic) metabolites (Taiz, 1992). The vacuolar membrane contains ATPase and pyrophosphatase proton pumps that actively acidify the vacuole, whereas the pH of the cytoplasm is kept approximately neutral. The resulting electrochemical gradient serves as a driving force for the uptake of various compounds by transporters and channels in the vacuolar membrane. Despite the physiological importance of vacuolar acidification, little is known about the mechanisms that regulate this process.
Because anthocyanins accumulate in vacuoles and because their absorption spectrum depends on pH, they provide a natural indicator of vacuolar pH (Mol et al., 1998). For example, the change from purple to blue during the development of Ipomoea flowers correlates with an increase in vacuolar pH (Yoshida et al., 1995) and requires the Na+/H+ exchanger PURPLE (Fukada-Tanaka et al., 2000), which is thought to consume the existing pH gradient, resulting in alkalinization of the vacuolar content.
Petunia flowers normally do not turn blue upon opening, and their vacuolar pH stays on the reddish (low-pH) side of the anthocyanin color spectrum. By genetic analyses, seven loci, designated PH1 to PH7, were identified that mutate to flowers with a more bluish color (de Vlaming et al., 1983; van Houwelingen et al., 1998). Because homogenates prepared from such flowers have higher pH, it is thought that these loci are required for the acidification of the vacuole (de Vlaming et al., 1983). Only one of these PH genes has been isolated (PH6; Chuck et al., 1993), but because no cDNA clones were isolated, the nature of the gene product remained unknown.
In this article, we show that AN1, AN2, and AN11 control, in addition to anthocyanin synthesis, the vacuolar pH in petal cells and the morphology of the seed coat epidermis. Our finding that ph6 mutants do not define a separate locus, as was thought previously, but represent specific alleles of AN1 indicates that AN1 controls anthocyanin synthesis and vacuolar pH by distinct mechanisms.
RESULTS
Mutations at AN1, AN2, and AN11 Increase the pH of Flower Homogenates
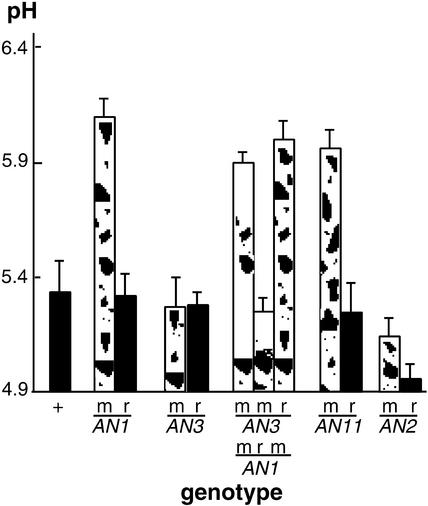
The petunia line W138 arose from the parental line R27 by the insertion of a dTph1 transposon in the AN1 gene and bears white flowers with red and pink revertant spots. During the phenotypic analysis of some new ph mutants that were identified among W138 progeny, we noticed that the an1-W138 mutation increased the pH of petal limb homogenates to a similar extent as did mutations in PH3 and PH4 (Quattrocchio, 1994). To establish the significance of this observation, we measured the pH of petal limb homogenates for a range of genotypes.
Figure 1 shows that extracts from an1-W138 homozygous flowers have a pH value that is ∼0.7 units higher than flowers homozygous for the AN1-R27 progenitor allele or from independently isolated AN1/an1-W138 revertant plants. To determine whether this pH shift is attributable to the absence of anthocyanins, we measured extracts of flowers harboring the unstable an3-R134 allele.
Figure 1.
Mutations in Regulatory Anthocyanin Genes Increase the pH of Petal Homogenates.
pH values (means ± sd; n ≥ 4) were measured in petal homogenates of various genotypes, as indicated on the horizontal axis: +, wild-type allele(s); m, mutable (unstable) alleles; r, derived full-revertant alleles.
AN3 encodes the enzyme flavanone 3β-hydroxylase (F3H), which catalyzes the conversion of flavanones to the corresponding dihydroflavonols, and an3 mutations block anthocyanin synthesis well before the an1 block (van Houwelingen et al., 1998). Extracts of an3-R134 and AN3 revertant flowers had pH values comparable to that of the wild-type progenitor (R27). Similar results were obtained for mutants containing two other an3 alleles (an3-S205 and an3-S206; data not shown) and for transgenic plants in which CHS genes, which encode chalcone synthase, the first enzyme of flavonoid metabolism, were cosuppressed.
Because the an3 mutation blocks the anthocyanin pathway at an earlier step than an1 mutations, an3 and an1 an3 double mutants accumulate the same flavonoids (eriodictyol in the R27 background). Even in an an3 background, the effect of the an1-W138 mutation on flower extract pH was observed (Figure 1; compare an1 an3 and AN1revan3). Thus, the pH shift observed in an1-W138 flowers cannot be explained by the absence of anthocyanins.
To determine whether AN2 and AN11 also are involved in pH control, we analyzed unstable mutants and revertants for these loci. Flowers harboring the unstable an11-W137 allele (de Vetten et al., 1997) in the R27 background had an increased pH value similar to that of an1-W138 flowers, whereas in AN11 revertant flowers, the pH shifted back to wild-type values (Figure 1). For the AN2 gene, we measured pH values in the line W82, which harbors an unstable an2 allele, and used isogenic AN2 revertants as a control. This showed that the an2-W82 mutation increased the pH value by 0.3.
Similar results were obtained when we compared pH values in a stable recessive an2 mutant and isogenic AN2 plants in which the mutation was complemented by a 35S:AN2 transgene (Quattrocchio et al., 1998). The limited effect of an2 on anthocyanin synthesis and flower extract pH presumably is caused by the partial redundancy of AN2 function (Quattrocchio et al., 1998, 1999).
Analysis of Functional AN1 Domains by in Vivo Mutagenesis
To identify functional domains in the AN1 protein, we exploited transposons to generate mutant an1 alleles by in vivo mutagenesis. The AN1 gene appears to be a focus for transposon insertions, which enabled us to isolate >30 independent unstable an1 alleles in random and directed transposon mutagenesis experiments by screening for mutants in which flower pigmentation was altered (van Houwelingen et al., 1998; this work).
We analyzed these unstable an1 alleles for: (1) molecular alterations in the structure and expression of an1, (2) their capacity to drive anthocyanin synthesis and the expression of the structural anthocyanin gene DFR, and (3) their capacity to regulate intracellular pH and drive the expression of PAT1, a gene of unknown function that is downregulated in ph4 and ph3 mutants (F. Quattrocchio, I. Roobeek, W. Verweij, C. Spelt, J. Mol, and R. Koes, unpublished data). The results for the most informative alleles are presented below.
The Wild-Type AN1 Allele Expresses Distinct mRNA Species That Result from Alternative Polyadenylation
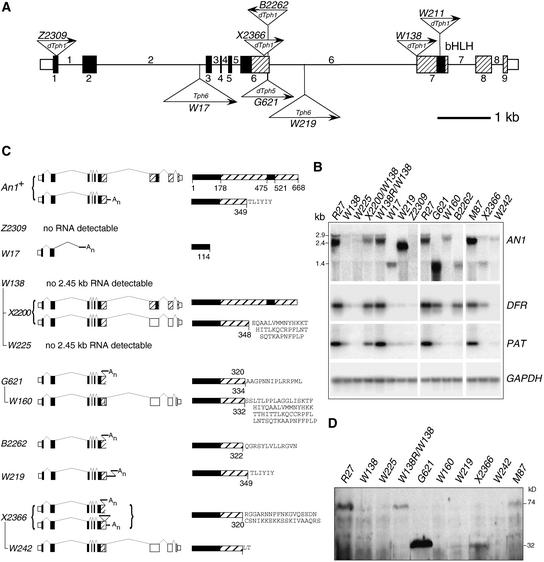
Line R27 bears full-colored (red) flowers with a pH of 5.4 ± 0.1 (Figure 2A) and contains a functional AN1 gene consisting of nine exons (Figure 3A). RNA gel blot analysis showed that R27 petal limbs contain a major AN1 mRNA of 2.45 kb (Figure 3B) that encodes a 668–amino acid protein with two well-conserved domains: (1) an ∼170–amino acid domain at the N terminus that is conserved in a number of plant bHLH proteins, including R, DEL, and JAF13; and (2) a bHLH domain in the C-terminal half of the protein (Spelt et al., 2000) (Figure 3C). In addition, R27 expresses small amounts of AN1 transcripts of ∼2.9 and ∼1.4 kb; the latter transcript was particularly evident in reverse transcriptase–mediated (RT)-PCR products because it is amplified more efficiently than the larger transcripts.
Figure 2.
Phenotypes of Flowers Harboring Different an1 Alleles.
Each image shows part of a flower and the pH value of its petal extracts at right (means ± sd; n ≥ 3). The an1 alleles present in each flower are indicated in italic type at top. For flowers that are homozygous for an an1 allele, only a single allele number is given. For flowers that are heterozygous, the numbers of both an1 alleles are given separated by a slash.
Figure 3.
Molecular Analysis of an1 Alleles.
(A) Structure of the AN1 gene and mutant alleles. Exons are indicated by rectangles, and introns are indicated by a horizontal line. Exonic regions that encode the conserved N-terminal domain of AN1 and the bHLH region are shaded black; regions encoding less conserved parts of AN1 are indicated by hatching. Exon regions that are not translated are indicated by rectangles of half the height. Triangles (not drawn to scale) denote transposon insertions in the indicated alleles; their orientations are indicated with arrows (arrows pointing right indicate that the orientation of the transposon relative to AN1 is the same as the sequences in the corresponding GenBank accessions).
(B) Analysis of RNAs expressed in AN1 and an1 flowers. RNAs of several genes (indicated at right) were detected by RNA gel blot analysis (AN1 mRNA) or RT-PCR (DFR, PAT, and GAPDH mRNA). For all alleles (indicated above the lanes) homozygous flowers were used, except for X2200 and W138R, which were heterozygous over the parental W138 allele.
(C) Structure of mRNAs and proteins expressed by an1 alleles. The numbers of the alleles are indicated at left, the structure of their mRNAs are indicated in the middle and the encoded proteins are indicated at right. Exons and transposon sequences are drawn as in (A); protein-coding sequences that are not translated in the mutant mRNA are shaded white. The mRNA splicing patterns are indicated by thin lines. The lines connecting the allele numbers indicate how distinct excision alleles derived from the original transposon insertion alleles.
(D) Gel blot analysis of AN1 proteins expressed in an1 mutants. The flowers analyzed were homozygous for the alleles indicated above the lanes, except for W138R, which was heterozygous over the parental mutable allele W138.
Rapid amplification of cDNA ends (Frohman et al., 1988) showed that the 1.4-kb an1 transcript resulted from premature polyadenylation in intron 6 and encodes a truncated AN1 protein (Figure 3C). Given that the removal of intron 7 is relatively slow (Spelt et al., 2000), we assume that the 2.9-kb an1 RNAs represent incompletely spliced transcripts.
an1 Alleles That Disrupt Both Short and Long AN1 mRNAs
To identify null alleles, we analyzed an1 alleles with mutations in the 5′ half of the gene, because such mutations affect both the major 2.45-kb mRNA and the minor 1.4-kb mRNA. The unstable allele an1-Z2309 contains a dTph1 insertion in exon 1 and represents the most 5′ disruption of the an1 gene that we found (Figure 3A). RNA analyses showed that an1-Z2309 flowers are completely devoid of detectable an1 transcripts (Figure 3B), indicating that it is a null allele. an1-Z2309 flowers are white (and dotted with red revertant spots) and have a high petal extract pH (Figure 2B), which correlates with a strong reduction of DFR and PAT1 mRNA levels (Figure 3B).
The an1-W17 allele, which is present in the R27 background, contains an insertion of a novel 3800-bp petunia transposon of the CACTA family, designated Tph6 (a more detailed characterization of Tph6 will be published elsewhere), that blocks expression of the wild-type 1.4- and 2.45-kb mRNAs and results in the expression of a 1.3-kb AN1 mRNA (Figures 3A and 3C). RT-PCR and 3′ rapid amplification of cDNA ends analyses indicated that this RNA arose by premature polyadenylation within Tph6, followed by the splicing of exon 2 to a cryptic splice site in Tph6; it encodes a truncated AN1 protein of 114 amino acids (Figure 3C). Flowers homozygous for an1-W17 are white (and contain few revertant spots), have a high petal pH (Figure 2C), and express little or no DFR and PAT mRNA (Figure 3B), indicating that the W17 mutation is similar in effect to Z2309 and is essentially a null allele.
The Long AN1 mRNA Is Essential to Direct Anthocyanin Synthesis and Vacuolar Acidification
To assess the contribution of the different AN1 mRNA species to flower pigmentation, we analyzed mutations that specifically affected the accumulation of the long 2.45-kb mRNA. The unstable allele an1-W138 specifies white flowers with a high petal pH that are dotted with pink and red spots (Figure 2D) as a result of a dTph1 insertion in the splice acceptor site of exon 7 (Spelt et al., 2000) (Figure 3A). This insertion did not interfere with the expression of the small 1.4-kb an1 mRNA (which is polyadenylated prematurely at a site upstream of the mutation), but it completely abolished the full-size 2.45-kb AN1 mRNA. This resulted in a strong reduction of both DFR and PAT1 expression (Figure 3B).
The dTph1 insertion in an1-W138 duplicated the acceptor splice of exon 7, and the derived excision alleles contain footprints that contain zero, one, two, or three potential acceptor splice sites at this position (Spelt et al., 2000). Full AN1 revertants (an1-W138R) contain a single splice site that results in the formation of a wild-type amount of the 2.45-kb AN1 mRNA (Figure 3B) with wild-type sequence (data not shown) (Spelt et al., 2000). By contrast, the excision allele an1-W225 harbors a 7-bp footprint that contains three potential splice sites. Apparently, most, if not all, of the long an1 mRNAs are misspliced on one of the alternative splice sites and degraded subsequently, because very low amounts of 2.45-kb an1 mRNAs were detected in W225 flowers, besides the normal amount of 1.4-kb prematurely polyadenylated transcripts (Figure 3B).
an1-W225 specifies white flowers with an increased petal pH and strongly reduced DFR and PAT1 mRNA levels (Figures 2E and 3B), similar to the null alleles Z2309 and W17. Because an1-W138 and an1-W225 still express the 1.4-kb AN1 mRNA but nevertheless have a null phenotype, we infer that the 1.4-kb AN1 mRNA has, at the levels at which it is expressed normally, little or no effect on the synthesis of anthocyanins and vacuolar acidification.
To examine how a reduction of the 2.45-kb an1 mRNA influenced anthocyanin synthesis and vacuolar acidification, we analyzed the partial revertants of an1-W138. Partial revertant alleles of an1-W138, such as an1-X2200, contain both the original and a potential alternative acceptor splice site (Spelt et al., 2000). RNA analysis showed that an1-X2200/an1-W138 heterozygous flowers express an amount of 2.45-kb mRNAs that is reduced approximately twofold compared with that expressed by full AN1/an1-W138 revertants (Figure 3B). Direct sequencing of RT-PCR products showed that these transcripts are an ∼1:1 mixture of two mRNAs, one in which exons 6 and 7 are spliced in frame on the original splice site and a second in which exons 6 and 7 are spliced out of frame on the alternative splice site (Figure 3C). Thus, in X2200 flowers, the amount of wild-type AN1 mRNA is reduced approximately fourfold.
Interestingly, an1-X2200 encodes a pink (i.e., light red) flower color, indicating that anthocyanin synthesis is diminished, whereas the pH value is similar to that of wild-type R27 (Figure 2F). Thus, anthocyanin synthesis is affected more strongly by reduced AN1 expression than is vacuolar pH, even though the amount of DFR and PAT mRNA is reduced equally (approximately twofold).
Mutant an1 Alleles That Partially Separate the Anthocyanin and pH Function
In the early 1980s, an F2 progeny of two red-flowering petunia lines (family G621) segregated 25% mutants with a purplish flower color. Because these mutants had an increased pH of petal homogenates and complemented mutants for ph1 to ph5, the mutation was assumed to define a new PH locus, which was designated PH6 (P. de Vlaming, unpublished results). Among G621 progeny, a new white-flowering mutant arose, which founded line W160. Allelism tests showed that W160 contained a stable recessive an1 allele, an1-W160; therefore, it was assumed to be an an1 ph6 double mutant (P. de Vlaming, unpublished data).
More than a decade later, Chuck et al. (1993) generated by mutagenesis with the maize transposon ACTIVATOR (AC) an unstable ph mutation that was allelic to that in W160 and G621 plants and concluded that they had tagged the PH6 locus. Using this AC-tagged ph6-m1 allele, Chuck et al. (1993) isolated a fragment of the PH6 gene, but no information on the encoded gene product was provided. However, when we received the sequence of a partial PH6 cDNA (T. Holton and H. Dooner, unpublished data) during the course of this research, we found to our surprise that it was nearly identical to the sequence that we had determined for AN1 (Spelt et al., 2000), except for a few sequence alterations that might be attributed to the different lines used for the cDNA cloning.
To determine whether ph6 mutants represent specific alleles of AN1, we analyzed the structure of the AN1 gene in progeny of the original G621 plants. The progeny of two of these mutants consisted of 34 plants that all bore purplish flowers with red spots (Figure 2G). DNA analysis indicated that they harbored a mutant an1 allele (an1-G621) that contained near the end of exon 6 an insertion of a new 873-bp transposon (Figure 3A) that we designated dTph5 (details on dTph5 will be published elsewhere). Molecular analysis of the stable an1-W160 allele, which arose in G621 progeny, showed that the dTph5 element had excised and produced an 8-bp footprint that disrupts the AN1 coding sequence (data not shown).
Because these results suggested that the G621 mutant represents a dTph5 insertion allele of AN1, rather than an allele of a separate PH6 locus, we performed complementation experiments. Crosses between G621 mutants and null an1 mutants (an1-W225 and an1-W138) all yielded progeny with a (purplish) ph mutant flower color rather than a (red) wild-type color, indicating that the G621 mutation was allelic to an1 (Figure 2H). Therefore, we concluded that ph6 mutants do not define a separate locus; rather, they represent particular alleles of AN1 that lost the control of vacuolar pH but retained the capacity to direct the nearly normal synthesis of anthocyanins. Consistent with this conclusion, DFR mRNA expression was reduced approximately twofold in an1-G621 petals, whereas PAT1 mRNA was abolished almost completely (Figure 3B). In derived an1-W160 mutants (Figure 2I), both DFR and PAT1 were downregulated strongly (Figure 3B).
RNA analysis showed that an1-G621 expresses a 1.4-kb an1 transcript that is polyadenylated prematurely in the dTph5 sequence (Figure 3C). This truncated transcript appears to be relatively stable, because it accumulates at threefold higher levels than the wild-type 2.45-kb an1 mRNA (Figure 3B). By contrast, the excision allele W160 expresses a reduced amount (∼30% of that in the wild-type line R27) of a 2.45-kb mRNA harboring an 8-bp footprint that shifts the reading frame (Figures 3B and 3C).
From the transposon mutagenesis screens, we isolated two additional an1 alleles, B3196 and B2262, with a strong effect on pH control and a mild effect on anthocyanin synthesis (Figure 2J and data not shown), which is reflected by a mild reduction of DFR mRNA and an almost complete reduction of PAT1 mRNA (Figure 3B). B2262 and B3196 contain a dTph1 insertion in exon 6 in the inverted orientation (Figure 3A and data not shown). This results in premature polyadenylation of the an1 mRNA in dTph1 sequences and the formation of a small amount of mRNAs encoding a truncated AN1 protein (Figures 3B and 3C). The AC-induced ph6-m1 allele also expresses a truncated transcript, but its structure and the sequence of the encoded protein are not known (Chuck et al., 1993).
Mutations That Truncate an1 mRNA and Reduce Both Anthocyanin Synthesis and Vacuolar Acidification
Among the an1 alleles isolated were several in which the AN1 coding sequence was truncated at a similar point as in the ph6 alleles but that nevertheless resulted in a strong reduction of anthocyanin synthesis. The allele an1-W219 had been isolated in progeny of W17 and encodes white flowers with red spots and an increased petal pH (Figure 2K). DNA analyses showed that an1-W219 contains a Tph6 insertion in intron 6 (Figure 3A), suggesting that it arose from an1-W17 by intragenic transposition. an1-W219 petals contain an1 transcripts of ∼2.1 kb that result from premature polyadenylation within Tph6 and that encode a truncated protein (Figures 3B and 3C).
Among the progeny of line M87 (Figure 2L), we found the unstable allele an1-X2366 containing a dTph1 insertion in exon 6 (Figure 3A). an1-X2366 clearly is not a null allele, because the flowers have a “blush” of variable intensity, dotted with full-colored revertant spots (Figure 2M). This incomplete block in anthocyanin synthesis is reflected by the relatively large amount of residual DFR mRNAs in X2366 homozygous flowers.
Strikingly, the X2366 mutation has a much stronger effect on PAT1 expression, because PAT1 mRNA is abolished almost completely (Figure 3B). X2366 petals express a small amount (∼20% of the amount in the isogenic wild-type line M87) of two truncated an1 transcripts that result from premature polyadenylation at two different sites: one in the dTph1 element and a site downstream of dTph1 within the intron. Both mRNAs encode an identical 345–amino acid protein (Figure 3C).
Excision of dTph1 from X2366 produced a stable recessive allele, an1-W242, that is stronger than the parental X2366 allele, because it encodes completely white flowers with a slightly higher petal pH (Figure 2N) and a complete reduction of both DFR and PAT1 expression (Figure 3B). an1-W242 expresses a small amount of full-size an1 transcript (∼10% of the amount in the isogenic An1+ line M87) (Figure 3B) that, as a result of the frameshift caused by the 7-bp footprint, encodes a truncated protein of 322 amino acids (Figure 3C).
Analysis of Mutant AN1 Proteins
Among the an1 alleles isolated were several that contained mutations that truncated the C-terminal end of AN1 in approximately the same region. These alleles all fail to drive vacuolar acidification and expression of PAT1 but show extensive differences in their capacity to activate anthocyanin synthesis and DFR expression in the order (from high to low): G621/B2262 > X2366 > W219/W160/W242. To solve this apparent discrepancy, we raised antibodies to AN1 and analyzed the AN1 proteins that accumulated in the mutants.
On protein blots, the anti-AN1 serum detected several bands (Figure 3D). The ∼74-kD protein detected in AN1 petals of R27, M87, and a full-colored revertant of W138 is the AN1 protein, because it is of the expected size and because this protein is missing in all an1 mutants. The other bands that were detected in these AN1 lines are not derived from AN1, because they were detected in all mutants, including those that lack AN1 mRNA.
In an1-G621 petals, we detected a new 32-kD protein, a size expected on the basis of the an1-G621 mRNA sequence (Figure 3D). Apparently, this mutant AN1 protein is very stable, because it is accumulated at ∼30-fold higher levels than wild-type AN1. A similarly sized protein was detected in X2366 and W219 petals, but at much lower levels (∼1-fold and 0.1-fold the amount of wild-type protein), but not in W242 and W160 petals (Figure 3D). Thus, the different capacities of these an1 alleles to induce anthocyanin synthesis and DFR expression, which is in the order G621 > X2366 > W219 > W160/W242, correlate perfectly with the amounts of truncated AN1 protein that are accumulated in petal cells.
Transient Expression of an1 Alleles
Because the protein analyses described above indicated that the different effects on anthocyanin synthesis of the mutant proteins G621 and X2366 are caused by differences in the stability (and hence the steady state levels) of these truncated proteins, rather than by differences in specific activity, we suspected that in vivo the AN1-G621 protein has a reduced capacity to activate anthocyanin synthesis that is compensated for by its ∼30-fold overaccumulation.
To reduce the contribution of the high protein stability and the consequent high protein levels to the activities that are measured, we expressed mutant an1 alleles from the strong and nearly constitutive 35S promoter of Cauliflower mosaic virus and assayed the activation of the DFR promoter in transient expression assays. The rationale behind these experiments is that the 35S promoter causes strong expression of all alleles, whereas the short time span of the experiment (24 h, compared with several days for the development of a mature flower on the plant) reduces the large differences in protein levels that result from differences in protein stability.
To generate 35S-driven gene constructs for distinct an1 alleles, we placed cDNA fragments amplified from mutant petals between the 35S promoter and the 3′ end of the NOPALINE SYNTHASE gene (see Methods). Each construct was introduced into leaf cells, in combination with a 35S-driven AN2 gene (35S:AN2), a LUCIFERASE reporter gene driven by the DFR promoter (DFR:LUC), and a constitutively expressed control gene (35S:β-GLUCURONIDASE) to correct for variations in transformation efficiency.
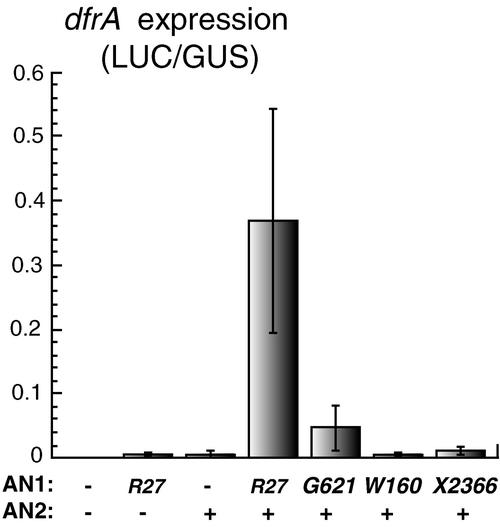
Figure 4 shows that coexpression of AN1 and AN2 strongly induced DFR:LUC expression in leaf cells, whereas either factor alone did not, consistent with previous results (Quattrocchio et al., 1998, 1999; Spelt et al., 2000). When we expressed, instead of the wild-type AN1 mRNA, the truncated mRNA encoded by an1-G621, the induction of DFR:LUC diminished ∼10-fold, indicating that AN1-G621 induces the DFR promoter less efficiently than wild-type AN1 does. Expression of AN1-X2366 induced the DFR promoter even more weakly, whereas AN1-W160 had no detectable activity at all.
Figure 4.
Transient Expression Assays of Mutant an1 Alleles.
The bars denote luciferase (LUC) activity expressed from a DFR:LUC reporter gene, normalized for β-glucuronidase (GUS) activity expressed from a codelivered 35S:GUS gene (means ± se; n = 5) in particle-bombarded leaves. Both reporters were codelivered with effector genes expressing AN2 and different mutant AN1 proteins from the 35S promoter, as indicated on the horizontal axis. A minus sign indicates that the corresponding effector was omitted.
These results indicate that AN1-G621 has a reduced capacity to induce the DFR promoter, which in vivo apparently is compensated for by the ∼30-fold overexpression of the protein, resulting in nearly normal DFR mRNA expression levels.
An Allelic Series Resulting from Alterations in the bHLH Domain
To assess the importance of the bHLH domain for AN1 function in greater detail, we exploited transposon insertions in the region encoding this domain. Among the many new pigmentation mutants found in W138 progeny were several that specified a reduced number of revertant spots that was clearly different from the pattern seen in the W138 parent. Genetic evidence (i.e., the segregation of spotting patterns in the original families and the results from allelism tests) showed that these new phenotypes were attributable to the intragenic transposition of the dTph1 insertion in the an1-W138 allele.
PCR and sequence analyses of one of these alleles, an1-W211, showed that the dTph1 element had excised from the original position at the border of intron 6 and exon 7 and created a footprint that would have restored an1 function (Figure 2T) if a dTph1 insertion had not disrupted the region encoding the first helix of the bHLH domain. This bHLH insertion apparently creates a null allele, because an1-W211 homozygous petal limbs lack anthocyanins, have increased pH (Figure 2O), and do not express DFR and PAT1 mRNA (Figure 5).
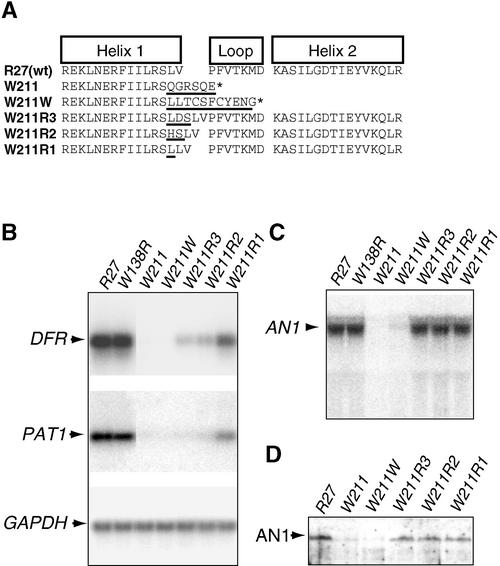
Figure 5.
Molecular Analysis of bHLH Domain Mutants of AN1.
(A) Sequences of the bHLH domains encoded by the wild-type (wt) allele R27, the transposon insertion allele W211, and derived excision alleles (W211R1 to W221W). Regions in which the protein differs from the wild type are underlined. The asterisk denotes the end of the protein.
(B) RT-PCR analysis of mRNAs expressed from the AN1-controlled genes DFR and PAT1 and a housekeeping gene (GAPDH) in the corolla limbs of an1 mutants. The analyzed corolla limbs were homozygous for alleles R27 and W211. Alleles W211W to W211R3 were heterozygous over the parental mutable allele W211, whereas the full revertant allele, W138R, was heterozygous over the parental allele W138.
(C) Analysis of the an1 transcripts expressed in the corolla limbs described in (B). The arrowhead indicates the wild-type 2.45-kb AN1 mRNA.
(D) Gel blot analysis of AN1 proteins expressed in corolla limbs of the mutants described in (B).
Somatic and germinal excisions of dTph1 from an1-W211 produced a series of four new stable an1 alleles with activities ranging from null to nearly wild type (Figures 2P to 2S). The an1-W211W allele specifies white flowers that completely lack DFR and PAT mRNA and a petal homogenate pH that is increased by 0.6 (Figure 2P), indicating that this is a null allele. PCR and sequence analysis showed that this allele contains a 7-bp footprint that results in a shift of the AN1 reading frame. The an1-W211R3 allele has some remaining activity and specifies light pink flowers in which the amount of DFR and PAT mRNA is reduced strongly (to 10% of the wild-type level); it also produces homogenates in which the pH is increased by 0.4 (Figures 2Q and 5B).
DNA analysis showed that an1-W211R3 arose from an1-W211 by excision of the dTph1 element, which resulted in the formation of a 9-bp footprint and the insertion of three amino acids in the bHLH domain (Figure 5A). The allele an1-W211R2 specifies pink flowers that contain 20% DFR mRNA (relative to the wild type) and produce extracts in which the pH is increased by 0.2 (Figure 2R). PCR and sequence analysis showed that this allele contained a 6-bp footprint resulting in the insertion of two amino acids in AN1. The an1-W211R1 allele, which specifies dark pink flowers with wild-type pH (Figure 2S) and nearly wild-type levels of DFR mRNA (∼70% of the amount in the wild type), contains a 3-bp footprint resulting in the insertion of one extra amino acid in AN1 (Figures 5A and 5B).
RNA gel blot analyses (Figure 5C), RT-PCR experiments (data not shown), and protein gel blot experiments (Figure 5D) showed that the three in-frame excision alleles (W211R1, W211R2, and W211R3) all produce wild-type amounts of an1 mRNA and AN1 protein, indicating that the different phenotypes result from a differences in the activity, not the amount, of the encoded AN1 protein. The dTph1 insertion in an1-W211 and the derived frameshift in an1-W211W result in strongly reduced amounts of an1 mRNA and AN1 protein, which explains why these are null alleles.
Regulatory Anthocyanin Genes Control Cell Shape in the Seed Coat
Earlier work indicated that AN1 controls the coloration not only of floral organs but also of seeds (Chuck et al., 1993; Quattrocchio et al., 1993). However, we noticed that the an1 and an11 seed coats also were more fragile (i.e., they tear and fall off regularly) than those of the wild type. To determine the reason for this, we subjected seeds to scanning electron microscopy.
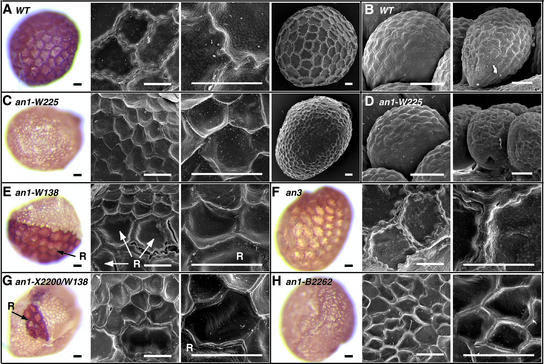
Figure 6A shows that the seed coat epidermis of mature wild-type seeds has a brown color and consists of relatively large cells with a rough, “bubbly” surface and thick, serrated crests where they are attached to neighboring cells. These cells derive from the outer layer of the single petunia integument, which is formed during ovule development, by an extensive differentiation process that involves cell enlargement and thickening of tangential walls (Colombo et al., 1997). However, the epidermal cells do not appear to divide during seed development, because the circumferences of mature seeds and unfertilized ovules contain a similar number of cells (Figure 6B).
Figure 6.
Effect of an1 Mutations on the Morphology of Seed Coat Cells.
(A) Seed coats of the wild-type (WT) line R27.
(B) Ovules of the wild-type (WT) line R27.
(C) Seed coats of line W225, which is homozygous for the stable null allele an1-W225.
(D) Ovules of line W225 (an1-W225).
(E) Seed coats of line W138, which is homozygous for the mutable allele an1-W138. Note the large revertant sector of large brown seed coat cells (marked R) at left. Revertant cells in the scanning electron micrographs (middle and right) are marked R.
(F) Seed coats of line W234, which is homozygous for a wild-type AN1 allele and a stable recessive an3 allele (an3-W1006) containing a large deletion (van Houwelingen et al., 1998).
(G) Seed coats of a plant heterozygous for the intermediate allele X2200 and the mutable allele W138. Revertant cells are indicated with R.
(H) Seed coats of a plant homozygous for an1-B2262.
The color photographs show untreated mature seeds, and the black-and-white images are scanning electron micrographs. Bars = 50 μm.
The seed coat epidermis of an1 seeds has a very different morphology: it is yellow and consists of relatively small cells that have a smooth surface and thinner, less serrated crests (Figure 6C). Because the reduced cell size is compensated for by an increase in cell number, an1 seeds are enclosed within a complete epidermis. Analysis of an1 ovules showed that the epidermis of the integument contained a similar number of cells than that of wild-type ovules (Figure 6D). This finding is consistent with expression data showing that AN1 is expressed in the integument endothelium but not in the epidermis (C. Spelt, M. Bliek, and R. Koes, unpublished data). Thus, the supernumerary cells in an1 seed coats arise from cell divisions that occur after fertilization, during development of the seed.
To determine whether AN1 acts cell autonomously during seed coat development, we analyzed seeds from unstable mutants. The epidermis of W138 seeds (and other unstable an1 mutants) mostly consists of small yellow cells, but some seeds contain patches of revertant cells with a wild-type phenotype (Figure 6E). At the borders of revertant sectors, the relatively large AN1 cells contact a larger than usual number of the much smaller neighboring mutant cells (Figure 6C). The crest between AN1 and an1 cells seems to consist of two halves with a wild-type and a mutant appearance, respectively, and crests between an1 cells have a mutant phenotype even when they contact an AN1 revertant cell on one end. Thus, the role of AN1 during the morphogenesis of seed coat cells is highly cell autonomous.
Seed coats of an an11 line had the same morphology as an1 seeds, whereas seed coats of an2, an4, and an2 an4 double mutants had a wild-type morphology (data not shown). Given the restricted expression domains of AN2 and AN4, it is possible that seed coat morphogenesis requires a distinct member of this gene family.
To determine whether the altered morphology of an1 and an11 seeds is caused by the block in flavonoid synthesis, we analyzed seeds in which the expression of flavonoid biosynthetic enzymes is blocked by either mutation or cosuppression. The epidermis of an3 seeds has a yellow color that apparently is caused by the absence of flavonoid pigments, but the size and the structure of the cells are similar to those of the wild type (Figure 6F). Similar results were obtained for seed coats of petunia lines in which the expression of CHS was blocked by cosuppression (data not shown). Thus, the altered morphology of an1 and an11 seed coat cells cannot be explained by the absence of flavonoid compounds.
To determine whether the AN1 domains involved in the specification of seed coat morphology were separable from those regulating vacuolar pH or anthocyanin synthesis, we analyzed the seed coats from various an1 mutants. Plants heterozygous for the an1 alleles X2200 and W138 express, in petals, an approximately fourfold reduced amount of wild-type an1 mRNA. The seed coats of such plants, however, appear fully mutant, because they consist of small yellow cells with occasional patches of revertant cells, which presumably result from reversion of the W138 allele (Figure 6G). Also, seeds that are homozygous for an1-B2262 or an1-G621, alleles that strongly affect PAT expression and pH control but that have little or no effect on DFR expression and anthocyanin synthesis, appear to have a full mutant phenotype (Figure 6H).
DISCUSSION
In this article, we show that AN1 of petunia acts on at least two and possibly more pathways: the well-defined anthocyanin biosynthesis pathway, and one or more poorly defined pathways involved in the acidification of the vacuole and the morphogenesis of the seed coat epidermis.
Control of Intracellular pH
In an1, an2, and an11 petals, the loss of anthocyanin pigments is associated with an increase in vacuolar pH. Because inactivation of enzymes of the anthocyanin pathway, such as F3H (in an3 mutants) or CHS (in cosuppressed lines), affects anthocyanin synthesis but not vacuolar pH, it is unlikely that anthocyanins have an effect on the pH or buffering capacity of the vacuole. On the basis of this result, and the finding that some an1 mutations (alleles previously known as ph6) differentially affect anthocyanin synthesis and vacuolar pH, we hypothesize that the effect of AN1 (and AN2 and AN11) on anthocyanin synthesis and vacuolar pH is mediated by distinct pathways.
AN1 is required for the expression of genes that encode at least eight different enzymes of the anthocyanin pathway (reviewed by Mol et al., 1998) and the glutathione S-transferase AN9, which is thought to play a role in the uptake of anthocyanins into the vacuole (Alfenito et al., 1998). Because AN1 controls pigmentation in a cell-autonomous manner but AN9 does not, it is thought that AN1 controls the expression of a protein that acts after AN9, possibly the pump that transports the anthocyanin into the vacuole (Alfenito et al., 1998). If this is the case, it would explain why full AN1 revertant spots on weakly colored flowers are surrounded by a “halo” of white anthocyaninless cells (Figure 2F).
High anthocyanin pumping activity in the revertant cells would result in rapid vacuolar sequestration of anthocyanins, including those that arrive by diffusion from neighboring mutant cells with weak AN1 and pump activity, resulting in depletion of anthocyanins in those cells. Consistent with this idea, the halos are several cells wide (large revertant sectors have wider halos than small revertant spots) and have a sharp inner edge with white cells neighboring the full-colored revertant cells, whereas the outer edge consists of a gradient of cells with colors ranging from white to the normal mutant color. If this idea is true, it suggests that the unknown transporter also is downregulated in an1-G621 and ph6-m1 mutants, because An1+ revertant spots on such flowers also are surrounded by a white halo (Figures 2G and 2H) (Chuck et al., 1993).
To date, two different transporters have been implicated in the transport of flavonoids: (1) the MATE-type (for multidrug and toxic compound extrusion) protein TT12, which is required for vacuolar localization of proanthocyanidins in the Arabidopsis seed coat (Debeaujon et al., 2001), and (2) two MRP-type (for multidrug-related protein) glutathione pumps that could transport anthocyanins when expressed in yeast (Lu et al., 1997). However, because it is not known whether these transporters translocate protons and whether their expression is controlled by TT8, the Arabidopsis AN1 homolog (Nesi et al., 2000), it remains to be determined whether anthocyanin transport and vacuolar acidification are linked directly.
Two genes have been identified that are coregulated by AN1 and one or more PH genes. One of them, MYB27, encodes a MYB domain protein of unknown function that is regulated by AN1 and PH4 (Mur, 1995; Spelt et al., 2000). The gene used in this study, PAT1, is downregulated in an1, an11, ph3, and ph4 mutants (F. Quattrocchio, I. Roobeek, W. Verweij, C. Spelt, J. Mol, and R. Koes, unpublished data) as well as in an1 mutants with specific defects in pH control (Figure 3B), suggesting that it operates in an AN1/PH3/PH4-controlled pH pathway. The further elucidation of this pathway will have to await the characterization of PAT1 and the identification of additional genes that function in the same pathway.
Control of Seed Coat Cell Morphology
Earlier work indicated that AN1 controls the coloration not only of floral organs but also of seeds (Chuck et al., 1993; Quattrocchio et al., 1993). The color of the seed coat epidermis depends on a flavonoid-derived pigment, presumably a proanthocyanidin, that requires the activity of enzymes such as CHS, F3H/AN3, and DFR (Figure 6) (Koes et al., 1990; Quattrocchio et al., 1993), similar to the situation in Arabidopsis (Debeaujon et al., 2000; Sagasser et al., 2002). Because ovules are uncolored, the synthesis of this pigment occurs after fertilization and coincides with a strong increase of DFR expression (Huits et al., 1994). Strikingly, AN1 and AN11 affect not only the color but also the morphogenesis of cells in the seed coat epidermis (Figure 6).
After fertilization, the seed increases severalfold in size, which requires that the epidermis extends proportionally. This extension seems primarily the result of cell expansion, not cell division, because the epidermis of the ovule integument and the mature seed coat consist of a similar number of cells (Figure 6). Cell expansion is accompanied by other morphological changes, such as extensive thickening of the cell wall and formation of starch granules (Colombo et al., 1997; Western et al., 2000; Windsor et al., 2000). One or more of these processes appears to be disrupted in an1 (and an11) seeds, because their epidermis consists of an increased number of smaller cells.
At least two explanations can account for this phenotype, and they are not mutually exclusive. First, the primary role of AN1 may be to prevent the growing epidermal cells from undergoing cell division. Alternatively, the primary role of AN1 may be in cell enlargement, and the induction of cell division in an1 seeds may be a secondary effect that is induced indirectly, for instance, by the more rapid expansion of underlying cell layers.
Although the mechanism by which AN1 controls seed coat development is unclear, some possibilities can be excluded. First, alterations in the flavonoid metabolism of an1 seeds cannot account for the observed morphological changes, because these are not seen in seeds in which the expression of the enzyme F3H (an3 mutants) or CHS (cosuppressed lines) is blocked. The synthesis of flavonoid (polymers) apparently is not required for the enlargement of these epidermal cells or for the thickening of their walls, because these processes show little or no alterations in an3 mutants or chs cosuppressed lines.
Because the mutations an1-G621 and an1-B2262, which in the flower primarily affect vacuolar acidification, have strong effects on the size and shape of seed coat cells, one may argue that this is an indirect result of the misregulation of vacuolar pH. However, we consider this possibility unlikely, because the seed coats of the known ph mutants consist of cells of nearly normal size and shape, even though some of the mutations affect seed color. Therefore, we favor the hypothesis that AN1 may control a third set of genes, in addition to those involved in anthocyanin synthesis and pH control, that is required for the normal morphology of seed coat cells.
Role of the bHLH Domain
In mammalian transcription factors, the bHLH domain enables these proteins to form homodimers and heterodimers through the HLH domain and to contact DNA through the basic domain (Massari and Murre, 2000). Although its conservation indicates an equally important role for the bHLH domain in plant proteins, its precise function remains unclear.
Transient expression experiments showed that deletion of the bHLH domain of B or R diminished the transcriptional activation of the BRONZE1 target promoter by only 50%, suggesting that the bHLH domain has a marginal role (Goff et al., 1992; Liu et al., 1998). By contrast, insertions of two, three, or seven amino acids into the bHLH region abolished the transcriptional activation of anthocyanin genes almost completely, in vivo as well as in transient expression assays, even though protein stability and nuclear import were unaffected (Liu et al., 1998). Although it remains unclear why deletion of the bHLH domain had less effect on the activity of R/B than small insertions, this finding raises the question of why the bHLH domain has been conserved over such a long period of evolution.
Mutational analyses of AN1, which is functionally and evolutionarily distinct from but structurally related to R, shed more light on this issue. Insertions of one, two, or three amino acids into the AN1 bHLH domain (as in the mutants W211R1 to W211R3) result in a progressive reduction of the capacity (but not in the amount) of AN1 to induce the expression of PAT1 and DFR and to drive anthocyanin synthesis and vacuolar acidification. This indicates that the bHLH domain is involved in the activation of anthocyanin synthesis and vacuolar acidification.
The removal of the C-terminal half of AN1, including the bHLH domain, while the amount of AN1 is kept approximately normal (as in an1-X2366 mutants) reduces anthocyanin synthesis and DFR expression strongly (to approximately the same extent as the insertion of two or three amino acids in the HLH region), but not completely. Although this result seems to indicate that AN1 is more sensitive to C-terminal deletions than is R or B, this is not certain. First, we cannot exclude the possibility that the transcription activation capacity of AN1-X2366 is underestimated as a result of difficulties with nuclear import. Second, because the bHLH deletion derivatives of R and B were analyzed in transient assays only, the stability and the specific activity of mutant R/B proteins are unknown.
Strikingly, removal of the C-terminal part of AN1 affects vacuolar acidification, seed coat morphogenesis, and PAT1 expression much stronger than anthocyanin synthesis or DFR expression. When overexpressed at levels ∼30-fold higher than those in the wild type (as in an1-G621 mutants), such a truncated AN1 protein drives nearly normal levels of DFR expression and anthocyanin synthesis but cannot drive vacuolar acidification, seed coat morphogenesis, or PAT1 expression.
This fact indicates that the C-terminal region of AN1, including the bHLH domain, plays a more prominent role in vacuolar acidification and seed coat morphogenesis than it does in anthocyanin synthesis, which may be the reason for the evolutionary conservation of this domain. Whether this is because AN1 regulates these different processes by interactions with different protein partners or via different cis-acting elements in the target gene promoters is the subject of further research.
Evolutionary Aspects
Based on comparative studies in petunia and maize (Quattrocchio et al., 1993, 1998, 1999) and the strong conservation of the WD repeat protein AN11 even in animals and yeast (de Vetten et al., 1997), we speculated that when the anthocyanin pathway arose during evolution, the newborn structural genes were linked to preexisting regulators, the ancestors of the Arabidopsis, petunia, and maize regulators that are studied today (Koes et al., 1994). The original function of the anthocyanin regulators has remained elusive, however. Whether one or more of the additional functions of anthocyanin regulators that have been revealed in Arabidopsis and petunia represents the hypothetical ancient function is difficult to determine, especially because these additional functions do not appear to be as widespread among plant species as anthocyanin synthesis.
In Arabidopsis, TTG controls hair cell fate by interaction with MYB proteins (GLABROUS1 [GL1] and WEREWOLF [WER]; Lee and Schiefelbein, 1999, 2001) and bHLH proteins (GL3; Payne et al., 2000) that are distinct from the anthocyanin-specific MYB (TT2, PAP1, and PAP2) and bHLH (TT8) proteins. Moreover, the genetic and physical interactions between TTG, GL1, WER, and GL3 (reviewed by Szymanski et al., 2000) closely resemble those between the anthocyanin regulators AN1, AN2, AN11, and JAF13 (A. Kroon and R. Koes, unpublished data) and C1 and B (Goff et al., 1992). This finding indicates that the mechanisms that specify hair cell fate in Arabidopsis are essentially similar to the mechanisms that drive anthocyanin synthesis in a wide variety of species.
However, the multicellular trichomes of Solanaceae appear to be specified by regulators that are distinct from those that specify the fate of the unicellular Arabidopsis hairs. First, neither mutations nor ectopic overexpression of AN1, AN2, and AN11 results in an obvious trichome phenotype (Quattrocchio et al., 1998; Spelt et al., 2000; this paper). Given that AN11 is a single gene in petunia (de Vetten et al., 1997), it is unlikely that the absence of an an11 trichome phenotype is attributable to genetic redundancy. Second, the ectopic expression of maize R induces trichome formation in Arabidopsis but not in tobacco or petunia (Lloyd et al., 1992; Quattrocchio et al., 1993). Third, trichome formation in tobacco and Arabidopsis involves MYB factors that are distinct from GL1 (Payne et al., 1999). Thus, the role of the WD repeat proteins TTG/AN11 in the specification of hairs appears to be less widespread than their role in anthocyanin synthesis.
Similarly the function of AN1 and AN11 in seed coat morphogenesis appears to be less widespread than their function in anthocyanin synthesis, because mutations in the homologous Arabidopsis genes seem to have little effect on the size and shape of seed coat epidermal cells (Debeaujon et al., 2000). The seed coat defects seen in tt1 mutants involve the differentiation of the seed coat endothelium (Sagasser et al., 2002), not the epidermis, as in an1 and an11 mutants. Whether anthocyanin regulators also control vacuolar acidification in species other than petunia remains unknown, because, to our knowledge, this question has not been analyzed in any other species.
METHODS
Petunia Lines and Mutants
Unless stated otherwise, all mutant lines used in this study were generated in the genetic background of the wild-type Petunia hybrida line R27. The alleles an1-W17 (formerly an1s/-+) and an1-W138 (formerly an1s/p-+) were isolated in progeny of R27 (Bianchi et al., 1978; Doodeman et al., 1984). an1-W219 was isolated in progeny of an1-W17 (T. Gerats and P. de Vlaming, unpublished data). The insertion alleles Z2083, B2261, and W2175 were identified among progeny of an1-W138 (van Houwelingen et al., 1998; this work). The excision allele X2200, derived from an1-W138, has been described previously (Spelt et al., 2000). The insertion alleles G621 (P. de Vlaming, unpublished data) and X2366 (van Houwelingen et al., 1998) arose in genetic backgrounds that were not related directly to R27.
Scanning Electron Microscopy
Mature seeds were fixed, critical point dried, and analyzed by scanning electron microscopy as described previously (Souer et al., 1996).
pH of Petal Extracts
The petal limbs of two flowers were ground with a pestle and mortar in 4 mL of distilled water, and the pH of the homogenate was measured immediately (to avoid alkalinization by uptake of atmospheric carbon dioxide) with a normal pH electrode. Although the differences in pH between petal extracts from different genotypes were always reproducible, the actual pH values varied between experiments, probably as a result of variations in environmental conditions. This variation is reflected by the small quantitative differences between the data sets shown in Figures 1 and 2, because these derive from distinct sets of plants grown at different periods.
Molecular Analysis of Mutant an1 Alleles
To determine the lesions in various mutant an1 alleles, we first determined the site and the origin of transposon insertions found in various unstable an1 alleles. Therefore, genomic DNA digested with different combinations of restriction enzymes was subjected to DNA gel blot analysis using fragments of the wild-type locus as a hybridization probe to visualize specific parts of the mutant locus. Once the insertions were mapped approximately, they were amplified by PCR together with the flanking an1 sequences using primers complementary to an1 and sequenced directly or first cloned in plasmid vectors and then sequenced. Lesions in derived stable recessive or revertant alleles were analyzed by PCR amplification and sequence analysis of the region surrounding the transposon insertion in the parental allele.
Transient Expression Assays
To generate 35S-driven constructs for distinct an1 alleles, we amplified, by reverse transcriptase–mediated (RT)-PCR on RNA isolated from petals containing the appropriate mutant an1 alleles, a cDNA fragment spanning the region from bp 1010 (the position in wild-type cDNA) to the poly(A) tail. RT-PCR products were digested with SacII (which cuts at position 1065) and XhoI (which cuts in the rapid amplification of cDNA ends primer) and used to replace the corresponding region of wild-type AN1 in the 35S:AN1 construct described by Spelt et al. (2000). The inserts of the resulting 35S:an1 constructs were resequenced to exclude PCR errors or cloning artifacts that had occurred.
Details regarding the fusion genes 35S:AN2, DFR:LUCIFERASE, and 35S:β-GLUCURONIDASE and procedures for the transient transformation of petunia W115 leaves are described elsewhere (de Vetten et al., 1997; Quattrocchio et al., 1998).
RNA Analyses
RNA isolation, RNA gel blot analyses, and RT-PCR were performed as described previously (de Vetten et al., 1997; Quattrocchio et al., 1998; Spelt et al., 2000). RT-PCR products were detected by gel blot hybridization and quantified by phosphor imaging. To analyze mutant an1 transcripts, they were first examined by RNA gel blot analysis to determine their size and quantity. To determine the structure in greater detail, specific parts of these an1 transcripts were amplified by PCR using either a combination of two different an1-specific primers or one an1-specific primer in combination with the 3′ rapid amplification of cDNA ends primer (Frohman et al., 1988). Products that had been amplified with “informative” primer pairs were cloned in plasmid vectors and sequenced or sequenced directly.
Expression of AN1 in Escherichia coli and Immunological Methods
Several AN1 protein fragments were expressed in E. coli as His6-tagged fusions and purified on nickel–nitrilotriacetic acid agarose columns (Qiagen, Valencia, CA) as described (de Vetten et al., 1997). One mouse was immunized with an AN1 fragment from amino acids 140 to 390 (AN140-390) and a second mouse with a 1:1 mixture of AN140-390 and the full AN1 protein as described previously (de Vetten et al., 1997). The protein blots shown in Figures 3D and 5D were made with the first antiserum; the other serum gave essentially the same results, but the background was higher.
Distribution of Biomaterials
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Accession Numbers
The GenBank accession numbers for the genes mentioned in this article are AF260918 (AN1), AF260921 (Tph6), and AF260920 (dTph5).
Acknowledgments
We thank Saskia Kars for help with scanning electron microscopy, Daisy Kloos for help with the genetic experiments, and Pieter Hoogeveen and Martina Meesters for their care of the plants.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.003772.
References
- Alfenito, M.R., Souer, E., Goodman, C.D., Buell, R., Mol, J., Koes, R., and Walbot, V. (1998). Functional complementation of anthocyanin sequestration in the vacuole by widely divergent glutathione S-transferases. Plant Cell 10, 1135–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi, F., Cornelissen, P.J.T., Gerats, A.G.M., and Hogervorst, J.M.W. (1978). Regulation of gene action in Petunia hybrida: Unstable alleles of a gene for flower colour. Theor. Appl. Genet. 53, 157–167. [DOI] [PubMed] [Google Scholar]
- Borevitz, J.O., Xia, Y., Blount, J., Dixon, R.A., and Lamb, C. (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12, 2383–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce, W., Folkerts, O., Garnaat, C., Crasta, O., Roth, B., and Bowen, B. (2000). Expression profiling of the maize flavonoid pathway genes controlled by estradiol-inducible transcription factors CRC and P. Plant Cell 12, 65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck, G., Robbins, T., Nijjar, C., Ralston, E., Courtney-Gutterson, N., and Dooner, H.K. (1993). Tagging and cloning of a petunia flower color gene with the maize transposable element Activator. Plant Cell 5, 371–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo, L., Franken, J., Van der Krol, A.R., Wittich, P.E., Dons, H.J., and Angenent, G.C. (1997). Downregulation of ovule-specific MADS box genes from petunia results in maternally controlled defects in seed development. Plant Cell 9, 703–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon, I., Leon-Kloosterziel, K.M., and Koornneef, M. (2000). Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol. 122, 403–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon, I., Peeters, A.J., Leon-Kloosterziel, K.M., and Koornneef, M. (2001). The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell 13, 853–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vetten, N., Quattrocchio, F., Mol, J., and Koes, R. (1997). The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants and animals. Genes Dev. 11, 1422–1434. [DOI] [PubMed] [Google Scholar]
- de Vlaming, P., Schram, A.W., and Wiering, H. (1983). Genes affecting flower colour and pH of flower limb homogenates in Petunia hybrida. Theor. Appl. Genet. 66, 271–278. [DOI] [PubMed] [Google Scholar]
- Doodeman, M., Boersma, E.A., Koomen, W., and Bianchi, F. (1984). Genetic analysis of instability in Petunia hybrida. 1. A highly unstable mutation induced by a transposable element inserted at the An1 locus for flower colour. Theor. Appl. Genet. 67, 345–355. [DOI] [PubMed] [Google Scholar]
- Frohman, M.A., Dush, M.K., and Martin, G.R. (1988). Rapid production of full-length cDNAs from rare transcripts: Amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85, 8998–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukada-Tanaka, S., Inagaki, Y., Yamaguchi, T., Saito, N., and Iida, S. (2000). Colour-enhancing protein in blue petals. Nature 407, 581. [DOI] [PubMed] [Google Scholar]
- Galway, M.E., Masucci, J.D., Lloyd, A.M., Walbot, V., Davis, R.W., and Schiefelbein, J.W. (1994). The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev. Biol. 166, 740–754. [DOI] [PubMed] [Google Scholar]
- Goff, S.A., Cone, K.C., and Chandler, V.L. (1992). Functional analysis of the transcription activator encoded by the maize B-gene: Evidence for a direct functional interaction between two classes of regulatory proteins. Genes Dev. 6, 864–875. [DOI] [PubMed] [Google Scholar]
- Holton, T.A., and Cornish, E.C. (1995). Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7, 1071–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huits, H.S.M., Gerats, A.G.M., Kreike, M.M., Mol, J.N.M., and Koes, R.E. (1994). Genetic control of dihydroflavonol 4-reductase gene expression in Petunia hybrida. Plant J. 6, 295–310. [DOI] [PubMed] [Google Scholar]
- Koes, R.E., Quattrocchio, F., and Mol, J.N.M. (1994). The flavonoid biosynthetic pathway in plants: Function and evolution. Bioessays 16, 123–132. [Google Scholar]
- Koes, R.E., Van Blokland, R., Quattrocchio, F., Van Tunen, A.J., and Mol, J.N.M. (1990). Chalcone synthase promoters in petunia are active in pigmented and unpigmented cell types. Plant Cell 2, 379–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo, H., Peeters, A.J., Aarts, M.G., Pereira, A., and Koornneef, M. (1999). ANTHOCYANINLESS2, a homeobox gene affecting anthocyanin distribution and root development in Arabidopsis. Plant Cell 11, 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M.M., and Schiefelbein, J. (1999). WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99, 473–483. [DOI] [PubMed] [Google Scholar]
- Lee, M.M., and Schiefelbein, J. (2001). Developmentally distinct MYB genes encode functionally equivalent proteins in Arabidopsis. Development 128, 1539–1546. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Wang, L., Kermicle, J.L., and Wessler, S.R. (1998). Molecular consequences of Ds insertion into and excision from the helix-loop-helix domain of the maize R gene. Genetics 150, 1639–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, A.M., Schena, M., Walbot, V., and Davis, R. (1994). Epidermal cell fate determination in Arabidopsis: Patterns defined by a steroid-inducible regulator. Science 266, 436–439. [DOI] [PubMed] [Google Scholar]
- Lloyd, A.M., Walbot, V., and Davis, R.W. (1992). Arabidopsis and Nicotiana anthocyanin production activated by maize regulators R and C1. Science 258, 1773–1775. [DOI] [PubMed] [Google Scholar]
- Lu, Y.-P., Li, Z.-S., and Rea, P.A. (1997). AtMRP1 gene of Arabidopsis encodes a glutathione S-conjugate pump: Isolation and functional definition of a plant ATP-binding cassette transporter gene. Proc. Natl. Acad. Sci. USA 94, 8243–8248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari, M.E., and Murre, C. (2000). Helix-loop-helix proteins: Regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20, 429–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol, J., Grotewold, E., and Koes, R. (1998). How genes paint flowers and seeds. Trends Plant Sci. 3, 212–217. [Google Scholar]
- Mur, L. (1995). Characterization of Members of the myb Gene Family of Transcription Factors from Petunia hybrida. PhD dissertation (Amsterdam: Vrije Universiteit).
- Nesi, N., Debeaujon, I., Jond, C., Pelletier, G., Caboche, M., and Lepiniec, L. (2000). The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 12, 1863–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesi, N., Jond, C., Debeaujon, I., Caboche, M., and Lepiniec, L. (2001). The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13, 2099–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne, C.T., Zhang, F., and Lloyd, A.M. (2000). GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156, 1349–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne, T., Clement, J., Arnold, D., and Lloyd, A. (1999). Heterologous myb genes distinct from GL1 enhance trichome production when overexpressed in Nicotiana tabacum. Development 126, 671–682. [DOI] [PubMed] [Google Scholar]
- Quattrocchio, F. (1994). Regulatory Genes Controlling Flower Pigmentation in Petunia hybrida. PhD thesis (Amsterdam: Vrije Universiteit).
- Quattrocchio, F., Wing, J., van der Woude, K., Souer, E., de Vetten, N., Mol, J., and Koes, R. (1999). Molecular analysis of the anthocyanin2 gene of Petunia and its role in the evolution of flower color. Plant Cell 11, 1433–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio, F., Wing, J.F., Leppen, H.T.C., Mol, J.N.M., and Koes, R.E. (1993). Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. Plant Cell 5, 1497–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio, F., Wing, J.F., van der Woude, K., Mol, J.N.M., and Koes, R. (1998). Analysis of bHLH and MYB-domain proteins: Species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J. 13, 475–488. [DOI] [PubMed] [Google Scholar]
- Sagasser, M., Lu, G.H., Hahlbrock, K., and Weisshaar, B. (2002). A. thaliana TRANSPARENT TESTA 1 is involved in seed coat development and defines the WIP subfamily of plant zinc finger proteins. Genes Dev. 16, 138–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souer, E., van Houwelingen, A., Kloos, D., Mol, J.N.M., and Koes, R. (1996). The no apical meristem gene of petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85, 159–170. [DOI] [PubMed] [Google Scholar]
- Spelt, C., Quattrocchio, F., Mol, J., and Koes, R. (2000). anthocyanin1 of petunia encodes a basic-helix loop helix protein that directly activates structural anthocyanin genes. Plant Cell 12, 1619–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski, D.B., Lloyd, A.M., and Marks, M.D. (2000). Progress in the molecular genetic analysis of trichome initiation and morphogenesis in Arabidopsis. Trends Plant Sci. 5, 214–219. [DOI] [PubMed] [Google Scholar]
- Taiz, L. (1992). The plant vacuole. J. Exp. Biol. 172, 113–122. [DOI] [PubMed] [Google Scholar]
- van Houwelingen, A., Souer, E., Spelt, C., Kloos, D., Mol, J., and Koes, R. (1998). Analysis of flower pigmentation mutants generated by random transposon mutagenesis in Petunia hybrida. Plant J. 13, 39–50. [DOI] [PubMed] [Google Scholar]
- Walker, A.R., Davison, P.A., Bolognesi-Winfield, A.C., James, C.M., Srinivasan, N., Blundell, T.L., Esch, J.J., Marks, M.D., and Gray, J.C. (1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11, 1337–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western, T.L., Skinner, D.J., and Haughn, G.W. (2000). Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiol. 122, 345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windsor, J.B., Symonds, V.V., Mendenhall, J., and Lloyd, A.M. (2000). Arabidopsis seed coat development: Morphological differentiation of the outer integument. Plant J. 22, 483–493. [DOI] [PubMed] [Google Scholar]
- Winkel-Shirley, B. (2001). Flavonoid biosynthesis: A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126, 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, K., Kondo, T., Okazaki, Y., and Katou, K. (1995). Cause of blue petal colour. Nature 373, 291. [Google Scholar]