Abstract
The ubiquitin-related protein RUB/Nedd8 is conjugated to members of the cullin family of proteins in plants, animals, and fungi. In Arabidopsis, the RUB conjugation pathway consists of a heterodimeric E1 (AXR1-ECR1) and a RUB-E2 called RCE1. The cullin CUL1 is a subunit in SCF-type ubiquitin protein ligases (E3s), including the SCFTIR1 complex, which is required for response to the plant hormone auxin. Our previous studies showed that conjugation of RUB to CUL1 is required for normal SCFTIR1 function. The RING-H2 finger protein RBX1 is a subunit of SCF complexes in fungi and animals. The function of RBX1 is to bind the ubiquitin-conjugating enzyme E2 and bring it into close proximity with the E3 substrate. We have identified two Arabidopsis genes encoding RING-H2 proteins related to human RBX1. Studies of one of these proteins indicate that, as in animals and fungi, Arabidopsis RBX1 is an SCF subunit. Reduced RBX1 levels result in severe defects in growth and development. Overexpression of RBX1 increases RUB modification of CUL1. This effect is associated with reduced auxin response and severe growth defects similar to those observed in axr1 mutants. As in the axr1 mutants, RBX1 overexpression stabilizes the SCFTIR1 substrate AXR2/IAA7. The RBX1 protein is a component of SCF complexes in Arabidopsis. In addition to its direct role in SCF E3 ligase activity, RBX1 promotes the RUB modification of CUL1 and probably functions as an E3 ligase in the RUB pathway. Hypermodification of CUL1 disrupts SCFTIR1 function, suggesting that cycles of RUB conjugation and removal are important for SCF activity.
INTRODUCTION
The ubiquitin-proteasome pathway has been implicated in diverse aspects of cellular regulation in plants, animals, and fungi (Hershko and Ciechanover, 1998; Callis and Vierstra, 2000). Ubiquitination of a target protein involves the sequential activity of three enzymes: a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin-protein ligase (E3). Initially, ubiquitin is activated by E1 through the formation of a thiol-ester linkage between the C terminus of ubiquitin and an internal Cys within the E1 enzyme. Ubiquitin then is transferred to an E2 enzyme, again by the formation of a thiol ester with an internal Cys. The function of the E3 is to bind the E2 and the target protein and facilitate the transfer of the ubiquitin molecule to the target.
The E3 proteins are diverse, but they can be divided into two main groups based on their mechanisms of action. Members of one group, the HECT domain E3s, form a thiol-ester intermediate with ubiquitin before they are transferred to the target protein (Hershko and Ciechanover, 1998). The second large group, the RING E3s, all contain a RING-H2 finger domain (Freemont, 2000). Many RING E3s consist of a single protein containing the RING-H2 finger domain. In the case of the SCF, VHL, and APC E3s, the RING finger protein is a subunit in a multiple-subunit complex. Unlike the HECT domain, RING E3s do not form a thiol-ester intermediate with ubiquitin. Instead, they appear to function as a platform for bringing the E2 and the substrate together.
The core module of the SCF and VHL complexes consists of a member of the cullin family and a RING-H2 finger protein called variously RBX, ROC, or Hrt (Kamura et al., 1999b; Ohta et al., 1999; Seol et al., 1999; Tan et al., 1999). One function of the RING finger protein is to bind E2, and in several instances, RBX-cullin complexes have been shown to have E3 activity in vitro (Ohta et al., 1999; Seol et al., 1999). There is no evidence that the RING protein participates more directly in ubiquitin transfer.
In the case of SCF E3s, a RBX1-cullin core binds a second module consisting of SKP1 and a member of a large family of F-box proteins (Deshaies, 1999; Freemont, 2000). The F-box proteins typically have additional protein–protein interaction motifs such as WD40 or Leu-rich repeats and are responsible for interacting with target proteins (del Pozo and Estelle, 2000; Kipreos and Pagano, 2000).
In Arabidopsis, SCF E3s have a particularly important role in regulating response to the hormone auxin (Gray et al., 1999, 2001; del Pozo and Estelle, 2000). This hormone is involved in many different aspects of plant growth and development ranging from embryogenesis to senescence. Recent studies indicate that auxin response depends on the degradation of a large family of transcriptional repressors called the Aux/IAA proteins (Ramos et al., 2001; Zenser et al., 2001). In response to auxin, these proteins are recognized by an E3 called SCFTIR1, ubiquitinated, and degraded (Gray et al., 2001).
Studies in Arabidopsis, as well as in animal and fungal systems, indicate that SCF function depends on modification of the cullin component of the complex by the ubiquitin-related protein RUB (Nedd8 in some species). RUB is conjugated to target proteins in a manner that is analogous to ubiquitin (Hochstrasser, 2000). In Arabidopsis, RUB is activated by a heterodimeric E1 enzyme composed of the AXR1 and ECR1 proteins and transferred to a RUB E2 enzyme called RCE1 (del Pozo and Estelle, 1999). Mutations in AXR1 result in reduced levels of RUB-CUL1, stabilization of Aux/IAA proteins, and decreased auxin response (Gray et al., 2001; del Pozo et al., 2002).
A unique RUB E3 activity has not been identified. One possibility is that RBX1 functions in this capacity. This hypothesis is supported by results showing that RBX1 promotes RUB modification of the budding yeast cullin Cdc53 and human Cul2 (Kamura et al., 1999a). In this study, we show that Arabidopsis RBX1 is associated with SCFTIR1. Furthermore, we demonstrate that overexpression of RBX1 in plants results in an increase in the relative level of RUB-CUL1. Surprisingly, this change is associated with stabilization of at least one SCF substrate, AXR2/IAA7, and a defect in auxin response.
RESULTS
The RBX Genes in Arabidopsis
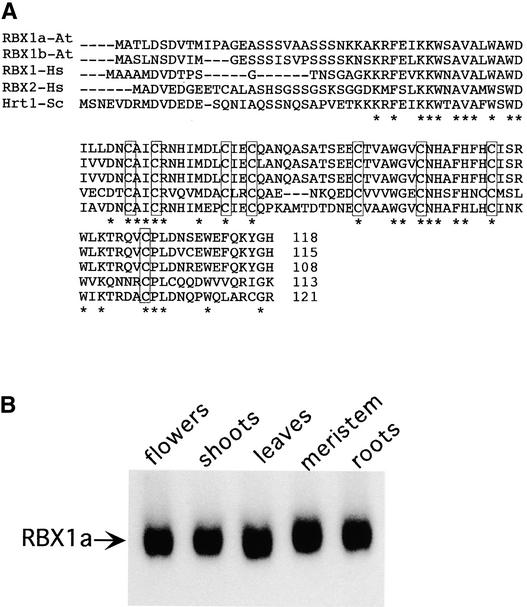
A search of Arabidopsis sequences in GenBank for genes related to human RBX1 revealed the presence of two RBX-like genes in the Arabidopsis genome. The putative proteins encoded by these genes (At3g42830 and At5g20570) are 83% identical. Both Arabidopsis proteins are 75% identical to human RBX1 and ∼50% identical to the yeast Hrt1 protein. Most of the differences among these proteins are found in the N-terminal 25 to 30 residues. Each of the predicted zinc binding residues is conserved (Figure 1A). We named these two genes RBX1a (At5g20570) and RBX1b (At3g42830). There are four EST sequences corresponding to RBX1a in GenBank and none corresponding to RBX1b. An RNA gel blot experiment indicated that RBX1a is expressed at significant levels throughout the plant (Figure 1B). For these reasons, we chose to focus our studies on RBX1a.
Figure 1.
RBX Genes in Arabidopsis.
(A) Alignment of RBX proteins in Arabidopsis (At) with human (Hs) and budding yeast (Sc) orthologs. Arabidopsis RBX1a is encoded by At5g20570 and RBX1b is encoded by At3g42830. Identical residues in all five proteins are indicated with asterisks. The predicted zinc binding Cys residues are boxed.
(B) RNA gel blot showing RBX1a expression in all tissues examined. Fifteen micrograms of total RNA from 30-day-old soil-grown plants was loaded in each lane.
RBX1 Is a Subunit of SCFTIR1
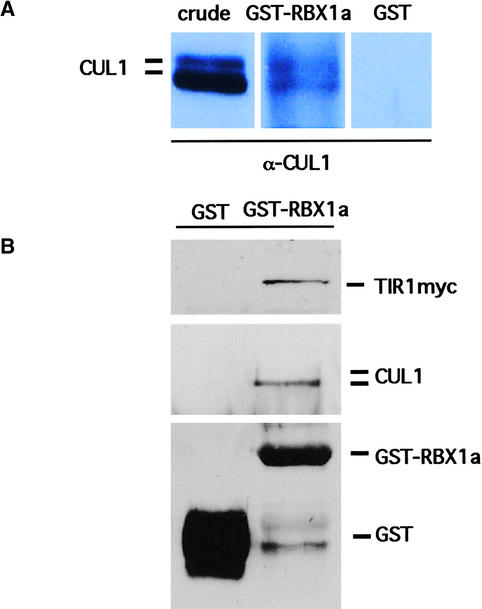
Studies in yeast and mammalian cells have shown that RBX1 and related RING-H2 proteins are components of SCF-type ubiquitin protein ligases. To determine if this is the case in plants as well, we tested for interaction between RBX1 and components of SCFTIR1. A glutathione S-transferase (GST)–RBX1a fusion protein was synthesized in Escherichia coli and added to protein extracts prepared from Arabidopsis seedlings. GST-RBX1a was recovered on agarose beads, and the pulldown was examined by protein gel blot analysis.
Figure 2A shows that both CUL1 and RUB1-modified CUL1 were recovered with GST-RBX1a. To show that RBX1 interacts with an intact SCFTIR1, the GST-RBX1a pulldown experiment was performed using a line carrying a TIR1-myc transgene. The results shown in Figure 2B indicate that GST-RBX1a interacts with a complex containing TIR1. As in Figure 2A, both CUL1 and RUB1-CUL1 were recovered in the pulldown. In this case, the modified CUL1 was visible in a longer exposure. These results confirm that Arabidopsis RBX1 binds SCF components, and they strongly suggest that, as in fungi and animals, RBX1 is a subunit of the SCF complex.
Figure 2.
RBX1a Is a Component of the SCF Complex in Arabidopsis.
(A) Recombinant GST-RBX1a or GST was added to extracts prepared from Arabidopsis seedlings. The GST pulldowns were examined for the presence of CUL1 by protein gel blot analysis using α-CUL1 antibody. The two CUL1 species are unmodified and RUB-modified protein. The three lanes are from the same autoradiogram.
(B) As in (A) except that extract was prepared from a tir1 line expressing TIR1-myc. CUL1, TIR1-myc, and GST-RBX1a were detected in the pulldown by protein gel blot analysis. The left lane is a control pulldown using GST alone.
Changes in RBX1 Levels Result in Dramatic Changes in Plant Morphology and Defects in Auxin Response
To investigate the physiological role of RBX1, we generated Arabidopsis lines with decreased and increased levels of RBX1a expression. To reduce RBX1a levels, we introduced an antisense construct into wild-type plants. Most of the transgenic plants recovered in this experiment died as young seedlings. These plants had purple cotyledons and elongated hypocotyls (Figure 3A). Surviving seedlings grew into small dark green rosettes with multiple inflorescences (Figure 3C). Most inflorescences stopped elongating after growing a few centimeters and terminated with compact clusters of dozens of flowers (Figure 3D). Occasionally, these plants also would produce an elongate inflorescence with normal fertile flowers. Although these defects were observed in several independent transgenic plants, we were unable to recover similarly affected plants in the T2 generation.
Figure 3.
Reduced Expression of RBX1 Results in Severe Growth Defects.
(A) Nine-day-old Col(α-RBX1) seedling.
(B) Nine-day-old Col(pROKII) seedling.
(C) Thirty-five-day-old Col(α-RBX1). Bar = 1 cm.
(D) Higher magnification of the inflorescence of a Col(α-RBX1) plant showing a floral cluster.
To observe the effects of increased RBX1a levels on growth and development, we introduced the RBX1a cDNA under the control of the 35S promoter into wild-type plants. Multiple independent transgenic lines exhibited the same phenotype. RNA gel blot analysis on two lines confirmed that the phenotype was associated with high levels of RBX1a expression (Figure 4A). Dark-grown seedlings displayed a partially deetiolated phenotype, including a reduction in hypocotyl length and loss of the apical hook (Figures 4B to 4D). The hypocotyls of the Col(35S::RBX1)1a line were 18.3 ± 1.8 mm compared with 22.3 ± 1.4 mm for the wild type. This phenotype is very similar to that of the axr1-12 mutant (Lincoln et al., 1990; Cernac et al., 1997).
Figure 4.
Effects of RBX1a Overexpression on Plant Growth and Development.
(A) RNA gel blot showing increased RBX1a expression in transgenic lines. RNA was extracted from 7-day-old Columbia (lane 1), Col(35S::RBX1)2d (lane 2), and Col(35S::RBX1)1a (lane 3) seedlings.
(B) to (D) Five-day-old dark-grown seedlings of Columbia (B), Col(35S::RBX1)1a (C), and Col(35S::RBX1)2d (D).
(E) to (H) Twenty-three-day-old plants of Columbia (E), axr1-12 (F), Col(35S::RBX1)1a (G), and Col(35S::RBX1)2b (H).
(I) to (K) Thirty-five-day-old plants of Columbia (I), Col(35S::RBX1)1a (J), and Col(35S::RBX1)2d (K).
(L) Morphology of (from left to right) Columbia, axr1-12, Col (35S::RBX1)1a, and Col(35S::RBX1)2b flowers.
(M) Morphology of (from left to right) Columbia, Col(35S::RBX1)1a, and Col(35S::RBX1)2b siliques.
(N) Forty-day-old plants of axr1-12 (left) and axr1-12(35S::RBX1)1a (right).
Bars = 1 cm.
When grown in the light, the transgenic plants had smaller cotyledons and produced fewer lateral roots than wild-type plants. At day 11 after germination, wild-type seedlings (n = 12) had 12.3 ± 1.7 lateral roots, whereas Col(35S::RBX1)2b seedlings (n = 11) had 4.1 ± 2.3 lateral roots. The rosette leaves of the 35S::RBX1 lines were smaller and misshapen compared with those of the wild type (Figures 4E to 4H) and were quite similar to those of axr1-12 (Figure 4F). After flowering, the transgenic plants were short and bushy and produced small flowers (Figures 4I to 4M).
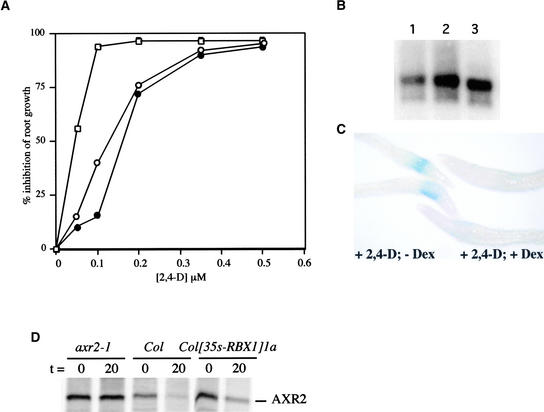
Many of the defects observed in the 35S::RBX1 lines were similar to those exhibited by the axr1 mutants, suggesting that an increase in RBX1a levels inhibits auxin response (cf. Figures 4F with 4G and 4H and Figures 4J and 4K with 4N; see also Figure 4L) (Lincoln et al., 1990; Cernac et al., 1997). To explore this possibility, we measured the effects of auxin on root elongation in two lines. The roots of two independent 35S::RBX1 lines were significantly less sensitive to the effects of auxin on root elongation than those of wild-type seedlings (Figure 5A).
Figure 5.
RBX1a Overexpression Results in Reduced Auxin Response and Stabilization of AXR2/IAA7.
(A) Response of seedling roots to auxin. Columbia (open squares), Col(35S::RBX1)1a (open circles), and Col(35S::RBX1)2e (closed circles). sd was <20% for each data point.
(B) RNA gel blot showing DEX-regulated accumulation of RBX1a RNA. RNA was isolated from 7-day-old seedlings treated with DEX for 12 h. Lane 1, Col(BA3::GUS); lane 2, Col(BA3::GUS,GVG::RBX1)a; lane 3, Col(BA3::GUS,GVG::RBX1)b.
(C) Seven-day-old Col(BA3::GUS,GVG::RBX1)a seedlings were pretreated with either Arabidopsis thaliana medium + sucrose buffer (left) or DEX (right) for 12 h followed by treatment with 0.2 μM 2,4-D for 8 h and staining for GUS activity.
(D) Pulse-chase analysis of AXR2/IAA7. Seedlings were treated as described in Methods. Labeled AXR2/IAA7 was immunoprecipitated as described by Gray et al. (2001). The half-life of AXR2/IAA7, based on four independent experiments, was 11.3 ± 1.4 min in wild-type and 14.7 ± 2.0 min in Col(35S::RBX1)1a seedlings.
To further document the effects of increased RBX1a expression on auxin response, we used the BA3::β-glucuronidase (GUS) auxin-responsive reporter gene (Oono et al., 1998). In this case, the RBX1a cDNA was cloned into the pTA7002 plasmid, allowing dexamethasone (DEX)-regulated expression of RBX1a (Aoyama and Chau, 1997). This transgene then was transformed directly into plants carrying the BA3::GUS gene. Seven-day-old seedlings were mock treated or pretreated with DEX for 12 h followed by an 8-h treatment with 0.2 μM 2,4-D and then stained for GUS.
Treatment with DEX resulted in significant accumulation of RBX1a RNA (Figure 5B). In the absence of DEX-induced RBX1a expression, GUS staining was observed in the elongation zone of the root (Figure 5C). No staining was observed in the DEX-treated seedlings, indicating that RBX1a expression inhibited auxin-regulated expression of the reporter gene. Analysis of BA3::GUS seedlings harboring the pTA7002 control vector revealed normal staining patterns, confirming that the observed reduction in GUS staining was attributable to expression of the RBX1a transgene (data not shown).
In previous studies, we showed that decreased auxin response in the axr1-12 and tir1 mutants is associated with stabilization of the Aux/IAA proteins (Gray et al., 2001). To determine if this is the case for the 35S::RBX1 lines, we performed a pulse-chase experiment to measure the half-life of AXR2 (IAA7) in wild-type and Col(35S::RBX1)1a seedlings. Representative data are shown in Figure 5D. AXR2 had a half-life of 11.3 ± 1.4 min (n = 4) in the wild type and 14.7 ± 2.0 min (n = 4) in the transgenic lines. A Student's t test confirmed that these two values were statistically different (P < 0.005). Thus, RBX1a overexpression is associated with increased stability of IAA7.
Overexpression of RBX1 Promotes RUB Modification of CUL1
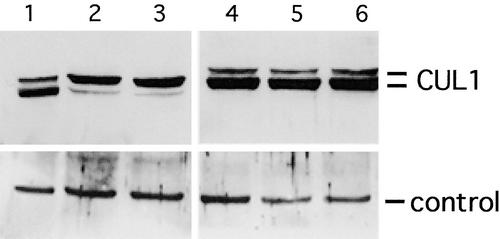
There are a number of possible explanations for the effects of 35S::RBX1 on auxin response. For example, increased levels of RBX1a may stimulate the degradation of a positive regulator of auxin response. Alternatively, studies in budding yeast and in mammals demonstrate that RBX1 promotes RUB/NEDD8 modification of cullins (Kamura et al., 1999b). Thus, increased RBX1 levels may affect the RUB conjugation pathway. Examination of CUL1 from wild-type and 35S::RBX1 seedlings by protein gel blot analysis indicates that this is the case.
In 35S-RBX1 seedlings, almost all of the CUL1 is in the modified form (Figure 6). AXR1 encodes one subunit of the RUB-activating enzyme, and the axr1 mutants have reduced levels of RUB-CUL1. When the (35S::RBX1)1a transgene was crossed into the axr1-12 background, the levels of RUB-CUL1 were reduced, indicating that the effects of increased RBX1a on RUB-CUL1 levels are dependent on the RUB protein conjugation pathway.
Figure 6.
Increased RUB-CUL1 Formation in 35S::RBX1 Lines Is AXR1 Dependent.
Extracts were prepared from 7-day-old seedlings and examined by protein blot analysis using α-CUL1 antiserum. Lane 1, Columbia; lanes 2 and 3, Col(35S-RBX1)1a; lane 4, axr1-12; lanes 5 and 6, axr1-12 (35S-RBX1)1a. The lines indicate CUL1 and RUB-CUL1. The control panels (bottom) show levels of an unknown cross-reacting protein.
Strikingly, axr1-12 and axr1-12(35S::RBX1)1a were indistinguishable, indicating that the severe dwarf phenotype exhibited by the Col(35S::RBX1)1a line is partially ameliorated by the axr1-12 mutation (Figure 4N). These results suggest that the phenotypic effects of RBX1a expression are largely the result of increased RUB conjugation and are not related to a direct role of RBX1a in SCF function.
DISCUSSION
Studies in animal and fungal systems have shown that the RING-H2 finger protein RBX is an integral component of SCF ubiquitin-protein ligases. In Arabidopsis, there are two closely related RBX orthologs that we have named RBX1a and RBX1b. RBX1a is expressed in all tissues of the plant and is represented multiple times in dBEST, suggesting that it has a general function in ubiquitin-mediated processes.
GST-RBX1a pulldown experiments indicate that RBX1a interacts with SCFTIR1. As in other species, we believe that this interaction occurs through the cullin component of the complex. In human Cul1, the sequence ILLQYN near the C terminus of the protein has been implicated in RBX1/ROC1 binding. In Arabidopsis CUL1, the corresponding sequence is the related VLLLFN, suggesting a similar function.
In both budding and filamentous yeast, RBX1 is essential for viability. In Arabidopsis, the introduction of an antisense RBX1 construct into plants caused a severe dwarf phenotype. These plants produced a limited number of seeds, but the resulting plants were always wild type in appearance, suggesting either that antisense suppression was unstable or that one of the effects of decreased RBX1 levels is gametophytic or embryonic lethality. Regardless of the explanation, it is clear that reduced RBX1 levels have a dramatic effect on plant growth and development, consistent with the central role of the protein in SCF function.
Overexpression of RBX1 also results in dramatic effects on plant morphology. These plants exhibit an increase in the amount of RUB-modified CUL1, suggesting that RBX1 promotes RUB modification of CUL1. This is consistent with earlier studies in vertebrate cells and in budding yeast (Kamura et al., 1999a). In the context of SCF E3 activity, the function of RBX1 is to bind the E2 protein and bring it into close proximity with the substrate. The fact that increased RBX1 levels promote RUB modification of CUL1 suggest that RBX1 also binds the RUB E2 enzyme.
Indeed, in separate studies of the RUB E2 RCE1, we found that RBX1 binds RCE1 (S. Dharmasiri and M. Estelle, unpublished data). However, we cannot exclude the possibility that the accumulation of RBX1 affects the level of RUB-CUL1 in some other way. For example, RBX1 has been shown to interact with two subunits of the COP9 signalosome (CSN) (Schwechheimer et al., 2001). Because the CSN is required for the removal of RUB from CUL1 (see below), it is possible that excess RBX1 disrupts this activity. Alternatively, RBX1 may titrate an unknown factor required for SCF function.
Surprisingly, RBX1 overexpression causes a phenotype that is very similar to the axr1 phenotype, including stabilization of the Aux/IAA protein AXR2 and reduced auxin response. These effects are best explained by a defect in SCFTIR1 function. Thus, both increased and decreased RUB-CUL1 levels appear to reduce the activity of SCF. These results are consistent with those obtained in a recent study of the CSN in Arabidopsis. A reduction in CSN levels results in the stabilization of Aux/IAA proteins, decreased auxin response, and increased levels of RUB-CUL1 (Schwechheimer et al., 2001). In this case, the change in RUB-CUL1 levels is caused by the loss of RUB-deconjugating activity associated with the CSN.
The precise role of the RUB modification remains unclear. Several possibilities have been suggested, including a role in the recruitment of F-box proteins or the E2 to the core SCF complex. In Arabidopsis, a reduction in CSN levels is associated with the accumulation of ubiquitinated proteins (Peng et al., 2001a, 2001b). However, it is not known if this defect is related directly to increased levels of RUB-CUL1 or to some other function of the CSN in protein degradation. Whatever the case, our results, together with those of Schwechheimer et al. (2001) and Lyapina et al. (2001), suggest that RUB conjugation is dynamic and that this is important for SCF function.
Our studies have shown that the RUB conjugation pathway is extremely important for the function of SCFTIR1. Is RUB modification also important for other SCFs? This seems likely, because RUB modification appears to be a general characteristic of all cullin proteins in diverse species (Lammer et al., 1998; Osaka et al., 1998; Hori et al., 1999; Wada et al., 1999; del Pozo and Estelle, 2000; Osaka et al., 2000). Recent estimates indicate that there are ∼700 F-box proteins encoded by the Arabidopsis genome (Gagne et al., 2002). However, only a small number have been assigned a biological role, making it difficult to assess the general role of CUL1 modification in SCF function.
Apart from TIR1, the best characterized of these is the COI1 protein, which is required for the response to jasmonic acid (JA). We and others have found that mutations in AXR1 have a moderate effect on JA response (Xu et al, 2002; S. Dharmasiri and M. Estelle, unpublished data; Tiryaki and Staswick, 2002). By contrast, the 35S::RBX1 lines have a normal JA response (data not shown). This difference may reflect differences in the severity of effects on SCF function. Further studies, including genomic and proteomic approaches to the identification of SCF substrates, will provide a clearer picture of the overall function of RUB modification.
METHODS
Plant Material and Growth Conditions
All mutants and transgenic Arabidopsis thaliana lines used in this study were in the Columbia ecotype. Seedlings were grown under long-day conditions on sterile, vertically oriented Arabidopsis thaliana medium + sucrose (ATS) plates (Lincoln et al., 1990). Seedlings used for protein extractions were grown for 5 to 7 days in liquid ATS medium.
To determine the response of seedling roots to auxin, Columbia and Col(35S::RBX1) seeds were sterilized and germinated on ATS nutrient medium. Five-day-old seedlings were transferred to ATS plates containing increasing concentrations of the synthetic auxin 2,4-D. Root growth was measured 5 days after transfer and expressed relative to the growth of roots on medium without auxin. Data points represent the average growth of 10 seedlings.
RBX1 Constructs
The RBX1 coding sequence from the Arabidopsis Stock Center EST clone T42038 was cloned into the plant expression vector pROK II in both the sense and antisense orientations. The dexamethasone-inducible derivative was constructed by introducing the RBX1 coding sequence into the pTA7002 expression vector (Aoyama and Chau, 1997). Constructs were introduced into Arabidopsis by Agrobacterium tumefaciens–mediated transformation as described previously (Bechtold et al., 1993).
The glutathione S-transferase (GST)–RBX1 expression vector was generated by cloning the RBX1 coding sequence into pGEX-2TK. GST-RBX1 was expressed and purified from Escherichia coli using standard techniques.
Antibodies and GST Pulldown Assays
Monoclonal α-myc was purchased from BabCo (Richmond, CA). The CUL1 antibody has been described previously (Gray et al., 1999). Pulldown assays were performed with 4 μg of purified GST or GST-RBX1 fusion protein and 4 mg of crude seedling extract prepared as described previously (Gray et al., 1999). After a 3-h incubation at 4°C, the fusion protein was purified from the seedling extract, washed extensively, and analyzed by SDS-PAGE and immunoblotting.
Pulse-Chase Analysis
Seven-day-old seedlings were transferred to 4 mL of ATS medium containing 200 μCi of 35S-Trans label (ICN, Costa Mesa, CA) and grown for 3.5 h. Labeled seedlings were washed, and proteins were extracted immediately or after a 30-min chase in medium containing 1 mM Met/Cys and 100 μg/mL cycloheximide. AXR2 was immunoprecipitated with affinity-purified α-AXR2 antibody. AXR2 half-life was calculated using the formula t1/2 = (0.693 × time in minutes)/ln(N0/Nx). Values presented are averages of three independent experiments ± sd.
RNA Gel Blot Hybridization
Total RNA was isolated from Arabidopsis seedlings or adult tissue and blotted using standard techniques. Seedlings were induced with 0.03 mM dexamethasone for 12 h where indicated.
Acknowledgments
This work was supported by National Institutes of Health Grant RO1-GM43644 and National Science Foundation Grant 0115870 to M.E. H.H. was supported by a grant from the Deutsche Forschungsgemeinschaft (HE 3224/1-1).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.003178.
References
- Aoyama, T., and Chau, N.H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11, 605–612. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris Life Sci. 316, 15–18. [Google Scholar]
- Callis, J., and Vierstra, R.D. (2000). Protein degradation in signaling. Curr. Opin. Plant Biol. 3, 381–386. [DOI] [PubMed] [Google Scholar]
- Cernac, A., Lincoln, C., Lammer, D., and Estelle, M. (1997). The SAR1 gene of Arabidopsis acts downstream of the AXR1 gene in auxin response. Development 124, 1583–1591. [DOI] [PubMed] [Google Scholar]
- del Pozo, J.C., Dharmasiri, S., Hellmann, H., Walker, L., Gray, W.M., and Estelle, M. (2002). AXR1-ECR1–dependent conjugation of RUB1 to the Arabidopsis cullin AtCUL1 is required for auxin response. Plant Cell 14, 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo, J.C., and Estelle, M. (1999). The Arabidopsis cullin AtCUL1 is modified by the ubiquitin-related protein RUB1. Proc. Natl. Acad. Sci. USA 96, 15342–15347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo, J.C., and Estelle, M. (2000). F-box proteins and protein degradation: An emerging theme in cellular regulation. Plant Mol. Biol. 44, 123–128. [DOI] [PubMed] [Google Scholar]
- Deshaies, R.J. (1999). SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15, 435–467. [DOI] [PubMed] [Google Scholar]
- Freemont, P.S. (2000). RING for destruction? Curr. Biol. 10, R84–R87. [DOI] [PubMed] [Google Scholar]
- Gagne, J.M., Downes, B.P., Shiu, S.H., Durski, A.M., and Vierstra, R.D. (2002). The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 99, 11519–11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., del Pozo, J.C., Walker, L., Hobbie, L., Risseeuw, E., Banks, T., Crosby, W.L., Yang, M., Ma, H., and Estelle, M. (1999). Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13, 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., Kepinski, S., Rouse, D., Leyser, O., and Estelle, M. (2001). Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414, 271–276. [DOI] [PubMed] [Google Scholar]
- Hershko, A., and Ciechanover, A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Hochstrasser, M. (2000). Evolution and function of ubiquitin-like protein-conjugation systems. Nat. Cell Biol. 2, E153–E157. [DOI] [PubMed] [Google Scholar]
- Hori, T., Osaka, F., Chiba, T., Miyamoto, C., Okabayashi, K., Shimbara, N., Kato, S., and Tanaka, K. (1999). Covalent modification of all members of human cullin family proteins by NEDD8. Oncogene 18, 6829–6834. [DOI] [PubMed] [Google Scholar]
- Kamura, T., Conrad, M.N., Yan, Q., Conaway, R.C., and Conaway, J.W. (1999. a). The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 13, 2928–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura, T., Koepp, D.M., Conrad, M.N., Skowyra, D., Moreland, R.J., Iliopoulos, O., Lane, W.S., Kaelin, W.G., Jr., Elledge, S.J., Conaway, R.C., Harper, J.W., and Conaway, J.W. (1999. b). Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284, 657–661. [DOI] [PubMed] [Google Scholar]
- Kipreos, E.T., and Pagano, M. (2000). The F-box protein family. Genome Biol. 1, 3002.3001–3002.3007. [DOI] [PMC free article] [PubMed]
- Lammer, D., Mathias, N., Laplaza, J.M., Jiang, W., Liu, Y., Callis, J., Goebl, M., and Estelle, M. (1998). Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes Dev. 12, 914–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln, C., Britton, J.H., and Estelle, M. (1990). Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2, 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina, S., Cope, G., Shevchenko, A., Serino, G., Tsuge, T., Zhou, C., Wolf, D.A., Wei, N., Shevchenko, A., and Deshaies, R.J. (2001). Promotion of NEDD8-CUL1 conjugate cleavage by COP9 signalosome. Science 292, 1382–1385. [DOI] [PubMed] [Google Scholar]
- Ohta, T., Michel, J.J., Schottelius, A.J., and Xiong, Y. (1999). ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell 3, 535–541. [DOI] [PubMed] [Google Scholar]
- Oono, Y., Chen, Q.G., Overvoorde, P.J., Kohler, C., and Theologis, A. (1998). age mutants of Arabidopsis exhibit altered auxin-regulated gene expression. Plant Cell 10, 1649–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaka, F., Kawasaki, H., Aida, N., Saeki, M., Chiba, T., Kawashima, S., Tanaka, K., and Kato, S. (1998). A new NEDD8-ligating system for cullin-4A. Genes Dev. 12, 2263–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaka, F., Saeki, M., Katayama, S., Aida, N., Toh, E.A., Kominami, K., Toda, T., Suzuki, T., Chiba, T., Tanaka, K., and Kato, S. (2000). Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J. 19, 3475–3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Z., Serino, G., and Deng, X.W. (2001. a). Molecular characterization of subunit 6 of the COP9 signalosome and its role in multifaceted developmental processes in Arabidopsis. Plant Cell 13, 2393–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, Z., Serino, G., and Deng, X.W. (2001. b). A role of Arabidopsis COP9 signalosome in multifaceted developmental processes revealed by the characterization of its subunit 3. Development 128, 4277–4288. [DOI] [PubMed] [Google Scholar]
- Ramos, J.A., Zenser, N., Leyser, H.M., and Callis, J. (2001). Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 13, 2349–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer, C., Serino, G., Callis, J., Crosby, W.L., Lyapina, S., Deshaies, R.J., Gray, W.M., Estelle, M., and Deng, X.W. (2001). Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIR1 in mediating auxin response. Science 292, 1379–1382. [DOI] [PubMed] [Google Scholar]
- Seol, J.H., Feldman, R.M., Zachariae, W., Shevchenko, A., Correll, C.C., Lyapina, S., Chi, Y., Galova, M., Claypool, J., Sandmeyer, S., Nasmyth, K., and Deshaies, R.J. (1999). Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 13, 1614–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, P., Fuchs, S.Y., Chen, A., Wu, K., Gomez, C., Ronai, Z., and Pan, Z.Q. (1999). Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of I kappa B alpha. Mol. Cell 3, 527–533. [DOI] [PubMed] [Google Scholar]
- Tiryaki, I., and Staswick, P. (2002). An Arabidopsis thaliana mutant defective in jasmonate response is allelic to the auxin signaling mutant axr1. Plant Physiol., in press. [DOI] [PMC free article] [PubMed]
- Wada, H., Yeh, E.T., and Kamitani, T. (1999). Identification of NEDD8-conjugation site in human cullin-2. Biochem. Biophys. Res. Commun. 257, 100–105. [DOI] [PubMed] [Google Scholar]
- Xu, L., Liu, F., Lechnerc, E., Genschick, P., Crosby, W.L., Ma, H., Peng, W., Huang, D., and Xie, D. (2002). The SCFCOI1 ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14, 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenser, N., Ellsmore, A., Leasure, C., and Callis, J. (2001). Auxin modulates the degradation rate of Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 98, 11795–11800. [DOI] [PMC free article] [PubMed] [Google Scholar]