Abstract
It has long been hypothesized that cortical microtubules (MTs) control the orientation of cellulose microfibril deposition, but no mutants with alterations of MT orientation have been shown to affect this process. We have shown previously that in Arabidopsis, the fra2 mutation causes aberrant cortical MT orientation and reduced cell elongation, and the gene responsible for the fra2 mutation encodes a katanin-like protein. In this study, using field emission scanning electron microscopy, we found that the fra2 mutation altered the normal orientation of cellulose microfibrils in walls of expanding cells. Although cellulose microfibrils in walls of wild-type cells were oriented transversely along the elongation axis, cellulose microfibrils in walls of fra2 cells often formed bands and ran in different directions. The fra2 mutation also caused aberrant deposition of cellulose microfibrils in secondary walls of fiber cells. The aberrant orientation of cellulose microfibrils was shown to be correlated with disorganized cortical MTs in several cell types examined. In addition, the thickness of both primary and secondary cell walls was reduced significantly in the fra2 mutant. These results indicate that the katanin-like protein is essential for oriented cellulose microfibril deposition and normal cell wall biosynthesis. We further demonstrated that the Arabidopsis katanin-like protein possessed MT-severing activity in vitro; thus, it is an ortholog of animal katanin. We propose that the aberrant MT orientation caused by the mutation of katanin results in the distorted deposition of cellulose microfibrils, which in turn leads to a defect in cell elongation. These findings strongly support the hypothesis that cortical MTs regulate the oriented deposition of cellulose microfibrils that determines the direction of cell elongation.
INTRODUCTION
Newly divided cells in plants undergo significant elongation before they mature into different cell types, such as fiber cells, tracheary elements, and sieve elements. The direction and degree of elongation determine the morphology of cells; thus, dissecting the molecular mechanisms that regulate cell elongation will help us understand cellular morphogenesis. Because plant cells are encased in rigid cell walls made of networks of cellulose, hemicellulose, and other matrix components, the patterned deposition of cell walls and the subsequent loosening of the cell wall networks are critical for the directional elongation of cells.
Cellulose microfibrils, which are deposited in a transverse direction along the axis of elongating cells, have been proposed to control cellular morphogenesis (Green, 1962). Disruption of the normal deposition of cellulose microfibrils by mutation of a cellulose synthase gene in the temperature-sensitive rsw1 mutant is correlated with the alteration of cell elongation (Sugimoto et al., 2001). The transverse deposition of cellulose microfibrils along the axis of elongating cells presumably allows directional expansion through loosening of the cellulose and hemicellulose networks. Enzymes such as expansins are proposed to be involved in the cell wall–loosening process during cell elongation (Cosgrove, 1998).
The transverse deposition of cellulose microfibrils along the long axis of elongating cells has long been proposed to be controlled by the transversely oriented cortical microtubules (MTs) lying underneath the plasma membrane, a hypothesis called MT/microfibril parallelism. Two lines of evidence support this hypothesis. First, cortical MTs often align in parallel with cellulose microfibrils in elongating cells. Since the first observation by Ledbetter and Porter (1963), the coalignment of MTs and microfibrils has been confirmed in both algae and land plants (Giddings and Staehelin, 1991; Baskin, 2001). Second, alteration of the orientation of cortical MTs by drugs has been shown to cause changes in the oriented deposition of cellulose microfibrils (Giddings and Staehelin, 1991; Baskin, 2001).
Treatment with MT-depolymerizing drugs, such as colchicine and amiprophos-methyl, causes randomization of both MTs and newly deposited cellulose microfibrils. Treatment with taxol, a MT-stabilizing drug, results in the stabilization of MT patterns and the concomitant coalignment of cellulose microfibrils. The coalignment of MTs and microfibrils has been demonstrated in both elongating cells and cells undergoing secondary wall thickening (Baskin, 2001).
Although it seems certain that cortical MTs control the orientation of cellulose microfibril deposition, which determines the direction of cell elongation, there are some exceptions that appear not to favor MT/microfibril parallelism (Preston, 1988). It has been observed that cellulose microfibrils appear to be oriented transversely earlier before the onset of cell elongation than do cortical MTs, which raises questions about the obligate nature of MT/microfibril parallelism (Sugimoto et al., 2000; Wasteneys, 2000). In addition, cells with tip growth, such as pollen tubes and root hairs, obviously do not exhibit the coalignment of MTs and cellulose microfibrils, indicating that cellulose microfibril deposition in tip-growing cells is independent of MT orientation (Emons et al., 1992).
An alternative hypothesis has been proposed whereby the coalignment of MTs and cellulose microfibrils might be determined by cellular geometry rather than by MT control of microfibril order (Emons and Mulder, 1998). It also has been suggested that simultaneous alteration of the orientations of MT and cellulose microfibrils by pharmacological drugs could be caused by a disruption of cell growth, which could evoke a second parameter that is responsible for the observed effect (Emons et al., 1992). As a result of these exceptions, it is necessary to use genetic tools to further investigate the phenomenon of MT/microfibril parallelism.
The use of mutants with direct alteration of cortical MTs or cellulose microfibril patterns could eliminate possible nonspecific effects that may occur with the use of pharmacological drugs. Cellulose microfibrils have been proposed to be able to impose the orientation of MTs (Williamson, 1990; Emons et al., 1992), and this is substantiated by pharmacological studies showing that drug inhibition of cellulose synthesis could alter the pattern of MTs (Fisher and Cyr, 1998). However, study of the rsw1 mutant, which has a defect in cellulose biosynthesis, demonstrates that although the orientation of cellulose microfibrils in the mutant is distorted dramatically, the cortical MT pattern retains the wild-type appearance (Sugimoto et al., 2001). Therefore, it was proposed that there is no direct control of MT orientation by cellulose microfibrils.
Several Arabidopsis mutants, such as fass (Torres-Ruiz and Jürgens, 1994; McClinton and Sung, 1997), ton1 and ton2 (Traas et al., 1995; Thion et al., 1998), cho (Mayer et al., 1999), hal (Mayer et al., 1999), pfi (Mayer et al., 1999), por (Mayer et al., 1999), fra2/bot1 (Bichet et al., 2001; Burk et al., 2001), and mor1 (Whittington et al., 2001), have been shown to alter cortical MT patterns and affect cellular morphology. The genes for two of these mutants have been cloned and characterized.
The MOR1 gene encodes a large MT-associated protein that is required for MT stability, and mutation of the MOR1 gene causes the fragmentation of cortical MTs and swollen cells (Whittington et al., 2001). The gene responsible for the fra2 mutation encodes a katanin-like protein (abbreviated AtKTN1) that is essential for the normal cortical MT patterns during the initiation and continuation of cell elongation (Burk et al., 2001). The katanin-like protein, which also is abbreviated AtKSS, appears to be localized in the cytoplasm and around the nuclear envelope (McClinton et al., 2001). Because MOR1 and katanin-like protein are involved directly in the regulation of cortical MT patterns, these mutants offer ideal opportunities for testing the roles of MTs in the control of oriented cellulose microfibril deposition.
Mutation of the katanin-like protein in the fra2 mutant caused a dramatic reduction in the length and an increase in the width of all cell types examined except tip-growing cells. Examination of the MT organization in roots showed that the fra2 mutation caused delays in the disassembly of perinuclear MT arrays and in the assembly of cortical MT arrays in expanding cells. As a result, cortical MTs in expanding cells of fra2 did not show the uniform transverse orientation that was seen in the wild type (Burk et al., 2001). Defects in cell elongation and MT pattern also have been demonstrated in the bot1 mutant (Bichet et al., 2001), which is believed to be allelic to the fra2 mutant. The correlation between the disruption of normal cortical MT pattern and the reduction in cell elongation in the fra2/bot1 mutants suggests a direct role of MTs in the control of cell elongation.
In this study, we investigate whether the altered MT pattern caused by the fra2 mutation affects the oriented deposition of cellulose microfibrils. Using field emission scanning electron microscopy, we show that the fra2 mutation dramatically alters the orientation of cellulose microfibrils in expanding cells. Although cellulose microfibrils in elongating cells of the wild type run in a transverse direction along the long axis, microfibrils in expanding cells of fra2 often form bands and run in different directions. We also show that cortical MTs in cells of fra2 stems and petioles are severely disorganized. In addition, the primary and secondary walls of fra2 cells are significantly thinner than those of the wild type. We demonstrate that the Arabidopsis katanin-like protein possesses in vitro MT-severing activity. These results suggest that the katanin-like protein, by influencing MT patterns, affects the oriented deposition of cellulose microfibrils, cell wall biosynthesis, and directional cell elongation.
RESULTS
Orientation of Cellulose Microfibrils in Elongating Root Cells
Arabidopsis primary roots are an ideal source for the examination of cells at different phases of cell elongation. It has been shown that cells at the early-elongating, rapidly elongating, and late-elongating phases all have cortical MTs aligned transversely along the axis of elongation (Sugimoto et al., 2000). In parallel with the MT orientation, cellulose microfibrils at the innermost layer of the cell wall also are positioned in a transverse pattern (Sugimoto et al., 2000) (Figure 1). This indicates that Arabidopsis roots are an excellent system in which to investigate the roles of MTs in the oriented deposition of microfibrils and cell elongation.
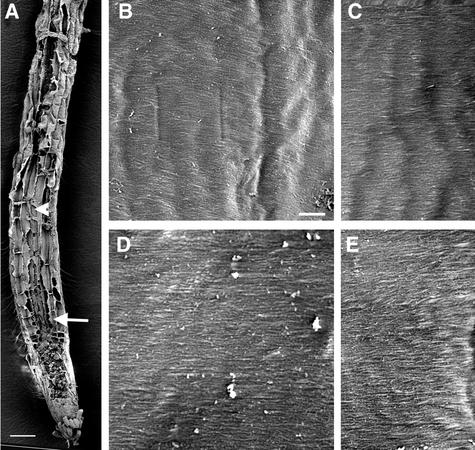
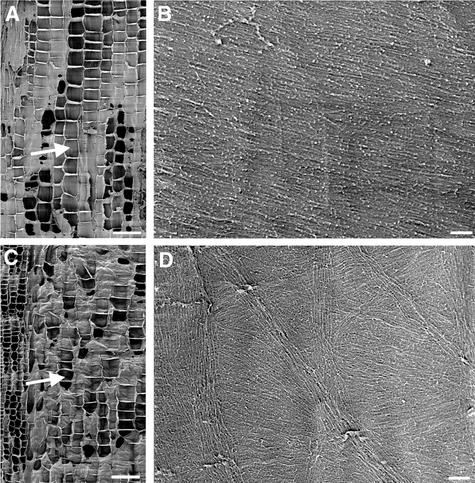
Figure 1.
Visualization of Cellulose Microfibrils in the Innermost Layer of Wild-Type Root Cell Walls.
Roots of 3-day-old wild-type seedlings were sectioned longitudinally, and the cellulose microfibrils in the innermost layer of cell walls were observed using field emission scanning electron microscopy. Individual microfibrils are seen as distinct lines. The vertical direction of the cellulose microfibril images in all figures corresponds to the elongation axis. Square marks, if present, at the centers of the images in all figures are the result of beam focusing.
(A) Longitudinal section of a root showing early-elongating (arrow) and rapidly elongating (arrowhead) cells.
(B) and (C) Cellulose microfibrils of rapidly elongating cells showing a transverse orientation along the elongation axis.
(D) and (E) Cellulose microfibrils of early-elongating cells showing a transverse orientation along the elongation axis.
Bar in (A) = 50 μm; bar in (B) = 0.5 μm for (B) to (E).
We showed previously that mutation of a katanin-like protein in the fra2 mutant causes alterations in cortical MT orientation and cell elongation (Burk et al., 2001). To investigate whether the fra2 mutation reduces cell elongation by altering cellulose microfibril deposition, we examined the orientation of cellulose microfibrils in elongating root cells of the wild type and the fra2 mutant (Figures 1 and 2). Using field emission scanning electron microscopy, we visualized cellulose microfibrils in the innermost layer of cell walls, which represent the most recently deposited microfibrils.
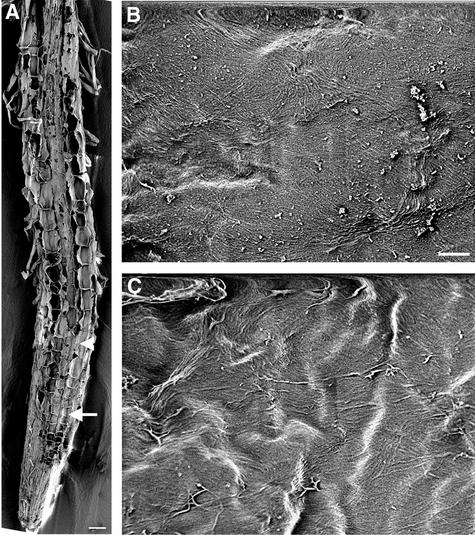
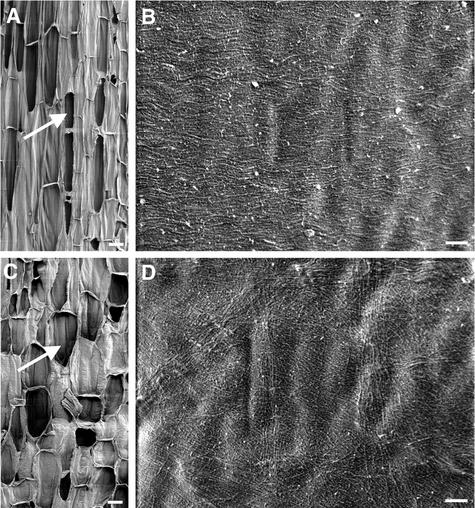
Figure 2.
Visualization of Cellulose Microfibrils in the Innermost Layer of fra2 Root Cell Walls.
(A) Longitudinal section of a root showing early-expanding (arrow) and rapidly expanding (arrowhead) cells.
(B) and (C) Cellulose microfibrils of rapidly expanding (B) and early-expanding (C) cells showing distorted orientations. Note that bands of microfibrils run in different directions.
Bar in (A) = 50 μm; bar in (B) = 0.5 μm for (B) and (C).
After exiting the apical meristem zone, cortical cells of fra2 roots (Figure 2A) underwent much less elongation compared with the wild type (Figure 1A). Cellulose microfibrils in walls of cortical cells from expanding regions of fra2 roots (Figures 2B and 2C) were distorted dramatically in orientation compared with the uniform transverse orientation seen in the wild type (Figures 1B to 1E). Some microfibrils appeared to form bands in groups of 8 to 12 individuals and ran in different directions with twists and turns. It was noted that some microfibril bands made a 180° turn within a small area (Figures 2B and 2C). These results clearly indicated that the fra2 mutation altered the oriented deposition of cellulose microfibrils in elongating root cells.
Orientation of Cortical MTs in Elongating Root Cells
Previous work has shown that the cortical MT orientation in root cells of fra2/bot1 is altered (Bichet et al., 2001; Burk et al., 2001), but no clear, detailed images of cortical MT pattern in cells from early-expanding and rapidly expanding regions of fra2/bot1 roots have been shown. To investigate in detail the cortical MT pattern in elongating cells of the wild type and the fra2 mutant, we performed immunofluorescence labeling of MTs in root cells.
In wild-type roots (Figure 3A), cortical MTs were oriented transversely along the long axis of both early-elongating and rapidly elongating cells (Figures 3B and 3C). By contrast, in fra2 roots (Figure 4A), cortical MTs in early-expanding cells were severely disoriented (Figures 4B to 4E) and ran in different directions (Figures 4F to 4H). It was noted that cortical MTs in some regions of cells aligned in a nearly transverse orientation (Figures 4B and 4C). It was evident that many cortical MTs converged at some common sites in both early-expanding (Figures 4D and 4E) and rapidly expanding (Figure 4H) cells, a pattern similar to that of the aster-like MTs seen around the perinuclear envelope of fra2 cells during the initiation of cell expansion (Burk et al., 2001).
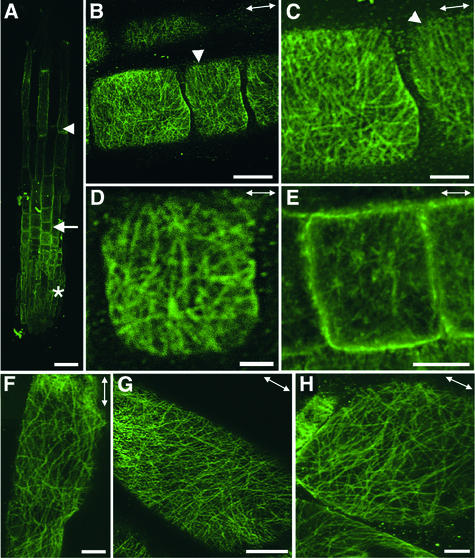
Figure 3.
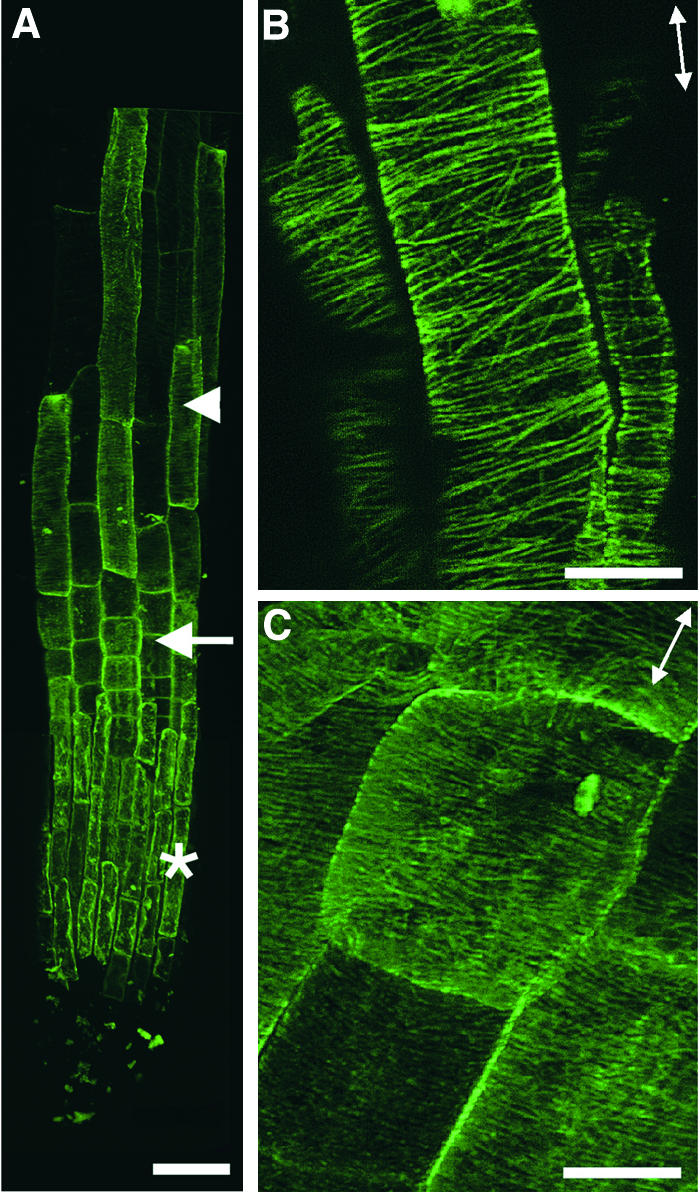
Visualization of Cortical MTs in Wild-Type Root Epidermal Cells.
Roots of 3-day-old seedlings were used for MT labeling with a monoclonal antibody against α-tubulin. Antibody-labeled MTs were detected with fluorescein isothiocyanate–conjugated secondary antibodies, and the signals of fluorescent MTs (green) were visualized using a confocal microscope. Double-headed arrows indicate the elongation axis.
(A) View of a whole root tip showing root cap cells (asterisk), early-elongating cells (arrow), and rapidly elongating cells (arrowhead).
(B) Rapidly elongating cells showing the transverse orientation of cortical MTs.
(C) Early-elongating cells showing the transverse orientation of dense cortical MTs.
Bar in (A) = 40 μm; bars in (B) and (C) = 8 μm.
Figure 4.
Visualization of Cortical MTs in fra2 Root Epidermal Cells.
Roots of 3-day-old seedlings were used for immunofluorescent labeling of MTs, and the signals of fluorescent MTs (green) were visualized using a confocal microscope. Double-headed arrows indicate the elongation axis.
(A) View of a whole root tip showing root cap cells (asterisk), early-expanding cells (arrow), and rapidly expanding cells (arrowhead).
(B) to (E) Early-expanding cells showing aberrant orientations of cortical MTs. Note that many MTs appeared to converge at some common sites. Also note that some regions of cells had cortical MTs aligned in a nearly transverse orientation (arrowheads).
(F) to (H) Rapidly expanding cells showing aberrant orientation of cortical MTs. Note that many MTs in (H) appeared to converge at some common sites.
Bar in (A) = 40 μm; bars in (B), (E), and (G) = 8 μm; bars in (C), (F), and (H) = 4 μm; bar in (D) = 2 μm.
Optical sectioning through the nuclear envelope did not show any apparent convergent MT sites around the nuclear envelope in expanding fra2 cells (data not shown). It appeared that the elongation of epidermal cells (Figure 4A) in fra2 roots was less affected than that of cortical cells (Figure 2A). These results indicated that the fra2 mutation caused a dramatic alteration in the orientations of both cortical MTs and cellulose microfibrils during different stages of cell elongation in roots.
Orientation of Cellulose Microfibrils in Elongating Cells of Hypocotyls, Stems, and Petioles
Because the fra2/bot1 mutations cause a dramatic reduction in cell length in all organs, such as roots, hypocotyls, stems, leaf blades, petioles, and floral organs (Bichet et al., 2001; Burk et al., 2001), we investigated whether an alteration in oriented cellulose microfibril deposition occurred in other organs in addition to roots. In walls of elongating cortical cells of wild-type hypocotyls (Figure 5A), cellulose microfibrils were oriented transversely along the axis of elongation (Figure 5B). By contrast, the microfibril orientation apparently was disorganized in the walls of expanding cortical cells of fra2 hypocotyls (Figures 5C and 5D). Like the microfibrils in the walls of expanding root cells, it appeared that groups of 6 to 12 microfibrils often formed bands that ran in different directions.
Figure 5.
Visualization of Cellulose Microfibrils in the Innermost Layer of Elongating Hypocotyl Cell Walls.
(A) Longitudinal section of a wild-type hypocotyl showing elongating cortical cells (arrow).
(B) Cellulose microfibrils of a wild-type elongating cortical cell showing a transverse orientation.
(C) Longitudinal section of an elongating fra2 hypocotyl showing cortical cells (arrow).
(D) Cellulose microfibrils of a fra2 cortical cell showing bands of microfibrils running in different directions.
Bars in (A) and (C) = 50 μm; bars in (B) and (D) = 0.5 μm.
The cell types affected most severely by the fra2 mutation were pith cells in inflorescence stems and parenchyma cells in petioles (Burk et al., 2001). These cells often were isodiametric in the mutant instead of cylindrical, as seen in the wild type. Some of the fra2 pith cells were reduced so severely in length that the cell width was longer than the length (Burk et al., 2001). Field emission scanning electron microscopy showed that the microfibrils in the walls of fra2 pith and petiole cells also were affected.
In wild-type elongating pith cells (Figure 6A), cellulose microfibrils ran in parallel and were oriented in a transverse direction (data not shown) or with a small angle relative to the transverse direction (Figure 6B). In expanding pith cells of fra2 (Figure 6C), the cellulose microfibrils often formed bands and were oriented in different directions (Figure 6D). Similarly, cellulose microfibrils in walls of parenchyma cells of petioles were oriented transversely along the elongating axis in the wild type (Figures 7A and 7B), whereas bands of microfibrils ran in various directions in the fra2 mutant (Figures 7C and 7D). Together, these results indicated that the fra2 mutation dramatically altered the cellulose microfibril deposition pattern in primary walls of elongating cells from various organs.
Figure 6.
Visualization of Cellulose Microfibrils in the Innermost Layer of Elongating Pith Cell Walls.
(A) Longitudinal section of the elongating region of a wild-type stem showing pith cells (arrow).
(B) Cellulose microfibrils in wild-type pith cell walls showing a transverse orientation.
(C) Longitudinal section of the elongating region of a fra2 stem showing pith cells (arrow).
(D) Cellulose microfibrils in fra2 pith cell walls showing various orientations.
Bars in (A) and (C) = 50 μm; bars in (B) and (D) = 0.25 μm.
Figure 7.
Visualization of Cellulose Microfibrils in the Innermost Layer of Elongating Petiole Cell Walls.
(A) Longitudinal section of an elongating wild-type petiole showing parenchyma cells (arrow).
(B) Cellulose microfibrils in wild-type parenchyma cell walls showing a transverse orientation.
(C) Longitudinal section of an elongating fra2 petiole showing parenchyma cells (arrow).
(D) Cellulose microfibrils in fra2 parenchyma cell walls showing a disoriented pattern.
Bars in (A) and (C) = 50 μm; bars in (B) and (D) = 0.25 μm.
Orientation of Cellulose Microfibrils in the Secondary Walls of Fiber Cells
Arabidopsis inflorescence stems develop interfascicular fiber cells next to the endodermis (Zhong et al., 1997, 2001). Interfascicular fiber cells in the fra2 mutant have shortened length and reduced thickness of secondary cell walls (Burk et al., 2001). To investigate whether the fra2 mutation affects the oriented cellulose microfibril deposition during secondary wall formation, we examined the orientation of cellulose microfibrils in the innermost layer of walls of mature fiber cells.
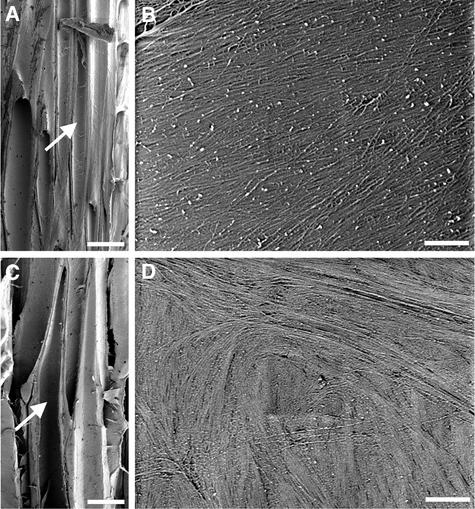
Cellulose microfibrils in the middle part of wild-type fiber cells ran uniformly in the same direction with an angle of 15 to 25° relative to the transverse orientation (Figures 8A and 8B). At the tip of fiber cells, cellulose microfibrils ran in parallel with the elongation axis (data not shown). By contrast, the orientation of cellulose microfibrils in secondary walls of fra2 fiber cells was severely distorted (Figures 8C and 8D). Cellulose microfibrils often formed bands and ran in different directions in a swirl-like pattern. This indicated that the fra2 mutation also altered the pattern of cellulose microfibril deposition in secondary walls of fiber cells.
Figure 8.
Visualization of Cellulose Microfibrils in the Innermost Layer of Fiber Cell Walls.
(A) Longitudinal section of the nonelongating region of a wild-type stem showing interfascicular fiber cells (arrow).
(B) Cellulose microfibrils in the middle part of a wild-type fiber cell. Note that microfibrils run in parallel at an angle of 15 to 25° relative to the transverse orientation.
(C) Longitudinal section of the nonelongating region of a fra2 stem showing interfascicular fiber cells (arrow).
(D) Cellulose microfibrils in the middle part of a fra2 fiber cell showing a disoriented pattern.
Bars in (A) and (C) = 25 μm; bars in (B) and (D) = 0.5 μm.
Quantitative Analysis of Cellulose Microfibril Orientations
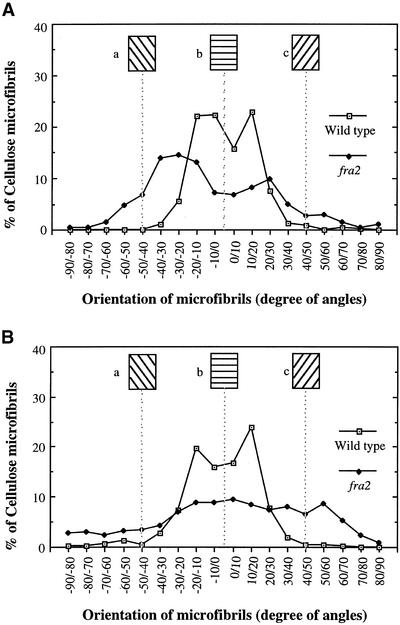
The results described above show clearly that the orientation of cellulose microfibrils in fra2 cell walls was altered compared with that in wild-type cell walls (Figures 1, 2, and 5 to 8). To quantify the degree of distortion of cellulose microfibril orientation in fra2 cell walls, we measured the angles of individual cellulose microfibrils. Most microfibrils in the walls of wild-type root (Figure 9A) and pith (Figure 9B) cells were oriented transversely, with <30° deviation from the transverse direction. Cellulose microfibrils in walls of fra2 root (Figure 9A) and pith (Figure 9B) cells had a much wider angle of deviation from the transverse orientation, especially those in walls of pith cells that lacked predominant angle peaks. This finding indicated that cellulose microfibrils in fra2 cells were oriented more randomly than those in wild-type cells.
Figure 9.
Quantitative Analysis of the Orientation of Cellulose Microfibrils.
Angles of 450 individual cellulose microfibrils from field emission scanning electron micrographs of the innermost layer of cell walls were measured and are represented in angles at 10° increments. Cellulose microfibrils at different angles were calculated as a percentage of total microfibrils measured. The transverse direction relative to the elongation axis was arbitrarily defined as 0°, and angles deviating from the transverse orientation are represented by positive or negative numbers of degrees. Insets (a), (b), and (c) show the directions of microfibrils corresponding to the defined angles.
(A) Cellulose microfibrils in walls of root cortical cells. Cellulose microfibrils in wild-type cell walls were oriented mostly in a transverse direction with a slight deviation, whereas the angles of microfibrils in fra2 cell walls had a much wider distribution.
(B) Cellulose microfibrils in walls of pith cells. Cellulose microfibrils in wild-type cell walls showed a prominent transverse orientation with <30° deviation, whereas the angles of microfibrils in fra2 cell walls had a much wider distribution.
Orientation of Cortical MTs in Elongating Cells of Stems and Petioles
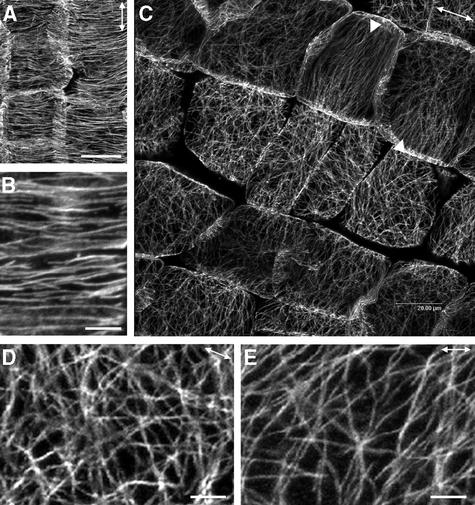
Because we have demonstrated that the aberrant cellulose microfibril deposition in root cells was correlated with disorganized cortical MTs in the fra2 mutant, we next investigated whether the altered cellulose microfibril deposition pattern in cells of other organs also was accompanied by a disrupted pattern of cortical MTs. We performed immunolocalization of cortical MTs in stems and petioles. In elongating pith cells of the wild type, cortical MTs were oriented transversely along the elongation axis (Figure 10A). A close-up of wild-type cells showed that cortical MTs were aligned in parallel with a small degree of deviation (Figure 10B). Visualization of cortical MTs in fra2 pith cells showed a sharp contrast to those in wild-type cells. Cortical MTs in fra2 cells were oriented aberrantly and lost their typical parallel alignment pattern (Figure 10C). It was noted that a few cells had cortical MTs oriented in a nearly transverse pattern. A close-up of MTs in fra2 cells clearly showed the aberrant organization of cortical MTs (Figures 10D and 10E) compared with the wild type (Figure 10B). Furthermore, many cortical MTs in fra2 cells appeared to converge into some common sites. Quantitative analysis of cortical MT angles showed that most cortical MTs in wild-type pith cells were oriented transversely, with <20° deviation from the transverse direction, whereas those in fra2 pith cells were oriented in various angles with a wide deviation from the transverse direction (Figure 11).
Figure 10.
Visualization of Cortical MTs in Pith Cells of Stems.
Pith cells from elongating internodes were used for MT labeling with a monoclonal antibody against α-tubulin. Antibody-labeled MTs were detected with fluorescein isothiocyanate–conjugated secondary antibodies, and the signals of fluorescent MTs were visualized with a confocal microscope. Double-headed arrows indicate the elongation axis.
(A) Wild-type pith cells showing the transversely oriented cortical MTs.
(B) High-magnification image of cortical MTs in the wild type showing their transverse alignment.
(C) fra2 pith cells showing aberrantly oriented cortical MTs. Note that in a few cells, cortical MTs aligned in a nearly transverse orientation (arrowheads).
(D) and (E) High-magnification image of cortical MTs in the fra2 mutant showing their aberrant patterns and converging sites.
Bars in (A) and (C) = 20 μm; bars in (B), (D), and (E) = 4 μm.
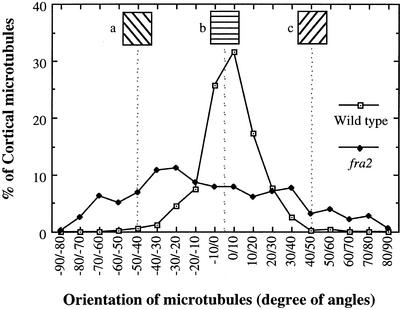
Figure 11.
Quantitative Analysis of the Orientation of Cortical MTs.
Angles of 535 individual cortical MTs from pith cells of the wild type and fra2 were measured and are represented in angles at 10° increments. The number of cortical MTs in a certain angle range was calculated as a percentage of total MTs measured. The transverse direction relative to the elongation axis was arbitrarily defined as 0°, and angles deviating from the transverse direction are represented by positive or negative numbers of degrees. Insets (a), (b), and (c) show the directions of cortical MTs corresponding to the defined angles. Cortical MTs in wild-type cells showed a prominent transverse orientation with <20° deviation, whereas the angles of cortical MTs in fra2 cells had a much wider distribution.
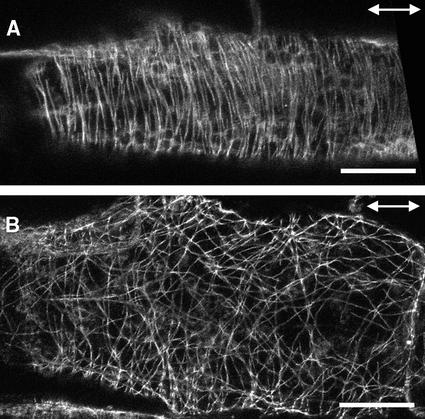
Immunolocalization of cortical MTs in petioles of the wild type and fra2 showed patterns similar to those in stems (Figure 12). Although cortical MTs in wild-type cells were aligned transversely along the elongation axis (Figure 12A), those in fra2 cells apparently were disorganized and often converged to some common sites (Figure 12B). These results demonstrate that the katanin-like protein is required for the normal transverse parallel alignment of cortical MTs, and the abnormal MT pattern is correlated directly with the aberrant deposition of cellulose microfibrils in the fra2 mutant.
Figure 12.
Visualization of Cortical MTs in Elongating Petiole Cells.
Parenchyma cells from elongating petioles were used for immunofluorescent labeling of MTs. Double-headed arrows indicate the elongation axis.
(A) A wild-type petiole parenchyma cell showing the transversely oriented cortical MTs.
(B) A fra2 petiole parenchyma cell showing aberrantly oriented cortical MTs.
Bars = 20 μm.
Anatomical Structure of Primary and Secondary Walls
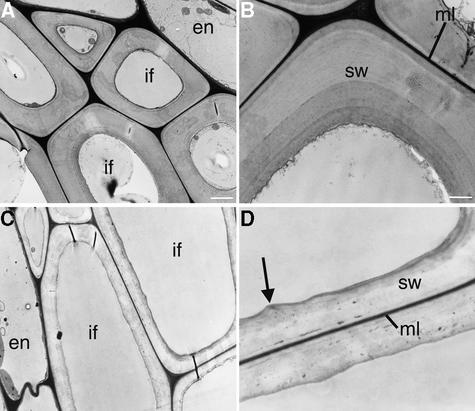
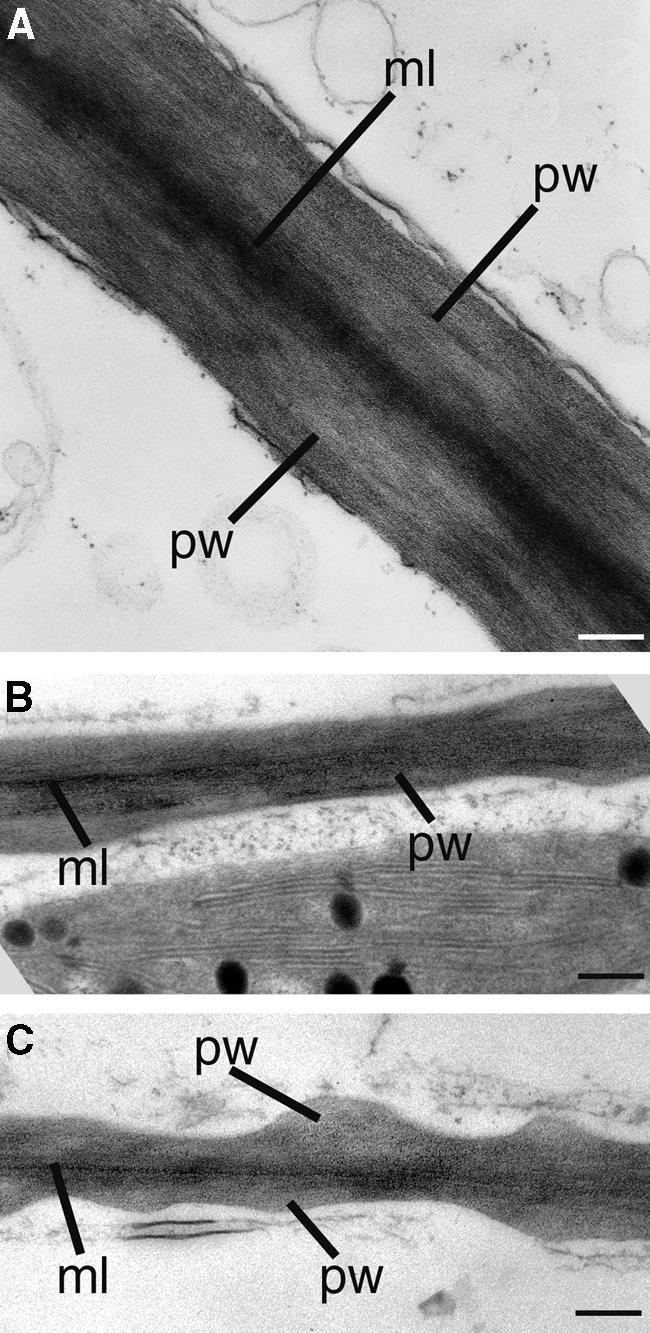
To investigate whether the distorted orientation of cellulose microfibrils affected cell wall morphology, we examined the ultrastructure of cell walls in the fra2 mutant. Transmission electron microscopy revealed that interfascicular fiber cells in the fra2 mutant were much larger in the radial dimension (Figure 13C) and that their walls were reduced dramatically in thickness (Figure 13D) compared with the same features in wild-type cells (Figures 13A and 13B).
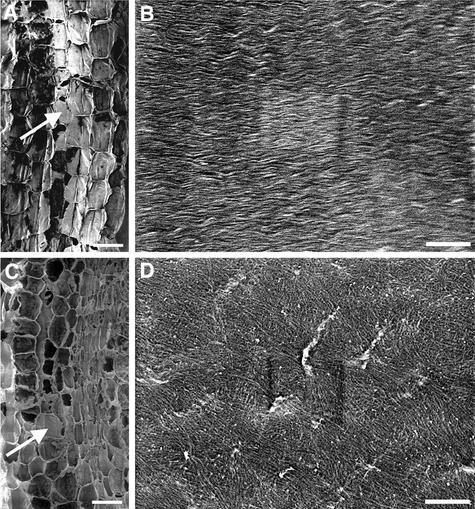
Figure 13.
Structure of Fiber Cell Walls in the Wild Type and the fra2 Mutant.
Nonelongating regions of stems of 8-week-old plants were cross-sectioned, and the interfascicular regions in the ultrathin sections were observed with a transmission electron microscope.
(A) Interfascicular fiber cells of the wild type showing thick cell walls and small intracellular space.
(B) High-magnification image of the secondary wall of a wild-type fiber cell showing clear layers and a smooth inner wall surface.
(C) Interfascicular fiber cells of the fra2 mutant showing much thinner cell walls and larger intracellular areas compared with those in the wild type (A).
(D) High-magnification image of the secondary wall of a fra2 fiber cell showing no apparent layers and a wavy inner wall surface (arrow).
En, endodermal cells; if, interfascicular fiber cells; ml, middle lamella; sw, secondary wall. Bars in (A) and (C) = 2.5 μm; bars in (B) and (D) = 1.1 μm.
In addition, although secondary walls of wild-type fiber cells were well stained and had distinct layers of wall thickening (Figure 13B), secondary walls of fra2 fiber cells stained less and lacked clear layers of wall thickening (Figure 13D). The middle lamella of fiber cell walls in both the wild type and fra2 were well stained (Figures 13B and 13D). Additionally, the inner wall surface of wild-type fiber cells was smooth (Figure 13B), whereas the inner wall surface of fra2 fiber cells appeared undulated (Figure 13D).
Similarly, examination of pith cells in inflorescence stems showed that the wall thickness was reduced significantly and that the inner wall surface was uneven in the fra2 mutant (Figures 14B and 14C) compared with the wild type (Figure 14A). Quantitative analysis showed that the wall thickness of fra2 pith cells and fiber cells was reduced to 63 and 58% of that in the wild type, respectively (Table 1). Longitudinal sections of fiber and pith cells did not show dramatic alterations in surface area of these cells between the wild type and the fra2 mutant (Burk et al., 2001). This finding indicated that the fra2 mutation dramatically affected the wall thickening of both pith cells and fiber cells.
Figure 14.
Structure of Pith Cell Walls in the Wild Type and the fra2 Mutant.
Nonelongating regions of stems of 8-week-old plants were cross-sectioned, and the pith cells in the ultrathin sections were observed with a transmission electron microscope.
(A) Walls of wild-type pith cells. Note the dark line of the middle lamella.
(B) and (C) Walls of fra2 pith cells showing thin walls and wavy inner wall surfaces.
ml, middle lamella; pw, primary wall. Bars = 0.21 μm.
Table 1.
Wall Thickness of Pith Cells and Fiber Cells in the Wild Type and the fra2 Mutant
| Wall Thickness (μm)a
|
||
|---|---|---|
| Cell Type | Pith Cellb | Fiber Cellc |
| Wild type | 0.35 ± 0.09 | 2.09 ± 0.50 |
| fra2 | 0.22 ± 0.07 | 1.21 ± 0.41 |
Wall thickness was measured from transmission electron micrographs of cells. Data shown are means ± se from 15 to 20 cells.
Pith cells from nonelongating internodes of inflorescence stems were used for the measurement of wall thickness.
Interfascicular fiber cells from nonelongating internodes of inflorescence stems were used for the measurement of wall thickness.
The Arabidopsis Katanin-Like Protein Possesses in Vitro MT-Severing Activity
The Arabidopsis katanin-like protein shows high sequence similarity to katanin from sea urchin, a protein known to sever MTs in vitro in an ATP-dependent manner (Hartman et al., 1998). Our finding that mutation of the katanin-like protein in the fra2 mutant disrupts the normal pattern of cortical MTs in elongating cells supports the roles of katanin in regulating MT organization. To confirm that the Arabidopsis katanin-like protein is a MT-severing protein, we tested its ability to sever MTs in vitro.
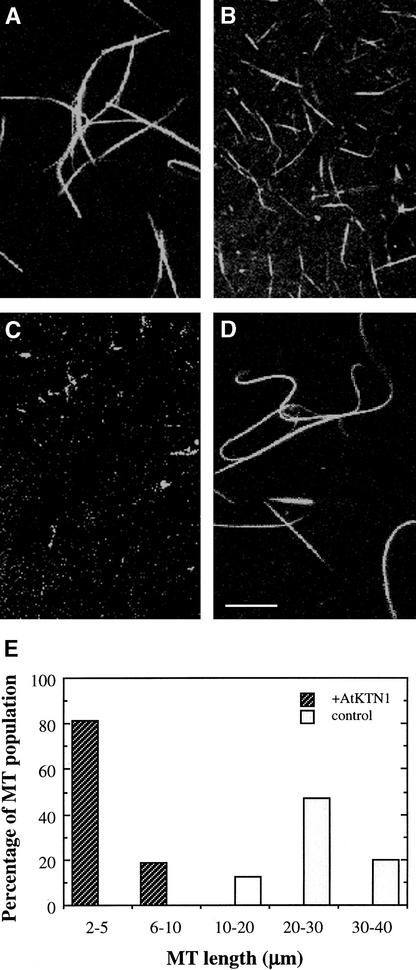
Recombinant Arabidopsis katanin-like protein expressed in insect cells was able to sever MTs (Figure 15). Taxol-stabilized, rhodamine-labeled MTs were fragmented into much shorter lengths after 5 min of incubation with the recombinant protein (Figure 15B). The length of MTs was reduced from the original average of 28 μm to an average of 4 μm (Figures 15B and 15E). After 10 min of incubation, most MTs disappeared, although a few very short MTs (<2 μm) were still visible (Figure 15C). When MTs were incubated with a control protein, they remained intact, with an average length of 28 μm (Figures 15D and 15E). These results demonstrate that the Arabidopsis katanin-like protein does possess MT-severing activity in vitro. They suggest that this protein is an ortholog of animal katanin and that it might affect MT patterns by controlling the MT disassembly process.
Figure 15.
MT-Severing Activity of Recombinant Arabidopsis Katanin.
Recombinant Arabidopsis katanin was expressed in insect cells and purified for assay of MT-severing activity. The purified protein was incubated with taxol-stabilized, rhodamine-labeled MTs, and the lengths of MTs were visualized with a confocal microscope. Typical fields of view are shown.
(A) MTs before incubation with recombinant katanin.
(B) MTs after incubation with recombinant katanin for 5 min. Note that MTs were cut into short fragments.
(C) MTs after incubation with recombinant katanin for 10 min. Note that some very short MTs (<2 μm) were still visible.
(D) MTs after incubation with a control protein for 10 min. Bar in (D) = 10 μm for (A) to (D).
(E) Distribution of MT lengths. The lengths of MTs were measured 5 min after treatment with a control protein (control) or recombinant Arabidopsis katanin (+AtKTN1). The distribution of MT lengths was expressed as a percentage of the total number of MTs measured (n = 150).
DISCUSSION
It has been proposed that cortical MTs undergo constant disassembly and assembly during cell elongation and that their overall transverse orientation along the elongation axis regulates the transverse pattern of cellulose microfibril deposition (Cyr and Palevitz, 1995; Baskin, 2001). Our findings demonstrate that the Arabidopsis katanin is essential for normal cortical MT patterns and the oriented deposition of cellulose microfibrils during cell elongation. These results provide genetic evidence in support of the hypothesis that cortical MTs control the pattern of cellulose microfibril deposition (Baskin, 2001).
Arabidopsis Katanin Is Essential for the Transverse Orientation of Cortical MTs
After cells exit from cytokinesis, MTs first appear around the nuclear envelope as a perinuclear MT array. MTs extend from the perinuclear area into the cortical region during the initial stage of cell expansion. It has been proposed that the MT nucleation sites are located around the nuclear envelope and that cortical MTs originate from these nucleation sites (Hasezawa and Nagata, 1991; Nagata et al., 1994; Hasezawa et al., 2000). During cell elongation, cortical MTs must adjust their orientation constantly to keep up with the increasing cell length. This constant change in MT organization, or MT dynamics, was proposed to be performed by the rapid disassembly, assembly, and translocation of MTs (Cyr and Palevitz, 1995).
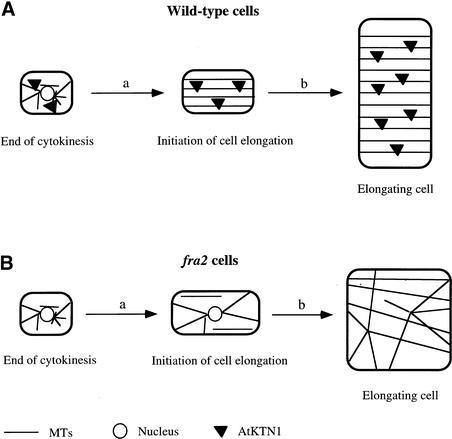
The findings that the fra2 mutation delays the disappearance of the perinuclear MT array during the initiation of cell elongation (Burk et al., 2001) and alters the normal cortical MT pattern throughout cell elongation (Figure 4) indicate that the katanin-like protein is essential for the normal MT patterns found during both the initiation and the continuation of cell elongation. The Arabidopsis katanin-like protein has been shown to possess MT-severing activity (Figure 15); thus, it is an ortholog of animal katanin and it most likely functions in the disassembly of the perinuclear MT array during the initiation of cell elongation (Figure 16A).
Figure 16.
Model of the Roles of Katanin (AtKTN1) in the Regulation of the Organization of Cortical MTs, Cellulose Microfibrils, and Directional Cell Elongation.
(A) In cells immediately after cytokinesis, AtKTN1 accelerates the formation of cortical MTs by disassembling perinuclear MTs from the nuclear envelope (a). During cell elongation (b), AtKTN1 mediates MT dynamic changes that are required for the transverse orientation of MTs along the elongation axis. The transversely oriented MTs regulate the parallel deposition of cellulose microfibrils, which determines the direction of cell elongation.
(B) Lack of AtKTN1 MT-severing activity in the fra2 mutant causes a delay in the disassembly of perinuclear MTs and a concomitant delay of cortical MT formation (a). During cell elongation (b), a decrease in MT dynamics caused by the lack of AtKTN1 MT-severing activity results in aberrant orientations of cortical MTs, which in turn leads to altered cellulose microfibril deposition and a reduction of directional cell elongation.
Disassembly of the perinuclear MT array by Arabidopsis katanin could be mediated by releasing perinuclear MTs that are then translocated to the cortical region or by severing perinuclear MTs into dimers that are then reassembled into MTs in the cortical region. During cell elongation, katanin is likely involved in the disassembly of cortical MTs that is essential to establish a transverse orientation (Figure 16A). In our model, lack of katanin MT-severing activity causes a delay in the disassembly of perinuclear MTs and, consequently, a delay in the appearance of cortical MTs during the initiation of cell elongation (Figure 16B). During cell elongation, lack of katanin MT-severing activity causes an alteration in normal cortical MT dynamics that in turn results in aberrant orientations of cortical MTs (Figure 16B).
It is important to note that, in addition to the aster-like MT pattern seen around the nuclear envelope during the initiation of cell expansion in the fra2 mutant (Burk et al., 2001), many cortical MTs also appeared to converge at some common sites in the cortical region of expanding fra2 cells (Figures 4, 10, and 12). It has been proposed that cortical MTs are likely nucleated at the cortical region (Vaughn and Harper, 1998); thus, these converged MT sites at the cortical region might be nucleation sites of cortical MTs during cell elongation. In the wild type, the katanin MT-severing activity might constantly release MTs from these sites, so that the converged MTs are not detectable. Lack of katanin MT-severing activity in the fra2 mutant might cause a delay in the release of MTs from these sites, enabling the converged MTs to be visualized at the cortical region.
It is tempting to propose that a timely release of MTs from these converged sites by katanin is essential for the formation of transversely oriented cortical MTs. A role of katanin MT-severing activity in the release of MTs from putative nucleation sites at the nuclear envelope and the cortical region is similar to the proposed roles of katanin MT-severing proteins in animal cells. Katanins in sea urchin embryonic cells (McNally et al., 1996; Hartman et al., 1998) and rat sympathetic neurons (Ahmad et al., 1999) are localized in centrosomes that are MT organization centers. Inactivation of katanin in neurons by microinjection of the katanin antibody resulted in a dramatic accumulation of MTs at the centrosome, indicating roles for katanin in the release of MTs from the centrosome (Ahmad et al., 1999). It will be very interesting to further investigate the nature of these converged MT sites in the fra2 mutant.
Despite the observation that the lack of katanin MT-severing activity in the Arabidopsis fra2 mutant causes a delay in MT dynamics, cortical MTs ultimately appear in the cortical region, although in aberrant orientations. This fact indicates that katanin MT-severing activity is not the only mechanism involved in the regulation of MT dynamics. Other mechanisms, such as dynamic instability and treadmilling (Walczak, 2000), also are likely involved in the regulation of MT dynamics in plants. However, these other mechanisms apparently could not compensate fully for the loss of katanin MT-severing activity in Arabidopsis.
Arabidopsis Katanin Is Required for Oriented Cellulose Microfibril Deposition and Cell Elongation
The fra2/bot1 mutations cause a dramatic reduction in cell length and an increase in cell width (Bichet et al., 2001; Burk et al., 2001). Because cellular morphology is determined by the ordered deposition of cellulose microfibrils, which is thought to be regulated by the pattern of cortical MTs (Baskin, 2001; Wasteneys, 2000), it is conceivable that the altered cellular morphology caused by the fra2/bot1 mutations could be a result of altered cellulose microfibril deposition. This notion was confirmed by field emission scanning electron microscopy showing the distorted orientation of cellulose microfibrils in walls of expanding cells of fra2.
Because Arabidopsis katanin is essential for normal MT patterns, we propose that katanin facilitates the transverse orientation of cortical MTs, which in turn dictates the ordered deposition of cellulose microfibrils during cell elongation (Figure 16A). Lack of katanin MT-severing activity in the fra2 mutant causes the formation of abnormal cortical MT patterns, which in turn results in a distorted orientation of cellulose microfibrils, leading to a dramatic reduction in cell elongation (Figure 16B).
Our finding that mutation of the katanin gene in the fra2 mutant alters the orientations of both cortical MTs and cellulose microfibrils provides genetic evidence of the roles that MTs play in the control of oriented cellulose microfibril deposition. Although previous studies have shown the coalignment of MTs and cellulose microfibrils and have documented the simultaneous alteration of their orientations by drugs, the hypothesis of MT and cellulose microfibril parallelism remains controversial (Wasteneys, 2000). Because animal katanin has been shown to target specifically to MTs (Hartman et al., 1998; Quarmby, 2000), it is unlikely that a lack of katanin MT-severing activity in plants would directly affect the movement of the cellulose synthase complex in the plasma membrane. Instead, the altered orientation of cellulose microfibrils in the fra2 mutant is most likely a result of the aberrant orientation of cortical MTs caused by a lack of katanin MT-severing activity.
Therefore, our results support the hypothesis that cortical MTs regulate the ordered deposition of cellulose microfibrils during cell elongation (Baskin, 2001). However, we could not completely exclude the possibility that the aberrant deposition of cellulose microfibrils in the fra2 mutant could be caused by the same mechanisms that account for the disorientation of cellulose microfibrils in the rsw1 mutant (Sugimoto et al., 2001), although it is not known how the rsw1 mutation causes aberrant orientations of cellulose microfibrils.
It is interesting that, unlike the uniformly aligned microfibrils in walls of wild-type cells, microfibrils in walls of fra2 mutant cells often form bands and run in different directions. It was proposed that cortical MTs might regulate the oriented deposition of cellulose microfibrils by delimiting the path of cellulose synthase movement (Herth, 1980; Giddings and Staehelin, 1991). If this is the case, the aberrantly oriented MTs in fra2 cells could cause abnormal delimitation of the path of cellulose synthase movement, which would lead to the formation of bands of microfibrils running in different directions.
In another model, it was hypothesized that cellulose synthase complexes might move directly along cortical MTs located underneath the plasma membrane. This model was first proposed by Heath (1974) and was supported experimentally by Hasezawa and Nozaki (1999). If this is the case, the aberrantly oriented and more stabilized cortical MTs in fra2 cells could provide prolonged tracks for the movement of cellulose synthase complexes and thus cause the formation of bands of microfibrils. In addition, it has been proposed that the movement of cellulose synthase complexes in the plasma membrane also might be constrained by cellular geometry (Emons and Mulder, 1998). This could explain why the orientations of cellulose microfibrils in fra2 cells are abnormal but not completely random, although the aberrantly oriented MTs in fra2 cells might lose their control of the oriented deposition of cellulose microfibrils. The formation of aberrant cellulose microfibril bands is likely the cause of the uneven inner wall surfaces seen in both primary and secondary walls of fra2 cells (Figures 13 and 14). This indicates that an ordered pattern of cortical MTs is not only essential for the oriented deposition of cellulose microfibrils but also important for the uniform thickening of cell walls around the cells.
In addition to its involvement in cell elongation, Arabidopsis katanin apparently is required for the normal branching of trichomes. In the fra2 mutant, most trichomes have one branch point instead of two branch points seen in the wild type (Burk et al., 2001). Because katanin is essential for the normal organization of MTs, this indicates that the normal patterns of cortical MTs are important in branch initiation during trichome morphogenesis. The roles of cortical MTs in trichome branching have been demonstrated in drug inhibitor experiments (Mathur and Chua, 2000) and in fass mutants, which are known to have altered MT patterns (Torres-Ruiz and Jürgens, 1994; McClinton and Sung, 1997). Because trichome branching is an anisotropic growth process that needs localized loosening of cellulose microfibrils, the reduced branch formation in the fra2 mutant might be caused by the altered deposition of cellulose microfibrils. It will be interesting to determine whether the patterns of MTs and cellulose microfibrils are altered in trichomes of the fra2 mutant.
Arabidopsis Katanin Is Required for Oriented Cellulose Microfibril Deposition during Secondary Wall Thickening
In addition to a role in regulating cellulose microfibril deposition in primary walls of elongating cells, cortical MTs are proposed to regulate cellulose microfibril deposition during secondary wall thickening (Baskin, 2001). This notion is supported by our observation that mutation of the katanin gene in the fra2 mutant causes aberrant orientations of cellulose microfibrils in fiber cell walls (Figure 8). Secondary walls of tracheids and fibers typically are composed of three distinct layers of cellulose microfibrils: S1, S2, and S3. The S1 and S3 layers have cellulose microfibrils in a flat helix, and the S2 layer has cellulose microfibrils in a steep helix (Harada and Coté, 1985). Cellulose microfibrils in secondary walls have been shown to be aligned in parallel with underlying cortical MTs (Robards and Kidwai, 1972; Seagull, 1992; Abe et al., 1994, 1995; Prodhan et al., 1995), suggesting that the cortical MTs undergo dynamic changes to form three distinctly oriented patterns during different stages of secondary wall thickening. Our finding that secondary walls of fra2 fiber cells lack distinct layers indicates that katanin is essential for the formation of distinct layers of cellulose microfibrils during secondary wall thickening.
MT Dynamics Is Required for Normal Cell Wall Biosynthesis
Although cortical MTs are believed to regulate the orientation of cellulose microfibrils, it was proposed that they do not affect the biosynthesis of cellulose and other cell wall components (Seagull and Falconer, 1991). This idea was based on early pharmacological studies showing that a disturbance of cortical MT organization alters the pattern of cellulose microfibril deposition but does not affect cellulose biosynthesis. However, the effects of cortical MT alterations on cell wall biosynthesis could not be determined easily in drug-treated cells. The fra2 mutation was shown to reduce the amount of cellulose and hemicellulose and to increase the condensation of lignin in total cell wall extracts of stems (Burk et al., 2001). The overall reduction of cell wall components apparently occurs in both primary and secondary walls, as revealed by transmission electron microscopy (Figures 13 and 14). This finding indicates that the normal MT pattern is essential not only for the oriented deposition of cellulose microfibrils but also for normal cell wall biosynthesis during primary and secondary wall formation. It is likely that the aberrant MT pattern in fra2 cells causes a decrease in the rate of cellulose synthase movement, which leads to a reduced amount of cellulose synthesis.
METHODS
Materials
Wild-type and fra2 Arabidopsis thaliana seeds were germinated on Murashige and Skoog (1962) medium on vertically placed plates and grown in a growth chamber at 24°C under a 16-h-light/8-h-dark photoperiod. Hypocotyls and roots from 3-day-old seedlings were used for the visualization of microtubules (MTs) or cellulose microfibrils. Stems and petioles used for the visualization of cellulose microfibrils were from plants grown in soil in a growth room at 22°C under a 12-h-light/12-h-dark photoperiod. The first emerging elongating internodes of inflorescence stems were used for the examination of MT and cellulose microfibrils in pith cells. The basal nonelongating internodes of inflorescence stems of 8-week-old plants were used for the examination of cellulose microfibrils in fiber cells.
Field Emission Scanning Electron Microscopy of Cellulose Microfibrils
Cellulose microfibrils at the innermost layer of cell walls were visualized using field emission scanning electron microscopy according to Sugimoto et al. (2000). Whole roots and hypocotyls were fixed directly, and elongating stems and petioles were cut longitudinally using a double-edge razor blade before fixation. Mature stems were cut longitudinally through the interfascicular fiber region using a dissection microscope before fixation. Roots and hypocotyls were sectioned longitudinally using a cryoultramicrotome (model MT6000-XL with CR2000 attachment; RMC Inc., Tucson, AZ).
After treatment, tissues were dried in a Samdri critical point dryer (Tousimis, Rockville, MD) before being mounted on stubs with carbon paste. Samples were coated with platinum using an Edwards 306 vacuum evaporator (Edwards High Vacuum International, Wilmington, MA) and viewed for cellulose microfibrils using a LEO 982 FE scanning electron microscope (LEO, Thornwood, NY).
Immunolocalization of MTs
MTs were visualized by immunolabeling α-tubulins according to Sugimoto et al. (2000). Roots from 3-day-old seedlings and young elongating petioles and stems were used for MT localization. Samples were probed first with mouse monoclonal antibody against chicken α-tubulin (1:800 dilution; Sigma) and then incubated with fluorescein isothiocyanate–conjugated goat antibody against mouse IgG (1:800 dilution; Sigma). MTs were visualized using a Leica TCS SP2 spectral confocal microscope (Leica Microsystems, Heidelberg, Germany). Images were saved and processed with Adobe Photoshop version 5.0 software (Adobe, San Jose, CA).
Transmission Electron Microscopy
Tissues were fixed in 2% (v/v) glutaraldehyde in phosphate buffer (50 mM, pH 7.2) at 4°C overnight. After fixation, tissues were postfixed in 2% (v/v) OsO4 for 2 h. After being washed in phosphate buffer, tissues were dehydrated in ethanol, infiltrated with Araldite/Embed 812 resin (Electron Microscopy Sciences, Fort Washington, PA), and finally polymerized in Araldite/Embed resin. For electron microscopy, 90-nm-thick sections were cut with a Reichert-Jung ultrathin microtome (C. Reichert Optische Werke AG, Vienna, Austria), mounted on formvar-coated gold slot grids, and poststained with uranyl acetate and lead citrate. Cell wall structure was visualized with a Zeiss EM 902A electron microscope (Jena, Germany).
Quantitative Analysis
Images of cellulose microfibrils at the innermost layer of the cell wall were obtained using a field emission scanning electron microscope and used for the measurements of cellulose microfibril angles. Briefly, three to four representative images of wild-type and fra2 cell walls from different cells were reduced to a fixed size, and a grid was overlaid on the image so that 130 evenly spaced grid points were available for analysis. At each grid point, the angle of the single microfibril that passed through or was closest to the point was measured using the ruler tool of Adobe Photoshop 5.0.
The microfibril was traced along its length to a fixed distance ∼0.24 μm from the grid point, and the angle was noted. If at any grid point the microfibrils were not resolved sufficiently for tracing, that point was discarded. All angles were measured relative to the axis perpendicular to the long axis of the cell. This perpendicular axis was arbitrarily assigned an angle of 0°. Using this method, microfibrils can approach angles of ±90° and correspond to fibrils approaching a longitudinal orientation in a left- or right-handed helix.
Images of cortical MTs were obtained using a confocal microscope and used for the measurement of cortical MTs. Representative images of wild-type and fra2 cells were overlaid with a grid so that at least 50 evenly spaced grid points were available for analysis within each cell. At selected grid points, the angle of the single MT that passed through or was closest to the point was measured using the ruler tool of Adobe Photoshop 5.0. The MT was traced along its length to a fixed distance ∼1 μm from the grid point, and the angle was noted. All angles were measured relative to the axis perpendicular to the long axis of the cell, which is consistent with the measurement technique used for cellulose microfibril angle determination.
Insect Cell Expression and Purification of Recombinant Arabidopsis Katanin
The expression of His-tagged Arabidopsis katanin was performed with an InsectSelect System kit (Invitrogen, Carlsbad, CA). Sf9 cells were grown and maintained as a monolayer at 27°C in TNM-FH medium plus 10% fetal bovine serum. Full-length Arabidopsis katanin cDNA containing a Kozak translation initiation sequence was cloned into BamHI and XbaI sites of the insect expression vector pIZ/V5-His to generate pIZ/AtKTN1-His. pIZ/V5-His vector contains the baculovirus immediate-early promoter OpIE2 that uses the host cell transcription machinery and does not require viral factor for the activation and expression of recombinant protein. pIZ/AtKTN1-His plasmid DNA was transfected into Sf9 cells with Insectin-Plus Liposomes (Invitrogen), and stable cell lines were selected and maintained in zeocin-containing TNM-FH medium (Invitrogen) plus 10% fetal bovine serum.
Cells expressing recombinant Arabidopsis katanin were suspended in lysis buffer (50 mM Na-phosphate, pH 8.0, 300 mM NaCl, 2 mM MgCl2, 20 mM imidazole, 0.05% [v/v] Nonidet P-40, 1 μg/mL aprotinin, 1 μg/mL leupeptin, and 0.1 mM phenylmethylsulfonyl fluoride) and freeze-thawed five times in a dry ice/ethanol bath and cold water. The cell lysate was centrifuged at 100,000g for 30 min to remove cell debris. His-tagged recombinant protein was purified on nickel–nitrilotriacetic acid agarose resin (Qiagen, Chatsworth, CA), and aliquots were frozen at −80°C until use.
MT-Severing Activity Assay
Rhodamine-labeled MTs were prepared by incubation of 5 mg/mL unlabeled tubulin and 5 mg/mL rhodamine-labeled tubulin (Cytoskeleton, Denver, CO) in PEM buffer (80 mM Na-Pipes, pH 6.9, 1 mM MgCl2, and 1 mM EGTA) containing 1 mM GTP and 5% glycerol at 37°C for 50 min. The MTs were stabilized by adding an equal volume of PEM buffer containing 40 μM taxol and incubated for 10 min at 37°C. Taxol-stabilized, rhodamine-labeled MTs were kept at room temperature in the dark for 24 h before being used for the severing assay. In our preparations, the MTs were 20 to 40 μm in length.
MT-severing assays were performed as described by Vale (1991). Briefly, purified recombinant Arabidopsis katanin was diluted 10-fold in assay buffer (20 mM Na-Hepes, pH 7.5, 1 mM MgCl2, 0.1 M EGTA, 1 mM ATP, and 20 μM taxol) and combined with fluorescent MTs in a tube by gentle pipetting with a cut tip. Two microliters of the mixture was gently pipetted with a cut tip onto the surfaces of glass slides and incubated at room temperature. Reactions were stopped at various times by adding 2 μL of 2% glutaraldehyde in PEM buffer containing 20 μM taxol onto the assay mixture and gently covering with a cover slip.
The length of MTs was viewed with a confocal microscope. The MT-severing assays were repeated three times, and similar results were obtained. Typical fields of view for each time point were recorded and measured for MT length distribution. Consistent with previous observations (Vale, 1991), this assay procedure did not cause shearing of long MTs. Although bundling of some long MTs occurred as described (Vale, 1991), the MT severing by recombinant Arabidopsis katanin was obvious. A control protein, His-tagged AtCTL1, a chitinase-like protein (Zhong et al., 2002) expressed in the Sf9 insect cells, was used as a control in the MT-severing activity assay.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Acknowledgments
We thank R. Zhong for her assistance with recombinant katanin expression and the MT-severing activity assay, J. Shields and M. Farmer for their help with scanning electron microscopy, E. Richardson and G. Freshour for their assistance with transmission electron microscopy, B. Palevitz for his helpful suggestions, and the coeditor and reviewer for their suggestions. D.H.B. was supported by a Plant Evolution Training Grant from the National Science Foundation. This work was supported by the Cooperative State Research, Education, and Extension Service of the U.S. Department of Agriculture.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.003947.
References
- Abe, H., Funada, R., Imaizumi, H., Ohtani, J., and Fukazawa, K. (1995). Dynamic changes in the arrangement of cortical microtubules in conifer tracheids during differentiation. Planta 197, 418–421. [Google Scholar]
- Abe, H., Ohtani, J., and Fukazawa, K. (1994). A scanning electron microscopic study of changes in microtubule distributions during secondary wall formation in tracheids. IAWA J. 15, 185–189. [Google Scholar]
- Ahmad, F.J., Yu, W., McNally, F.J., and Baas, P.W. (1999). An essential role for katanin in severing microtubules in the neuron. J. Cell Biol. 145, 305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskin, T.I. (2001). On the alignment of cellulose microfibrils by cortical microtubules: A review and a model. Protoplasma 215, 150–171. [DOI] [PubMed] [Google Scholar]
- Bichet, A., Desnos, T., Turner, S., Grandjean, O., and Höfte, H. (2001). BOTERO1 is required for normal orientation of cortical microtubules and anisotropic cell expansion in Arabidopsis. Plant J. 25, 137–148. [DOI] [PubMed] [Google Scholar]
- Burk, D.H., Liu, B., Zhong, R., Morrison, W.H., and Ye, Z.-H. (2001). A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell 13, 807–827. [PMC free article] [PubMed] [Google Scholar]
- Cosgrove, D.J. (1998). Cell wall loosening by expansins. Plant Physiol. 118, 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr, R.J., and Palevitz, B.A. (1995). Organization of cortical microtubules in plant cells. Curr. Opin. Cell Biol. 7, 65–71. [DOI] [PubMed] [Google Scholar]
- Emons, A.M.C., Derksen, J., and Sassen, M.M.A. (1992). Do microtubules orient plant cell wall microtubules? Physiol. Plant. 84, 486–493. [Google Scholar]
- Emons, A.M.C., and Mulder, B.M. (1998). The making of the architecture of the plant cell wall: How cells exploit geometry. Proc. Natl. Acad. Sci. USA 95, 7215–7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, D.D., and Cyr, R.J. (1998). Extending the microtubule/microfibril paradigm: Cellulose synthesis is required for normal cortical microtubule alignment in elongating cells. Plant Physiol. 116, 1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddings, T.H., and Staehelin, L.A. (1991). Microtubule-mediated control of microfibril deposition: A re-examination of the hypothesis. In The Cytoskeletal Basis of Plant Growth and Form, C.W. Lloyd, ed (San Diego, CA: Academic Press), pp. 85–99.
- Green, P.B. (1962). Mechanism for plant cellular morphogenesis. Science 138, 1404–1405. [DOI] [PubMed] [Google Scholar]
- Harada, T., and Coté, W.A. (1985). Structure of wood. In Biosynthesis and Biodegradation of Wood Components, T. Higuchi, ed (San Diego, CA: Academic Press), pp. 1–42.
- Hartman, J.J., Mahr, J., McNally, K., Okawa, K., Iwamatsu, A., Thomas, S., Cheesman, S., Heuser, J., Vale, R.D., and McNally, F.J. (1998). Katanin, a microtubule-severing protein, is a novel AAA ATPase that targets to the centrosome using a WD40-containing subunit. Cell 93, 277–287. [DOI] [PubMed] [Google Scholar]
- Hasezawa, S., and Nagata, T. (1991). Dynamic organization of plant microtubules at the three distinct transition points during the cell cycle progression of synchronized tobacco BY-2 cells. Bot. Acta 104, 206–211. [Google Scholar]
- Hasezawa, S., and Nozaki, H. (1999). Role of cortical microtubules in the orientation of cellulose microfibril deposition in higher-plant cells. Protoplasma 209, 98–104. [DOI] [PubMed] [Google Scholar]
- Hasezawa, S., Ueda, K., and Kumagai, F. (2000). Time-sequence observation of microtubule dynamics throughout mitosis in living cell suspensions of stable transgenic Arabidopsis: Direct evidence for the origin of cortical microtubules at M/G1 interface. Plant Cell Physiol. 41, 244–250. [DOI] [PubMed] [Google Scholar]
- Heath, I.B. (1974). A unified hypothesis for the role of membrane bound enzyme complexes and microtubules in plant cell wall synthesis. J. Theor. Biol. 48, 445–449. [DOI] [PubMed] [Google Scholar]
- Herth, W. (1980). Calcofluor white and Congo red inhibit chitin microfibril assembly of Poterioochromonas: Evidence for a gap between polymerization and microfibril formation. J. Cell Biol. 87, 442–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledbetter, M.C., and Porter, K.R. (1963). A “microtubule” in plant cell fine structure. J. Cell Biol. 19, 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur, J., and Chua, N.-H. (2000). Microtubule stabilization leads to growth reorientation in Arabidopsis trichomes. Plant Cell 12, 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, U., Herzog, U., Berger, F., Inzé, D., and Jürgens, G. (1999). Mutations in the PILZ group genes disrupt the microtubule cytoskeleton and uncouple cell cycle progression from cell division in Arabidopsis embryo and endosperm. Eur. J. Cell Biol. 78, 100–108. [DOI] [PubMed] [Google Scholar]
- McClinton, R.S., Chandler, J.S., and Callis, J. (2001). cDNA isolation, characterization, and protein intracellular localization of a katanin-like p60 subunit from Arabidopsis thaliana. Protoplasma 216, 181–190. [DOI] [PubMed] [Google Scholar]
- McClinton, R.S., and Sung, Z.R. (1997). Organization of cortical microtubules at the plasma membrane in Arabidopsis. Planta 201, 252–260. [DOI] [PubMed] [Google Scholar]
- McNally, F.J., Okawa, K., Iwamatsu, A., and Vale, R.D. (1996). Katanin, the microtubule-severing ATPase, is concentrated at centrosomes. J. Cell Sci. 109, 561–567. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nagata, T., Kumagai, F., and Hasezawa, S. (1994). The origin and organization of cortical microtubules during the transition be-tween M and G1 phases of the cell cycle as observed in highly synchronized cells of tobacco BY-2. Planta 193, 567–572. [Google Scholar]
- Preston, R.D. (1988). Cellulose-microfibril-orienting mechanisms in plant cells walls. Planta 174, 67–74. [DOI] [PubMed] [Google Scholar]
- Prodhan, A.K.M.A., Funada, R., Ohtani, J., Abe, H., and Fukazawa, K. (1995). Orientation of microfibrils and microtubules in developing tension-wood fibers of Japanese ash (Fraxinus mandshurica var. japonica). Planta 196, 577–585. [Google Scholar]
- Quarmby, L. (2000). Cellular samurai: Katanin and the severing of microtubules. J. Cell Sci. 113, 2821–2827. [DOI] [PubMed] [Google Scholar]
- Robards, A.W., and Kidwai, P. (1972). Microtubules and microfibrils in xylem fibers during secondary wall formation. Cytobiologie 6, 1–21. [Google Scholar]
- Seagull, R.W. (1992). A quantitative electron microscopic study of changes in microtubule arrays and wall microfibril orientation during in vitro cotton fiber development. J. Cell Sci. 101, 561–577. [Google Scholar]
- Seagull, R.W., and Falconer, M.M. (1991). In vitro xylogenesis. In The Cytoskeletal Basis of Plant Growth and Form, C.W. Lloyd, ed (San Diego, CA: Academic Press), pp. 183–194.
- Sugimoto, K., Williamson, R.E., and Wasteneys, G.O. (2000). New techniques enable comparative analysis of microtubule orientation, wall texture, and growth rate in intact roots of Arabidopsis. Plant Physiol. 124, 1493–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto, K., Williamson, R.E., and Wasteneys, G.O. (2001). Wall architecture in the cellulose-deficient rsw1 mutant of Arabidopsis thaliana: Microfibrils but not microtubules lose their transverse alignment before microfibrils become unrecognizable in the mitotic and elongation zones of roots. Protoplasma 215, 172–183. [DOI] [PubMed] [Google Scholar]
- Thion, L., Mazars, C., Nacry, P., Bouchez, D., Moreau, M., Ranjeva, R., and Thuleau, P. (1998). Plasma membrane depolarization-activated calcium channels, stimulated by microtubule-depolymerizing drugs in wild-type Arabidopsis thaliana protoplasts, display constitutively large activities and a longer half-life in ton2 mutant cells affected in the organization of cortical microtubules. Plant J. 13, 603–610. [DOI] [PubMed] [Google Scholar]
- Torres-Ruiz, R.A., and Jürgens, G. (1994). Mutations in the FASS gene uncouple pattern formation and morphogenesis in Arabidopsis development. Development 120, 2967–2978. [DOI] [PubMed] [Google Scholar]
- Traas, J., Bellini, C., Nacry, P., Kronenberger, J., Bouchez, D., and Caboche, M. (1995). Normal differentiation patterns in plants lacking microtubular preprophase bands. Nature 375, 676–677. [Google Scholar]
- Vale, R.D. (1991). Severing of stable microtubules by a mitotically activated protein in Xenopus egg extracts. Cell 64, 827–839. [DOI] [PubMed] [Google Scholar]
- Vaughn, K.C., and Harper, J.D.I. (1998). Microtubule-organization centers and nucleating sites in land plants. Int. Rev. Cytol. 181, 75–149. [DOI] [PubMed] [Google Scholar]
- Walczak, C.E. (2000). Microtubule dynamics and tubulin interacting proteins. Curr. Opin. Cell Biol. 12, 52–56. [DOI] [PubMed] [Google Scholar]
- Wasteneys, G.O. (2000). The cytoskeleton and growth polarity. Curr. Opin. Plant Biol. 3, 503–511. [DOI] [PubMed] [Google Scholar]
- Whittington, A.T., Vugrek, O., Wei, K.J., Hasenbein, N.G., Sugimoto, K., Rashbrooke, M.C., and Wasteneys, G.O. (2001). MOR1 is essential for organizing cortical microtubules in plants. Nature 411, 610–613. [DOI] [PubMed] [Google Scholar]
- Williamson, R.E. (1990). Alignment of cortical microtubules by anisotropic wall stresses. Aust. J. Plant Physiol. 17, 601–613. [Google Scholar]
- Zhong, R., Burk, D.H., and Ye, Z.-H. (2001). Fibers: A model for studying cell differentiation, cell elongation, and cell wall biosynthesis. Plant Physiol. 126, 477–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Stanley, J.K., Schroeder, B.P., and Ye, Z.-H. (2002). Mutation of a chitinase-like protein causes ectopic deposition of lignin, aberrant cell shapes, and overproduction of ethylene. Plant Cell 14, 165–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Taylor, J.J., and Ye, Z.-H. (1997). Disruption of interfascicular fiber differentiation in an Arabidopsis mutant. Plant Cell 9, 2159–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]