Abstract
The degradation of storage compounds just after germination is essential to plant development, providing energy and molecules necessary for the building of a photosynthetic apparatus and allowing autotrophic growth. We identified à bout de souffle (bou), a new Arabidopsis mutation. Mutant plants stopped developing after germination and degraded storage lipids, but they did not proceed to autotrophic growth. Neither leaves nor roots developed in the mutant. However, externally added sugar or germination in the dark could bypass this developmental block and allowed mutant plants to develop. The mutated gene was cloned using the transposon Dissociation as a molecular tag. The gene coding sequence showed similarity to those of the mitochondrial carnitine acyl carriers (CACs) or CAC-like proteins. In animals and yeast, these transmembrane proteins are involved in the transport of lipid-derived molecules across mitochondrial membranes for energy and carbon supply. The data presented here suggest that BOU identifies a novel mitochondrial pathway that is necessary to seedling development in the light. The BOU pathway would be an alternative to the well-known glyoxylate pathway.
INTRODUCTION
From germination until the establishment of autotrophic growth (referred to here as postgerminative growth), young seedlings rely on nutrients that were stored during embryogenesis and seed development. The degradation of storage molecules provides energy and materials for the building of photosynthetic chloroplasts that, in turn, produce sugars, which are a source of carbon and energy. Oilseeds, such as Arabidopsis, store part of the energy required for postgerminative growth as lipids in the form of triacylglycerides (TAG) that are contained in oil bodies within the cotyledon cells of the embryo. When oilseeds germinate, glyoxysomes, a specialized form of peroxisome, differentiate, with TAG degradation occurring concomitantly (for review, see Olsen and Harada, 1995; Olsen, 1998). Plant glyoxysomes have been shown to contain the enzymes necessary for the complete degradation of the fatty acids through the process of β-oxidation (Kleiter and Gerhardt, 1998; Hayashi et al., 1999).
The pathway and location of fatty acid degradation in the plant cell differ from those in animal cells. In animal cells, the β-oxidation pathway is split between peroxisomes and mitochondria. Fatty acids are activated as acyl-CoA. Very-long-chain fatty acids are first shortened within the peroxisome to medium-chain acyl-CoA. The acyl group is conjugated to carnitine forming acylcarnitine. Acylcarnitine is transported across the inner mitochondrial membrane. In the mitochondrial matrix, the fatty acid moiety is reassociated with CoA to regenerate acyl-CoA, which is degraded through β-oxidation. The animal carnitine acyl carrier (CAC) is the transmembrane carrier of acylcarnitine molecules. CAC was shown in vitro to catalyze the import of acylcarnitine into liposomes (Indiveri et al., 1997, 1998).
Several CAC protein mutants have been identified in animals, including human, fruit fly, and worm. In Caenorhabditis elegans, the DIF mutation affects embryos before tissue differentiation (Ahringer, 1995). In the Drosophila melanogaster colt mutant, the primary defect is the collapse of tracheae tissues (Hartenstein et al., 1997). These examples illustrate the vital role of lipid metabolism and transport in animal cells.
By contrast, in yeast and other fungi, fatty acid breakdown takes place exclusively in peroxisomes. In the yeast Saccharomyces cerevisiae, acetyl-CoA produced by the β-oxidation of oleate acts as a source of succinate through the glyoxylate cycle or is conjugated to carnitine by the peroxisomal carnitine acetyl transferase (CAT2) enzyme (Elgersma et al., 1995). The resulting acetylcarnitine is transported to the mitochondria. Both the succinate and the acetylcarnitine pathways sustain yeast growth when oleate is the sole carbon source (van Roermund et al., 1995, 1999). It is necessary to disrupt both pathways to prevent growth on oleate. A single mutation in the peroxisomal CAT2 or the peroxisomal citrate synthase (CIT2), a key enzyme of the glyoxylate cycle, does not affect growth on oleate. However, the double mutant in which both CAT2 and CIT2 are inactivated cannot grow on oleate (van Roermund et al., 1995, 1999).
It has been shown that in oleate-grown S. cerevisiae, acetylcarnitine is imported into the mitochondria by a CAC-like protein, CRC-1, which is present in the mitochondrial membrane. Mutation of the CRC-1 locus prevents growth on oleate when the citrate synthase locus also is deleted (van Roermund et al., 1999). In Aspergillus nidulans, the acuH locus, which has been identified from oleate-grown mutants, encodes a CAC-like protein. Similarly, its suggested function would be that of a mitochondrial acetylcarnitine transporter (DeLucas et al., 1999).
There are many reports of the presence of carnitine in plant cells and of its role in plant metabolism, including the transport of acyl molecules across the mitochondrial membranes (for review, see Wood et al., 1992). A CAT has been purified from pumpkin mitochondria (Schwadbedissen-Gerbling and Gerhardt, 1995). However, the biological role of carnitine-dependent transport in fatty acid degradation and during oilseed germination is not clear.
During oilseed germination, β-oxidation of fatty acids is known to occur in the glyoxysome (Kleiter and Gerhardt, 1998; Hayashi et al., 1999). Acetyl-CoA that is produced by the degradation of fatty acids is converted into succinate by the glyoxylate cycle. Succinate then is transported into the mitochondria (ap Rees, 1987; Gerhardt, 1993). Succinate is a substrate for respiratory ATP production and is converted to malate, which can be exported to the cytosol for sugar synthesis by gluconeogenesis. However, it was shown recently that a mutant (icl) that is impaired in the succinate-producing glyoxylate cycle enzyme isocitrate lyase (ICL) shows an impaired-growth phenotype when grown in the dark but not when grown in the light (Eastmond et al., 2000). It has been suggested that an alternative to the glyoxylate pathway must exist to account for the icl mutant's use of carbon-containing compounds produced after fatty acid β-oxidation (Eastmond and Graham, 2001).
We identified à bout de souffle (bou), an Arabidopsis mutation that results in a defective postgerminative phenotype that is light dependent. When grown in the light, mutant plants were unable to proceed to autotrophic growth. However, when seedlings were germinated in the dark, or when sugar was added to the medium, they proceeded to develop. We identified the BOU gene, which in the mutant was disrupted by a transposon. The BOU coding sequence showed homology with the CAC-like protein family that is involved in the translocation of the carnitine ester of fatty acids (acylcarnitine) or acetate (acetylcarnitine) into the mitochondria. The BOU protein is located in mitochondria. We measured storage lipid degradation in bou seedlings and BOU gene expression. Our results suggest that BOU identifies a light-regulated pathway that could be an alternative to the glyoxylate cycle during postgerminative growth.
RESULTS
Characterization of the Mutant Phenotype
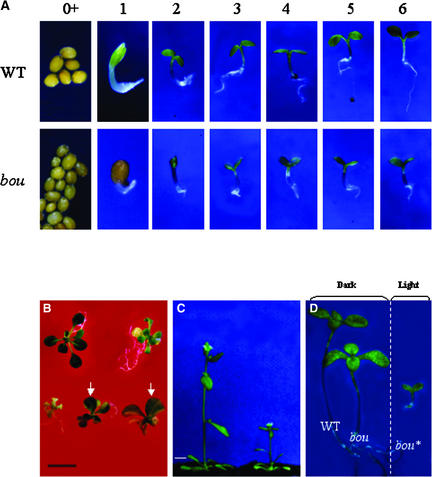
Using a modified Dissociation element [Ds(HYG)] that was introduced into Arabidopsis for insertional mutagenesis (Coupland, 1992; Long et al., 1993), we isolated a mutant on the basis of its pale-green adult phenotype. When mutant seeds germinated on a medium deprived of sugar, they remained arrested at the cotyledon stage, unable to produce the first pair of leaves, and did not develop any root (Figure 1A).
Figure 1.
Phenotype of the bou Mutant.
(A) The bou phenotype is apparent after germination on mineral medium. 0+, seeds that had imbibed (magnification ×30); 1, 1 day of growth (magnification ×40); 2 to 6, 2 to 6 days of growth (magnification ×15). Top row, wild-type (WT) seedlings; bottom row, bou seedlings.
(B) Top row, wild-type plants (left) and mutant plants (right) grown for 20 days on sugar-containing medium. Bottom row, stable bou mutant (left) and two bou mutants showing an unstable phenotype with wild-type sectors (right). The arrows indicate the presence of a revertant sector seen as dark green on a pale green background. Bar = 1 cm.
(C) Wild-type (left) and bou (right) plants grown for 2 weeks on Murashige and Skoog (1962) medium and then on soil for 1 more week. Bar = 1 cm.
(D) Dark indicates seedlings germinated in the dark for 4 days and then transferred to long-day conditions for 15 days. Light indicates mutants kept in long-day conditions for 17 days (magnification ×7.5).
Arrested mutant seedlings that were transferred to a medium containing sugar resumed their development: the root elongated and leaves formed, although they were paler than those of the wild type (Figure 1B). When mutant plants grown on sugar were transferred to soil at the juvenile stage, with one pair of leaves, they grew and set seed. Mutant plants, however, were smaller than wild-type plants (Figure 1C), and their development was slower than that of the wild type, although organ formation was not altered. The progeny of mutant plants had a phenotype identical to that of the parent.
From these observations, we conclude that the primary defect of the mutant is in the transition from the embryonic stage to the juvenile autotrophic stage. The small adult phenotype suggests that BOU also is required for normal adult development. Because mutant seedlings stop developing unless sugar is added, we named the mutant à bout de souffle (which means “out of breath”).
Sensitivity to 2,4-Dichlorophenoxybutyric Acid
The postgerminative phenotype observed in bou was similar to that of peroxisome-deficient (ped) mutants, which are deficient in the glyoxysome fatty acid β-oxidation thiolase protein. ped mutants are unable to grow after germination unless sugar is present in the medium (Hayashi et al., 1998). It is thought that this arrested development is the direct consequence of their deficiency in fatty acid degradation. ped mutants were selected on medium containing sugar and 2,4-dichlorophenoxybutyric acid (2,4-DB), an inactive precursor of the 2,4-D growth factor (an analog of the endogenous hormone auxin). 2,4-DB is converted to the active 2,4-D via β-oxidation of its butyrate moiety.
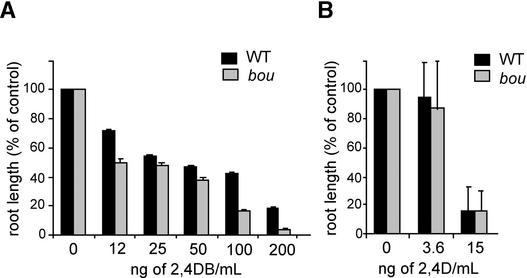
Wild-type plants germinated on 2,4-DB showed arrested development, because the 2,4-D produced caused abnormal cell growth and division. In ped mutants, glyoxysomal β-oxidation activity was inefficient, 2,4-DB was not converted to 2,4-D, and plants displayed resistant growth compared with that of the wild type. When assayed for their root growth response to 2,4-DB, bou mutant plants exhibited a sensitivity to 2,4-DB that was comparable to that of wild-type seedlings, showing that β-oxidation is not affected in bou (Figure 2A). Sensitivity to 2,4-D was comparable between wild-type and bou seedlings (Figure 2B).
Figure 2.
Root Sensitivity to 2,4-DB.
(A) Root elongation of plants (n = 10) grown on ATG medium containing a range of concentrations of 2,4-DB.
(B) Root elongation of plants (n = 10) grown on ATG medium containing a range of concentrations of the active compound 2,4-D.
Plants were grown for 1 week on a vertically orientated plate before root length measurement. Root length is expressed as a percentage of the length of wild-type and bou roots grown in the absence of 2,4-DB and 2,4-D, respectively. bou, mutant plants; WT, wild-type plants. Bars represent the standard deviation.
Lipid Degradation
Analysis of the lipid content of wild-type and bou seeds showed similar composition of total fatty acids from TAG (data not shown). TAG represented 90% of the lipid content in dry seeds. Seeds that were harvested from soil-grown bou plants had approximately half of the wild-type lipid content. When seeds were harvested from a mutant bou plant grown on sugar-supplemented medium, the lipid content was similar to that of the wild type (Table 1). Whatever their seed lipid content, all bou seedlings showed the arrested phenotype.
Table 1.
Lipid Content in Mutant and Wild-Type Seeds
| Sample | Total Lipids | TAG |
|---|---|---|
| Wild type | 5.5 ± 0.9 | 5.0 ± 0.7 |
| bou (sugar grown) | 4.2 ± 0.7 | 3.95 ± 0.3 |
| bou (soil grown) | 2.6 ± 0.8 | 2.3 ± 0.6 |
The total lipid content and the purified TAG were quantified in wild-type seeds and two types of mutant seeds. bou (sugar grown) refers to seeds from a mutant plant grown on medium supplemented with 1% Suc. Compared with bou (soil grown), the lipid content (total or TAG) in the sugar-grown sample was not significantly different from that in the wild-type sample.
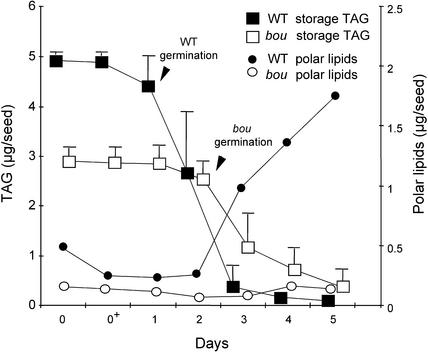
In the wild type, storage lipid degradation was low during the first day of growth, when germination occurred, but increased dramatically thereafter, at the postgermination stage. Less than 2% of the TAG remained after 5 days, indicating the mobilization of storage lipids from the storage oil bodies. In mutant seedlings, storage lipids were degraded more slowly (Figure 3). In wild-type plants, polar lipids, which are present in membranes, were being synthesized when TAG were being degraded after germination was completed. In mutant seedlings, very few polar lipids were synthesized (Figure 3).
Figure 3.
Lipid Degradation and Polar Lipid Accumulation during Early Arabidopsis Seedling Development.
Storage lipids (TAG) and polar lipids were measured each day during seed germination. The arrows indicate the apparent completion of germination (protrusion of the radicle from the seed coat). The germination of bou was delayed by 1 day compared with that of the wild type (WT). Seeds were germinated in vitro on ATG medium (without sugar). 0, dry seeds; 0+, seeds allowed to imbibe overnight at 4°C before germination; 1 to 5, days of culture at 24°C in the light. Bars indicate the standard deviation from an average of three experiments (n = 10). Polar lipids were measured from a pool of 20 seedlings.
Because TAG degradation and β-oxidation occur in mutant seedlings, it is likely that the primary defect in bou is not in the degradation of lipid storage molecules. It is more likely that bou seedlings are defective in the use of the products of fatty acid degradation.
Genetic Characterization of the bou Mutation
The Ds-modified element used to produce bou carried a hygromycin resistance gene. From the progeny of a heterozygous (Ds/+) plant, we tested 180 lines heterozygous for their resistance to hygromycin [Ds(HYG)/+]. All of these lines showed a segregation of the bou phenotype in their progeny. This finding suggests that all of the Ds-containing lines also are heterozygous for bou (bou/+). After crossing heterozygous hygromycin-resistant plants to wild-type Arabidopsis, half (n = 40) of the F1 plants were found to be resistant to hygromycin. The progeny of resistant plants showed a segregation for the mutant phenotype.
It is clear from these experiments that the Ds(HYG) transposon is linked tightly to the mutation. Furthermore, the bou phenotype results from a recessive monogenic trait that is associated with the Ds(HYG) transposon and is most likely caused by the transposon. Heterozygous plants, which carry an active transposase gene, were screened for the loss of the mutant phenotype. An average of 2% of the mutant plants showed a sectored phenotype in which wild-type leaf tissue developed on a mutant background (Figure 1B). Six of the mutants showed a reversion phenotype that encompassed the floral meristem, giving rise to a revertant floral branch. The progeny of the revertant branches segregated for wild-type plants. This finding shows that the phenotypic reversion is transmissible genetically and is not merely a phenotypic variation. This behavior is typical of a transposon-caused mutation.
Cloning of BOU
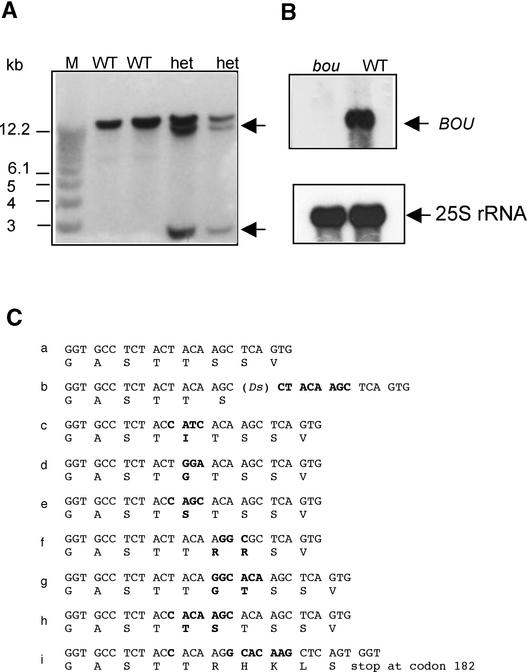
A 200-bp genomic DNA fragment flanking the Ds(HYG) transposon was obtained by inverse PCR. Its sequence allowed the identification of a putative Arabidopsis gene in the database that is part of the BAC clone MZA15 mapped on Arabidopsis chromosome V. DNA gel blot hybridization showed that BOU gene organization was disrupted in plants carrying the bou mutation. Heterozygous plants showed both a wild-type copy and an altered version of the gene (Figure 4A). The presence of the transposon within the BOU gene was confirmed by PCR (data not shown). Total RNA probed with the BOU cDNA identified a transcript of ∼1.2 kb in the wild-type sample that was undetectable in RNA extracted from mutant plants (Figure 4B). It is likely that the bou phenotype is caused by the insertion of Ds(HYG), which interrupts the transcription of the BOU gene.
Figure 4.
Molecular Analysis of the bou Mutation.
(A) DNA gel blot analysis of the bou mutation. DNA extracted from wild-type (WT) and heterozygous (het) hygromycin-resistant lines was digested with EcoRI, transferred to a nitrocellulose membrane, and probed with the BOU cDNA. Two extra genomic DNA fragments were revealed (arrows) in the heterozygote samples. M, molecular mass markers.
(B) RNA gel blot showing a BOU transcript in wild-type seedlings (arrow) that was absent from the mutant sample. RNA extracted from 2-week-old seedlings was probed with the BOU cDNA. Hybridization to the 25S rRNA was used as a loading control.
(C) Sequence analysis of the transposon insertion. The nucleotides and the deduced amino acids that differed from the wild-type sequence are shown in boldface.
(a) Relevant sequence of the BOU gene.
(b) Sequence of the insertion allele.
(c) to (h) Sequences of six revertant alleles.
(i) Sequence of a stable mutant with an insertion resulting in frameshifting and a truncated reading frame.
The sequence of the mutated bou gene showed an 8-bp footprint at the Ds(HYG) insertion site, as expected from a Ds family transposable element (Figure 4C, sequence b). We determined the BOU sequences of 10 revertant plants of wild-type phenotype that were obtained either from revertant branches (as described above) or from the progeny of mutants in which revertants occur at low frequency. In all cases, Ds was not found. The sequences of four independent revertants showed a wild-type DNA sequence at the point of insertion. The sequences of six other independent revertants showed a footprint of 3 to 6 bp at the insertion site, restoring the open reading frame (Figure 4C, sequences c to h).
The footprint did not seem to be a remnant of the 8-bp duplication created upon the insertion of the transposon; rather, it appeared to be a rearrangement of the DNA upon transposon excision. A similar effect has been observed in Arabidopsis and tomato (Keddie et al., 1996; Carol et al., 1999). It seems that a strong bias for amino acids compatible with an active gene product would select for DNA repair at the insertion site, creating acceptable codons for biological activity.
In addition, we obtained a new mutant allele in which the Ds upon excision left a footprint causing the disruption of the reading frame (Figure 4C, sequence i). The unstable mutant phenotype we observed, and the restoration of the gene sequence in revertant alleles, confirmed that the bou mutation is caused by the Ds(HYG) element.
BOU Gene Expression
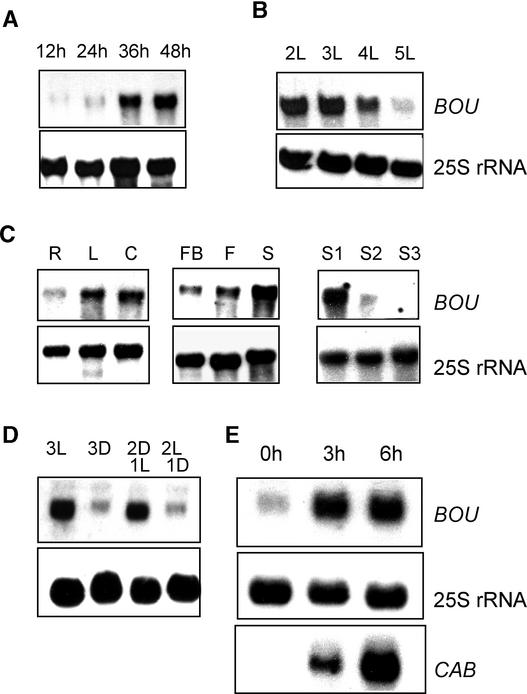
We investigated the expression of the BOU gene during the germination of Arabidopsis seedlings (Figure 5). For the first 12 to 24 h, which corresponds to germination itself, BOU mRNA was barely detectable. Germination ended with the protrusion of the radicle after 24 h. BOU mRNA accumulated to a high level after 36 h (Figure 5A) and remained so for up to 2 more days. The signal decreased thereafter (Figure 5B).
Figure 5.
RNA Gel Blot Analysis of BOU Expression.
(A) Detection of BOU mRNA during seed germination from 12 to 48 h.
(B) Detection of BOU mRNA after germination in seedlings grown in the light for 2 to 5 days.
(C) Detection of BOU mRNA in Arabidopsis organs. R, roots; L, leaves; C, 3-day-old cotyledons; FB, flower buds; F, flowers; S, siliques; S1, immature developing siliques (up to 7 days after fertilization); S2, developing siliques (1 to 2 weeks after fertilization); S3, maturing siliques (2 to 3 weeks after fertilization).
(D) Detection of BOU mRNA in light-grown seedlings. 3L, 3 days in the light; 3D, 3 days in the dark; 2D1L, 2 days in the dark followed by 1 day in the light; 2L1D, 2 days in the light followed by 1 day in the dark.
(E) Effect of light treatment (0, 3, or 6 h) on BOU RNA accumulation in 3-day-old dark-grown seedlings. The CAB gene was used as a control for light-induced gene expression.
In all experiments, the loading control was the 25S rRNA hybridization signal.
In adult plants, BOU was expressed more highly in leaves and flowers and developing fruits (siliques) than in roots and maturing siliques (Figure 5C). The expression profile of BOU mRNA with higher levels in green (photosynthetic) tissues, which are known for light-regulated gene expression, suggested that BOU mRNA level could be modulated by light. A strong BOU mRNA signal was detected in light-grown seedlings, whereas dark-grown seedlings showed much less BOU mRNA (Figure 5D). When dark-grown seedlings were placed in light growth conditions, a rapid increase in BOU mRNA was detected (Figure 5E).
Light Modulates the bou Phenotype
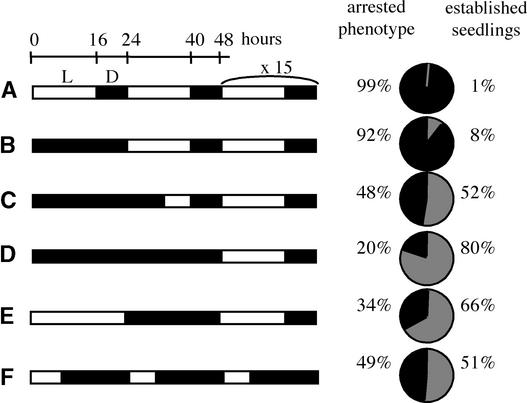
These results indicate that BOU has a light-regulated function that is important after the seed has germinated. To assess the importance of BOU in the absence of light, when its expression is low, bou seedlings were germinated in the dark for 4 days before being transferred to the light. Dark-treated seedlings did not show the usual arrested phenotype after germination (Figure 1D), because the seedlings became established and autotrophic. The quantification of the development and establishment of seedlings in various light and dark growth conditions is shown in Figure 6.
Figure 6.
Scheme of the Light-Dependent bou Phenotype.
Before growth, bou seeds that had imbibed were germinated for 2 days in the light or in the dark for various time periods. The time scale represents the germination of bou seedlings, starting at 0 h. Germinated seedlings were grown for 15 days, and the phenotype was scored as arrested if the seedlings had not developed and as established if the seedlings had developed roots and leaves. At left ([A] to [F]), the light periods (L) are shown with open boxes, and the dark periods (D) are shown with black boxes. At right, for each light regime, the percentage of seedlings that were arrested (black) is compared with the percentage of established seedlings (gray). Results shown are averages of two independent experiments with 100 seeds each. A control sample of wild-type seeds always showed normal development (100% established) in all of the described conditions.
In long-day conditions (Figure 6A), seedlings showed the arrested phenotype. A few seedlings (8%; Figure 6B) became established when germinated for 24 h in the dark. Lengthening of the dark period increased the percentage of established seedlings (Figure 6C). Up to 80% of the seedlings developed and became established when seeds were germinated for 48 h in the dark (Figure 6D). Fewer seedlings (66%) became established if a 24-h light period was applied during the first day of germination (Figure 6E). A short-day light regime during and after germination allowed the establishment of approximately half of the seedlings (Figure 6F). In conclusion, the presence or absence of light during and just after germination modulated the bou phenotype. Therefore, it seems that BOU function, which is vital for the establishment of seedlings in the light, is not necessary for seedling establishment in the dark.
BOU Shows Similarity to CAC
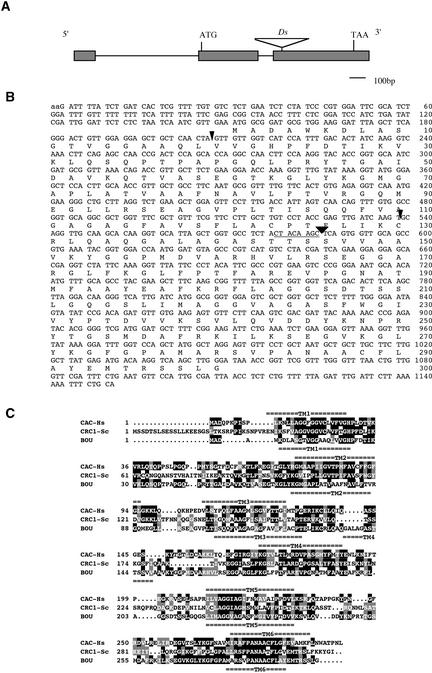
The sequence flanking the Ds(HYG) insertion in bou corresponded to that of the DIF-like gene, which was identified by computer analysis of the chromosome V clone MZA15. We identified an EST cDNA that allowed us to identify a coding sequence and the intron-exon locations within the gene. The BOU coding sequence encompasses 2 kb of genomic DNA comprising three exons and two introns (Figure 7A). The cDNA contains an open reading frame of 300 amino acids, starting with a Met in a plant translation initiation consensus (Figure 7B). Database searches revealed homology with mitochondrial transmembrane carrier proteins of the CAC protein family from mammals, the DIF protein from C. elegans, and the COLT protein from Drosophila. This similarity extends to the yeast CRC1 acetylcarnitine carrier protein and other fungi CAC-like proteins.
Figure 7.
The BOU Gene and Its Coding Sequence.
(A) Structure of the BOU gene. Exons are represented as boxes, and introns are represented as a line. The relative position of the Ds transposon is indicated. Bar = 100 bp.
(B) Nucleotide and amino acid sequences of BOU cDNA. The arrowheads indicate the positions of the introns in the gene. The triangle indicates the insertion point of the Ds transposon. The 8-bp sequence duplicated upon Ds insertion is underlined.
(C) Comparison of the BOU protein with CAC-like proteins. CAC-Hs, CAC from human; CRC1-Sc, acetylcarnitine carrier CRC-1 from S. cerevisiae. Identical amino acids are shaded in black, and similar amino acids are shaded in gray. Putative transmembrane domains (TM1 to TM6) are underlined. The sequences were aligned using the Genetics Computer Group computer package version 8.1 (Madison, WI). The transmembrane domains were predicted using TMpred (Hofmann and Stoffel, 1993) (http://www.ch.embnet.org/).
The comparison of BOU with the human acylcarnitine carrier and the yeast acetylcarnitine carrier proteins, whose functions are known, revealed similar features (Figure 7C). The CAC-like proteins have six transmembrane domains (TM). Comparing the predicted TM relative spacing of CAC and BOU, we found that five of six TM are colinear, but TM4 is predicted to be at a different position in BOU. The predicted relocation of TM4 corresponds to an amino acid insertion sequence unique to BOU that is not found in other CAC-related proteins.
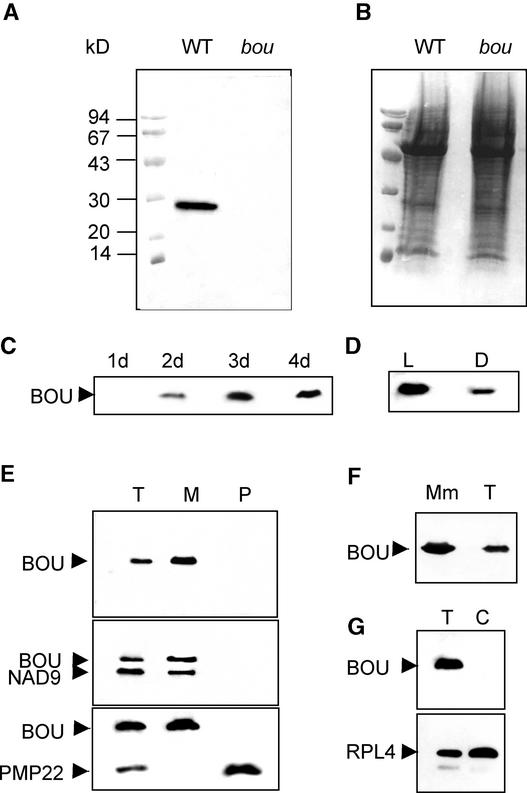
Location of BOU
BOU was detected in wild-type Arabidopsis as a 28-kD protein that was not found in bou mutant plants (Figure 8A). After germination, BOU was detected in 2-day-old seedlings (Figure 8C) and was present at high levels in light-grown seedlings (Figure 8D). Immunodetection of BOU in Arabidopsis organelles isolated from leaves showed a strong signal in mitochondria-containing fractions. No signal was found in peroxisomes or chloroplasts (Figures 8E and 8F). Furthermore, BOU was detected in purified mitochondrial membranes from cultured Arabidopsis cells (Figure 8G). A BOU-related protein of 30 kD also was immunodetected in the mitochondrial fraction purified from rapeseed seedlings (data not shown). Therefore, the cellular location of BOU is likely to be the mitochondrial membrane.
Figure 8.
Immunodetection of BOU, a Mitochondrial Protein.
(A) to (D) Immunodetection of BOU in total plant protein extracts. M, molecular mass markers.
(A) Immunodetection of BOU in wild-type (WT) and mutant (bou) protein extracts.
(B) Coomassie blue staining of the samples.
(C) Immunodetection of BOU during seed germination, from 1 to 4 days of seedling development.
(D) Immunodetection of BOU in light-grown (L) and dark-grown (D) 3-day-old seedlings.
(E) and (F) Immunodetection of BOU in Arabidopsis organelles.
(E) Comparison of total (T) proteins and mitochondrial (M) and peroxisome (P) fractions from leaf tissue of 3-week-old Arabidopsis. NAD9 was used as a mitochondria-specific control, and PMP22 was used as a peroxisome-specific control.
(F) Immunodetection of BOU in purified mitochondrial membrane (MM) and total proteins (T) from Arabidopsis cells.
(G) Comparison of total proteins (T) and chloroplast fraction (C) from leaf tissue of 3-week-old Arabidopsis. The detection of RPL4 chloroplast protein was used as a control.
DISCUSSION
bou and Peroxisome Function
The primary phenotype associated with bou was the inability to proceed from embryonic to juvenile autotrophic growth. This phenotype is similar to that of ped mutant plants, which are defective in glyoxysomal thiolase activity and are unable to produce acetate through the β-oxidation of fatty acids. In Arabidopsis, up to 5 μg of TAG was stored in the seed. Quantification of TAG throughout seedling development showed that TAG remained for 24 h, when germination occurred, before being degraded completely during the subsequent 2 days. We conclude that lipid degradation is not used for germination per se. It is likely that the energy and molecules generated by lipid degradation are used at the postgerminative stage, when the seedling embryonic structure develops photosynthetic tissues, leading to an autotrophic plant.
bou mutant seedlings displayed an arrested growth phenotype whether the seeds had a normal complement of storage lipids (the progeny of mutant plants grown on sugar) or half of the wild-type lipid content (the progeny of a soil-grown mutant plant). The mutant phenotype also was observed in the progeny of heterozygous plants. We found that bou mutants could degrade storage lipids. This was confirmed by the biological assay for β-oxidation activity in germinating seedlings, in which mutant seedlings behaved similarly to wild-type seedlings. This result clearly shows that the glyoxysomal β-oxidation of fatty acid is functional in the mutant.
Mutant plants that remained arrested for a few days were able to resume development when moved to sugar-containing medium. Sugar was needed until the first leaves appeared. Thereafter, mutant plants could sustain autotrophic growth. From these observations, we conclude that the defect in bou mutants affects neither germination nor storage lipid degradation. Most likely, the bou mutation prevents postgerminative plant development or the use of storage compound degradation products. The germination of mutant seedlings in the dark alleviated the developmental block. This light-dependent phenotype suggests that BOU performs a function that is necessary only in the light, and another different pathway is sufficient for development in the dark.
BOU Expression
We detected more BOU mRNA and its protein product in light growth conditions, suggesting that dark development does not require a high level of BOU expression. An explanation for this finding might be that BOU is part of a metabolic pathway that is associated with photomorphogenesis. In light conditions, in which chloroplasts differentiate, BOU would be needed. In dark conditions, in which chloroplasts do not differentiate, BOU would be downregulated. This is consistent with the low level of BOU mRNA detected in roots, which are nonphotosynthetic.
During seedling development, when storage lipids sustain cell growth and differentiation, embryonic cotyledons undergo a metabolic transition from storage degradation to photosynthetic tissue. BOU expression was light induced in these developing seedlings. Later, after 4 days, when cotyledons became senescent, there was a decrease of BOU expression. BOU was detected again in newly formed photosynthetic leaves. Although BOU mRNA was detected first after 36 h and reached a maximum after 2 days of growth, BOU protein was detected after 2 days and reached a maximum at 3 days of growth. It is possible that a translational mechanism regulates BOU production.
The accumulation of BOU in the wild type coincided with the stage at which the bou phenotype was first detected in the mutant (i.e., after germination). BOU gene expression also was correlated with the light-dependent phenotype of the mutant. The bou phenotype appeared in the light growth conditions that corresponded to the highest BOU accumulation in the wild type. In the dark, when BOU was expressed at a low level, the function was not necessary for seedling growth, because the mutants were able to develop.
Putative Transport Function of BOU
The amino acid similarity between BOU, mammalian CAC, and yeast CAC-like protein suggests that they are members of the same protein family. The presence of transmembrane domains at the same positions in both BOU and CAC suggests similar secondary and tertiary structures. The demonstrated function of CAC in mammals is the translocation of acylcarnitine (fatty acid esterified to carnitine) molecules across the membrane of mitochondria, in which the fatty acid moiety is degraded to provide energy through the mechanism of β-oxidation (Indiveri et al., 1998). In yeast, the CAC-like protein CRC-1, which is located in the mitochondrial membrane, is responsible for the transport of acetylcarnitine (acetate esterified to carnitine), which is produced by the degradation of fatty acid in the peroxisomes. It is transported to the mitochondria to feed into the tricarboxylic acid pathway (Palmieri et al., 1999; van Roermund et al., 1999).
The transport function of BOU remains to be assessed. Plant cells do contain carnitine (for review, see Wood et al., 1992). The carnitine-dependent transport of fatty acid molecules into pea mitochondria has been reported (Wood et al., 1984; Thomas et al., 1988), and acetylcarnitine transferase activity has been purified from pumpkin mitochondria (Schwadbedissen-Gerbling and Gerhardt, 1995). However, the presence of acetylcarnitine has yet to be correlated with Arabidopsis postgerminative growth and glyoxysomal fatty acid β-oxidation.
Hypothesizing an Alternative to the Glyoxylate Cycle in Plants
In plants, acetate (in the form of acetyl-CoA) that is produced through fatty acid degradation enters the glyoxylate cycle, providing the mitochondria with succinate. This pathway has several functions: to produce NADH for energy, to compensate for the constant loss of organic molecules entering various biochemical pathways (which is the anaplerotic role of the glyoxylate cycle), and to produce sugar molecules through the conversion of succinate to malate and then oxaloacetate after export to the cytoplasm.
Surprisingly, it has been shown that Arabidopsis icl mutants, which are defective for the ICL gene coding for the enzyme ICL from the glyoxylate cycle, are able to germinate and develop into autotrophic photosynthetic plants in the light (Eastmond et al., 2000). Therefore, the glyoxylate cycle is dispensable for seedling establishment, and another pathway must compensate. However, in dark-grown plants, the glyoxylate pathway is important and is not compensated for, because icl mutants fail to develop properly when germinated in the dark.
This is similar to the metabolism of yeast, in which fatty acid β-oxidation produces acetate that is exported to the mitochondria either as succinate via the glyoxylate cycle or directly as acetylcarnitine by CRC-1 CAC-like protein. In yeast, these two pathways are redundant, because a mutation in either of them does not prevent growth on oleate fatty acid, but when both pathways are inactivated, yeast mutants fail to grow on oleate fatty acid (van Roermund et al., 1999).
The possible occurrence of an acetylcarnitine pathway in Arabidopsis has been speculated (Eastmond and Graham, 2001). Based on the amino acid sequence and protein localization similarity between BOU and the CAC-like protein, it can be speculated that in the light, plant mitochondria could use BOU transport function as an alternative or a complement to the well-known succinate-producing glyoxylate cycle. In dark growth conditions, and in the root, BOU is expressed at a low level. Furthermore, bou mutants can be rescued when grown in the dark, a condition in which the glyoxylate cycle is functional and important. The glyoxylate cycle or other pathways may compensate for bou in dark-grown seedlings.
In addition to its role during seedling development, BOU also is required during the entire plant life cycle. Adult bou plants are shorter and paler green than wild-type plants, and they accumulate fewer storage fatty acids. It can be speculated that the BOU transport function is not involved directly in lipid biosynthesis, because it has been shown that the pool of acetate is not a primary source of carbon for lipid biosynthesis in leaves (Bao et al., 2000). In addition, in leaf mitochondria, molecules that might be transported via BOU may play a role in other mitochondrial functions, especially those associated with photosynthetic tissue development and metabolic functions associated with photosynthesis.
METHODS
Plant Growth Conditions
The Arabidopsis thaliana Landsberg erecta Ds(Hyg)-containing line has been described previously (Coupland, 1992; Long et al., 1993), and the T-DNA insertion carrying the Ds(Hyg) transposon was mapped on chromosome V (Long et al., 1997). Detection of plants carrying the transposase construct was performed by monitoring the β-glucuronidase activity associated with 35S::β-glucuronidase (Long et al., 1993). Antibiotic selection of Ds(Hyg), namely hygromycin resistance and streptomycin resistance (associated with Ds excision from the T-DNA), was described by Long et al. (1993). The Arabidopsis Landsberg erecta line carrying the Activator transposase gene fused to the 35S promoter of Cauliflower mosaic virus (35S::TPase) has been described previously (Swinburne et al., 1992).
Plants were grown on standard Murashige and Skoog (1962) medium (Sigma) with or without the addition of 1% Suc before sterilization. Seeds were surface-sterilized with a mix of commercial bleach in ethanol (1:3) for 10 min, rinsed once in ethanol, and allowed to dry. Seeds were allowed to imbibe overnight on Murashige and Skoog (1962) medium at 4°C before transfer to long-day conditions (16 h of light) in a growth chamber at 24°C. Typically, germination was completed 1 day after transfer (Figure 1).
Lipid Analysis
Seeds were boiled for 10 min in 200 μL of water to inactivate any lipase activity, and lipids (total extract) were extracted from 10 seeds as described by Christie (1993). Neutral lipids were separated on a silica thin layer chromatography plate (Si60; Merck) developed with petroleum ether:diethylether:acetic acid (80:20:1, v/v). Lipids were visualized under UV light after spraying with 8-anilinonaphthalene-1-sulfonic acid (0.2 g in 100 mL of methanol). For fatty acid analysis, lipids were transesterified using methanol:sulfuric acid (100:2.5, v/v) at 100°C for 2 h. Fatty acid methyl esters were extracted with hexane.
The total fatty acids of seeds were analyzed by gas chromatography. Thereafter, triacylglycerides (TAG) were computed from the measured C20:1 value in a total lipid extract. Because C20:1 is a typical storage fatty acid found in TAG, it can be used as a marker to determine the overall content of TAG in Arabidopsis seedlings, because the overall fatty acid composition of TAG fatty acids does not vary over 4 days after germination (data not shown). Heneicosanoic acid (C21:0) was used as an internal standard. Gas chromatography was performed using a capillary column (BPX70; Scientific Glass Engineering, Victoria, Australia).
Cloning of BOU
DNA from plants heterozygous for the bou mutation was cleaved with BstYI. Inverse PCR was used to isolate DNA flanking the Ds(Hyg) element according to Long et al. (1993). The primers used to amplify the Ds flanking sequence described in this study were DL6 and D71 (Long et al., 1993). The PCR product was gel purified and sequenced.
DNA and RNA Analysis
Isolation of Arabidopsis DNA was performed as described elsewhere (Sundaresan et al., 1995). The DNA gel blot transfer was probed with the full-length cDNA radioactively labeled by random priming using 32P-dCTP (Sambrook et al., 1989) and exposed to Kodak MS-1 x-ray film (Rochester, NY). Plant RNA was extracted from 200 mg of fresh tissues ground in Eppendorf tubes as described elsewhere (Cowling et al., 1998), separated by agarose-formaldehyde gel electrophoresis, transferred to Optran nitrocellulose membranes (Schleicher & Schuell), and hybridized to probes radioactively labeled by random priming using 32P-dCTP (Sambrook et al., 1989).
To isolate DNA from revertants, PCR was performed using the primers H (5′-TCAAATCTAAAAACAGAGAGG-3′) and C (5′-GAA-ATGGCGGATGCGTGG-3′) using 10 ng of plant DNA as a template, followed by 4 min of denaturation at 94°C and 30 cycles of amplification (1 min at 94°C, 1 min at 55°C, and 1 min at 72°C). The PCR product was purified and sequenced using the PCR sequencing method with primers M (5′-GCTTGTCCTACCGAGTTG-3′) and N (5′-TTC-ACGGGCGAATGTAGG-3′).
Immunoblot Analysis
The complete BOU coding sequence was cloned in pQE30 plasmid (Qiagen, Hilden, Germany) to overproduce a His-tagged BOU protein. After affinity purification on an agarose-nickel column, the purified BOU protein was used to immunize a rabbit. This anti-BOU serum was used to detect the BOU protein in plant protein extracts. Total protein extracts from plants and from cells were purified according to Hurkman and Tanaka (1986) and then dissolved in loading buffer. Purified organelles were resuspended directly in concentrated (two times) loading buffer. An estimated 60 μg of total proteins and 2 to 10 μg of organellar proteins were used for SDS-PAGE and blotting onto nylon Immobilon-P membranes (Millipore, Bedford, MA).
After transfer, the filters were blocked in TBS (20 mM Tris-HCl, pH 7.6, and 137 mM NaCl) with 5% nonfat powdered milk, and the primary antibody was incubated overnight at 4°C in TBS. The filters were rinsed three times in TBS before incubating with the secondary antibody anti-rabbit goat IgG-peroxidase (Bio-Rad, Hercules, CA) for 1 h at room temperature. The signal was developed using the enhanced chemiluminescence kit (Amersham Pharmacia Biotech).
Isolation of Organelles
Plant peroxisomes and mitochondria were purified from 3-week-old Arabidopsis plants and from 5-day-old rapeseed (Brassica napus) grown in the light. A total of 100 g of plant tissue was homogenized in 400 mL of extraction buffer (10 mM Tricine-KOH, pH 7.5, 10 mM KCl, 1 mM MgCl2, 1 mM EDTA, 1 M Suc, 0.3% BSA, 5 mM DTT, 0.1 mM phenylmethylsulfonyl fluoride, and 0.1 mM benzamidine). Chloroplasts were eliminated by centrifugation at 4000g for 10 min. Organelles were pelleted for 20 min at 11,000g, resuspended in extraction buffer, and then loaded on top of a multilayer Suc density gradient (57, 52, 47, 42, and 35% [w/v] Suc in 10 mM EDTA, pH 7.4). The gradient was centrifuged at 72,000g for 3 h.
Peroxisomes were collected from the top of the 57 and 52% layers and mitochondria were collected from the top of the 42% layer. The organelles were fractionated once more using the same gradient and centrifugation conditions, diluted in extraction buffer, and pelleted for 2 h at 72,000g. All of the steps were performed at 4°C. A catalase activity assay (the decrease of H2O2 by absorbance at 240 nm in 0.1 M phosphate buffer, pH 7, 10% Triton X-100, and 0.01% H2O2) confirmed the identification of the peroxisome fraction.
Mitochondria were identified by immunoreaction with anti-NAD9 serum (Lamattina et al., 1993), and peroxisomes were identified with anti-PMP22 serum (Tugal et al., 1999). Chloroplasts were purified separately on Percoll gradients and characterized using anti-RPL4 antibody as described by Trifa et al. (1998). Mitochondrial membranes that were purified from Arabidopsis cells were kindly provided by Jacques Bourguignon from the Laboratoire de Physiologie des Plantes (Grenoble, France; Bourguignon et al., 1988).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Accession Numbers
The GenBank accession numbers for the proteins/genes mentioned in this article are dbj|ABO16882|gi|3449323| (MZA15), W43756 (clone H4G5T7), AJ277732 (BOU cDNA), CAB55356 (CAC from H. sapiens), and AJ250124 (CRC-1 from S. cerevisiae).
Acknowledgments
The authors thank Jean-Pierre Alcaraz for the DNA sequence data, Gérard Clabault for his help with computers, Mireille Jaubert for her help with the photographs, and Daniel Salvi for his help with lipid analysis. Alison Baker, Florence Courtois, Roland Douce, and Ian Graham are thanked for their helpful discussion on the metabolism of glyoxysomes. We thank Jacques Bourguignon for his kind gift of pure Arabidopsis mitochondrial membranes and Rachel Carol for correcting the manuscript.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.002485.
References
- Ahringer, J. (1995). Embryonic tissue differentiation in Caenorhabditis elegans requires dif-1, a gene homologous to mitochondrial solute carriers. EMBO J. 14, 2307–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ap Rees, T. (1987). Compartmentation of plant metabolism. In The Biochemistry of Plants: A Comprehensive Treatise, Vol. 12, P.K. Stumpf and E.E. Conn, eds (New York: Academic Press), pp. 87–115.
- Bao, X., Focke, M., Pollard, M., and Ohlrogge, J. (2000). Understanding in vivo carbon precursor for fatty acid synthesis in leaf tissue. Plant J. 22, 39–50. [DOI] [PubMed] [Google Scholar]
- Bourguignon, J., Neuburger, M., and Douce, R. (1988). Resolution and characterization of the glycine-cleavage reaction in pea leaf mitochondria: Properties of the forward reaction catalysed by glycine decarboxylase and serine hydroxymethyltransferase. Biochem. J. 255, 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carol, P., Stevenson, D., Bisanz, C., Breitenbach, J., Sandmann, G., Coupland, G., Mache, R., and Kuntz, M. (1999). Mutations in the Arabidopsis gene IMMUTANS cause a variegated phenotype by inactivating a chloroplast terminal oxidase associated with phytoene desaturation. Plant Cell 11, 57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie, W.W. (1993). Preparation of ester derivatives of fatty acids for chromatographic analysis. In Advances in Lipid Methodology II, W.W. Christie, ed (Dundee, UK: Oily Press), pp. 69–111.
- Coupland, G. (1992). Transposon tagging in Arabidopsis. In Methods in Arabidopsis Research, C. Koncz, N.H. Chua, and J. Schell, eds (Singapore: World Scientific), pp. 290–310.
- Cowling, R.J., Kamiya, Y., Seto, H., and Harberd, N.P. (1998). Gibberellin dose-response regulation of GA4 gene transcript levels in Arabidopsis thaliana. Plant Physiol. 117, 1195–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLucas, J.R., Dominguez, A.I., Valenciano, S., Turner, G., and Laborda, F. (1999). The acuH gene of Aspergillus nidulans, required for growth on acetate and long chain fatty acids, encodes a putative homologue of the mammalian carnitine/acylcarnitine carrier. Arch. Microbiol. 171, 386–396. [DOI] [PubMed] [Google Scholar]
- Eastmond, P.J., Germain, V., Longe, P.R., Bryce, J.H., Smith, S.M., and Graham, I.A. (2000). Postgerminative growth and lipid catabolism in oilseeds lacking the glyoxylate cycle. Proc. Natl. Acad. Sci. USA 97, 5669–5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond, P.J., and Graham, I.A. (2001). Re-examining the role of the glyoxylate cycle in oilseeds. Trends Plant Sci. 6, 72–77. [DOI] [PubMed] [Google Scholar]
- Elgersma, Y., van Roermund, C.W.T., Wanders, R.J.A., and Tabak, H.F. (1995). Peroxisomal and mitochondrial acetyltransferase of Saccharomyces cerevisiae are encoded by a single gene. EMBO J. 14, 3472–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt, B. (1993). Catabolism of fatty acid α and β-oxidation. In Lipid Metabolism in Plants, T.M. Moore, ed (Boca Raton, FL: CRC Press, pp. 527–565).
- Hartenstein, K., Sinha, P., Mishra, A., Schenkel, H., Török, I., and Mechler, B.M. (1997). The congested-like tracheae gene of Drosophila melanogaster encodes a member of the mitochondrial carrier family required for gas-filling of the tracheal system and expansion of the wings after eclosion. Genetics 147, 1755–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, H., De Bellis, L., Cirurli, A., Kondo, M., Hayashi, M., and Nishimura, M. (1999). A novel acyl-CoA oxidase that can oxidize short-chain acyl-CoA in plant peroxisomes. J. Biol. Chem. 274, 12715–12721. [DOI] [PubMed] [Google Scholar]
- Hayashi, M., Toriyama, K., Kondo, M., and Nishimura, M. (1998). 2,4—Dichlorophenoxybutyric acid–resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid β-oxidation. Plant Cell 10, 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann, K., and Stoffel, W. (1993). TMbase: A database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 347, 166. [Google Scholar]
- Hurkman, W.J., and Tanaka, C.K. (1986). Solubilisation of plant membrane proteins for analysis by two dimensional gel electrophoresis. Plant Physiol. 81, 802–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indiveri, C., Iacobazzi, V., Giangregorio, N., and Palmieri, F. (1997). The mitochondrial carnitine carrier protein: cDNA cloning, primary structure and comparison with other mitochondrial transport proteins. Biochem. J. 321, 713–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indiveri, C., Iacobazzi, V., Giangregorio, N., and Palmieri, F. (1998). Bacterial overexpression, purification and reconstitution of the carnitine/acylcarnitine carrier from rat liver. Biochem. Biophys. Res. Commun. 249, 589–594. [DOI] [PubMed] [Google Scholar]
- Keddie, J.S., Carroll, B., Jones, J.D., and Gruissem, W. (1996). The DCL gene of tomato is required for chloroplast development and palisade cell morphogenesis in leaves. EMBO J. 15, 4208–4217. [PMC free article] [PubMed] [Google Scholar]
- Kleiter, A.E., and Gerhardt, B. (1998). Glyoxysomal β-oxidation of long-chain fatty acids: Completeness of degradation. Planta 206, 125–130. [Google Scholar]
- Lamattina, L., Gonzalez, D., Gualberto, J.M., and Grienenberger, J.M. (1993). Higher plant mitochondria encode a homologue of the nuclear-encoded 30 kDa subunit of bovine mitochondrial complex I. Eur. J. Biochem. 217, 831–838. [DOI] [PubMed] [Google Scholar]
- Long, D., Goodrich, J., Wilson, K., Sundberg, E., Martin, M., Puangsomlee, P., and Coupland, G. (1997). Ds elements on all five Arabidopsis chromosomes and assessment of their utility for transposon tagging. Plant J. 11, 145–148. [DOI] [PubMed] [Google Scholar]
- Long, D., Martin, M., Sundberg, E., Swinburne, J., Puangsomlee, P., and Coupland, G. (1993). The maize transposable element system Ac/Ds as a mutagen in Arabidopsis: Identification of an albino mutation induced by Ds insertion. Proc. Natl. Acad. Sci. USA 90, 10370–10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Olsen, L.J. (1998). The surprising complexity of peroxisome biogenesis. Plant Mol. Biol. 38, 163–189. [PubMed] [Google Scholar]
- Olsen, L.J., and Harada, J.J. (1995). Peroxisomes and their assembly in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 123–146. [Google Scholar]
- Palmieri, L., Lasorsa, F.M., Iacobazzi, V., Runswick, M.J., Palmieri, F., and Walker, J.E. (1999). Identification of the mitochondrial carnitine carrier in Saccharomyces cerevisiae. FEBS Lett. 462, 472–476. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schwadbedissen-Gerbling, H., and Gerhardt, B. (1995). Purification and characterization of carnitine acyltransferase from higher plant mitochondria. Phytochemistry 39, 39–44. [Google Scholar]
- Sundaresan, V., Springer, P., Volpe, T., Haward, S., Jones, J.D., Dean, C., Ma, H., and Martienssen, R. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9, 1797–1810. [DOI] [PubMed] [Google Scholar]
- Swinburne, J., Balcells, L., Scofield, S., Jones, J., and Coupland, G. (1992). Elevated levels of Activator transposase mRNA are associated with high frequencies of Dissociation excision in Arabidopsis. Plant Cell 4, 583–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, D.R., Wood, C., and Masterson, C. (1988). Long-chain acyl CoA synthase, carnitine and β-oxidation in pea-seed mitochondrion. Planta 173, 263–266. [DOI] [PubMed] [Google Scholar]
- Trifa, Y., Privat, I., Gagnon, J., Baeza, L., and Lerbs-Mache, S. (1998). The nuclear RPL4 gene encodes a chloroplast protein that co-purifies with the T7-like transcription complex as well as plastid ribosome. J. Biol. Chem. 273, 3980–3985. [DOI] [PubMed] [Google Scholar]
- Tugal, H.B., Pool, M., and Baker, A. (1999). Arabidopsis 22-kilodalton peroxisomal membrane protein: Nucleotide sequence analysis and biochemical characterization. Plant Physiol. 120, 309.–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Roermund, C.W.T., Elgersma, Y., Singh, N., Wanders, R.J.A., and Tabak, H.F. (1995). The membrane of peroxisomes in Saccharomyces cerevisiae is impermeable to NAD(H) and acetyl-CoA under in vivo conditions. EMBO J. 14, 3480–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Roermund, C.W.T., Hettema, E.W., van den Berg, M., Tabak, H.F., and Wanders, R.J.A. (1999). Molecular characterization of carnitine-dependent transport of acetyl-CoA from peroxisome to mitochondria in Saccharomyces cerevisiae and identification of a plasma membrane carnitine transporter, Agp2p. EMBO J. 18, 5843–5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, C., Hj Jalil, N., McLaren, I., Yong, B.C.S., Ariffin, A., McNeil, P.H., Burgess, N., and Thomas, D.R. (1984). Carnitine long-chain acyltransferase and oxidation of palmitate, palmitoyl coenzyme A and palmitoylcarnitine by pea mitochondrial preparations. Planta 161, 255–260. [DOI] [PubMed] [Google Scholar]
- Wood, C., Masterson, C., and Thomas, D.R. (1992). The role of carnitine in plant cell metabolism. In Plant Organelles, Society for Experimental Biology Seminar Series 50, A.K. Tobin, ed (Cambridge, UK: Cambridge University Press), pp. 229–263.