Abstract
Little is known about the molecular processes that govern female gametophyte (FG) development and function, and few FG-expressed genes have been identified. We report the identification and phenotypic analysis of 31 new FG mutants in Arabidopsis. These mutants have defects throughout development, indicating that FG-expressed genes govern essentially every step of FG development. To identify genes involved in cell death during FG development, we analyzed this mutant collection for lines with cell death defects. From this analysis, we identified one mutant, gfa2, with a defect in synergid cell death. Additionally, the gfa2 mutant has a defect in fusion of the polar nuclei. We isolated the GFA2 gene and show that it encodes a J-domain–containing protein. Of the J-domain–containing proteins in Saccharomyces cerevisiae (budding yeast), GFA2 is most similar to Mdj1p, which functions as a chaperone in the mitochondrial matrix. GFA2 is targeted to mitochondria in Arabidopsis and partially complements a yeast mdj1 mutant, suggesting that GFA2 is the Arabidopsis ortholog of yeast Mdj1p. These data suggest a role for mitochondria in cell death in plants.
INTRODUCTION
The female gametophyte (FG) is a haploid structure that plays an integral role in the angiosperm life cycle. It develops within ovules and most often consists of an egg cell, a central cell, two synergid cells, and three antipodal cells (Figure 1). The FG is important for many aspects of the angiosperm reproductive process: in addition to giving rise to the embryo and endosperm of the seed, the FG plays a role in pollen tube guidance (Hülskamp et al., 1995; Ray et al., 1997; Shimizu and Okada, 2000; Higashiyama et al., 2001), fertilization (Russell, 1992, 1996), and the induction of seed development (Ohad et al., 1996; Chaudhury et al., 1997; Grossniklaus et al., 1998).
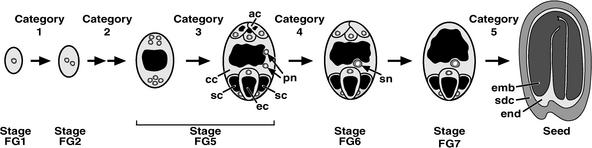
Figure 1.
Developmental Stages Affected in the FG Mutants.
Megagametogenesis has been described previously and divided into seven stages (Christensen et al., 1997). The haploid megaspore defines stage FG1. The megaspore undergoes three rounds of mitosis without cytokinesis, giving rise to an eight-nucleate syncytium (early-stage FG5). Immediately after the third mitosis, cell walls form (late-stage FG5). The central cell inherits two nuclei, called the polar nuclei, which fuse to form the homodiploid secondary nucleus (stage FG6). Finally, the antipodal cells degenerate, producing a mature FG (stage FG7). Early during the fertilization process, one of the synergid cells is induced to undergo cell death (data not shown). Category designations show the developmental stages affected in the FG mutants. In this figure, cytoplasm is shown in light gray, vacuoles are shown in black, nuclei are shown in dark gray, and nucleoli are represented as the white areas within the nuclei. ac, antipodal cell; cc, central cell; ec, egg cell; emb, embryo; end, endosperm; pn, polar nucleus; sc, synergid cell; sdc, seed coat; sn, secondary nucleus.
FG development requires many fundamental cellular processes, including mitosis, vacuole formation, cell wall formation, nuclear migration, nuclear fusion, and cell death (Figure 1). Thus, mutations that affect these processes are likely to affect the FG, exhibit reduced transmission through the FG, and appear at reduced frequency in the sporophyte generation. Consequently, genetic analysis of these cellular processes is likely to require gametophytic screens and analysis of the gametophyte generation. An example of this is cell death, which is important in the diploid sporophyte generation for tissue development, senescence, and plant defense (Martienssen, 1997; Buckner et al., 1998; Richberg et al., 1998; Lam et al., 1999). However, because cell death occurs during the gametophytic phase (Figure 1), cell death mutants may not be identified readily in sporophyte-oriented screens (Martienssen, 1997).
Little is known about the molecular and genetic processes that govern FG development and the FG's reproductive functions. The number and identities of genes expressed in the FG, and the proportion of these that are unique to the FG, are unknown. These genes presumably regulate and mediate FG development and function. However, for all but a few of these genes, specific functions in the FG are unknown. Also, little is known about FG physiology. The physiological pathways that exist in the FG, and the extent to which FG physiology is independent from that of the surrounding sporophytic tissue, are unknown. Thus, it is not clear whether FG development and physiology are governed primarily by the haploid or the diploid genome.
As a step toward addressing these issues, several groups have performed screens for gametophytic mutants in Arabidopsis and maize, and a number of FG mutations have been identified (Bonhomme et al., 1998; Christensen et al., 1998; Drews et al., 1998; Grossniklaus et al., 1998; Howden et al., 1998; Grini et al., 1999; Shimizu and Okada, 2000; Evans and Kermicle, 2001). However, relatively few of these mutations have been characterized at the phenotypic and molecular levels, and a specific function in the FG is known for only few genes.
The present study had two main objectives. The first was to determine the extent to which FG development and function are controlled/mediated by FG-expressed genes and to identify specific genes required by these developmental/reproductive steps. To address these issues, we have identified and characterized a large collection of FG mutants. The second objective was to initiate a molecular genetic analysis of cell death during FG development. To this end, we identified and characterized a mutant, gfa2, that has a defect in synergid cell death during the reproductive process. Our analysis of the GFA2 gene suggests a role for mitochondria in cell death in plants.
RESULTS
Analysis of 39 Female Gametophyte Mutants
To gain insight into the steps of FG development mediated by FG-expressed genes, we identified and analyzed a large collection of FG mutants. We identified 31 new FG mutants (fem5 to fem38) from T-DNA– and transposon-mutagenized lines (see Methods) and analyzed these along with the previously identified ctr1 mutant (Kieber and Ecker, 1994). These mutants are summarized in Table 1. Because the FG is haploid, complementation tests could not be performed to determine whether any of the FG mutations are allelic. However, preliminary molecular analysis of these and seven previously identified mutants (gfa2 to gfa5 and fem2 to fem4; Table 1) has shown that for all mutants analyzed to date (22 of 39 mutants), the T-DNA/transposon insertion sites fall within different genomic regions (C.A. Christensen, R.H. Brown, and G.N. Drews, unpublished data), suggesting that these mutations are not allelic.
Table 1.
Mutants Discussed in This Article
| Mutant | Isolate No. | Mutagen | Ecotype | Source |
|---|---|---|---|---|
| constitutive triple response (ctr1) | – | X-ray | L-er | Kieber and Ecker, 1994 |
| female gametophyte2 (fem2) | 2462-5 | T-DNA | Ws | Christensen et al., 1998 |
| female gametophyte3 (fem3) | 2465-35 | T-DNA | Ws | Christensen et al., 1998 |
| female gametophyte4 (fem4) | TJ160 | T-DNA | Col | Christensen et al., 1998 |
| female gametophyte5 (fem5) | TJ13 | T-DNA | Col | This article |
| female gametophyte6 (fem6) | TJ1335 | T-DNA | Col | This article |
| female gametophyte7 (fem7) | TJ1350 | T-DNA | Col | This article |
| female gametophyte8 (fem8) | TJ1507 | T-DNA | Col | This article |
| female gametophyte9 (fem9) | TJ2111 | T-DNA | Col | This article |
| female gametophyte10 (fem10) | TJ2118 | T-DNA | Col | This article |
| female gametophyte11 (fem11) | a308 | Transposon | L-er | This article |
| female gametophyte12 (fem12) | a466 | Transposon | L-er | This article |
| female gametophyte13 (fem13) | B271 | Transposon | L-er | This article |
| female gametophyte14 (fem14) | A839 | Transposon | L-er | This article |
| female gametophyte15 (fem15) | A845 | Transposon | L-er | This article |
| female gametophyte16 (fem16) | E160 | Transposon | L-er | This article |
| female gametophyte17 (fem17) | TJ761 | T-DNA | Col | This article |
| female gametophyte18 (fem18) | RB46 | T-DNA | Col | This article |
| female gametophyte19 (fem19) | RB81 | T-DNA | Col | This article |
| female gametophyte20 (fem20) | RB158 | T-DNA | Col | This article |
| female gametophyte21 (fem21) | RB160 | T-DNA | Col | This article |
| female gametophyte22 (fem22) | RB299 | T-DNA | Col | This article |
| female gametophyte23 (fem23) | RB302 | T-DNA | Col | This article |
| female gametophyte24 (fem24) | RB309 | T-DNA | Col | This article |
| female gametophyte25 (fem25) | RB312 | T-DNA | Col | This article |
| female gametophyte26 (fem26) | RB353 | T-DNA | Col | This article |
| female gametophyte29 (fem29) | RB117 | T-DNA | Col | This article |
| female gametophyte30 (fem30) | RB140 | T-DNA | Col | This article |
| female gametophyte31 (fem31) | RB184 | T-DNA | Col | This article |
| female gametophyte33 (fem33) | RB273 | T-DNA | Col | This article |
| female gametophyte34 (fem34) | RB279 | T-DNA | Col | This article |
| female gametophyte35 (fem35) | RB341 | T-DNA | Col | This article |
| female gametophyte36 (fem36) | RB405 | T-DNA | Col | This article |
| female gametophyte37 (fem37) | RB428 | T-DNA | Col | This article |
| female gametophyte38 (fem38) | RB433 | T-DNA | Col | This article |
| gametophytic factor2 (gfa2) | 102 | T-DNA | Ws | Feldmann et al., 1997 |
| gametophytic factor3 (gfa3) | 13/21 | T-DNA | Ws | Feldmann et al., 1997 |
| gametophytic factor4 (gfa4) | 84 | T-DNA | Ws | Feldmann et al., 1997 |
| gametophytic factor5 (gfa5) | 114 | T-DNA | Ws | Feldmann et al., 1997 |
Col, Columbia; L-er, Landsberg erecta; Ws, Wassilewskija.
To determine the penetrance of these mutations in the FG (i.e., the proportion of genotypically mutant FGs that fail to transmit the mutant allele), we crossed heterozygous mutants as females to wild-type males and scored the number of heterozygous (kanamycin-resistant) and homozygous wild-type (kanamycin-sensitive) progeny (Table 2). To determine whether these mutations also affect the male gametophyte, we crossed heterozygous mutants as males to wild-type females and scored the number of heterozygous (kanamycin-resistant) and homozygous wild-type (kanamycin-sensitive) progeny (Table 2). As shown in Table 2, of the new mutations, fem8, fem9, fem17, and fem20 appear to affect the FG specifically.
Table 2.
Penetrance, Female Gametophyte Specificity, and Phenotype
| Mutant | Penetrance in FGa | Penetrance in MGa | Terminal Phenotypeb |
|---|---|---|---|
| ctr1-2 | 58%c | 0%c | Wild type (5) |
| fem2 | 44%d | 0%d | Stage FG1d (1) |
| fem3 | 42%d | 44%d | Stage FG1d (1) |
| fem4 | 100%d | 42%d | Abnormal cellular morphology, stage FG5/6/7 (3) |
| fem5 | 87% (685) | 72% (1799) | Abnormal nuclear number, size, and position (2) |
| fem6 | 93% (455) | 86% (811) | Abnormal cellular morphology (3) |
| fem7 | 81% (831) | 30% (614) | Abnormal cellular morphology (3) |
| fem8 | 67% (441) | 9%e (623) | Abnormal cellular morphology (3) |
| fem9 | 42% (431) | 12%e (857) | Stage FG1 (1) |
| fem10 | 93% (1275) | 92% (1347) | Abnormal nuclear number and position (2) |
| fem11 | 54% (624) | 100% (100) | Stage FG1 to stage FG6 (2) |
| fem12 | 73% (1111) | 100% (403) | Stage FG1 (1) |
| fem13 | 84% (406) | 100% (423) | Abnormal nuclear number, cellularization; twinning (2) |
| fem14 | 79% (105) | 100% (218) | Abnormal nuclear number, position; collapse (2) |
| fem15 | 100% (99) | 64% (64) | Abnormal nuclear number and cellularization (2) |
| fem16 | 66% (206) | 71% (298) | No FG (1) |
| fem17 | 98% (66) | 10%e (133) | Wild type (5) |
| fem18 | 39% (305) | 65% (694) | Stage FG1 to stage FG2 (1) |
| fem19 | 100% (306) | 100% (459) | Abnormal nuclear number and position (2) |
| fem20 | 34% (154) | 0% (413) | Stage FG1, stage FG5 (2) |
| fem21 | 100% (108) | 100% (376) | Stage FG3 to stage FG5 (2) |
| fem22 | 100% (710) | 99% (601) | Stage FG4 to stage FG5 (2) |
| fem23 | 98% (826) | 43% (1024) | Stage FG3 to stage FG5 (2) |
| fem24 | 83% (504) | 16% (726) | Stage FG1, stage FG3 to stage FG5 (2) |
| fem25 | 86% (342) | 99% (847) | Abnormal nuclear number, position; stage FG1 to FG2 (2) |
| fem26 | 99% (260) | 17% (796) | Stage FG1 (1) |
| fem29 | 17% (387) | 71% (535) | Stage FG1 (1) |
| fem30 | 15% (183) | 97% (774) | Stage FG1 to stage FG5; abnormal nuclear number (2) |
| fem31 | 66% (506) | 88% (256) | Stage FG1, stage FG3 (1) |
| fem33 | 94% (174) | 70% (214) | Stage FG1, stage FG3 to stage FG4 (2) |
| fem34 | 100% (300) | 98% (360) | Stage FG3 (2) |
| fem35 | 35% (295) | 31% (602) | Stage FG1 (1) |
| fem36 | 99% (315) | 92% (246) | Stage FG1, stage FG3 to stage FG4 (2) |
| fem37 | 42% (328) | 16% (668) | Stage FG1 (1) |
| fem38 | 51% (330) | 53% (355) | Stage FG1 (1) |
| gfa2 | 99% (571) | 86% (704) | Unfused polar nucleid (4) |
| gfa3 | 82% (281) | 99% (365) | Unfused polar nuclei, cellularization failured (4) |
| gfa4 | 61% (1252) | 100% (908) | Stage FG1d (1) |
| gfa5 | 79% (441) | 84% (649) | Stage FG1d (1) |
FG, female gametophyte; MG, male gametophyte. The number of progeny scored is in parentheses.
Phenotypic category is in parentheses.
Previously described by Kieber and Ecker (1994).
Previously described by Christensen et al. (1998).
Not significantly different from 0 at P < 0.05.
To determine the steps of FG development affected by the mutations, we analyzed the terminal phenotypes using confocal laser scanning microscopy (CLSM) (Christensen et al., 1997, 1998) and divided the mutants into phenotypic categories. To comprehensively define the spectrum of possible phenotypes, we included seven previously analyzed mutants (gfa2 to gfa5 and fem2 to fem4; Table 1) in our categorization scheme. Together, the 39 FG mutants fall into five phenotypic categories (Table 2). These phenotypic categories along with wild-type development are summarized in Figure 1. Confocal images of the FG mutants are shown in Figure 2.
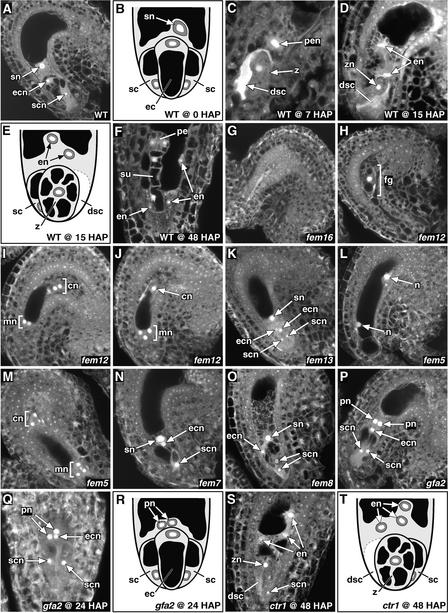
Figure 2.
Phenotypic Analysis of the FG Mutants.
(A) and (B) Wild-type FG at the terminal developmental stage (stage FG7). The egg is pear shaped, has a nucleus at its chalazal end, and has a single large vacuole occupying the remainder of the cell. Both synergid cells are present, and the secondary nucleus is undivided.
(C) Wild-type (ecotype Ws) seed at 7 HAP containing a single-celled zygote, an undivided endosperm nucleus (primary endosperm nucleus), one degenerated synergid cell, and one persistent synergid cell (data not shown).
(D) and (E) Wild-type (ecotype Ws) seed at 15 HAP. The zygote is relatively round, has several small vacuoles, and has a centrally located nucleus. One of the synergids is degenerate, and the primary endosperm nucleus has divided.
(F) Wild-type (ecotype Ws) seed at 48 HAP containing a four-celled proembryo (two cells are not projected in this image) and multiple peripheral endosperm nuclei.
(G) fem16 FG that has collapsed.
(H) fem12 FG arrested at the one-nucleate stage (stage FG1).
(I) fem12 FG arrested at the four-nucleate stage.
(J) fem12 FG containing three micropylar nuclei and one chalazal nucleus.
(K) fem13 FG containing two nuclei in the position of the egg nucleus.
(L) fem5 FG containing two large nuclei.
(M) fem5 FG containing four micropylar nuclei and two chalazal nuclei.
(N) fem7 FG with reduced cytoplasm associated with the nuclei and fuzzy nuclear margins. The second synergid was present but is not projected in this image.
(O) fem8 FG containing a large number of small vacuoles in the central cell cytoplasm and a disordered egg apparatus.
(P) gfa2 FG with unfused polar nuclei.
(Q) and (R) gfa2 FG at 24 HAP. Both synergids are intact, the egg cell appears to be unfertilized (it is pear shaped, has a nucleus at its chalazal end, and has a single large vacuole), and the polar nuclei appear to be unfertilized (they are unfused and remain near the chalazal end of the egg cell). gfa2 embryo sacs at 7 to 15 HAP have similar morphology.
(S) and (T) ctr1-2 seed at 48 HAP showing early signs of embryogenesis. This seed has three endosperm nuclei, a single-celled zygote, a degenerated synergid, and a persistent synergid.
(A), (C), (D), (F) to (Q), and (S) are CLSM images. (B), (E), (R), and (T) are depictions. All CLSM images are projections of several optical sections taken at the terminal developmental stage or at the HAP indicated. In the CLSM images, cytoplasm is shown in gray, vacuoles are shown in black, and nucleoli are shown in white. In the depictions, cytoplasm is shown in light gray, vacuoles are shown in black, nuclei are shown in dark gray, and nucleoli are shown in white. In all images, the micropylar pole is at the bottom and the chalazal pole is at the top. cn, chalazal nuclei; dsc, degenerate synergid cell; ec, egg cell; ecn, egg cell nucleus; en, endosperm nucleus; fg, female gametophyte; mn, micropylar nuclei; n, nucleus; pe, proembryo; pen, primary endosperm nucleus; pn, polar nucleus; sc, synergid cell; scn, synergid cell nucleus; sn, secondary nucleus; su, suspensor; WT, wild type; z, zygote; zn, zygote nucleus.
Category 1 and category 2 mutants had defects during the nuclear division phase of megagametogenesis (stages FG1 to early FG5) and failed to cellularize. With category 1 mutants (14 of 39 mutants), all or most (>60%) of the mutant FGs arrested at stage FG1. Arrested FGs either persisted (Figure 2H) or degenerated (Figure 2G) during ovule development. Mutant FGs that progressed beyond stage FG1 exhibited category 2 defects. With category 2 mutants (17 of 39 mutants), all or most (>60%) of the mutant FGs progressed beyond stage FG1. Category 2 defects included abnormal nuclear numbers and positions (Figures 2J to 2M) and developmental arrest at stages FG2 to early FG5 (Figure 2I).
Category 3 and category 4 mutants became cellularized and had defects during the later phases of megagametogenesis. Category 3 mutants (4 of 39 mutants) had defects in cellular morphology, including abnormal nuclear positions within cells, misshapen cells, and unusual cell features. With fem4, the defects were quite pronounced and included irregular cell shapes and polarities of the egg and synergid cells (Christensen et al., 1998). With the other three category 3 mutants, FG morphology was almost wild type: fem7 (Figure 2N) and fem9 (data not shown) FGs had slightly reduced cytoplasm and slightly abnormal cell shape, and fem8 FGs had many small vacuoles in the central cell cytoplasm (Figure 2O). Category 4 mutants (2 of 39 mutants) had defects in fusion of the polar nuclei. With both mutants, the polar nuclei migrated properly, came to lie side by side, but failed to fuse. gfa2 affected nuclear fusion only (Figure 2P), whereas gfa3 affected both nuclear fusion (predominantly) and cellularization (Christensen et al., 1998).
Category 5 mutants (2 of 39 mutants) appeared normal at the terminal developmental stage (stage FG7), suggesting that megagametogenesis was not affected. This finding, together with the reduced transmission of these mutations through the FG (Table 2), suggests that category 5 mutations affect one of the FG's reproductive functions (pollen tube guidance, fertilization, etc.). Molecular analysis of the T-DNA insertion site in fem17 (C.A. Christensen, R.H. Brown, and G.N. Drews, unpublished data) suggests that the disrupted gene corresponds to the FIS2 gene (Luo et al., 1999), which is required to repress endosperm development before fertilization (Chaudhury et al., 1997).
To investigate the role of CTR1, we analyzed the postpollination phenotypes of ctr1. In ctr1/ctr1 siliques, 92% (45 of 49) of ovules contained a pollen tube in the micropyle (97% in the wild-type control), indicating that pollen tube guidance was not affected significantly. We next examined fertilized ctr1 embryo sacs at 15 to 48 h after pollination (HAP) with wild-type pollen. ctr1 seeds at 48 HAP (Figures 2S and 2T) resembled wild-type seeds at 15 HAP (Figures 2D and 2E): the embryo was single celled and relatively round and contained a central nucleus and many small vacuoles; the endosperm contained two to four nuclei; and one synergid was degenerated. These data suggest that ctr1 FGs become fertilized but arrest very early in seed development.
Together, these data suggest that most (∼95%) FG-expressed genes are required for megagametogenesis and that these genes play a role in specific developmental steps (e.g., cell formation) and/or specific cellular processes (e.g., nuclear fusion). Furthermore, these data suggest that a proportion (∼5%) of FG-expressed genes play no role in megagametogenesis and instead are required for one of the FG's reproductive functions.
The gfa2 Mutation Affects Synergid Cell Death
In an effort to identify the genes involved in cell death during FG development, we analyzed our mutant collection for lines that display cell death defects. Cell death occurs three times during FG development: first, immediately after meiosis, three of the megaspores undergo cell death; second, late during megagametogenesis, the three antipodal cells degenerate (Figure 1); third, in response to pollination, one of the synergid cells undergoes cell death (Christensen et al., 1997). As discussed above, the analysis of megagametogenesis in each of our FG mutants did not reveal mutants with defects in megaspore or antipodal cell death.
We also asked whether synergid cell death after pollination is affected in our FG mutants. We focused on those mutants in which the egg apparatus (egg and synergid cells) is morphologically normal: fem17/fis2 (discussed above), ctr1 (Figures 2S and 2T), and gfa2 (Figure 2P). As discussed above (Figures 2S and 2T) and by others (Chaudhury et al., 1997), the fertilization process appears to be normal in the ctr1 and fem17/fis2 mutants, indicating that synergid cell death is not affected in these mutants.
To determine whether the gfa2 mutation affects synergid cell death, we pollinated gfa2/GFA2 pistils with wild-type pollen and examined gfa2 embryo sacs (those with unfused polar nuclei) at 7 to 24 HAP. In the wild type (Wassilewskija [Ws-2] ecotype), one of the synergid cells was degenerated by 7 HAP (Figure 2C) in essentially all (40 of 41) ovules. In contrast to the wild type, with most (44 of 55) gfa2 FGs, both synergids were intact and showed no signs of degeneration (Figures 2Q and 2R). All gfa2 embryo sacs that failed to undergo synergid cell death (i.e., those with two intact synergids) showed no signs of fertilization (cf. Figures 2A to 2E with Figures 2Q and 2R).
The synergid-cell-death defect of gfa2 could be a secondary consequence of a failure to attract pollen tubes. To address this issue, we pollinated gfa2/GFA2 pistils with wild-type pollen and scored the number of ovules with pollen tubes in the micropyle. In gfa2/GFA2 siliques, >99% (148 of 149) of ovules had a pollen tube in the micropyle (100% in the wild-type control), indicating that the gfa2 mutation does not affect pollen tube guidance. Together, these data indicate that the GFA2 gene product is required for synergid cell death during the fertilization process.
GFA2 Encodes a Member of the DnaJ Protein Family
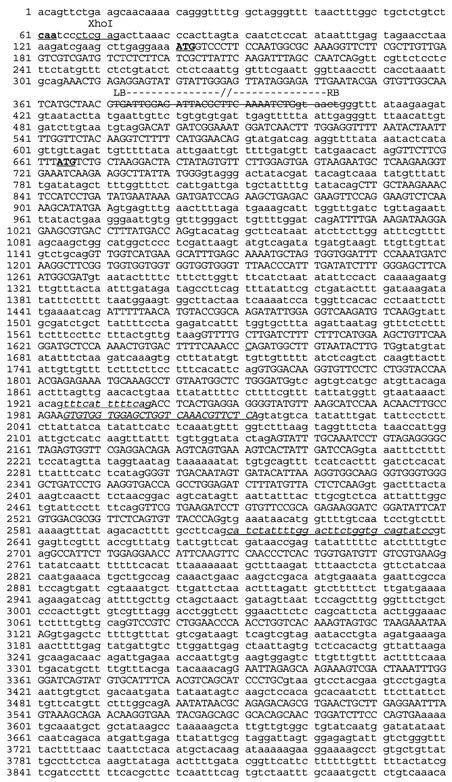
We used inverse PCR (Triglia et al., 1988) to identify the T-DNA insertion site in gfa2 mutants. The insertion site corresponded to locus At5 g48030 from BAC clone MDN11 on chromosome 5. Introducing a wild-type copy of this gene into the gfa2 mutant restored transmission of the gfa2 allele through the FG and rescued the nuclear fusion defect (data not shown), indicating that locus At5 g48030 corresponds to the GFA2 gene. To determine GFA2 gene structure, we isolated a GFA2 cDNA clone and compared its sequence with that of the genomic sequence. As shown in Figure 3, the GFA2 gene consists of 18 exons, has an open reading frame of 1368 bp, and is predicted to encode a protein of 456 amino acids. The T-DNA insert in the gfa2 mutant fell within the second exon, likely creating a null allele (Figure 3).
Figure 3.
GFA2 Gene Sequence.
Exons are indicated with uppercase letters. A 33-bp deletion associated with the site of the T-DNA insert is indicated by strikethrough. The position of the T-DNA left border is indicated above the strikethrough. Plant DNA on the other side of the T-DNA insert is adjacent to the internal T-DNA sequence. The predicted GFA2 protein contains two in-frame ATG codons (marked in boldface and underlined) in its N terminus. The first Met codon is expected to initiate translation because it satisfies the consensus sequence criteria for a translation initiation codon (Kozak, 1991). Some of the exon-intron boundaries derived from the cDNA sequence differ from the annotation of the At5 g48030 locus shown in the MIPS Arabidopsis thaliana database (MATDB; http://mips.gsf.de/proj/thal/db/index.html). Miscalled exons and splice sites relative to the annotation for At5 g48030 are underlined. The underlined regions are indicated as exons or introns according the structure of the GFA2 cDNA.
BLAST searches against the SWISS-PROT database using the predicted GFA2 protein sequence revealed homology with bacterial DnaJ (HSP40) proteins. As shown in Figure 4, within the region of homology (residues 90 to 456), GFA2 is 37% identical and 53% similar to Escherichia coli DnaJ. DnaJ proteins are defined by the presence of an ∼70–amino acid J-domain, which contains the highly conserved HPD tripeptide (Kelley, 1998). GFA2 has this domain and, like many other DnaJ family members, also contains a Gly/Phe-rich domain, a Cys-rich zinc-finger domain, and a less well-conserved C-terminal domain (Figure 4). The J-domain and the Gly/Phe-rich domain are thought to mediate the interactions between DnaJ and its cochaperone DnaK, and the zinc-finger and C-terminal domains are thought to mediate the interactions between DnaJ and its substrate proteins (Hartl, 1996; Bukau and Horwich, 1998; Kelley, 1998; Fink, 1999).
Figure 4.
Alignment of the Predicted Protein Sequences for Arabidopsis GFA2, Yeast Mdj1p, and E. coli DnaJ.
The J domain (residues 90 to 156) and the zinc-finger domain (residues 238 to 294) are double underlined. The Gly/Phe-rich domain (residues 157 to 237) lies between these two domains. Sequence similarity is indicated by shading. Relative to DnaJ, the predicted GFA2 sequence contains an 89-residue extension at the N terminus that has several characteristics of a mitochondrial targeting sequence: an overrepresentation of Arg, Ala, and Ser residues (25 of 89 residues); a paucity of Asp and Glu residues (3 of 89 residues); and an ability to form an amphiphilic α-helix (residues 7 to 16 may form an amphiphilic α-helix region) (Gavel et al., 1988; Roise, 1997).
The GFA2 Gene Is Expressed Throughout the Plant
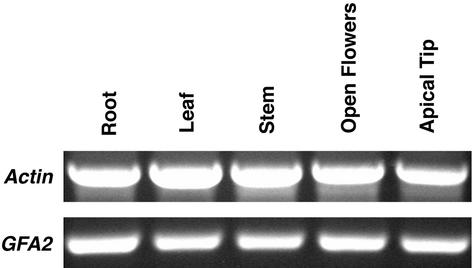
We performed two assays to determine where the GFA2 gene is expressed within the plant. First, we used reverse transcriptase–PCR to assay the presence of GFA2 RNA in various plant organs. Total RNA purified from roots, rosette leaves, stems, open flowers, and apical tips (including unopened flowers) was used as a template to make single-stranded cDNA by reverse transcription, and primers specific for GFA2 and actin were used to amplify cDNAs from each of the RNA populations. As shown in Figure 5, PCR bands representing the spliced transcripts of both genes were amplified from each RNA population, indicating that GFA2 RNA is present in each organ.
Figure 5.
Reverse Transcriptase –PCR Analysis of GFA2 Expression.
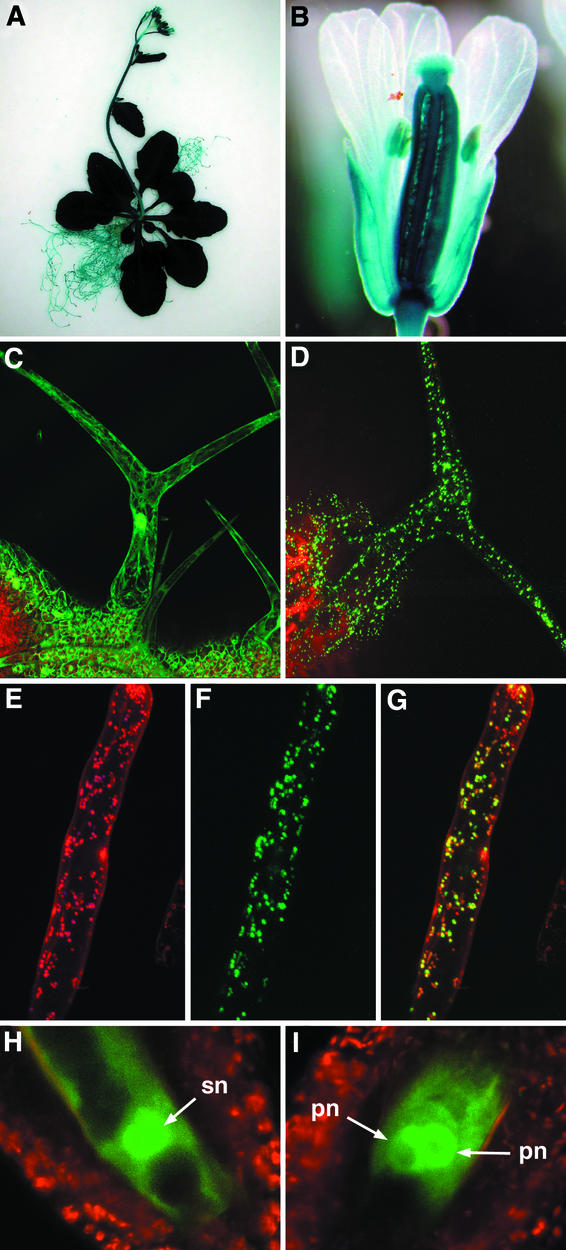
Second, we transformed plants with a GFA2::β-glucuronidase (GUS) reporter construct and analyzed GUS expression in seedlings and whole flowering plants. As shown in Figures 6A and 6B, strong GUS activity was detected in all organs. GUS activity also was detected in ovules during FG development (Figure 6B). Together, these data indicate that the GFA2 gene is expressed throughout the plant.
Figure 6.
GFA2::GUS Expression in the Wild Type, GFA2-GFP Protein Localization in the Wild Type, and FIE::FIE-sGFP Expression in the gfa2 Mutant.
(A) and (B) Histochemical assay for the expression of the GFA2::GUS transgene in the whole plant (A) and the flower (B). Longer histochemical staining showed expression in the petals.
(C) Trichome from a transgenic plant expressing 35S::GFP.
(D) Trichome from a transgenic plant expressing 35S::GFA2-GFP.
(E) to (G) Mitotracker-stained root hair from a transgenic plant expressing 35S::GFA2-GFP.
(E) Mitotracker fluorescence.
(F) GFP fluorescence.
(G) Merger of red and green channel fluorescence.
(H) and (I) Expression of the FIE::FIE-GFP construct in the wild type (H) and the gfa2 mutant (I).
pn, polar nucleus; sn, secondary nucleus.
GFA2 Is Similar to Yeast Mdj1p and Localizes to Mitochondria
We compared GFA2 to the J-domain proteins from Saccharomyces cerevisiae and found that Mdj1p is the yeast protein most similar to GFA2 (data not shown). Mdj1p is a mitochondria-targeted DnaJ family member that functions as a chaperone in the mitochondrial matrix (Rowley et al., 1994; Westermann et al., 1996; Duchniewicz et al., 1999). This sequence similarity suggests that GFA2 might be a mitochondria-targeted protein. Furthermore, the GFA2 N terminus has the characteristics of a mitochondrial targeting sequence (Figure 4) (Gavel et al., 1988; Roise, 1997).
To determine whether GFA2 is targeted to mitochondria, we transformed plants with a GFA2–green fluorescent protein (GFP) translational fusion construct driven by the 35S promoter of Cauliflower mosaic virus (35S::GFA2-GFP). As a control, we also transformed plants with a GFP that lacks a targeting sequence (35S::GFP) (von Arnim et al., 1998). With both constructs, transgenic plants appeared normal. Control plants containing the 35S::GFP construct exhibited GFP fluorescence in the cytoplasm and nucleus (Figure 6C), in agreement with previous studies (Grebenok et al., 1997; von Arnim et al., 1998). By contrast, in plants containing the 35S::GFA2-GFP construct, GFP fluorescence was detected in multiple small intracellular compartments (Figure 6D). This pattern was seen in all tissues examined and colocalized with a mitochondrion-specific dye, Mitotracker (Figures 6E to 6G). Together, these data indicate that the GFA2 protein localizes to mitochondria.
GFA2 Partially Complements a Yeast mdj1 Deletion
The sequence similarity and mitochondrial localization suggest that GFA2 is the Arabidopsis ortholog of yeast MDJ1. To test this possibility further, we determined whether GFA2 protein can substitute for Mdj1p in yeast by expressing GFA2 in an mdj1 yeast strain. The yeast strains used in this study are listed in Table 3. To generate an MDJ1 null allele, we replaced the entire MDJ1 coding region with the HIS3 gene. We refer to this allele as mdj1::HIS3. As expected from previous studies (Rowley et al., 1994; Duchniewicz et al., 1999), mdj1::HIS3 cells at 25°C failed to grow on the nonfermentable carbon source, glycerol, but grew slowly on the fermentable carbon source, dextrose. This petite phenotype is consistent with a defect in mitochondrial function. In addition, at 37°C, mdj1::HIS3 cells failed to grow on both carbon sources, indicating that Mdj1p function also is required for survival at high temperature (Duchniewicz et al., 1999).
Table 3.
Yeast Strains Used in This Study
| Strain Name | Genotype |
|---|---|
| JSY4037 | MATa/α ura3-52/ura3-52 his3Δ200/his3Δ200 leu2Δ1/leu2Δ1 trp1Δ63/trp1Δ63 lys2Δ202/lys2Δ202 mdj1::HIS3/MDJ1 with pRS416-MET25-GFA2 |
| JSY4042 | MATa/α ura3-52/ura3-52 his3Δ200/his3Δ200 leu2Δ1/leu2Δ1 trp1Δ63/trp1Δ63 lys2Δ202/lys2Δ202 mdj1::HIS3/MDJ1 with pRS416-MET25 |
| JSY4046 | MATa ura3-52 his3Δ200 leu2Δ1 trp1Δ63 lys2Δ202 mdj1::HIS3 with pRS416-MET25-GFA2 |
| JSY4048 | MATa ura3-52 his3Δ200 leu2Δ1 trp1Δ63 lys2Δ202 MDJ1 with pRS416-MET25-GFA2 |
| JSY4058 | MATa ura3-52 his3Δ200 leu2Δ1 trp1Δ63 lys2Δ202 MDJ1 with pRS416-MET25 |
| JSY4059 | MATa ura3-52 his3Δ200 leu2Δ1 trp1Δ63 lys2Δ202 mdj1::HIS3 with pRS416-MET25 |
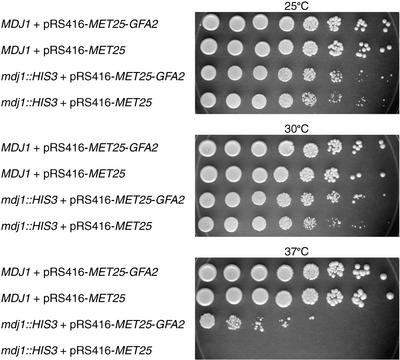
We introduced an expression vector containing the GFA2 cDNA (pRS416-MET25-GFA2) into heterozygous yeast cells (MDJ1/mdj1::HIS3). As a control, we transformed heterozygous cells (MDJ1/mdj1::HIS3) with the pRS416-MET25 vector alone. Transformed heterozygous diploids then were sporulated, and tetrads were dissected onto glycerol- or dextrose-containing plates and grown at 25, 30, and 37°C. As shown in Figure 7, mdj1::HIS3 cells containing the GFA2 expression vector grew slightly faster than mdj1::HIS3 cells containing the empty vector at 25°C (cf. rows 3 and 4) and 30°C (cf. rows 7 and 8).
Figure 7.
GFA2 Complementation of the Yeast mdj1 Mutant Growth Defect.
Cultures of the four possible combinations of the MDJ1 or mdj1::HIS3 allele and the expression vectors pRS416-MET25-GFA2 or pRS416-MET25 were diluted serially (1:10) and grown at 25, 30, and 37°C on dextrose.
In addition, the presence of GFA2 rescued the lethality of mdj1::HIS3 at 37°C (cf. rows 11 and 12). However, the presence of the GFA2 expression vector did not restore the ability of mdj1::HIS3 cells to grow on glycerol (data not shown). These data (summarized in Table 4) indicate that Arabidopsis GFA2 can partially substitute for Mdj1p in yeast. These data, along with the sequence similarity and mitochondrial localization, suggest very strongly that GFA2 is the ortholog of yeast MDJ1 and that GFA2 protein functions as a chaperone in the mitochondrial matrix.
Table 4.
Complementation of Yeast mdj1::HIS3 Growth Defects by Arabidopsis GFA2
| 25°C
|
30°C
|
37°C
|
||||
|---|---|---|---|---|---|---|
| Construct | Glycerol | Dextrose | Glycerol | Dextrose | Glycerol | Dextrose |
| MDJ1 + pRS416-MET25 | ++ | +++ | ++ | +++ | ++ | +++ |
| MDJ1 + pRS416-MET25-GFA2 | ++ | +++ | ++ | +++ | ++ | +++ |
| mdj1::HIS3 + pRS416-MET25 | − | ++ | − | + | − | − |
| mdj1::HIS3 + pRS416-MET25-GFA2 | − | ++ | − | ++ | − | ++ |
The gfa2 Mutation Does Not Affect Viability
The data presented above suggest that the gfa2 mutation causes defects in mitochondrial function. Thus, the gfa2 defects could be a secondary consequence of decreased metabolic activity. To assess this possibility, we asked whether gfa2 FGs express genes associated with central cell differentiation. We analyzed the expression of a FIE reporter gene construct (FIE::GFP) whose expression is initiated in the central cell at approximately the time of polar nuclei fusion (Yadegari et al., 2000). We examined ovules from plants that were heterozygous for the gfa2 mutation and homozygous for FIE::GFP.
As expected, approximately half (39 of 70) of the ovules contained a FG with unfused polar nuclei. All of the ovules examined (n = 98) expressed FIE::GFP in the central cell (data not shown). We also examined ovules from plants heterozygous for the gfa2 mutation and containing a FIE-GFP fusion protein construct (FIE::FIE-GFP). GFP expressed from this construct is localized in the central cell nucleus (Kinoshita et al., 2001). As shown in Figures 6H and 6I, the FIE::FIE-GFP construct is localized to the nucleus and is expressed at the same level in mutant and wild-type FGs.
gfa2 FGs exhibited several other indicators of cell viability and metabolic activity. First, the failure of polar nuclei fusion was the only defect observed during megagametogenesis (Figure 2P). In the wild type, many other energy-requiring steps (e.g., cell wall formation) occurred at approximately the same time as nuclear fusion. Yet, none of these steps appeared to be affected in the gfa2 mutant (Figure 2P). Furthermore, the nuclei, vacuoles, and cell membranes in gfa2 FGs were intact and showed no signs of degradation (Figures 2P and data not shown). Second, as discussed above, gfa2 FGs attracted pollen tubes. The presence of a functional FG is essential for pollen tube attraction to an ovule (Hülskamp et al., 1995; Ray et al., 1997; Shimizu and Okada, 2000; Higashiyama et al., 2001). Third, within emasculated flowers, gfa2 FGs persisted as long as wild-type FGs (data not shown). Together, these data suggest that gfa2 FGs are viable and metabolically active.
DISCUSSION
FG-Expressed Genes are Required throughout FG Development
One of the objectives of this study was to identify and characterize a large collection of mutants so that we could infer the developmental and reproductive steps regulated and mediated by FG-expressed genes. Our analysis has shown that a large number of FG mutants can be recovered (discussed below) and that these mutants have defects throughout development. Many of these mutations appear to affect specific processes and can be ordered within the FG developmental pathway (Figure 1). These data indicate that megagametogenesis is controlled/mediated extensively by FG-expressed genes and that FG-expressed genes control/mediate essentially every step of FG development. The one developmental step for which no mutants were recovered is degeneration of the antipodal cells, possibly because mutations that affect this step occur infrequently or do not affect FG viability.
In addition, we identified a mutant class that includes ctr1 and fem17, in which megagametogenesis is not affected detectably. This, together with the fact that these mutations exhibit reduced transmission through the FG, suggests that the affected genes are required for one of the FG's reproductive functions. Consistent with this notion, the T-DNA insertion in fem17 (C.A. Christensen, R.H. Brown, and G.N. Drews, unpublished data) disrupts the previously identified FIS2 gene (Luo et al., 1999), which plays a role in the induction of endosperm development (Chaudhury et al., 1997).
The CTR1 gene encodes a Raf Ser/Thr protein kinase that is involved in ethylene signal transduction (Kieber et al., 1993). Seed development becomes arrested soon after the fertilization of ctr1 FGs with wild-type pollen (Figure 2S). These data suggest either that maternal expression (i.e., FG expression) of the CTR1 gene is required for embryo and/or endosperm development after fertilization or that the CTR1 gene is imprinted paternally and required after fertilization for embryo and/or endosperm development (Chaudhury and Berger, 2001).
In the three independent screens we performed, mutant frequency was 0.5 to 0.7% among T-DNA/transposon lines (see Methods). However, this number is an underestimate because lines containing multiple independently segregating inserts were not considered in segregation distortion screens. In our T-DNA screens, approximately one-third of the lines contained multiple inserts; thus, the corrected frequency of FG mutants was ∼1.0%. Mutant frequencies similar to this have been reported by other groups (Moore et al., 1997; Bonhomme et al., 1998; Howden et al., 1998). In a saturation screen, 180,000 T-DNA insertions must be screened to achieve a 95% probability of mutating any gene of median size (Krysan et al., 1999), and approximately three alleles per gene are isolated on average. Using these three numbers, we estimate that ∼600 (0.01 × 180,000 × 0.33) independent FG loci could be identified via mutation.
The gfa2 Mutation Affects Cell Death and Nuclear Fusion
A second objective of this study was to initiate a genetic analysis of cell death during FG development. Cell death plays a prominent role in FG development: soon after meiosis, three of the meiotic products (megaspores) undergo cell death; late during megagametogenesis, the three antidopdal cells degenerate; and soon after pollination, one of the synergid cells undergoes cell death. Although we did not identify mutants with defects in megaspore or antipodal cell death, one mutant, gfa2, exhibited a defect in synergid cell death. In the wild type, most FGs contain a degenerating synergid cell at 7 HAP (Figure 2C), whereas in most gfa2 FGs, a degenerating synergid cell is not observed even at 24 HAP (Figure 2Q). Megaspore and antipodal cell death both occur normally in gfa2 mutants, suggesting that synergid cell death may occur via a different mechanism than megaspore and antipodal cell death.
The synergid cell death process has been described in a number of species. An exact time course has not been reported; however, the process invariably involves a dramatic decrease in cell volume, collapse of the vacuoles, and complete disintegration of the plasma membrane and most organelles (Jensen and Fisher, 1968; Cass and Jensen, 1970; van Went, 1970; Mogensen, 1972; van Went and Cresti, 1988; Huang et al., 1993). Synergid cell death has not been described in Arabidopsis. Thus, the extent to which synergid cell death in Arabidopsis resembles synergid cell death in other species remains unknown. It also is not clear which step of synergid cell death is affected in gfa2 mutants. To resolve these issues, we are analyzing synergid cell death in wild-type and gfa2 FGs using transmission electron microscopy.
The gfa2 mutation also affects the fusion of the polar nuclei during megagametogenesis (Figure 2P). Fusion of the polar nuclei begins with contact of the endoplasmic reticulum (ER) membranes that are continuous with the outer nuclear membranes of the two nuclei. Fusion of the ER membranes results in outer nuclear membranes that are continuous. Finally, the inner nuclear membranes come into contact and merge (Jensen, 1964; Schulz and Jensen, 1973; Sumner and Van Caeseele, 1990; Huang and Russell, 1992a). In gfa2 mutants, the outer nuclear membranes of the polar nuclei come into contact but do not fuse (Figure 2P and data not shown), suggesting that the GFA2 gene product is required for the membrane fusion step.
GFA2 Is a Mitochondrial DnaJ Protein
GFA2 encodes a member of the DnaJ protein family, which function as chaperones alone or in association with DnaK (HSP70) partners. Chaperone activity prevents the aggregation or misfolding of nascent polypeptides, assists in the assembly/disassembly of protein complexes, prevents nonproductive interactions with other cellular components, and prevents protein aggregation during stress (Hartl, 1996; Bukau and Horwich, 1998; Kelley, 1998; Fink, 1999). Of the yeast DnaJ family members, GFA2 is most similar to mitochondrially localized Mdj1p. GFA2 protein localized to mitochondria (Figures 6E to 6G), and the GFA2 gene partially complemented a yeast mdj1 deletion (Figure 7, Table 4). Together, these data suggest very strongly that GFA2 is the Arabidopsis ortholog of MDJ1.
Disruption of the MDJ1 gene leads to mitochondrial genome loss and reduced oxidative phosphorylation and ATP production by mitochondria (Rowley et al., 1994; Prip-Buus et al., 1996; Westermann et al., 1996; Duchniewicz et al., 1999). The gfa2 mutation most likely causes defects in mitochondrial function similar to those caused by the mdj1 mutation (Rowley et al., 1994; Prip-Buus et al., 1996; Westermann et al., 1996; Duchniewicz et al., 1999). Thus, the gfa2 defects could be a secondary consequence of reduced metabolic activity.
Several lines of evidence argue against this possibility. First, the nuclear fusion defect is the only defect detected during megagametogenesis (Figures 2P to 2R) (Christensen et al., 1998). Nuclear division, nuclear migration, cell wall formation, and cell differentiation all occur normally. If gfa2 FGs were energy deficient, pleiotropic effects on these other energy-requiring processes would be expected. Second, gfa2 FGs do not appear degraded and persist within unfertilized ovules as long as wild-type FGs. Third, gfa2 FGs are metabolically active: they attract pollen tubes and express genes at the same level as wild-type cells (Figures 6H and 6I). Decreased metabolic activity would be expected if gfa2 FGs were energy deficient. Together, these observations suggest that the cell death and nuclear fusion defects are not attributable simply to a lack of respiratory activity and reduced ATP levels.
These observations also suggest that the surrounding sporophytic tissue provides metabolites to the FG. As discussed above, oxidative phosphorylation and ATP production most likely are reduced in gfa2 FGs. If this is the case, then the apparently normal metabolic activity of gfa2 FGs suggests that this energy deficiency is rescued by the provision of metabolites by the surrounding sporophytic tissue.
Role of GFA2 in Cell Death
In animals, mitochondria play a central role in cell death (Green and Reed, 1998; Wang, 2001). This is established most clearly in mammals, in which the mitochondrial intermembrane space contains several cell death activators (e.g., cytochrome c) and effectors (e.g., nucleases). In response to a variety of cell death stimuli, the cell death activators/effectors are released from mitochondria (Hengartner, 2000; Wang, 2001). The cell death activators promote cell death by activating caspases that are largely responsible for the morphological changes associated with cell death (Hengartner, 2000). Much less is known about cell death in plants, and a role for mitochondria is not firmly established (Lam et al., 1999; Jones, 2000).
In this study, we show that the GFA2 gene is required for synergid cell death and that the absence of GFA2 protein most likely compromises mitochondrial function. These data suggest very strongly that mitochondria play a role in cell death in plants. Several other studies support this conclusion.
First, in several experimental systems, the release of cytochrome c from mitochondria has been shown to precede cell death (Balk et al., 1999; Stein and Hansen, 1999; Sun et al., 1999; Balk and Leaver, 2001).
Second, Bax, a cell death–promoting member of the Bcl-2 protein family, triggers cell death when expressed in plant cells. Deletion of the C-terminal hydrophobic tail (transmembrane domain) eliminates this activity, and fusion of the transmembrane domain to GFP targets GFP to mitochondria, suggesting that mitochondrial targeting of Bax is necessary for its death-promoting activity in plants (Lacomme and Santa Cruz, 1999).
Third, victorin, a fungal toxin that binds mitochondrial Gly decarboxylase, induces cell death in treated tissue (Navarre and Wolpert, 1999).
Finally, mutants or experimental manipulations that lead to increased levels of reactive oxygen species, which are produced primarily in the mitochondrion, can induce cell death or the expression of genes associated with cell death (Hu et al., 1998; Maxwell et al., 1999; Molina et al., 1999).
Given that GFA2 functions as a chaperone within the mitochondrial matrix, it is unlikely to play a direct role in cell death. More likely, the gfa2 mutation affects cell death indirectly by compromising the folding of proteins that are involved directly in the cell death process. For example, GFA2-deficient mitochondria may fail to undergo the morphological and biochemical changes required to release cytochrome c in response to cell death stimuli. To investigate these issues further, we are analyzing mitochondrial morphology and cytochrome c localization during synergid cell death in wild-type and gfa2 FGs.
Role of GFA2 in Nuclear Fusion
Essentially nothing is known about the molecular aspects of nuclear fusion in plants. However, genetic and biochemical studies in yeast have identified a number of proteins required for nuclear fusion during karyogamy, which occurs via a process similar to nuclear fusion in plants (Rose, 1996). All of the identified proteins localize to the ER, and most of them are components of the translocon (Rose, 1996; Brizzio et al., 1999). Although a direct requirement for these translocon components in membrane fusion has been demonstrated, a specific role of the translocon in nuclear fusion remains unclear.
The mitochondrial localization of GFA2 raises the question of what role this protein plays in nuclear fusion. As discussed above, the nuclear fusion defect does not appear to be attributable simply to a lack of respiratory activity. One possibility is that functional mitochondria are required for nuclear fusion in plants for reasons other than energy production. For example, nuclear membrane fusion may require diffusible factors derived from one of the mitochondrion's other metabolic activities, including hemes, cytochromes, carbon skeletons, and thymidylates (Mackenzie and McIntosh, 1999; Kushnir et al., 2001).
Alternatively, mitochondria may provide factors via physical contact with the outer nuclear membrane (Lichtscheidl et al., 1990). For example, lipid composition, which is important for membrane trafficking within the secretory pathway (Burger, 2000; Huijbregts et al., 2000; Huttner and Schmidt, 2000), may be an important factor in membrane fusion, and mitochondrial-nuclear contact may be required to shuttle lipids between the two compartments (Staehelin, 1997). The analysis of additional nuclear fusion mutants as well as mitochondrial mutants in Arabidopsis and yeast should help to clarify the possible role of the mitochondrion in nuclear fusion in these two organisms.
Synergid Cell Death in Arabidopsis
The timing and induction of synergid cell death appears to be variable among species. In some species, synergid cell death requires pollination and does not occur if pollination is prevented (Christensen et al., 1997; Huang and Russell, 1992b; Jensen et al., 1983), which suggests that synergid degeneration is not a feature of the megagametogenesis developmental program in these species. By contrast, in other species, synergid cell death occurs before pollination and clearly is not dependent on pollination, which suggests that synergid degeneration is an integral aspect of the megagametogenesis developmental program in these species (Huang and Russell, 1992b). Thus, different species may use different mechanisms to induce synergid cell death.
In Arabidopsis, synergid cell death does not occur in the absence of pollination (Christensen et al., 1997), and a degenerating synergid cell is apparent soon after pollination (within 7 HAP; Figure 2C). These observations suggest very strongly that synergid cell death is induced by pollination or the presence of pollen tubes within the female tissue (Christensen et al., 1997). At present, it is not known whether synergid degeneration in Arabidopsis occurs before or upon pollen tube arrival at the FG. Thus, it is unclear whether synergid cell death is triggered by a long-range signal or upon FG–pollen tube contact.
In some species, the synergid degenerates only after pollen tube arrival, and degeneration is thought to occur via a mechanical process: the release of pollen tube contents into the synergid cell may cause a massive increase in volume and pressure, resulting in a bursting of the synergid membrane (van Went and Willemse, 1984; Willemse and van Went, 1984; Russell, 1992; Higashiyama et al., 2000). The gfa2 mutation should not affect a mechanical process such as this, suggesting that synergid degeneration in Arabidopsis entails a physiological response by the synergid cell.
The gfa2 phenotype suggests that synergid degeneration is not required for pollen tube attraction. The synergid cells are the source of a pollen tube attractant (Higashiyama et al., 2001), and synergid cell death could, in principle, be important for attractant release (van Went and Willemse, 1984; Willemse and van Went, 1984; Russell, 1992; Cheung, 1996). In gfa2 FGs, synergid cell death failed to occur and ovules containing gfa2 FGs attracted pollen tubes, suggesting that the attractant was released by intact synergid cells. These data are consistent with those showing that at least one intact synergid is required for pollen tube attraction (Higashiyama et al., 2001).
Summary
We have shown that screens for FG mutants can identify genes important for many developmental and cellular processes, including cellularization, nuclear fusion, cell death, and the induction of seed development. One example of this is GFA2, which is required for cell death of the synergid cell. We have shown that GFA2 protein localized to mitochondria and that the gfa2 mutation most likely affected mitochondrial function. These data suggest very strongly that mitochondria are involved in cell death in plants. Thus, the role of mitochondria as important regulators of cell death appears to be conserved evolutionarily in plants, mammals (Green and Reed, 1998), and Caenorhabditis elegans (Chen et al., 2000; Parrish et al., 2001).
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana growth conditions were as described previously, except that plants were grown in Scott's Terra-lite soil mix (Maysville, OH) (Christensen et al., 1997). We obtained seeds from transgenic plants for two FIE reporter construct lines. Line 24 contains 4.9 kb of FIE promoter fused to sGFP (FIE::sGFP) (Yadegari et al., 2000), and line 11 contains 1.6 kb of FIE promoter fused to the FIE cDNA and sGFP (FIE::FIE-sGFP) (Kinoshita et al., 2001).
Plant Transformation
T-DNA constructs were introduced into Agrobacterium tumefaciens strain ASE by freeze-thaw transformation (Chen et al., 1994). Arabidopsis plants were transformed by the floral dip method (Clough and Bent, 1998). Transformed progeny were selected by sowing surface-sterilized T1 seeds on 50 μg/mL kanamycin in Murashige and Skoog (1962) (MS) basal medium with Suc and agar (M-9274; Sigma).
Mutant Screen
Mutagenized individuals were transferred to soil, grown, and screened for reduced seed set or the presence of abnormal seeds (white or brown seeds). Selfed seeds were collected from plants exhibiting reduced seed set, and the ratio of kanamycin-resistant to kanamycin-sensitive progeny was determined. With lines exhibiting a resistant-to-sensitive ratio of ≤1.5:1, ∼20 individuals were transferred to soil and screened to determine if reduced seed set cosegregated with kanamycin resistance. Two individuals were used in reciprocal crosses with the wild type, and lines exhibiting significantly reduced transmission of kanamycin through the female gametophyte (FG) were selected for further analysis. Backcrossed individuals were transplanted to soil for repeated reciprocal crosses (2 individuals), repeated cosegregation analysis (50 individuals), and analysis of the FG terminal phenotype (2 individuals). The theoretical aspects of this screen have been described previously (Christensen et al., 1998; Drews et al., 1998).
We performed three independent screens. In the first screen, we identified seven mutants (fem5 to fem10 and fem17) from 1228 T-DNA–mutagenized lines generated with the pD991 vector (Campisi et al., 1999); in the second screen, we identified six mutants (fem11 to fem16) from 819 transposon-mutagenized lines generated with the DsG transposon (Sundaresan et al., 1995); and in the third screen, we identified 18 mutants (fem18 to fem38) from 3426 T-DNA–mutagenized lines generated with the pCGN1547 vector (McBride and Summerfelt, 1990). The mutant frequency from these three screens was 0.6, 0.7, and 0.5%, respectively.
Phenotypic Analysis of Mutants
Confocal laser scanning microscopy was performed as described previously (Christensen et al., 1997, 1998). For analysis of pollen tube guidance, siliques at 24 h after pollination were fixed in 10% glacial acetic acid, 30% chloroform, and 60% absolute ethanol for ∼1.5 h; rinsed with tap water; soaked in 4 N NaOH at 65°C for 15 to 20 min; rinsed with tap water; and then stained in 0.1% aniline blue (Smith and McCully, 1978) and 0.1 M K3PO4 overnight. Stained tissue was mounted in a drop of glycerol, pressed with a cover slip, and examined using a fluorescence microscope. In all cases, terminal phenotypes were analyzed. To obtain ovules of the appropriate stage, we emasculated flowers of stage 12c, waited 24 to 48 h, and processed the tissue for confocal analysis (Christensen et al., 1997).
Sequence Analysis
We used GENSCANW (http://CCR-081.mit.edu/cgi-bin/genscanw.cgi) to identify genomic sequence features such as exon/intron boundaries and poly(A) addition sites (Vector NTI Suite version 5.3 for Macintosh; Informax Inc., Bethesda, MD) for manipulation of plasmid constructs, primer design, and analysis of protein sequences. We used the AlignX module of Vector NTI suite with a BLOSUM matrix and the neighbor-joining algorithm (Saitou and Nei, 1987) for the alignment of protein sequences.
GFA2 Gene Cloning and gfa2 Complementation
We used inverse PCR to identify the plant sequence flanking the T-DNA border in the gfa2 mutant (Triglia et al., 1988). We used the HindIII restriction enzyme with the LB1 (5′-CATCTCATCGATGCT-TGGTAATAATTG-3′) and LB2 (5′-GAGCTATTGGCACACGAAGAATGGT-3′) primers. Approximately 600 bp was amplified that contained T-DNA left border sequence and 240 bp of plant DNA.
The construct for molecular complementation of the gfa2 mutation consisted of the GFA2 promoter fused to the GFA2 cDNA (see below). A 3654-bp XhoI fragment from a partial genomic subclone of BAC MDN11 (plasmid pBSII-C/P5′GFA2-8) was subcloned into the XhoI site in the pCRII-6/14GFA2c plasmid, resulting in plasmid pCR-GFA2c/g. To add a transcription termination sequence, the primers 3′NOS-F (SacI; 5′-GCGCGAGCTCTGAATCCTGTTGCCGGTCTTG-3′) and 3′NOS-R (PstI-SacI; 5′-GCGCGAGCTCTGCAGCGATCTAGT-AACATAGATGACAC-3′) were used to amplify and introduce restriction sites into the 3′ nopaline synthase (NOS) termination sequence from pGEM NOS-1.
This product was digested with SacI and ligated into the SacI site of pCR-GFA2c/g. A 5513-bp SphI-PstI fragment corresponding to the rescue construct (GFA2::GFA2-NOS) then was isolated from the resultant plasmid, pCR-GFA2c/g-NOS, blunted, and subcloned into the unique PmeI restriction site of the T-DNA vector pCGNlox2b (Sieburth et al., 1998), resulting in transformation vector plox-GFA2c/g-NOS. Transgenic progeny from wild-type plants transformed with plox-GFA2c/g-NOS were selected and crossed with the gfa2 mutant. By crossing and screening with PCR, we generated lines that were heterozygous for the gfa2 mutation and homozygous for the GFA2 transgene. In such lines, FG morphology was analyzed, and transmission of the gfa2 allele through the FG and male gametophyte was tested.
Reverse Transcriptase PCR and cDNA Cloning
RNA was isolated using the Plant RNeasy purification kit (Qiagen, Valencia, CA) from ecotype Columbia. First-strand cDNA synthesis was performed using the SuperScript Preamplification System for First Strand cDNA Synthesis (Gibco BRL). Subsequent amplification of the first-strand cDNA was performed using Biolase polymerase (Bioline, Reno, NV), and all thermal cycling was performed on an MJ Research PTC-200 thermal cycler. For tissue-specific reverse transcriptase–PCR, we used primer sets gfa2-11 (5′-CAACCCTCACTGGTGATGTTGTCGTGAA-3′) and gfa2-14 (5′-GTAGAATTAGTTAAAAGTAAAACCCAGACA-3′) along with ActConF (5′-GATTTG-GCATCACACTTTCTACAATG-3′) and ActConR (5′-GTTCCACCACTG-AGCACAATG-3′) to assay the presence of spliced cDNAs from the GFA2 and actin genes, respectively.
The fully processed cDNA fragments amplified by these primer sets were 385 bp for GFA2 and 650 bp for actin. To amplify a cDNA encompassing the entire open reading frame of GFA2, we used primer set gfa2-6 (5′-TAGGGTTTTAACTTTGGCTGCTCTGTCTCAA-3′) and gfa2-14. The cDNA was cloned into the pCRII-TOPO vector using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA), resulting in plasmid pCRII-6/14GFA2c.
GFA2::β-Glucuronidase Reporter Construct
To make the β-glucuronidase (GUS) reporter plasmid, we started with pBSII-mGFP5-ter that was prepared by amplifying the mGFP5 coding region and the transcription termination region from pAVA393 (von Arnim et al., 1998) using mGFP5-F (5′-GGGGCTCGAGCC-ATGGGTAAAGGAGAACTTTTCACT-3′) and mGFP5-R (5′-GGG-GGGTACCAGGTCACTGGATTTTGGTTTTAGGAA-3′), which introduced XhoI and KpnI sites, respectively, and cloned this product in pBluescript II KS−. The uidA coding sequence from transgenic GUS-expressing plants (transformed with the pD991 enhancer trap vector [Campisi et al., 1999]) was amplified using primers GUS-F (5′-GGGGCCATGGCATTACGTCCTGTAGAAACCCCAA-3′) and GUS-R (5′-GGGGTCTAGAGTTGTTTGCCTCCCTGCTGCGGTT-3′), which introduced a NcoI site at the uidA start codon and an XbaI site at the uidA stop codon.
The PCR product was digested with NcoI and XbaI and used to replace the mGFP5 coding region in pBSII-mGFP5-ter, resulting in plasmid pBSII-GUS-ter. The GFA2 upstream regulatory sequences and the first three exons were obtained by PCR amplification using the gfa2-17 (5′-GGGGCTGCAGCAGATCAGTCGACGTTCATGCCA-TCTCAT-3′) and gfa2-18 (5′-GGGGCCATGGCAAACGAAGAACCTGTTCCATGAAAAGACCT-3′) primers to introduce PstI and NcoI sites, respectively. The resulting PCR product was cloned into pBSII-GUS-ter using the PstI and NcoI sites, resulting in plasmid pBSII-GFA2pro-GUS. The GFA2::GUS-ter cassette was liberated by BssHII digestion and blunting and subcloned into the blunted XbaI site of pCGN1547, resulting in plasmid pCGN-GFA2pro-GUS. This construct includes 2993 bp of sequence upstream of the translational start codon and the first three exons of GFA2.
Progeny from plants transformed with pCGN-GFA2pro-GUS were selected on 50 μg/mL kanamycin MS medium. After ∼10 days of growth, seedlings were transplanted to Magenta boxes (Gibco BRL) containing only MS medium and allowed to develop until 5 to 10 flowers were present. Plants were stained in 50-mL conical tubes containing 25 mL of 1 mM 5-bromo-4-chloro-3-indolyl-β-glucuronic acid (U.S. Biological, Swampscott, MA), 1 mM potassium ferrocyanide, 1 mM potassium ferricyanide, 0.1% Triton X-100, and 50 mM sodium phosphate buffer, pH 7.0. Holes were punched in the lid of the tube to allow air exchange.
A house vacuum was used for infiltration during the first 10 to 20 min of staining. Tissue was incubated in stain at 37°C overnight and then cleared in 70% ethanol. Flowers and siliques were stained in the same way, except that they were removed from the plants and the carpels were dissected using a 30-gauge syringe needle and stained in 1.5-mL tubes. Images of GUS-stained whole plants were taken with a hand-held Nikon Coolpix 990 digital camera (Tokyo, Japan). Images of flowers were taken with an Olympus SZx12 dissecting microscope equipped with the same camera (Tokyo, Japan).
GFA2-mGFP5 Protein Localization Construct
We used the plasmid pAVA393 (von Arnim et al., 1998) to construct a C-terminal fusion of mGFP5 to the GFA2 protein. We amplified the GFA2 open reading frame from plasmid pCR-GFA2c/g using primers gfa2-19 (5′-GGGGCCATGGTCCCTTCCAATGGCGCAAAGGTT-3′) and gfa2-20 (5′-CCCCCCATGGCTCCGCCACCTCCGCCACCC-TGGGAAGATCCAGTTGCTGTGCGCT-3′), which introduce a NcoI site at the 5′ and 3′ ends of the gene and introduce six Gly codons at the 3′ end of the gene. The gfa2-19/gfa2-20 PCR product was digested with NcoI and cloned into the NcoI site of pAVA393, resulting in plasmid pAVA393-GFA2-mGFP5(6G).
The 35S Cauliflower mosaic virus::GFA2-6G-mGFP5 fusion protein construct in this plasmid was moved into pCGN1547 using BamHI and HindIII restriction sites, resulting in plasmid pCGN-35S::GFA2-mGFP5(6G). As a control for protein localization, the 35S Cauliflower mosaic virus::mGFP5 cassette from pAVA393 also was cloned into pCGN1547 using the BamHI and HindIII restriction sites, resulting in plasmid pCGN-35S::mGFP5.
Progeny from pCGN-35S::GFA2-mGFP5(6G) and pCGN-35S:: mGFP5 transformants were selected on 50 μg/mL kanamycin MS medium. After 5 to 10 days of growth, seedlings were harvested into a freshly made staining solution of 500 nM CM-H2XRos (Mitotracker Red; Molecular Probes, Eugene, OR) in a 1 × MS salts solution (11117-058; Gibco BRL) and allowed to stain for 15 min at room temperature. After staining, the seedlings were washed three times in MS salts solution for ∼10 min (Hedtke et al., 1999).
Roots were mounted under a cover slip in a drop of 1% low-melting-point agarose stored at 37°C (Gibco BRL). Images of mGFP5 fluorescence and Mitotracker fluorescence were captured using a Leica LSM510 confocal laser scanning microscope (Wetzlar, Germany). The two channels were excited separately by 488-nm (mGFP5) and 543-nm (Mitotracker) laser lines, and fluorescent emissions were gathered using the rhodamine/fluorescein isothiocyanate filter set. Images of trichomes from plants transformed with pCGN-35S::GFA2-mGFP5(6G) and pCGN-35S::mGFP5 were taken in the same manner, except that whole seedlings were mounted in 1% low melting point agarose.
Analysis of FIE::sGFP Expression in the gfa2 Mutant
These lines were crossed with gfa2, and progeny that carried both the GFP construct and the gfa2 mutation were selected by GFP fluorescence and a PCR assay for the gfa2 mutation. We emasculated flowers from these plants, waited 2 days, and then fixed half of them for phenotypic examination and mounted the other half for GFP expression analysis. Before mounting the pistil in 1% low-melting-point agarose (Gibco BRL), sepals, petals, and anthers were removed and the carpel walls were dissected using a 30-gauge syringe needle. Images of gfa2 mutants expressing FIE::FIE-sGFP were taken as described above for GFA2-mGFP5 protein localization.
Yeast Growth
Standard methods were used for the growth, transformation, and genetic manipulation of Saccharomyces cerevisiae in rich medium (yeast extract plus peptone containing either dextrose or glycerol) and synthetic medium (synthetic dextrose) as described (Sherman, 1986).
MDJ1 Disruption and Complementation by GFA2
An mdj1::HIS3 disruption that replaced the MDJ1 coding region was generated by gene replacement in a diploid strain as described (Baudin et al., 1993). The resulting heterozygous diploid was sporulated, and the tetrads were dissected to obtain the mdj1::HIS3 haploid strains. To make the GFA2 complementation construct, a 1.716-kb BamHI-XbaI fragment from pCRII-6/14GFA2c that contained the GFA2 open reading frame was cloned into the BamHI-XbaI sites in the polylinker of the yeast expression vector pRS416-MET25, resulting in plasmid pRS416-MET25-GFA2. The pRS416-MET25 and pRS416-MET25-GFA2 vectors were transformed into the heterozygous diploid mdj1::HIS3/MDJ1 before sporulation and dissection.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Accession Numbers
The GenBank accession number for the GFA2 cDNA sequence is AY103490. The GFA2 gene corresponds to locus At5 g48030 from BAC clone MDN11 on chromosome 5 (GenBank accession number AB017064).
Acknowledgments
We thank the ABRC for the MDN11 BAC clones; Albrecht von Arnim (University of Tennessee) for the pAVA393 (35S::mGFP5) plasmid; Ramin Yadegari, Tetsu Kinoshita, and Bob Fischer (University of California, Berkeley) for seeds from transgenic plants expressing FIE::sGFP and FIE::FIE-sGFP; and F.B. Pickett for GUS staining suggestions. We also thank Ramin Yadegari and John Harada for critical review of the manuscript. This work was funded in part by a National Institutes of Health Developmental Biology Training Grant appointment to C.A.C. and by grants from the National Science Foundation (IBN-96-30371) and Ceres, Inc., to G.N.D.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.002170.
References
- Balk, J., and Leaver, C.J. (2001). The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. Plant Cell 13, 1803–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk, J., Leaver, C.J., and McCabe, P.F. (1999). Translocation of cytochrome c from the mitochondria to the cytosol occurs during heat-induced programmed cell death in cucumber plants. FEBS Lett. 463, 151–154. [DOI] [PubMed] [Google Scholar]
- Baudin, A., Ozier-Kalogeropoulos, O., Denouel, A., Lacroute, F., and Cullin, C. (1993). A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21, 3329–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhomme, S., Horlow, C., Vezon, D., de Laissardiere, S., Guyon, A., Ferault, M., Marchand, M., Bechtold, N., and Pelletier, G. (1998). T-DNA mediated disruption of essential gametophytic genes in Arabidopsis is unexpectedly rare and cannot be inferred from segregation distortion alone. Mol. Gen. Genet. 260, 444–452. [DOI] [PubMed] [Google Scholar]
- Brizzio, V., Khalfan, W., Huddler, D., Beh, C.T., Andersen, S.S., Latterich, M., and Rose, M.D. (1999). Genetic interactions between KAR7/SEC71, KAR8/JEM1, KAR5, and KAR2 during nuclear fusion in Saccharomyces cerevisiae. Mol. Biol. Cell 10, 609–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner, B., Janick-Buckner, D., Gray, J., and Johal, G.S. (1998). Cell-death mechanisms in maize. Trends Plant Sci. 3, 218–223. [Google Scholar]
- Bukau, B., and Horwich, A.L. (1998). The Hsp70 and Hsp60 chaperone machines. Cell 92, 351–366. [DOI] [PubMed] [Google Scholar]
- Burger, K.N. (2000). Greasing membrane fusion and fission machineries. Traffic 1, 605–613. [DOI] [PubMed] [Google Scholar]
- Campisi, L., Yang, Y., Yi, Y., Heilig, E., Herman, B., Cassista, A.J., Allen, D.W., Xiang, H., and Jack, T. (1999). Generation of enhancer trap lines in Arabidopsis and characterization of expression patterns in the inflorescence. Plant J. 17, 699–707. [DOI] [PubMed] [Google Scholar]
- Cass, D.D., and Jensen, W.A. (1970). Fertilization in barley. Am. J. Bot. 57, 62–70. [Google Scholar]
- Chaudhury, A.M., and Berger, F. (2001). Maternal control of seed development. Semin. Cell Dev. Biol. 12, 381–386. [DOI] [PubMed] [Google Scholar]
- Chaudhury, A.M., Ming, L., Miller, C., Craig, S., Dennis, E.S., and Peacock, W.J. (1997). Fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94, 4223–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, F., Hersh, B.M., Conradt, B., Zhou, Z., Riemer, D., Gruenbaum, Y., and Horvitz, H.R. (2000). Translocation of C. elegans CED-4 to nuclear membranes during programmed cell death. Science 287, 1485–1489. [DOI] [PubMed] [Google Scholar]
- Chen, H., Nelson, R.S., and Sherwood, J.L. (1994). Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Biotechniques 16, 664–668, 670 [PubMed] [Google Scholar]
- Cheung, A.Y. (1996). The pollen tube growth pathway: Its molecular and biochemical contributions and responses to pollination. Sex. Plant Reprod. 9, 330–336. [Google Scholar]
- Christensen, C.A., King, E.J., Jordan, J.R., and Drews, G.N. (1997). Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex. Plant Reprod. 10, 49–64. [Google Scholar]
- Christensen, C.A., Subramanian, S., and Drews, G.N. (1998). Identification of gametophytic mutations affecting female gametophyte development in Arabidopsis. Dev. Biol. 202, 136–151. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Drews, G.N., Lee, D., and Christensen, C.A. (1998). Genetic control of female gametophyte development and function. Plant Cell 10, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchniewicz, M., Germaniuk, A., Westermann, B., Neupert, W., Schwarz, E., and Marszalek, J. (1999). Dual role of the mitochondrial chaperone Mdj1p in inheritance of mitochondrial DNA in yeast. Mol. Cell. Biol. 19, 8201–8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, M.M., and Kermicle, J.L. (2001). Interaction between maternal effect and zygotic effect mutations during maize seed development. Genetics 159, 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann, K.A., Coury, D.A., and Christianson, M.L. (1997). Exceptional segregation of a selectable marker (KanR) in Arabidopsis identifies genes important for gametophytic growth and development. Genetics 147, 1411–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink, A.L. (1999). Chaperone-mediated protein folding. Physiol. Rev. 79, 425–449. [DOI] [PubMed] [Google Scholar]
- Gavel, Y., Nilsson, L., and von Heijne, G. (1988). Mitochondrial targeting sequences: Why “non-amphiphilic” peptides may still be amphiphilic. FEBS Lett. 235, 173–177. [DOI] [PubMed] [Google Scholar]
- Grebenok, R.J., Pierson, E., Lambert, G.M., Gong, F.C., Afonso, C.L., Haldeman-Cahill, R., Carrington, J.C., and Galbraith, D.W. (1997). Green-fluorescent protein fusions for efficient characterization of nuclear targeting. Plant J. 11, 573–586. [DOI] [PubMed] [Google Scholar]
- Green, D.R., and Reed, J.C. (1998). Mitochondria and apoptosis. Science 281, 1309–1312. [DOI] [PubMed] [Google Scholar]
- Grini, P.E., Schnittger, A., Schwarz, H., Zimmermann, I., Schwab, B., Jürgens, G., and Hülskamp, M. (1999). Isolation of ethyl methanesulfonate-induced gametophytic mutants in Arabidopsis thaliana by a segregation distortion assay using the multimarker chromosome 1. Genetics 151, 849–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus, U., Vielle-Calzada, J.-P., Hoeppner, M.A., and Gagliano, W.B. (1998). Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280, 446–450. [DOI] [PubMed] [Google Scholar]
- Hartl, F.U. (1996). Molecular chaperones in cellular protein folding. Nature 381, 571–579. [DOI] [PubMed] [Google Scholar]
- Hedtke, B., Meixner, M., Gillandt, S., Richter, E., Borner, T., and Weihe, A. (1999). Green fluorescent protein as a marker to investigate targeting of organellar RNA polymerases of higher plants in vivo. Plant J. 17, 557–561. [DOI] [PubMed] [Google Scholar]
- Hengartner, M.O. (2000). The biochemistry of apoptosis. Nature 407, 770–776. [DOI] [PubMed] [Google Scholar]
- Higashiyama, T., Kuroiwa, H., Kawano, S., and Kuroiwa, T. (2000). Explosive discharge of pollen tube contents in Torenia fournieri. Plant Physiol. 122, 11–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashiyama, T., Yabe, S., Sasaki, N., Nishimura, Y., Miyagishima, S., Kuroiwa, H., and Kuroiwa, T. (2001). Pollen tube attraction by the synergid cell. Science 293, 1480–1483. [DOI] [PubMed] [Google Scholar]
- Howden, R., Park, S.K., Moore, J.M., Orme, J., Grossniklaus, U., and Twell, D. (1998). Selection of T-DNA-tagged male and female gametophytic mutants by segregation distortion in Arabidopsis. Genetics 149, 621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, G., Yalpani, N., Briggs, S.P., and Johal, G.S. (1998). A porphyrin pathway impairment is responsible for the phenotype of a dominant disease lesion mimic mutant of maize. Plant Cell 10, 1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, B.-Q., Pierson, E.S., Russell, S.D., Tiezzi, A., and Cresti, M. (1993). Cytoskeletal organization and modification during pollen tube arrival, gamete delivery and fertilization in Plumbago zeylanica. Zygote 1, 143–154. [DOI] [PubMed] [Google Scholar]
- Huang, B.-Q., and Russell, S.D. (1992. a). Female germ unit: Organization, isolation, and function. Int. Rev. Cytol. 140, 233–292. [Google Scholar]
- Huang, B.-Q., and Russell, S.D. (1992. b). Synergid degeneration in Nicotiana: A quantitative, fluorochromatic and chlorotetracycline study. Sex. Plant Reprod. 5, 151–155. [Google Scholar]
- Huijbregts, R.P., Topalof, L., and Bankaitis, V.A. (2000). Lipid metabolism and regulation of membrane trafficking. Traffic 1, 195–202. [DOI] [PubMed] [Google Scholar]
- Hülskamp, M., Schneitz, K., and Pruitt, R.E. (1995). Genetic evidence for a long-range activity that directs pollen tube guidance in Arabidopsis. Plant Cell 7, 57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttner, W.B., and Schmidt, A. (2000). Lipids, lipid modification and lipid-protein interaction in membrane budding and fission: Insights from the roles of endophilin A1 and synaptophysin in synaptic vesicle endocytosis. Curr. Opin. Neurobiol. 10, 543–551. [DOI] [PubMed] [Google Scholar]
- Jensen, W.A. (1964). Observations on the fusion of nuclei in plants. J. Cell Biol. 23, 669–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, W.A., Ashton, M.E., and Beasley, C.A. (1983). Pollen tube-embryo sac interaction in cotton. In Pollen: Biology and Implications for Plant Breeding, D.L. Mulcahy and E. Ottaviano, eds (New York: Elsevier Science Publishing), pp. 67–72.
- Jensen, W.A., and Fisher, D.B. (1968). Cotton embryogenesis: The entrance and discharge of the pollen tube in the embryo sac. Planta 78, 158–183. [DOI] [PubMed] [Google Scholar]
- Jones, A. (2000). Does the plant mitochondrion integrate cellular stress and regulate programmed cell death? Trends Plant Sci. 5, 225–230. [DOI] [PubMed] [Google Scholar]
- Kelley, W.L. (1998). The J-domain family and the recruitment of chaperone power. Trends Biochem. Sci. 23, 222–227. [DOI] [PubMed] [Google Scholar]
- Kieber, J.J., and Ecker, J.R. (1994). Molecular and genetic analysis of the constitutive ethylene response mutation ctr1. In Plant Molecular Biology: Molecular Genetic Analysis of Plant Development and Metabolism, Vol. H81, G. Coruzzi and P. Puigdomenech, eds (Berlin: Springer-Verlag), pp. 193–201.
- Kieber, J.J., Rothenberg, M., Roman, G., Feldman, K.A., and Ecker, J.R. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell 72, 427–441. [DOI] [PubMed] [Google Scholar]
- Kinoshita, T., Harada, J.J., Goldberg, R.B., and Fischer, R.L. (2001). Polycomb repression of flowering during early plant development. Proc. Natl. Acad. Sci. USA 98, 14156–14161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak, M. (1991). Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 266, 19867–19870. [PubMed] [Google Scholar]
- Krysan, P.J., Young, J.C., and Sussman, M.R. (1999). T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11, 2283–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnir, S., et al. (2001). A mutation of the mitochondrial ABC transporter Sta1 leads to dwarfism and chlorosis in the Arabidopsis mutant starik. Plant Cell 13, 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacomme, C., and Santa Cruz, S. (1999). Bax-induced cell death in tobacco is similar to the hypersensitive response. Proc. Natl. Acad. Sci. USA 96, 7956–7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, E., Pontier, D., and del Pozo, O. (1999). Die and let live: Programmed cell death in plants. Curr. Opin. Plant Biol. 2, 502–507. [DOI] [PubMed] [Google Scholar]
- Lichtscheidl, I.K., Lancelle, S.A., and Hepler, P.K. (1990). Actin-endoplasmic reticulum complexes in Drosera: Their structural relationship with the plasmalemma, nucleus, and organelles in cells prepared by high pressure freezing. Protoplasma 155, 116–126. [Google Scholar]
- Luo, M., Bilodeau, P., Koltunow, A., Dennis, E.S., Peacock, W.J., and Chaudhury, A.M. (1999). Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96, 296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie, S., and McIntosh, L. (1999). Higher plant mitochondria. Plant Cell 11, 571–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen, R. (1997). Cell death: Fatal induction in plants. Curr. Biol. 7, R534–R537. [DOI] [PubMed] [Google Scholar]
- Maxwell, D.P., Wang, Y., and McIntosh, L. (1999). The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc. Natl. Acad. Sci. USA 96, 8271–8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride, K.E., and Summerfelt, K.R. (1990). Improved binary vectors for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 14, 269–276. [DOI] [PubMed] [Google Scholar]
- Mogensen, H.L. (1972). Fine structure and composition of the egg apparatus before and after fertilization in Quercus gambelii: The functional ovule. Am. J. Bot. 59, 931–941. [Google Scholar]
- Molina, A., Volrath, S., Guyer, D., Maleck, K., Ryals, J., and Ward, E. (1999). Inhibition of protoporphyrinogen oxidase expression in Arabidopsis causes a lesion-mimic phenotype that induces systemic acquired resistance. Plant J. 17, 667–678. [DOI] [PubMed] [Google Scholar]
- Moore, J.M., Calzada, J.-P.V., Gagliano, W., and Grossniklaus, U. (1997). Genetic characterization of hadad, a mutant disrupting female gametogenesis in Arabidopsis thaliana. Cold Spring Harbor Symp. Quant. Biol. 62, 35–47. [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Navarre, D.A., and Wolpert, T.J. (1999). Victorin induction of an apoptotic/senescence-like response in oats. Plant Cell 11, 237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad, N., Margossian, L., Hsu, Y.-C., Williams, C., Repetti, P., and Fischer, R.L. (1996). A mutation that allows endosperm development without fertilization. Proc. Natl. Acad. Sci. USA 93, 5319–5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish, J., Li, L., Klotz, K., Ledwich, D., Wang, X., and Xue, D. (2001). Mitochondrial endonuclease G is important for apoptosis in C. elegans. Nature 412, 90–94. [DOI] [PubMed] [Google Scholar]
- Prip-Buus, C., Westerman, B., Schmitt, M., Langer, T., Neupert, W., and Schwarz, E. (1996). Role of the mitochondrial DnaJ homologue, Mdj1p, in the prevention of heat-induced protein aggregation. FEBS Lett. 380, 142–146. [DOI] [PubMed] [Google Scholar]
- Ray, S., Park, S.-S., and Ray, A. (1997). Pollen tube guidance by the female gametophyte. Development 124, 2489–2498. [DOI] [PubMed] [Google Scholar]
- Richberg, M.H., Aviv, D.H., and Dangl, J.L. (1998). Dead cells do tell tales. Curr. Opin. Plant Biol. 1, 480–485. [DOI] [PubMed] [Google Scholar]
- Roise, D. (1997). Recognition and binding of mitochondrial presequences during import of proteins into mitochondria. J. Bioenerg. Biomembr. 29, 19–27. [DOI] [PubMed] [Google Scholar]
- Rose, M.D. (1996). Nuclear fusion in the yeast Saccharomyces cerevisiae. Annu. Rev. Cell Dev. Biol. 12, 663–695. [DOI] [PubMed] [Google Scholar]
- Rowley, N., Prip-Buus, C., Westermann, B., Brown, C., Schwarz, E., Barrell, B., and Neupert, W. (1994). Mdj1p, a novel chaperone of the DnaJ family, is involved in mitochondrial biogenesis and protein folding. Cell 77, 249–259. [DOI] [PubMed] [Google Scholar]
- Russell, S.D. (1992). Double fertilization. Int. Rev. Cytol. 140, 357–388. [Google Scholar]
- Russell, S.D. (1996). Attraction and transport of male gametes for fertilization. Sex. Plant Reprod. 9, 337–342. [Google Scholar]
- Saitou, N., and Nei, M. (1987). The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Schulz, P., and Jensen, W.A. (1973). Capsella embryogenesis: The central cell. J. Cell Sci. 12, 741–763. [DOI] [PubMed] [Google Scholar]
- Sherman, F. (1986). Translation, post-translational processing, and mitochondrial translocation of yeast iso-1-cytochrome c. Basic Life Sci. 40, 533–544. [DOI] [PubMed] [Google Scholar]
- Shimizu, K.K., and Okada, K. (2000). Attractive and repulsive interactions between female and male gametophytes in Arabidopsis pollen tube guidance. Development 127, 4511–4518. [DOI] [PubMed] [Google Scholar]
- Sieburth, L.E., Drews, G.N., and Meyerowitz, E.M. (1998). Non-autonomy of AGAMOUS function in flower development: Use of a Cre/loxP method for mosaic analysis in Arabidopsis. Development 125, 4303–4312. [DOI] [PubMed] [Google Scholar]
- Smith, M.M., and McCully, M.E. (1978). A critical evaluation of the specificity of aniline blue induced fluorescence. Protoplasma 95, 229–254. [Google Scholar]
- Staehelin, L.A. (1997). The plant ER: A dynamic organelle composed of a large number of discrete functional domains. Plant J. 11, 1151–1165. [DOI] [PubMed] [Google Scholar]
- Stein, J.C., and Hansen, G. (1999). Mannose induces an endonuclease responsible for DNA laddering in plant cells. Plant Physiol. 121, 71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner, M.J., and Van Caeseele, L. (1990). The development of the central cell of Brassica campestris prior to fertilization. Can. J. Bot. 68, 2553–2563. [Google Scholar]
- Sun, Y.L., Zhao, Y., Hong, X., and Zhai, Z.H. (1999). Cytochrome c release and caspase activation during menadione-induced apoptosis in plants. FEBS Lett. 462, 317–321. [DOI] [PubMed] [Google Scholar]
- Sundaresan, V., Springer, P., Volpe, T., Haward, S., Jones, J.D.G., Dean, C., Ma, H., and Martienssen, R. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9, 1797–1810. [DOI] [PubMed] [Google Scholar]
- Triglia, T., Peterson, M.G., and Kemp, D.J. (1988). A procedure for in vivo amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 16, 8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Went, J., and Cresti, M. (1988). Pre-fertilization degeneration of both synergids in Brassica campestris ovules. Sex. Plant Reprod. 1, 208–216. [Google Scholar]
- van Went, J.L. (1970). The ultrastructure of the fertilized embryo sac of petunia. Acta Bot. Neerl. 19, 468–474. [Google Scholar]
- van Went, J.L., and Willemse, M.T.M. (1984). Fertilization. In Embryology of Angiosperms, B.M. Johri, ed (Berlin: Springer-Verlag), pp. 273–317.
- von Arnim, A.G., Deng, X.W., and Stacey, M.G. (1998). Cloning vectors for the expression of green fluorescent protein fusion proteins in transgenic plants. Gene 221, 35–43. [DOI] [PubMed] [Google Scholar]
- Wang, X. (2001). The expanding role of mitochondria in apoptosis. Genes Dev. 15, 2922–2933. [PubMed] [Google Scholar]
- Westermann, B., Gaume, B., Herrmann, J.M., Neupert, W., and Schwarz, E. (1996). Role of the mitochondrial DnaJ homolog Mdj1p as a chaperone for mitochondrially synthesized and imported proteins. Mol. Cell. Biol. 16, 7063–7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemse, M.T.M., and van Went, J.L. (1984). The female gametophyte. In Embryology of Angiosperms, B.M. Johri, ed (Berlin: Springer-Verlag), pp. 159–196.
- Yadegari, R., et al. (2000). Mutations in the FIE and MEA genes that encode interacting polycomb proteins cause parent-of-origin effects on seed development by distinct mechanisms. Plant Cell 12, 2367–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]