Abstract
Pollen germination and pollen tube growth are thought to require extracellular cues, but how these cues are perceived and transduced remains largely unknown. Pollen receptor kinases are plausible candidates for this role; they might bind extracellular ligands and thereby mediate cytoplasmic events required for pollen germination and pollen tube growth. To search for pollen-expressed ligands for pollen receptor kinases, we used the extracellular domains of three pollen-specific receptor kinases of tomato (LePRK1, LePRK2, and LePRK3) as baits in a yeast two-hybrid screen. We identified numerous secreted or plasma membrane–bound candidate ligands. One of these, the Cys-rich protein LAT52, was known to be essential during pollen hydration and pollen tube growth. We used in vivo coimmunoprecipitation to demonstrate that LAT52 was capable of forming a complex with LePRK2 in pollen and to show that the extracellular domain of LePRK2 was sufficient for the interaction. Soluble LAT52 can exist in differently sized forms, but only the larger form can interact with LePRK2. We propose that LAT52 might be a ligand for LePRK2.
INTRODUCTION
Pollen germination and pollen tube growth are not only crucial for the successful fertilization of plants but also offer an excellent model for studying cell signaling (reviewed by Wheeler et al., 2001). In the process of pollination, mature pollen grains are released from the anther in a partially dehydrated state and rehydrate after they contact the female pistil. The pollen tube emerges from one of the apertures and grows by tip growth through the extracellular matrix of the pistil. The pollen tube eventually reaches the ovule and delivers the sperm cells to the embryo sac.
This complicated and tightly controlled process presumably involves signaling between the pollen tube and the pistil: pollen tubes may adhere to the extracellular matrix and follow guidance cues as they traverse different types of tissues en route to an ovule. Recent evidence suggests that, in the pistil, the arabinogalactan protein TTS (Cheung et al., 1995; Wu et al., 2000), lipid-transfer protein (Park et al., 2000), and stylar pectin (Mollet et al., 2000) might serve as extracellular signaling cues. Although pollen tube growth occurs normally in the female tissue, pollen of many species also can germinate and grow pollen tubes when placed in a simple medium (Taylor and Hepler, 1997). This in vitro growth suggests that signaling molecules that regulate tube growth also might be produced by pollen itself.
Extracellular signaling molecules from pollen, if they exist, also might contribute to the pollen tube–pollen tube adhesion sometimes seen in vitro (Lush et al., 1997) and in vivo (Park et al., 2000). Somehow, these guidance cues must be transduced to the pollen cytoplasm, allowing the cytoskeletal changes (Franklin-Tong et al., 1999) and other cytoplasmic events required for tip growth. Whether guidance cues originate from pollen, from pistil, or from both, receptor kinases are plausible candidates to perceive and transduce these diverse signals to the pollen cytoplasm.
Leu-rich repeat receptor kinases (LRR kinases) constitute a large gene family in higher plants (Shiu and Bleecker, 2001). Ligands are known for only a few of the members of the LRR kinase family. For example, a small peptide, CLV3, is the ligand for the LRR kinase CLV1 (Trotochaud et al., 2000). A brassinosteroid is the ligand for the LRR kinase BRI1 (He et al., 2000). Flagellin, a bacterial elicitor, is the ligand for the LRR kinase FLS2 (Gomez-Gomez et al., 2001). In pollen, the best characterized LRR kinases have an extracellular domain composed of five or six LRRs, a transmembrane domain, a variable domain, and a cytoplasmic kinase domain (Mu et al., 1994; Muschietti et al., 1998).
In tomato, three pollen-specific receptor kinases (LePRK1, LePRK2, and LePRK3) localize on the pollen tube wall, with distinct but overlapping patterns (Muschietti et al., 1998; Kim et al., 2002). Furthermore, LePRK2 is phosphorylated in pollen membranes and dephosphorylated specifically after the membranes are incubated with style extracts (Muschietti et al., 1998), strongly suggesting that at least LePRK2 is involved in pollen–pistil interactions. However, the ligands for the pollen LRR kinases are unknown. If the LePRKs mediate signaling during pollen tube growth, their overlapping localization patterns suggest that they might bind the same ligands (albeit perhaps with different affinities). However, the extracellular domains of LePRKs are rather dissimilar (Muschietti et al., 1998; Kim et al., 2002), and these differences suggest that they might bind different ligands.
It is typical that ligands are produced and secreted by cells other than those that express the receptor. However, in autocrine signaling systems, both the ligand and its receptor are produced by the same cell (DeWitt et al., 2001). Here, we focus on the signaling pathway potentially mediated by LePRKs and the candidate ligands expressed by pollen itself. To identify candidate ligands, we used a yeast two-hybrid screen with the extracellular domains of the LePRKs as bait and a pollen cDNA library as prey. We identified numerous secreted or plasma membrane–bound candidates, including some small Cys-rich proteins, some cell wall–remodeling proteins, a LRR protein, a protein with EF-hand motifs, and several unknown proteins.
One of the small Cys-rich proteins, LAT52, was of special interest. Previously, we showed that LAT52 is essential during pollen hydration and pollen tube growth, because pollen expressing antisense LAT52 RNA hydrates and germinates abnormally and cannot achieve fertilization (Muschietti et al., 1994). We used coimmunoprecipitation to demonstrate that the interaction between LAT52 and LePRK2 occurs in pollen. The LAT52 protein binds only weakly to LePRK1 and not at all to LePRK3, showing that the LePRK2–LAT52 interaction in pollen is specific. Moreover, the extracellular domain of LePRK2 is sufficient to bind LAT52. Using size fractionation, we show that native LAT52 in extracts of mature pollen is a component of a larger complex whose formation is independent of its interaction with LePRK2. Under certain conditions, the LAT52 protein is not present in the larger form and cannot interact with LePRK2.
RESULTS
Yeast Two-Hybrid Screens Identify Extracellular Binding Partners for LePRKs
To identify pollen proteins that interact with the extracellular domains of the pollen kinases, we used a yeast two-hybrid screen. Although the yeast two-hybrid screen requires that proteins interact in the yeast nucleus, this approach has been successful occasionally (Ozenberger and Young, 1995; Zhu and Kahn, 1997; Park et al., 2001) in identifying interacting proteins that are located normally in the plasma membrane or the extracellular space. A cDNA library prepared from mRNA of mature pollen of tomato was fused to the GAL4 activation domain. This library was screened separately against three bait constructs, each encoding an extracellular domain (ECD) of a LePRK fused to the GAL4 DNA binding domain. Perhaps surprisingly, a remarkable portion (∼60%) of the positive cDNA clones from each screen encoded bona-fide secreted proteins or proteins containing a predicted N-terminal signal peptide and therefore were candidate ligands (see supplementary data online for details).
These candidate ligands fall into five groups. The first group comprises diverse Cys-rich proteins (molecular mass ranging from 6 to 16 kD). This group was especially interesting because another small Cys-rich protein, SCR, found on the pollen coat in Brassica species, was shown recently to be the ligand for the stigma-expressed S-locus receptor kinase SRK (Schopfer et al., 1999; Kachroo et al., 2001; Takayama et al., 2001). However, SRK is not an LRR kinase, and the arrangement of Cys residues is not conserved between SCR and the Cys-rich proteins we identified from the yeast two-hybrid screen. The second group includes various cell wall–remodeling proteins. This group was interesting because some cell wall components, such as pectins, are modified during different phases of pollen tube growth (reviewed by Lord, 2000).
The third group is represented by a protein containing LRRs. Because LRRs mediate protein–protein interactions (Kobe and Deisenhofer, 1994), it seemed reasonable that the extracellular domain of LePRKs might interact with another LRR protein. The fourth group is represented by a protein containing EF-hand (calcium binding) motifs. This candidate seemed interesting because of the well-known role of calcium in regulating pollen tube growth (Steer and Steer, 1989). The fifth group comprises proteins with no significant similarity to proteins with known functions.
The previously characterized LAT52 gene (Twell et al., 1989, 1990) encodes one of the small Cys-rich proteins that was isolated with the LePRK2 bait. LAT52 has an N-terminal signal peptide and is secreted when expressed from a baculovirus expression system (Muschietti et al., 1994). LAT52 was isolated four times from the yeast two-hybrid screen with the ECD of LePRK2 but not at all from the equivalently large screens with the extracellular domains of LePRK1 (ECD1) or LePRK3 (ECD3). One of the LAT52 clones from the LePRK2 screen is full length (161 amino acids), whereas the others have the N terminus truncated by 1, 30, or 39 amino acids. This delimits the interaction region to the C-terminal 122 amino acids of LAT52.
To determine whether LAT52 could interact with the other LePRKs, the full-length LAT52 in the prey vector (pAD-LAT52) or the empty prey vector (pAD) were retransformed into yeast cells harboring bait plasmids (pBD) of the extracellular domains of LePRK1, LePRK2, or LePRK3 or the cytoplasmic domain of LePRK2 (control). Table 1 shows that only the transformants harboring both pAD-LAT52 and pBD-ECD2 grew on selection plates. The β-galactosidase activity of yeast cells harboring both pAD-LAT52 and pBD-ECD2 was 10 times higher than that of any other combination. These results showed that in the yeast two-hybrid system, LAT52 interacts reproducibly and specifically with ECD2 but not with ECD1 or ECD3. Additional candidate ligands also were confirmed in the yeast two-hybrid specificity test (see supplementary data online for details).
Table 1.
LAT52 Interacts Specifically with the Extracellular Domain of LePRK2 in Yeast
| pAD
|
pAD-LAT52
|
|||||
|---|---|---|---|---|---|---|
| Sample | +His, +Adea | −His, −Ade, +3ATb | β-Gal Activityc(Miller units) | +His, +Adea | −His, −Ade, +3ATb | β-Gal Activityc(Miller units) |
| pBD-ECD1 | + | − | 0.4 ± 0.3 | + | − | 1.0 ± 0.1 |
| pBD-ECD2 | + | − | 1.8 ± 0.2 | + | + | 30.8 ± 9.5 |
| pBD-ECD3 | + | − | 1.2 ± 0.3 | + | − | 1.5 ± 0.6 |
| pBD-CD2 | + | − | 0.6 ± 0.5 | + | − | 1.4 ± 0.4 |
| pBD | + | − | 2.3 ± 0.4 | + | − | 2.7 ± 0.8 |
ECD1, ECD2, and ECD3 refer to the extracellular domains of LePRK1, LePRK2, and LePRK3, respectively.
CD2 refers to the cytoplasmic domain of LePRK2. + and − indicate that cells could and could not grow on the indicated medium.
The yeast transformants were streaked on medium supplied with His (+His) and adenine (+Ade).
The yeast transformants were streaked on medium lacking His (−His) and adenine (−Ade) but supplied with 3 mM 3-amino-1,2,4-triazole (+3-AT) to monitor the interaction.
β-Galactosidase activity also was assayed. Values shown are from three independent β-galactosidase liquid assays using o-nitrophenyl-β-d-galactopyranoside as a substrate.
In Vivo Coimmunoprecipitation Demonstrates That LAT52 and LePRK2 Are in the Same Complex in Mature Pollen Extracts
Because pollen expressing antisense LAT52 RNA hydrates and germinates abnormally (Muschietti et al., 1994), LAT52 seemed the most promising candidate ligand for further characterization. We had antibodies (Figure 1A) for both LAT52 (Muschietti et al., 1994) and LePRK2 (Muschietti et al., 1998), so we were able to test whether LAT52 and LePRK2 interact with each other in pollen, using a coimmunoprecipitation assay. When equivalent amounts of total protein (S10) extracts from mature pollen were immunoprecipitated with anti-LePRK2 or with normal mouse serum, LAT52 protein was detected only in the immunopellet of anti-LePRK2 (Figure 1B). This result shows that LAT52 and LePRK2 are in the same complex in total protein extracts from mature pollen and confirms that LAT52 and LePRK2 interact with each other in pollen.
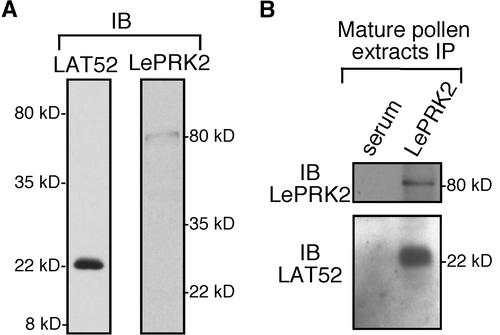
Figure 1.
LAT52 and LePRK2 Are in the Same Complex in Mature Pollen Extracts.
(A) Immunoblot (IB) analysis of mature pollen extracts (S10) with anti-LAT52 (left) or anti-LePRK2 (right) antibody.
(B) Coimmunoprecipitation (IP) of LAT52 with anti-LePRK2, followed by separate immunoblot analysis with anti-LePRK2 (top) or anti-LAT52 (bottom). Mature pollen extracts (S10; 1 mg each) in PGM buffer were immunoprecipitated separately with mouse serum (left) or anti-LePRK2 antibody (right). Molecular mass markers are indicated.
In the yeast two-hybrid system, LAT52 interacted specifically with LePRK2 but not with LePRK1 or LePRK3. We tested whether this specificity could be demonstrated in pollen extracts, using antibodies raised against the extracellular domains of the LePRKs (Figure 2A). In three different experiments, equivalent amounts of total protein extracts from mature pollen were immunoprecipitated separately with antibodies against the extracellular domains of LePRK1, LePRK2, and LePRK3. In all three experiments, a significant amount of LAT52 protein was present in the immunopellet of anti-LePRK2 (Figure 2B), but LAT52 protein was not present in the immunopellet of anti-LePRK3.
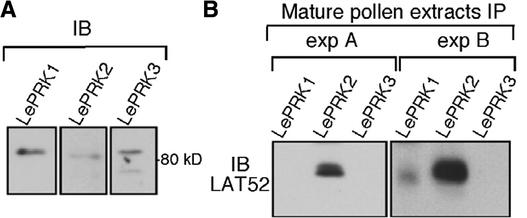
Figure 2.
Interaction Specificity of LAT52 with LePRKs in Pollen.
(A) Immunoblot (IB) analysis of mature pollen extracts (S10) with anti-LePRK1, anti-LePRK2, or anti-LePRK3 antibody.
(B) Mature pollen extracts (S10; 1 mg each) in PGM buffer were immunoprecipitated (IP) with anti-LePRK1, anti-LePRK2, or anti-LePRK3 antibody. Immunoprecipitated proteins were subjected to immunoblot analysis with anti-LAT52 antibody. Two independent experiments (A and B) are shown.
In one of three experiments, a small amount of LAT52 was present in the immunopellet of anti-LePRK1 (Figure 2B, experiment B), whereas in the other two experiments, no LAT52 was present in the anti-LePRK1 immunopellet (Figure 2B, experiment A). Thus, in pollen, LAT52 does interact with LePRK2, does not interact with LePRK3, and does not interact significantly with LePRK1. We could not perform the reciprocal coimmunoprecipitation assay with anti-LAT52 because it does not recognize native LAT52 protein in pollen extracts, although it recognizes denatured LAT52 protein and is useful for immunoblot analysis (Muschietti et al., 1994).
The Presence of EDTA Abolishes the Interaction between LAT52 and LePRK2
In the process of optimizing the coimmunoprecipitation assays (Figure 2), we noticed that LAT52 was not coimmunoprecipitated with the anti-LePRK2 antibody if TED buffer (Tris, EDTA, and DTT), pH 7.6, was used (Figure 3). However, when we used a buffer designed to mimic conditions optimized for pollen tube growth, coimmunoprecipitation was successful (Figures 1 to 3). The pollen germination mimic (PGM) buffer contains Mes, pH 6.0, boric acid (BA), MgSO4, Ca(NO3)2, and KCl, with concentrations of each component identical to those used for in vitro germination (see Methods). The TED buffer and PGM buffer differ in their pH and in whether EDTA, DTT, BA, and KCl are present.
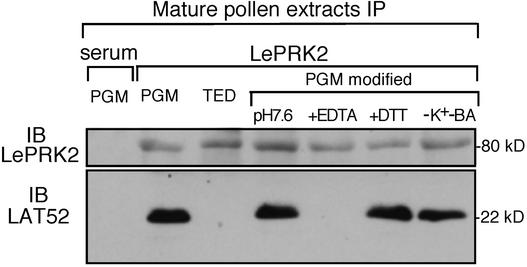
Figure 3.
EDTA Abolishes the Interaction of LAT52 and LePRK2 in Mature Pollen Extracts.
Coimmunoprecipitation (IP) of LAT52 from mature pollen extracts (1 mg each) in TED buffer, PGM buffer, or modified versions of PGM buffer (PGM modified). The modified versions of PGM buffer were as follows: pH 7.6, 50 mM Tris-Cl, pH 7.6, instead of Mes, pH 6.0; +EDTA, addition of 1 mM EDTA but omission of MgSO4 and Ca(NO3)2; +DTT, addition of 1 mM DTT; −K+−BA, omission of KCl and boric acid. The left lane was immunoprecipitated with normal mouse serum. All other lanes were immunoprecipitated with anti-LePRK2 antibody. The immunoprecipitated proteins were subjected to immunoblot analysis (IB) with anti-LePRK2 (top) or anti-LAT52 (bottom) antibody. The gel at top shows a loading control and demonstrates that similar amounts of LePRK2 were precipitated with anti-LePRK2 in the different buffers. Molecular mass markers are indicated.
To determine what component of the two buffers might account for the difference in coimmunoprecipitation success, we tested modified PGM buffers. Figure 3 shows that changing the pH did not affect the amount of coimmunoprecipitated LAT52 significantly. The addition of DTT to PGM buffer did not affect coimmunoprecipitation, suggesting that disulfide bonds are not required for the interaction. Nor did the removal of BA and K+ from PGM buffer reduce the amount of coimmunoprecipitated LAT52. However, the addition of EDTA and the omission of MgSO4 and Ca(NO3)2 completely prevented the coimmunoprecipitation of LAT52. Because EDTA is a chelator for metal ions such as Ca2+ and Mg2+, this result suggests that a metal ion (Ca2+ and/or Mg2+) is required for the LAT52–LePRK2 interaction.
The Extracellular Portion of LePRK2 Is Sufficient for Its Interaction with LAT52
Binding of an extracellular protein to the extracellular domain of a receptor kinase can be independent of the cytoplasmic domain (Kachroo et al., 2001) or dependent on the cytoplasmic kinase activity (Trotochaud et al., 2000). Only the extracellular domain of LePRK2 is required for the interaction with LAT52 in the yeast two-hybrid system, but in the coimmunoprecipitation experiments (Figures 1 to 3), the entire LePRK2 protein was immunoprecipitated. To determine whether the ECD of LePRK2 is sufficient for the interaction in pollen, we used a His-tagged version of ECD2.
The Escherichia coli–expressed His-ECD2 fusion protein was purified under native conditions (Figure 4A). Total protein extracts from mature pollen were incubated with the His-ECD2 protein, and then LAT52 was coimmunoprecipitated. The purified ECD2 can be recognized by both anti-His and anti-LePRK2 antibodies, but the endogenous LePRK2 can be recognized only by the anti-LePRK2 antibody. We first determined the amounts of LAT52 and LePRK2 in the soluble (S100) and membrane-associated (P100) fractions of mature pollen extracts by immunoblot analysis (Figure 4B). The LAT52 protein was found mostly in the soluble protein fraction. The residual amount of LAT52 detected in the membrane-associated fraction might be attributable to interaction with membrane-associated components or to incomplete fractionation.
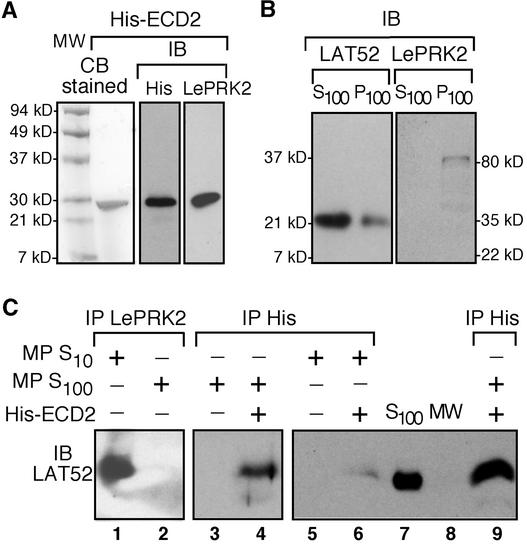
Figure 4.
The Extracellular Domain of LePRK2 Is Sufficient for Interaction with LAT52.
(A) Expression and purification of the extracellular domain of LePRK2 fused with a His tag (His-ECD2). His-ECD2 protein was expressed and purified by nickel–nitrilotriacetic acid agarose and separated by SDS-PAGE. Gels were stained with Coomassie blue (CB stained) or immunoblotted (IB) with anti-His or anti-LePRK2 antibody. His-ECD2 was used in the subsequent semi-in vivo coimmunoprecipitation assays.
(B) Subcellular location of LAT52 and LePRK2. Immunoblots of soluble (S100) or membrane-associated (P100) fractions of mature pollen extracts are shown. The S100 fraction was used in the subsequent semi-in vivo coimmunoprecipitation assays.
(C) Coimmunoprecipitation of LAT52 with either endogenous LePRK2 from mature pollen (MP) extracts (IP LePRK2) or exogenous ECD2 protein (IP His). Lanes 1 and 2, mature pollen extracts (S10 or S100) were immunoprecipitated with anti-LePRK2 antibody. Lanes 3 and 5, mature pollen extracts (S10 or S100) were immunoprecipitated with anti-His antibody. Lanes 4, 6, and 9, mature pollen extracts (S10 or S100) were incubated with 5 μg of purified His-ECD2 protein and immunoprecipitated with anti-His antibody. Immunoprecipitated proteins were subjected to immunoblot analysis with anti-LAT52 antibody. Lane 7, mature pollen extract (S100) as a control. Lane 8, molecular mass marker (MW).
As we already knew, LePRK2 can be detected only in the membrane-associated fraction. Consistent with these locations, Figure 4C (lanes 1 and 2) shows that LAT52 was coimmunoprecipitated only with anti-LePRK2 antibody from total protein extracts (S10) from mature pollen, but not from the S100 fraction in which LAT52 was enriched but from which the endogenous LePRK2 was depleted. In the semi-in vivo immunoprecipitation assay, the LAT52 in the mature pollen S100 fraction was able to be coimmunoprecipitated with anti-His antibody if exogenous His-ECD2 protein was added (Figure 4C, compare lanes 3 and 4). When the His-ECD2 protein was added to total protein extracts (S10) of mature pollen, LAT52 was coimmunoprecipitated (Figure 4C, lane 6), but the signal was much weaker than if the coimmunoprecipitation was from a similar amount of S100 fraction (Figure 4C, lane 9). We presume that this difference is caused by competition from the endogenous LePRK2 that was present in the S10 extract.
The results showed that LAT52 in pollen extracts also could form a complex with the His-ECD2 protein (Figure 4C). Thus, the ECD of LePRK2 is sufficient for this interaction, and the cytoplasmic portion of LePRK2, including the kinase domain, is not necessary for binding to LAT52. This experiment also indicates that the LAT52 protein in the mature pollen S100 fraction, although temporarily not in the same complex with LePRK2, remains capable of forming a complex with LePRK2.
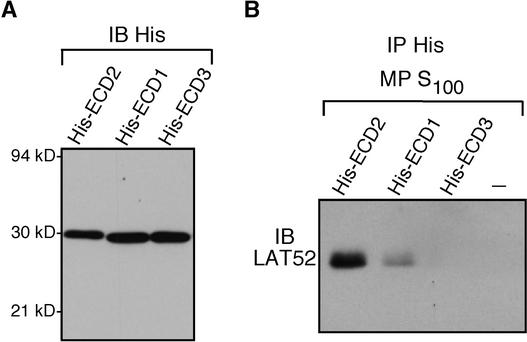
We used the His-tagged fusion proteins (Figure 5A) to determine if LAT52 and the extracellular domains of the other two LePRKs could interact. Figure 5B shows that LAT52 was not precipitated with anti-His antibody when His-ECD3 was incubated with pollen extracts; it also shows that a small amount of LAT52 was detected in the precipitate after His-ECD1 was incubated with pollen extracts. These results are essentially consistent with the in vivo coimmunoprecipitation results (Figure 2).
Figure 5.
Interaction Specificity of LAT52 with His-ECD Fusion Proteins.
(A) The extracellular domains of LePRK2, LePRK1, and LePRK3 fused with a His tag (His-ECD2, His-ECD1, and His-ECD3) were purified and immunoblotted with anti-His antibody (IB His). Molecular mass markers are indicated.
(B) Mature pollen (MP) S100 fractions with the addition of His-ECD2, His-ECD1, or His-ECD3 (5 μg each), or without the addition of a fusion protein (−), were immunoprecipitated with anti-His antibody (IP His). Immunoprecipitated proteins were subjected to immunoblot analysis with anti-LAT52 antibody.
Pollen Germination or Heating Abolishes the Interaction of LAT52 with LePRK2
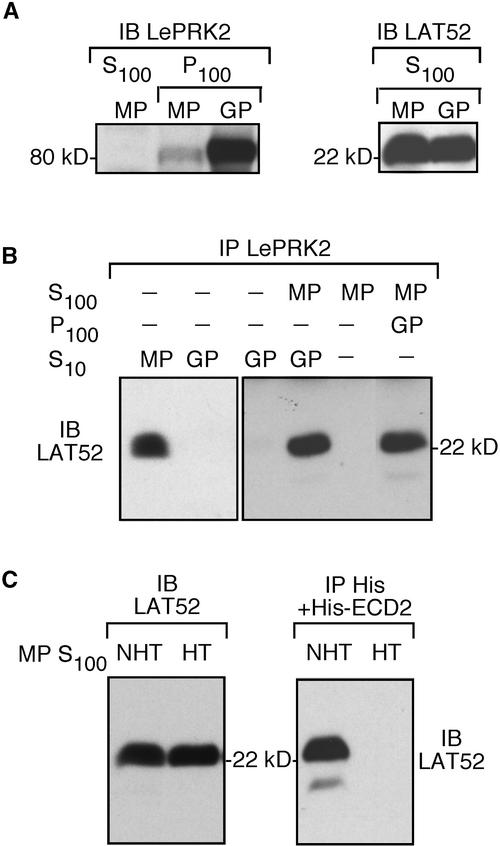
We know that LAT52 and LePRK2 can be found in the same complex when protein extracts are prepared from mature pollen under conditions appropriate for pollen germination. We determined if LAT52 and LePRK2 also interact with each other after pollen germination. Both LAT52 and LePRK2 were present in pollen that had been germinated in vitro (Figure 6A), and as shown previously (Muschietti et al., 1998), the amount of LePRK2 was increased dramatically after germination. To our surprise, LAT52 was not coimmunoprecipitated with anti-LePRK2 from pollen that had been germinated in vitro for 4 h (data not shown) or 16 h (Figure 6B, left).
Figure 6.
Under Certain Conditions, LAT52 Cannot Interact with LePRK2.
(A) Detection of LePRK2 (left) and LAT52 (right) by immunoblot (IB) analysis of mature pollen (MP) or germinated pollen (GP) extracts. P100, membrane-associated fraction; S100, soluble fraction.
(B) Coimmunoprecipitation (IP) of LAT52 with LePRK2 from mature pollen extracts, from germinated pollen extracts, or from mixtures, as indicated. At left, mature pollen and pollen germinated in vitro (S10; 1 mg each) were immunoprecipitated with anti-LePRK2 antibody. At right, the germinated pollen extract (S10 or P100, as indicated) was incubated with 500 μg of S100 from mature pollen and immunoprecipitated with anti-LePRK2 antibody. As a control, 500 μg of the S100 from mature pollen also was immunoprecipitated with anti-LePRK2. All immunoprecipitated proteins were subjected to immunoblot analysis with anti-LAT52 antibody. Molecular mass markers are indicated.
(C) Coimmunoprecipitation of heat-treated LAT52 with the extracellular domain of LePRK2. One of two identical aliquots of mature pollen extracts (5 μg/μL) was heated (70°C for 2 min) to obtain a heat-treated (HT) S100, whereas the other aliquot, which was not heat treated (NHT), was held on ice. At left, 2 μL of each aliquot was immunoblotted with anti-LAT52 antibody. At right, the NHT and HT S100 aliquots (200 μL each) were incubated with His-ECD2 protein (5 μg) and precipitated with anti-His antibody. Immunoprecipitated proteins were subjected to immunoblot analysis with anti-LAT52 antibody. Molecular mass markers are indicated.
We surmised that either LAT52 or LePRK2 had changed after germination, although no obvious size difference for either protein was apparent from immunoblot analysis (Figure 6A). Because we knew that LAT52 in the S100 fraction from mature pollen remains capable of forming a complex with LePRK2 (Figure 4C), we tested whether LAT52 or LePRK2 had changed after germination. We added the S100 fraction from mature pollen before germination, which contains LAT52, to either the S10 or P100 protein that had been extracted from pollen germinated in vitro, which contains LePRK2. Figure 6B (right) shows that under these conditions, LAT52 was coimmunoprecipitated with anti-LePRK2 antibody. Therefore, LAT52 before germination can form a complex with LePRK2 after germination. This result indicates that it is LAT52, and not LePRK2, that is changed after germination.
We showed previously that LAT52 is a heat-stable protein (Muschietti et al., 1994). When the protein extracted from pollen is heat treated (2 min at 90°C), most proteins are denatured and are precipitated, but LAT52 is soluble and remains in the supernatant fraction. Figure 6C (left) shows that LAT52 also remains soluble after a less severe heat treatment (70°C). However, we found that when the 70°C-treated S100 fraction from mature pollen was incubated with the His-ECD2 protein, LAT52 was not coimmunoprecipitated with anti-His antibody (Figure 6C, right). Nonetheless, Figure 6C shows that the mass and amount of LAT52 are similar before and after heat treatment. Apparently, heat treatment alters the conformation of LAT52 (or denatures another protein that might be required for the complex) so that LAT52 cannot interact with His-ECD2.
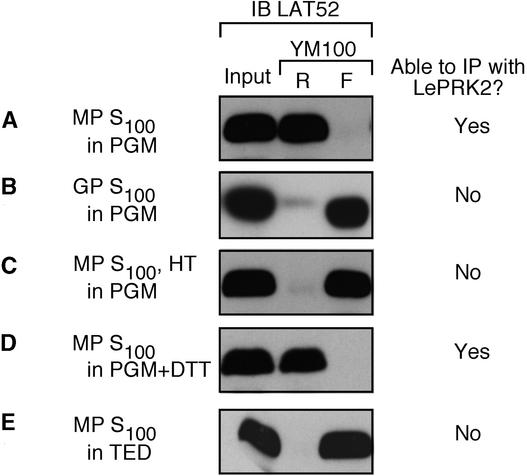
Thus, there are two conditions under which LAT52 cannot interact with LePRK2: after heat treatment and after pollen germination. To help determine what might account for this difference, we used crude size fractionation. The LAT52 gene encodes a protein of 161 amino acids, but after processing of the signal peptide, the calculated molecular mass of the mature LAT52 protein is 16 kD. Because LAT52 is glycosylated (Muschietti et al., 1994), its apparent molecular mass after SDS-PAGE is ∼20 kD. Therefore, we expected that LAT52 would not be retained on centrifugal filters (YM50 and YM100) that retain molecules of >50 kD and 100 kD but allow smaller molecules to flow through. To our surprise, LAT52 from mature pollen prepared in the PGM buffer (nondenaturing conditions, S100) was retained on both the YM50 (data not shown) and YM100 (Figure 7A) filters.
Figure 7.
LAT52 Protein Exists in Differently Sized Forms under Different Conditions.
Mature pollen (MP) or germinated pollen (GP) S100, in PGM or TED buffers, as noted, were loaded on YM100 centrifugal filters (cutoff molecular mass of 100 kD). In (C), the S100 was first subjected to 70°C for 2 min (HT). After centrifugation, the retentate (R), the flow-through (F), and the input sample were subjected to immunoblot (IB) analysis with anti-LAT52 antibody. Whether LAT52 was coimmunoprecipitated (IP) with LePRK2 or ECD2 (data from Figures 4, 5, and 6) is noted for reference.
We conclude that the soluble LAT52 extracted from mature pollen in PGM buffer exists in a form whose molecular mass obviously is larger than that of the LAT52 monomer. Because the soluble fraction is depleted for most of the membrane-associated LePRK2, this complex does not include LePRK2 (Figure 6A). We then used the YM100 filter to determine the size of LAT52 in the heat-treated S100 protein faction from mature pollen and in the S100 fraction from pollen germinated in vitro. Figures 7B and 7C show that LAT52 was not retained on the YM100 filter in either case, and after heat treatment it was not retained on the YM50 filter (data not shown). These results indicate that there are at least two differently sized forms possible for soluble LAT52. Furthermore, when LAT52 cannot be retained on the YM100 filter, it also is not able to interact with LePRK2.
When 10 mM DTT was added to mature pollen extracts, LAT52 was retained on the YM100 filter (Figure 7D), and we showed (Figure 3) that DTT did not prevent the coimmunoprecipitation of LAT52 with anti-LePRK2 from mature pollen. Furthermore, 1 mM EDTA prevented LAT52 from being retained on the YM100 filter (Figure 7E) and prevented it from interacting with LePRK2 (Figure 3). Together, these results suggest that the larger form is required for LAT52 to interact with LePRK2.
DISCUSSION
Despite anecdotal reports that yeast two-hybrid screens for extracellular proteins are fraught with difficulty, our results show that screens with the extracellular domains of the LePRKs were successful in identifying several interacting proteins that also are extracellular. In this regard, it might be significant that the extracellular domains of the LePRKs are small (∼200 amino acids, with five to six LRRs). By contrast, the extracellular domains of many other LRR receptor kinases (Shiu and Bleecker, 2001) are large (20 or more LRRs) and might not be well expressed or might not fold properly in yeast.
Even though we identified numerous extracellular proteins, they still might be false positives. It is always necessary to use independent biochemical assays to confirm candidates obtained from two-hybrid screens. Therefore, we used coimmunoprecipitation from pollen extracts to demonstrate that one of these extracellular proteins, LAT52, does interact with the extracellular domain of LePRK2 in pollen. It will require further work to determine if LAT52 is in fact a ligand whose binding activates the receptor.
Of the many interacting proteins obtained from the yeast two-hybrid screen, we focused on characterizing the LAT52–LePRK2 interaction, because we had shown previously a role for LAT52 during pollen germination (Muschietti et al., 1994). Pollen expressing antisense LAT52 appears normal at the mature pollen stage. Pollen expressing antisense LAT52 can hydrate normally when placed in germination medium without polyethylene glycol, but it cannot hydrate normally in vitro when placed in germination medium with polyethylene glycol (which is thought to slow water uptake). Pollen expressing antisense LAT52 also has a mutant phenotype on the pistil: many grains did not germinate, and those that did grew twisted pollen tubes and did not achieve fertilization (Muschietti et al., 1994). From these results, it seemed that LAT52 played a role at two phases: during hydration/germination and during tube growth.
LAT52 is highly expressed in mature pollen, and LAT52 transcripts still are detected after 18 h of in vitro germination (Ursin et al., 1989). LAT52 protein is present in mature pollen and at equivalent levels in extracts of pollen after 16 h of in vitro germination, presumably from continued translation. LePRK2 also is present in mature pollen, but its level increases ∼10-fold after germination. Immunolocalization with antibodies raised against the extracellular domain of LePRK2 showed that it is localized along the entire tube, but in addition it forms a collar (Kim et al., 2002) near the grain–tube interface. These differences in protein levels and localization of LePRK2 also suggest two roles for LePRK2, one at the beginning stage of germination and one later during tube growth. To determine if the LAT52–LePRK2 interaction would affect either one or both phases of the antisense LAT52 pollen phenotype, it was necessary to determine if they interact with each other during the two phases.
The complex containing both LAT52 and LePRK2 was detected in the total proteins of mature pollen extracted in PGM buffer (Figures 1 to 3). In fact, when placed in buffer, pollen hydrated immediately, so it is more accurate to say that our coimmunoprecipitation results represent interactions in hydrated pollen, the beginning stage of pollen germination. Because LAT52 and LePRK2 are in a complex at this first stage, the abnormality in hydration/germination in the antisense LAT52–expressing pollen could be attributable to the absence of this complex. However, we have shown that the LAT52–LePRK2 complex cannot be detected in the total extracts of pollen germinated in vitro (Figure 6B), suggesting that LAT52 and LePRK2 dissociate as in vitro germination proceeds.
In the course of determining why the LAT52–LePRK2 interaction is different after in vitro germination, we found that LAT52 can exist in differently sized forms (Figure 7) and that soluble LAT52 from pollen germinated in vitro does not exist in a large form (Figure 7B). Because we could reconstitute the LAT52–LePRK2 complex using LePRK2 from germinated pollen and soluble LAT52 from mature pollen (Figure 6B), the binding characteristics of LePRK2 do not change after germination. This finding suggests that only the large form of soluble LAT52 interacts with LePRK2.
In some receptor–ligand interactions in animals, ligand multimers are required for receptor binding. For example, ephrin must dimerize (Toth et al., 2001), and trimerization of tumor necrosis factor receptor–associated factors is required for high-affinity interactions with the CD40 receptor (Pullen et al., 1999). However, in some other cases, the monomer is the active ligand (Qiu et al., 2000). We do not know if the large form of soluble LAT52 is a heteromultimer or a homomultimer, but it appears that disulfide bonds are not required for its formation (Figures 3 and 7).
Why would pollen receptors bind to extracellular molecules produced by pollen itself? Autocrine signaling, in which cells secrete soluble ligands that bind to receptors on their own surfaces, has been characterized for several animal growth factors and their receptors. This type of intracellular signaling allows the cell to monitor itself, and in some cases, how the ligand is delivered can have significant effects on the response. For example, when epidermal growth factor (EGF) was delivered to the EGF receptor from the same epithelial cell expressing the receptor, cell migration was prolonged and directional, whereas when EGF was delivered exogenously, cells migrated randomly (Maheshwari et al., 2001). It is possible that pollen is monitoring its own germination status when LePRK2 interacts with LAT52, because LAT52 exists in differently sized forms before and after germination.
The demonstration that LAT52 interacts with LePRK2 is only a first step toward understanding the signaling pathway mediated by pollen receptor kinases and their extracellular binding partner(s). Interactions of receptors with extracellular binding partners often cause receptor dimerization and phosphorylation, and we know that LePRK1 and LePRK2 can form heterodimers in pollen and when expressed in yeast (I. Valsecchi, M.L. Cabanas, D. Wengier, W. Tang, S. McCormick, and J. Muschietti, unpublished data). Furthermore, the phosphorylation status of LePRK2 changes upon incubation with style extracts (Muschietti et al., 1998). It is possible that the large form of LAT52 is in a complex with LePRK2 before germination and is displaced upon germination, so that the extracellular domain of LePRK2 can bind to other proteins from the pistil (or to other proteins from pollen).
It is worth noting that the failure of the smaller form of LAT52 to interact with LePRK2 does not exclude the possibility of their continued interaction, perhaps with the participation of other molecules from the pistil. In fact, on the pistil, the pollen expressing antisense LAT52 showed a less severe phenotype than when germinated in vitro (Muschietti et al., 1994), suggesting that a molecule in the pistil might be capable of substituting for the lack of LAT52, at least in the beginning stages of germination.
There are at least three LePRKs expressed at similar stages and locations (Kim et al., 2002). Our yeast two-hybrid screens identified many other candidates that might bind to the extracellular domains of the LePRKs in vivo (see supplementary data online). A yeast two-hybrid screen with a pistil library (I. Ezcurra, R. Cotter, and S. McCormick, unpublished data) yielded pistil-expressed members of the Cys-rich and cell wall–remodeling groups of candidates. It is interesting that in the yeast two-hybrid screens, there were both distinct and overlapping interaction patterns; for example, some candidates interacted only with LePRK1 and LePRK2, and others interacted only with LePRK2.
Although all three LePRKs have extracellular domains composed of LRRs, LePRK3 did not interact with LAT52 under any condition we tested (Figure 2). The weaker interaction between LePRK1 and LAT52 might be the result of the heterodimerization of LePRK1 and LePRK2 or of a low affinity of LAT52 for LePRK1. The apparent higher affinity of LAT52 for LePRK2 rather than LePRK1 illustrates the potential complexity of signaling: LePRKs may form homodimers or heterodimers and interact with different extracellular binding partners along the path of pollen tube growth through the pistil. The challenge now is to test this hypothesis by demonstrating that additional candidates do in fact bind in vivo to the LePRKs and by demonstrating that the binding of LAT52 (or other candidates) initiates the signal transduction pathways required for pollen germination and pollen tube growth.
METHODS
Plant Material
Mature pollen from tomato (Lycopersicon esculentum cv VF36) was obtained by vibrating the anthers of open flowers and was stored immediately at −80°C or used directly for germination experiments. Pollen was germinated in vitro for up to 16 h in pollen germination medium [20 mM Mes, pH 6.0, 3 mM Ca(NO3)2, 1 mM KCl, 0.8 mM MgSO4, 1.6 mM boric acid, 2.5% (w/v) Suc, and 24% (w/v) polyethylene glycol 4000], as described by Muschietti et al. (1994).
Yeast Two-Hybrid Screens
The phagemid vectors pBD-GAL4 Cam and pAD-GAL4-2.1 (Stratagene, La Jolla, CA) were used to construct the baits or the prey cDNA library. The cDNAs encoding the mature extracellular domains of LePRK1, LePRK2, and LePRK3 (i.e., without signal peptides) were ligated separately to the SmaI site of pBD-GAL4 to generate in-frame fusions between the sequence of the DNA binding domain of GAL4 and those of the extracellular domains (pBD-ECD1, pBD-ECD2, and pBD-ECD3). All constructs were sequenced to confirm the orientation. Poly(A)+ RNA from mature pollen was prepared using the PolyATtract System IV (Promega, Madison, WI). The cDNA was synthesized with a Stratagene cDNA synthesis kit and ligated to the HybriZAP-2.1 two-hybrid vector (Stratagene) to generate the GAL4-AD–cDNA fusion library.
The yeast strain PJ69-4A was used as the host strain (James et al., 1996). A modified lithium acetate method (Gietz et al., 1992) was used in all yeast transformations. PJ69-4A cells were transformed with bait plasmids pBD-ECD1, pBD-ECD2, and pBD-ECD3 separately, and the transformants were transformed subsequently with DNA from the pAD-GAL4 pollen library. More than 1 million transformants were screened for each bait. Initial selection was on synthetic complete (SC) medium lacking Leu, Trp, and adenine. Colonies were tested subsequently for growth on SC medium supplemented with 3 mM 3-amino-1,2,4-triazole and lacking Leu, Trp, and His, and those that grew were assayed using an X-gal filter lift method (Breeden and Nasmyth, 1985). Colonies that passed all three tests were analyzed further.
Yeast DNA was extracted and transformed into Escherichia coli XL1Blue to amplify pAD-GAL4 plasmids with cDNA inserts. The plasmids then were retransformed separately into PJ69-4A cells harboring the pBD-GAL4, pBD-ECD1, pBD-ECD2, or pBD-ECD3 plasmid. All transformants then were plated on SC medium supplemented with 3 mM 3-amino-1,2,4-triazole and lacking Leu, Trp, Ade, and His. β-Galactosidase activity was assayed quantitatively after growth in liquid culture using o-nitrophenyl β-d-galactopyranoside as a substrate according to Clontech's yeast protocol (Palo Alto, CA).
DNA Sequence Analysis
The cDNA inserts in the positive colonies were sequenced. Database searches were conducted with the BLAST program at The Arabidopsis Information Resource (www.arabidopsis.org) and at the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov). Cellular locations were predicted using PSORT (psort.nibb.ac.jp) and SignalP (www.cbs.dtu.dk/services/SignalP).
Protein Extraction
Protein extracts and fractions were prepared as described by Muschietti et al. (1998) with the following change. Proteins were extracted using TED buffer (50 mM Tris-HCl, pH 7.6, 1 mM EDTA, 1 mM DTT, and 1 × Protease Inhibitor Cocktail Complete [Boehringer Mannheim]) or pollen germination condition mimic (PGM) buffer [20 mM Mes, pH 6.0, 3 mM Ca(NO3)2, 1 mM KCl, 0.8 mM MgSO4, 1.6 mM boric acid, and 1 × Protease Inhibitor Cocktail Complete].
Pollen homogenates were centrifuged at 10,000g for 20 min at 4°C, and the supernatants (S10, or total protein extracts) were used directly in immunoprecipitation experiments or reserved for further fractionation. The S10 was fractionated by centrifugation at 100,000g for 3 h at 4°C. The resulting supernatant was the S100 (soluble) protein fraction. The pellet was resuspended in PGM buffer with the addition of 0.1% Triton X-100 to yield the P100 (membrane-associated) protein fraction. Protein samples were frozen quickly in liquid nitrogen and stored at −80°C. The protein concentration of each sample was determined with the bicinchoninic acid protein assay kit (Pierce).
Expression of Extracellular Domains of LePRKs and Polyclonal Antibodies
The extracellular domains of LePRK1, LePRK2, and LePRK3 (termed ECD1, ECD2, and ECD3) were fused with a His tag and expressed as described by Muschietti et al. (1998) and Kim et al. (2002). The extracellular domain–His fusion proteins were purified further under native conditions with nickel–nitrilotriacetic acid agarose spin columns (Qiagen, Valencia, CA), and then the imidazole was depleted by centrifugation with a Microcon YM3 filter (Millipore, Bedford, MA) four times with the addition of PGM buffer. Protein samples were frozen quickly in liquid nitrogen and stored at −80°C. Polyclonal anti-ECD1 and anti-ECD2 antibodies (Muschietti et al., 1998), anti-ECD3 antibody (Kim et al., 2002), and anti-LAT52 antibody (Muschietti et al., 1994) were characterized previously.
Immunoprecipitations
In Vivo Coimmunoprecipitation
For each sample, 1 mg of protein extract (S10) in 200 μL of PGM buffer (or other buffer, as indicated), 4 μL of an anti-ECD antibody, and 200 μL of immunoprecipitation buffer (50 mM Tris-HCl, pH 7.6, 0.5% Nonidet P-40 [Sigma], 100 mM NaCl, and 1 × Protease Inhibitor Cocktail Complete [Boehringer Mannheim]) were mixed, adjusted to 0.5% Nonidet P-40, and incubated for 2 h at 4°C. Then, 20 μL of 50% protein A–agarose (preequilibrated with immunoprecipitation buffer; Santa Cruz Biotechnology, Santa Cruz, CA) was added and incubated for another 30 min.
After brief centrifugation to precipitate the agarose beads, immunopellets were washed three times with 1 mL of wash buffer (50 mM Tris-HCl, pH 7.6, 0.2% Nonidet P-40, and 100 mM NaCl). The pellet was resuspended in 30 μL of 1 × Laemmli SDS-PAGE loading buffer, mixed vigorously (using a Vortex mixer, Scientific Industries, Bohemia, NY) for 1 min, and boiled for 1 min. After brief centrifugation, the immunoprecipitated proteins were separated by 15% SDS-PAGE, transferred to a nitrocellulose membrane, and subjected to immunoblot analysis with anti-LAT52 antibody. As a control, duplicate samples were subjected to immunoblot analysis with anti-ECD2 antibody.
Semi-In Vivo Immunoprecipitation
A total of 800 μg of S100 protein from mature pollen was incubated in 200 μL of PGM buffer with the corresponding purified extracellular domain–His fusion protein (∼5 μg) for 1 h at 4°C. Then, 3 μL of anti-RGSHis antibody (Qiagen) and 200 μL of immunoprecipitation buffer were added, and the samples were processed as described for in vivo coimmunoprecipitation.
SDS-PAGE and Immunoblot Analysis
Protein samples were first separated by SDS-PAGE and then transferred to a Hybond enhanced chemiluminescence nitrocellulose membrane (Amersham Pharmacia Biotech). To detect LAT52 or extracellular domain–His fusion proteins, 15% SDS–polyacrylamide gels were used; to detect LePRK proteins, 4 to 20% gradient SDS–polyacrylamide gels (ISC BioExpress, Kaysville, UT) were used. The membranes were immunoblotted with the corresponding primary antibody (anti-LAT52, 1:1200 dilution; anti-RGSHis, 1:1500 dilution; anti-ECD2, 1:1000 dilution) and then with a sheep anti-mouse secondary antibody conjugated with horseradish peroxidase (Amersham Pharmacia Biotech). Finally, the membrane was developed with the enhanced chemiluminescence protein gel blot detection kit (Amersham Pharmacia Biotech).
Protein Partitioning under Native Conditions
For each sample, 520 μg of S100 protein in TED buffer, in PGM buffer, or in modified PGM buffer, as described above, was adjusted to a final volume of 520 μL. After removing 20 μL (input control), the remaining 500 μL was loaded onto the sample reservoir of a Microcon YM100 centrifugal filter device (cutoff molecular mass of 100 kD). The filters were centrifuged at 4°C and 10,000g until the volume retained in the sample reservoir was ∼50 μL or less and the flow-through in the bottom of the vial was ∼450 μL.
The sample reservoir was inverted and recentrifuged briefly to recover the retentate, and the volume was measured. The ∼450-μL flow-through was concentrated to ∼50 μL with an Amicon YM3 centrifugal filter device (cutoff molecular mass of 3 kD; Millipore), and the volume was measured precisely. The input control (10 μL), one-fifth volume of the retentate, and one-fifth volume of the flow-through were processed for immunoblot analysis with anti-LAT52.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Supplementary Material
Acknowledgments
We thank Robyn Cotter for protocols and advice for the yeast two-hybrid screens, Roy Nattiv, a University of California, Berkeley, undergraduate apprentice, for help in characterizing the many positive yeast clones, and David Hantz for excellent care of the greenhouse plants. We thank Michele Engel, Jenn Fletcher, and Paul Herzmark for comments on the manuscript, and we thank all members of our laboratory for useful discussions throughout the course of this work. This work was supported by U.S. Department of Agriculture Grant CRIS 5335-21000-011-00D.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.003103.
Footnotes
Online version contains Web-only data.
References
- Breeden, L., and Nasmyth, K. (1985). Regulation of the yeast HO gene. Cold Spring Harbor Symp. Quant. Biol. 50, 643–650. [DOI] [PubMed] [Google Scholar]
- Cheung, A.Y., Wang, H., and Wu, H.M. (1995). A floral transmitting tissue-specific glycoprotein attracts pollen tubes and stimulates their growth. Cell 82, 383–393. [DOI] [PubMed] [Google Scholar]
- DeWitt, A.E., Dong, J.Y., Wiley, H.S., Lauffenburger, D.A., Guhaniyogi, J., and Brewer, G. (2001). Quantitative analysis of the EGF receptor autocrine system reveals cryptic regulation of cell response by ligand capture. J. Cell Sci. 114, 2301–2313. [DOI] [PubMed] [Google Scholar]
- Franklin-Tong, V.E., Yang, Z., Feijo, J.A., Sainhas, J., Holdaway-Clarke, T., Cordeiro, M.S., Kunkel, J.G., and Hepler, P.K. (1999). Signaling and the modulation of pollen tube growth. Plant Cell 11, 727–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, D., St. Jean, A., Woods, R.A., and Schiestl, R.H. (1992). Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gomez, L., Bauer, Z., Boller, T., Guhaniyogi, J., and Brewer, G. (2001). Both the extracellular leucine-rich repeat domain and the kinase activity of FLS2 are required for flagellin binding and signaling in Arabidopsis. Plant Cell 13, 1155–1163. [PMC free article] [PubMed] [Google Scholar]
- He, Z., Wang, Z.Y., Li, J., Zhu, Q., Lamb, C., Ronald, P., and Chory, J. (2000). Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science 288, 2360–2363. [DOI] [PubMed] [Google Scholar]
- James, P., Halladay, J., and Craig, E.A. (1996). Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo, A., Schopfer, C.R., Nasrallah, M.E., and Nasrallah, J.B. (2001). Allele-specific receptor-ligand interactions in Brassica self-incompatibility. Science 293, 1824–1826. [DOI] [PubMed] [Google Scholar]
- Kim, H.U., Cotter, R., Johnson, S., Senda, M., Dodds, P., Kulikauskas, R., Tang, W., Ezcurra, I., Herzmark, P., and McCormick, S. (2002). New pollen-specific receptor kinases identified in tomato, maize and Arabidopsis: The tomato kinases show overlapping but distinct localization patterns on pollen tubes. Plant Mol. Biol. 50, 1–16. [DOI] [PubMed] [Google Scholar]
- Kobe, B., and Deisenhofer, J. (1994). The leucine-rich repeat: A versatile binding motif. Trends Biochem. Sci. 19, 415–421. [DOI] [PubMed] [Google Scholar]
- Lord, E. (2000). Adhesion and cell movement during pollination: Cherchez la femme. Trends Plant Sci. 5, 368–373. [DOI] [PubMed] [Google Scholar]
- Lush, M.W., Opat, A.S., Nie, F., and Clark, A.E. (1997). An in vitro assay for assessing the effects of growth factors on Nicotiana alata pollen tubes. Sex. Plant Reprod. 10, 351–357. [Google Scholar]
- Maheshwari, G., Wiley, H.S., Lauffenburger, D.A., Guhaniyogi, J., and Brewer, G. (2001). Autocrine epidermal growth factor signaling stimulates directionally persistent mammary epithelial cell migration. J. Cell Biol. 155, 1123–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollet, J.C., Park, S.Y., Nothnagel, E.A., and Lord, E.M. (2000). A lily stylar pectin is necessary for pollen tube adhesion to an in vitro stylar matrix. Plant Cell 12, 1737–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu, J.H., Lee, H.S., and Kao, T.H. (1994). Characterization of a pollen-expressed receptor-like kinase gene of Petunia inflata and the activity of its encoded kinase. Plant Cell 6, 709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschietti, J., Dircks, L., Vancanneyt, G., and McCormick, S. (1994). LAT52 protein is essential for tomato pollen development: Pollen expressing antisense LAT52 RNA hydrates and germinates abnormally and cannot achieve fertilization. Plant J. 6, 321–338. [DOI] [PubMed] [Google Scholar]
- Muschietti, J., Eyal, Y., and McCormick, S. (1998). Pollen tube localization implies a role in pollen-pistil interactions for the tomato receptor-like protein kinases LePRK1 and LePRK2. Plant Cell 10, 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozenberger, B.A., and Young, K.H. (1995). Functional interaction of ligands and receptors of the hematopoietic superfamily in yeast. Mol. Endocrinol. 9, 1321–1329. [DOI] [PubMed] [Google Scholar]
- Park, A.R., Cho, S.K., Yun, U.J., Jin, M.Y., Lee, S.H., Sachetto-Martins, G., and Park, O.K. (2001). Interaction of the Arabidopsis receptor protein kinase Wak1 with a glycine-rich protein, AtGRP-3. J. Biol. Chem. 276, 26688–26693. [DOI] [PubMed] [Google Scholar]
- Park, S.Y., Jauh, G.Y., Mollet, J.C., Eckard, K.J., Nothnagel, E.A., Walling, L.L., and Lord, E.M. (2000). A lipid transfer-like protein is necessary for lily pollen tube adhesion to an in vitro stylar matrix. Plant Cell 12, 151–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullen, S.S., Labadia, M.E., Ingraham, R.H., McWhirter, S.M., Everdeen, D.S., Alber, T., Crute, J.J., and Kehry, M.R. (1999). High-affinity interactions of tumor necrosis factor receptor-associated factors (TRAFs) and CD40 require TRAF trimerization and CD40 multimerization. Biochemistry 38, 10168–10177. [DOI] [PubMed] [Google Scholar]
- Qiu, L., Escalante, C.R., Aggarwal, A.K., Wilson, P.D., and Burrow, C.R. (2000). Monomeric midkine induces tumor cell proliferation in the absence of cell-surface proteoglycan binding. Biochemistry 39, 5977–5987. [DOI] [PubMed] [Google Scholar]
- Schopfer, C.R., Nasrallah, M.E., and Nasrallah, J.B. (1999). The male determinant of self-incompatibility in Brassica. Science 286, 1697–1700. [DOI] [PubMed] [Google Scholar]
- Shiu, S.H., and Bleecker, A.B. (2001). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 98, 10763–10768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steer, M., and Steer, J. (1989). Pollen tube tip growth. New Phytol. 111, 323–358. [DOI] [PubMed] [Google Scholar]
- Takayama, S., Shimosato, H., Shiba, H., Funato, M., Che, F.S., Watanabe, M., Iwano, M., and Isogai, A. (2001). Direct ligand-receptor complex interaction controls Brassica self-incompatibility. Nature 413, 534–538. [DOI] [PubMed] [Google Scholar]
- Taylor, L.P., and Hepler, P.K. (1997). Pollen germination and tube growth. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 461–491. [DOI] [PubMed] [Google Scholar]
- Toth, J., Cutforth, T., Gelinas, A.D., Bethoney, K.A., Bard, J., and Harrison, C.J. (2001). Crystal structure of an ephrin ectodomain. Dev. Cell 1, 83–92. [DOI] [PubMed] [Google Scholar]
- Trotochaud, A.E., Jeong, S., and Clark, S.E. (2000). CLAVATA3, a multimeric ligand for the CLAVATA1 receptor-kinase. Science 289, 613–617. [DOI] [PubMed] [Google Scholar]
- Twell, D., Wing, R., Yamaguchi, J., and McCormick, S. (1989). Isolation and expression of an anther-specific gene from tomato. Mol. Gen. Genet. 217, 240–245. [DOI] [PubMed] [Google Scholar]
- Twell, D., Yamaguchi, J., and McCormick, S. (1990). Pollen-specific gene expression in transgenic plants: Coordinate regulation of two different tomato gene promoters during microsporogenesis. Development 109, 705–713. [DOI] [PubMed] [Google Scholar]
- Ursin, V.M., Yamaguchi, J., and McCormick, S. (1989). Gametophytic and sporophytic expression of anther-specific genes in developing tomato anthers. Plant Cell 1, 727–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler, M.J., Franklin-Tong, V.E., and Franklin, F.C.H. (2001). The molecular and genetic basis of pollen–pistil interactions. New Phytol. 151, 565–584. [DOI] [PubMed] [Google Scholar]
- Wu, H.M., Wong, E., Ogdahl, J., and Cheung, A.Y. (2000). A pollen tube growth-promoting arabinogalactan protein from Nicotiana alata is similar to the tobacco TTS protein. Plant J. 22, 165–176. [DOI] [PubMed] [Google Scholar]
- Zhu, J., and Kahn, C.R. (1997). Analysis of a peptide hormone-receptor interaction in the yeast two-hybrid system. Proc. Natl. Acad. Sci. USA 94, 13063–13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.