Abstract
The interaction between two phytohormones, gibberellins (GA) and abscisic acid (ABA), is an important factor regulating the developmental transition from seed dormancy to germination. In cereal aleurone tissue, GA induces and ABA suppresses the expression of α-amylases that are essential for the utilization of starch stored in the endosperm. In this work, the signaling pathways mediated by these hormones were investigated in the aleurone cells of barley seeds using double-stranded RNA interference (RNAi) technology. In this tissue, double-stranded RNA molecules generated from the transient expression of DNA templates caused a sequence-specific suppression of the target genes. We demonstrate that the transcription factor, GAMyb, is not only sufficient but also necessary for the GA induction of α-amylase. Another regulatory protein, SLN1, is shown to be a repressor of GA action, and the use of RNAi technology to inhibit the synthesis of SLN1 led to derepression of α-amylase even in the absence of GA. However, this effect still was suppressed by ABA. Although the ABA-induced Ser/Thr protein kinase, PKABA1, is known to suppress GA-induced α-amylase expression, PKABA1 RNAi did not hamper the inhibitory effect of ABA on the expression of α-amylase, indicating that a PKABA1-independent signaling pathway also may exist. We suggest that the generation of specific RNAi in a transient expression approach is a useful technique for elucidating the role of regulatory molecules in biological systems in which conventional mutational studies cannot be performed easily.
INTRODUCTION
The regulation of metabolism and development by hormone interactions is a recurrent theme in biology. In insects, the proper balance between juvenile hormones and ecdysteroids is essential to properly coordinate the larva-to-adult transformation (Gilbert et al., 1996). In plants, the auxin/cytokinin ratio is essential for root and shoot initiation (Inoue et al., 2001), cell division and elongation, and apical dominance (Tamas, 1995). The developmental transition from seed dormancy to germination is regulated by the interaction between gibberellins (GA) and abscisic acid (ABA) (Chrispeels and Varner, 1966; Koornneef et al., 1982; Jacobsen et al., 1995). Barley aleurone cells have provided an amenable system to study the GA/ABA regulation of gene expression. These highly specialized cells are dedicated to the secretion of hydrolytic enzymes that are necessary for the breakdown and use of reserves stored in the endosperm (Yomo and Varner, 1971; Jacobsen et al., 1995).
The involvement of GA as a stimulator of this process is well documented (Bethke et al., 1997; Ritchie and Gilroy, 1998, and references therein). ABA, on the other hand, is capable of blocking the action of GA. Early studies have determined that the GA/ABA antagonism does not follow strictly competitive kinetics, suggesting that GA and ABA do not compete for the same binding site in the cell (Chrispeels and Varner, 1966). More recently, it has been shown that the molecular interaction between the signaling cascades mediated by GA and ABA occurs midway in the pathway that leads to the synthesis of hydrolytic enzymes (Gómez-Cadenas et al., 2001).
Early responses to GA include the activation of G-proteins (Jones et al., 1998; Ueguchi-Tanaka et al., 2000) and increased cGMP (Penson et al., 1996) and cytoplasmic Ca2+ (Gilroy, 1996) concentrations, which culminate in the induction of a transcription factor, GAMyb (Gubler et al., 1995). GAMyb promotes the expression of genes, such as high- and low-pI α-amylases, proteinases, and β-glucanases, by binding directly to the promoter GA response element, which has been shown to be essential for the GA induction of these genes (Lanahan et al., 1992; Rogers and Rogers, 1992; Cercós et al., 1999; Gubler et al., 1999).
Another important regulator in the GA signaling cascade in cereal aleurone cells is the slender gene product. The slender mutants of barley and rice express α-amylase in aleurone cells even in the absence of GA, and their shoots elongate as if they were growing under saturating concentrations of GA (Chandler, 1988; Lanahan and Ho, 1988; Ikeda et al., 2001). Interestingly, the constitutive GA response in a barley slender mutant can be blocked by ABA (Lanahan and Ho, 1988). Recently, the slender gene of rice (SLR1) was cloned and found to be an ortholog of wheat Rht1, maize D8, and Arabidopsis GAI and RGA (Peng et al., 1997, 1999; Silverstone et al., 1998; Ikeda et al., 2001). In transgenic rice, overexpression of SLR1 resulted in a dwarfing phenotype, and GA treatment of plants expressing an SLR1–green fluorescent protein (GFP) fusion protein caused the disappearance of the protein from nuclei (Itoh et al., 2002).
The discovery of an ABA-inducible protein kinase, PKABA1 (Anderberg and Walker-Simmons, 1992), provides the opportunity to study the effect of ABA and this kinase on the GA signaling pathway. Recent studies have shown that ABA blocks the GA signal transduction cascade before the formation of active GAMyb. PKABA1 is able to mimic the effect of ABA, suggesting a role for this kinase in the ABA-mediated inhibition of GA signaling (Gómez-Cadenas et al., 1999, 2001).
The evidence obtained to date indicates that GAMyb, SLN1, and PKABA1 play pivotal roles in the GA/ABA signaling cascades; however, it is not clear whether they are absolutely necessary for these pathways to function. Functional redundancy is not uncommon among similar regulatory proteins, which could hamper studies of the roles of these proteins. For instance, the Arabidopsis proteins GAI and RGA control overlapping processes and a strong phenotype becomes evident only when both gene products are absent (Dill and Sun, 2001; King et al., 2001).
The “gain-of-function” approach has been used successfully to address the question of whether a particular factor, such as a hormone, signaling molecule, or regulator of transcription, is involved in gene expression (Gubler et al., 1995, 1999; Shen et al., 1996; Cercós et al., 1999; Gómez-Cadenas et al., 1999, 2001). However, “loss-of-function” experiments are more difficult to implement, especially in transient expression systems. Antisense technology is not always effective, and approaches such as overexpression of dominant-negative mutant proteins often are unavailable. Random insertional mutagenesis followed by systematic screening of gene knockouts has been very successful for the isolation of mutants in specific genes, but this technique is better suited for species with relatively small genomes, such as Arabidopsis and rice. The barley genome (4.9 × 109 bp) is ∼10 times larger than that of rice (4.3 × 108 bp) (Dean and Schmidt, 1995), which would make such a screen much more laborious.
The recent discovery that double-stranded RNA (dsRNA) effectively interferes with gene expression in a sequence-specific manner (Fire et al., 1998) and the successful utilization of this technology in Arabidopsis (Chuang and Meyerowitz, 2000) opened the possibility of using a similar approach to nullify the expression of important components of the GA and ABA signaling cascades in barley and to assess their contribution in regulating GA/ABA-controlled processes.
In this report, we show that dsRNA interference (RNAi) can be used successfully in transient expression experiments in highly specialized and terminally differentiated aleurone tissue. We demonstrate that this technique can be adapted to study induced and preexisting factors such as GAMyb and SLN1, respectively. GAMyb RNAi hampered the GA induction of α-amylase, and blocking the expression of SLN1 by RNAi allowed the derepression of α-amylase even in the absence of GA, which demonstrates the central role of GAMyb and SLN1 in the GA signaling pathway. Finally, based on our observations with PKABA1 RNAi, we propose the existence of an alternative ABA signaling cascade independent of PKABA1.
RESULTS
Transient Expression of RNAi Specifically Inactivates Target Genes
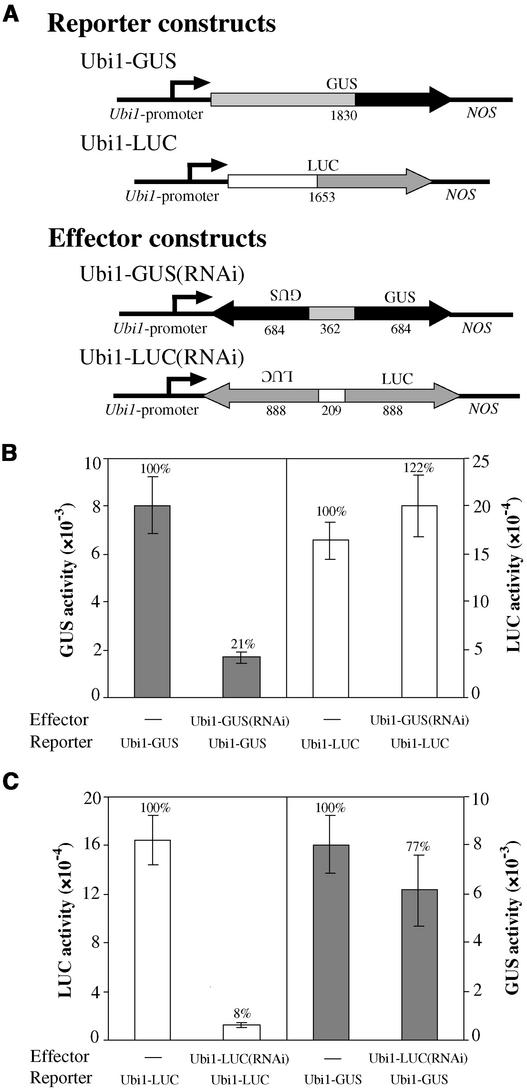
The first question addressed in this work was whether RNAi technology could be used in our experimental system, which is based on the transient transformation of highly differentiated aleurone cells. Our initial attempts focused on interfering with the expression of reporter genes that encode either β-glucuronidase (GUS) or luciferase (LUC). RNAi effector constructs were prepared using inverted repeats of portions of each target gene under the control of the maize constitutive Ubi1 promoter (Figure 1A; see Methods). Extracts from transformed aleurones were assayed for the accumulation of the respective reporter enzymes. Because transformation effectiveness varies from sample to sample, we usually normalized reporter enzyme activity with the enzyme activity of an internal transformation control.
Figure 1.
Transient Transformation with RNAi-Generating Gene Constructs Interferes with the Expression of Target Genes.
(A) Schemes of gene constructs. Different box shades indicate regions of the respective reporter genes used to create each effector construct. Arrowheads indicate the orientation of each gene or gene fragment. Numbers below the boxes represent the size (in bp) of each segment or entire gene (not drawn to scale).
(B) Reporter construct Ubi1-GUS and internal transformation control Ubi1-LUC were cobombarded with either the Ubi1-Empty vector (−) or the effector construct Ubi1-GUS(RNAi).
(C) Reporter construct Ubi1-LUC and internal control Ubi1-GUS were cotransformed with either the Ubi1-Empty vector (−) or the effector construct Ubi1-LUC(RNAi).
For (B) and (C), all plasmids were mixed at a 1:1:1 ratio (reporter:effector:internal control). Closed and open bars represent GUS and LUC activities, respectively, ± se (n = 6).
For clarity, in Figure 1, we present enzyme activities before normalization to show that the effector constructs did not have any significant effect on the expression of the internal controls. At left in Figures 1B and 1C, we show that GUS and LUC activities were reduced strongly by the introduction of the respective RNAi effector constructs. At right in Figures 1B and 1C, we show that enzyme activities from the constructs Ubi1-LUC and Ubi1-GUS, which were used as internal controls for transformation, were not changed significantly by the presence of the RNAi constructs. These results demonstrate the specificity of each RNAi construct for its target gene.
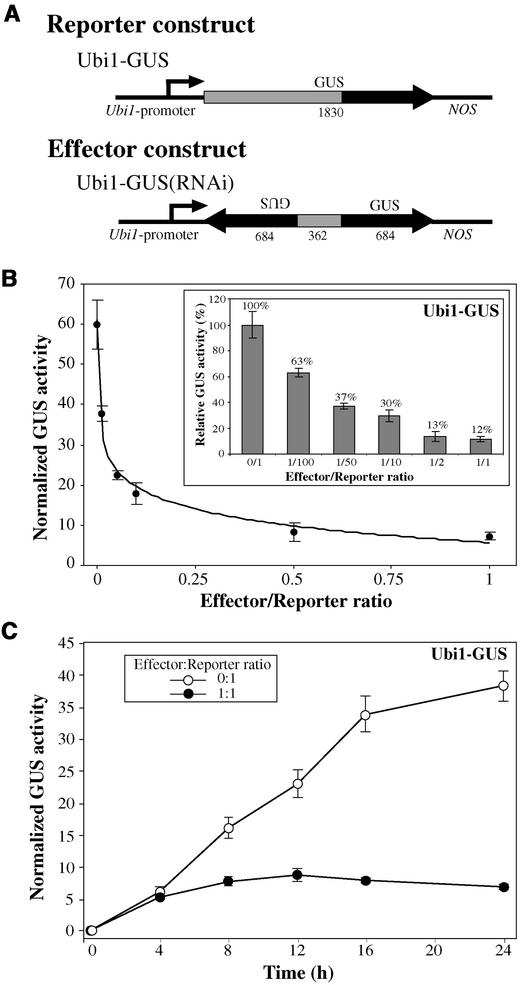
RNAi Is Effective Even at Very Low Effector-to-Reporter Construct Ratios
To determine the effectiveness of RNAi, a dosage-response curve was established with effector-to-reporter ratios ranging from 0.01:1 to 1:1. As shown in Figure 2B, when one-tenth of the amount of RNAi construct versus reporter construct (0.1:1 ratio) was used, GUS activities were approximately one-third of what was observed when no Ubi1-GUS(RNAi) was included (0:1 ratio). When the ratio was decreased to 0.01:1, GUS activities were ∼75% of those of the control (0:1). These results led us to conclude that the effect of Ubi1-GUS(RNAi) is dose dependent and that GUS activity is repressed progressively with increasing concentrations of effector construct. It is interesting that, at least with these constructs, the RNAi effect almost reached a plateau at a ratio of 0.5:1 with respect to the target construct. With this in mind, all of the subsequent experiments were performed at a ratio of 1:1, unless indicated otherwise.
Figure 2.
Response of Aleurone Cells to Increasing Amounts of RNAi, and Time Course of Induction of the RNAi Mechanism.
(A) Schemes of gene constructs. Different box shades indicate regions of the GUS gene used to create the effector construct. Arrowheads indicate the orientation of the gene or gene fragment. Numbers below the boxes represent the size (in bp) of every segment or entire gene (not drawn to scale).
(B) The reporter construct Ubi1-GUS was cobombarded with increasing amounts of effector Ubi1-GUS(RNAi). Decreasing amounts of Ubi1-Empty vector were used to maintain the DNA ratio at 1:1 ([effector + empty vector]:reporter). The curve y = −6.016ln(x) + 5.448 was fitted to the data. The inset represents relative GUS activities at different effector:reporter ratios, with 0:1 representing 100% activity.
(C) The reporter construct Ubi1-GUS was cobombarded with the Ubi1-Empty vector (open circles) or the effector construct Ubi1-GUS(RNAi) (closed circles) at a 1:1 ratio.
For (B) and (C), data points represent normalized GUS activities ± se. For (B), n = 4; for (C), n = 6.
The RNAi Effect Takes Place within 4 h after Transformation
GUS activity in tissue cotransformed with Ubi1-GUS and Ubi1-GUS(RNAi) constructs at a ratio of 1:1 was ∼12% of that of the control after a 24-h incubation period (Figure 2B). However, it was unclear how the level of GUS activity accumulated over time. At least two possibilities exist. (1) GUS activity accumulates in a linear fashion, indicating that the effect of Ubi1-GUS(RNAi) is constant from the beginning of the experiment; in this case, ∼10% of the mRNA would be translated at all times, indicating a certain degree of leakiness of the system. (2) GUS activity accumulates at the same rate as in control samples until the interference machinery is activated, at which time GUS accumulation levels off.
In the former case, we would expect a nearly linear accumulation of GUS over time; in the latter possibility, a constant accumulation rate would occur in the early hours of the experiment, and then GUS activity would cease to increase at some point. As shown in Figure 2C, our results fit better with the second possibility. GUS accumulation occurred normally during the first 4 h of the experiment, but then GUS levels stopped accumulating and remained steady for the next 20 h. Thus, expression of the target gene was repressed nearly 100% after the RNAi mechanism was established in 4 h.
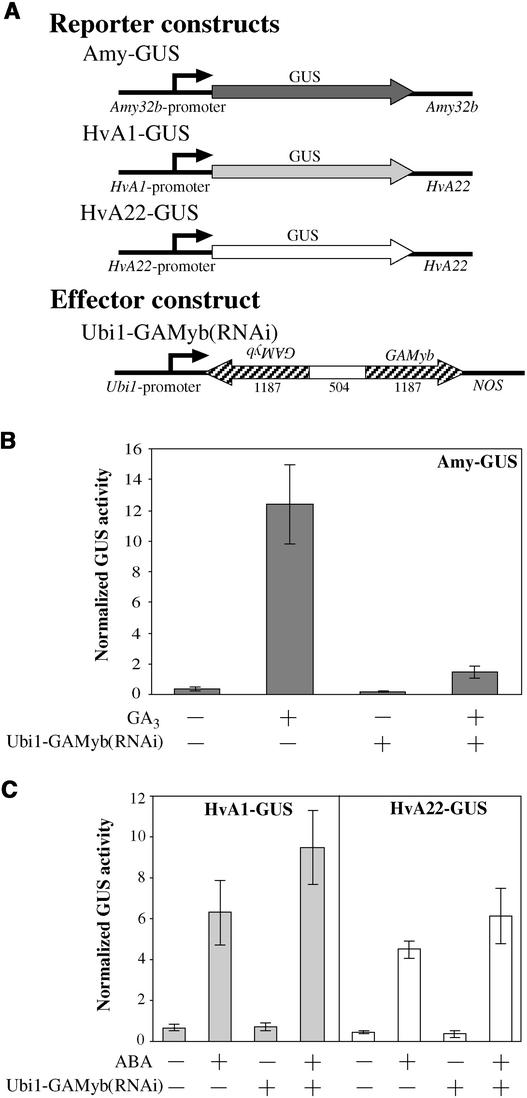
Interference of GAMyb Expression Reduces the GA Induction of α-Amylase
It has been shown that the transcription factor GAMyb binds and activates the promoter of α-amylase genes by gel-shift and transient expression assays (Gubler et al., 1995, 1999). In addition, blockage of the GA response element binding site with a truncated version of GAMyb that lacks the transactivation domain of the protein strongly reduced α-amylase expression (Gómez-Cadenas et al., 2001). However, it has not been resolved whether GAMyb is absolutely necessary to induce α-amylase expression in aleurone cells. Because GA induces GAMyb transcription, its expression is likely to be susceptible to RNAi. By specifically targeting GAMyb transcripts, we monitored the GA-mediated induction of an α-amylase promoter–GUS reporter construct (Amy-GUS).
The effector construct Ubi1-GAMyb(RNAi) was generated using a region of the GAMyb gene that codes for the transactivation domain (Figure 3A). We purposely excluded the conserved Myb-like DNA binding domain to reduce any possible cross-interference with other Myb factors. The results shown in Figure 3B indicate that blocking the expression of GAMyb led to a dramatic reduction of the GA induction of α-amylase. Other genes not regulated by GA, such as the ABA-induced HvA1 and HvA22 genes, were not affected by the presence of the Ubi1-GAMyb(RNAi) construct (Figure 3C). In addition, the same RNAi construct showed no apparent effect on the expression of the internal transformation control Ubi1-LUC (data not shown). These results indicate that the Ubi1-GAMyb(RNAi) construct is specific and that the expression of GAMyb is required for the GA induction of α-amylase.
Figure 3.
GAMyb RNAi Specifically Reduces the GA-Mediated Induction of α-Amylase.
(A) Schemes of gene constructs. The sequence of the effector construct is identical to that of a part of the endogenous GAMyb gene. Arrowheads indicate the orientation of every gene or gene fragment. Numbers below the effector construct represent the size (in bp) of every segment (not drawn to scale).
(B) The GA-inducible reporter construct Amy-GUS was cotransformed with the Ubi1-Empty vector (−) or the effector construct Ubi1-GAMyb(RNAi) at a 1:1 ratio. Embryoless half-seeds then were incubated for 24 h with (+) or without (−) 1 μM GA3.
(C) An ABA-inducible construct, HvA1-GUS (left) or HvA22-GUS (right), was cotransformed with the Ubi1-Empty vector (−) or with Ubi1-GAMyb(RNAi) at a 1:1 ratio. Embryoless half-seeds then were incubated for 24 h with (+) or without (−) 20 μM ABA.
For (B) and (C), bars represent normalized GUS activities ± se (n = 4).
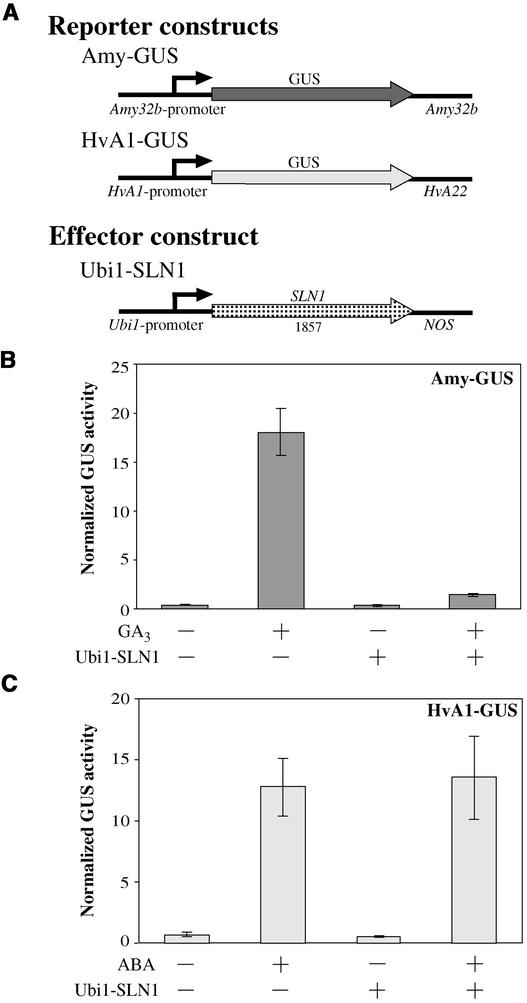
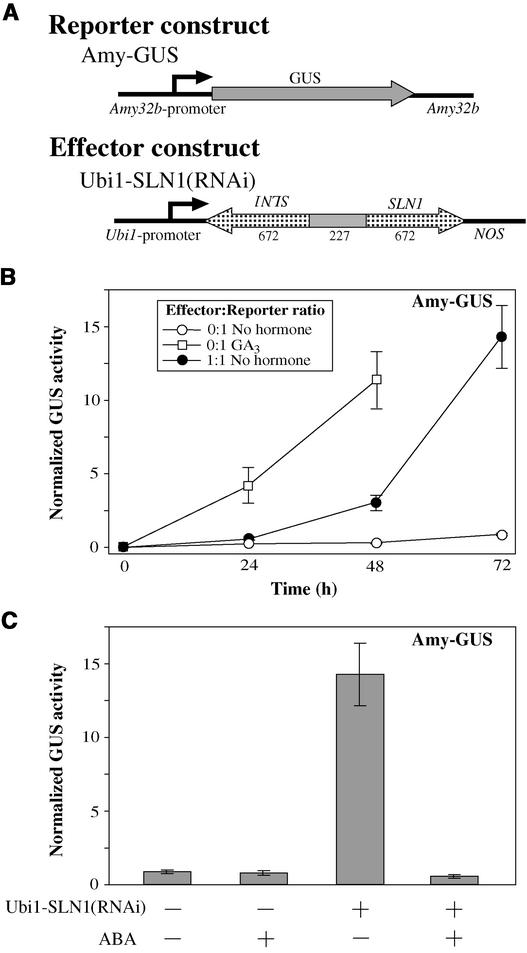
SLN1 Is a Strong Repressor of α-Amylase Expression, and Its Removal Causes Derepression of α-Amylase in the Absence of GA
The recent discovery that several proteins belonging to the GRAS family of transcriptional regulators are involved in GA signaling (Peng et al., 1999; Pysh et al., 1999; Silverstone and Sun, 2000) prompted us to test their involvement in the expression of GA-controlled genes in aleurone cells. A pair of primers was designed based on a DNA sequence alignment of the cereal genes SLR1 (rice), Rht-1 (wheat), and D8 (maize). A PCR product of a conserved region was obtained and used to screen barley genomic and aleurone cDNA libraries.
The longest cDNA clone recovered lacked the first 10 amino acids at the N terminus of the protein compared with a genomic clone. The sequence is practically identical to the SLN1 gene sequence deposited in GenBank by Chandler et al. (accession number AF460219), with the exception of a few bases that cause changes in three amino acids. When the reconstituted open reading frame (ORF) of 1857 bp was overexpressed in aleurone cells, the GA-induced expression of α-amylase was inhibited strongly (Figure 4B). On the other hand, Ubi1-SLN1 had no effect on the ABA induction of the HvA1-GUS reporter construct (Figure 4C). These results indicate that barley SLN1 is a specific repressor of the GA.
Figure 4.
SLN1 Is a Repressor of the GA-Mediated Expression of α-Amylase.
(A) Schemes of gene constructs. Arrowheads represent the ORF of each gene. Numbers below the effector construct represent the size (in bp) of the ORF (not drawn to scale).
(B) The GA-inducible reporter construct Amy-GUS was cotransformed with either the Ubi1-Empty vector (−) or the effector construct Ubi1-SLN1 at a 1:1 ratio. Embryoless half-seeds then were incubated for 24 h with (+) or without (−) 1 μM GA3.
(C) The ABA-inducible construct HvA1-GUS was cotransformed with the Ubi1-Empty vector (−) or the effector construct Ubi1-SLN1 at a 1:1 ratio and incubated for 24 h with (+) or without (−) 20 μM ABA.
For (B) and (C), bars represent normalized GUS activities ± se (n = 4).
When a Ubi1-SLN1(RNAi) construct was tested, a gradual derepression of α-amylase occurred over time (Figure 5B). In contrast to the response observed with GAMyb RNAi, the effect of SLN1 RNAi was not evident at 24 h. However, after 48 h of incubation in the absence of GA, the levels of α-amylase almost reached those observed at 24 h in GA-treated samples transformed with the reporter construct and an empty effector construct (Figure 5B). At 72 h after transformation in the absence of GA, the levels of α-amylase expression were similar to those of samples treated with GA for 48 h (Figure 5B).
Figure 5.
SLN1-RNAi Causes Derepression of α-Amylase in the Absence of GA.
(A) Schemes of gene constructs. The effector construct shares 100% sequence identity with part of the endogenous SLN1 transcript. Arrowheads indicate the orientation of the gene or gene fragment. Numbers below the effector construct represent the size (in bp) of every segment (not drawn to scale).
(B) The GA-inducible reporter construct Amy-GUS was cotransformed with either the Ubi1-Empty (open symbols) or the effector construct Ubi1-SLN1(RNAi) (closed circles) at a 1:1 mass ratio. Embryoless half-seeds then were incubated for 24, 48, or 72 h in the absence of hormones (closed and open circles) or in the presence of 1 μM GA3 (open squares).
(C) The reporter construct Amy-GUS was cobombarded with either the Ubi1-Empty vector (−) or the effector construct Ubi1-SLN1(RNAi) at a 1:1 ratio. Embryoless half-seeds then were incubated for 72 h with (+) or without (−) 20 μM ABA.
Data points in (B) and bars in (C) represent normalized GUS activities ± se (n = 6).
These results indicate that the endogenous SLN1 represses α-amylase expression and that by interfering with the expression of SLN1, α-amylase is derepressed, even in the absence of GA. The delay in the response to SLN1 RNAi can be attributed to the high stability of preexisting SLN1 protein. We also observed that the derepressed expression of α-amylase was blocked effectively by the addition of ABA (Figure 5C), which confirms our own observations that SLN1 acts upstream of the point of ABA action (Gómez-Cadenas et al., 2001).
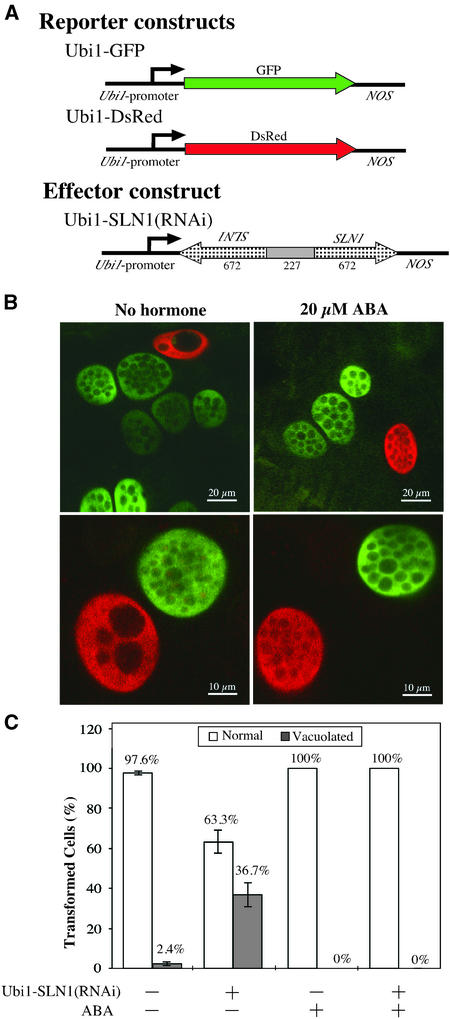
SLN1 Regulates General GA Responses in Aleurone Cells
In addition to the derepression of the α-amylase–GUS construct, we determined if the Ubi1-SLN1(RNAi) caused the pleiotropic effect observed in aleurone cells treated with GA. One well-characterized morphological change of aleurone cells is the vacuolation process that takes place when cells are engaged in GA responses (Bethke et al., 1998). As cells progress in the GA response, a dramatic reconfiguration of the cellular interior takes place to switch from a storage function to secretory machinery. Upon GA treatment, the endoplasmic reticulum and the Golgi apparatus are developed and the synthesis and secretion of hydrolytic enzymes occur. As the contents in protein storage vacuoles are degraded to provide amino acids for protein synthesis, these vacuoles fuse, enlarge, and become less dense.
An experiment was devised to distinguish between cells transformed with the Ubi1-Empty vector and those transformed with the Ubi1-SLN1(RNAi) construct in the same sample. In previous microscopic observations, we noted that the fluorescent proteins GFP and DsRed were localized to cytoplasm and nucleus but unable to penetrate the vacuolar membrane of aleurone cells. Thus, vacuoles appeared as dark intracellular bodies whose size and morphology could be assessed easily. A Ubi1-GFP construct was mixed with Ubi1-Empty and cotransformed into the aleurone of embryoless half-seeds. A second particle bombardment event was performed with the same half-seeds, delivering a mixture of Ubi1-DsRed and Ubi1-SLN1(RNAi) constructs.
Two representative confocal microscopy images of the vacuolation effect at 48 h after transformation are presented in Figure 6B (left). The green cells cotransformed with the Ubi1-Empty vector show the distinct internal morphology of cells not engaged in GA responses; protein storage vacuoles in the range of 2 to 5 μm are easily distinguished. Red cells carrying the Ubi1-SLN1(RNAi) construct developed larger vacuoles (>10 μm) typical of a GA response.
Figure 6.
SLN1 RNAi Triggers a General GA-Like Response in Aleurone Cells in the Absence of GA.
(A) Schemes of gene constructs. The effector construct shares 100% sequence identity with part of the endogenous SLN1 transcript. Arrowheads indicate the orientation of the gene segment with respect to the sense strand of the gene. Numbers under the effector construct represent the size (in bp) of every segment (not drawn to scale).
(B) Composite images of green and red channels of confocal micrographs at lower (top) and higher (bottom) magnifications. Embryoless half-seeds were cotransformed with Ubi1-GFP and Ubi1-Empty vectors (green cells), or with Ubi1-DsRed and Ubi1-SLN1(RNAi) (red cells), at a 1:2 mass ratio. Samples were examined 48 h after incubation in the presence (right) or absence (left) of 20 μM ABA.
(C) Transformation conditions were the same as in (B). Red and green cells that vacuolated (closed bars) or did not vacuolate (open bars) were scored. The numbers from each aleurone layer were counted as one sample. Bars represent the percentage of transformed cells ± se (n = 6). The experiment was repeated three times with similar results.
In an attempt to quantify this process, green and red cells, either vacuolated or nonvacuolated, were counted. More than 1000 cells of each color were evaluated. Although not all of the red cells showed extensive vacuolation at 48 h, ∼37% clearly were vacuolated (Figure 6C). By contrast, the majority of the green cells did not develop large vacuoles. Only 3% of the green cells showed a GA-like response, of which approximately one-third were doubly transformed, containing both GFP and DsRed. Vacuolation of both green and red cells was inhibited completely if the half-seeds were incubated in the presence of 20 μM ABA (Figure 6B).
It is important to note that when the reporter constructs were exchanged [i.e., Ubi1-GFP mixed with Ubi1-SLN1(RNAi) and Ubi1-DsRed mixed with Ubi1-Empty], the same outcome was observed (i.e., green cells became vacuolated). These observations demonstrate that SLN1 not only regulates the expression of GAMyb-induced genes but also is involved in general GA responses in aleurone cells. Furthermore, the diverse effects of SLN1-RNAi can be blocked completely by ABA.
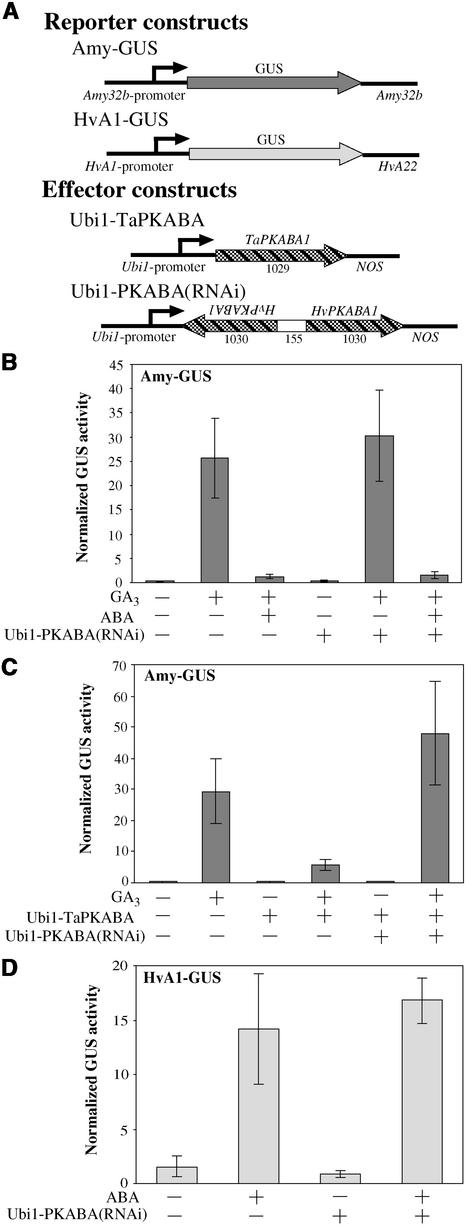
Interference of PKABA1 Expression Does Not Affect the ABA Repression of α-Amylase
It has been reported that overexpression of the wheat ABA-induced protein kinase, TaPKABA1, but not of other protein kinases, greatly reduces the GA-mediated expression of α-amylase, mimicking the effect of ABA (Gómez-Cadenas et al., 1999). We have observed that the barley HvPKABA1 is equally effective (Yamauchi et al., 2002). We decided to use RNAi to block the expression of HvPKABA1 and observe the effect on the ABA signaling cascade involved in the repression of α-amylase. In Figure 7B, we show that ABA is an effective antagonist of the GA induction of α-amylase (third bar from left); however, when we introduced the Ubi1-PKABA(RNAi) construct, it had no effect on the antagonistic pathway mediated by ABA (Figure 7B, sixth bar from left).
Figure 7.
PKABA1 RNAi Does Not Affect the Antagonistic Effect of ABA on the GA-Mediated Induction of α-Amylase.
(A) Schemes of gene constructs. The effector construct shares 100% sequence identity with the endogenous gene HvPKABA1 throughout the entire inverted repeats and shares 94% identity with the wheat TaPKABA1. Arrowheads indicate the orientation of the gene or gene fragment. Numbers below the effector constructs represent the size (in bp) of every segment or the entire ORF (not drawn to scale).
(B) The reporter construct Amy-GUS was cobombarded with either the Ubi1-Empty vector (−) or the effector construct Ubi1-PKABA(RNAi) at a 1:1 ratio. Embryoless half-seeds then were incubated with (+) or without (−) 1 μM GA3 and/or 20 μM ABA for 24 h.
(C) The GA-inducible reporter construct Amy-GUS was cotransformed with the Ubi1-TaPKABA and/or Ubi1-PKABA(RNAi) effector constructs. The Ubi1-Empty vector (−) replaced either or both effector constructs to account for a 1:1:1 ratio. Embryoless half-seeds were incubated for 24 h with (+) or without (−) 1 μM GA3.
(D) The ABA-inducible reporter construct HvA1-GUS was cotransformed with the Ubi1-Empty vector (−) or the effector construct Ubi1-PKABA(RNAi) at a 1:1 ratio. Embryoless half-seeds then were incubated for 24 h with (+) or without (−) 20 μM ABA.
For (B) to (D), bars represent normalized GUS activities ± se (n = 4).
To determine whether Ubi1-PKABA(RNAi) was a functional construct, we tested its effect on overexpressed TaPKABA1. As expected, Ubi1-TaPKABA was able to repress the GA induction of α-amylase (Figure 7C, fourth bar from left). When the Ubi1-PKABA(RNAi) construct was included in the transformation mix, it counteracted the inhibitory effect of Ubi1-TaPKABA (Figure 7C, sixth bar from left). We also determined if PKABA1 RNAi affected the expression of ABA-induced genes. We cobombarded the ABA-responsive construct HvA1-GUS with the Ubi1-PKABA(RNAi) construct. As shown in Figure 7D, ABA still induced HvA1 expression, even when PKABA1 expression was suppressed. These results indicate that PKABA1 is not absolutely necessary for the ABA suppression of α-amylase or for the induction of HvA1.
DISCUSSION
In this work, we demonstrate that RNAi technology can be used in transient expression experiments to specifically suppress the expression of target genes in barley aleurone cells in which traditional mutational studies are difficult. The evidence presented here shows that RNAi technology works within 4 h of the introduction of the effector construct. Currently, barley is not an amenable genetic system for specific gene knockouts, and screens for mutants in specific genes can be very laborious and time consuming. Although transposon tagging and Agrobacterium tumefaciens–mediated transformation have been developed (Horvath et al., 2000; Koprek et al., 2001; Scholz et al., 2001), these methodologies still require a great deal of effort and time.
Considering that the genome size of barley is >10 times larger than that of rice and ∼40 times larger than that of Arabidopsis, the effort invested in screening for insertion mutants could be enormous. The recent development of targeting of induced local lesions in genomes as a way to screen for mutations in specific genes is very promising (McCallum et al., 2000), but the establishment of a barley mutant population for systematic screening requires extensive facilities. Moreover, certain genes may be lethal in hemizygous or homozygous states, which could pose an even greater difficulty in obtaining mutants for a particular gene. RNAi has been used successfully as a genetic tool in several organisms, including protozoa, fungi, mammals, and plants (Bosher and Labouesse, 2000; Chuang and Meyerowitz, 2000; Sharp, 2001), and it can reveal valuable information even if a stable knockout line cannot be generated.
It has been shown that RNAi can occur in epidermal cells of maize, wheat, and barley. In fact, both dsRNA and a DNA construct with a basic structure similar to those used in this work interfered with the expression of target genes (Schweizer et al., 2000). However, early observations in the worm Caenorhabditis elegans showed that certain cell types might be less susceptible to dsRNAi (Fire et al., 1998). Consequently, it was difficult to predict the outcome in triploid, highly differentiated, and specialized cereal aleurone cells (Ritchie and Gilroy, 1998).
The evidence obtained in this work suggests that the interference mechanism in aleurone cells is similar to RNAi observed in other organisms. With the aid of reporter genes encoding GUS and LUC, we have shown that the accumulation of these enzymes can be reduced by up to 90% when both the RNAi and the target gene constructs are coexpressed under the control of strong constitutive promoters (Figure 1). A 10 to 15% residual activity was always observed, probably because both constructs are under the control of the same Ubi1 promoter and the reporter construct has enough time to be expressed (∼4 h) before the RNAi mechanism is fully functional (Figure 2C). We suggest that the use of inducible or weaker promoters to drive the expression of the reporter gene would allow more time for the RNAi construct to be expressed; thus, the interference mechanism could be fully active before the transcription of the reporter construct begins.
We used the promoter of an α-amylase gene fused to GUS to monitor the activity of the GA signaling pathways upon different hormonal treatments. There is ample evidence to suggest that GAMyb is sufficient to transactivate the expression of α-amylase (Gubler et al., 1995; Gómez-Cadenas et al., 1999, 2001); however, whether GAMyb is necessary for GA induction is unclear, mainly because no GAMyb mutant has been reported in any plant. Gómez-Cadenas et al. (2001) showed that a truncated GAMyb protein containing only the DNA binding domain, but lacking the transactivation domain, was capable of blocking the expression of an α-amylase reporter construct in the presence of GA. Presumably, this truncated protein also binds the GA response element sequence, thus competing with the endogenous full-length GAMyb. However, the possibility of functional redundancy among similar Myb proteins or the existence of a different transcription factor that could bind to nearby DNA elements in the α-amylase promoter and activate transcription cannot be excluded. In this work, we demonstrate through the inactivation of GAMyb expression that there are no other factors that can substitute for the GAMyb function in aleurone cells (Figure 3B). One might argue that it is still possible that our RNAi construct could interfere with the expression of other transcription factors that share sequence similarity with GAMyb. We were careful to choose a region outside of the Myb-like binding domain that showed no similarity to any other sequence in the database other than to homologous genes in other species. Also, we observed that GAMyb RNAi did not affect the expression of other genes, that is, the ABA induction of HvA1 and HvA22 (Figure 3C) or the constitutive expression of the Ubi1-LUC internal control (data not shown).
The importance of SLN1 as a repressor of GA action has been demonstrated in barley and other plant species. The recessive slender mutant of barley shows constitutive GA-like responses throughout the plant, including extraordinary shoot elongation and α-amylase expression in seeds, even in the presence of GA biosynthesis inhibitors (Chandler, 1988; Lanahan and Ho, 1988). These observations suggest that this factor probably works as a repressor, except when GA responses occur. When SLN1 was overexpressed, the GA-induced expression of α-amylase was blocked dramatically, which confirms the role of SLN1 protein as a repressor of GA action (Figure 4B).
Considering that SLN1 protein likely preexists in aleurone cells, as is the case in vegetative tissues of rice (Itoh et al., 2002), we realized that the effect of SLN1 RNAi might not be detectable immediately after the RNAi construct is introduced into the cells. In fact, our first experiments were inconclusive when samples were analyzed 24 h after transformation (data not shown). The success of the RNAi experiments depended on the stability and turnover rate of the target gene products. We reasoned that if enough time was allowed for SLN1 to decay naturally and at the same time SLN1 expression was blocked by RNAi, then we would see α-amylase expression even in the absence of GA, similar to what is observed in the barley slender mutant. Indeed, 48 h after transformation provided enough time to derepress α-amylase expression almost fully in the absence of GA (Figure 5B).
Independent results obtained with rice SLR1 and Arabidopsis RGA suggest that the stability of these proteins is decreased when GA is applied to these plants (Dill et al., 2001; Itoh et al., 2002). Sequences at the N terminus seem to be important in destabilizing these proteins, in particular the DELLA domain, a highly conserved sequence present in all members of the GRAS family that are involved in GA responses (Pysh et al., 1999; Silverstone and Sun, 2000). We believe that a similar destabilizing process occurs in barley aleurone cells, because the effect of SLN1 RNAi was observed 48 h after transformation, whereas the effect of GA treatment was observed at 24 h (Figure 5B). Another possibility is that the kinetics of RNAi observed with transiently expressed genes is not the same as that for endogenous genes.
Another aspect of the GA response is the vacuolation process, which is characteristic of aleurone cells engaged in GA responses (Bethke et al., 1998). We observed that SLN1 RNAi also stimulated the vacuolation of aleurone cells (Figures 6B and 6C), which indicates that SLN1 regulates GA action in general rather than just in genes responsive to GAMyb. As in the case of α-amylase expression, vacuolation caused by SLN1 RNAi was delayed by ∼24 h. Normally, vacuolation of GA-treated aleurone cells is observed within 24 to 36 h (data not shown) (Bethke et al., 1998). However, SLN1 RNAi promoted this process 48 h after transformation in the absence of GA. It is important to note that all of the changes promoted by the SLN1 RNAi were effectively blocked by ABA (Figures 5C, 6B, and 6C). This finding is consistent with the suggestion that ABA blocks the GA signaling pathway downstream of SLN1 (Figure 8) (Gómez-Cadenas et al., 2001).
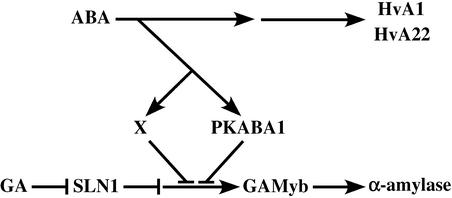
Figure 8.
Important Steps Involved in the Hormonal Regulation of α-Amylase in Aleurone Cells.
A repressor, SLN1, and an activator, GAMyb, are essential components in the GA signaling pathway leading to the induction of α-amylase. An alternative route from ABA to the site of interaction with the GA signaling cascade (represented by X) is proposed to indicate that the transcriptional induction of PKABA1 is sufficient, but not necessary, to block α-amylase expression.
The expression of PKABA1 has been shown to be upregulated by ABA treatment (Anderberg and Walker-Simmons, 1992; Gómez-Cadenas et al., 1999), and overexpression of this protein kinase represses the GA-mediated expression of α-amylase (Gómez-Cadenas et al., 1999). Because PKABA1 is induced transcriptionally by ABA, it is a good target for RNAi to determine whether PKABA1 is not only sufficient but also necessary for the ABA suppression of α-amylase. Our results indicate that PKABA1 RNAi does not hamper the antagonistic effect that ABA has on the GA induction of α-amylase. However, it is able to block the repressive effect of a transgene, TaPKABA1, on the expression of α-amylase.
One interpretation of these observations is that there are at least two independent ABA signaling pathways that lead to the suppression of α-amylase, one dependent and another independent of PKABA1. Another possibility is that PKABA1 preexists in aleurone cells; therefore, transcription of this gene would not be a prerequisite for its action. However, the latter possibility seems to be contradictory to the observation that the expression of PKABA1 is induced by ABA treatment (Anderberg and Walker-Simmons, 1992; Gómez-Cadenas et al., 1999; Yamauchi et al., 2002). In addition, we have explored time courses of up to 72 h of preincubation in the presence of PKABA1 RNAi before the addition of hormones. Our results provide no indication that PKABA1 stability is the reason why PKABA1 RNAi does not inhibit the repressive effect of ABA. Nonetheless, we cannot exclude the possibility of PKABA1 being stable for >3 days without significant turnover. A third possibility is that PKABA1 is not normally involved in the control of ABA processes, but overexpression of this protein kinase blocks GA action by inactivating a component of the GA signaling cascade in a manner similar to ABA. Contrary to what occurs with GAMyb and SLN1, there are sequences similar to PKABA1 in the genome of some cereals that might share a redundant role with this kinase.
In support of this argument, Holappa and Walker-Simmons (1997) have reported the existence of TaPK3, protein kinase highly similar to PKABA1. However, the expression of TaPK3 does not seem to be regulated by ABA, and its transcripts are more abundant in greening tissues. Furthermore, Huttly and Phillips (1995) detected by reverse transcriptase–mediated PCR the expression of 10 different protein kinases in oat aleurone cells, and at least 2 of them shared sequence similarity to PKABA1 and at least one of them, Aspk4, is expressed constitutively in this tissue.
A model of the signaling network that incorporates the observations from this work is presented in Figure 8. Whereas the components of the GA signaling pathway are absolutely necessary to regulate the expression of α-amylase, the ABA pathway involved in the suppression of α-amylase is bifurcated into two routes, one dependent on and another independent of PKABA1.
In summary, we have demonstrated that RNAi technology can be used successfully in transient expression experiments, especially when traditional genetic approaches are not successful in generating mutants in genes of interest. Transiently expressed RNAi can be a complement to the widely used overexpression approach to better understand the function of a gene in a short period of time. The use of this technology allowed us to answer questions that otherwise could not have been resolved in our experimental system.
METHODS
Plant Material
Barley seeds (Hordeum vulgare cv Himalaya), harvest 1991, were obtained from the Department of Agronomy and Soils at Washington State University (Pullman, WA). Embryoless half-seeds were used in all experiments. Half-seeds were prepared according to Gómez-Cadenas et al. (2001).
Preparation of DNA Constructs
To obtain a SLN1 clone, a pair of primers (5′-CTACCTCAAGTTCGCGCACTT-3′ and 5′-CTCGCCTGCTTGTAGGCATT-3′) was designed based on an alignment of rice SLR1, wheat Rht1, and maize D8 genes. A PCR product was amplified, with a barley (cv Himalaya) aleurone cDNA library obtained from Stratagene (La Jolla, CA) used as a template. The ∼800-bp fragment was cloned and sequenced. It was used to screen the same cDNA library as well as a barley (cv Igri) genomic library, also obtained from Stratagene. The longest cDNA had an incomplete open reading frame (ORF), which was restored by PCR using the genomic clone as a template.
Primers 5′-ATCGGATCCACCATGAAGCGCGAGTACCAGGA-3′ and 5′-ATCGAATTCAAAACTCGCGATCACGG-3′ were used to introduce BamHI and EcoRI sites at the 5′ and 3′ ends of the gene, respectively, in two independent PCR procedures. The region between the SphI and BglII sites was replaced with the cDNA to eliminate possible PCR errors, and the entire ORF was sequenced to ensure accuracy.
Reporter Constructs
For Ubi1-GUS, the Escherichia coli UidA gene coding for β-glucuronidase (GUS), with a modified ATG initiation codon (Lanahan et al., 1992), was cloned into the BamHI and KpnI sites of a vector containing the promoter and the first intron of the maize Ubi1 gene and the Agrobacterium tumefaciens NOS gene terminator (Christensen and Quail, 1996). Ubi1-LUC is a modified version of pAHC18 (Christensen and Quail, 1996) in which the 5′ untranslated region of the firefly luciferase (LUC) gene and the EcoRI sites were eliminated. Amy-GUS is equivalent to MBL022 described by Lanahan et al. (1992), and HvA1-GUS and HvA22-GUS are the same as constructs C1 and C17 reported by Shen et al. (1996). Ubi1-GFP was constructed by replacing the LUC ORF in Ubi1-LUC with the green fluorescent protein (GFP) ORF in clone CD3-327 (Davis and Vierstra, 1998) obtained from the ABRC (Ohio State University, Columbus). Ubi1-DsRed was made by replacing the LUC ORF in Ubi1-LUC with the DsRed ORF in plasmid pDsRed2-C1 (Clontech, Palo Alto, CA).
Effector Constructs
In cases in which double-stranded RNA was the intended transcription product, two fragments of the gene of interest were selected, one being a longer version of the other with extra sequence at the 5′ end [except for Ubi1-GAMyb(RNAi); see below]. These fragments were cloned in reverse orientation, generating two inverted repeats separated by a “stuffer” region. All RNA interference (RNAi) constructs were driven by the maize Ubi1 promoter and terminated by the NOS poly(A) addition signal, and both sequences were taken from pAHC17 or its derivative constructs (Christensen and Quail, 1996). Ubi1-GUS(RNAi) was generated by cutting Ubi1-GUS with BamHI and EcoRV enzymes, eliminating more than half of the coding region at the 5′ end of GUS. A PCR product of the 3′ end of GUS was obtained using primers 5′-GAAGGCCTCAGCAAGCGCACTTAC-AGG-3′ and 5′-CGGGATCCATGGTGCGCCAGGAGAGTTGT-3′. The PCR product was treated with BamHI and StuI and then ligated to the cut Ubi1-GUS plasmid in reverse orientation.
For Ubi1-LUC(RNAi), the last ∼900 bp of the LUC gene was subcloned by cutting at the internal SapI site, followed by blunt ending with Klenow enzyme and a second cut with BamHI near the 3′ end of the gene beyond the stop codon. This fragment was linked in reverse orientation into Ubi1-LUC, which was cut previously with EcoRI, blunt ended with Klenow, and cut again with BamHI.
For Ubi1-GAMyb(RNAi), a PCR product of the last ∼1200 bp of the ORF was obtained using primers 5′-TATGATATCAATCTTCAAATGAA-3′ and 5′-CCGCTCGAGTCATTTGAATTCCTCCGA-3′ and cloned into the EcoRV site in pBluescript KS+ (Stratagene). The DNA binding domain of the yeast protein GAL4 was subcloned at the 5′ end of the GAMyb PCR product and served as stuffer in the final construct. This PCR product was subcloned in reverse orientation before the GAL4 binding domain. This construct then was transferred to a derivative of pAHC17 as a BamHI-KpnI insert. Ubi1-SLN1 was generated by subcloning the ORF of SLN1 into a derivative of pAHC17 as a BamHI-EcoRI insert. To construct Ubi1-SLN1(RNAi), a BglII-KpnI fragment that includes the 3′ end of the SLN1 cDNA and part of the polylinker was cloned in reverse orientation into a plasmid containing a longer piece of the 3′ end of SLN1, from BssHII to the end of the cDNA. This intermediary construct was transferred to a pAHC17 derivative as a KpnI-SacI insert.
Ubi1-PKABA(RNAi) was generated using the barley HvPKABA1 cDNA (Yamauchi et al., 2002), which is 94.3% identical to the equivalent portion of the wheat cDNA. The entire ORF of HvPKABA1 was amplified by PCR using primers 5′-CGCGGATCCAAGGATGGATCGGTACGAGGTGG-3′ and 5′-CGGGGTACCTCAAAGCTTCAACGGGCACACGAAGTCCCC-3′. The PCR product was cloned into pBluescript SK+ as a BamHI-KpnI fragment. The new plasmid then was cut with BamHI, treated with Klenow enzyme, and cut again with SpeI, and the original cDNA containing both the 5′ and 3′ untranslated regions was subcloned in reverse orientation as a BamHI-SmaI fragment. The 5′ untranslated region of the cDNA became the stuffer between the two inverted repeats. The insert containing the inverted segments was transferred to a derivative of pAHC17 as a BamHI-KpnI fragment.
Construct Ubi1-TaPKABA is identical to construct UBI-PKABA described by Gómez-Cadenas et al. (1999). Ubi1-Empty is the same as pAHC17 (Christensen and Quail, 1996).
Particle Bombardment and Enzyme Assays
A detailed description of the particle bombardment procedure has been reported elsewhere (Lanahan et al., 1992; Shen et al., 1993). Effector, reporter, and internal control plasmid mixtures are referred to as mass ratios and are described in the figure legends. For normalization purposes, an internal control for transformation (Ubi1-LUC) was always included at a 1:1 ratio with the reporter plasmid. After bombardment and incubation for 24 h (unless noted otherwise), in the presence or absence of 1 μM GA3 or 20 μM abscisic acid, each sample of four half-seeds was processed, and LUC and GUS activities were measured as follows.
The half-seeds were homogenized in 1 mL of grinding buffer (Shen et al., 1993). After clarification at 12,000g for 15 min, 100 μL of supernatant was used for LUC assays using a luminometer (Moonlight 2010; Analytical Luminescence Laboratory, San Diego, CA). For GUS assays, 50 μL of supernatant was mixed with 200 μL of GUS assay buffer (Shen et al., 1993) and incubated for 20 h at 37°C; then, 50 μL of reaction mixture was diluted in 2 mL of 200 mM sodium carbonate. GUS activity was measured with a fluorometer (model 450; Sequoia-Turner, Mountain View, CA) calibrated to 1000 units with a standard made by diluting 50 μL of 1 μM 4-methylumbelliferone in 2 mL of 200 mM sodium carbonate. Normalized GUS activity were calculated by dividing GUS activity by LUC activity of the respective sample and multiplying by a constant of 2000. All experiments consisted of at least four replicates, and the entire experiment was repeated a minimum of two times with similar results.
Samples used for confocal microscopy imaging were transformed once with the Ubi1-SLN1(RNAi):Ubi1-DsRed DNA mixture and a second time with the Ubi1-Empty:Ubi1-GFP DNA mixture at a ratio of 2:1. The embryoless half-seeds then were incubated for 48 h in the presence or absence of hormones.
Confocal Microscopy Imaging
After the 48-h incubation period, the aleurone layers were isolated, removing as much starchy endosperm as possible. To accommodate the samples, two cover slides were glued onto a microscope slide with a space left between them. An aleurone layer was placed on the microscope slide between the two cover slides. Then, a third cover slide was placed on top of the sample to form a glass bridge. Samples were imaged using Leica Confocal System TCS2 (Leica Microsystems, Heidelberg, Germany). Imaging of GFP and DsRed was sequential. To image GFP, an argon laser beam (488-nm excitation) was used, capturing emission in the 510- to 550-nm range. For DsRed, a krypton laser beam (568-nm excitation) was used, capturing emission in the 580- to 650-nm range.
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
NOTE ADDED IN PROOF
Two recent papers (Gubler, F., Chandler, P.M., White, R.G., Llewellyn, D.J., and Jacobsen, J.V. [2002]. Gibberellin signaling in barley aleurone cells: Control of SLN1 and GAMYB expression. Plant Physiol. 129, 191–200, and Chandler, P.M., Marion-Poll, A., Ellis, M., and Gubler, F. [2002]. Mutants at the Slender1 locus of barley cv Himalaya: Molecular and physiological characterization. Plant Physiol. 129, 181–190) also address the role of SLN1 in GA signaling in barley.
Acknowledgments
We thank A.H. Christensen and P.H. Quail for providing pAHC18 and M.K. Walker-Simmons for providing Ubi1-TaPKABA1. Technical assistance from Mike Veith at the Microscopy Facility, Department of Biology, Washington University, was greatly appreciated. This work was supported by grants from the National Science Foundation (IBN-9983126) and Monsanto Co. (St. Louis, MO) to T.-H.D.H. Support for R.Z. by the Consejo Nacional de Ciencia y Tecnológia, Mexico, and for D.Y. from the Itoh Science Foundation, Japan, also are acknowledged.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.003376.
References
- Anderberg, R.J., and Walker-Simmons, M.K. (1992). Isolation of a wheat cDNA clone for an abscisic acid-inducible transcript with homology to protein kinases. Proc. Natl. Acad. Sci. USA 89, 10183–10187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke, P.C., Schuurink, R., and Jones, R.L. (1997). Hormonal signalling in cereal aleurone. J. Exp. Bot. 48, 1337–1356. [Google Scholar]
- Bethke, P.C., Swanson, S.J., Hillmer, S., and Jones, R.L. (1998). From storage compartment to lytic organelle: The metamorphosis of the aleurone protein storage vacuole. Ann. Bot. 82, 399–412. [Google Scholar]
- Bosher, J.M., and Labouesse, M. (2000). RNA interference: Genetic wand and genetic watchdog. Nat. Cell Biol. 2, E31–E36. [DOI] [PubMed] [Google Scholar]
- Cercós, M., Gómez-Cadenas, A., and Ho, T.-H.D. (1999). Hormonal regulation of a cysteine proteinase gene, EPB-1, in barley aleurone layers: Cis- and trans-acting elements involved in the co-ordinated gene expression regulated by gibberellins and abscisic acid. Plant J. 19, 107–118. [DOI] [PubMed] [Google Scholar]
- Chandler, P.M. (1988). Hormonal regulation of gene expression in the “slender” mutant of barley (Hordeum vulgare L.). Planta 175, 115–120. [DOI] [PubMed] [Google Scholar]
- Chrispeels, M., and Varner, J. (1966). Inhibition of gibberellic acid induced formation of alpha-amylase by abscisin II. Nature 212, 1066–1067. [Google Scholar]
- Christensen, A.H., and Quail, P.H. (1996). Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 5, 213–218. [DOI] [PubMed] [Google Scholar]
- Chuang, C.F., and Meyerowitz, E.M. (2000). Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97, 4985–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, S.J., and Vierstra, R.D. (1998). Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol. Biol. 36, 521–528. [DOI] [PubMed] [Google Scholar]
- Dean, C., and Schmidt, R. (1995). Plant genomes: A current molecular description. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 395–418. [Google Scholar]
- Dill, A., Jung, H.S., and Sun, T.P. (2001). The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc. Natl. Acad. Sci. USA 98, 14162–14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill, A., and Sun, T. (2001). Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159, 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Gilbert, L.I., Tata, J.R., and Atkinson, B.G. (1996). Metamorphosis: Postembryonic Reprogramming of Gene Expression in Amphibian and Insect Cells. (San Diego, CA: Academic Press).
- Gilroy, S. (1996). Signal transduction in barley aleurone protoplasts is calcium dependent and independent. Plant Cell 8, 2193–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Cadenas, A., Verhey, S.D., Holappa, L.D., Shen, Q., Ho, T.-H.D., and Walker-Simmons, M.K. (1999). An abscisic acid-induced protein kinase, PKABA1, mediates abscisic acid-suppressed gene expression in barley aleurone layers. Proc. Natl. Acad. Sci. USA 96, 1767–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Cadenas, A., Zentella, R., Walker-Simmons, M.K., and Ho, T.-H.D. (2001). Gibberellin/abscisic acid antagonism in barley aleurone cells: Site of action of the protein kinase PKABA1 in relation to gibberellin signaling molecules. Plant Cell 13, 667–679. [PMC free article] [PubMed] [Google Scholar]
- Gubler, F., Kalla, R., Roberts, J.K., and Jacobsen, J.V. (1995). Gibberellin-regulated expression of a myb gene in barley aleurone cells: Evidence for Myb transactivation of a high-pI α-amylase gene promoter. Plant Cell 7, 1879–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler, F., Raventos, D., Keys, M., Watts, R., Mundy, J., and Jacobsen, J.V. (1999). Target genes and regulatory domains of the GAMYB transcriptional activator in cereal aleurone. Plant J. 17, 1–9. [DOI] [PubMed] [Google Scholar]
- Holappa, L.D., and Walker-Simmons, M.K. (1997). The wheat protein kinase gene, TaPK3, of the PKABA1 subfamily is differentially regulated in greening wheat seedlings. Plant Mol. Biol. 33, 935–941. [DOI] [PubMed] [Google Scholar]
- Horvath, H., Huang, J., Wong, O., Kohl, E., Okita, T., Kannangara, C.G., and von Wettstein, D. (2000). The production of recombinant proteins in transgenic barley grains. Proc. Natl. Acad. Sci. USA 97, 1914–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttly, A.K., and Phillips, A.L. (1995). Gibberellin-regulated expression in oat aleurone cells of two kinases that show homology to MAP kinase and ribosomal protein kinase. Plant Mol. Biol. 27, 1043–1052. [DOI] [PubMed] [Google Scholar]
- Ikeda, A., Ueguchi-Tanaka, M., Sonoda, Y., Kitano, H., Koshioka, M., Futsuhara, Y., Matsuoka, M., and Yamaguchi, J. (2001). slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13, 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, T., Higuchi, M., Hashimoto, Y., Seki, M., Kobayashi, M., Kato, T., Tabata, S., Shinozaki, K., and Kakimoto, T. (2001). Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409, 1060–1063. [DOI] [PubMed] [Google Scholar]
- Itoh, H., Ueguchi-Tanaka, M., Sato, Y., Ashikari, M., and Matsuoka, M. (2002). The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14, 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen, J.V., Gubler, F., and Chandler, P.M. (1995). Gibberellin action in germinated cereal grains. In Plant Hormones: Physiology, Biochemistry, and Molecular Biology, P.J. Davies, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- Jones, H.D., Smith, S.J., Desikan, R., Plakidou-Dymock, S., Lovegrove, A., and Hooley, R. (1998). Heterotrimeric G proteins are implicated in gibberellin induction of α-amylase gene expression in wild oat aleurone. Plant Cell 10, 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, K.E., Moritz, T., and Harberd, N.P. (2001). Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159, 767–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., Jorna, M.L., Brinkhorst-van der Swan, D.L.C., and Karssen, C.M. (1982). The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana L. Heynh. Theor. Appl. Genet. 61, 385–393. [DOI] [PubMed] [Google Scholar]
- Koprek, T., Rangel, S., McElroy, D., Louwerse, J.D., Williams-Carrier, R.E., and Lemaux, P.G. (2001). Transposon-mediated single-copy gene delivery leads to increased transgene expression stability in barley. Plant Physiol. 125, 1354–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanahan, M.B., and Ho, T.-H.D. (1988). Slender barley: A constitutive gibberellin-response mutant. Planta 175, 107–114. [DOI] [PubMed] [Google Scholar]
- Lanahan, M.B., Ho, T.-H.D., Rogers, S.W., and Rogers, J.C. (1992). A gibberellin response complex in cereal α-amylase gene promoters. Plant Cell 4, 203–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum, C.M., Comai, L., Greene, E.A., and Henikoff, S. (2000). Targeting induced local lesions in genomes (TILLING) for plant functional genomics. Plant Physiol. 123, 439–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., Carol, P., Richards, D.E., King, K.E., Cowling, R.J., Murphy, G.P., and Harberd, N.P. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11, 3194–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, J., et al. (1999). ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400, 256–261. [DOI] [PubMed] [Google Scholar]
- Penson, S.P., Schuurink, R.C., Fath, A., Gubler, F., Jacobsen, J.V., and Jones, R.L. (1996). cGMP is required for gibberellic acid-induced gene expression in barley aleurone. Plant Cell 8, 2325–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pysh, L.D., Wysocka-Diller, J.W., Camilleri, C., Bouchez, D., and Benfey, P.N. (1999). The GRAS gene family in Arabidopsis: Sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 18, 111–119. [DOI] [PubMed] [Google Scholar]
- Ritchie, S., and Gilroy, S. (1998). Tansley Review No. 100. Gibberellins: Regulating genes and germination. New Phytol. 140, 363–383. [DOI] [PubMed] [Google Scholar]
- Rogers, J.C., and Rogers, S.W. (1992). Definition and functional implications of gibberellin and abscisic acid cis-acting hormone response complexes. Plant Cell 4, 1443–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz, S., Lorz, H., and Lutticke, S. (2001). Transposition of the maize transposable element Ac in barley (Hordeum vulgare L.). Mol. Gen. Genet. 264, 653–661. [DOI] [PubMed] [Google Scholar]
- Schweizer, P., Pokorny, J., Schulze-Lefert, P., and Dudler, R. (2000). Technical Advance. Double-stranded RNA interferes with gene function at the single-cell level in cereals. Plant J. 24, 895–903. [DOI] [PubMed] [Google Scholar]
- Sharp, P.A. (2001). RNA interference-2001. Genes Dev. 15, 485–490. [DOI] [PubMed] [Google Scholar]
- Shen, Q., Uknes, S.J., and Ho, T.-H.D. (1993). Hormone response complex of a novel abscisic acid and cycloheximide inducible barley gene. J. Biol. Chem. 268, 23652–23660. [PubMed] [Google Scholar]
- Shen, Q., Zhang, P., and Ho, T.-H.D. (1996). Modular nature of abscisic acid (ABA) response complexes: Composite promoter units that are necessary and sufficient for ABA induction of gene expression in barley. Plant Cell 8, 1107–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone, A.L., Ciampaglio, C.N., and Sun, T. (1998). The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10, 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone, A.L., and Sun, T. (2000). Gibberellins and the green revolution. Trends Plant Sci. 5, 1–2. [DOI] [PubMed] [Google Scholar]
- Tamas, I.A. (1995). Hormonal regulation of apical dominance. In Plant Hormones: Physiology, Biochemistry, and Molecular Biology, P.J. Davies, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 572–597.
- Ueguchi-Tanaka, M., Fujisawa, Y., Kobayashi, M., Ashikari, M., Iwasaki, Y., Kitano, H., and Matsuoka, M. (2000). Rice dwarf mutant d1, which is defective in the alpha subunit of the heterotrimeric G protein, affects gibberellin signal transduction. Proc. Natl. Acad. Sci. USA 97, 11638–11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi, D., Zentella, R., and Ho, T.-H.D. (2002). Molecular analysis of the barley (Hordeum vulgare L.) gene encoding the protein kinase PKABA1 capable of suppressing gibberellin action in aleurone layers. Planta, 215, 319–326. [DOI] [PubMed] [Google Scholar]
- Yomo, H., and Varner, J.E. (1971). Hormonal control of a secretory tissue. In Current Topics in Developmental Biology, A.A. Moscona and A. Monroy, eds (New York: Academic Press), pp. 111–144. [DOI] [PubMed]