Abstract
Caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase (COMT) from alfalfa is an S-adenosyl-l-Met–dependent O-methyltransferase involved in lignin biosynthesis. COMT methylates caffeoyl- and 5-hydroxyferuloyl–containing acids, aldehydes, and alcohols in vitro while displaying a kinetic preference for the alcohols and aldehydes over the free acids. The 2.2-Å crystal structure of COMT in complex with S-adenosyl-l-homocysteine (SAH) and ferulic acid (ferulate form), as well as the 2.4-Å crystal structure of COMT in complex with SAH and 5-hydroxyconiferaldehyde, provide a structural understanding of the observed substrate preferences. These crystal structures identify residues lining the active site surface that contact the substrates. Structurally guided site-directed mutagenesis of active site residues was performed with the goal of altering the kinetic preferences for physiological substrates. The kinetic parameters of the COMT mutants versus wild-type enzyme are presented, and coupled with the high-resolution crystal structures, they will serve as a starting point for the in vivo manipulation of lignin monomers in transgenic plants. Ultimately, this structurally based approach to metabolic engineering will allow the further alteration of the lignin biosynthetic pathway in agronomically important plants. This approach will lead to a better understanding of the in vivo operation of the potential metabolic grid for monolignol biosynthesis.
INTRODUCTION
With the advent of large-scale genomic sequencing, the necessity of gene product characterization increases concomitantly. Structural biology provides an important tool for the detailed characterization of proteins at the atomic level. This molecular approach can lead to a more complete understanding not only of a single enzymatic reaction, for example, but also helps characterize complex biosynthetic pathways by elucidating the mechanisms of individual biosynthetic reactions. The biosynthetic pathway resulting in distinct lignin monomers of varying O-methylation patterns provides a useful example of the utility of this architectural information. This complex metabolic grid involves multiple methylation reactions of polyhydroxylated phenylpropanoids. High-resolution x-ray crystal structures of a methyltransferase enzyme integral to this pathway provide a more complete picture of monolignol biosynthesis and offer a useful template for predictably altering substrate specificity and turnover at several critical steps in the lignin metabolic grid.
Lignin is a principal structural component of cell walls in higher terrestrial plants and, after cellulose, the second most abundant plant polymer. The lignin polymer is composed of phenylpropane units oxidatively coupled through ether and carbon-carbon linkages. This natural polymer can function as a genetically inducible physical barrier in response to microbial attack (Lawton and Lamb, 1987; Jaeck et al., 1992; Ni et al., 1996; Baldridge et al., 1998; Hatfield and Vermerris, 2001). In addition to structural support and pathogen defense, lignin functions in water transport as a hydrophobic constituent of vascular phloem and xylem cells (Ros Barcelo, 1997; Inoue et al., 1998).
In angiosperms, lignin is composed of two major monomeric phenolic constituents: guaiacyl (G) and syringyl (S) units. The G unit is singly methylated on the 3-hydroxyl group, whereas the S subunit is methylated on both the 3- and 5-hydroxyl moieties. The ratio of S-to-G subunits dictates the degree of lignin condensation by allowing for different types of polymeric linkages. Increased G content leads to more highly condensed lignin composed of a greater proportion of biphenyl and other carbon-carbon linkages, whereas S subunits are commonly linked through more labile ether bonds at the 4-hydroxyl position (Dixon et al., 1996; Li et al., 2000; Guo et al., 2001).
The type and extent of lignin polymerization has profound industrial, environmental, and agricultural consequences. Industrial processes such as Kraft pulping for wood processing necessitate the delignification of cellulose fibers. Procedurally, the presence of more chemically labile S lignin is desirable to reduce the monetary and environmental impact of wood processing (Lapierre et al., 1999; Jouanin et al., 2000). Agriculturally, lignin affects the digestibility of important forage crops such as alfalfa (Inoue et al., 1998; Pincon et al., 2001). As plants mature, their lignin content increases, particularly S subunits. Unlike their reactivity during chemical delignification, S subunits may negatively affect the digestibility and ultimately the nutrient availability of alfalfa and other forage grasses (Pond et al., 1987).
The final lignin polymer composition and the extent of cross-linking are determined by the hydroxylation and methylation state of the individual lignin monomers. The biosynthetic route to these monolignols is not understood clearly. A number of possible hydroxylated substrates exist, and the in vivo flux of these substrates through the lignin pathway needs to be clarified. The structure determinations of methyltransferase enzymes, such as caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase (COMT), provide a structural basis for understanding substrate discrimination and enable the introduction of targeted mutations into COMT, allowing for the modulation of methyltransferase activity and lignin monomer methylation.
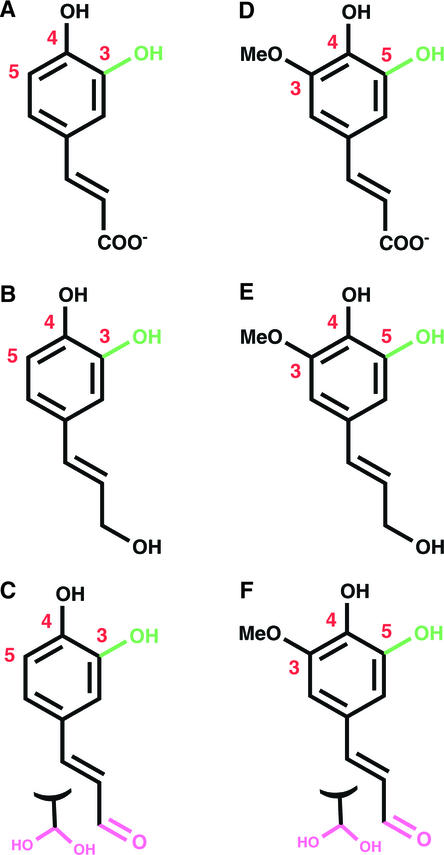
COMT from alfalfa is an S-adenosyl-l-Met (SAM)–dependent methyltransferase that methylates 3-hydroxyl– and 5-hydroxyl–containing phenylpropanoid-derived lignin precursors (Edwards and Dixon, 1991). In vitro kinetic analysis of COMT demonstrates methylation of caffeate, 5-hydroxyferulate, caffeoyl aldehyde, caffeoyl alcohol, 5-hydroxyconiferaldehyde, and 5-hydroxyconiferyl alcohol (Figure 1), yielding the corresponding methylated phenyl ethers and S-adenosyl-l-homocysteine (SAH) as products. Whereas the in vivo preferences of COMT toward these substrates is not fully defined, recent evidence suggests that the enzyme exhibits a kinetic preference for the alcohols and aldehydes over the free acids (Osakabe et al., 1999; Li et al., 2000; Parvathi et al., 2001). The 2.2- to 2.4-Å crystal structures of COMT in complex with the products SAH and ferulic acid in the form of deprotonated ferulate, as well as the product SAH and the substrate 5-hydroxyconiferaldehyde, afford a mechanistic understanding of kinetic discrimination between chemically similar monomeric lignin precursors.
Figure 1.
Phenylpropanoid-Derived Substrates of COMT.
The hydroxyl site that is methylated is shown in green. The 3, 4, and 5 positions of the phenyl ring are numbered. Aldehydes (pink) are shown as hydrates that likely form in aqueous solutions.
(A) Caffeate.
(B) Caffeoyl alcohol.
(C) Caffeoyl aldehyde.
(D) 5-Hydroxyferulate.
(E) 5-Hydroxyconiferyl alcohol.
(F) 5-Hydroxyconiferaldehyde.
RESULTS
Structural Elucidation by Protein X-Ray Crystallography
Recombinant COMT was expressed in Escherichia coli as an N-terminal polyhistidine-tagged protein and purified by nickel–nitrilotriacetic acid agarose affinity and gel filtration chromatography (Zubieta et al., 2001). Recombinant COMT possesses activity comparable to that of the native enzyme when assayed with SAM and caffeate, 5-hydroxyferulate, alcohols, and aldehydes (Parvathi et al., 2001). COMT was crystallized from polyethylene glycol solutions containing a 2-fold molar excess of the reaction product SAH (2 mM) and a 1.5-fold molar excess of ferulic acid or 5-hydroxyconiferaldehyde (1.5 mM). Initial phasing was accomplished with selenomethionine (SeMet)-substituted protein crystallized in the presence of SAM, and a partial model was built using these phases. The SeMet-substituted crystal was highly disordered in that two of the three monomers in the asymmetric unit were poorly defined. Therefore, the SeMet structure was never refined fully. However, the partial model derived from this phase set provided an adequate search model for performing molecular replacement of COMT complexed with ferulic acid and 5-hydroxyconiferaldehyde (Table 1).
Table 1.
Crystallographic Data for the COMT SeMet-Substituted Protein and the Ferulic Acid/SAH and 5-Hydroxyconiferaldehyde/SAH Complexes
| λ1 | λ2 | λ3 | FAa | 5-OH CALDb | |
|---|---|---|---|---|---|
| Wavelength (Å) | 0.9746 | 0.9785 | 0.9797 | 1.08 | 1.08 |
| Resolution (Å) | 50 to 2.2 a = 103.8 |
50 to 2.2 a = 103.8 |
50 to 2.2 a = 103.8 |
99 to 2.1 a = 104.6 |
90 to 2.4 a = 104.5 |
| Cell dimensions (a, b, and c = Å, β = °) |
b = 61.6 c = 110.4 β = 112.4 |
b = 61.6 c = 110.4 β = 112.4 |
b = 61.6 c = 110.4 β = 112.4 |
b = 61.9 c = 112.2 β = 111.3 |
b = 62.0 c = 112.2 β = 111.3 |
| Total observations | 124,360 | 124,421 | 124,700 | 388,692 | 207,847 |
| Unique reflections | 65,808 | 65,822 | 65,953 | 88,232 | 53,959 |
| Completenessc | 97 (97) | 97 (97) | 97 (97) | 98 (99) | 89 (43) |
| Rsymc,d | 3.2 (31) | 3.3 (33) | 3.3 (29) | 4.9 (51) | 4.7 (42) |
| No. of selenium sites | 14 | 14 | 14 | – | – |
| PPisoe (acentric/centric) | – | 3.0/2.2 | 2.2/1.6 | – | – |
| PPanoe | 2.2 | 2.7 | 2.4 | – | – |
| No. of water molecules | – | – | – | 264 | 225 |
| Rcrystf | – | – | – | 23.6 | 20.5 |
| Rfreeg | – | – | – | 27.4 | 25.9 |
| R.m.s.d.h bonds (Å) | – | – | – | 0.006 | 0.009 |
| R.m.s.d. angles (°) | – | – | – | 1.2 | 1.4 |
Ferulic acid.
5-Hydroxyconiferaldehyde.
Numbers in parentheses refer to the highest shell.
Rsym=Σ|Ih − <Ih>|/ΣIh, where <Ih> is the average intensity over symmetry equivalent reflections.
Phasing power = <|FH(calc)|/|E|, where |FH(calc)| is the calculated difference and E is the lack of closure.
Rcryst = Σ|Fobs − Fcalc|/ΣFobs, where summation is over the data used for refinement.
Rfree was calculated as for Rcryst using 5% of the data that was excluded from refinement.
Root mean square deviations.
Three-Dimensional Architecture
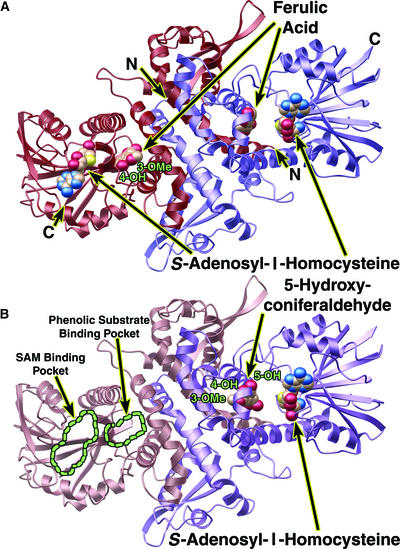
The tertiary and quaternary structures of COMT are similar to those of previously characterized plant phenolic O-methyltransferases from alfalfa (Zubieta et al., 2001). The 43-kD protein forms a dimer in solution, and this dimeric arrangement of the enzyme is critical for its activity. The dimer excludes solvent from the active site and is necessary for the formation of the substrate binding cavity. Furthermore, gel filtration and crystallographic studies provide no evidence for monomeric forms of COMT. The COMT structure consists of a large C-terminal catalytic domain involved in SAM/SAH and phenolic substrate binding and a smaller N-terminal domain involved primarily in mediating dimer formation (Figures 2A and 2B).
Figure 2.
Ribbon Diagrams of the COMT Dimer Protein Backbone.
Ferulic acid (ferulate), 5-hydroxyconiferaldehyde, and SAH are shown as color-coded van der Waals spheres in which carbon is beige, nitrogen is blue, oxygen is red, and sulfur is yellow. Ribbon diagrams were produced with MOLSCRIPT (Kraulis, 1991) and rendered with POV-RAY (http://www.povray.org).
(A) Ribbon diagram of the COMT dimer in complex with ferulic acid (ferulate) and SAH. One monomer is shown in red (left), and the other monomer is shown in purple (right). The N and C termini are labeled N and C, respectively. The 3-methoxy and 4-hydroxyl sites of ferulate bound to the red monomer (left) are labeled.
(B) Ribbon diagram of the COMT dimer in complex with 5-hydroxyconiferaldehyde and SAH. One monomer is shown in rose (left), and the other monomer is shown in purple (right). The rose monomer lacked noticeable electron density for SAM/SAH or a phenylpropanoid substrate. The SAM/SAH and the phenolic substrate binding sites are outlined in green. The 5-hydroxyconiferaldehyde molecule bound to the purple monomer appears to be hydrated; thus, it was interpreted and modeled as a diol.
Each asymmetric unit in the crystals of COMT contains three polypeptide chains. Two of these chains organize as a homodimer around a noncrystallographic twofold axis, and the third polypeptide chain forms an identical homodimer with a neighboring polypeptide chain in a second asymmetric unit by organizing around a crystallographic twofold axis. All of the COMT molecules in the ferulic acid (ferulate) complexes contain SAH and ferulic acid likely in the ferulate form. The 5-hydroxyconiferaldehyde complex exhibited electron density for SAH in two of the monomers in the asymmetric unit and reliable density affording modeling and refinement for 5-hydroxyconiferaldehyde in only one of the three monomers. Low root mean square deviations (<0.5 Å2) between the backbone atoms of all three monomers in the asymmetric unit imply a fairly preorganized active site cavity and therefore do not suggest that a dramatic conformational rearrangement accompanies substrate binding.
The catalytic C-terminal domain consists of a core α/β Rossmann fold commonly found in nucleotide binding proteins (Rossmann et al., 1974). This extensive β-sheet motif is shared by all structurally characterized SAM-dependent methyltransferases. Phenolic substrate binding also occurs in the C-terminal domain and is mediated by the presence of the partner monomer, which helps enclose the active site and creates a more solvent-excluded region for the sequestration of phenylpropanoid-derived substrates.
The N-terminal domain facilitates dimerization through electrostatic interactions and complementary van der Waals interactions between opposing α-helices of each monomer. These interactions form a broad dimer interface, which buries ∼30% of the total surface area of the dimer. The N-terminal domain also contributes to the formation of the active site by providing residues that line the back wall of the substrate binding cavity and help enclose the recognition surface. Specifically, Ser-28 protrudes into the active site of its partner monomer, helping to exclude solvent and form the propanoid tail binding pocket.
SAM/SAH Binding Pocket
Recognition and sequestration of SAM/SAH is highly conserved in all SAM-dependent methyltransferases, and the residues involved form a readily recognizable signature motif (Ibrahim et al., 1998; Joshi and Chiang, 1998). Extensive hydrogen bonding interactions sequester the cosubstrate methyl donor and position the methyl group near hydroxylated acceptor moieties for transmethylation (Figures 3A and 3B). The carboxylate group of SAH forms an electrostatic interaction with Lys-265, and the terminal amino group participates in a hydrogen bonding network with Asp-206 and a water molecule. The ribose hydroxyls are tethered by hydrogen bonds to Asp-231, whereas the exocyclic amino group of the adenine ring forms a hydrogen bond with Asp-251. As seen in sequence alignments of representative methyltransferases, the hydrogen bonding partners of SAM/SAH are shared universally (Figure 4).
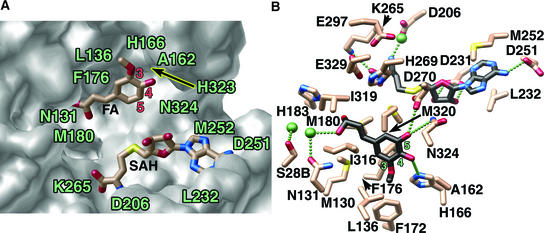
Figure 3.
Close-Up Views of the COMT Active Site.
(A) Surface representation of the active site cavity for the COMT–SAH–ferulate (FA) complex illustrating the complementary shape and size to ferulate and SAH. The accessible surface was calculated with GRASP (Nicholls et al., 1991) and rendered with POV-RAY (http://www.povray.org). SAH and ferulate are depicted as half-colored bonds and are color coded as in Figure 2.
(B) Active site arrangement for the COMT–SAH–5-hydroxyconiferaldehyde complex. Bonds of the COMT active site residues are color coded as in Figure 2. Carbon atoms of SAH and 5-hydroxyconiferaldehyde are shown in black to distinguish them from the active site residues. Water molecules are shown as green spheres, and putative hydrogen bonds are shown as a linear series of small green spheres. This image was produced with MOLSCRIPT (Kraulis, 1991) and rendered with POV-RAY (http://www.povray.org).
Figure 4.
Sequence Alignment of Four COMT Enzymes and Three Representative Plant O-Methyltransferases.
Shown are primary sequences of COMT from alfalfa, COMT from Clarkia (fairy fans), COMT from Pinus pinaster (pine), COMT from Chrysosplenium americanum (saxifrage), IEMT from Clarkia (fairy fans), chalcone O-methyltransferase (ChOMT; Zubieta et al., 2001) from alfalfa, and isoflavone O-methyltransferase (IOMT; Zubieta et al., 2001) from alfalfa.
The aromatic adenine ring is sequestered by a number of hydrophobic residues, with Met-252 and Leu-232 bracketing the ring and Trp-271 and Phe-253 (data not shown) involved in favorable edge-to-face interactions. Although the hydrogen bonding partners of SAM/SAH are highly conserved, the hydrophobic and aromatic residues interacting with SAM/SAH vary among methyltransferases, likely the result of several energetically favorable sets of van der Waals interactions with the adenine ring.
Lignin Monomer Binding Interactions
Ferulic acid (ferulate) and 5-hydroxyconiferaldehyde bind in a similar manner to the active sites in the two complexes characterized crystallographically (Figures 3A and 3B). In both cases, the hydrophobic residues Phe-176, Met-130, Met-320, and Met-180 sequester the phenyl ring that presents the reactive hydroxyl group to SAM. Hydrogen bonding of His-166 with the 3-methoxy group of ferulic acid helps orient the substrate for preferential methylation of the 5-hydroxyl position (Figure 3A). A slightly altered substrate orientation is observed for binding of 5-hydroxyconiferaldehyde. His-166 forms a hydrogen bond with the hydroxyl moiety at the 4 position of the substrate, and additional hydrogen bonds form between Asp-270 and Asn-324 and the 5-hydroxyl group (Figure 3B). The orientation observed for 5-hydroxyconiferaldehyde presents the 5-hydroxyl moiety for methylation. Moreover, the methoxy groups of both ferulic acid and 5-hydroxyconiferaldehyde reside in a complementary hydrophobic methyl binding pocket consisting of Leu-136, Phe-172, Phe-176, and Ala-162. This conserved pattern of favorable van der Waals interactions most likely explains the kinetic preferences of COMT for the 3-methoxy-4,5-dihydroxyl–substituted substrates over the 3,4-dihydroxyl–substituted substrates.
The propanoid tails of the potential COMT substrates include carboxylate, aldehyde, and alcohol functionalities. To accept different substrates, the active site must be spacious and versatile enough to allow the binding of these different molecules. The environment for binding the propanoid terminus consists of two water molecules and the hydrophilic residues His-183 and Asn-131. The water molecules mediate binding interactions between His-183, Asn-131, and the hydrophilic tail of the substrates. The same hydrogen bonding pattern and position of the substrate tail were observed for the aldehyde- and carboxylate-containing substrates. In addition to the hydrophilic residues, the propanoid tail is surrounded by a number of hydrophobic groups, including Met-180, Met-130, Ile-316, and Ile-319 (Figures 3A and 3B). The structure and relative hydrophobicity of this propanoid tail binding pocket suggests that selectivity for neutral aldehydes and alcohols over negatively charged carboxylate groups is conferred by this region of the COMT active site.
Catalytic Mechanism
The catalytic mechanism for COMT is comparable to that of previously characterized plant O-methyltransferases (Zubieta et al., 2001) and PRMT3, a protein Arg N-methyltransferase from rat (Zhang et al., 2000), all of which use a catalytic His as a general base. In COMT, the 3- or 5-hydroxyl group is deprotonated by His-269, facilitating the transfer of the reactive methyl group of SAM to the newly formed phenolate anion. This general base is held in a catalytically productive position by hydrogen bonds with Glu-329. Glu-297 and Asp-270 are adjacent to the His and most likely contribute to the orientation of His-269 and the pKa of the neighboring hydroxyl moiety poised for transmethylation. As was the case for chalcone O-methyltransferase and isoflavone O-methyltransferase (Zubieta et al., 2001), mutations of His-269 in COMT to Leu, Asn, and Gln result in enzymes that lack methyltransferase activity, further supporting the role of His-269 as the essential catalytic base (data not shown).
Interestingly, unlike the previously characterized plant O-methyltransferases, the positions of phenol-containing substrates in the COMT active site near His-269 and SAM exhibit many interatomic distances larger then those observed previously (Zubieta et al., 2001). This more spacious phenylpropanoid binding architecture supports the observed activity of COMT toward several different phenol-containing substrates. The local environment around the reactive hydroxyl group depends less on precise positioning than on the general chemical nature of the active site surface. In turn, this rather spacious active site environment affords a level of substrate promiscuity that may be important for in vivo biological activity.
Structurally Guided Alteration of COMT's Kinetic Discrimination among Putative Lignin Monomers
To help determine the structural and catalytic roles of residues lining the active site and their relative importance in kinetic discrimination among potential substrates, a series of mutations were designed, expressed, and kinetically evaluated based on the structures of the COMT complexes described above (Figures 5A and 5B). Wild-type COMT exhibited a twofold preference (Vmax/Km) for 3- and 5-substituted phenylpropanoids over the equivalent compounds substituted at the 3 position alone. Additionally, the aldehydes were the preferred substrates over the alcohols and free acids. Site-directed mutations were introduced to modulate this in vitro selectivity, with potentially similar results for in vivo activity, and to determine which residues contributed most strongly to substrate discrimination.
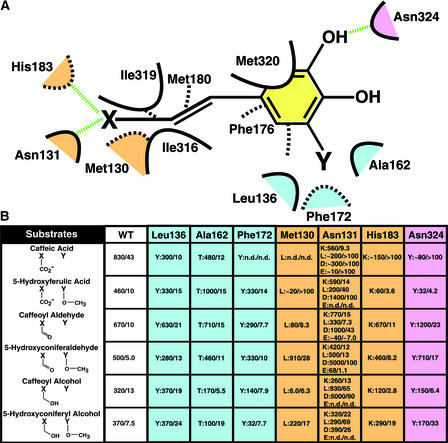
Figure 5.
Scheme and Mutagenesis Study of the Lignin Monomer Binding Site of COMT.
(A) Scheme of the lignin monomer binding site of COMT. van der Waals interactions are shown as dashed or solid curves. Hydrogen bonds are shown as dashed green lines. The spatial orientation of residues is approximate, with solid curves representing residues in the foreground and dashed curves representing residues in the background. Residues that are color coded as light blue (methoxy binding pocket), pink (hydroxyl group binding), and beige (propanoid tail binding region) constitute COMT residues selected for mutagenesis and further kinetic analysis. X represents a carboxylate moiety, a hydroxyl group, or an aldehyde moiety. Y signifies a methoxy group or the absence of any functional moiety on the phenyl ring of the putative COMT substrates.
(B) Kinetic analysis of a series of wild-type (WT) and mutant forms of COMT. The positions chosen for mutagenic replacement are color coded and correspond to the color scheme in (A). The mutations are given as single-letter amino acid codes. Each mutant is shown as mutation:Vmax/Km. All assays were performed in quadruplicate. Vmax values are given as pkat/mg COMT, and Km values are expressed in μM. n.d., no activity determined.
Although the in vivo significance of the steady state kinetic parameters shown in Figure 5B is being evaluated at present, a subset of COMT mutants provides a tantalizing glimpse of future utility in plants. For instance, F172Y displayed a complete loss of caffeate binding and turnover, a severe loss in 5-hydroxyconiferyl alcohol turnover, but little kinetic effect on the remaining lignin monomers. The severe decrease in the Vmax for the 5-hydroxyconiferyl alcohol substrate coupled with little change in Km is attributable most likely to an increased affinity for the product, mediated by the formation of a new hydrogen bond between the side chain hydroxyl moiety of Tyr-172 and the methoxy group of the sinapyl alcohol product. Subtle binding differences between the alcohol/acid propanoid tails and the aldehyde tail containing substrates most likely moderate this discriminatory kinetic effect when using the 5-hydroxyconiferyl alcohol substrate.
L136Y showed little substrate discrimination with a moderate level of caffeate specificity. M130L displayed a complete loss of caffeate binding and turnover with strong selectivity in terms of both Km and Vmax for 5-hydroxyconiferaldehyde and 5-hydroxyconiferyl alcohol. A162T, like the aforementioned L136Y mutant, showed a loss of kinetic discrimination among all known COMT substrates. N131K behaved kinetically like L136Y and A162T, but unlike these mutants, it presented a mutant side chain near the propanoid terminus. Finally, although N131D displayed statistically higher turnover against several putative COMT sub-strates, this mutant exhibited an across-the-board increase in Km for all tested substrates. These kinetic properties may prove essential in properly evaluating the physiological significance of in vitro steady state kinetic profiles. Given the possible existence of substrate channeling in loosely associated multienzyme complexes, steady state kinetic parameters such as Vmax and Km may not reflect the local conditions that affect turnover in plant cells as metabolic flux is directed down highly bifurcating metabolic pathways. In addition, further structural studies of these and other mutants with a complementary set of substrates and/or products bound will provide additional information regarding the apparent plasticity exhibited by COMT during substrate recognition and turnover.
DISCUSSION
Compared with isoflavone O-methyltransferase and chalcone O-methyltransferase (Zubieta et al., 2001), the active site of COMT is more spacious and allows the facile accommodation of 3- and 5-substituted phenolic substrates (Figures 3A and 3B). As shown by the broader range of acceptable COMT substrates, the size of the active site affords turnover of a series of monomeric lignin precursors. In both complexes described here, the substrate hydroxyl group resides 7 to 9 Å away from the donor methyl moiety of SAM based on the position of SAH. Because of the tight constraints on SAH/SAM binding conferred by extensive hydrogen bonding and van der Waals interactions, it is probable that the phenolic substrates, and not SAM, undergo dynamic shifts as transmethylation proceeds. The largely hydrophobic region of the active site, which binds the phenyl portion of the substrate through a set of shape-complementary sandwiching interactions, allows a high degree of substrate sliding.
Unlike previous structurally characterized O-methyltransferases, which demonstrated a high degree of substrate specificity using an extensive repertoire of hydrophilic and hydrophobic binding interactions, COMT exhibits broader substrate tolerances and relatively few hydrogen bonding interactions. The majority of phenolic substrate–protein interactions appear to include extensive van der Waals forces that conform to the general shape of the phenylpropanoid skeleton. Adaptive processes have given rise to a COMT active site architecture in which substrate molecules access a number of energetically similar binding arrangements. These structures provide visual clarity for at least two slightly different binding modes for ferulic acid (ferulate) and 5-hydroxyconiferaldehyde. With the addition of the cosubstrate SAM and its positively charged sulfonium center, as opposed to the demethylated and uncharged product SAH, additional substrate movement is probable as the phenoxide anion positions itself near SAM.
In vitro studies have shown that COMTs from a number of different plant species are positionally selective for hydroxyl groups at the 3- and 5-hydroxyl positions around the phenyl ring, with preference for the 5-hydroxyl moiety in 3-methoxy–containing substrates (Bugos et al., 1992; Inoue et al., 2000; Dixon et al., 2001; Parvathi et al., 2001). Putative hydrogen bonding partners, specifically Asp-270, Asn-324, and His-166, which can interact with hydroxyl groups or methoxy moieties, and the COMT methoxy binding pocket, specifically Leu-136, Phe-172, Phe-176, and Ala-162, most likely are responsible for the demonstrated preference of COMT for 5-hydroxyconiferaldehyde over caffeoyl aldehyde, 5-hydroxyconiferyl alcohol over caffeoyl alcohol, and 5-hydroxyferulate over caffeate. Hydrogen bonding of the 5-hydroxyl group sequesters the substrate in close proximity to the catalytic machinery of the enzyme. Thus, the 5-hydroxyl moiety is in a more favorable conformation for deprotonation and eventual nucleophilic attack on the reactive methyl group of SAM.
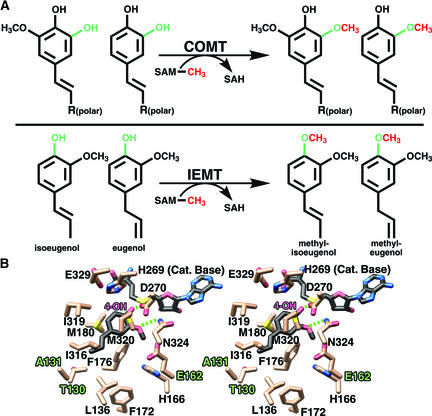
The propanoid tail binding region of the active site functions to prevent methylation at the 4-hydroxyl position. Moreover, this region confers some degree of substrate specificity in the order aldehydes > alcohols > acids. Previous studies have shown that mutations in the propanoid tail recognition site were sufficient to convert COMT from Clarkia breweri to an (iso)eugenol O-methyltransferase (IEMT) (Wang and Pichersky, 1999). IEMT methylates at the 4-hydroxyl (para) position of eugenol and isoeugenol to form methyl-eugenol and methyl-isoeugenol, respectively, whereas COMT is specific for methylation at the 3- and 5-hydroxyl positions of caffeoyl and 5-hydroxyferuloyl-containing substrates (Figure 6A). Based on sequence alignments of IEMT and COMT from Clarkia and the three-dimensional structure of COMT from alfalfa (Figure 4), the high conservation of residues lining the active site between the two COMTs implies a similar active site topology. Mutations within the propanoid tail binding pocket of Clarkia COMT conferred the most significant degree of substrate specificity for the allyl versus the free acid propanoid tail and ultimately influenced the ability of the mutant Clarkia COMT enzyme to methylate at the 4-hydroxyl position.
Figure 6.
Modeling of the Putative IEMT Active Site and the Molecular Basis for Substrate Discrimination and 4-Hydroxyl Methylation.
(A) Comparison of the COMT- and IEMT-catalyzed reactions. Hydroxyl groups subject to SAM-mediated transmethylation are shown in green, and the transferred methyl group is shown in red.
(B) Stereo view of the putative (iso)eugenol binding site of IEMT based on the x-ray crystal structure of alfalfa COMT. Bonds are colored by atom type, with dashed lines representing possible hydrogen bonds. Single-letter amino acid designations are used. Eugenol and SAH carbons are shown in black, and protein carbon atoms are shown in beige. The numbering scheme corresponds to that of alfalfa COMT used in the course of this structural and kinetic study. Three key amino acid differences in the phenylpropanoid binding pocket of IEMT likely result in repositioning of the propanoid tail of (iso)eugenol, resulting in the methylation of the 4-hydroxyl moiety rather then the 3- or 5-hydroxyl group. Met-130 is replaced by Thr, Asn-131 is replaced by Ala, and Ala-162 is replaced by Glu in IEMT. This image was produced with MOLSCRIPT (Kraulis, 1991) and rendered with POV-RAY (http://www.povray.org).
Modeling studies of IEMT based on the structure of COMT explain these results and further clarify the role of active site residues in substrate binding. The shorter hydrophobic tails of eugenol and isoeugenol cannot form hydrogen bonds with residues in IEMT. The M130T and N131A mutants of COMT enlarge the propanoid tail binding pocket and allow the allylic propanoid tail of eugenol and isoeugenol to shift positions, with a corresponding reorientation of their respective phenolic rings. Additional mutations near the phenolic ring further allow eugenol and isoeugenol to adopt conformations more amenable to 4-hydroxyl methylation. Specifically, the Ala-162 change to Glu (Figure 4) most likely assists in the repositioning of the phenolic ring closer to the SAM molecule and in the positioning of the 4-hydroxyl moiety for transmethylation (Figure 6B).
In native COMT, the necessity of accommodating the hydrophilic tail of the free acid, aldehyde, or alcohol substrates in a more hydrophilic environment, as well as the addition of hydrogen bonding interactions and steric bulk, helps prevent repositioning of the substrate and thus precludes access to the 4-hydroxyl group for methylation. The structural roots of this transformation are evident from the crystal structures presented here. Although the active site of alfalfa COMT, and most likely the majority of other COMTs, is rather promiscuous and able to accommodate a variety of substrates, methylation is regiospecific for the 3- and 5-hydroxyl positions as a result of anchoring of the hydrophilic propanoid tail in a complementary binding pocket within the active site.
Finally, the mutagenesis studies provide a starting point for the targeted alteration of lignin monomer selectivity in the lignin biosynthetic pathway. Manipulation of enzymes such as COMT will help modulate flux through the lignin biosynthetic pathway and thus modify the type and extent of lignin cross-links. Furthermore, structural studies of COMT facilitate the production of active site–directed mutant libraries. The limitations of generating point mutants and measuring their effects can be overcome by the structurally guided production of a more broad-based mutant library. Coupled with both in vitro and in vivo screening, this rather straightforward approach should provide important information regarding the control of lignin biosynthesis. As seen in the kinetic constants measured, structurally guided mutations affect enzymatic efficiency and substrate selectivity.
These studies highlight the importance of unequivocal identification of residues lining the active site cavity and suggest additional residues for inclusion in more diverse mutant libraries. Moreover, selected point mutants with very different kinetic profiles and selectivities versus wild-type COMT (summarized in Figure 5B) are being examined in vivo using plants downregulated for COMT activity. These studies provide a novel approach for the modification of lignin content and composition, both of which are important factors that affect improved approaches for modulating forage digestibility and paper pulping. Furthermore, these types of structure/function analyses offer a direct means of assessing the extent to which the biosynthesis of monolignols in vivo uses the full “metabolic grid potential” suggested by the promiscuous substrate specificity of COMT in vitro.
METHODS
Materials
The pET-15b expression vector and the Escherichia coli strain BL21 (DE3) were purchased from Novagen (Madison, WI). Nickel–nitrilotriacetic acid agarose (Ni2+-NTA) resin was purchased from Qiagen (Valencia, CA). Benzamidine-Sepharose and Superdex-200 fast protein liquid chromatography columns were obtained from Amersham Biosciences (Piscataway, NJ). Selenomethionine was obtained from Sigma (St. Louis, MO). Cryoloops for crystal freezing were purchased from Hampton Research (Laguna Niguel, CA). Caffeoyl and 5-hydroxyconiferaldehydes and alcohols were synthesized as described previously (Chen et al., 2001).
Caffeic Acid/5-Hydroxyferulic Acid 3/5-O-Methyltransferase Expression, Purification, and Mutagenesis
Alfalfa (Medicago sativa) caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase (COMT) was subcloned into the E. coli expression vector pET-15b (Inoue et al., 1998). The COMT construct was transformed into E. coli BL21 (DE3). Transformed E. coli were grown at 37°C in terrific broth containing 100 μg/mL ampicillin until A600 reached a value of 1.0. After induction with 0.5 mM isopropyl 1-thio-β-galactopyranoside, the cultures were grown for 6 h at 25°C. Cells were pelleted, harvested, and resuspended in lysis buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 20 mM imidazole, pH 8.0, 20 mM β-mercaptoethanol, 10% [v/v] glycerol, and 1% [v/v] Tween 20). After sonication and centrifugation, the supernatant was passed over a Ni2+-NTA column, washed with 10 bed volumes of lysis buffer and 10 bed volumes of wash buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 20 mM imidazole, pH 8.0, 20 mM β-mercaptoethanol, and 10% [v/v] glycerol), and the His-tagged protein was eluted with elution buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 250 mM imidazole, pH 8.0, 20 mM β-mercaptoethanol, and 10% [v/v] glycerol).
Incubation with thrombin during dialysis for 24 h at 4°C against 25 mM HEPES-Na+, pH 7.5, 100 mM NaCl, and 1 mM DTT removed the N-terminal His tag. Dialyzed protein was reloaded onto a Ni2+-NTA column to remove the cleaved His tag followed by thrombin depletion using a benzamidine-Sepharose column. Gel filtration chromatography on a Superdex-200 column equilibrated with 25 mM HEPES-Na+, pH 7.5, 100 mM NaCl, and 1 mM DTT afforded purified protein of >99% homogeneity. Fractions containing the protein of interest were pooled and concentrated to ∼25 mg/mL and stored at −80°C. Selenomethionine (SeMet)-substituted protein was obtained from E. coli grown in minimal medium with appropriate amino acids and added SeMet (Doublie, 1997). Expression and purification steps were performed as described above. Site-directed mutants were generated using the QuikChange protocol (Stratagene, La Jolla, CA) and purified from E. coli cultures as described previously.
Kinetic Analysis
COMT activities were assayed as described previously (Gowri et al., 1991; Ni et al., 1996; Inoue et al., 1998, 2000). For the determination of Vmax and apparent Km, caffeic acid (5 to 100 μM except for mutants L136Y, A162T, and N131K, which had 0.25 to 100 μM), 5-hydroxy ferulic acid (0.25 to 100 μM), caffeoyl aldehyde (0.25 to 100 μM), caffeoyl alcohol (0.25 to 100 μM), 5-hydroxyconiferaldehyde (0.25 to 100 μM), and 5-hydroxyconiferyl alcohol (0.25 to 100 μM) were used as substrates in the COMT reaction. Vmax and Km values were determined from Lineweaver-Burk plots.
Crystallographic Structure Determinations
Crystals of COMT were grown by vapor diffusion in hanging drops containing a 1:1 mixture of protein and crystallization buffer (12% [w/v] polyethylene Glycol 8000, 0.05 M TAPS-Na+, pH 8.5, 0.25 M calcium acetate, and 2 mM DTT at 4°C). Crystals grew in space group P2 with three molecules per asymmetric unit. Unit cell dimensions for COMT SeMet derivative crystallized in the presence of 1 mM S-adenosyl-l-Met (SAM) were a = 103.8Å, b = 61.6Å, c = 110.4Å, and β = 112.4°; dimensions for the ferulic acid/S-adenosyl-l-homocysteine (SAH) complex were a = 104.6Å, b = 61.9Å, c = 112.2Å, β = 111.3°. The COMT 5-hydroxyconiferaldehyde/SAH complex crystallized with similar unit cell dimensions of a = 104.5Å, b = 62.0Å, c = 112.2Å, and β = 111.3°. Diffraction data were collected from single crystals mounted in a cryoloop and flash frozen in a nitrogen stream at 105K. All diffraction data were collected at the Stanford Synchrotron Radiation Laboratory (Stanford, CA), beam line 9-2 (COMT SeMet/SAM complex) on a Quantum 4 charge-coupled device detector and beam line 7-1 (COMT ferulic acid and 5-hydroxyconiferaldehyde complexes) using a 30-cm imaging plate. All images were indexed and scaled using DENZO (Otwinowski and Minor, 1997), and the reflections were merged with SCALEPACK (Otwinowski and Minor, 1997).
The COMT SeMet structure was determined using multiple wavelength anomalous dispersion phasing. Initial heavy-atom sites were found with SOLVE (Terwilliger and Berendzen, 1999). SHARP (de la Fourtelle and Bricogne, 1997) was used to refine the initial selenium sites and to locate additional sites. Wavelength anomalous dispersion phases were improved with SOLOMON (Abrahams and Leslie, 1996). Multiple rounds of noncrystallographic averaging and manual rebuilding of the SeMet/SAM model resulted in a nearly complete backbone trace and a majority of side chains positioned. However, the quality of the electron-density maps did not permit the rapid convergence of a complete COMT model. Therefore, refinement of the SAM complex was halted, and subsequent complexes were solved by molecular replacement based on this partial SeMet-derived structure using rigid body refinement in CNS (Brunger et al., 1998). All subsequent refinements were performed using CNS.
During the initial stages of refinement, noncrystallographic symmetry restraints were used to improve phases and electron-density map quality. As refinement and manual rebuilding progressed significantly, these noncrystallographic restraints were gradually released and ultimately discarded to prevent the inclusion of model bias, particularly with regard to side chain positions and substrate/product binding mode elucidation. During refinements, structure factors obtained from intensity data were used to generate |2FO − FC| and |FO − FC| SIGMAA-weighted electron-density maps with phases calculated from the structure of the in-progress model. Inspection of the electron-density maps and model building were performed in O (Jones et al., 1991).
Comparisons of the backbone trace of the partial SeMet COMT and the two reported complexes displayed nearly identical architecture and no evidence for conformational differences between the various apo and complex forms of COMT. Therefore, refinement of the original SeMet COMT structure was not continued. The quality of the resultant models was assessed with the program PROCHECK (Laskowski et al., 1996). The COMT–ferulic acid/SAH complex had 89.4, 9.5, and 1.2% of the residues in the most favored, allowed, and generously allowed regions of the Ramachandran plot, respectively. The COMT–5-hydroxyconiferaldehyde/SAH complex had 89.5% of the residues in the most favored, 9.0% of the residues in the allowed, and 1.1% of the residues in the generously allowed regions of the Ramachandran plot, with 0.4% of the residues in the disallowed region. The atomic coordinates (1KYW–5-hydroxyconiferaldehyde/SAH COMT complex and 1KYZ–ferulic acid/SAH COMT complex) have been deposited in the Protein Data Bank.
Accession Numbers
The accession numbers for the sequences shown in Figure 4 are as follows: COMT from alfalfa (AAB46623), COMT from Clarkia (fairy fans; AAB71141), COMT from Pinus pinaster (pine; CAC21601), COMT from Chrysosplenium americanum (saxifrage; AAA86982), IEMT from Clarkia (fairy fans; AAC01533), chalcone O-methyltransferase (ChOMT) from alfalfa (AAB48059), and isoflavone O-methyltransferase (IOMT) from alfalfa (AAC49927).
Acknowledgments
We thank the staff of the Stanford Synchrotron Radiation Laboratory macromolecular crystallography team for technical assistance. This work was supported by an innovation grant awarded to J.P.N. by the Salk Institute for Biological Studies. The Samuel Roberts Noble Foundation supported work conducted by P.K. and R.A.D. The National Institutes of Health molecular biophysics training grant administered by the University of California, San Diego, supported C.Z. The Stanford Synchrotron Radiation Laboratory Biotechnology Program is supported by the National Institutes of Health, the National Center for Research Resources, the Biomedical Technology Program, and the Department of Energy, Office of Biological and Environmental Research.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.001412.
References
- Abrahams, J.P., and Leslie, A.G.W. (1996). Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D Biol. Crystallogr. 49, 148–157. [DOI] [PubMed] [Google Scholar]
- Baldridge, G.D., O'Neill, N.R., and Samac, D.A. (1998). Alfalfa (Medicago sativa L.) resistance to the root-lesion nematode, Pratylenchus penetrans: Defense-response gene mRNA and isoflavonoid phytoalexin levels in roots. Plant Mol. Biol. 38, 999–1010. [DOI] [PubMed] [Google Scholar]
- Brunger, A.T., et al. (1998). Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Bugos, R.C., Chiang, V.L., and Campbell, W.H. (1992). Characterization of bispecific caffeic acid/5-hydroxyferulic acid O-methyltransferase from aspen. Phytochemistry 31, 1495–1498. [DOI] [PubMed] [Google Scholar]
- Chen, F., Parvathi, K., Blount, J.W., and Dixon, R.A. (2001). Chemical synthesis of caffeoyl and 5-OH coniferyl aldehydes and alcohols and the determination of lignin O-methyltransferase activities in dicot and monocot species. Phytochemistry 58, 1035–1042. [DOI] [PubMed] [Google Scholar]
- de la Fourtelle, E., and Bricogne, G. (1997). Maximum likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 276, 472–494. [DOI] [PubMed] [Google Scholar]
- Dixon, R.A., Chen, F., Guo, D., and Parvathi, K. (2001). The biosynthesis of monolignols: A “metabolic grid”, or independent pathways to guaiacyl and syringyl units? Phytochemistry 57, 1069–1084. [DOI] [PubMed] [Google Scholar]
- Dixon, R.A., Lamb, C.J., Masoud, S., Sewalt, V.J., and Paiva, N.L. (1996). Metabolic engineering: Prospects for crop improvement through the genetic manipulation of phenylpropanoid biosynthesis and defense responses. A review. Gene 179, 61–71. [DOI] [PubMed] [Google Scholar]
- Doublie, S. (1997). Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 276, 523–530. [PubMed] [Google Scholar]
- Edwards, R., and Dixon, R.A. (1991). Purification and characterization of S-adenosyl-l-methionine:caffeic acid 3-O-methyltransferase from suspension cultures of alfalfa (Medicago sativa L.). Arch. Biochem. Biophys. 287, 372–379. [DOI] [PubMed] [Google Scholar]
- Gowri, G., Paiva, N.L., and Dixon, R.A. (1991). Stress responses in alfalfa (Medicago sativa L.). 12. Sequence analysis of phenylalanine ammonia-lyase (PAL) cDNA clones and appearance of PAL transcripts in elicitor-treated cell cultures and developing plants. Plant Mol. Biol. 17, 415–429. [DOI] [PubMed] [Google Scholar]
- Guo, D., Chen, F., Inoue, K., Blount, J., and Dixon, R.A. (2001). Downregulation of caffeic acid 3-O-methyltransferase and caffeoyl CoA 3-O-methyltransferase in transgenic alfalfa: Impacts on lignin structure and implications for the biosynthesis of G and S lignin. Plant Cell 13, 73–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield, R., and Vermerris, W. (2001). Lignin formation in plants: The dilemma of linkage specificity. Plant Physiol. 126, 1351–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim, R.K., Bruneau, A., and Bantignies, B. (1998). Plant O-methyltransferases: Molecular analysis, common signature and classification. Plant Mol. Biol. 36, 1–10. [DOI] [PubMed] [Google Scholar]
- Inoue, K., Parvathi, K., and Dixon, R.A. (2000). Substrate preferences of caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferases in developing stems of alfalfa (Medicago sativa L.). Arch. Biochem. Biophys. 375, 175–182. [DOI] [PubMed] [Google Scholar]
- Inoue, K., Sewalt, V.J., Murray, G.B., Ni, W., Sturzer, C., and Dixon, R.A. (1998). Developmental expression and substrate specificities of alfalfa caffeic acid 3-O-methyltransferase and caffeoyl coenzyme A 3-O-methyltransferase in relation to lignification. Plant Physiol. 117, 761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeck, E., Dumas, B., Geoffroy, P., Favet, N., Inze, D., Van Montagu, M., Fritig, B., and Legrand, M. (1992). Regulation of enzymes involved in lignin biosynthesis: Induction of O-methyltransferase mRNAs during the hypersensitive reaction of tobacco to tobacco mosaic virus. Mol. Plant-Microbe Interact. 5, 294–300. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. (1991). Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Joshi, C.P., and Chiang, V.L. (1998). Conserved sequence motifs in plant S-adenosyl-l-methionine-dependent methyltransferases. Plant Mol. Biol. 37, 663–674. [DOI] [PubMed] [Google Scholar]
- Jouanin, L., Goujon, T., de Nadai, V., Martin, M.T., Mila, I., Vallet, C., Pollet, B., Yoshinaga, A., Chabbert, B., Petit-Conil, M., and Lapierre, C. (2000). Lignification in transgenic poplars with extremely reduced caffeic acid O-methyltransferase activity. Plant Physiol. 123, 1363–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraulis, P.J. (1991). MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24, 946–950. [Google Scholar]
- Lapierre, C., Pollet, B., Petit-Conil, M., Toval, G., Romero, J., Pilate, G., Leple, J.C., Boerjan, W., Ferret, V.V., De Nadai, V., and Jouanin, L. (1999). Structural alterations of lignins in transgenic poplars with depressed cinnamyl alcohol dehydrogenase or caffeic acid O-methyltransferase activity have an opposite impact on the efficiency of industrial Kraft pulping. Plant Physiol. 119, 153–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski, R.A., Rullmannn, J.A., MacArthur, M.W., Kaptein, R., and Thornton, J.M. (1996). AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486. [DOI] [PubMed] [Google Scholar]
- Lawton, M.A., and Lamb, C.J. (1987). Transcriptional activation of plant defense genes by fungal elicitor, wounding, and infection. Mol. Cell. Biol. 7, 335–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., Popko, J.L., Umezawa, T., and Chiang, V.L. (2000). 5-Hydroxyconiferyl aldehyde modulates enzymatic methylation for syringyl monolignol formation, a new view of monolignol biosynthesis in angiosperms. J. Biol. Chem. 275, 6537–6545. [DOI] [PubMed] [Google Scholar]
- Ni, W., Fahrendorf, T., Ballance, G.M., Lamb, C.J., and Dixon, R.A. (1996). Stress responses in alfalfa (Medicago sativa L.). XX. Transcriptional activation of phenylpropanoid pathway genes in elicitor-induced cell suspension cultures. Plant Mol. Biol. 30, 427–438. [DOI] [PubMed] [Google Scholar]
- Nicholls, A., Charp, K., and Honig, B. (1991). Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Protein Struct. Funct. Genet. 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Osakabe, K., Tsao, C.C., Li, L., Popko, J.L., Umezawa, T., Carraway, D.T., Smeltzer, R.H., Joshi, C.P., and Chiang, V.L. (1999). Coniferyl aldehyde 5-hydroxylation and methylation direct syringyl lignin biosynthesis in angiosperms. Proc. Natl. Acad. Sci. USA 96, 8955–8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski, Z., and Minor, W. (1997). Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Parvathi, K., Chen, F., Guo, D., Blount, J.W., and Dixon, R.A. (2001). Substrate preferences of O-methyltransferases in alfalfa suggest new pathways for 3-O-methylation of monolignols. Plant J. 25, 193–202. [DOI] [PubMed] [Google Scholar]
- Pincon, G., Maury, S., Hoffmann, L., Geoffroy, P., Lapierre, C., Pollet, B., and Legrand, M. (2001). Repression of O-methyltransferase genes in transgenic tobacco affects lignin synthesis and plant growth. Phytochemistry 57, 1167–1176. [DOI] [PubMed] [Google Scholar]
- Pond, K.R., Ellis, W.C., Lascano, C.E., and Akin, D.E. (1987). Fragmentation and flow of grazed coastal Bermudagrass through the digestive tract of cattle. J. Anim. Sci. 65, 609–618. [DOI] [PubMed] [Google Scholar]
- Ros Barcelo, A. (1997). Lignification in plant cell walls. Int. Rev. Cytol. 176, 87–132. [PubMed] [Google Scholar]
- Rossmann, M.G., Moras, D., and Olsen, K.W. (1974). Chemical and biological evolution of nucleotide-binding protein. Nature 250, 194–199. [DOI] [PubMed] [Google Scholar]
- Terwilliger, T.C., and Berendzen, J. (1999). Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., and Pichersky, E. (1999). Identification of specific residues involved in substrate discrimination in two plant O-methyltransferases. Arch. Biochem. Biophys. 368, 172–180. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Zhou, L., and Cheng, X. (2000). Crystal structure of the conserved core of protein arginine methyltransferase PRMT3. EMBO J. 19, 3509–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubieta, C., He, X.-Z., Dixon, R.A., and Noel, J.P. (2001). Structures of two natural product methyltransferases reveal the basis for substrate specificity in plant O-methyltransferases. Nat. Struct. Biol. 8, 271–279. [DOI] [PubMed] [Google Scholar]