Abstract
Iron, an essential nutrient, is not readily available to plants because of its low solubility. In addition, iron is toxic in excess, catalyzing the formation of hydroxyl radicals that can damage cellular constituents. Consequently, plants must carefully regulate iron uptake so that iron homeostasis is maintained. The Arabidopsis IRT1 gene is the major transporter responsible for high-affinity iron uptake from the soil. Here, we show that the steady state level of IRT1 mRNA was induced within 24 h after transfer of plants to iron-deficient conditions, with protein levels peaking 72 h after transfer. IRT1 mRNA and protein were undetectable 12 h after plants were shifted back to iron-sufficient conditions. Overexpression of IRT1 did not confer dominant gain-of-function enhancement of metal uptake. Analysis of 35S-IRT1 transgenic plants revealed that although IRT1 mRNA was expressed constitutively in these plants, IRT1 protein was present only in the roots when iron is limiting. Under these conditions, plants that overexpressed IRT1 accumulated higher levels of cadmium and zinc than wild-type plants, indicating that IRT1 is responsible for the uptake of these metals and that IRT1 protein levels are indeed increased in these plants. Our results suggest that the expression of IRT1 is controlled by two distinct mechanisms that provide an effective means of regulating metal transport in response to changing environmental conditions.
INTRODUCTION
Improving the mineral content of plants so that they can serve as sources of the 14 minerals required in the human diet presents researchers with a number of challenges. In the case of iron, these include the facts that iron is not readily available in the rhizosphere, often limiting plant growth, and that iron can be toxic if present in excess, forcing organisms to carefully regulate its uptake (Eide et al., 1996; Robinson et al., 1999) and storage (Lescure et al., 1991; Briat and Lobréaux, 1997; Wei and Theil, 2000). Because iron deficiency is the leading human nutritional disorder in the world today (World Health Organization, 2002) and because plants serve as the primary source of dietary iron for most of the world's population, we clearly need to understand iron homeostasis in plants if we wish to improve the iron content of food.
Work in our laboratory has focused on iron uptake from the soil into the plant root. After Fe(III) chelates are reduced at the cell membrane (Robinson et al., 1999), iron is transported into the Arabidopsis root via IRT1 (Eide et al., 1996; Vert et al., 2002). IRT1 is one of three founding members of the ZIP (for ZRT-IRT–like proteins) family of transporters that function in metal transport in a diverse array of eukaryotic organisms (Guerinot, 2000). ZIP family members characterized to date function in the transport of iron, zinc, and/or manganese in bacteria (Grass et al., 2002), yeast (Zhao and Eide, 1996a, 1996b; MacDiarmid et al., 2000), humans (Gaither and Eide, 2000, 2001), and plants (Eide et al., 1996; Grotz et al., 1998; Pence et al., 2000; Assuncao et al., 2001; Eckhardt et al., 2001; Vert et al., 2001). When expressed in yeast, IRT1 itself mediates the uptake of iron (Eide et al., 1996), zinc, and manganese (Korshunova et al., 1999). Cadmium inhibits the uptake of these metals by IRT1 (Eide et al., 1996), and expression of IRT1 in yeast results in increased sensitivity to cadmium (Rogers et al., 2000), suggesting that cadmium also is transported by IRT1.
Here, we report on the regulation of expression of the Arabidopsis metal transporter IRT1. Previous work demonstrated that IRT1 mRNA accumulates preferentially in the roots of iron-deficient plants (Eide et al., 1996). We hypothesized that overexpression of IRT1 in transgenic plants might lead to enhanced accumulation of iron. Using this approach, we discovered post-transcriptional regulation of IRT1: IRT1 protein accumulated only in the roots of iron-starved transgenic 35S-IRT1 plants, despite the fact that IRT1 mRNA was expressed constitutively in the same plants. 35S-IRT1 transgenic plants showed enhanced sensitivity to cadmium only when grown on iron-deficient medium, as a result of increased levels of IRT1 protein in the roots of iron-deficient transgenic plants. Thus, overexpression of IRT1 protein was permitted only when plants were iron starved. Furthermore, our results show that expression of IRT1 was regulated at the level of transcript accumulation in response to iron, zinc, and cadmium and at the level of protein accumulation in response to iron and zinc. The fact that the accumulation of IRT1 was controlled at multiple levels serves to emphasize the importance of maintaining metal homeostasis within cells.
RESULTS
Time Course of IRT1 Induction and Turnover
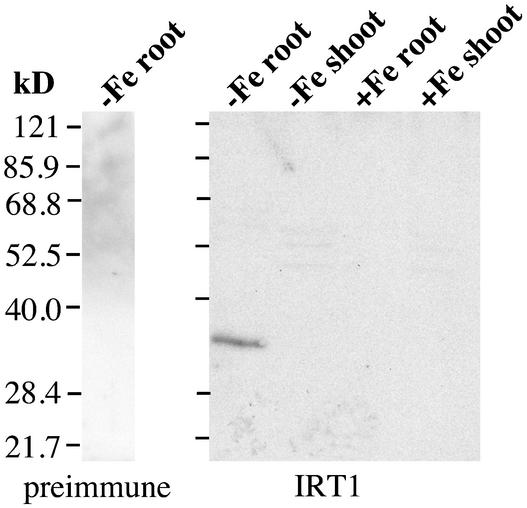
IRT1 mRNA is expressed in the roots of iron-starved Arabidopsis plants 3 days after transfer of the plants to iron-deficient growth conditions (Eide et al., 1996). To examine the kinetics of the induction of IRT1 expression, a time-course experiment was performed in which the levels of both IRT1 mRNA and protein were evaluated. IRT1 antiserum was raised against a synthetic peptide corresponding to a portion of the variable region between transmembrane domains III and IV. The antiserum detected a protein of ∼35 kD in the roots of iron-starved plants (Figure 1), corresponding well with the predicted molecular mass of the processed protein (35.9 kD). RNA gel blot analysis using the IRT1 cDNA as a hybridization probe showed that IRT1 RNA was detectable in the roots of plants 24 h after transfer of plants to iron-deficient medium (Figure 2A). IRT1 steady state RNA levels peaked 72 h after the transfer of plants to iron-deficient medium. Immunoblot analysis showed that IRT1 protein was detectable 48 h after the transfer of plants to iron-deficient conditions and that protein levels were highest 72 h after the transfer of plants to iron-deficient medium (Figure 2A).
Figure 1.
Characterization of IRT1 Antiserum.
Wild-type plants were grown for 2 weeks on B5 medium and transferred to either iron-deficient or iron-sufficient medium, and roots and shoots were harvested at 3 days after the transfer. Protein samples were prepared from each tissue sample and used to prepare protein gel blots. IRT1 protein was detected using affinity-purified antiserum raised against a synthetic peptide. The reaction of proteins prepared from iron-deficient roots with preimmune serum is shown as a control. Numbers at left indicate molecular mass (kD).
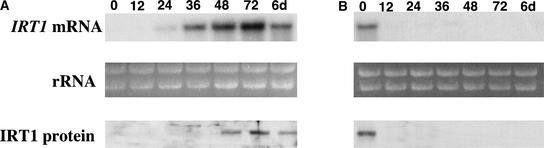
Figure 2.
Time Course of IRT1 mRNA and Protein Abundance Patterns in Response to Iron-Deficient and Iron-Sufficient Growth Conditions.
(A) Wild-type plants were grown for 2 weeks on B5 medium and transferred to iron-deficient medium, and roots were harvested at 0, 12, 24, 36, 48, and 72 h and 6 days after the transfer. RNA and protein samples were prepared from each tissue sample and used to prepare RNA and protein gel blots. The IRT1 cDNA was used to probe the RNA gel blot. Ethidium bromide–stained rRNA is shown as a control for loading. IRT1 protein was detected using an IRT1 affinity-purified peptide antibody.
(B) Wild-type plants were grown for 2 weeks on B5 medium, transferred to iron-deficient medium for 3 days, and transferred a second time to iron-sufficient medium. Roots were harvested at various times as indicated, and RNA gel blot and immunoblot analyses were performed as described above.
In a complementary experiment, we examined how quickly IRT1 mRNA and protein levels change in response to the presence of iron. IRT1 mRNA was present at time 0 but was undetectable 12 h after transfer of plants from iron-deficient to iron-sufficient conditions (Figure 2B). Immunoblot analysis showed that IRT1 protein also was undetectable 12 h after transfer of plants to iron-sufficient conditions (Figure 2B). These results indicate that steady state levels of both IRT1 mRNA and protein change rapidly after transfer of plants to iron-sufficient conditions, demonstrating that the expression of IRT1 is tightly regulated. To control for potential changes in IRT1 mRNA or protein abundance in response to the physical transfer of plants, we performed an experiment in which plants were first transferred from B5 medium to iron-deficient medium for 3 days and then transferred a second time to iron-deficient medium. Roots were harvested at 0, 12, 24, 36, 48, and 72 h as well as 6 days after the transfer. RNA gel blot and protein gel blot analysis revealed that the transfer did not affect the abundance of either mRNA or protein (data not shown).
Post-Transcriptional Regulation of IRT1 by Iron
Expression of IRT1 in yeast leads to the uptake of iron, manganese, zinc, and cadmium (Eide et al., 1996; Korshunova et al., 1999; Rogers et al., 2000). We reasoned that overexpression of IRT1 in Arabidopsis might lead to enhanced metal uptake. We constructed 35S-IRT1 transgenic plants and identified six independent, homozygous, single-insertion transgenic lines based on segregation ratios and genomic DNA gel blot analysis (data not shown). The 35S-IRT1 transgenic lines showed no visible morphological phenotype when grown on soil or on B5 plates under standard growth conditions (data not shown).
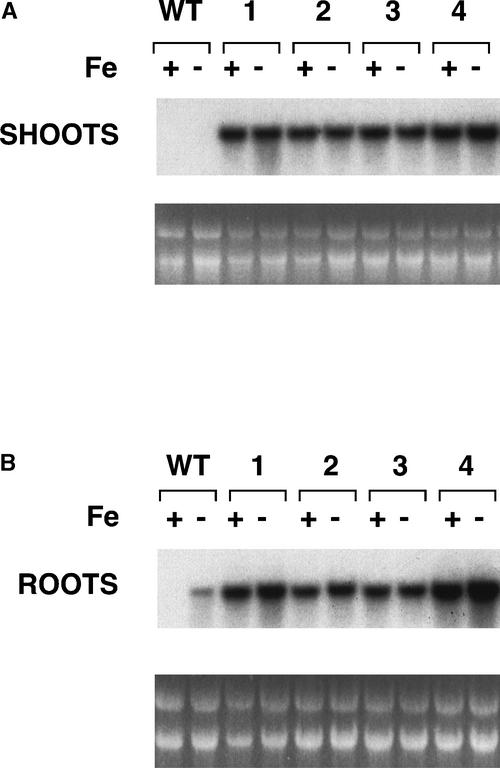
To examine steady state IRT1 mRNA levels in the transgenic plants, we performed RNA gel blot analysis on four of the 35S-IRT1 lines using the IRT1 cDNA as a probe. RNA was isolated from the roots and shoots of T4 plants grown on either iron-sufficient or iron-deficient medium. The plants were grown for 3 days under these conditions because IRT1 mRNA was abundant 3 days after the transfer to iron-deficient medium (Figure 2A) and because IRT1 mRNA was undetectable 3 days after the transfer from iron-deficient to iron-sufficient medium (Figure 2B). In wild-type plants, IRT1 mRNA was not expressed in the shoots of plants grown with or without iron (Figure 3A) but was expressed exclusively in the roots of iron-starved plants (Figure 3B), as has been shown (Eide et al., 1996). As expected, IRT1 mRNA was expressed at high levels in both the shoots (Figure 3A) and roots (Figure 3B) of all four transgenic lines regardless of the iron status of the plants.
Figure 3.
RNA Gel Blot Analysis of Wild-Type and Transgenic 35S-IRT1 Plants.
The IRT1 cDNA was used to probe a RNA gel blot containing RNA prepared from the shoots (A) and roots (B) of plants grown for 3 days on either iron-sufficient (+) or iron-deficient (−) medium. RNA from wild-type plants (WT) was electrophoresed next to RNA from the transgenic lines (lines 1 to 4). Ethidium bromide–stained rRNA is shown as a control for loading.
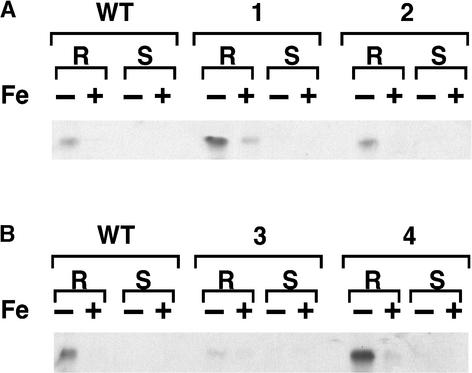
Despite the fact that IRT1 mRNA was present in the transgenic lines under all conditions examined, IRT1 protein only accumulated to high levels in iron-deficient roots (Figure 4). These results demonstrate that IRT1 is subject to an additional level of regulation that occurs post-transcriptionally in response to iron. Consequently, iron-deficient roots are considered the “permissive” condition for the expression of IRT1. It is important to note, however, that IRT1 protein levels were increased in the roots of two transgenic lines (lines 1 and 4) compared with wild-type plants when plants were iron deficient.
Figure 4.
Protein Gel Blot Analysis of Wild-Type and Transgenic 35S-IRT1 Plants.
IRT1 protein was detected using the affinity-purified IRT1 peptide antibody. Each lane contained 10 μg of protein extracted from roots (R) or shoots (S) of plants grown for 3 days on either iron-sufficient (+) or iron-deficient (−) medium. Protein extracted from wild-type (WT) plants was electrophoresed next to protein extracted from transgenic lines 1 and 2 (A) and transgenic lines 3 and 4 (B).
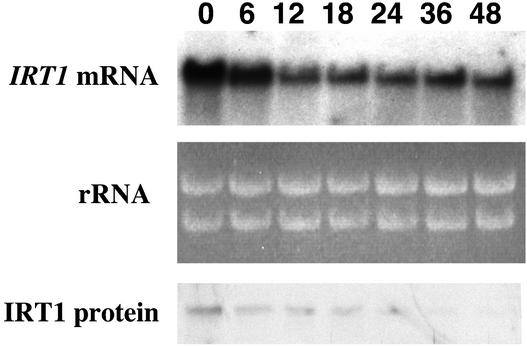
Next, we performed a time-course analysis using transgenic 35S-IRT1 (line 4) plants that were grown for 3 days on iron-deficient medium and then transferred to iron-sufficient medium. IRT1 steady state RNA levels remained high after the transfer of plants from iron-deficient to iron-sufficient medium (Figure 5). The IRT1 RNA detected corresponds to expression of the endogenous IRT1 gene as well as that of the IRT1 transgene. As a result, IRT1 steady state RNA levels were higher at 0 and 6 h than they were at later times because of expression of the endogenous IRT1 gene (for comparison, see Figure 2B). In contrast, IRT1 protein levels declined to very low levels by 36 h after the transfer of transgenic plants from iron-deficient to iron-sufficient medium (Figure 5).
Figure 5.
Time Course of IRT1 mRNA and Protein Abundance Patterns in 35S-IRT1 Plants.
35S-IRT1 plants (line 4) were grown on B5 plates for 2 weeks, transferred to iron-deficient plates for 3 days, and transferred again to iron-sufficient plates. Roots were harvested at 0, 6, 12, 18, 24, 36, and 48 h after the second transfer. RNA and protein samples were prepared from each tissue sample and used to prepare RNA and protein gel blots. The IRT1 cDNA was used to probe the RNA gel blot. Ethidium bromide–stained rRNA is shown as a control for loading. IRT1 protein was detected using the IRT1 affinity-purified peptide antibody.
Regulation of IRT1 Expression by Zinc
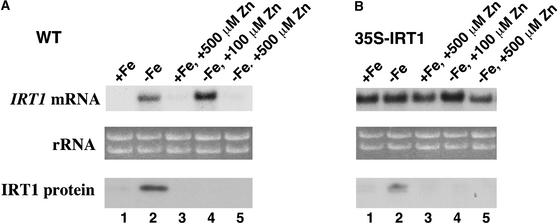
Several experiments have shown that IRT1 transports zinc. First, zinc inhibits IRT1-dependent iron uptake in yeast (Eide et al., 1996). Second, expression of IRT1 complements the zinc-limited growth defect of a yeast zinc uptake mutant (zrt1 zrt2) (Korshunova et al., 1999). Finally, IRT1 mediates the uptake of radiolabeled zinc in yeast (Korshunova et al., 1999). Although the growth of plants on zinc-deficient medium does not induce the expression of IRT1 (our unpublished data), we wanted to determine whether added zinc could affect the expression of IRT1. When wild-type plants were grown for 3 days on iron-deficient plates containing 100 μM zinc, IRT1 mRNA was detected in the roots (Figure 6A). However, IRT1 protein was undetectable in the roots of the same plants despite the presence of high levels of IRT1 mRNA, indicating that 100 μM zinc results in post-transcriptional regulation of IRT1 (Figure 6A). IRT1 protein was undetectable in the roots of 35S-IRT1 transgenic plants grown on iron-deficient medium containing zinc (Figure 6B), although IRT1 mRNA was abundant in the same tissue (Figure 6B), confirming that zinc causes post-transcriptional regulation of IRT1.
Figure 6.
Zinc Affects the Abundance of IRT1 mRNA and Protein.
Wild-type (WT [A]) and 35S-IRT1 (line 4) transgenic (B) plants were grown for 2 weeks on B5 plates. Seedlings were transferred subsequently and grown for 3 days on iron-sufficient plates (lane 1), iron-deficient plates (lane 2), iron-sufficient plates supplemented with 500 μM zinc (lane 3), iron-deficient plates supplemented with 100 μM zinc (lane 4), or iron-deficient plates supplemented with 500 μM zinc (lane 5). RNA and protein samples were prepared from each root sample and used to prepare RNA and protein gel blots. The IRT1 cDNA was used to probe the RNA gel blots. Ethidium bromide–stained rRNA is shown as a control for loading. IRT1 protein was detected using the IRT1 affinity-purified peptide antibody.
When wild-type plants were grown on iron-deficient medium containing 500 μM zinc, neither IRT1 mRNA nor IRT1 protein could be detected in the roots. This result demonstrates that zinc additionally mediates the regulation of IRT1 at the level of transcript accumulation when it is present in the growth medium at high levels. Arabidopsis plants grown on medium containing 500 μM zinc showed zinc toxicity symptoms, including the arrest of root growth (data not shown). As a result, the absence of IRT1 RNA in plants grown on medium containing 500 μM zinc may be attributable simply to the toxicity of zinc. However, IRT1 RNA was present in the roots of 35S-IRT1 transgenic plants grown on iron-deficient medium supplemented with 500 μM zinc (Figure 6B), supporting the idea that the repression of IRT1 RNA levels in wild-type plants in response to high zinc is mediated through the IRT1 promoter.
To determine how quickly IRT1 protein levels decline after the addition of 100 μM zinc to the medium, a time-course analysis was performed on wild-type plants that were grown on iron-deficient medium for 3 days and then transferred to iron-deficient medium containing 100 μM zinc. Although IRT1 steady state RNA levels remained high after the addition of 100 μM zinc, IRT1 protein levels declined rapidly after plants were exposed to 100 μM zinc (Figure 7). IRT1 protein was undetectable 6 h after the transfer of plants to medium containing 100 μM zinc. Growth of wild-type plants on medium containing 100 μM zinc resulted in a slight reduction in root growth over time, indicating that this level of zinc is somewhat toxic for Arabidopsis (data not shown).
Figure 7.
Time Course of IRT1 mRNA and Protein Abundance Patterns in Wild-Type Plants Grown on Plates Containing Zinc.
Wild-type plants were grown on B5 plates for 2 weeks, transferred to iron-deficient plates for 3 days, and transferred again to iron-deficient medium supplemented with 100 μM zinc. Roots were harvested at 0, 6, 12, 24, 36, 48, and 72 h after the second transfer. RNA and protein samples were prepared from each tissue sample and used to prepare RNA and protein gel blots. The IRT1 cDNA was used to probe the RNA gel blot. Ethidium bromide–stained rRNA is shown as a control for loading. IRT1 protein was detected using the IRT1 affinity-purified peptide antibody.
Regulation of IRT1 Expression by Cadmium
Cadmium is not essential for plant growth and is known to cause phytotoxicity at relatively low concentrations (Sanita di Toppi and Gabbrielli, 1999). Cadmium is able to compete for IRT1-dependent iron uptake in yeast (Eide et al., 1996). In addition, yeast expressing IRT1 show enhanced sensitivity to cadmium (Rogers et al., 2000). These studies suggest that IRT1 mediates the uptake of cadmium in addition to iron, zinc, and manganese.
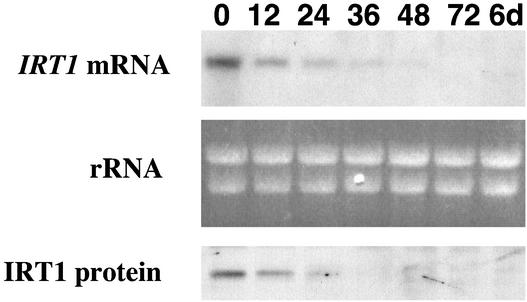
We examined the effect of cadmium on the expression of IRT1. Wild-type plants were grown for 3 days on iron-deficient medium and then transferred to iron-deficient medium supplemented with 90 μM CdSO4. We chose 90 μM as the cadmium concentration for these experiments because previous studies showed that this concentration of cadmium inhibited root growth of wild-type Arabidopsis plants (Howden and Cobbett, 1992). IRT1 RNA was abundant at the time of the transfer (0 h) and declined steadily until it became undetectable 72 h later. IRT1 protein levels also declined when cadmium was present in the medium (Figure 8). Arabidopsis plants grown on plates containing 90 μM cadmium exhibited cadmium toxicity symptoms. It is possible that the decrease in IRT1 mRNA and protein abundance in response to cadmium was attributable simply to the toxicity of cadmium. As a control, we hybridized the RNA gel blot shown in Figure 8 with a gene-specific probe corresponding to the 5′ untranslated region of the Arabidopsis H+ ATPase 2 (AHA2) gene. AHA2 mRNA levels were unchanged in response to cadmium (data not shown).
Figure 8.
Time Course of IRT1 mRNA and Protein Abundance Patterns in Wild-Type Plants Grown on Plates Containing Cadmium.
Plants were grown on B5 plates for 2 weeks, transferred to iron-deficient plates for 3 days, and transferred again to iron-deficient medium supplemented with 90 μM CdSO4. Roots were harvested at 0, 12, 24, 36, 48, and 72 h and 6 days after the second transfer. RNA and protein samples were prepared from each tissue sample and used to prepare RNA and protein gel blots. The IRT1 cDNA was used to probe the RNA gel blot. Ethidium bromide–stained rRNA is shown as a control for loading. IRT1 protein was detected using the IRT1 affinity-purified peptide antibody.
35S-IRT1 Plants Are Sensitive to Cadmium
To examine the effects of cadmium on the growth of the 35S-IRT1 transgenic lines, we measured the root growth of wild-type and 35S-IRT1 transgenic plants on iron-deficient plates that contained various concentrations of cadmium (0 to 500 μM). The plates were placed in the growth chamber in the vertical orientation so that the roots would grow down along the surface of the agar, and root growth was measured every 24 h. Wild-type roots grew well on iron-deficient plates supplemented with 50 μM cadmium. In contrast, the growth of the roots of transgenic plants was arrested completely on iron-deficient plates supplemented with 50 μM cadmium (Figure 9).
Figure 9.
Root Growth of 35S-IRT1 Transgenic Plants on Plates Containing Cadmium.
Seedlings (wild type [WT] and transgenic line 4) were grown on B5 plates for 8 days before being transferred to iron-deficient plates that contained 50 μM CdSO4. Plates were placed in the growth chamber in the vertical orientation such that root growth occurred along the surface of the agar, and root growth was measured every 24 h. Results are means of six independent measurements, and bars indicate standard error of mean. The experiment was performed twice.
In addition, the transgenic lines showed sensitivity to cadmium at much lower concentrations than the wild type, presumably as a result of the expression of IRT1. Root growth of transgenic plants was inhibited by 0.01 μM cadmium, whereas the root growth of wild-type plants was not inhibited by growth on cadmium levels <10 μM; the transgenic plants were 1000 times more sensitive to cadmium than the wild-type plants (data not shown). Finally, the sensitivity of the 35S-IRT1 lines to growth in the presence of cadmium was greatest when the plants were grown without iron.
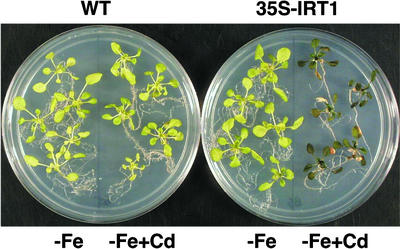
Although root growth was inhibited by lower concentrations of cadmium, the visible effects of cadmium on the aerial portions of the plant were best seen using higher concentrations of cadmium (90 μM). Wild-type plants grown without iron for 6 days appeared chlorotic (Figure 10). Wild-type plants grown on iron-deficient plates that contained cadmium resembled plants grown on iron-deficient plates without cadmium. 35S-IRT1 plants resembled wild-type plants when grown on iron-deficient plates. However, when the transgenic plants were grown on iron-deficient plates that contained cadmium, they showed a severe cadmium sensitivity phenotype. Compared with wild-type plants, the transgenic plants were smaller, and some leaves were bleached and necrotic when grown on this medium. In addition, the leaves of the transgenic plants were purple, presumably because of the accumulation of the pigment anthocyanin (Figure 10).
Figure 10.
Sensitivity of 35S-IRT1 Transgenic Plants to Cadmium.
Seedlings were grown for 2 weeks on B5 plates before being transferred to plates that were either iron deficient (−) or iron deficient plus 90 μM CdSO4. Seedlings were allowed to grow for 6 days more before being transferred for photography. Wild-type (WT) plants are shown next to transgenic line 4 plants.
The roots of the transgenic plants were brown and root growth was arrested completely when grown on iron-deficient plates that contained 90 μM cadmium. The transgenic plants were not dead, however, because they recovered when transferred to iron-sufficient plates without cadmium (data not shown). All four transgenic lines tested showed enhanced sensitivity to cadmium to varying degrees, with transgenic line 4 showing the most severe phenotype; this line also appeared to have the highest levels of IRT1 protein in iron-deficient roots, as detected by immunoblot analysis (Figure 4). Together, these results demonstrate that overexpression of IRT1 in transgenic Arabidopsis results in enhanced sensitivity to cadmium.
Elemental Analysis of 35S-IRT1 Plants
Finally, we wanted to determine whether or not the transgenic plants take up and accumulate increased levels of metals when grown under the permissive condition for the expression of IRT1 (iron deficiency). We performed elemental analysis on the roots and shoots of wild-type and 35S-IRT1 plants that were grown without iron for 6 days. Zinc levels in the 35S-IRT1 plants were significantly different from the levels in wild-type plants (P < 0.05) (Table 1). 35S-IRT1 plants contained less zinc in the shoots and more zinc in the roots than the wild type. We found that iron and manganese levels in the 35S-IRT1 plants were not significantly different from the levels in wild-type plants (Table 1). It is not surprising that iron levels were unchanged in the transgenic plants compared with the wild type because the plants were grown on iron-deficient medium. However, we expected that manganese levels would be increased in the transgenic plants compared with wild-type plants. It is possible that we would be able to detect increased manganese levels in the transgenic plants if we allowed the plants to grow on iron-deficient medium for a longer period of time.
Table 1.
Elemental Analysis of Wild-Type and 35S-IRT1 Plants
| Genotype | Tissue | Iron | Zinca | Manganese |
|---|---|---|---|---|
| Wild type | Shoot | 77.3 ± 22 | 122 ± 12.1 | 99.5 ± 25.6 |
| 35S-IRT1 | Shoot | 66.2 ± 8.4 | 68 ± 13.6 | 154 ± 24.7 |
| Wild type | Root | 314 ± 61.3 | 512 ± 134 | 64.5 ± 8.5 |
| 35S-IRT1 | Root | 200 ± 26.1 | 779 ± 178 | 59.2 ± 16.2 |
Values shown are μg/g dry weight ± se.
Means for zinc in wild-type and 35S-IRT1 plants are significantly different (P < 0.05).
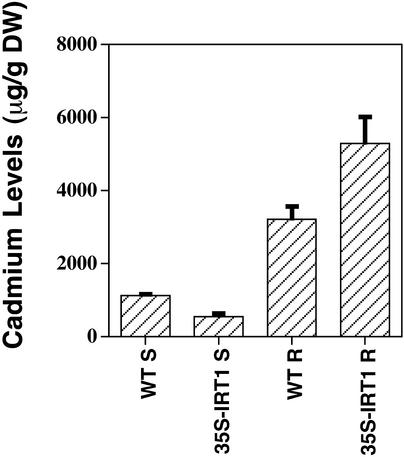
In addition, we performed elemental analysis on the roots and shoots of wild-type and 35S-IRT1 plants that were grown on plates that were iron deficient and contained cadmium. The results showed that cadmium levels in the 35S-IRT1 plants were significantly different from the levels in wild-type plants (P < 0.05); the roots of transgenic plants contained ∼2000 μg/g dry weight more cadmium than the roots of wild-type plants (Figure 11). Interestingly, the transgenic plants accumulated less cadmium in their shoots than wild-type plants. Presumably, increased levels of cadmium in the roots cause a stress response that results in more cadmium being sequestered in the root and less cadmium being mobilized to the shoot (Sanita di Toppi and Gabbrielli, 1999). A similar hypothesis would explain the zinc data described above.
Figure 11.
Elemental Analysis of Wild-Type and 35S-IRT1 Transgenic Plants Grown on Plates Containing Cadmium.
Plants were grown on B5 plates before being transferred to iron-deficient medium that contained 90 μM CdSO4. Plants were allowed to grow for 6 days, at which time the roots and shoots were harvested separately and subjected to elemental analysis. Approximately 50 plants were pooled for each experiment, and results are means of three (WT) or six (35S-IRT1) independent experiments. DW, dry weight. Bars indicate standard error of mean.
DISCUSSION
In this report, we have demonstrated that the abundance of the Arabidopsis metal transporter IRT1 is controlled at the levels of transcript and protein accumulation. Iron deficiency resulted in an induction of IRT1 transcript accumulation, whereas iron sufficiency resulted in a reduction in IRT1 transcript levels. High levels of zinc or cadmium also caused a reduction in IRT1 transcript levels. Interestingly, iron and zinc also caused post-transcriptional regulation of IRT1 such that IRT1 protein did not accumulate when these metals were present in the growth medium. It is logical that the uptake of these metals is tightly controlled because they are both essential and potentially toxic.
Zinc uptake in Saccharomyces cerevisiae is known to be tightly regulated (Guerinot and Eide, 1999). ZRT1 and ZRT2, the yeast high- and low-affinity zinc transporters (Zhao and Eide, 1996a, 1996b), are regulated at the level of transcription by the ZAP1 transcription factor (Zhao and Eide, 1997; Zhao et al., 1998). When zinc is limiting, ZAP1 induces the expression of ZRT1 and ZRT2. ZRT1 activity is subject to a second level of regulation in response to zinc availability; high zinc causes the removal of the protein from the plasma membrane (Gitan et al., 1998). High zinc concentrations trigger the conjugation of ubiquitin to ZRT1, which in turn induces endocytosis of the transporter (Gitan and Eide, 2000). This regulatory system allows the rapid turnover of the transporter when cells come in contact with high levels of zinc, thus preventing the overaccumulation of zinc, which can be toxic at high levels. ZRT2 also is subject to zinc-induced endocytosis, although this regulation has not been well characterized (D.J. Eide, unpublished data).
Recent studies in yeast have described the post-translational regulation of several plasma membrane metal transporters in response to changing environmental conditions. These include the SMF1 protein, a Nramp family member involved in metal uptake (Liu and Culotta, 1999), and the CTR1 protein, a copper transporter (Ooi et al., 1996). In addition, MAC1p, a copper-sensing transcription factor, also is subject to degradation in a copper-specific manner (Zhu et al., 1998). There also is evidence for post-translational control of metal transporters in mammalian cells. DMT1, a divalent metal transporter involved in iron transport across the apical membrane of human intestinal enterocytes, changes its distribution between the plasma membrane and the cytosol in response to iron levels (Sharp et al., 2002).
ZRT1 and ZRT2 are ZIP family members and thus show a high degree of sequence similarity to IRT1. Most ZIP family members contain eight potential membrane-spanning domains and have a similar predicted membrane topology in which the N and C termini are predicted to reside on the outside surface of the plasma membrane. ZIPs also have an intracellular loop between transmembrane domains III and IV. This intracellular loop has been termed the variable region because it is variable in both length and sequence (Guerinot and Eide, 1999). The variable region is of interest here for two reasons.
First, it is the site of a His-rich repeat that may be involved in metal binding and/or sensing. In IRT1, the sequence of the repeat is HGHGHGH (Eide et al., 1996), and in ZRT1, the sequence of the repeat is HDHTHDE (Zhao and Eide, 1996a). Second, mutation of a specific Lys residue in ZRT1 blocks zinc-regulated ubiquitination and endocytosis of ZRT1; this critical Lys residue (Lys-195) resides in the variable region (Gitan and Eide, 2000). Based on our knowledge of the regulation of ZRT1, we hypothesize that the decline in IRT1 protein abundance in response to iron and zinc is mediated post-translationally via a mechanism analogous to the post-translational mechanism that controls ZRT1 abundance in yeast. Indeed, there are two Lys residues present within the variable region of the IRT1 protein that may serve as ubiquitin attachment sites.
The permissive condition for the expression of IRT1 is iron deficiency; IRT1 is not expressed in zinc-deficient roots (data not shown). However, when plants are grown without iron, high zinc causes post-transcriptional regulation of IRT1. It is possible that post-transcriptional regulation of IRT1 by zinc is mediated via either a direct or an indirect mechanism. If the regulation is mediated via a direct mechanism, then the sensing machinery would respond to either high intracellular iron or zinc levels to activate post-transcriptional regulation of IRT1. In contrast, it is possible that high zinc levels alter iron pools within the cell, resulting in indirect activation of the post-transcriptional regulation of IRT1. For example, it is possible that high zinc competes with iron for ferrochelatase in the production of heme. As a result, lower use of iron would lead to higher iron pools, which in turn would affect IRT1 protein accumulation.
The observation that 35S-IRT1 transgenic plants are sensitive to cadmium when grown on iron-deficient medium supports the hypothesis that IRT1 mediates the transport of this metal. Cadmium is an important environmental pollutant; it is released into the environment by a number of anthropogenic activities, including the use of power stations and waste incinerators (Sanita di Toppi and Gabbrielli, 1999). Plants take up cadmium and thus serve as the entry point for cadmium into the food chain, where it poses a threat to human health. In recent years, phytoremediation, the use of plants to clean up contaminated areas, has been proposed as an environmentally friendly, inexpensive way to remove toxic metals such as cadmium from the environment. Because IRT1 mediates the transport of cadmium, it may be possible to engineer plants that specifically hyperaccumulate cadmium, thus removing it from contaminated soils. Indeed, recent work showed that single amino acid changes in IRT1 result in altered selectivity of transport and is promising for this application (Rogers et al., 2000). However, to design plants capable of hyperaccumulating cadmium, we first must elucidate the mechanism of post-transcriptional regulation in response to metals.
Together, our results demonstrate that metal uptake via IRT1 is a carefully regulated process that is controlled at the levels of transcript and protein accumulation, thereby providing a means to take up sufficient amounts of essential metals while preventing their accumulation to potentially toxic levels. Studies of the iron-storage protein ferritin in the maize ys1 mutant have shown that it too is subject to post-transcriptional regulation in response to iron (Fobis-Loisy et al., 1996). ys1 accumulates less iron than wild-type plants, and it is known that ferritin protein abundance correlates with iron loading. Fobis-Loisy and coworkers showed that ferritin mRNA but not protein accumulates in the maize ys1 mutant when treated with iron.
Thus, the use of the ys1 mutant allowed the uncoupling of ferritin mRNA and protein accumulation. Our study represents a new example of metal-dependent post-transcriptional regulation of a metal transporter in plants. Moreover, this result demonstrates that the targeted overexpression of IRT1 is not sufficient to confer dominant gain-of-function enhancement of metal uptake. It seems that it will not be a straightforward matter to engineer plants that accumulate iron; currently, we are working to identify mutations within IRT1 that render it insensitive to post-transcriptional regulation. Overexpression of such IRT1 alleles may lead to increased iron accumulation in plants, particularly in conjunction with the overexpression of other proteins necessary for the transport and storage of iron in plants.
METHODS
Plant Growth Conditions
Wild-type seeds of Arabidopsis thaliana (ecotype Columbia gl-1) and transgenic 35S-IRT1 seeds were surface-sterilized, placed in the dark at 4°C for 2 days, and then sown on plates of Gamborg's B5 medium (Sigma-Aldrich, St. Louis, MO) supplemented with 2% sucrose, 1 mM Mes, and 0.6% agar, pH 5.8. Transgenic plants were grown on plates supplemented with kanamycin (50 μg/mL). Plates were incubated at 21°C under constant illumination (∼90 μE·m−2·s−1) for 12 to 14 days until they reached the four- to six-true-leaf stage. Plants were grown under a yellow filter (acrylic yellow-2208; Cadillac Plastic and Chemical, Pittsburgh, PA) to prevent the photochemical degradation of Fe(III)-EDTA (Hangarter and Stasinopoulos, 1991).
Seedlings were transferred to plates that were either iron sufficient [50 μM Fe(III)-EDTA] or iron deficient {300 μM FerroZine [3- (2-pyridyl)-5,6-diphenyl-1,2,4-triazine sulfonate]; HACH Chemical, Ames, IA}. The medium contained macronutrients and micronutrients (Marschner et al., 1982), 0.6% agar, and 1 mM Mes, pH 6.0. In some cases, plants were grown on either iron-sufficient or iron-deficient plates that were supplemented with 100 μM ZnSO4, 500 μM ZnSO4, 50 μM CdSO4, or 90 μM CdSO4. Plants were incubated for various times after transfer in a growth chamber as described.
Construction of the Chimeric 35S-IRT1 Gene of Cauliflower mosaic virus
The Arabidopsis IRT1 cDNA was cloned into the BamHI site of pCGN18, creating pIRT1 sense. pCGN18 is derived from pCGN1547 (McBride and Summerfelt, 1990). A HindIII-KpnI fragment containing the 35S promoter of Cauliflower mosaic virus and 3′ nopaline synthase terminator was cloned into the HindIII-KpnI sites of pCGN1547 to create pCGN18 (a kind gift of Dr. T. Jack, Department of Biological Sciences, Dartmouth College, Hanover, NH). The promoter and terminator are separated by a single BamHI site.
Plant Transformation
The pIRT1 sense construct was used to transform Agrobacterium tumefaciens strain ASE (Rogers et al., 1988), and transformants were selected on medium containing kanamycin (50 μg/mL) and gentamycin (30 μg/mL). Agrobacterium-mediated transformation of wild-type Arabidopsis plants (Columbia gl-1) was accomplished using vacuum infiltration (Bent et al., 1994). T1 seeds obtained from self-fertilization of the primary transformants were surface-sterilized and sown on Gamborg's B5 medium supplemented with kanamycin. Kanamycin-resistant plants were transferred to soil, and the T2 seeds resulting from self-fertilization were collected.
The T2 seeds were surface-sterilized, plated on the same medium, and scored for resistance to the antibiotic. Transgenic lines that displayed 3:1 segregation for kanamycin resistance to kanamycin sensitivity in the T2 generation and that were 100% kanamycin resistant in the T3 generation were selected for further analysis. Six independent, single-insertion transgenic lines were isolated in this way. Single insertions were verified using genomic DNA gel blot analysis. All further experiments were performed using T4 or T5 seeds.
Isolation of RNA and RNA Gel Blot Analysis
Total RNA was prepared (Verwoerd et al., 1989) from the roots and shoots of plants grown axenically on plates that were either iron deficient or iron sufficient. Plates were supplemented with 100 μM ZnSO4, 500 μM ZnSO4, or 90 μM CdSO4 as noted. RNA samples (10 μg) were modified covalently by treatment with glyoxal (McMaster and Carmichael, 1977), separated on a 1.2% agarose gel containing 10 mM NaPO4, pH 6.5, transferred to a nylon membrane, and bound to the membrane by UV cross-linking (Stratalinker; Stratagene, La Jolla, CA). Hybridizations were performed in 50% formamide at 42°C using standard procedures (Ausubel et al., 2002). Membranes were washed twice for 15 min at room temperature in 1 × SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.1% SDS, followed by two 15-min washes in 0.1 × SSC and 0.1% SDS at 65°C. The 1.4-kb NotI fragment from pIRT-1 containing the IRT1 cDNA was used as a probe for RNA gel blot analysis (Eide et al., 1996). The probe is specific for IRT1, because no signal is detected on a RNA gel blot of RNA prepared from an IRT1 knockout plant (Vert et al., 2002). DNA fragments used as probes were radiolabeled according to the random-primer method (Feinberg and Vogelstein, 1984).
Isolation of Protein and Immunoblot Analysis
Total protein was prepared from the roots and shoots of plants grown axenically on plates that were either iron deficient or iron sufficient. Extracts were prepared by grinding tissue (2 mL of buffer per 1 g of wet tissue) on ice in extraction buffer (50 mM Tris, pH 8.0, 5% glycerol, 4% SDS, 1% polyvinylpolypyrrolidone, and 1 mM phenylmethylsulfonyl fluoride) followed by centrifugation at 4°C for 15 min at 14,000g. The supernatant was recovered, and total protein was estimated using the bicinchoninic acid protein assay (Pierce, Rockford, IL). Samples for SDS-PAGE were diluted with an equal volume of 2 × sample prep buffer (Ausubel et al., 2002) and boiled for 2 min.
Total protein (10 μg) was separated by SDS-PAGE (Laemmli, 1970) and transferred to polyvinylidene fluoride membranes by electroblotting (Towbin et al., 1979). Membranes were blocked in 1 × PBST (0.1% Tween 20 in 1× PBS) with 5% nonfat dry milk for 3 h at 37°C and then washed two times in 1 × PBST for 5 min each. The membranes then were incubated overnight at 4°C with affinity-purified IRT1 peptide antibody (1:1000 dilution in 1× PBST and 1% nonfat dry milk). The IRT1 peptide antibody was raised against a synthetic peptide (PANDVTLPIKEDDSSN) that corresponds to amino acids 162 to 177 of the IRT1 deduced protein sequence and is unique to IRT1 (Quality Controlled Biochemicals, Hopkinton, MA). The antibody is specific for IRT1, because no antigen is detected in extracts from an IRT1 knockout line (Vert et al., 2002). Next, the membranes were washed in 1 × PBST four times for 15 min each. Membranes then were incubated for 1 h with goat anti-rabbit IgG conjugated to horseradish peroxidase (1:5000 dilution in 1× PBST and 1% nonfat dry milk) followed by four washes for 15 min each in 1 × PBST. Chemiluminescence was performed using the Renaissance protein gel blot chemiluminescence reagent according to the directions of the manufacturer (DuPont–New England Nuclear, Boston, MA).
Elemental Analysis
Mineral concentrations in the shoots and roots of wild-type and transgenic plants grown on plates were determined. Plants were germinated on Gamborg's B5 medium and were transferred at the four- to six-true-leaf stage to plates that were either iron sufficient or iron deficient either with or without added cadmium (90 μM CdSO4). After 6 days, plants were harvested, and the roots and shoots were separated and dried overnight in a 65°C oven. Approximately 50 plants were pooled for each sample. Elemental analysis was performed using inductively coupled argon plasma spectrometry at the Soil and Plant Tissue Testing Laboratory at the University of Massachusetts (Amherst). The data were analyzed using the multivariate analysis of variance test.
Root Growth Sensitivity to Cadmium
Seedlings (Columbia gl-1 and transgenic line 4) were grown for 8 days as described above. After 8 days, plants were transferred to plates that were either iron sufficient [50 μM Fe(III)-EDTA] or iron deficient (300 μM FerroZine) and contained 0, 0.01, 0.1, 1, 10, 50, 100, 250, or 500 μM CdSO4. Plants were placed on the plates such that their roots extended in as straight a line as possible across the surface of the agar. Plates were placed in the growth chamber in a vertical orientation so that the roots grew down along the surface of the agar. Root length was measured at days 0, 1, 2, 3, 4, 5, and 6.
Accession Number
The GenBank accession number for the IRT1 cDNA is U27590.
Acknowledgments
We are grateful to Blair Seidler for technical assistance and to Mark McPeek for assistance with statistical analysis. We thank Rob McClung and David Eide for critical reading of the manuscript. This work was supported by Department of Energy Grant 07-97ER20292 to M.L.G. and U.S. Department of Agriculture National Research Initiative Competitive Grants Program Grant 9900598 to E.L.C.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.001263.
References
- Assuncao, A.G.L., Martins, P., De Folter, S., Vooijs, R., Schat, H., and Aarts, M.G.G. (2001). Elevated expression of metal transporter genes in three accessions of the metal hyperaccumulator Thlaspi caerulescens. Plant Cell Environ. 24, 217–226. [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (2002). Current Protocols in Molecular Biology. (New York: John Wiley & Sons).
- Bent, A.F., Kunkel, B.N., Dahlbeck, D., Brown, K.L., Schmidt, R., Giraudat, J., Leung, J., and Staskawicz, B.J. (1994). RPS2 of Arabidopsis thaliana: A leucine-rich repeat class of plant disease resistance genes. Science 265, 1856–1860. [DOI] [PubMed] [Google Scholar]
- Briat, J.-F., and Lobréaux, S. (1997). Iron transport and storage in plants. Trends Plant Sci. 2, 187–193. [Google Scholar]
- Eckhardt, U., Marques, A.M., and Buckout, T.J. (2001). Two iron-regulated cation transporters from tomato complement metal uptake-deficient yeast mutants. Plant Mol. Biol. 45, 437–448. [DOI] [PubMed] [Google Scholar]
- Eide, D., Broderius, M., Fett, J., and Guerinot, M.L. (1996). A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. USA 93, 5624–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg, A.P., and Vogelstein, B. (1984). A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 137, 266–267. [DOI] [PubMed] [Google Scholar]
- Fobis-Loisy, I., Aussel, L., and Briat, J.-F. (1996). Post-transcriptional regulation of plant ferritin accumulation in response to iron as observed in the maize mutant ys1. FEBS Lett. 397, 149–154. [DOI] [PubMed] [Google Scholar]
- Gaither, L.A., and Eide, D.J. (2000). Functional expression of the human hZIP2 zinc transporter. J. Biol. Chem. 275, 5560–5564. [DOI] [PubMed] [Google Scholar]
- Gaither, L.A., and Eide, D.J. (2001). The human ZIP1 transporter mediates zinc uptake in human K562 erythroleukemia cells. J. Biol. Chem. 276, 22258–22264. [DOI] [PubMed] [Google Scholar]
- Gitan, R.S., and Eide, D.J. (2000). Zinc-regulated ubiquitin conjugation signals endocytosis of the yeast ZRT1 zinc transporter. Biochem. J. 346, 329–336. [PMC free article] [PubMed] [Google Scholar]
- Gitan, R.S., Luo, H., Rodgers, J., Broderius, M., and Eide, D. (1998). Zinc-induced inactivation of the yeast ZRT1 zinc transporter occurs through endocytosis and vacuolar degradation. J. Biol. Chem. 273, 28617–28624. [DOI] [PubMed] [Google Scholar]
- Grass, G., Wong, M.D., Rosen, B.P., Smith, R.L., and Rensing, C. (2002). ZupT is a Zn(II) uptake system in E. coli. J. Bacteriol. 184, 864–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotz, N., Fox, T., Connolly, E.L., Park, W., Guerinot, M.L., and Eide, D. (1998). Identification of a family of zinc transporter genes from Arabidopsis that respond to zinc deficiency. Proc. Natl. Acad. Sci. USA 95, 7220–7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerinot, M.L. (2000). The ZIP family of metal transporters. Biochim. Biophys. Acta 1465, 190–198. [DOI] [PubMed] [Google Scholar]
- Guerinot, M.L., and Eide, D. (1999). Zeroing in on zinc uptake in yeast and plants. Curr. Opin. Plant Biol. 2, 244–249. [DOI] [PubMed] [Google Scholar]
- Hangarter, R.P., and Stasinopoulos, T.C. (1991). Effect of Fe-catalyzed photooxidation of EDTA on root growth in plant culture media. Plant Physiol. 96, 843–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden, R., and Cobbett, C.S. (1992). Cadmium-sensitive mutants of Arabidopsis thaliana. Plant Physiol. 99, 100–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korshunova, Y.O., Eide, D., Clark, W.G., Guerinot, M.L., and Pakrasi, H.B. (1999). The IRT1 protein from Arabidopsis thaliana is a metal transporter with broad specificity. Plant Mol. Biol. 40, 37–44. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lescure, A.-M., Proudhon, D., Pesey, H., Ragland, M., Theil, E.C., and Briat, J.-F. (1991). Ferritin gene transcription is regulated by iron in soybean cell cultures. Proc. Natl. Acad. Sci. USA 88, 8222–8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X.F., and Culotta, V.C. (1999). Post-translation control of Nramp metal transport in yeast: Role of metal ions and the BSD2 gene. J. Biol. Chem. 274, 4863–4868. [DOI] [PubMed] [Google Scholar]
- MacDiarmid, C.W., Gaither, L.A., and Eide, D.J. (2000). Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J. 19, 2845–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner, H., Römheld, V., and Ossenberg-Neuhaus, H. (1982). Rapid method for measuring changes in pH and reducing processes along roots of intact plants. Z. Pflanzenphysiol. 105, 407–416. [Google Scholar]
- McBride, K.E., and Summerfelt, K.R. (1990). Improved binary vectors for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 14, 269–276. [DOI] [PubMed] [Google Scholar]
- McMaster, G.K., and Carmichael, G.G. (1977). Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels using glyoxal and acridine orange. Proc. Natl. Acad. Sci. USA 74, 4835–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi, C.E., Rabinovich, E., Dancis, A., Bonifacino, J.S., and Klausner, R.D. (1996). Copper-dependent degradation of the Saccharomyces cerevisiae plasma membrane copper transporter Ctr1p in the apparent absence of endocytosis. EMBO J. 15, 3515–3523. [PMC free article] [PubMed] [Google Scholar]
- Pence, N.S., Larsen, P.B., Ebbs, S.D., Letham, D.L.D., Lasat, M.M., Garvin, D.F., Eide, D., and Kochian, L.V. (2000). The molecular physiology of heavy metal transport in the Zn/Cd hyperaccumulator Thlaspi caerulescens. Proc. Natl. Acad. Sci. USA 97, 4956–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, N.J., Proctor, C.M., Connolly, E.L., and Guerinot, M.L. (1999). A ferric-chelate reductase for iron uptake from soils. Nature 397, 694–697. [DOI] [PubMed] [Google Scholar]
- Rogers, E.E., Eide, D.J., and Guerinot, M.L. (2000). Altered selectivity in an Arabidopsis metal transporter. Proc. Natl. Acad. Sci. USA 97, 12356–12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, S.G., Klee, H.J., Horsch, R.B., and Fraley, R.T. (1988). Improved vectors for plant transformation: Expression cassette vectors and new selectable markers. Methods Enzymol. 253, 253–277. [Google Scholar]
- Sanita di Toppi, L., and Gabbrielli, R. (1999). Response to cadmium in higher plants. Environ. Exp. Bot. 41, 105–130. [Google Scholar]
- Sharp, P., Tandy, S., Yamaji, S., Tennant, J., Williams, M., and Srai, S.K.S. (2002). Rapid regulation of divalent metal transporter (DMT1) protein but not mRNA expression by non-haem iron in human intestinal Caco-2 cells. FEBS Lett. 510, 71–76. [DOI] [PubMed] [Google Scholar]
- Towbin, H., Staehelin, T., and Gordon, J. (1979). Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA 76, 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert, G., Briat, J.-F., and Curie, C. (2001). Arabidopsis IRT2 gene encodes a root-periphery transporter. Plant J. 26, 181–189. [DOI] [PubMed] [Google Scholar]
- Vert, G., Grotz, N., Dédaldéchamp, F., Gaymard, F., Guerinot, M.L., Briat, J.-F., and Curie, C. (2002). IRT1, an Arabidopsis transporter essential for iron uptake from the soil and plant growth. Plant Cell 14, 1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwoerd, T.C., Dekker, B.M.M., and Hoekema, A. (1989). A small scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res. 17, 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, J., and Theil, E.C. (2000). Identification and characterization of the iron regulatory element in the ferritin gene of a plant (soybean). J. Biol. Chem. 275, 17488–17493. [DOI] [PubMed] [Google Scholar]
- World Health Organization. (2002). Available at http://www.who.int/nut/ida.htm. Micronutrient deficiencies. June 9, 2002. [Google Scholar]
- Zhao, H., Butler, E., Rodgers, J., Spizzo, T., Duesterhoeft, S., and Eide, D. (1998). Regulation of zinc homeostasis in yeast by binding of the ZAP1 transcriptional activator to zinc-responsive promoter elements. J. Biol. Chem. 273, 28713–28720. [DOI] [PubMed] [Google Scholar]
- Zhao, H., and Eide, D. (1996. a). The yeast ZRT1 gene encodes the zinc transporter of a high affinity uptake system induced by zinc limitation. Proc. Natl. Acad. Sci. USA 93, 2454–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, H., and Eide, D. (1996. b). The ZRT2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J. Biol. Chem. 271, 23203–23210. [DOI] [PubMed] [Google Scholar]
- Zhao, H., and Eide, D. (1997). Zap1p, a metalloregulatory protein involved in zinc-responsive transcriptional regulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 17, 5044–5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, Z., Labbe, S., Pena, M.M.O., and Thiele, D.J. (1998). Copper differentially regulates the activity and degradation of yeast Mac1 transcription factor. J. Biol. Chem. 273, 1277–1280. [DOI] [PubMed] [Google Scholar]