Abstract
The Arabidopsis NPR1 protein is a key regulator of salicylic acid (SA)–mediated gene expression in systemic acquired resistance. Based on yeast two-hybrid analysis, NPR1 has been suggested to interact with members of the TGA family of transcription factors, including TGA2 (AHBP-1b). However, genetic evidence demonstrating that the NPR1–TGA interaction occurs in planta is still lacking, and the role of this interaction in SA-mediated gene activation has yet to be determined. In this study, we expressed a truncated form of TGA2 in Arabidopsis and found that the resulting transgenic lines displayed phenotypes similar to those of npr1 mutants. This dominant-negative effect of the TGA2 mutant shows that TGA2 and NPR1 interact in planta. We also present biochemical evidence indicating that this interaction is specific and enhanced by SA treatment. Moreover, using a chimera reporter system, we found that a chimeric TGA2GAL4 transcription factor activated a UASGAL::GUS reporter gene in response to SA and that this activation was abolished in the npr1 mutant. NPR1 is required for the DNA binding activity of the transcription factor. These genetic data clearly demonstrate that TGA2 is a SA-responsive and NPR1-dependent transcription activator.
INTRODUCTION
Systemic acquired resistance (SAR) is a plant defense response induced after a local hypersensitive response to avirulent pathogens or by treatment with signal molecules such as salicylic acid (SA), 2,6-dichloroisonicotinic acid (INA), and benzothiadiazole (Ryals et al., 1996). Induction of SAR involves the activation of many pathogenesis-related (PR) genes, which function in concert to confer resistance against a broad spectrum of pathogens (Ward et al., 1991; Uknes et al., 1992). The signal transduction pathway leading to SAR has been studied using both genetic and molecular approaches. The Arabidopsis npr1 (nonexpresser of PR genes) mutants (also known as nim1 and sai1) (Cao et al., 1994; Delaney et al., 1995; Glazebrook et al., 1996; Shah et al., 1997) were identified by their loss of PR gene induction and disease resistance under SAR-activating conditions.
The positive role of NPR1 in SAR, suggested by the phenotype of these recessive npr1 mutants, was further demonstrated by overexpression experiments. Overexpression of the NPR1 gene in Arabidopsis and rice rendered the transgenic plants more resistant to various pathogens in the absence of a SAR inducer or after treatment with lower-than-normal concentrations of the inducer (Cao et al., 1998; Chern et al., 2001; Friedrich et al., 2001). Interestingly, overexpression of NPR1 did not result in constitutive PR gene expression before pathogen challenge, indicating that the NPR1 protein requires activation, perhaps by SA, to be functional in PR gene activation. The derived amino acid sequence of the NPR1 protein (Cao et al., 1997; Ryals et al., 1997) has provided some hints about its molecular function.
A bipartite nuclear localization sequence at the carboxyl end of NPR1 mediates its nuclear localization, which is required for the induction of PR genes (Kinkema et al., 2000). NPR1 also contains two protein–protein interaction domains: a BTB/POZ domain (Aravind and Koonin, 1999) at the N-terminal end and an ankyrin-repeat domain (ARD) in the center of the protein (Cao et al., 1997). Although many proteins contain either BTB/POZ or ARD domains, NPR1 belongs to a unique family of proteins that carry both domains. The functional importance of these protein–protein interaction domains is highlighted by the various npr1 mutants identified with amino acid changes in the consensus of these domains (Cao et al., 1997; Ryals et al., 1997).
The presence of two protein–protein interaction domains but the lack of a DNA binding domain suggest that NPR1 may exert its regulatory role in PR gene expression through interaction with transcription factors. Indeed, using yeast two-hybrid screens, we and others found that NPR1 interacts with the TGA subclass of basic Leu zipper (bZIP) transcription factors (Zhang et al., 1999; Després et al., 2000; Niggeweg et al., 2000b; Zhou et al., 2000; Chern et al., 2001), suggesting that TGA factors could be the missing link between NPR1 and its target genes. This notion is supported by several studies in which the binding sites for TGA transcription factors (e.g., the as-1 element) in PR gene promoters were found to be responsible for SA-mediated gene induction (Qin et al., 1994; Shah and Klessig, 1996; Lebel et al., 1998).
Although the seven known Arabidopsis TGA transcription factors have high degrees of amino acid sequence identity and similarity, they have different affinities toward NPR1 in the yeast two-hybrid assay, with TGA2 (first known as AHBP-1b), TGA3, and TGA6 showing the strongest binding (Zhang et al., 1999; Després et al., 2000; Niggeweg et al., 2000b; Zhou et al., 2000). The TGA factors also differ in their binding affinity and specificity to the as-1 element (Miao et al., 1994; Lam and Lam, 1995; Xiang et al., 1997). These results suggest that some TGA factors may have redundant or overlapping functions, whereas others may play different roles in regulating genes in plant defense and other biological processes. Moreover, TGA factors can form homodimers and heterodimers through their highly conserved bZIP domains, further enhancing the versatility of these transcription factors (Foster et al., 1994; Lam and Lam, 1995).
However, these characteristics make it difficult to define the function of any specific TGA factor. Indeed, examination of Arabidopsis mutants in the TGA2, TGA3, and TGA6 genes have yet to reveal a detectable phenotype (M. Kesarwani and X. Dong, unpublished data), suggesting that these NPR1-interacting TGA factors are at least partially redundant in function. In addition to the TGA knockout mutants, trans-dominant TGA mutants compromised in their ability to bind to the as-1 element have been created and characterized in tobacco. Transgenic plants overexpressing these TGA mutants showed different phenotypes. In one study, overexpression of a tobacco TGA2.2 mutant (a homolog of Arabidopsis TGA2) diminished SA- and auxin-inducible PR gene induction (Niggeweg et al., 2000a). In another study, in which an Arabidopsis TGA2 mutant was overexpressed, PR gene induction was enhanced further (Pontier et al., 2001).
Because both mutants used in these studies were defective only in the basic domain involved in DNA binding, they could interact not only with NPR1 but also with other TGA factors. The complexity of the interactions made it difficult to determine the exact cause of the observed phenotypes and to define the specific functions of these TGA transcription factors. Therefore, genetic evidence demonstrating that the NPR1–TGA interaction occurs in planta is still lacking, and the role of this interaction in SA-mediated gene activation has yet to be established.
In this study, we present both genetic and biochemical evidence demonstrating that TGA2 and NPR1 interact specifically in planta and that this interaction is enhanced by SA treatment. Moreover, we provide clear evidence showing that TGA2 is a SA-responsive and NPR1-dependent transcription activator. Based on these new findings, a working model is proposed to explain the molecular mechanism by which NPR1 regulates SAR-related gene expression.
RESULTS
Construction of the Dominant-Negative Mutant of TGA2
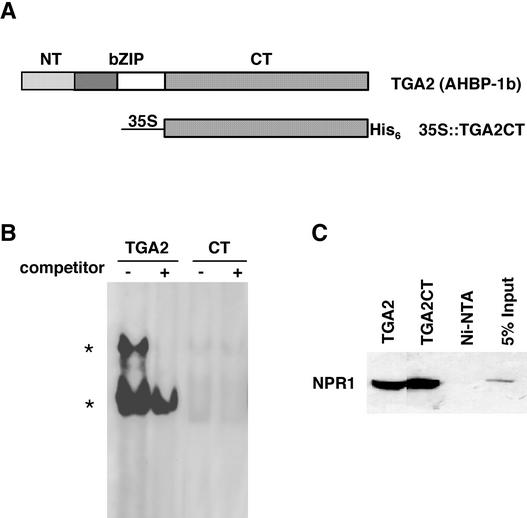
Based on their amino acid sequence similarities, Arabidopsis TGA transcription factors can be further divided into several subgroups, with TGA2, OBF5, and TGA6 belonging to one subgroup (Niggeweg et al., 2000b). Both TGA2 and TGA6 interact strongly with NPR1 in yeast two-hybrid analysis, whereas the interaction between OBF5 and NPR1 is significantly weaker (Zhang et al., 1999). In this study, we focus on understanding the function of TGA2. As shown in Figure 1A, the TGA2 transcription factor consists of three distinct domains: the N-terminal domain (NT; amino acids 1 to 46), which affects the stability (Zhou et al., 2000) and possibly the transactivation activity of the protein (Pascuzzi et al., 1998); the bZIP domain (bZIP; amino acids 47 to 94), which is involved in DNA binding and dimerization (Katagiri et al., 1992); and the C-terminal domain (CT; amino acids 95 to 330), which is sufficient for interaction with NPR1 in yeast (Zhang et al., 1999; Zhou et al., 2000). Crystal structure studies of other bZIP transcription factors suggest that each of these domains is independent in folding and function (O'Shea et al., 1991; Ellenberger et al., 1992). Therefore, it is possible to remove or replace a specific domain in TGA2 without significantly affecting the structure and function of other domains.
Figure 1.
In Vitro Characterization of TGA2CT.
(A) TGA2 and 35S::TGA2CT. The TGA2 transcription factor consists of the NT domain (amino acids 1 to 46), the bZIP domain (amino acids 47 to 94), and the CT domain (amino acids 95 to 330). TGA2CT was put under the control of the 35S CaMV, and the resulting protein was tagged with a His6 tag at the C terminus.
(B) GMSA. The recombinant proteins were purified from E. coli. The proteins (0.1 μg of TGA2 or 1 μg of TGA2CT [CT]) were incubated with 32P-labeled as-1 element (4 × 104 cpm) in the presence (+) or absence (−) of 5 ng of unlabeled probe as a competitor. The TGA2–as-1 complexes are indicated by asterisks.
(C) In vitro copurification analysis. Purified recombinant TGA proteins (5 μg) were mixed with 20 μL of protein extract from an NPR1-expressing insect cell line (Zhang et al., 1999) and loaded onto Ni-NTA columns. The His-tagged TGA proteins and their interactors were eluted, and the eluates were run on an SDS-PAGE gel. As a control, the NPR1 extract alone was loaded onto an Ni-NTA column, and the eluate was loaded onto the gel. As an additional control, 1 μL of the NPR1 extract (5% input) was applied directly to the gel. The NPR1 protein that coeluted with the TGA factors was detected by protein gel blot analysis using an antibody against NPR1 (Cao et al., 1998).
To examine the activity of TGA2 in the regulation of PR genes in Arabidopsis, we generated a truncated TGA2 construct, TGA2CT, in which the coding regions of NT and the bZIP domains were deleted, leaving only that of the CT domain (Figure 1A). To facilitate the detection of this mutant protein, the sequence encoding a polyhistidine (His6) tag was added to the 3′ end of the gene. As shown in Figure 1A, the in planta expression of this recombinant gene is controlled by a modified 35S promoter of Cauliflower mosaic virus (35S CaMV) (Mindrinos et al., 1994). The functionality of the mutant protein was examined first using in vitro analysis. The His-tagged TGA2CT protein was produced in Escherichia coli and purified using a nickel–nitrilotriacetic acid agarose (Ni-NTA) column (Zhang et al., 1999). The TGA2CT mutant could no longer bind to the as-1 element, as demonstrated by a gel mobility shift assay (GMSA) (Figure 1B), but it did maintain the ability to interact with NPR1, as shown by the copurification experiment (Figure 1C) (Zhang et al., 1999).
Expression of TGA2CT in Wild-Type Plants Renders npr1 Mutant-Like Phenotypes
After the in vitro tests had verified the expected properties of the TGA2CT protein, 35S::TGA2CT was transformed into both wild-type and npr1-2 mutant plants. In the wild-type background, 23 transformants were analyzed. As shown by the protein gel blots in Figure 2A, TGA2CT accumulated to high levels in lines 5, 6, and 17. The effect of the TGA2CT protein then was investigated by examining the expression of PR genes in response to SAR induction. As shown in Figure 2B, the INA-induced PR-1 expression was reduced significantly in lines with high levels of TGA2CT (lines 5, 6, and 17). This reduction was much less prominent in lines with low levels of TGA2CT (such as line 18). Expression of another PR gene, BGL2 (also known as PR-2), was affected similarly (data not shown). These results indicated that the accumulation of TGA2CT in a wild-type background caused a defect in SAR gene expression, a phenotype similar to that of the npr1 mutants.
Figure 2.
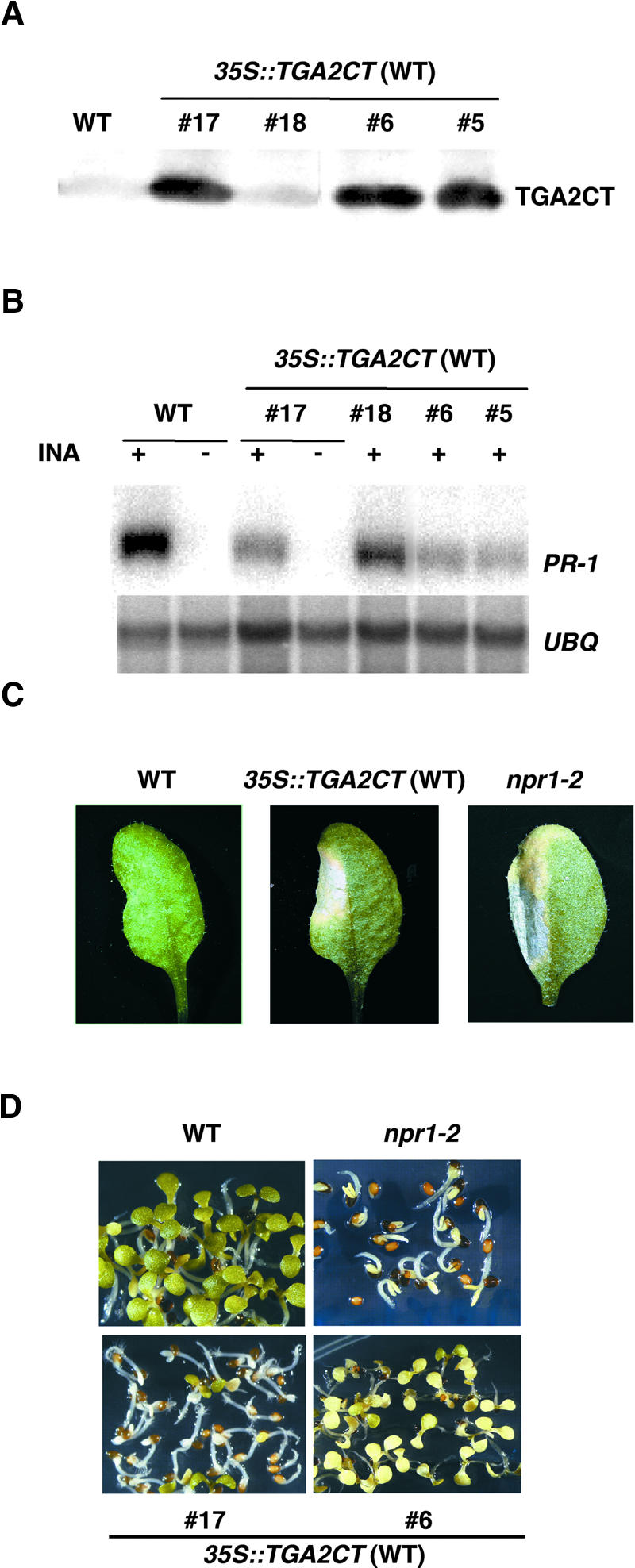
Overexpression of TGA2CT in Wild-Type Plants Results in an npr1-Like Phenotype.
(A) Protein gel blot analysis of the TGA2CT protein in 35S::TGA2CT transformants. Protein extracts were made from 2-week-old plants. Independent 35S::TGA2CT transformants (5, 6, 17, and 18) in the wild-type background (35S::TGA2CT [WT]) were compared with untransformed wild-type plants (WT). The His6-tagged TGA2CT was detected using an antibody against the His tag.
(B) RNA gel blot analysis of PR-1 gene expression. Plants were grown for 2 weeks on MS medium with (+) or without (−) 20 μM INA. Total RNA (10 μg) was used to make the blot, which was probed for the PR-1 and UBQ5 mRNAs.
(C) Disease symptoms caused by P. syringae pv maculicola ES4326. Leaves from 4-week-old wild-type, 35S::TGA2CT (WT) (line 17), and npr1-2 plants were infiltrated with a P. syringae pv maculicola ES4326 suspension of OD600 = 0.0001. Photographs were taken of representative leaves 4 days after infection.
(D) Growth of plants on MS medium containing 0.5 mM SA. The photographs were taken 7 days after germination.
To further characterize the dominant-negative effect of TGA2CT, we examined the transgenic lines for other phenotypes associated with a deficiency in NPR1 function. As seen in all npr1 mutants, infection of 35S::TGA2CT transformants (such as line 17) with a low titer (OD600 = 0.0001) of the bacterial pathogen Pseudomonas syringae pv maculicola ES4326 resulted in enhanced disease symptoms compared with the wild-type control (Figure 2C). We also examined the growth and development of the transgenic lines in the presence of a high concentration of SA (0.5 mM) and found that they were unable to develop beyond the cotyledon stage (Figure 2D). This reduced tolerance to SA has been observed in all npr1 mutants, indicating that the wild-type NPR1 protein is not only required for SA signal transduction but also is involved in the feedback regulation of SA accumulation (Cao et al., 1997).
Dominant-Negative Effect of TGA2CT Is Abolished in the npr1-2 Mutant Background
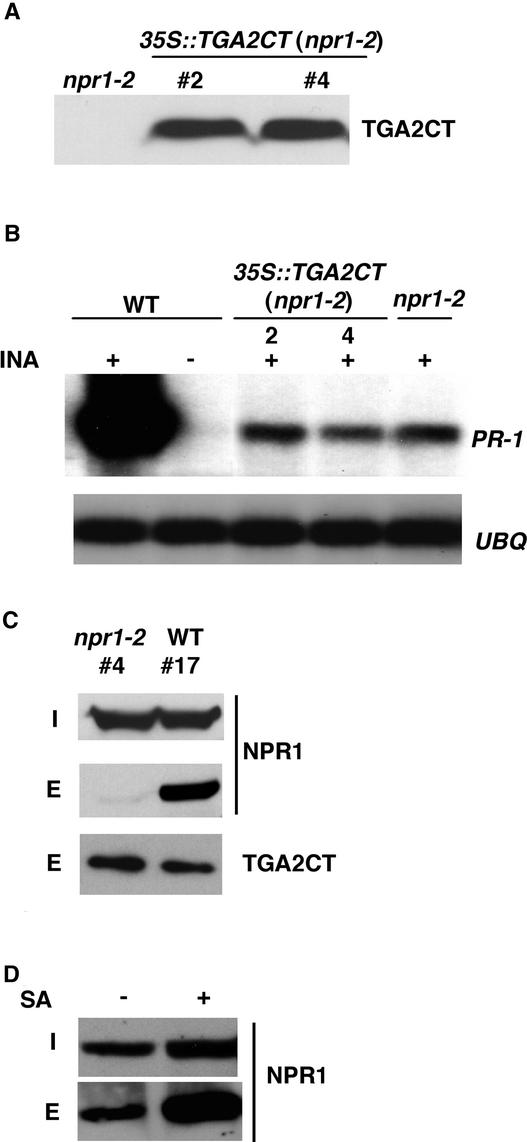
The accumulation of TGA2CT in wild-type plants resulted in mutant phenotypes that resembled those of a weak allele of npr1. If this dominant-negative effect was caused by in vivo interaction between TGA2CT and NPR1, then disruption of this interaction should alleviate the effect. From the yeast two-hybrid screens, we found that both the H334Y mutation in npr1-1 and the C150Y mutation in npr1-2 disrupt the interaction of NPR1 with TGA2 (Zhang et al., 1999; Zhou et al., 2000). Indeed, overexpression of TGA2CT in the npr1-2 mutant background, as shown in Figure 3A, did not cause any significant dominant-negative effect. The residual PR-1 gene induction normally observed in the leaky npr1-2 mutant was not affected by the accumulation of TGA2CT (Figure 3B).
Figure 3.
The Dominant-Negative Effect of TGA2CT Is Dependent on Interaction with NPR1.
(A) Protein gel blot analysis of the TGA2CT protein in independent 35S::TGA2CT transformants 2 and 4 in the npr1-2 mutant background. Protein extracts were made from 4-week-old npr1-2 control and 35S::TGA2CT transgenic lines. The amount of His6-tagged TGA2CT protein in each line was determined using an antibody against the His6 tag.
(B) RNA gel blot analysis of PR-1 gene expression in wild-type (WT), npr1-2, and 35S::TGA2CT (npr1-2) transformants. Plants were grown for 2 weeks on MS medium with (+) or without (−) 20 μM INA. Total RNA (10 μg) was used for RNA gel blot analysis using probes specific for PR-1 and UBQ5.
(C) Copurification of TGA2CT and NPR1 in 35S::TGA2CT transgenic plants. Equal amounts of protein extract from the wild type and npr1-2 transformed with 35S::TGA2CT (lines 4 and 17, respectively) were loaded onto Ni-NTA columns. The input protein extracts (I) and the eluates (E) were run on a 6 to 16% gradient SDS-PAGE gel. The presence of TGA2CT and NPR1 proteins was detected by protein gel blot analysis using antibodies against the His6 tag and NPR1, respectively.
(D) Effect of SA on TGA2CT-NPR1 copurification. Protein extracts were made from 35S::TGA2CT transformant line 17 in the wild-type background with (+) or without (−) a 24-h treatment of 1 mM SA. The amounts of the TGA2CT-NPR1 complex were measured as described in (C).
TGA2CT and NPR1 Form a Complex
To verify further that this lack of dominant-negative effect was attributable to a loss of NPR1–TGA2CT interaction, a copurification assay was performed. Total protein extracts from plants expressing similar levels of the His6-tagged TGA2CT (line 17 in the wild-type background and line 4 in the npr1-2 background) were loaded onto separate Ni-NTA columns. TGA2CT and any associated proteins in the extracts were bound to the columns and then eluted. Using protein gel blot analysis, the presence of wild-type NPR1 and mutant npr1-2 proteins was examined in the input protein extracts in the eluates. As shown in Figure 3C, equal amounts of NPR1 and npr1-2 proteins were detected in the input extracts, indicating that the npr1-2 mutation did not affect the synthesis and accumulation of the protein. However, only the wild-type NPR1 protein was found to coelute with TGA2CT. These results clearly indicate that TGA2CT and NPR1 form a complex in wild-type plants resulting in the dominant-negative effect of TGA2CT, and this complex is disrupted in the npr1-2 mutant, abolishing the dominant-negative effect.
NPR1-TGA2CT Complex Formation Increases in Response to SA Induction
We also investigated the effect of SA on the binding of TGA2CT to NPR1 in planta using the same copurification assay. Protein extracts were made from 35S::TGA2CT in the wild-type background with or without SA treatment and applied to separate Ni-NTA columns. As shown in Figure 3D, the input extract made from SA-treated plants showed a slightly higher level of the NPR1 protein. This finding is consistent with our previous observation that expression of the NPR1 gene is increased moderately during SAR induction (Kinkema et al., 2000). The amount of TGA2CT protein, on the other hand, was found to be constant after SA treatment (data not shown). After washing, the amounts of NPR1 that coeluted with TGA2CT were measured to determine its binding affinity to TGA2CT. In the absence of SA treatment, a significant amount of NPR1 was found in the eluate, suggesting that SA is not required for NPR1-TGA2 complex formation (Figure 3D). In the SA-treated sample, a noticeable increase in NPR1 was observed in the eluate, indicating that SA either enhances the NPR1–TGA2 interaction or, through an unknown mechanism, makes NPR1 more available for binding to TGA2.
Construction of the Chimera Reporter System
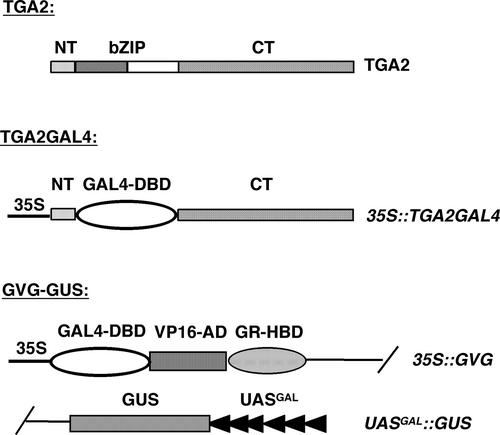
The dominant-negative effect of TGA2CT clearly demonstrated that NPR1 and TGA2 interact in vivo and that TGA2 may participate in SA- and NPR1-regulated gene expression. To observe the biological effect of the NPR1–TGA2 interaction on the transcriptional activity of TGA2 in planta, we constructed the chimera reporter system shown in Figure 4. The bZIP domain of TGA2 (amino acids 47 to 94), which is involved in DNA binding and homodimer and heterodimer formation, was replaced by the DNA binding domain of the yeast GAL4 transcription factor (amino acids 2 to 146) to create the chimeric transcription factor TGA2GAL4. TGA2GAL4 was put under the expression control of the constitutive 35S CaMV (Figure 4). We chose to replace the entire bZIP domain to prevent the formation of dimers between the chimeric TGA2GAL4 transcription factor and other bZIP factors in the cell, which could complicate data interpretation.
Figure 4.
Construction of the TGA2GAL4 Chimeric Transcription Factor and the UASGAL::GUS Reporter.
The bZIP domain of TGA2 (residues 47 to 94) was replaced by the DNA binding domain of the yeast GAL4 transcription factor (GAL4-DBD), and the resulting chimeric transcription factor was put under the control of the 35S promoter to generate 35S::TGA2GAL4. The GAL4-responsive reporter UASGAL::GUS was created by inserting the coding region of β-glucuronidase into plasmid pTA7001 (Aoyama and Chua, 1997) under the control of the synthetic promoter UASGAL, which contains the minimal 35S promoter sequence and six GAL4 binding sites. The plasmid (pGVG-GUS) carrying the UASGAL::GUS reporter also contains the chimeric transcription factor GVG, which consists of the DNA binding domain of GAL4 (GAL4-DBD), the activation domain of VP16 (VP16-AD), and the hormone binding domain of glucocorticoid receptor (GR-HBD).
A reporter for the chimeric transcription factor was constructed by putting the coding region of β-glucuronidase (GUS) under the control of a synthetic promoter consisting of a minimal 35S promoter sequence and six GAL4 binding sites (UASGAL) in the vector pTA7001 (Aoyama and Chua, 1997) (Figure 4). This vector also encodes a chimeric transcription factor, GAL4-VP16-GR (GVG). GVG is active in inducing the UASGAL::GUS reporter only in the presence of the steroid hormone dexamethasone (DEX), which is required for the nuclear translocation of this transcription factor (Aoyama and Chua, 1997). This 35S::GVG/UASGAL::GUS (GVG-GUS) construct (Figure 4) was made with the notion that TGA2 could be either a transcriptional activator or a repressor. To detect transcriptional repression, the UASGAL:: GUS reporter has to be transcribed actively, which can be achieved in this GVG-GUS system using DEX treatment.
The GVG-GUS construct was transformed into both wild-type and npr1-2 plants, and the reporter gene activity in each transformant was examined in the presence or absence of DEX (5 μM). Using this assay, we identified transformants that had low basal levels of GUS expression in the absence of DEX and showed clear induction of GUS in the presence of DEX. From these transformants, one homozygous line in the wild-type background (line 22-15) and one in the npr1-2 mutant (line 44-36) were chosen for their similar levels of GUS reporter gene expression in response to activation of GVG. The chimeric transcription factor construct 35S::TGA2GAL4 then was introduced into these two reporter lines through a second round of transformation. Multiple independent transformants were allowed to self-fertilize, and the progeny homozygous for both TGA2GAL4 and GVG-GUS were identified. These lines were selected for the analyses described below.
TGA2GAL4 Activates Reporter Gene Expression in Response to SAR Induction and Is Dependent on NPR1
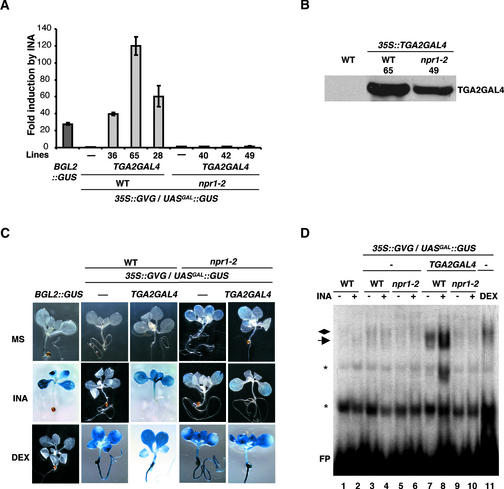
To examine the transactivation activity of TGA2GAL4, UASGAL::GUS reporter expression was measured through both GUS staining and the more quantitative fluorometric 4-methylumbelliferyl β-d-glucuronide assay (Jefferson, 1987). Transgenic lines carrying the TGA2GAL4/UASGAL::GUS chimera reporter system in the wild-type (lines 28, 36, and 65) and npr1-2 (lines 40, 42, and 49) backgrounds were grown in Murashige and Skoog (1962) (MS) medium with or without 100 μM SA or 50 μM INA. To test the integrity of the UASGAL::GUS reporter, some plants were treated with 1 μM DEX. As a control, we used the INA-responsive BGL2::GUS reporter line generated in our laboratory (Bowling et al., 1994; Cao et al., 1994) to monitor the effectiveness of the induction. The BGL2 promoter of this reporter contains as-1–like sequences, suggesting that it might be regulated by the TGA factors.
In the fluorometric GUS assay shown in Figure 5A, in the absence of an inducer, no significant expression of the UASGAL::GUS reporter was observed in either wild-type or npr1-2 mutant backgrounds, indicating that the TGA2GAL4 transcription factor is not autoactive. However, in response to INA induction, expression of the UASGAL::GUS reporter increased by 40- to 120-fold in the three independent lines tested in the wild-type background (Figure 5A). In comparison, the other SA- and INA-responsive reporter, BGL2::GUS, was induced 30-fold (Figure 5A). In the absence of TGA2GAL4, the GVG-GUS plants showed no detectable GUS activity in response to induction (Figure 5A), indicating that TGA2GAL4 is required for the INA induction of the UASGAL::GUS reporter. Treatment of the plants with SA produced similar results (data not shown).
Figure 5.
Characterization of TGA2GAL4 Transcriptional Activity in SAR Induction.
(A) Quantitative GUS assay of plants transformed with the chimera reporter system. Plants were grown on MS medium with and without 50 μM INA for 2 weeks, and GUS activities were measured using 4-methylumbelliferyl β-d-glucuronide as a substrate. The fold induction by INA was calculated for each genotype by comparing the GUS activity of INA-treated plants with that of untreated plants. The values represent averages of three replicates ±se. A transgenic line containing the INA-responsive BGL2::GUS reporter (Bowling et al., 1994; Cao et al., 1994) was used as a positive control. Three independent 35S::TGA2GAL4/UASGAL::GUS transformants in the wild-type background (WT; lines 28, 36, and 65) and three in the npr1-2 background (lines 40, 42, and 49) were examined.
(B) Protein gel blot analysis of the TGA2GAL4 protein in plants transformed with the chimera reporter system. Protein extracts were made from wild-type plants and wild-type and npr1-2 plants transformed with the chimera reporter system (lines 65 and 49, respectively). TGA2GAL4 and GVG were immunoprecipitated using a rabbit polyclonal antibody against GAL4-DBD (amino acids 1 to 147). The TGA2GAL4 protein then was detected using a mouse monoclonal antibody against residues 94 to 147 of GAL4-DBD. The GVG protein, which contains only amino acids 1 to 74 of GAL4-DBD, was not detected on this blot.
(C) Histochemical staining of GUS activities in plants transformed with the chimera reporter system. Plants were grown on MS medium (MS) or MS medium with 50 μM INA (INA) for 2 weeks. As controls, some plants for each genotype were taken from the MS plates, treated with 1 μM DEX in liquid MS medium for 24 h (DEX), and then stained for GUS.
(D) GMSA. Protein extracts were made from 2-week-old plants grown in MS medium with (+) or without (−) 50 μM INA or treated with DEX for 24 h (DEX). 35S::TGA2GAL4/UASGAL::GUS transgenic line 65 in the wild-type background and line 49 in the npr1-2 background were used. Each extract (20 μg) was incubated with a 32P-labeled probe (17-mer) containing UASGAL (4 × 104 cpm), and the mixtures were run on a 4% native PAGE gel. The protein-DNA complexes were detected by autoradiography. The arrow points to the TGA2GAL4-UASGAL complex, the diamond marks the GVG-UASGAL complex, and the asterisks indicate nonspecific complexes. FP, free probe.
These results indicated that TGA2GAL4 is a SA-responsive transcription activator. We further investigated whether the INA-induced activation of TGA2GAL4 is dependent on NPR1. We found that in the npr1-2 background, no induction of the UASGAL::GUS reporter was detected (Figure 5A). To confirm that this lack of reporter induction in the npr1-2 background was not caused by an absence of the TGA2GAL4 protein, immunoprecipitation and protein gel blot analysis were performed. As shown in Figure 5B, significant levels of TGA2GAL4 were detected in both the wild type and the npr1-2 mutant. Thus, we conclude that the transcriptional activity of TGA2GAL4 requires the function of NPR1.
We further examined the spatial expression pattern of the UASGAL::GUS reporter using histochemical GUS staining in transgenic line 65 in the wild-type background and in line 49 in the npr1-2 background. As shown in Figure 5C, the UASGAL::GUS reporter was activated by INA only in the presence of both TGA2GAL4 and NPR1. The expression pattern of UASGAL::GUS, which is controlled by an artificial promoter, was compared with that of BGL2::GUS, which is regulated by the entire promoter sequence of the PR gene, BGL2 (PR-2). In the BGL2::GUS transgenic plants, reporter gene expression was detected only in the aerial parts of the plants, not in roots (Figure 5C). Interestingly, TGA2GAL4-mediated UASGAL::GUS expression showed a similar tissue-specific staining pattern (Figure 5C), despite the ubiquitous presence of the TGA2GAL4 transcription factor.
Induction by SA produced the same result (data not shown). In the npr1-2 background, the reporter gene was not induced by INA (Figure 5C) or SA (data not shown). The tissue-specific and NPR1-dependent expression pattern observed in the wild-type background reflected the transcriptional activity of the TGA2GAL4 factor, not the responsiveness of the reporter gene, because the same UASGAL::GUS reporter showed a ubiquitous expression pattern in both shoots and roots and in both wild-type and npr1-2 backgrounds when it was activated by GVG after DEX treatment. The spatial activity of TGA2GAL4 is consistent with its role as an activator of PR gene expression.
The specificity of SA- and INA-induced TGA2GAL4 activity was investigated by treating the TGA2GAL4/GVG-GUS transformants with several other signal molecules that stimulate various plant biological processes. We found that although SA and INA induced strong GUS staining, no reporter expression was detected in response to stimuli such as auxins, 2,4-D (50 or 500 μM) and indole-3-acetic acid (10 or 200 μM); jasmonic acid (10 or 50 μM); a precursor of ethylene, 1-aminocyclopropane-1-carboxylic acid (50 μM); H2O2 (3 mM); and high salt (150 mM NaCl) (data not shown). These results indicate that TGA2GAL4 responds specifically to SAR inducers.
TGA2GAL4 DNA Binding Is Dependent on NPR1 and Enhanced by the SAR Inducer
To determine the mechanism by which NPR1 regulates TGA2GAL4, we examined the DNA binding activity of this chimeric transcription factor. GMSA was performed using a radiolabeled oligomer containing the GAL4 binding sites (UASGAL) as a probe and protein extracts made from various genotypes after different treatments. As shown in Figure 5D, protein extracts made from the wild-type control did not display any detectable DNA binding activity with or without INA induction (lanes 1 and 2), indicating that no transcription factors in the wild-type plants could bind to the GAL4 binding site. Similarly, no specific DNA binding was detected in plants transformed with GVG-GUS alone (lanes 3 to 6), indicating that GVG did not bind DNA without DEX induction.
DNA binding was observed for GVG only in extracts made from DEX-treated plants (lane 11). However, using the extracts prepared from TGA2GAL4 transgenic line 65 in the wild-type background, DNA binding activity was observed. The DNA-protein complex band was much more intense in samples from the INA-treated plants than in those from untreated plants (lanes 7 and 8). This binding activity was dependent on the function of NPR1, as shown by the diminished DNA binding activity in the npr1-2 background (line 49; lanes 9 and 10). Because the TGA2GAL4 protein accumulated in both the wild-type and npr1-2 backgrounds (Figure 5B), NPR1 must be required for TGA2GAL4 DNA binding but not for TGA2GAL4 stability. However, NPR1 is unlikely to be in the final protein-DNA complex, based on the results of supershift experiments. When two antibodies against different parts of NPR1 were added to the DNA binding reactions, neither of them produced a supershift in the mobility of the DNA-protein complex. However, an antibody against the GAL4 DNA binding domain produced a clear supershifted band in the GMSA (data not shown), indicating that the DNA-protein complex contained TGA2GAL4 but not NPR1. It is possible that the NPR1–TGA2GAL4 interaction is disrupted in the GMSA. It also is possible that upon DNA binding, NPR1 may be released from the complex.
DISCUSSION
TGA2 and other members of the TGA family of bZIP transcription factors, such as TGA3, OBF5, and TGA6, have been shown to interact with NPR1 by several independent yeast two-hybrid screens (Zhang et al., 1999; Després et al., 2000; Niggeweg et al., 2000b; Zhou et al., 2000; Chern et al., 2001). However, as a result of the artificial nature of the yeast two-hybrid system, it was unclear whether the TGA factors interact with NPR1 in planta and whether they take part in the SAR signaling pathway. It is difficult to use knockout mutants to study these highly conserved transcription factors, because they may have redundant or overlapping transcriptional activities. To remedy this problem, we used a dominant-negative mutant and a chimera reporter system to study the function of a specific TGA factor, TGA2. We chose to focus on the properties of TGA2 because it is one of the strong NPR1 interactors identified in the yeast two-hybrid screens (Zhang et al., 1999; Després et al., 2000; Zhou et al., 2000) as well as the major component of the as-1 binding activity in Arabidopsis (Lam and Lam, 1995).
To detect the TGA2–NPR1 interaction in planta, we cloned the CT domain–coding region of TGA2 under the control the 35S promoter and expressed the resulting 35S::TGA2CT gene in wild-type plants. We found that the accumulation of TGA2CT resulted in dosage-dependent phenotypes that resembled those of the npr1 mutants. We showed that this dominant-negative effect of TGA2CT was attributable to in planta complex formation between TGA2CT and NPR1; disruption of this complex formation in the npr1-2 mutant abolished the dominant-negative effect. Copurification experiments showed that formation of the TGA2CT-NPR1 complex did not require the presence of SA but was enhanced by it.
This finding was consistent with the results of a recent study using a transient expression system in tobacco and tomato protoplasts (Subramaniam et al., 2001). The mechanism by which SA enhances NPR1-TGA2 complex formation is unknown. SA may enhance the affinity of the NPR1–TGA2 interaction or increase the availability of NPR1 for TGA2 binding. In a previous experiment, we found that SA treatment increased the distribution of NPR1 protein to the nuclear fraction in which the TGA transcription factors reside (Kinkema et al., 2000). It also is possible that SA releases NPR1 from an inhibitory complex, allowing it to bind TGA2.
The TGA2GAL4/UASGAL::GUS chimera reporter system was created to make in planta observations of TGA2 transcriptional activity during SAR induction. Our data clearly demonstrate that TGA2 is a transcription activator and that its activity requires the presence of SA and a functional NPR1. GMSAs showed that SA and NPR1 both are required for the DNA binding activity of TGA2. This is consistent with previous findings that SA treatment causes an increase in as-1 binding activity in protein extracts (Jupin and Chua, 1996; Stange et al., 1997; Després et al., 2000) and that NPR1 could enhance TGA2 binding to the as-1 element in vitro (Després et al., 2000). Our results now show that changes in the DNA binding of TGA2 result in transcriptional activation in planta.
The TGA2 transcription factor alone is sufficient for SA-responsive, NPR1-dependent gene expression. The CT domain of TGA2 interacts with NPR1 to transduce the SA signal, resulting in enhanced affinity of the bZIP domain for DNA. The transactivation activity of TGA2 seems to reside in the NT domain of the protein, as is the case for the tobacco TGA1a transcription factor (Johnson et al., 2001). When this domain was deleted from TGA2GAL4, the resulting transcription factor no longer activated the UASGAL::GUS reporter, even though it still bound NPR1 (data not shown). It remains to be determined whether the NT domain alone is sufficient for TGA2 transactivation activity.
The chimera reporter system generated in this study allowed us to overcome the problem caused by functional redundancy among the TGA transcription factors and effectively determine the regulatory role of the TGA2 transcription factor in SAR-related gene expression. All TGA factors have the potential to bind to the as-1 element; thus, GMSA is unable to distinguish the binding activity of different TGA factors. Moreover, DNA binding does not necessarily result in transcription activation. In a GMSA conducted by Després et al. (2000) using endogenous TGA factors, an increase in as-1 binding activity was observed in the SAR-induced sample. Surprisingly, when extracts made from an npr1 mutant were used, constitutive as-1 binding activity was detected (Després et al., 2000).
These results indicate that under noninducing conditions, wild-type NPR1 may sequester some TGA factors (e.g., TGA2) from binding to the as-1 element through its association with these transcription factors. In the npr1 mutant, the NPR1–TGA interaction is inhibited; therefore, more TGA factors can bind to the as-1 element. Obviously, in the absence of a functional NPR1, this constitutive as-1 binding activity does not lead to transcription activation.
The endogenous PR gene promoters contain both positive and negative regulatory elements (Lebel et al., 1998). Therefore, regulation of these promoters is certainly more complex than that of the UASGAL::GUS reporter. When the bacterial pathogen P. syringae pv maculicola ES4326 was used to infect transgenic plants carrying the chimera reporter system, little UASGAL::GUS induction was observed (data not shown). The same pathogen infection can cause a significant induction of endogenous PR genes and BGL2:: GUS (Bowling et al., 1994; Cao et al., 1994). This finding indicates that other promoter elements besides as-1, or other transcription factors in addition to TGA2, are important for enhancing the sensitivity of the PR genes to induction. In addition to positive regulators, PR gene expression also is modulated by negative regulators. We found that a loss-of-function mutation in a SAR-suppressor gene, SNI1, resulted in increased levels of background PR gene expression and suppression of the npr1 mutant phenotype (Li et al., 1999).
The results of this study place TGA2 solidly in the SAR signaling pathway. However, the target genes for TGA2 have yet to be identified. The binding sites for the TGA factors exist in many genes, responding to diverse signals in pathogen defense, wounding, and xenobiotic stresses (Xiang et al., 1996; Pascuzzi et al., 1998; Chen and Singh, 1999). In tobacco, TGA1a is involved in regulating several auxin-induced glutathione S-transferase genes such as GNT1 and GNT35 (Pascuzzi et al., 1998; Johnson et al., 2001). In contrast, Arabidopsis TGA2 responds only to the SAR inducers INA and SA but not to any of the xenobiotic stresses tested. TGA2 and TGA1a differ in their interacting proteins, with TGA2 binding to NPR1, a positive transcriptional regulator, and TGA1a associating with a negative transcriptional regulator, p120 (Johnson et al., 2001). It is evident from these studies that the specific activity of a TGA factor is determined or modified by its interacting protein. In the case of TGA2, interaction with NPR1 enables it to activate SA-responsive gene expression. However, we cannot exclude the possibility that TGA2 functions as a repressor when it is associated with other TGA factors or regulators.
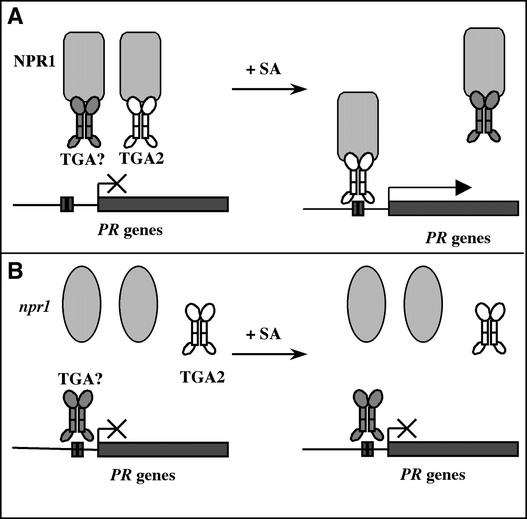
To summarize our current understanding of the regulatory mechanism of TGA2 in SAR-related gene expression, we propose the model shown in Figure 6. In wild-type plants, NPR1 forms complexes with TGA2 and other TGA factors. Under noninducing conditions, these complexes are unable to bind to DNA. Upon SAR induction, more NPR1 becomes available to form the NPR1-TGA2 complex. Through an unknown mechanism, NPR1 enhances the DNA binding activity of TGA2, leading to PR gene induction (Figure 6A). In an npr1 mutant (Figure 6B), the NPR1–TGA2 interaction is disrupted; thus, TGA2 cannot bind to DNA. However, some TGA factors other than TGA2 may be able to bind to DNA in the absence of NPR1, as observed by Després et al. (2000), but they are not active in inducing PR gene expression. This hypothesis can be tested by applying the chimera reporter system described in this work to other members of the TGA transcription factor family.
Figure 6.
Proposed Model for the Regulatory Mechanism of TGA2 in SAR-Related Gene Expression.
(A) In wild-type plants, NPR1 forms complexes with TGA2 and other TGA factors (TGA?). Under noninducing conditions, these complexes are unable to bind to DNA. Upon SAR induction (+SA), more NPR1 becomes available to form the NPR1-TGA2 complex. Through an unknown mechanism, NPR1 enhances the DNA binding activity of TGA2, leading to PR gene induction.
(B) In an npr1 mutant, the NPR1–TGA2 interaction is disrupted, so TGA2 cannot bind to DNA. Some TGA factors (TGA?) may be able to bind to DNA in the absence of NPR1, but they are not active in inducing PR gene expression.
METHODS
DNA Constructs
The TGA2 cDNA was cloned as described previously (Zhang et al., 1999). TGA2CT was cloned by PCR amplification of the DNA fragment encoding amino acids 96 to 330 of TGA2, with an ATG codon added at the 5′ end. A polyhistidine (His6) tag was added to TGA2CT through PCR, and the resulting fragment was cloned into the NdeI and XhoI sites in the Escherichia coli expression vector pET24c(+) (Novagen) (Zhang et al., 1999) and the SalI and BamHI sites in the binary vector pBI1.4t under the control of a modified 35S promoter of Cauliflower mosaic virus (Mindrinos et al., 1994). The 35S::TGA2GAL4 construct was made by substituting the DNA fragment encoding the basic Leu zipper domain (amino acids 47 to 94) with the coding sequence of the GAL4 DNA binding domain (amino acids 2 to 146) from plasmid pMA564 (Ma et al., 1988). The TGA2GAL4 fusion then was cloned into the plant transformation vector pBI1.4t under the control of the 35S promoter. To construct the UASGAL::β-glucuronidase (GUS) reporter, the uidA gene was amplified by PCR from plasmid pBI101 (Jefferson et al., 1986) and cloned into the XhoI and SpeI sites of plasmid pTA7001 (Aoyama and Chua, 1997). All of the clones were sequenced to ensure that no undesired mutations were introduced by PCR.
RNA Gel Blot Analysis of Transgenic Plants
Total RNA was extracted from 2-week-old Arabidopsis plants grown on Murashige and Skoog (1962) (MS) medium or MS medium with 2,6-dichloroisonicotinic acid (INA) or salicylic acid at the concentrations indicated in the figure legends (Cao et al., 1994). RNA samples (10 μg) were used for gel blot analysis as described previously (Cao et al., 1994). To detect the expression of TGA2 and TGA2CT, the region encoding residues 135 to 319 of TGA2 was amplified by PCR and used as a template to generate the gene-specific probe. Fragments used to generate probes specific to PR-1 and UBQ5 were described previously (Cao et al., 1994; Clarke et al., 1998). 32P labeling of the probes was performed using strand-biased PCR (Bowling et al., 1997; Clarke et al., 1998).
Extraction of Arabidopsis Proteins and Protein Gel Blot Analysis
Total protein extracts were made from Arabidopsis plants by grinding leaf tissue first in liquid nitrogen and then on ice in 2 volumes of whole cell extraction buffer (WCEB; 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Triton X-100, 0.2% Nonidet P-40, 6 mM β-mercaptoethanol, and 1 × plant protease inhibitor cocktail [Sigma]). The extracts were cleared of cell debris by centrifugation at 14,000 rpm in a desktop centrifuge at 4°C. Protein concentrations in the extracts were measured by the Bradford (1976) assay (Bio-Rad). A 200-μg aliquot of proteins from each extract was separated on a 12% SDS-PAGE gel (Laemmli, 1970) and then transferred onto a nitrocellulose membrane.
Subsequently, the blots were probed with an antibody against the His6 tag (Amersham Pharmacia Biotech), an antibody against NPR1 (Cao et al., 1998), or an antibody against the GAL4 DNA binding domain (mouse monoclonal anti-GAL4-BD; Santa Cruz Biotechnology, Santa Cruz, CA). The antibodies were diluted in blocking buffer (1× PBS containing 5% nonfat milk and 0.1% Tween 20) to 1:1000 (anti-His6 and anti-NPR1) or 1:500 (anti-GAL4-BD). After washing in 1 × PBS, the blots were probed with appropriate secondary antibodies conjugated with horseradish peroxidase. The antibody-bound proteins were detected by a chemiluminescence reaction using the SuperSignal Kit (Pierce).
Copurification of TGA2CT and NPR1
Total protein was extracted from 10 g of leaf tissue from 4-week-old soil-grown Arabidopsis plants using the method described above. The crude extracts were cleared of cell debris by centrifugation as described above and loaded onto columns containing 1 mL of nickel–nitrilotriacetic acid agarose (Qiagen, Valencia, CA). The His6-tagged TGA2CT protein and its interactors were retained on the columns. To remove any unbound proteins, the columns were washed with wash buffer (50 mM Tris-HCl, 150 mM NaCl, 6 mM β-mercaptoethanol, and 25 mM imidazole). Then, the proteins bound to the columns were eluted with 5 mL of wash buffer plus 150 mM imidazole. Fractions of 1 mL were collected, and the fraction with the highest protein concentration was saved for further analysis. The TGA2CT and NPR1 proteins in the eluates were examined using protein gel blot analysis as described above, except that a 6 to 16% gradient SDS-PAGE was used.
Quantitative GUS Assay
Plants were grown on MS medium or MS medium with 50 μM INA for 2 weeks and then collected. Quantitative GUS assays using the GUS substrate 4-methylumbelliferyl β-d-glucuronide were performed as described previously (Jefferson et al., 1987; Bowling et al., 1994). Three replicate samples were taken for each genotype and treatment. The GUS activity of each sample was specified as the rate of increase of the fluorescent product 4-methylumbelliferone to protein concentration (measured using the Bradford [1976] assay). The fold induction by INA for each genotype was determined by comparing the GUS activity of the INA-induced sample with that of the untreated sample.
Histochemical GUS Assay
Two-week-old plants were stained for GUS activity using a solution containing 0.5 mg/mL 5-bromo-4-chloro-3-indolyl glucuronide in 0.1 M Na2HPO4, pH 7.0, 10 mM EDTA, 0.5 mM potassium ferricyanide/ferrocyanide, and 0.06% Triton X-100 (Jefferson, 1987) at 37°C for 16 h. The staining solution was removed, and the samples were cleared of chlorophyll by sequential changes of 75 and 95% ethanol.
Immunoprecipitation
Arabidopsis proteins were extracted from 4-week-old soil-grown plants by grinding the tissue (2 g) on ice in 3 volumes of WCEB. Cell lysates (6 mL each) were cleared of debris by centrifugation at 14,000g for 15 min at 4°C. The protein level in each lysate was determined by the Bradford (1976) assay and then adjusted to the same concentration with WCEB. Equal volumes (6 mL) of lysates were transferred to new centrifuge tubes, to which 100 μL of protein A–agarose (Upstate, Lake Placid, NY) was added. After incubation at 4°C for 1 h, the protein A–agarose beads were precipitated by centrifugation at 10,000g for 20 s.
The supernatants were transferred to new tubes, and 12 μg of the antibody (rabbit polyclonal anti-GAL4-DBD; Santa Cruz Biotechnology) was added. After incubation at 4°C for 2 h, 100 μL of protein A–agarose was added to precipitate the antigen-antibody complex. The protein A–agarose beads were collected after 1 h of incubation at 4°C by centrifugation at 10,000g for 20 s. The beads then were washed three times with 5 volumes of WCEB. The antigen-antibody complex was eluted by boiling in SDS sample buffer (Laemmli, 1970) and subsequently run on a 10% SDS-PAGE gel.
Gel Mobility Shift Assay
Expression and purification of the recombinant TGA2 and TGA2CT proteins were conducted using previously described methods (Zhang et al., 1999). A 32P-labeled as-1 probe was used for the gel mobility shift assay (GMSA) of the recombinant proteins as described (Zhang et al., 1999). The oligonucleotide 5′-CTAGCGGAGGACTGTCCTCCG-3′ (17-mer) was used as a probe for GMSA of the proteins containing the GAL4 DNA binding domain. A 20-pmol aliquot of the 17-mer oligonucleotide was end labeled with γ-32P-ATP using T4 polynucleotide kinase (New England Biolabs, Beverly, MA). The labeled probe was purified using a Nucleotide Removal Kit (Qiagen) according to the manufacturer's instructions.
Cell extracts were prepared by grinding 2-week-old Arabidopsis plants on ice in 2 volumes of extraction buffer (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 2 mM MgCl2, 0.2 mM EDTA, 10% glycerol, 1 mM DTT, and protease inhibitor cocktail [Sigma]). The crude extracts were cleared of cell debris by centrifugation at 15,000g for 15 min at 4°C. Protein concentrations of the supernatants were determined by the Bradford (1976) assay. A 20-μg aliquot of the supernatant was used to incubate with 4 × 104 cpm of the 17-mer probe and was run subsequently on a 4% native PAGE gel in 0.5× Tris-Borate EDTA buffer. The gel was dried and exposed to a piece of x-ray film.
Acknowledgments
We thank Nam-Hai Chua for providing the pTA7001 vector and Jun Ma for the pMA564 vector. We also thank Wisuwat Songnuan and Lou Annie Yi for assistance with laboratory work and Wendy Durrant, Meenu Kesarwani, Rebecca Mosher, and James Siedow for critical reading of the manuscript. This work was supported by National Science Foundation Grant 0090887 and a grant from Monsanto to X.D.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.001628.
References
- Aoyama, T., and Chua, N.-H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11, 605–612. [DOI] [PubMed] [Google Scholar]
- Aravind, L., and Koonin, E.V. (1999). Fold prediction and evolutionary analysis of the POZ domain: Structural and evolutionary relationship with the potassium channel tetramerization domain. J. Mol. Biol. 285, 1353–1361. [DOI] [PubMed] [Google Scholar]
- Bowling, S.A., Clarke, J.D., Liu, Y., Klessig, D.F., and Dong, X. (1997). The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9, 1573–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling, S.A., Guo, A., Cao, H., Gordon, A.S., Klessig, D.F., and Dong, X. (1994). A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6, 1845–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Cao, H., Bowling, S.A., Gordon, S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, H., Glazebrook, J., Clark, J.D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–64. [DOI] [PubMed] [Google Scholar]
- Cao, H., Li, X., and Dong, X. (1998). Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc. Natl. Acad. Sci. USA 95, 6531–6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W., and Singh, K.B. (1999). The auxin, hydrogen peroxide and salicylic acid induced expression of the Arabidopsis GST6 promoter is mediated in part by an ocs element. Plant J. 19, 667–677. [DOI] [PubMed] [Google Scholar]
- Chern, M.S., Fitzgerald, H.A., Yadav, R.C., Canlas, P.E., Dong, X., and Ronald, P.C. (2001). Evidence for a disease-resistance pathway in rice similar to the NPR1-mediated signaling pathway in Arabidopsis. Plant J. 27, 101–113. [DOI] [PubMed] [Google Scholar]
- Clarke, J.D., Liu, Y., Klessig, D.F., and Dong, X. (1998). Uncoupling PR gene expression from NPR1 and bacterial resistance: Characterization of the dominant Arabidopsis cpr6-1 mutant. Plant Cell 10, 557–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney, T.P., Friedrich, L., and Ryals, J.A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA 92, 6602–6606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després, C., DeLong, C., Glaze, S., Liu, E., and Fobert, P.R. (2000). The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12, 279–290. [PMC free article] [PubMed] [Google Scholar]
- Ellenberger, T.E., Brandl, C.J., Struhl, K., and Harrison, S.C. (1992). The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: Crystal structure of the protein-DNA complex. Cell 71, 1223–1237. [DOI] [PubMed] [Google Scholar]
- Foster, R., Izawa, T., and Chua, N.H. (1994). Plant bZIP proteins gather at ACGT elements. FASEB J. 8, 192–200. [DOI] [PubMed] [Google Scholar]
- Friedrich, L., Lawton, K., Dietrich, R., Willits, M., Cade, R., and Ryals, J. (2001). NIM1 overexpression in Arabidopsis potentiates plant disease resistance and results in enhanced effectiveness of fungicides. Mol. Plant-Microbe Interact. 14, 1114–1124. [DOI] [PubMed] [Google Scholar]
- Glazebrook, J., Rogers, E.E., and Ausubel, F.M. (1996). Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics 143, 973–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R.A. (1987). Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 5, 387–405. [Google Scholar]
- Jefferson, R.A., Burgess, S.M., and Hirsh, D. (1986). β-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc. Natl. Acad. Sci. USA 83, 8447–8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, C., Glover, G., and Arias, J. (2001). Regulation of DNA binding and trans-activation by a xenobiotic stress-activated plant transcription factor. J. Biol. Chem. 276, 172–178. [DOI] [PubMed] [Google Scholar]
- Jupin, I., and Chua, N.H. (1996). Activation of the CaMV as-1 cis-element by salicylic acid: Differential DNA-binding of a factor related to TGA1a. EMBO J. 15, 5679–5689. [PMC free article] [PubMed] [Google Scholar]
- Katagiri, F., Seipel, K., and Chua, N.H. (1992). Identification of a novel dimer stabilization region in a plant bZIP transcription activator. Mol. Cell. Biol. 12, 4809–4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkema, M., Fan, W., and Dong, X. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12, 2339–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lam, E., and Lam, Y.K. (1995). Binding site requirements and differential representation of TGF factors in nuclear ASF-1 activity. Nucleic Acids Res. 23, 3778–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebel, E., Heifetz, P., Thorne, L., Uknes, S., Ryals, J., and Ward, E. (1998). Functional analysis of regulatory sequences controlling PR-1 gene expression in Arabidopsis. Plant J. 16, 223–233. [DOI] [PubMed] [Google Scholar]
- Li, X., Zhang, Y., Clarke, J.D., Li, Y., and Dong, X. (1999). Identification and cloning of a negative regulator of systemic acquired resistance, SNI1, through a screen for suppressors of npr1-1. Cell 98, 329–339. [DOI] [PubMed] [Google Scholar]
- Ma, J., Przibilla, E., Hu, J., Bogorad, L., and Ptashne, M. (1988). Yeast activators stimulate plant gene expression. Nature 334, 631–633. [DOI] [PubMed] [Google Scholar]
- Miao, Z.H., Liu, X., and Lam, E. (1994). TGA3 is a distinct member of the TGA family of bZIP transcription factors in Arabidopsis thaliana. Plant Mol. Biol. 25, 1–11. [DOI] [PubMed] [Google Scholar]
- Mindrinos, M., Katagiri, F., Yu, G.L., and Ausubel, F.M. (1994). The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78, 1089–1099. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Niggeweg, R., Thurow, C., Kegler, C., and Gatz, C. (2000. a). Tobacco transcription factor TGA2.2 is the main component of as-1-binding factor ASF-1 and is involved in salicylic acid- and auxin-inducible expression of as-1-containing target promoters. J. Biol. Chem. 275, 19897–19905. [DOI] [PubMed] [Google Scholar]
- Niggeweg, R., Thurow, C., Weigel, R., Pfitzner, U., and Gatz, C. (2000. b). Tobacco TGA factors differ with respect to interaction with NPR1, activation potential and DNA-binding properties. Plant Mol. Biol. 42, 775–788. [DOI] [PubMed] [Google Scholar]
- O'Shea, E.K., Klemm, J.D., Kim, P.S., and Alber, T. (1991). X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science 254, 539–544. [DOI] [PubMed] [Google Scholar]
- Pascuzzi, P., Hamilton, D., Bodily, K., and Arias, J. (1998). Auxin-induced stress potentiates trans-activation by a conserved plant basic/leucine-zipper factor. J. Biol. Chem. 273, 26631–26637. [DOI] [PubMed] [Google Scholar]
- Pontier, D., Miao, Z.H., and Lam, E. (2001). Trans-dominant suppression of plant TGA factors reveals their negative and positive roles in plant defense responses. Plant J. 27, 529–538. [DOI] [PubMed] [Google Scholar]
- Qin, X.-F., Holuige, L., Horvath, D.M., and Chua, N.-H. (1994). Immediate early transcription activation by salicylic acid via the cauliflower mosaic virus as-1 element. Plant Cell 6, 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals, J., Weymann, K., Lawton, K., Friedrich, L., Ellis, D., Steiner, H.-Y., Johnson, J., Delaney, T.P., Jesse, T., Vos, P., and Uknes, S. (1997). The Arabidopsis NIM1 protein shows homology to the mammalian transcription factor inhibitor IκB. Plant Cell 9, 425–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals, J.A., Neuenschwander, U.H., Willits, M.G., Molina, A., Steiner, H.Y., and Hunt, M.D. (1996). Systemic acquired resistance. Plant Cell 8, 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah, J., and Klessig, D.F. (1996). Identification of a salicylic acid-responsive element in the promoter of the tobacco pathogenesis-related beta-1,3-glucanase gene, PR-2d. Plant J. 10, 1089–1101. [DOI] [PubMed] [Google Scholar]
- Shah, J., Tsui, F., and Klessig, D.F. (1997). Characterization of a salicylic acid-insensitive mutant (sai1) of Arabidopsis thaliana identified in a selective screen utilizing the SA-inducible expression of the tms2 gene. Mol. Plant-Microbe Interact. 10, 69–78. [DOI] [PubMed] [Google Scholar]
- Stange, C., Ramirez, I., Gomez, I., Jordana, X., and Holuigue, L. (1997). Phosphorylation of nuclear proteins directs binding to salicylic acid-responsive elements. Plant J. 11, 1315–1324. [DOI] [PubMed] [Google Scholar]
- Subramaniam, R., Desveaux, D., Spickler, C., Michnick, S.W., and Brisson, N. (2001). Direct visualization of protein interactions in plant cells. Nat. Biotechnol. 19, 769–772. [DOI] [PubMed] [Google Scholar]
- Uknes, S., Mauch-Mani, B., Moyer, M., Potter, S., Williams, S., Dincher, S., Chandler, D., Slusarenko, A., Ward, E., and Ryals, J. (1992). Acquired resistance in Arabidopsis. Plant Cell 4, 645–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, E.R., Uknes, S.J., Williams, S.C., Dincher, S.S., Wiederhold, D.L., Alexander, D.C., Ahl-Goy, P., Métraux, J.-P., and Ryals, J.A. (1991). Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3, 1085–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, C., Miao, Z., and Lam, E. (1997). DNA-binding properties, genomic organization and expression pattern of TGA6, a new member of the TGA family of bZIP transcription factors in Arabidopsis thaliana. Plant Mol. Biol. 34, 403–415. [DOI] [PubMed] [Google Scholar]
- Xiang, C., Miao, Z.H., and Lam, E. (1996). Coordinated activation of as-1-type elements and a tobacco glutathione S-transferase gene by auxins, salicylic acid, methyl-jasmonate and hydrogen peroxide. Plant Mol. Biol. 32, 415–426. [DOI] [PubMed] [Google Scholar]
- Zhang, Y.L., Fan, W.H., Kinkema, M., Li, X., and Dong, X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. USA 96, 6523–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J.M., Trifa, Y., Silva, H., Pontier, D., Lam, E., Shah, J., and Klessig, D.F. (2000). NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol. Plant-Microbe Interact. 13, 191–202. [DOI] [PubMed] [Google Scholar]