Abstract
In Arabidopsis, the basic leucine zipper transcription factor ABI5 activates several late embryogenesis–abundant genes, including AtEm1 and AtEm6. However, the expression of many other seed maturation genes is independent of ABI5. We investigated the possibility that ABI5 homologs also participate in the regulation of gene expression during seed maturation. We identified 13 ABI5-related genes in the Arabidopsis genomic sequence. RNA gel blot analysis showed that seven of these genes are active during seed maturation and that they display distinct expression kinetics. We isolated and characterized two mutant alleles of one of these genes, AtbZIP12/EEL. Unlike abi5, the eel mutations did not inhibit the expression of any of the maturation marker genes that we monitored. On the contrary, the accumulation of the AtEm1 and AtEm6 mRNAs was enhanced in eel mutant seeds compared with wild-type seeds. Gel mobility shift assays, combined with analysis of the genetic interactions among the eel and abi5 mutations, indicated that ABI5 and EEL compete for the same binding sites within the AtEm1 promoter. This study illustrates how two homologous transcription factors can play antagonistic roles to fine-tune gene expression.
INTRODUCTION
In higher plants, seed development is divided roughly into two successive phases. Cell division and morphogenesis occur during the first phase. Once embryogenesis is completed, the seed enters a second phase called maturation, during which the embryo accumulates storage compounds, becomes dormant, and acquires desiccation tolerance (Wobus and Weber, 1999). All of these traits are of crucial importance for the survival of seed plant species, including Arabidopsis. At the molecular level, seed maturation is characterized by the coordinated expression of several classes of genes with distinct temporal patterns (Hughes and Galau, 1989; Parcy et al., 1994). The maturation MAT class includes genes that code for storage proteins such as 2S and 12S globulins in Arabidopsis (Pang et al., 1988; Guerche et al., 1990). The late embryogenesis–abundant (LEA) class comprises genes that code for LEA proteins that are thought to be involved in desiccation tolerance (Hoekstra et al., 2001).
Our knowledge of the mechanisms that control MAT and LEA gene expression is fragmentary. The phytohormone abscisic acid (ABA) accumulates during seed maturation and positively regulates the expression of genes in both the MAT and LEA classes (Koornneef et al., 1989; Parcy et al., 1994; Phillips et al., 1997). Several transcription factors have been identified that control different aspects of seed maturation (Wobus and Weber, 1999; Stone et al., 2001). In particular, ABA-INSENSITIVE3 (ABI3) is considered a global regulator of seed maturation because it is necessary for both the proper accumulation of storage proteins and the acquisition of desiccation tolerance (Nambara et al., 1992). In abi3 mutant seeds, the expression of multiple genes from both the MAT and LEA classes is reduced compared with that in wild-type seeds (Parcy et al., 1994; Nambara et al., 1995, 2000).
How the ABI3 protein regulates these genes is not known. The isolated basic domain B3 of the VP1 protein, the maize ortholog of ABI3, is able to bind DNA in vitro (Suzuki et al., 1997). However, DNA binding has never been observed with the full-length VP1 (Suzuki et al., 1997) or ABI3 proteins. This suggests that other proteins might be needed to recruit ABI3 to the promoter of its target genes. Consistent with this hypothesis, OsVP1, the ABI3/VP1 ortholog from rice, interacts physically with the basic leucine zipper (bZIP) transcription factor TRAB1, which binds the ABA response element (ABRE) CACGTG that is present in the promoter of LEA genes (Hobo et al., 1999a, 1999b).
Genetic evidence that a TRAB1-related bZIP protein is involved in the ABI3-dependent regulation of LEA genes came from the analysis of the abi5 mutant of Arabidopsis. The abi5 mutation leads to reduced ABA sensitivity during germination and to decreased expression of some LEA genes, including AtEm1 and AtEm6, during seed maturation (Finkelstein, 1993, 1994; Gaubier et al., 1993). Cloning of the ABI5 gene revealed that it encodes a bZIP protein from the same subfamily as TRAB1 (Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000). Moreover, ABI5 interacts with ABI3 in two-hybrid experiments (Nakamura et al., 2001). Hence, the current model for both monocotyledonous and dicotyledonous species is that the recruitment of ABI3 (or its ortholog) on the ABREs present in LEA promoters is mediated by bZIP proteins. This model also might apply to storage protein genes, because the promoters of these genes contain ABREs that are functionally important for expression during seed maturation (Chern et al., 1996; Ezcurra et al., 1999).
The abi5 mutant has a markedly milder phenotype than abi3. In contrast to abi3, abi5 seeds are tolerant to desiccation and have a storage protein content similar to that of wild-type seeds (Finkelstein, 1994). At the molecular level, the expression of several storage protein genes is reduced in abi3 but not in abi5 (Parcy et al., 1994; Nambara et al., 1995; Finkelstein and Lynch, 2000; Delseny et al., 2001). The abi3 mutation inhibits the expression of a wider range of individual LEA genes than abi5. Finally, the accumulation of the LEA mRNAs AtEm1 and AtEm6 is abolished in abi3, whereas it is only partially reduced in abi5 (Parcy et al., 1994; Finkelstein and Lynch, 2000; Delseny et al., 2001). These data indicate that ABI5 contributes to only a subpart of the various roles of ABI3 during seed maturation.
An attractive possibility, therefore, is that additional ABI5-related bZIP proteins contribute to the ABI3-dependent regulation of gene expression during seed maturation and mediate the recruitment of ABI3 to distinct promoters. For instance, at least three different ABI5-related bZIP genes (DPBF-1, DPBF-2, and DPBF-3) are expressed in sunflower seed tissue. These three DPBF proteins are able to bind the ABRE-like motifs present in the promoter of the LEA gene DC3 from carrot (Kim et al., 1997; Kim and Thomas, 1998). In Arabidopsis, several ABI5-related genes have been described: ABF1, ABF2/AREB1, ABF3, ABF4/AREB2, AREB3, and GBF4 (Menkens and Cashmore, 1994; Choi et al., 2000; Uno et al., 2000).
All of these genes are expressed in vegetative tissues, and at least several of them seem to be involved in ABA-dependent processes. ABF1, ABF2/AREB1, ABF3, and ABF4/AREB2 are induced differentially in response to abiotic stress or ABA treatment (Choi et al., 2000; Uno et al., 2000). ABF1 binds to the ACGTGT/GC motif in vitro (Choi et al., 2000), and two such sequence motifs are present in the stress- and ABA-inducible RD29B promoter, which can be activated by the transient expression of ABF2/AREB1 or ABF4/AREB2 in protoplasts (Uno et al., 2000). However, the possible involvement of these ABI5 homologs in seed maturation has not been tested.
In the present study, we performed an exhaustive search of the Arabidopsis genomic sequence and identified 13 ABI5-related genes, including 6 that had not been described previously. Seven of these bZIP genes are expressed during seed maturation. Two genes, including ABI5, displayed expression kinetics similar to that of the LEA genes. Remarkably, three other bZIP genes displayed expression kinetics similar to that of storage protein genes, suggesting that these bZIPs could regulate the expression of genes from the MAT class. We tested this hypothesis by isolating two insertional mutants for one of the three latter bZIP genes, AtbZIP12. The accumulation of storage protein mRNAs was not affected in the atbzip12 mutants. However, unexpectedly, the abundance of the LEA mRNAs AtEm1 and AtEm6 was increased in atbzip12, whereas the level of these same mRNAs was decreased in the abi5 mutant. We analyzed the genetic and molecular interactions between ABI5 and AtbZIP12 and propose a model of how these two homologous transcription factors play antagonistic roles to fine-tune LEA gene expression during Arabidopsis seed development.
RESULTS
The Arabidopsis ABI5/ABF/AREB Family Contains 13 Members
In the complete sequence of the Arabidopsis genome, we identified 13 genes that encode proteins belonging to the ABI5/ABF/AREB family. Seven of these genes had been characterized previously and had been named ABI5, ABF1, ABF2/AREB1, ABF3, ABF4/AREB2, AREB3, and GBF4 (Menkens and Cashmore, 1994; Choi et al., 2000; Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000; Uno et al., 2000). The six additional genes were designated AtbZIP12, AtbZIP13, AtbZIP14, AtbZIP15, AtbZIP27, and AtbZIP67, according to the unified nomenclature for AtbZIP genes proposed by Jacoby et al. (2002). The complete coding sequences of AtbZIP12, AtbZIP13, AtbZIP15, and AtbZIP67 were obtained by sequencing cDNA clones and assembling available EST sequences. In contrast, cDNA clones have not been identified for AtbZIP14 or AtbZIP27. Thus, the coding sequences proposed for these two genes are based solely on the results of gene-modeling programs and optimization of protein sequence alignments to other members of the ABI5/ABF/AREB family. Furthermore, because we were unable to detect their expression in the various tissues analyzed, we cannot exclude the possibility that AtbZIP14 and AtbZIP27 represent pseudogenes.
As shown in Figure 1, the amino acid sequence of the basic region is extremely conserved between the 13 members of this bZIP subfamily. This region contains, in particular, the motifs M/K-I-K and Q-A-Y/Q, which are not found in any other Arabidopsis bZIP outside of this subfamily. The high degree of conservation of this DNA binding region suggests that all members of this subfamily recognize similar cis elements. As noted previously, three additional conserved domains are found in the N-terminal half of the proteins, and a fourth is found at their C-terminal end (Choi et al., 2000; Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000; Uno et al., 2000). The C2 and C3 domains are present in all 13 members, although they are less well conserved in GBF4, AtbZIP13, AtbZIP14, and AtbZIP27. In addition, these four proteins do not have any recognizable C1 domain.
Figure 1.
Sequence Comparison of Arabidopsis bZIP Proteins of the ABI5/ABF/AREB Family.
Alignment of the predicted amino acid sequences of the 13 members of the ABI5/ABF/AREB family. Identical and conserved residues are shaded in black and gray, respectively. The positions of the C1 to C4 conserved domains and of the basic region are indicated by lines above the protein sequences. Potential phosphoresidues corresponding to consensus phosphorylation sites for Ca2+-dependent protein kinase (R/K-X-X-S/T), protein kinase C (S/T-X-K/R), cGMP-dependent protein kinase (K/R-X-X-X-S/T), or casein kinase II (S/T-X-X-D/E) are shown in lowercase letters, and their positions are indicated by asterisks. Residues from the basic region specific to this subfamily of bZIP are indicated by dots. The positions of the conserved Leu residues in the Leu zipper domain are indicated by z's. Protein sequence alignments were performed using ClustalW at the European Bioinformatics Institute (http://www.ebi.ac.uk/clustalw/) and edited using Macboxshade 2.11 software (Institute of Animal Health, Pirbright, Surrey, UK).
The C1, C2, and C3 domains share some sequence similarity: for instance, they all contain stretches of two acidic residues (D or E) surrounded by nonpolar residues (F, I, L, M, or V). Finally, there is a fourth conserved small domain (C4) located at the C-terminal end of the proteins. The C1 to C4 domains contain consensus phosphorylation sites for various protein kinases that are well conserved in the different proteins. Interestingly, the four consensus phosphorylation sites for Ca2+-dependent protein kinase (R/K-X-X-S/T) are all found two residues C terminal to a conserved Leu (L-X-R/K-X-X-S/T), suggesting that they might be recognized by similar kinases.
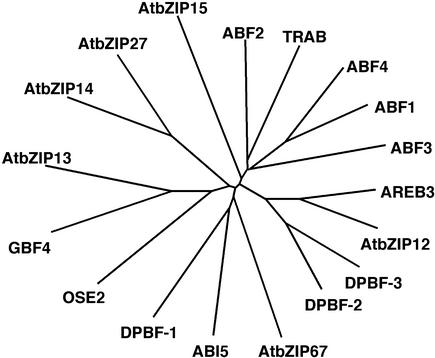
Figure 2 shows the degree of similarity between the various members of the Arabidopsis subfamily and related bZIP proteins from other plant species. It also illustrates the existence of several pairs of closely related Arabidopsis proteins that might have similar functional properties (ABF1 and ABF4/AREB2, AREB3 and AtbZIP12, GBF4 and AtbZIP13, and AtbZIP14 and AtbZIP27).
Figure 2.
Relation Tree of bZIP Proteins from Arabidopsis and Other Plant Species.
This tree includes the 13 Arabidopsis proteins shown in Figure 1, DPBF-1, DPBF-2, and DPBF-3 from sunflower, and TRAB1 and OSE2 from rice. Proteins were aligned using ClustalW, and the tree was constructed using Treeview software.
Expression Analysis during Seed Development
As a first step toward the identification of bZIP genes involved in seed maturation, we analyzed which of the bZIP genes described above are expressed during silique development. For this purpose, we generated a gene-specific probe for each of the bZIP genes. We used a maximum level of 70% nucleotide identity over 100 bp as a specificity criterion. This criterion, however, could not be met for a few pairs of closely related bZIP genes, and we had to accept up to 79% identity between the AtbZIP14 probe and the corresponding region of AtbZIP27. In all cases, we also verified that the probe does not share any significant sequence similarity with any other predicted coding sequence of the Arabidopsis genome.
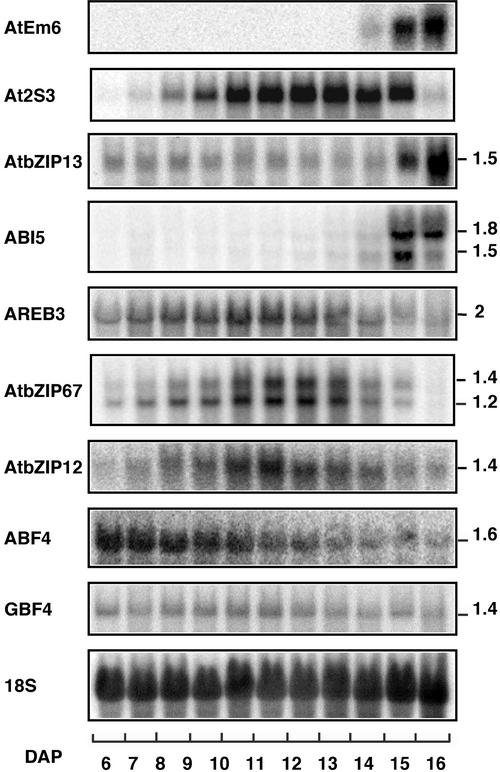
These probes were used to analyze the expression of the various bZIP genes during seed maturation. This developmental process is characterized by the existence of several temporal programs of gene expression. The storage protein At2S3 and LEA AtEm6 genes are representative members of the MAT and LEA programs, respectively (Parcy et al., 1994). These marker genes were included as temporal landmarks for our bZIP expression analysis. Figure 3 shows mRNA expression kinetics between 6 and 16 days after pollination (DAP), a period extending from the torpedo stage of embryo development to the dry seed stage. Upon RNA gel blot analysis, hybridization signals were detected for 7 of the 13 bZIP genes monitored. In agreement with previous reports (Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000), ABI5 mRNA accumulated during the last few days of seed development with an expression pattern similar to that of the LEA marker AtEm6.
Figure 3.
RNA Gel Blot Analysis of Gene Expression during Silique Development.
Each of the indicated probes was hybridized to 12 μg of total RNA isolated from Wassilewskija (Ws) wild-type siliques harvested between 6 and 16 DAP. An 18S rRNA probe was used as a control for equal amounts of RNA loading in the different lanes. Estimated bZIP transcript sizes (kb) are indicated at right.
AtbZIP13 mRNA was expressed at all stages analyzed, and like ABI5, its level increased sharply during the last 2 days of seed development. The AtbZIP12, AtbZIP67, and AREB3 mRNAs also were detectable as early as 6 to 7 DAP, but their abundance decreased toward the end of seed maturation. The expression kinetics of these three bZIPs is similar to that of the storage protein marker At2S3. It is unclear why two hybridizing mRNA bands were observed with the ABI5 and AtbZIP67 probes. Each of these probes shows a low level of sequence similarity to the other bZIP genes; thus, it is unlikely to cross-hybridize with another member of the gene family. Hence, the double bands may be indicative of alternative splicing in the ABI5 and AtbZIP67 genes. Finally, GBF4 mRNA was expressed uniformly throughout seed maturation, whereas ABF4 mRNA abundance was maximum at 6 DAP and then decreased progressively until the dry seed stage. Their distinctive expression kinetics suggest that the seven bZIP genes described above play different regulatory roles during seed maturation.
Isolation and Characterization of atbzip12/eel Mutants
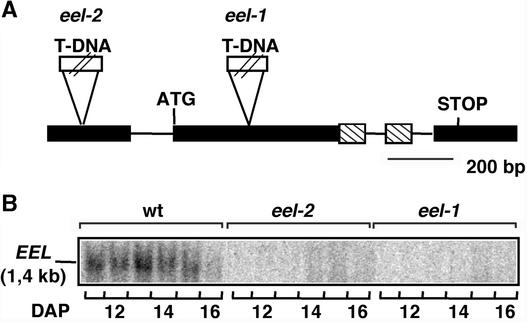
We have undertaken a systematic functional analysis of this bZIP subfamily. AtbZIP12 was the first member for which we identified knockout mutants. Two independent mutant alleles of AtbZIP12 were isolated by PCR screening of DNA pools from collections of T-DNA insertion lines. As described below, these two mutants displayed enhanced expression of the AtEm1 mRNA, and AtbZIP12 was thus renamed EEL (enhanced Em level). The eel-1 mutant contains a T-DNA insertion 5′ to the codon for residue Asp-85 of the EEL protein (Figure 4A). In eel-2, the T-DNA is inserted in the 5′ untranslated leader sequence of EEL (upstream of nucleotide 118). EEL mRNA was undetectable by RNA gel blot analysis in both mutant alleles (Figure 4B). The EEL coding sequence is not disrupted by the T-DNA insertion in eel-2. However, the EEL transcript was undetectable by reverse transcriptase–mediated PCR analysis in eel-2 mutant seeds (data not shown). Hence, eel-1 and eel-2 are null or strong hypomorphic mutant alleles.
Figure 4.
Insertion Mutants in AtbZIP12/EEL.
(A) Structure of the AtbZIP12/EEL gene. Lines indicate introns, and boxes represent exons. The gene region encoding the basic region of the EEL protein is indicated by hatched boxes. Positions of the T-DNA insertions in the eel mutants are shown.
(B) RNA gel blot analysis of EEL expression in the Ws wild type (wt) and eel-1 and eel-2 mutants. The EEL-specific probe was hybridized to 12 μg of total RNA from siliques of the indicated genotypes harvested between 11 and 16 DAP.
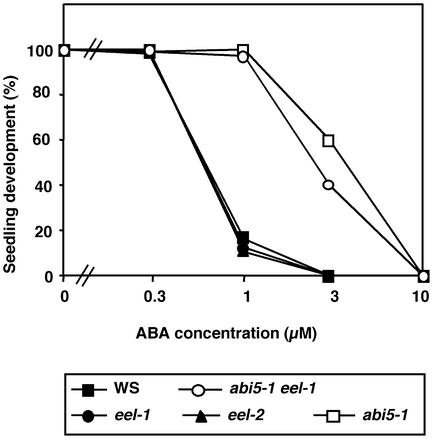
Seeds from the eel-1 and eel-2 mutants did not display any morphological alteration. Seed development, defined as the period extending from pollination to the dry seed stage, lasted the same amount of time (16 days) as in the wild type, and the eel mutant seeds were as dormant as wild-type seeds. The eel mutants also showed wild-type sensitivity to the ABA inhibition of seedling establishment, whereas, as described previously (Finkelstein, 1994; Lopez-Molina et al., 2001), the abi5-1 mutant showed reduced ABA sensitivity in this bioassay (Figure 5).
Figure 5.
ABA Inhibition of Seedling Establishment.
Seeds of Ws wild type (closed squares), eel-1 (closed circles), eel-2 (closed triangles), abi5-1 (open squares), and abi5-1 eel-1 (open circles) were plated on medium supplemented with the indicated concentrations of ABA, chilled for 4 days at 4°C in darkness, and incubated for 3 days at 21°C with a 16-h photoperiod. The number of seedlings with green cotyledons is expressed as the percentage of the total number of seeds plated (40 to 280 seeds).
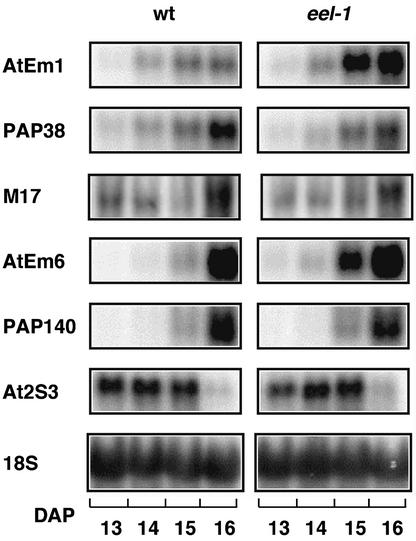
The abi5 mutation leads to decreased expression of some LEA genes during seed maturation (Finkelstein and Lynch, 2000). We initially compared the abundance of the storage protein At2S3 mRNA and a few LEA mRNAs (AtEm1, AtEm6, M17, PAP38, and PAP140) in wild-type and eel-1 seeds. For At2S3, M17, PAP38, and PAP140, we occasionally observed a slight difference in mRNA abundance between wild-type and mutant seeds. However, these fluctuations were observed only once within a given experiment and were not reproducible. Hence, such variations were considered to be within the experimental fluctuations. They most likely result, at least in part, from the fact that staging and harvesting of the siliques inevitably vary slightly from one genotype to another and from one experiment to another. In contrast, the abundance of the AtEm1 and AtEm6 mRNAs was reproducibly higher in eel-1 than in the wild type (Figure 6). This observation was particularly surprising because, in marked contrast to eel-1, the abi5 mutation inhibits the expression of AtEm1 and AtEm6 (Finkelstein and Lynch, 2000).
Figure 6.
Comparative Gel Blot Analysis of the Expression of Marker mRNAs during Wild-Type and eel-1 Silique Development.
The indicated gene-specific probes were hybridized to 12 μg of total RNA from siliques of the Ws wild type (wt) and the eel-1 mutant harvested between 13 and 16 DAP. An 18S rRNA probe was used as a control for equal amounts of RNA loading in the different lanes.
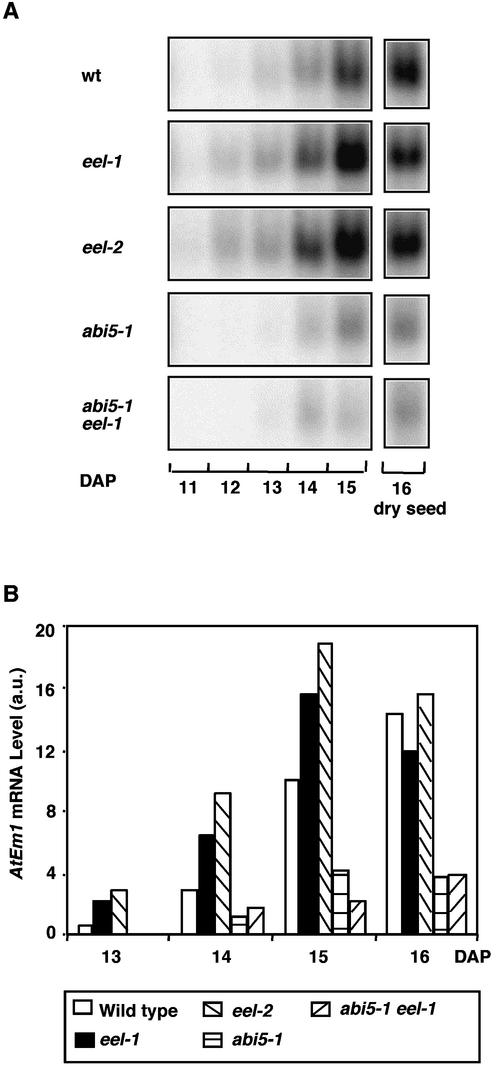
We analyzed in more detail the expression of AtEm1, for which the eel phenotype seemed the clearest. In wild-type seed, AtEm1 mRNA was expressed during the last 4 days of seed maturation, and its level reached a maximum in the desiccated seeds (Figure 7). The abundance of the AtEm1 mRNA was reduced approximately fourfold in abi5-1. In contrast, in eel-1 and eel-2, the AtEm1 mRNA level was approximately threefold higher than in the wild type, and the expression of this mRNA was detectable 1 day earlier than in the wild type (Figure 7B). Based on seven independent experiments (five with eel-1 and two with eel-2), the ratio between the AtEm1 level in eel and the wild type stands with 95% confidence between 3.3 and 6.5 at 13 DAP, between 2.2 and 4.7 at 14 DAP, and between 2.0 and 3.5 at 15 DAP. However, in dry seeds (16 DAP), in most cases the AtEm1 mRNA level was similar in the wild type and the eel mutants (Figure 7B).
Figure 7.
Gel Blot Analysis of AtEm1 mRNA Expression during Silique Development.
(A) Developmental time course of AtEm1 expression in the Ws wild type (wt), the eel-1, eel-2, and abi5-1 mutants, and the abi5-1 eel-1 double mutant. The AtEm1 probe was hybridized to 12 μg of total RNA from siliques of the indicated genotypes harvested between 11 and 16 DAP.
(B) Levels of AtEm1 mRNA. Hybridization signals shown in (A) were quantified using a Storm imager and were normalized using the signals obtained after stripping and hybridizing the blot with an 18S rRNA probe. AtEm1 mRNA levels are expressed in arbitrary units (a.u.).
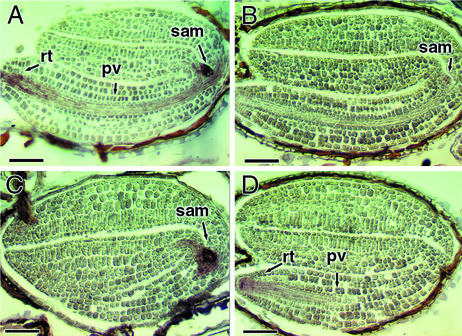
We analyzed the spatial pattern of AtEm1 mRNA expression by in situ hybridization. In agreement with previous reports (Gaubier et al., 1993; Vicient et al., 2000), AtEm1 expression in the wild-type seeds was restricted mostly to the provascular tissue, the root tip, and the shoot apical meristem of the embryo (Figure 8A). In abi5-1, AtEm1 mRNA was undetectable in most embryos (data not shown), but a faint hybridization signal was observed in a few cases (Figure 8B). In eel-1, the spatial distribution of AtEm1 mRNA was similar to that in wild-type seeds (Figures 8C and 8D). The increase in AtEm1 mRNA level detected by gel blot analysis was not apparent in the in situ hybridizations. Nevertheless, the combined results of gel blot and in situ hybridization analyses indicate that the eel mutations do not induce an ectopic accumulation of AtEm1 mRNA; rather, they increase the intensity of AtEm1 expression without altering its spatial distribution.
Figure 8.
In Situ Hybridization Analysis of AtEm1 expression.
In situ hybridization was performed on sections of Ws (A), abi5-1 (B), and eel-1 ([C] and [D]) embryos with an AtEm1-specific probe. In Ws and eel-1, the AtEm1 transcript (brown signal) was detected primarily in the provascular tissues (pv), the root tip (rt), and the shoot apical meristem (sam). In abi5-1 embryos, the AtEm1 signal was either absent or of reduced intensity. Embryos were harvested at 14 to 15 DAP. Bars = 50 μm.
Genetic Interaction between ABI5 and EEL
eel-1 and abi5-1 are null mutations that have opposite effects on AtEm1 expression. We tested the epistatic relationship between these mutations by analyzing AtEm1 expression in the abi5-1 eel-1 double mutant. The expression kinetics and abundance of the AtEm1 mRNA were similar in the abi5-1 eel-1 double mutant and the abi5-1 single mutant (Figure 7B). This result indicates that abi5 is epistatic to eel. In other words, the increased AtEm1 expression observed in the eel mutant is completely dependent on a functional ABI5 gene. A possible explanation for this finding could then be that EEL acts upstream of ABI5, for instance, by negatively regulating ABI5 expression. However, we analyzed the expression kinetics of the ABI5 mRNA from 11 to 16 DAP and found no obvious difference between wild-type and eel mutant seeds (data not shown). Another possibility is that EEL and ABI5 act at the same level to regulate AtEm1 expression. Consequently, we investigated whether these two transcription factors bind directly to the AtEm1 promoter.
ABI5 and EEL Bind to the Same Sites in the AtEm1 Promoter
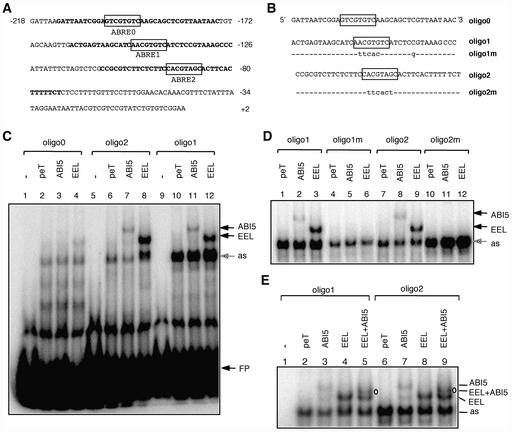
Deletion analysis of the AtEm1 promoter in transgenic Arabidopsis plants has shown that the 200 bp upstream of the transcription start are sufficient to confer the AtEm1 expression pattern to the β-glucuronidase reporter gene (Delseny et al., 2001). This 200-bp region contains three potential ABREs (Figure 9A). ABRE1 and ABRE2 contain an ACGT core. ABRE0 does not contain an ACGT core, but available evidence indicates that this sequence motif still might be recognized by members of the ABF/AREB family (Hobo et al., 1999a; Choi et al., 2000). We tested by electrophoretic mobility shift assay (EMSA) whether ABI5 and EEL were able to interact with double-stranded DNA oligonucleotides corresponding to AtEm1 promoter regions centered on these three ABREs (Figure 9B).
Figure 9.
ABI5 and EEL Interact with the Same ABREs in the AtEm1 Promoter.
(A) Sequence of the minimal AtEm1 promoter. Nucleotides are numbered relative to the transcription start site (+1). ABRE motifs are shown in boxes. Promoter regions used as probes for EMSA are indicated in boldface.
(B) Sequences of the wild-type (oligo0, oligo1, and oligo2) and mutated (oligo1m and oligo2m) oligonucleotides used as probes for EMSA. Mutated nucleotides are indicated in lowercase letters. Sequences are presented in the 5′ to 3′ orientation and are shown as single strand only.
(C) ABI5 and EEL bind to oligo1 and oligo2. Oligo0 (lanes 1 to 4), oligo2 (lanes 5 to 8), and oligo1 (lanes 9 to 12) were incubated without protein extract (free probe; lanes 1, 5, and 9) or with in vitro–transcribed and translated vector pET16b (negative control; lanes 2, 6, and 10), ABI5 (lanes 3, 7, and 11), or EEL (lanes 4, 8, and 12). Bands shifted by ABI5 or EEL are indicated by arrows. as, an aspecific band shifted by reticulocyte proteins; FP, free probe.
(D) Mutations in ABRE1 and ABRE2 abolish the binding of ABI5 and EEL. Oligo1 (lanes 1 to 3), oligo1m (lanes 4 to 6), oligo2 (lanes 7 to 9), or oligo2m (lanes 10 to 12) were incubated with in vitro–transcribed and translated vector pET16b (negative control; lanes 1, 4, 7, and 10), ABI5 (lanes 2, 5, 8, and 11), or EEL (lanes 3, 6, 9, and 12). In contrast to the wild-type oligo1 and oligo2, the mutant oligo1m and oligo2m were not shifted by ABI5 and EEL proteins.
(E) ABI5 and EEL bind to oligo1 and oligo2 as homodimers and as a heterodimer. Oligo1 (lanes 1 to 5) or oligo2 (lanes 6 to 9) were incubated without protein extract (free probe; lane 1) or with in vitro–transcribed and translated vector pET16b (negative control; lanes 2 and 6), ABI5 (lanes 3 and 7), EEL (lanes 4 and 8), or cotranslated ABI5 and EEL (lanes 5 and 9). The shifted band corresponding to the ABI5-EEL heterodimer is indicated (EEL + ABI5).
In vitro–produced ABI5 and EEL proteins bound weakly, if at all, to oligonucleotide 0 containing ABRE0 (Figure 9C). In contrast, both proteins bound clearly to oligonucleotides 1 and 2, which contain ABRE1 and ABRE2, respectively (Figure 9C). No binding was observed when the ABRE motifs were mutated (Figure 9D). Thus, these data indicate that both ABI5 and EEL are able to bind to ABRE1 and ABRE2. Because bZIP proteins bind DNA as dimers, the shifted bands observed in Figure 9C correspond to complexes between the oligonucleotide and homodimers of either ABI5 or EEL. When EMSA was performed with cotranslated ABI5 and EEL proteins, an additional shifted band was observed that had mobility intermediate between those of the bands observed with ABI5 alone or EEL alone (Figure 9E, lanes 5 and 9). This result indicates that an ABI5-EEL heterodimer can form and can bind to ABRE1 and ABRE2.
DISCUSSION
Seven members of the ABI5 subfamily of bZIP genes are expressed during seed maturation. These genes, however, display various expression kinetics, suggesting that they play different roles in the control of gene expression programs during seed maturation. The ABI5 mRNA accumulates during the later stages of seed development, and ABI5 controls the expression of some LEA genes (Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000). The expression kinetics of the AtbZIP12/EEL mRNA resembles that of storage protein mRNAs. Thus, we thought initially that AtbZIP12/EEL might be involved in regulating storage protein genes. However, using gel blot analysis and hybridization of macroarrays carrying 500 seed cDNA clones, we detected no effect of the eel mutations on the accumulation of napin and cruciferin mRNAs or of any other MAT mRNA. In contrast, the LEA mRNAs AtEm1 and AtEm6 were expressed at higher levels in eel mutant seeds than in wild-type seeds. Conversely, the AtEm1 and AtEm6 mRNA levels were lower in abi5 than in the wild type. Thus, genetically, these two homologous bZIPs have opposite roles in the control of AtEm1 and AtEm6: the expression of the AtEm genes is regulated positively by ABI5 and regulated negatively by EEL.
EEL function was investigated in more detail by studying AtEm1 expression. Double-mutant analysis showed that abi5 is epistatic to eel, indicating that the increased AtEm1 expression in eel mutant seeds is attributable to ABI5. Thus, genetically, EEL acts either upstream or at the same level as ABI5. It seems unlikely that EEL controls AtEm1 indirectly by inhibiting ABI5 expression, because we did not detect any increase in ABI5 mRNA level in the eel mutant. Moreover, overexpression of ABI5 results in supersensitivity to ABA inhibition of seed germination and seedling establishment (Lopez-Molina et al., 2001), whereas the eel mutants displayed a wild-type ABA sensitivity in this bioassay.
Our gel retardation experiments suggest that both ABI5 and EEL regulate AtEm1 expression by binding directly to the AtEm1 promoter. The decreased abundance of AtEm1 mRNA in the abi5 mutant suggests that ABI5 is a transcriptional activator of AtEm1. In contrast, EEL seems unable to activate AtEm1 transcription: the AtEm1 mRNA level was higher in eel than in the wild type. Moreover, the abi5 single mutant and the abi5 eel double mutant had similar levels of AtEm1 mRNA, indicating that EEL is not responsible for the residual AtEm1 expression observed in abi5. EEL may be unable to activate AtEm1 transcription because, unlike ABI5 (Nakamura et al., 2001), it cannot recruit the ABI3 coactivator. Also, unlike ABI5, EEL lacks the fourth consensus phosphorylation site in the C1 domain, which could be important for the functional regulation of this subfamily of bZIPs (Figures 1 and 2) (Uno et al., 2000; Lopez-Molina et al., 2001).
In vitro, the EEL and ABI5 homodimers as well as the EEL-ABI5 heterodimer bind to the ABRE1 and ABRE2 elements of the AtEm1 promoter. The eel and abi5 mutant phenotypes then can be explained by assuming that, in the seed, EEL and ABI5 compete for these binding sites on the AtEm1 promoter. In eel mutant seeds, the EEL protein is absent and ABI5 can freely activate AtEm1 expression to maximum levels. In wild-type seeds, the EEL-ABI5 heterodimer and the EEL homodimer compete with the ABI5 homodimer for binding to the AtEm1 promoter, preventing ABI5 from optimally activating AtEm1. Consequently, AtEm1 expression is lower in wild-type seeds than in eel. In abi5 and abi5 eel mutant seeds, the ABI5 protein is absent and AtEm1 expression is reduced further.
The regulation of Em genes in monocots also might involve bZIP proteins with antagonistic functions. The bZIP EMBP-1 from wheat, a putative positive regulator of Em, binds in vitro to ABREs present in the wheat Em promoter. In the presence of a rice bZIP, osZIP-2a or osZIP-2b, heterodimerization between EmBP-1 and osZIP-2 prevents EmBP-1 from binding to the ABRE (Nantel and Quatrano, 1996). However, in contrast to EEL, the osZIP-2 proteins are not able to bind to ABREs. Similarly, the ROM1 and ROM2 bZIPs have been proposed to play opposite roles in the regulation of MAT genes during seed development in bean (Chern et al., 1996).
Competition between ABI5 and EEL could be a mechanism adopted by plants to tightly control the expression of LEA genes during seed development. RNA gel blot analysis suggests that until ∼13 DAP, EEL expression is highest, whereas ABI5 mRNA is barely detectable. We believe that, as a result, the AtEm1 gene is not expressed during this period. From 13 DAP onward, ABI5 expression increases but EEL expression decreases. This may explain the quick upregulation of AtEm1. AtEm1 mRNA reaches its maximum level at the end of seed maturation, when ABI5 also is highly expressed and EEL expression decreases. ABI5 might even contribute to the downregulation of EEL expression, because EEL mRNA level was higher in abi5 than in wild-type seeds (data not shown). Therefore, seeds appear to use two transcription factors based on the same DNA binding module to fine-tune the expression of LEA genes.
The model described above for AtEm1 probably does not apply to all LEA genes. For instance, the abundance of the LEA mRNA M17 is increased (not decreased) in abi5 (Finkelstein and Lynch, 2000) and is not affected in the eel mutant. Moreover, several LEA mRNAs have similar expression levels in abi5 and wild-type seeds (Finkelstein and Lynch, 2000). It is noteworthy that AtbZIP13 and ABI5 have similar expression kinetics during seed development. Thus, AtbZIP13 may be involved in activating ABI5-independent LEA genes. It will be of particular interest to analyze LEA gene expression in an atbzip13 mutant or in an abi5 atbzip13 double mutant.
Our macroarray analysis did not reveal any effect of the eel mutation on the accumulation of MAT mRNAs. However, EEL (AtbZIP12), AtbZIP67, and AREB3 display similar expression patterns and may have overlapping roles in regulating MAT genes. Testing this hypothesis will require the isolation of plants that combine mutations in EEL, AREB3, and AtbZIP67. Sequencing of the Arabidopsis genome revealed that, in this model dicotyledonous species, transcription factors exist as large families (Riechmann et al., 2000). Each family contains recognizable subgroups of closely related proteins for which, in most cases, the biological function has yet to be established (Eulgem et al., 2000; Riechmann et al., 2000).
Mutant analysis of several subgroups of MADS box genes underscored the existence of functional overlap between homologous transcription factors that leads to partial genetic redundancy (Jack, 2001). In contrast, the MYB genes WER and GL1 demonstrate that two closely related proteins can have different biological functions simply as a result of their distinct expression patterns (Lee and Schiefelbein, 2001). We showed here that two related bZIP proteins play antagonistic roles in regulating common target genes. Future studies will reveal to which extent other members of the ABI5 subfamily of bZIP transcription factors act redundantly, antagonistically, or in distinct tissues.
METHODS
Plant Material and Culture Conditions
eel-1, eel-2, and abi5-1 mutants are derived from the Arabidopsis thaliana Wassilewskija accession. eel-1 (line COU38) was isolated from the Versailles collection of T-DNA insertion lines (Bechtold et al., 1993). eel-2 (line CS11393) was isolated from the Feldmann collection of T-DNA insertion lines available at the ABRC (Ohio State University, Columbus). abi5-1 seeds (Finkelstein, 1994) were obtained from Ruth Finkelstein (University of California, Santa Barbara).
Plants were grown routinely in a greenhouse (22°C with a 16-h photoperiod) on soil irrigated with mineral nutrients. For aseptic growth, seeds were surface-sterilized and plated on medium containing the inorganic salts of Murashige and Skoog (1962) at half concentrations, 100 mg/L myoinositol, 1 mg/L thiamine-HCl, 0.5 mg/L nicotinic acid, 0.5 mg/L pyridoxine-HCl, 0.8% agar, and 2.5 mM (2-[N-morpholino]ethanesulfonic acid)–KOH, pH 5.7. Abscisic acid (mixed isomers; Sigma) was diluted from 10-mM stock solutions prepared in methanol; equivalent volumes of methanol were included in the abscisic acid–free controls.
bZIP Sequences
The coding sequence of AtbZIP13 was determined by assembling available EST sequences. The coding sequence of AtbZIP12/EEL was determined by assembling the sequence of a partial cDNA obtained by reverse transcriptase–mediated PCR and available EST sequences. The coding sequences of AtbZIP15 and AtbZIP67 were determined by sequencing cDNA clones SQ099e02F (Asamizu et al., 2000) and 600039423R1 (White et al., 2000), respectively. The AtbZIP12/EEL coding sequence is identical to that of the recently deposited DPBF-4, whereas AtbZIP67 is very similar to DPBF-2. The coding sequences of AtbZIP14 and AtbZIP27 were predicted by gene modeling using Genemark.hmm (http://dixie.biology.gatech.edu/GeneMark/eukhmm.cgi) and Netgene2 (http://www.cbs.dtu.dk/services/NetGene2/) software as recommended by Pavy et al. (1999).
DNA regions used as gene-specific probes were selected by pair-wise comparison of the bZIP sequences using Bestfit (GCG package; Genetics Computer Group, Madison, WI) and by BlastN comparison against all predicted Arabidopsis coding sequences at TAIR (http://www.arabidopsis.org). Gene-specific probes were prepared by PCR amplification, and the amplified DNA was subcloned into PCR-TOPO2.1 (Invitrogen, La Jolla, CA) or pGEM-T easy (Promega).
RNA Gel Blot Analysis
Staging of the developing siliques was performed by tagging individual flowers on the day of pollination (Parcy et al., 1994). Siliques were pooled from several primary inflorescences of plants grown simultaneously and were frozen in liquid nitrogen. RNA extraction and gel blot analysis were performed as described by Parcy et al. (1994). 32P-labeled DNA probes were generated by random or reverse primer extension. The gene-specific probes used for the detection of At2S3, PAP38, M17, and PAP140 mRNAs have been described previously (Parcy et al., 1994). Gene-specific probes for AtEm1 and AtEm6 were prepared by PCR amplification with oligonucleotides Em1f (5′-GTGATAGTAGTTACGAGCTAC-3′)/Em1r (5′-CGGCAACCGCATTACATAAC-3′) and Em6f (5′-GGACCAAGACCTAAATCAAAC-3′)/Em6r (5′-CGAGACACTTTACGTAGACG-3′), respectively. The amplified DNA was subcloned into pGEM-T easy (Promega) to generate plasmids pGATEM1 (AtEm1 probe) and pGATEM6 (AtEm6 probe).
Mutant Isolation and Genotyping
Oligonucleotides used for PCR screening of the collections of T-DNA insertion lines were as follows: TAG3 (5′-CTGATACCAGACGTTGCCCGCATAA-3′) and TAG5 (5′-CTACAAATTGCCTTTTCTTATCGAC-3′) for the T-DNA in the Versailles collection; oFPLB (5′-GATGCACTCGAAATCAGCCAATTTTAGAC-3′) and oFPRB (5′-TCCTTCAATCGTTGCGGTTCTGTCAGTTC-3′) for the T-DNA in the Feldman collection; and oFP1085 (5′-GAGTCTTTAGGATCAGAGAGA-AGCAGAG-3′) and oFP1079 (5′-TCTGGAAAACCACTAGGAAGCAT-3′) for the EEL gene. The positions of the T-DNA insertions in eel-1 and eel-2 correspond to nucleotides 31,314 and 30,754, respectively, in BAC clone T3K9.
Plant genotyping for the eel-1 mutation was performed by PCR using oligonucleotides oSR505 (5′-GCCACCAGCTGAGGAAGGGCTT-3′), oSR506 (5′-GGCTGATGATGATACTCAACCCATT-3′), and TAG3 (see above). Plant genotyping for the eel-2 mutation was performed by PCR using oligonucleotides oFP1080 (5′-CTCCACAATCTCTCCAGCTACTC-3′), oFP1108 (5′-GTTCGAGTGTAACAGTTTCTA-AGGC-3′), and oFPLB (see above). Plant genotyping for the abi5-1 mutation was performed by PCR using oligonucleotides oFP1081 (5′-GGTTATTGTTGTGTATATGATGCAGTTG-3′) and oFP1082 (5′-CCACTACTCTTTTCCTTCCCC-3′) followed by digestion with AvaII. Plant DNA was extracted as described by Edwards et al. (1991).
In Situ Hybridization
Plants were fixed in 4% formaldehyde (fresh from paraformaldehyde) in PBS under vacuum for 2 × 20 min and left in fixative overnight. After fixation, plants were washed, dehydrated, and embedded in paraffin, essentially as described previously (Jackson et al., 1991). Paraffin sections (8 to 10 mm thick) were cut with a disposable metal knife and attached to precoated glass slides (Fisher). Sections were hybridized with digoxigenin (DIG)-labeled antisense AtEm1 RNAs. The antisense AtEm1 RNA was synthesized from the plasmid pGATEM1 using the SP6 RNA polymerase and DIG according to the manufacturer's instructions (Boehringer Mannheim). Immunodetection of the DIG-labeled probe was performed using an anti-DIG antibody coupled to alkaline phosphatase as described by the manufacturer (Boehringer Mannheim). Sections were photographed on a Nikon Microphot FXA microscope (Garden City, NY).
Electrophoretic Mobility Shift Assays
The pET16b-ABI5 construct, corresponding to the ABI5 coding sequence cloned in the pET16b vector, was kindly supplied by Cristel Carles (Perpignan, France). To build the pET16b-EEL construct, a partial EEL cDNA was amplified by reverse transcriptase–mediated PCR from seed RNA using oligonucleotides oFP1079 (5′-TCTGGAAAACCACTAGGAAGCAT-3′) and oFP1085 (5′-GAGTCTTTAGGATCAGAGAGAAGCAGAG-3′). This partial cDNA was completed with a fragment obtained by PCR amplification of genomic DNA with oligonucleotides oSB1009 (5′-ACTCTAGAGGATCCTATGGGTTCTATTAGAGGAAACATTG-3′) and oSR506 (5′-GGCTGATGATGATAC-TCAACCCATT-3′). This EEL cDNA was cloned into the BamHI site of the pET16b vector (Novagen, Darmstadt, Germany). pET16b-ABI5 and pET16b-EEL were used for in vitro transcription and translation in the TNT rabbit reticulocyte system (Promega) according to the manufacturer's instructions. These ABI5 and EEL proteins contain six additional His residues at their N termini.
Overlapping oligonucleotides were annealed in 50 mM Tris-HCl, pH 8, 100 mM NaCl, and 10 mM MgCl2 from 85°C to room temperature, filled by T4 DNA polymerase with α-32P-dCTP, and purified on a column of Sephadex G-50 fine (Pharmacia, Uppsala, Sweden). Binding reactions were performed on ice for 30 min in 10 μL (final volume) of binding buffer (25 mM Hepes, pH 7.6, 60 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 1 μM DTT, and 5% glycerol) (Kuras et al., 1996) with 2.5 μL of in vitro–translated product, 0.5 to 1 ng of radiolabeled probe, and 100 ng of poly(dIdC). Binding reaction products then were loaded on 6% polyacrylamide gels in 225 mM Tris-borate and 0.5 mM EDTA. Gels were run at 5 V/cm at 4°C for 2 h, dried on Whatman 3MM paper, and analyzed using a Storm imager (Molecular Dynamics, Sunnyvale, CA).
Accession Numbers
Accession numbers (indicated as protein identification number/Arabidopsis Genome Initiative identification number) of the sequences shown in Figures 1 and 2 are as follows: ABI5 (AAD21438.1/At2 g36270), ABF1 (AAF27179/At1 g49720), ABF2 (AAF27180), ABF3 (AAF27181/At4 g34000), ABF4 (AAF27182/At3 g19290), AREB3 (BAB12406/At3 g56850), AtbZIP12 (BN000024/At2 g41070), AtbZIP13 (BN000023/At5 g44080), AtbZIP14 (BN000021/At4 g35900), AtbZIP15 (AJ419599/At5 g42910), AtbZIP27 (BN000022/At2 g17770), AtbZIP67 (AJ419600/At3 g44460), GBF4 (P42777/At1 g03970), DPBF-1 (T12621), DPBF-2 (T12622), DPBF-3 (T12585), TRAB1 (BAA83740), and OSE2 (AAF65459).
Accession numbers for EST and other sequences mentioned in Methods are as follows: EST sequences for AtbZIP13 (AV527966, 202M14T7, and AV549823), AtbZIP12/EEL partial cDNA (AJ420881), EST sequences for AtbZIP12 (BE529226, BE524624, and AV561543), DPBF-4 (AF334209), DPBF-2 (AF334207), and BAC clone T3K9 (AC004261).
Accession numbers for the regions used as gene-specific probes are as follows: ABI5 (nucleotides 22 to 1074 in AF334206), ABF1 (1163 to 1428 in AF093544), ABF2 (550 to 1017 in AF093545), ABF3 (608 to 1053 in AF093546), ABF4 (1262 to 1592 in AF093547), AREB3 (63,805 to 64,439 in AL390921), AtbZIP12 (100 to 623 in AF334209), AtbZIP13 (69,270 to 69,846 in MRH10), AtbZIP14 (76,538 to 76,863 in AL161588), AtbZIP15 (545 to 1005 in AJ419599), AtbZIP27 (54,898 to 55,124 in AF024504), AtbZIP67 (154 to 626 in AJ419600), and GBF4 (70 to 370 in U01823).
Acknowledgments
We thank Ranjiv Khush and Michel Delseny for helpful comments on the manuscript, David Bouchez for access to the Versailles collection of T-DNA lines, Dominique Thomas for help with EMSA, Cristel Carles and Michel Delseny for the ABI5 expression clone, Monique Raynal for the M17 cDNA, Philippe Peynot for help with figures, Ruth Finkelstein for seeds, Erika Asamizu at Kazusa DNA Research Institute for cDNA clones, and the ABRC for seeds and DNA clones. This work was supported by the Centre National de la Recherche Scientifique and the European Community REGIA project (Grant QLG2-CT1999-00876). S.B. was the recipient of a fellowship from the Ministère de l'Education Nationale de la Recherche et de la Technologie.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.000869.
References
- Asamizu, E., Nakamura, Y., Sato, S., and Tabata, S. (2000). A large scale analysis of cDNA in Arabidopsis thaliana: Generation of 12,028 non-redundant expressed sequence tags from normalized and size-selected cDNA libraries. DNA Res. 7, 175–180. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. 316, 1194–1199. [DOI] [PubMed] [Google Scholar]
- Chern, M.S., Bobb, A.J., and Bustos, M.M. (1996). The regulator of MAT2 (ROM2) protein binds to early maturation promoters and represses PvALF-activated transcription. Plant Cell 8, 305–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, H., Hong, J., Ha, J., Kang, J., and Kim, S.Y. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275, 1723–1730. [DOI] [PubMed] [Google Scholar]
- Delseny, M., Bies-Etheve, N., Carles, C., Hull, G., Vicient, C.M., Raynal, M., Grellet, F., and Aspart, L. (2001). Late embryogenesis abundant (LEA) protein gene regulation during Arabidopsis seed maturation. J. Plant Physiol. 158, 419–427. [Google Scholar]
- Edwards, K., Johnstone, C., and Thompson, C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19, 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem, T., Rushton, P.J., Robatzek, S., and Somssich, I.E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. [DOI] [PubMed] [Google Scholar]
- Ezcurra, I., Ellerstrom, M., Wycliffe, P., Stalberg, K., and Rask, L. (1999). Interaction between composite elements in the napA promoter: Both the B-box ABA-responsive complex and the RY/G complex are necessary for seed-specific expression. Plant Mol. Biol. 40, 699–709. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R.R. (1993). Abscisic acid-insensitive mutations provide evidence for stage-specific signal pathways regulating expression of an Arabidopsis late embryogenesis-abundant (lea) gene. Mol. Gen. Genet. 238, 401–408. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R.R. (1994). Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J. 5, 765–771. [Google Scholar]
- Finkelstein, R.R., and Lynch, T.J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12, 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaubier, P., Raynal, M., Hull, G., Huestis, G.M., Grellet, F., Arenas, C., Pagès, M., and Delseny, M. (1993). Two different Em-like genes are expressed in Arabidopsis thaliana seeds during maturation. Mol. Gen. Genet. 238, 409–418. [DOI] [PubMed] [Google Scholar]
- Guerche, P., Tire, C., Grossi de Sa, F., De Clercq, A., Van Montagu, M., and Krebbers, E. (1990). Differential expression of the Arabidopsis 2S albumin genes and the effect of increasing gene family size. Plant Cell 2, 469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobo, T., Asada, M., Kowyama, Y., and Hattori, T. (1999. a). ACGT-containing abscisic acid response element (ABRE) and coupling element3 (CE3) are functionally equivalent. Plant J. 19, 679–689. [DOI] [PubMed] [Google Scholar]
- Hobo, T., Kowyama, Y., and Hattori, T. (1999. b). A bZIP factor, TRAB1, interacts with VP1 and mediates abscisic acid-induced transcription. Proc. Natl. Acad. Sci. USA 96, 15348–15353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra, F.A., Golovina, E.A., and Buitink, J. (2001). Mechanisms of plant desiccation tolerance. Trends Plant Sci. 6, 431–438. [DOI] [PubMed] [Google Scholar]
- Hughes, D.W., and Galau, G.A. (1989). Temporally modular gene expression during cotyledon development. Genes Dev. 3, 358–369. [DOI] [PubMed] [Google Scholar]
- Jack, T. (2001). Relearning our ABCs: New twists on an old model. Trends Plant Sci. 6, 310–316. [DOI] [PubMed] [Google Scholar]
- Jackson, D., Culianez-Macia, F., Prescott, A.G., Roberts, K., and Martin, C. (1991). Expression patterns of myb genes from Antirrhinum flowers. Plant Cell 3, 115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby, M., Weisshaar, B., Droege-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., and Parcy, F. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111. [DOI] [PubMed] [Google Scholar]
- Kim, S.Y., Chung, H.J., and Thomas, T.L. (1997). Isolation of a novel class of bZIP transcription factors that interact with ABA-responsive and embryo-specification elements in the Dc3 promoter using a modified yeast one-hybrid system. Plant J. 11, 1237–1251. [DOI] [PubMed] [Google Scholar]
- Kim, S.Y., and Thomas, T.L. (1998). A family of novel basic leucine zipper proteins binds to seed-specification elements in the carrot DC3 gene promoter. J. Plant Physiol. 152, 607–613. [Google Scholar]
- Koornneef, M., Hanhart, C.J., Hilhorst, H.W.M., and Karssen, C.M. (1989). In vivo inhibition of seed development and reserve protein accumulation in recombinants of abscisic acid biosynthesis and responsiveness mutants in Arabidopsis thaliana. Plant Physiol. 90, 463–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuras, L., Cherest, H., Surdin-Kerjan, Y., and Thomas, D. (1996). A heteromeric complex containing the centromere binding factor 1 and two basic leucine zipper factors, Met4 and Met28, mediates the transcription activation of yeast sulfur metabolism. EMBO J. 15, 2519–2529. [PMC free article] [PubMed] [Google Scholar]
- Lee, M.M., and Schiefelbein, J. (2001). Developmentally distinct MYB genes encode functionally equivalent proteins in Arabidopsis. Development 128, 1539–1546. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina, L., and Chua, N.H. (2000). A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol. 41, 541–547. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina, L., Mongrand, S., and Chua, N.H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 98, 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menkens, A.E., and Cashmore, A.R. (1994). Isolation and characterization of a fourth Arabidopsis thaliana G-box-binding factor, which has similarities to Fos oncoprotein. Proc. Natl. Acad. Sci. USA 91, 2522–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, S., Lynch, T.J., and Finkelstein, R.R. (2001). Physical interactions between ABA response loci of Arabidopsis. Plant J. 26, 627–635. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nambara, E., Hayama, R., Tsuchiya, Y., Nishimura, M., Kawaide, H., Kamiya, Y., and Naito, S. (2000). The role of ABI3 and FUS3 loci in Arabidopsis thaliana on phase transition from late embryo development to germination. Dev. Biol. 220, 412–423. [DOI] [PubMed] [Google Scholar]
- Nambara, E., Keith, K., McCourt, P., and Naito, S. (1995). A regulatory role for the ABI3 gene in the establishment of embryo maturation in Arabidopsis thaliana. Development 121, 629–636. [Google Scholar]
- Nambara, E., Naito, S., and McCourt, P. (1992). A mutant of Arabidopsis which is defective in seed development and storage protein accumulation is a new abi3 allele. Plant J. 2, 435–441. [Google Scholar]
- Nantel, A., and Quatrano, R.S. (1996). Characterization of three rice basic/leucine zipper factors, including two inhibitors of EmBP-1 DNA binding activity. J. Biol. Chem. 271, 31296–31305. [DOI] [PubMed] [Google Scholar]
- Pang, P.P., Pruitt, R.E., and Meyerowitz, E.M. (1988). Molecular cloning, genomic organization, expression and evolution of 12S seed storage protein genes of Arabidopsis thaliana. Plant Mol. Biol. 11, 805–820. [DOI] [PubMed] [Google Scholar]
- Parcy, F., Valon, C., Raynal, M., Gaubier-Comella, P., Delseny, M., and Giraudat, J. (1994). Regulation of gene expression programs during Arabidopsis seed development: Roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6, 1567–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavy, N., Rombauts, S., Dehais, P., Mathe, C., Ramana, D.V., Leroy, P., and Rouze, P. (1999). Evaluation of gene prediction software using a genomic data set: Application to Arabidopsis thaliana sequences. Bioinformatics 15, 887–899. [DOI] [PubMed] [Google Scholar]
- Phillips, J., Artsaenko, O., Fiedler, U., Horstmann, C., Mock, H.P., Muntz, K., and Conrad, U. (1997). Seed-specific immunomodulation of abscisic acid activity induces a developmental switch. EMBO J. 16, 4489–4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann, J.L., et al. (2000). Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. [DOI] [PubMed] [Google Scholar]
- Stone, S.L., Kwong, L.W., Yee, K.M., Pelletier, J., Lepiniec, L., Fischer, R.L., Goldberg, R.B., and Harada, J.J. (2001). LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. USA 98, 11806–11811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, M., Kao, C.Y., and McCarty, D.R. (1997). The conserved B3 domain of VIVIPAROUS1 has a cooperative DNA binding activity. Plant Cell 9, 799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno, Y., Furihata, T., Abe, H., Yoshida, R., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 97, 11632–11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicient, C.M., Hull, G., Guilleminot, J., Devic, M., and Delseny, M. (2000). Differential expression of the Arabidopsis genes coding for Em-like proteins. J. Exp. Bot. 51, 1211–1220. [PubMed] [Google Scholar]
- White, J.A., Todd, J., Newman, T., Focks, N., Girke, T., de Ilarduya, O.M., Jaworski, J.G., Ohlrogge, J.B., and Benning, C. (2000). A new set of Arabidopsis expressed sequence tags from developing seeds: The metabolic pathway from carbohydrates to seed oil. Plant Physiol. 124, 1582–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus, U., and Weber, H. (1999). Seed maturation: Genetic programmes and control signals. Curr. Opin. Plant Biol. 2, 33–38. [DOI] [PubMed] [Google Scholar]