Abstract
The chloroplastic drought-induced stress protein of 32 kD (CDSP32) is composed of two thioredoxin modules and is induced by environmental and oxidative stress conditions. We investigated whether the plastidic protein BAS1, which is related to eubacterial 2-Cys peroxiredoxin, is a target for CDSP32. Using a CDSP32 active-site mutant, we showed that the BAS1 and CDSP32 proteins form a mixed disulfide complex in vitro. Moreover, affinity chromatography indicated that BAS1 is a major target for CDSP32 in chloroplasts. CDSP32 was able to reduce BAS1 in vitro, and BAS1 displayed CDSP32-dependent peroxidase activity. The function of CDSP32 was investigated in transgenic potato lines without detectable levels of the protein as a result of cosuppression. Under conditions of photooxidative stress induced by incubation with either methyl viologen or t-butyl hydroperoxide or by exposure to low temperature under high light, plants lacking CDSP32 exhibited decreased maximal photosystem II photochemical efficiencies compared with the wild type and transgenic controls. In addition, plants without CDSP32 retained much less chlorophyll than controls under stress, indicating increased damage to photosynthetic membranes. We conclude that CDSP32 is a thioredoxin with a critical role in plastid defense against oxidative damage and that this role is related to its function as a physiological electron donor to the BAS1 peroxiredoxin.
INTRODUCTION
Thioredoxins are small proteins with a Cys-Gly-Pro-Cys active site domain that is able to reduce disulfide bridges on target proteins. They are found in all living cells and display a typical structure, called the thioredoxin fold, which is characterized by a central pleated β sheet composed of five strands surrounded by α helices (Eklund et al., 1991). In addition to a general function in the regulation of enzyme activity via thiol redox control, thioredoxins participate in many other processes, such as sulfate assimilation, phage assembly, DNA transcription, and cell apoptosis (Holmgren, 1985; Schenk et al., 1994; Saitoh et al., 1998). In bacteria, yeast, and animals, thioredoxins have been shown to be involved in oxidative stress responses. For example, the oxidative stress–induced expression of thioredoxin 2 in Escherichia coli is activated by the transcription factor OxyR (Ritz et al., 2000), and a thioredoxin-deficient yeast mutant is hypersensitive to H2O2 (Muller, 1991).
Evidence for a role for thioredoxins in H2O2 reduction has been obtained in yeast and mammals. This role is linked to the action of 2-Cys peroxiredoxins, a family of thioredoxin-dependent peroxidases similar to the bacterial subunit C of alkyl hydroperoxide reductase (AhpC). These peroxidases are able to reduce H2O2 or alkyl hydroperoxides by transferring electrons from sulfhydryl residues (Chae et al., 1994). In Salmonella typhimurium, in which the system has been best characterized, AhpC is reduced by AhpF, a flavin adenine dinucleotide–containing protein with a C-terminal domain similar to the E. coli 35-kD flavoprotein thioredoxin reductase Tr-R (Poole et al., 2000b). A recent study showed that the N-terminal domain of AhpF contains a tandem repeat of the thioredoxin fold. The second of these folds retains a redox-active disulfide center able to mediate electron transfer from the AhpF TrR-like domain to AhpC (Poole et al., 2000a).
Three types of thioredoxins (h, f, and m) have been identified in plants, each encoded by a multigene family (Meyer et al., 1999). Thioredoxin h is located in the cytosol and is suggested to participate in cell communication, seed germination, and reproduction (for review, see Schürmann and Jacquot, 2000). Thioredoxins f and m are present in chloroplasts, where they regulate, through conformational changes, the activity of certain enzymes involved in photosynthetic carbon metabolism, such as Fru-1,6-bisphosphatase and NADP malate dehydrogenase (Jacquot et al., 1997; Ruelland and Miginiac-Maslow, 1999). The role of thioredoxins in oxidative stress responses in plants is largely unexplored. Arabidopsis thioredoxins h3 and m have been reported to partially complement the H2O2 hypersensitivity of a yeast thioredoxin-deficient mutant (Mouaheb et al., 1998; Issakidis-Bourguet et al., 2001). An active-site mutant of Arabidopsis thioredoxin h3, when expressed in yeast, was found to interact with a peroxiredoxin (Verdoucq et al., 1999). The plant 2-Cys peroxiredoxin called BAS1, which is related to AhpC, has been found in chloroplasts, in which it exists as a homodimer associated with the stromal surface of the thylakoids (Baier and Dietz, 1999a). Baier and Dietz (1999b) showed that BAS1 protects the photosynthetic apparatus against oxidative damage during leaf development in Arabidopsis. To date, no electron donor reducing the plastidic 2-Cys peroxiredoxin has been identified.
We characterized a novel type of plant thioredoxin in potato called CDSP32 (chloroplastic drought-induced stress protein of 32 kD) (Pruvot et al., 1996). The CDSP32 protein, which is encoded in the nucleus and located in the stroma, is abundant during severe drought and oxidative stress conditions (Rey et al., 1998; Eymery and Rey, 1999; Broin et al., 2000). The mature CDSP32 protein (243 residues) exhibits structural features similar to those described for the AhpF N-terminal domain described previously. Each protein contains two thioredoxin domains, with the one closest to the C terminus retaining a redox-active disulfide (Rey et al., 1998; Poole et al., 2000a).
In the present study, based on the sequence similarities between AhpF and CDSP32 and between AhpC and BAS1, we investigated whether CDSP32 could act as an electron donor to BAS1 in a manner similar to that described for the bacterial AhpF/AhpC system. Because the thioredoxin reduction catalytic mechanism proceeds via the formation of a transient covalent complex with the substrate, through a disulfide bridge between the first Cys of the thioredoxin active site and the substrate Cys (Yang and Wells, 1991), we used a mutation strategy to modify the CDSP32 active site. Replacing the second Cys of the active site with a Ser (a nonreactive structural analog) results in stabilization of the complex (Wynn et al., 1995; Goyer et al., 1999; Verdoucq et al., 1999; Balmer and Schürmann, 2001). Using a CDSP32 active-site mutant, we show the formation of a CDSP32/BAS1 mixed disulfide complex in vitro. In addition, affinity chromatography indicates that BAS1 is a major target for CDSP32 in plant extracts. As a step toward clarifying CDSP32 function, we generated transgenic potato plants altered in CDSP32 expression. We report that cosuppressed lines lacking CDSP32 exhibit an increased susceptibility to photooxidative stress conditions. These data demonstrate the presence of a thioredoxin-dependent defense system that prevents oxidative damage in plants. We propose that the peroxiredoxin-dependent reduction of alkyl hydroperoxides in chloroplasts proceeds through CDSP32.
RESULTS
Cloning and Sequence Analysis of Arabidopsis CDSP32 cDNA
Using the potato CDSP32 (StCDSP32) cDNA as a probe, a full-length Arabidopsis cDNA, encoding a 302-residue protein, was isolated and designated AtCDSP32. The mature AtCDSP32 and StCDSP32 proteins (244 and 243 residues, respectively) were found to share 75% identity and 82% similarity (Figure 1). Interestingly, higher percentages of identity and similarity were noted in the half C-terminal domain (83 and 88%, respectively). As described for StCDSP32 (Rey et al., 1998), the AtCDSP32 half N- and C-terminal domains displayed 27% identity and 44% similarity, with no active site in the N-terminal part.
Figure 1.
Sequence Alignment of Arabidopsis (AtCDSP32) and Potato (StCDSP32) CDSP32 Precursor Proteins.
Identity and similarity of amino acids are indicated by colons and dots, respectively. The arrow indicates the putative transit peptide cleavage site, and the asterisk indicates the first residues of the C-terminal domains. The active sites are underlined, and residues important for preserving the thioredoxin fold (Eklund et al., 1991) appear in boldface type.
DNA gel blot data indicated that AtCDSP32 is very likely a single-copy gene in Arabidopsis (data not shown) as in potato (Rey et al., 1998). Sequence analysis using the BLAST algorithm (Altschul et al., 1990) revealed that two overlapping genomic fragments from Arabidopsis chromosome I contained AtCDSP32 in their common region, confirming the presence of one copy in this species.
Production, Integrity, and Activity of Recombinant Proteins
The Arabidopsis and potato cDNAs coding for mature CDSP32 proteins (AtCDSP32m and StCDSP32m, respectively), for mutated CDSP32 proteins, with the second Cys of the active site replaced by a Ser (AtCDSP32mC164S and StCDSP32mC163S), and for CDSP32 half C-terminal domains (AtCDSP32C-ter and StCDSP32C-ter) were cloned into pQE30 expression vectors. The recombinant proteins were produced in E. coli and purified in native conditions. Protein purity and identity were verified using SDS-PAGE (Figure 2) and immunoblot analysis (data not shown). Circular dichroism in the 190- to 260-nm range indicated correct folding for all proteins, with no difference between wild-type and mutant spectra (data not shown). Furthermore, the folding of AtCDSP32m and StCDSP32m was verified using two-dimensional NMR analysis. Preliminary interpretation of these data confirmed that both proteins were composed of two typical thioredoxin folds, as suggested by sequence analysis and predicted using the RasMol2 molecular graphics program (data not shown).
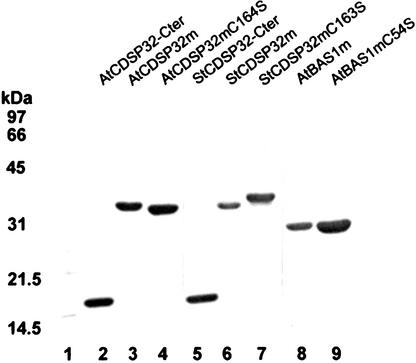
Figure 2.
Purification of Recombinant CDSP32 and BAS1 Proteins.
The proteins produced in E. coli with a six-His N-terminal tag were purified using a nickel affinity column, separated by SDS-PAGE, and stained with Coomassie blue. Lane 1, molecular mass markers; lane 2, AtCDSP32C-ter; lane 3, AtCDSP32m; lane 4, AtCDSP32mC164S; lane 5, StCDSP32C-ter; lane 6, StCDSP32m; lane 7, StCDSP32mC163S; lane 8, AtBAS1m; lane 9, AtBAS1mC54S.
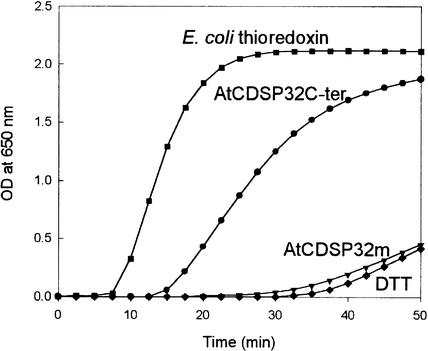
The thioredoxin activity of CDSP32 proteins was investigated by performing insulin reduction assays (Holmgren, 1979). As shown in Figure 3, DTT alone resulted in slow insulin precipitation after 32 min. When 5 μM E. coli thioredoxin was added as a positive control, insulin precipitation was observed after 7 min and occurred at a maximal rate of 0.199 OD unit/min. Using 5 μM AtCDSP32C-ter, insulin precipitation took place after 13 min with a lower rate (maximal rate of 0.090 OD unit/min). Similar results have been reported by Rey et al. (1998) with regard to the insulin reduction properties of StCDSP32C-ter. In contrast, the whole AtCDSP32m and StCDSP32m proteins did not reduce insulin at a faster rate than DTT alone (Figure 3 and data not shown). Because circular dichroism and NMR indicated correct folding for both proteins, we conclude that the mature CDSP32 proteins are not able to reduce insulin.
Figure 3.
Thioredoxin Activity of Arabidopsis CDSP32 Protein.
The reduction of insulin (1 mg/mL) by 5 μM AtCDSP32m (inverted triangles) or by 5 μM AtCDSP32C-ter (circles) was monitored as an increase in turbidity at 650 nm because of insulin precipitation. The reaction was started by DTT addition. DTT alone (diamonds) and E. coli thioredoxin at 5 μM (squares) were used as negative and positive controls, respectively.
In plant cells, the plastidic BAS1 2-Cys peroxiredoxin exists as a homodimer, with the two subunits linked through a disulfide bridge between Cys-54 of one subunit and Cys-175 of the other subunit (Baier and Dietz, 1997). Mutation of either Cys residue in the AhpC protein has been reported to prevent dimerization (Ellis and Poole, 1997). The Arabidopsis cDNAs that code for mature BAS1 (AtBAS1m) and for mutated BAS1, with the first Cys replaced by a Ser (AtBAS1mC54S), were cloned into the pQE32 expression vector. Purified recombinant proteins were analyzed using SDS-PAGE (Figure 2) and immunodetection (data not shown). On the basis of circular dichroism spectra in the 190- to 260-nm range, the proteins appeared to be folded correctly, with no difference between AtBAS1m and AtBAS1mC54S (data not shown).
Interaction in Vitro between a CDSP32 Active-Site Mutant and BAS1
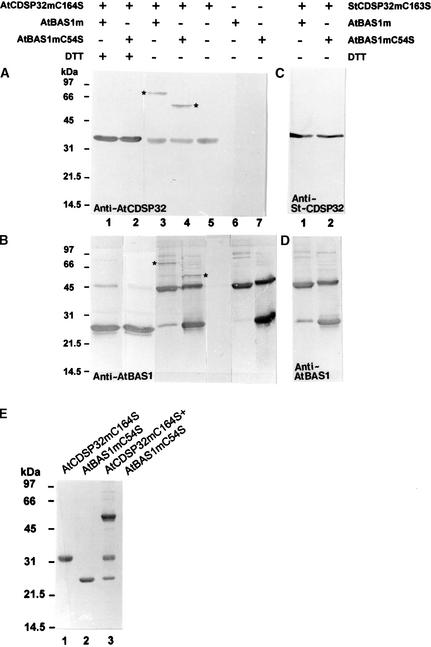
We analyzed the ability of CDSP32 to interact with BAS1 by incubating AtCDSP32mC164S or StCDSP32mC163S with AtBAS1m or AtBAS1mC54S. After incubation, the proteins were separated by SDS-PAGE in the presence or absence of a reducing agent (DTT) and analyzed by immunodetection using sera raised against CDSP32 and BAS1. When AtCDSP32mC164S was incubated alone and SDS-PAGE was run without DTT, a single 32-kD band was recognized by the anti-AtCDSP32 serum (Figure 4A, lane 5). When AtBAS1m and AtBAS1mC54S were treated similarly, 25- and 45-kD bands corresponding to BAS1 monomer and dimer, respectively, were revealed using the anti-AtBAS1m serum, the dimer being the major form with AtBAS1m (Figure 4B, lanes 6 and 7). These data indicate that dimerization of AtBAS1mC54S can occur through the other Cys (Cys-175) of each subunit. In addition, several faint bands likely corresponding to BAS1 multimers were detected in both extracts. In samples containing the CDSP32 and BAS1 proteins separated in the presence of DTT, the anti-AtCDSP32 serum revealed a 32-kD band corresponding to AtCDSP32mC164S (Figure 4A, lanes 1 and 2), whereas the serum raised against AtBAS1 revealed a main 25-kD band corresponding to a monomer of AtBAS1m and a faint 45-kD band corresponding to a dimer (Figure 4B, lanes 1 and 2). When mixed AtCDSP32mC164S and AtBAS1m were separated without DTT, an extra band of ∼66 kD was revealed using both anti-AtCDSP32 and anti-AtBAS1 sera (Figures 4A and 4B, lanes 3). The analysis of AtCDSP32mC164S and AtBAS1mC54S mixtures separated without DTT showed a supplementary 55-kD band detected using the anti-AtBAS1 and anti-AtCDSP32 sera (Figures 4A and 4B, lanes 4).
Figure 4.
In Vitro Analysis of the Interaction Products between CDSP32 and BAS1 Proteins.
(A) to (D) Immunoblot analysis of the interaction products between CDSP32 and BAS1 proteins. Recombinant AtBAS1m and AtBAS1mC54S were incubated in vitro with AtCDSP32mC164S ([A] and [B]) or StCDSP32mC163S ([C] and [D]) for 15 min at room temperature in phosphate buffer, separated using SDS-PAGE in the presence (+) or absence (−) of DTT in the loading buffer, and revealed by immunoblot analysis using the anti-AtCDSP32 (A), anti-StCDSP32 (C), and anti-AtBAS1 ([B] and [D]) sera. Asterisks indicate the bands recognized by both anti-AtCDSP32 and anti-AtBAS1 sera.
(E) SDS-PAGE analysis of the interaction product between AtBAS1mC54S-TNB and AtCDSP32mC164S. AtBAS1mC54S was reacted with dithiobisnitrobenzoic acid to form a AtBAS1mC54S-TNB mixed disulfide before incubation with AtCDSP32mC164S prereduced with DTT. The proteins were incubated in vitro for 15 min at room temperature in phosphate buffer, separated using SDS-PAGE in the absence of DTT, and revealed using Coomassie blue staining. Lane 1, AtCDSP32mC164S; lane 2, AtBAS1mC54S-TNB; lane 3, AtCDSP32mC164S + AtBAS1mC54S-TNB.
The 66- and 55-kD bands were not observed with only one protein type in the incubation mixture or when AtBAS1m and AtCDSP32m were mixed and separated in the presence of DTT. Because they are recognized by both anti-AtBAS1 and anti-AtCDSP32 sera, they correspond to 66- and 55-kD heterodimeric disulfide complexes composed of AtCDSP32mC164S and a dimer of AtBAS1m or a monomer of AtBAS1mC54S, respectively. No extra band was observed when wild-type AtCDSP32m was incubated with AtBAS1m or with AtBAS1mC54S (data not shown), clearly demonstrating the necessity of modifying the AtCDSP32m active site to stabilize the complex. Note also that no additional band was observed in mixtures containing StCDSP32mC163S and AtBAS1m or AtBAS1mC54S after separation without DTT (Figures 4C and 4D, lanes 1 and 2).
In these incubation experiments, only a low proportion of the AtBAS1 and AtCDSP32 pools appeared to form a stable complex. We investigated whether modifying the redox state of AtBAS1 and AtCDSP32 Cys residues could change the proportion of complexed proteins. To ensure full AtBAS1mC54S oxidation, the protein was reacted with dithiobisnitrobenzoic acid (Wang et al., 1996) to give a mixed disulfide derivative, AtBAS1mC54S-TNB. The C54S mutant was used rather than the wild-type protein to avoid reduction of the TNB derivative via dimerization. AtBAS1mC54S-TNB was incubated for 15 min with AtCDSP32mC164S prereduced with DTT. SDS-PAGE revealed that after incubation, >50% of AtBAS1C54S and AtCDSP32C164S were complexed as a stable mixed disulfide of 55 kD (Figure 4E). This experiment clearly demonstrates that AtBAS1 oxidation and AtCDSP32 reduction substantially increase the formation of the disulfide complex.
Affinity Chromatography Using AtCDSP32m and StCDSP32m as Bait
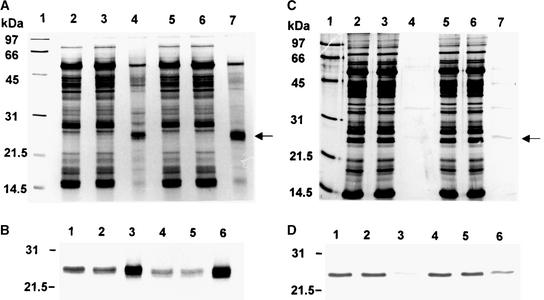
To investigate the specificity of the CDSP32–BAS1 interaction in native plant proteins, we performed affinity chromatography using CDSP32 mutant proteins as bait to analyze extracts of leaf and chloroplastic soluble proteins. After immobilization of either AtCDSP32mC164S or StCDSP32mC163S on Hi-Trap N-hydroxysuccinimide-activated columns, extracts from Arabidopsis leaves or potato chloroplasts, respectively, were loaded on the columns. After washing, the proteins that were retained specifically through a disulfide bridge to CDSP32 were eluted using DTT. Samples representative of each chromatography step were separated by SDS-PAGE and analyzed using silver staining and immunodetection with anti-AtBAS1 serum (Figure 5).
Figure 5.
Affinity Chromatography of Arabidopsis and Potato Plant Extracts Using Mutated CDSP32 as Bait.
After application of Arabidopsis leaf ([A] and [B]) and potato chloroplastic ([C] and [D]) soluble proteins on affinity columns retaining immobilized AtCDSP32mC164S ([A] and [B]) or StCDSP32mC163S ([C] and [D]), the proteins from each representative chromatography step were analyzed using SDS-PAGE and silver staining ([A] and [C]) or immunodetection with the anti-AtBAS1 serum ([B] and [D]).
(A) and (C) Lane 1, molecular mass markers; lane 2, crude extract; lane 3, unbound proteins; lane 4, DTT-eluted proteins; lane 5, unbound fraction oxidized with 100 μM tBOOH for 30 min at 4°C; lane 6, unbound proteins from the tBOOH-oxidized fraction; lane 7, DTT-eluted proteins from the tBOOH-oxidized fraction.
(B) and (D) Immunoblot analysis with the anti-AtBAS1 serum of the fractions corresponding to lanes 2 to 7 of (A) and (C), respectively.
For both Arabidopsis and potato extracts, few proteins were revealed in DTT-recovered fractions, and a 25-kD protein appeared to be a major band out of these proteins (Figures 5A and 5C, lanes 4). Immunoblot analysis clearly identified this band as BAS1 (Figures 5B and 5D, lanes 3) and also revealed a large BAS1 abundance in the unbound fractions (Figures 5B and 5D, lanes 2). The protein was not detected in washing fractions (data not shown). Because most other eluted bands were observed in washing fractions, they may correspond to contaminant proteins abundant in the crude extract rather than to proteins bound specifically to CDSP32 through a disulfide bridge. However, note that the eluted proteins of 50 and 15 kD were identified as ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) subunits by immunoblot analysis (data not shown) and that Rubisco small subunit also has been identified as a target for thioredoxin m by Motohashi et al. (2001). The physiological relevance of an interaction between thioredoxins and Rubisco remains to be explored.
To investigate whether the redox state of BAS1 alters its binding capacity, as observed in the incubation experiments described previously, the unbound fractions were treated using t-butyl hydroperoxide (tBOOH), a 2-Cys peroxiredoxin substrate, and reapplied to the columns. As shown in lanes 7 of Figures 5A and 5C and lanes 6 of Figures 5B and 5D, an increased BAS1 amount was retained and eluted using DTT in both species. These chromatography data clearly show that the native AtBAS1 and StBAS1 peroxiredoxins are major targets for the AtCDSP32 and StCDSP32 thioredoxins, respectively.
BAS1 Reduction by CDSP32
The AtBAS1m protein was engineered by linking a fluorophore (carboxyfluorescein succinimidyl ester; FAM-SE) via a disulfide bond to Cys-175 of the AtBAS1mC54S mutant. The C54S mutant was used to prevent reduction of a FAM derivative via dimerization. Such a strategy has been used to measure the reduction rate of AhpC, the bacterial homolog of BAS1, by AhpF (Poole et al., 2000a). The substrate reduction results in fluorophore release and can be monitored as an increase in fluorescence at 510 nm.
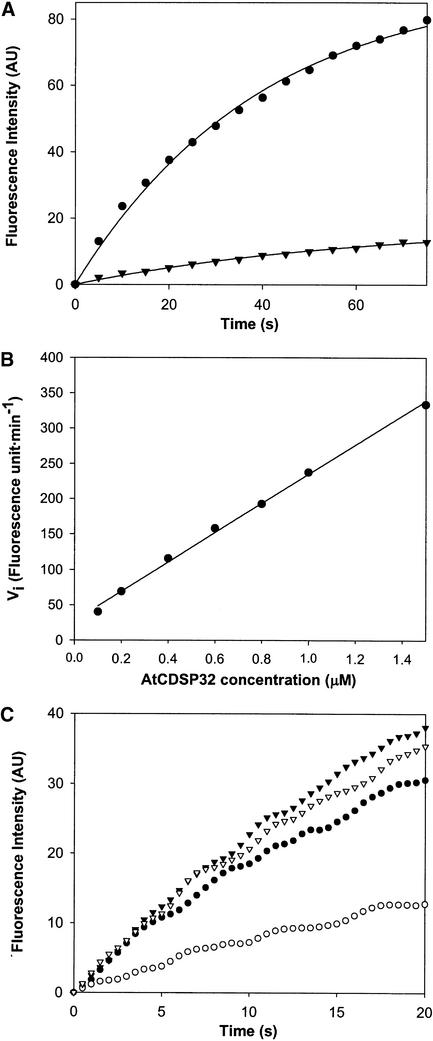
As shown in Figure 6A, a substantial increase in fluorescence was recorded after 75 s when using prereduced AtCDSP32m at 0.5 μM and AtBAS1m-FAM at 1 μM. A maximal linear increase (∼140 arbitrary units/min) was observed during the first 10 s. By contrast, almost no fluorescence change was noted using nonreduced AtCDSP32m (Figure 6A). These data clearly show the ability of AtCDSP32m to act as an electron donor to the AtBAS1 2-Cys peroxiredoxin.
Figure 6.
Reduction of AtBAS1m-FAM by CDSP32.
The reduction of AtBAS1m-FAM was monitored as a fluorescence increase at 510 nm generated by the release of FAM. The reactions were performed in phosphate buffer and started by adding CDSP32. AU, arbitrary units.
(A) Increase in fluorescence intensity in a reaction mixture containing 1 μM AtBAS1m-FAM and 0.5 μM reduced (closed circles) or nonreduced (closed inverted triangles) AtCDSP32m. The fluorescence intensity values were read every 5 s after starting the reaction.
(B) Initial rates of fluorescence increase measured in reaction mixtures containing 1 μM AtBAS1m-FAM and AtCDSP32m in a concentration range from 0.1 to 1.5 μM. Vi was calculated as the rate of increase in fluorescence intensity (AU) during a 6-s period.
(C) Increase in fluorescence intensity in reaction mixtures containing AtBAS1m-FAM and AtCDSP32m (closed circles), AtCDSP32C-ter (closed inverted triangles), StCDSP32m (open circles), or StCDSP32C-ter (open inverted triangles). A total of 1 μM AtBAS1m-FAM was mixed with 0.5 μM CDSP32 proteins. The fluorescence intensity values were recorded every 0.5 s for 20 s.
We then measured the maximal initial rates of AtBAS1m-FAM reduction (at 1 μM) during the first 6 s of the reaction using various AtCDSP32m concentrations. We observed that, during this period, the AtBAS1 reduction rate increased linearly with AtCDSP32m concentration in a range from 0.1 to 1.5 μM (Figure 6B). Finally, we compared the initial rates of AtBAS1m-FAM reduction (at 1 μM) by AtCDSP32m, AtCDSP32C-ter, StCDSP32m, and StCDSP32C-ter (at 0.5 μM) (Figure 6C). We observed that, whereas AtCDSP32m, AtCDSP32C-ter, and StCDSP32C-ter displayed close maximal linear rates during the first 6 s (∼130 arbitrary units/min), StCDSP32m reduced AtBAS1m in a substantially less efficient manner (maximal rate of ∼50 arbitrary units/min).
Peroxidase Activity of BAS1 in the Presence of CDSP32
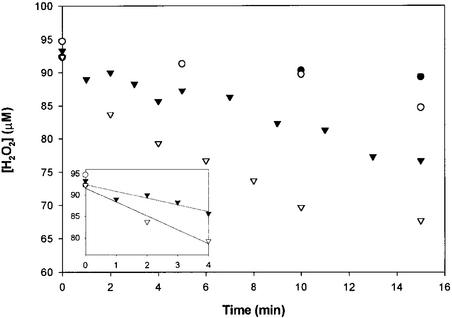
We investigated whether AtBAS1m displays CDSP32-dependent peroxidase activity by measuring the rate of H2O2 consumption in mixtures containing prereduced AtCDSP32m, AtBAS1m, and 100 μM H2O2 (Figure 7). In the absence of AtBAS1m, the H2O2 concentration remained almost stable. In the absence of AtCDSP32m, a slight decrease in H2O2 concentration, particularly during the first 10 min of the reaction, was noted. When 5 μM prereduced AtCDSP32m was mixed with 5 or 10 μM AtBAS1m, the H2O2 concentration decreased to 77 or 68 μM after 15 min, respectively. In both cases, the initial rate of H2O2 consumption, measured within the first 4 min, was ∼0.32 μmol H2O2·min−1·μmol−1 AtBAS1m (Figure 7, subfigure).
Figure 7.
Peroxidase Activity of AtBAS1m in the Presence of AtCDSP32m.
A total of 5 μM (closed inverted triangles) or 10 μM (open inverted triangles) AtBAS1m was mixed with 5 μM AtCDSP32m, prereduced with DTT, in phosphate buffer containing 100 μM H2O2. The reaction was started by adding AtBAS1m. The H2O2 concentration was determined in a 5-μL reaction mixture using the FOX1 reagent (Wolff, 1994). A total of 10 μM AtBAS1m (open circles) or 5 μM AtCDSP32m (closed circles) were used alone as controls. The subfigure shows the linear regression of H2O2 concentration as a function of time within the first 4 min of the reaction using 5 μM AtCDSP32m and 5 μM (closed inverted triangles) or 10 μM (open inverted triangles) AtBAS1m. The regression parameters were calculated using SigmaPlot, with a confidence interval of 95%. The calculated rates of H2O2 consumption were 1.59 μmol/min (r2 = 0.83) and 3.25 μmol/min (r2 = 0.97) using 5 and 10 μM AtBAS1, respectively.
These data indicate that AtCDSP32m provides electrons to AtBAS1m, which in turn can reduce H2O2. The peroxidase activity appears rather limited because of the absence of an electron donor to AtCDSP32m to sustain the reducing flux. Poole et al. (2000a) reported similar results when measuring the peroxidase activity of AhpC in the presence of a truncated AhpF protein corresponding to its double thioredoxin domain.
Generation of Transgenic Potato Plants with Modified CDSP32 Levels
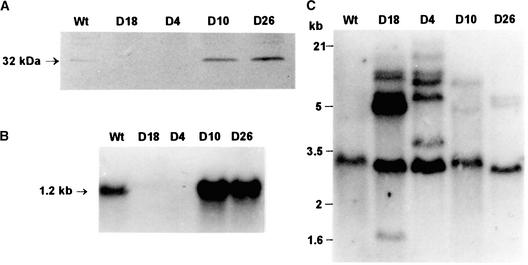
Potato cv Désirée plants were transformed with the whole StCDSP32 cDNA in the sense orientation under the control of the 35S promoter of Cauliflower mosaic virus. A total of 31 kanamycin-resistant plants were screened using immunoblot analysis for the CDSP32 abundance in leaves. Four lines (D4, D10, D18, and D26) with notably altered levels of CDSP32 were selected for further investigation. In lines D10 and D26, fivefold higher CDSP32 abundance was observed compared with that in the wild type (Figure 8A; image analysis performed using Genetools [Syngene, Cambridge, UK]) and in four independent control lines transformed with the vector carrying only the kanamycin resistance gene (data not shown). The stromal localization of the overexpressed protein was confirmed by immunoblot analysis of chloroplastic proteins (data not shown). By contrast, in lines D18 and D4, no protein was detected (Figure 8A), even when larger protein amounts were loaded and when proteins from younger or older leaves were analyzed (data not shown).
Figure 8.
Characterization of Transgenic Potato Lines with Modified CDSP32 Levels.
(A) Immunoblot analysis. Leaf soluble proteins (20 μg) from wild-type, D18, D4, D10, and D26 lines were analyzed using anti-StCDSP32 serum.
(B) RNA gel blot analysis. Total leaf RNA (6 μg) from the same lines were hybridized with a DIG-labeled StCDSP32 cDNA.
(C) DNA gel blot analysis. Genomic DNA (3 μg) from the same lines was digested with XbaI and hybridized with a DIG-labeled StCDSP32 cDNA.
Sizes of marker DNAs are shown at left. Wt, wild type.
RNA gel blot hybridization was performed to investigate the abundance of CDSP32 mRNA in leaves from transgenic lines. A single 1.2-kb mRNA band was revealed in the wild type (Figure 8B). Using the same exposure and revelation procedures, a much higher level of transcript was observed in lines D10 and D26, whereas the transcript was not detected in lines D18 and D4 (Figure 8B).
DNA gel blot analysis was performed on genomic DNA digested with XbaI, which does not cleave StCDSP32 cDNA. The probe hybridized one major band in wild-type DNA (Figure 8C), likely indicating the presence of one gene copy in cv Désirée, as reported for cv Haig (Rey et al., 1998). In digests from transformants, different patterns were observed, with approximately five additional bands for lines D18 and D4 (Figure 8C) and two supplementary bands for lines D10 and D26 (Figure 8C). Because lines D18 and D4 displayed the highest number of inserted CDSP32 copies and lacked the CDSP32 mRNA and the protein, we concluded that a cosuppression mechanism occurred at the transcript level in these lines.
Under standard phytotron conditions, no obvious phenotype was noted for the four independent transgenic lines, which exhibited growth and development characteristics similar to those of the wild type and transgenic controls (data not shown). Because lines D4 and D18, on the one hand, and lines D10 and D26, on the other hand, displayed similar characteristics with regard to stress response, additional experiments were performed using an equal number of plants from each line. Cosuppressed and overexpressing lines were designated CDSP32− and CDSP32+, respectively.
Photosynthetic Damage in Leaf Discs Subjected to Oxidative Treatments
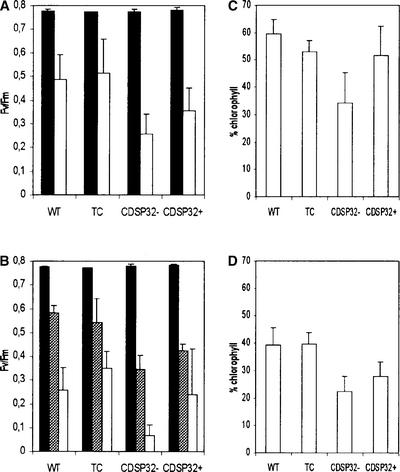
We investigated the characteristics of transgenic lines using leaf discs treated with either the photooxidizing photosystem (PS) I acceptor methyl viologen (Babbs et al., 1989) or the alkyl hydroperoxide tBOOH. We used PSII photochemical efficiency and chlorophyll content as indicators of photosynthetic performance to evaluate the resistance to oxidative stress. The maximal PSII photochemical efficiency was measured using the chlorophyll fluorescence parameter Fv/Fm (see Methods), which reflects the PSII capacity to reduce electron acceptors in the photosynthetic pathway (Maxwell and Johnson, 2000). In discs floated on 1 μM methyl viologen, a severe decrease in Fv/Fm was recorded in all plant types after 3 h in the light (Figure 9A). No decrease was noted in dark-incubated discs (data not shown).
Figure 9.
Maximal PSII Photochemical Efficiency and Chlorophyll Retention in Leaf Discs from Wild-Type, Transgenic Control, CDSP32-Cosuppressed, and CDSP32-Overexpressing Potato Plants Subjected to Photooxidative Treatments.
(A) Fv/Fm values measured in leaf discs floated on water (closed bars) or 1 μM methyl viologen (open bars) and exposed to light for 3 h after 16 h in the dark.
(B) Fv/Fm values measured in leaf discs floated on water (closed bars) or 2 mM tBOOH and exposed to light for 2.5 h (gray bars) or 4 h (open bars) after 16 h in the dark.
(C) and (D) Retention of chlorophyll in leaf discs floated on 1 μM methyl viologen (C) and 2 mM tBOOH (D) and exposed to light for 26 h under phytotron photoperiod conditions after 16 h in the dark.
For each treatment, data from 6 to 10 independent experiments are reported (one plant per experiment). The presented Fv/Fm values are means ± sd of the average values measured on 10 discs per plant. The percentage of chlorophyll retention was calculated on the basis of the content of untreated discs floated on water under the same light conditions. The presented percentage values are means ± sd of the average values determined on 10 discs from one plant. CDSP32−, CDSP32 cosuppressed; CDSP32+, CDSP32 overexpressing; TC, transgenic control; WT, wild type.
The discs from CDSP32− plants exhibited a much higher sensitivity to methyl viologen, with a Fv/Fm mean value (0.256) significantly lower than those of the wild type (0.484) and transgenic controls (0.512). Note that leaf discs from CDSP32+ plants showed reduced PSII efficiency compared with the wild type and transgenic controls (Figure 9A). The methyl viologen treatment, applied for 26 h under phytotron light conditions, resulted in a loss of chlorophyll and bleaching. The loss appeared to be much more severe in discs from cosuppressed plants (34% chlorophyll retained compared with that in untreated discs floated on water) than in the wild type and transgenic controls (59 and 53%, respectively) (Figure 9C). Discs from CDSP32+ plants were found to retain 51% chlorophyll under the photooxidative treatment (Figure 9C).
When discs from cosuppressed plants were floated on 2 mM tBOOH under light, the PSII photochemical efficiency decreased severely after 2.5 h (0.344) and was almost null after 4 h (0.068) (Figure 9B). In contrast, the Fv/Fm values were significantly higher in treated discs from the wild type (0.593 and 0.258 after 2.5 and 4 h, respectively) and from transgenic controls (0.542 and 0.350) (Figure 9B). Leaf discs from CDSP32+ lines were not less sensitive to tBOOH than those from plants with unmodified CDSP32 levels (Figure 9B). After exposure for 26 h, discs from wild-type and transgenic control plants retained ∼40% chlorophyll, whereas those from CDSP32− and CDSP32+ lines retained 22 and 28% chlorophyll, respectively (Figure 9D). Together, these data demonstrate a greater susceptibility to photooxidative stress among lines lacking CDSP32 with regard to the stability of photosynthetic structures.
PSII Efficiency in Plants Subjected to Photooxidative Stress
The sensitivity to photooxidative stress of whole transgenic plants was investigated under high light (1100 μmol·m−2·s−1) combined with low temperature (6°C), conditions that induce CDSP32 expression (Broin et al., 2000). After 25 h of treatment (including 12 h of night), young leaves with no bleaching symptoms were used for measurements of chlorophyll fluorescence. The treatment resulted in severe decreases in PSII photochemical efficiency, as indicated by Fv/Fm values <0.4 in all lines (Table 1). However, the CDSP32− plants displayed increased sensitivity to the treatment, with Fv/Fm mean values lower than those measured in the wild type (Table 1). The overexpressing lines also exhibited reduced PSII efficiency, although to a lesser extent than the cosuppressed lines.
Table 1.
Maximal PSII Photochemical Efficiency in Potato Plants with Modified CDSP32 Levels Subjected to Photooxidative Stress
| Plant | Experiment 1 | Experiment 2 | Experiment 3 |
|---|---|---|---|
| Wild type | 0.291 ± 0.020 (5) | 0.208 ± 0.019 (3) | 0.335 ± 0.076 (5) |
| CDSP32− | 0.217 ± 0.036 (6) | 0.158 ± 0.029 (4) | 0.281 ± 0.104 (6) |
| CDSP32+ | 0.222 ± 0.079 (6) | 0.182 ± 0.029 (4) | 0.318 ± 0.083 (6) |
Three-week-old potato plants were subjected to high light (1100 μmol·m−2·s−1) and low temperature (6/5°C day/night) for 25 h (including 12 h of night) before measuring Fv/Fm. The results from three independent experiments are reported, with the number of plants given in parentheses. The presented Fv/Fm values are means ± sd of the average values obtained from 10 replicates per plant.
DISCUSSION
In the present study, we show that the CDSP32 thioredoxin, carrying a Cys-to-Ser mutation in the active site, can form a heterodimer complex through a disulfide bridge with the plastidic 2-Cys peroxiredoxin BAS1. These data, revealing that BAS1 is a target for CDSP32, greatly improve our knowledge concerning the identity of CDSP32 substrate(s), because the thioredoxin activity of the protein had been shown only on its C-terminal domain using insulin (Rey et al., 1998). Because affinity chromatography revealed a highly specific interaction between the two proteins, we propose that the plant 2-Cys peroxiredoxin BAS1 is a major target for the CDSP32 thioredoxin.
Recently, Motohashi et al. (2001) identified >14 targets, including Rubisco activase, glutamine synthase, and different peroxiredoxins, for an immobilized m-type thioredoxin mutant from spinach. The 2-Cys peroxiredoxin type also was found to interact with thioredoxin m, but in a much less specific manner than in our experiments. Thus, the double-module CDSP32 thioredoxin appears to display a much greater specificity toward 2-Cys peroxiredoxin compared with a simple module such as the m type.
The mature CDSP32 protein is composed of two thioredoxin domains, with the one closest to the C terminus containing a redox disulfide active site. Both AtCDSP32m and StCDSP32m C-terminal domains were shown to reduce insulin, revealing the functionality of the active site (Figure 3) (Rey et al., 1998). By contrast, whole mature CDSP32 proteins were unable to reduce insulin. In these assays, protein misconformation was excluded because circular dichroism spectra as well as two-dimensional NMR prints revealed correct protein folding. Consequently, it can be hypothesized that the higher specificity of the double module is linked to the presence of the N-terminal domain. The formation of a complex composed of AtCDSP32mC164S and AtBAS1m was observed clearly in vitro, but no complex was noted when StCDSP32mC163S was mixed with AtBAS1m. These incubation assays confirm the role of the CDSP32 N-terminal domain in the interaction with the target, because most differences between AtCDSP32m and StCDSP32m are located in the N-terminal domain (AtCDSP32m and StCDSP32m C-terminal domains share 83% identity, and their N-terminal domains share 67% identity). Accordingly, whereas AtCDSP32m, AtCDSP32C-ter, and StCDSP32C-ter displayed close initial rates of AtBAS1 reduction, a notably lower rate was measured using StCDSP32m. Collectively, these data suggest that the N-terminal thioredoxin-like domain would allow the thioredoxin catalytic activity of CDSP32 to proceed via a role in the stability of the interaction with the substrate. Site-directed mutagenesis in the CDSP32 N-terminal domain would help identify the amino acids involved in the interaction with the target.
According to the endosymbiotic origin of chloroplast (Whatley, 1993), the plant gene BAS1, which is related to AhpC (Baier and Dietz, 1997), is likely to originate from the eubacteria-like genome. To date, no plant AhpF homolog has been found in either sequencing or proteomics projects. Like AhpF, CDSP32 exhibits a double thioredoxin module and has been described as most closely related to the plant h-type thioredoxin (Rey et al., 1998), which is believed to originate from the eukaryotic host (Meyer et al., 1999). Thus, whereas AhpF may have evolved directly from thioredoxin reductase in eubacteria (Poole et al., 2000b), CDSP32 could have appeared later in evolution from a eukaryotic thioredoxin gene duplication (StCDSP32 and AtCDSP32 N-terminal and C-terminal domains share ∼45% similarity).
To date, very few transgenic plants modified in the expression of thioredoxin genes have been reported. Only barley plants overexpressing a thioredoxin h in the grain endosperm have been described, using a seed-specific promoter (Cho et al., 1999). The absence of any other study concerned with modifying the level of thioredoxins in plants suggests that these proteins fulfill essential functions and that altering their expression results in severe perturbations that impair development. With regard to CDSP32, overexpressing and cosuppressed transformants were indistinguishable from the wild type and transgenic controls under standard growth conditions. These data indicate that the CDSP32 thioredoxin is dispensable for plant growth and development and that the protein is involved mainly in stress response, consistent with our previous reports showing CDSP32 induction under drought and photooxidative conditions (Rey et al., 1998; Broin et al., 2000).
Under photooxidative stress, wild-type and transgenic potato plants exhibited substantial damage to the photosynthetic apparatus, as indicated by PSII efficiency and chlorophyll content measurements. This decreased photosynthetic capacity likely originates from general membrane degradation, as reported for wheat leaves subjected to high light or high temperature (Mishra and Singhal, 1992) or for methyl viologen–treated tobacco leaf discs (Hideg et al., 2000). Our data clearly demonstrate that the lines lacking CDSP32 are more susceptible to photooxidative treatments, and they indicate that the thioredoxin fulfills an important function in the protection of the photosynthetic apparatus against oxidative damage. Note that the plants overexpressing CDSP32 were found to be more sensitive to photooxidative stress than the wild type and the transgenic controls, although to a lesser extent compared with the CDSP32− lines. A greater susceptibility to oxidative stress was noted when overexpressing γ-glutamylcysteine synthetase, an enzyme involved in glutathione synthesis in tobacco chloroplasts, probably as a result of a failure in the redox-sensing process (Creissen et al., 1999). Thus, we propose that the overexpression of a thioredoxin such as CDSP32 also could disturb the plastidic redox state. Because CDSP32 expression is induced under severe stress conditions in potato (Pruvot et al., 1996; Rey et al., 1998; Broin et al., 2000) and in Arabidopsis (data not shown), the high CDSP32 abundance in overexpressing plants under mild stress conditions could result in diverting the reducing power necessary for other protection systems.
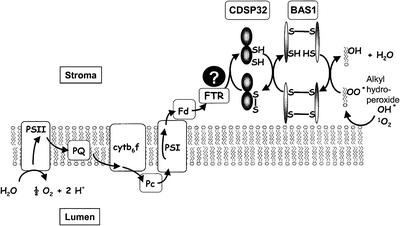
The results showing that CDSP32 downregulation provokes increased oxidative damage in potato chloroplasts indicate that CDSP32 participates in tolerance to stress. Because we identified BAS1 as a major target for CDSP32 in Arabidopsis and potato, and because AtBAS1 was found to display AtCDSP32-dependent peroxidase activity, we propose that the CDSP32 thioredoxin participates in the reduction of alkyl hydroperoxides by acting as a physiological electron donor to the BAS1 peroxiredoxin. The chloroplast, where CDSP32 and BAS1 are located, is a major site of active oxygen species formation in plants (Smirnoff, 1993; Asada, 1994). The functioning of the photosynthetic apparatus, particularly under abiotic constraints, gives rise to the formation of superoxide anion at the PSI level or to singlet oxygen in the light-harvesting antenna. Increased levels of these harmful species can lead to damage such as lipid peroxidation in the thylakoids (Mishra and Singhal, 1992). Consistent with our data, BAS1 was shown to exert a protective function on the photosynthetic apparatus against oxidative damage in Arabidopsis antisense mutants (Baier and Dietz, 1999a) and to form an integral part of the plastidic antioxidant network (Baier et al., 2000). Like the bacterial AhpF/AhpC system that participates in protection against osmotic and oxidative stress (Poole et al., 2000b), the CDSP32/BAS1 system could be essential to reduce alkyl hydroperoxides in plant chloroplasts during times of environmental constraint (Figure 10). In this system, the CDSP32 thioredoxin would be a critical component for transferring the reducing power, and the protein might be reduced by ferredoxin-thioredoxin reductase, which is known to mediate the flow of electrons from the photosynthetic electron chain to thioredoxins m and f (Jacquot et al., 1997).
Figure 10.
Putative Model for the Thioredoxin-Dependent Reduction of Alkyl Hydroperoxides in Chloroplast.
The electrons originating from the photosynthetic electron transfer chain may be transferred from ferredoxin to alkyl hydroperoxides through ferredoxin-thioredoxin reductase, CDSP32, and BAS1. cytb6f, cytochrome b6f; Fd, ferredoxin; FTR, ferredoxin-thioredoxin reductase; Pc, plastocyanin; PQ, plastoquinone; PSI and PSII, photosystems I and II.
METHODS
cDNA Cloning, Expression in Escherichia coli, and Purification of Recombinant Proteins
Arabidopsis thaliana CDSP32 cDNA was cloned using a probe corresponding to the full-length potato (Solanum tuberosum) CDSP32 cDNA to screen a λZAPII cDNA library (Stratagene) constructed from poly(A)+ RNA originating from 20-day-old Landsberg erecta plants (Parker et al., 1997). After blotting to Biodyne B (Pall Gelman Sciences, Ann Arbor, MI), hybridization to the random-primed probe labeled with α-32P-dATP (3000 Ci/mmol; Amersham-Pharmacia) was performed in 5 × SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate), 5 × Denhardt's solution (1× Denhardt's solution is 0.02% Ficoll, 0.02% polyvinylpyrrolidone, and 0.02% BSA), 0.1% SDS, and 100 μg/mL calf thymus DNA at 65°C for 16 h. Membranes were washed in 2 × SSC and 0.1% SDS successively at 25, 35, 50, and 60°C for 10 min each.
The cDNAs coding for mature CDSP32 proteins (AtCDSP32m and StCDSP32m) and for CDSP32 half C-terminal regions (AtCDSP32C-ter and StCDSP32C-ter) were amplified using PCR with primers containing 5′ extensions (BamHI site for the forward primer, HindIII site for the reverse primer) and cloned into the BamHI-HindIII sites of E. coli expression vector pQE30 (Qiagen, Valencia, CA). The cDNA coding for mature Arabidopsis BAS1 (AtBAS1m) was amplified from a cDNA library (Clontech, Palo Alto, CA) using primers containing 5′ extensions (BamHI site for the forward and reverse primers) and cloned into the BamHI site of expression vector pQE32 (Qiagen). The cDNAs coding for AtCDSP32mC164S, AtBAS1mC54S, and StCDSP32mC163S (Cys positions are given with respect to cleavage sites) were obtained by site-directed mutagenesis using a pair of complementary primers of 20 bp covering the mutation site in which the Cys codon has been replaced by a Ser codon, according to the “splicing by overlap extension” method described by Clackson et al. (1991). After cloning, all PCR-generated cDNAs were verified by sequencing.
Expression of recombinant proteins was performed in E. coli strain M15Rep4 grown at 37°C to an OD600 of 0.5 (Uvikon 942; Kontron, Plaisir, France) (0.5-μm disposable cuvettes, 1-mL culture) in 2XYT medium (16 g/L tryptone, 10 g/L yeast extract, 5 g/L NaCl). After induction by 100 μM isopropylthio-β-galactoside and overnight culture at room temperature, bacteria were pelleted by centrifugation and lysed using a French press (16,000 p.s.i.) in 100 mM Tris, pH 8.0, 500 mM NaCl, 10% glycerol, 25 mM imidazole, 1 mM phenylmethylsulfonyl fluoride, and 1 mM benzamidine. The supernatant recovered by centrifugation (12,000g for 20 min) was used to purify polyhistidine-tagged fusion proteins on a nickel affinity column (nickel–nitrilotriacetic acid agarose resin; Qiagen). After fixation and washing in lysis buffer, washing and elution of recombinant proteins were performed using 82.5 and 192.5 mM imidazole, respectively. Purified proteins were dialyzed against 50 mM phosphate buffer, pH 7.0, and protein concentration was measured spectrophotometrically at 280 nm using the molar extinction coefficient. Yields were between 10 and 15 mg/L for CDSP32 proteins and ∼30 mg/L for BAS1 proteins.
Incubation in Vitro, SDS-PAGE, and Immunoblot Analysis
For standard incubation, recombinant AtCDSP32mC164S or StCDSP32mC163S (0.25 μg/μL) was incubated in an equimolar ratio with AtBAS1m or AtBAS1mC54S in 40 μL of 50 mM potassium phosphate buffer, pH 7.0, for 15 min at room temperature. When using dithiobisnitrobenzoic acid, AtBAS1m was reacted with the reagent to form AtBAS1m-TNB complexes, as described by Wang et al. (1996). AtCDSP32mC164S was prereduced using DTT and reacted with AtBAS1m-TNB in an equimolar ratio in 50 mM potassium phosphate buffer, pH 7.0, for 15 min at room temperature.
Proteins were separated by SDS-PAGE in 13% acrylamide gels (Laemmli, 1970) and electroblotted onto 0.45 μM nitrocellulose (Pall Gelman Sciences) for immunoblot analysis (Rey et al., 1998). The serum against the StCDSP32m N-terminal region (Rey et al., 1998) was diluted 1:1500. The AtCDSP32m antiserum was raised using a synthetic peptide corresponding to a N-terminal part (NDEKVQKIHSGEEFD) coupled to keyhole limpet hemacyanin (Agro-Bio, Villeny, France) and used at a dilution of 1:1000. After cloning AtBAS1m cDNA in pQE32 (Qiagen), a rabbit serum was raised against the purified recombinant protein (Agro-Bio) and used at a dilution of 1:5000. Bound antibodies were detected using an anti-rabbit immunoglobulin G alkaline phosphatase conjugate at a dilution of 1:10,000 (Sigma).
Leaf and Chloroplast Soluble Protein Preparation and Affinity Chromatography Experiments
Arabidopsis cv Columbia and potato cv Désirée leaves were ground in liquid nitrogen using a mortar and pestle. The powder was resuspended in 50 mM Tris and 250 mM NaCl, pH 7.5, and filtered through 0.22-μm Miracloth (Calbiochem). After centrifugation (10,000g at 20 min), soluble proteins were collected in the supernatant. Protein concentration was determined using a modified Lowry procedure (Sigma). Intact chloroplasts from potato leaves were isolated at 4°C according to a modified method of Mills and Joy (1980) as described by Pruvot et al. (1996), except that 40% Percoll instead of 25% was used. Chlorophyll determination was performed according to Lichtenthaler and Wellburn (1983). After freezing and thawing, lysed chloroplasts were washed for 1 h at 4°C in 50 mM Tris-HCl and 250 mM NaCl, pH 7.5. Stromal proteins were collected in the supernatant after centrifugation (10,000g at 20 min) and filtration through a 0.45-μm filter (Millipore, Bedford, MA).
After desalting on 5-mL Hi-Trap desalting columns (Amersham Pharmacia Biotech), ∼7 mg of AtCDSP32mC164S or StCDSP32mC163S was coupled to the Hi-Trap NHS-activated 1-mL affinity column (Amersham Pharmacia Biotech) according to the manufacturer's procedure. Leaf or chloroplastic soluble proteins in 25 mM Tris, pH 7.5, and 250 mM NaCl (∼80 mg, 1 mg/mL) were applied to the column by injection at a flow rate of 0.5 mL/min. The column was washed successively with 25 mM Tris, pH 7.5, and buffers containing 250 mM NaCl (10 mL) and 500 mM NaCl (10 mL) at 0.5 mL/min. Elution was performed in 25 mM Tris, pH 7.5, and 250 mM NaCl supplemented with 2 mM DTT. Oxidation of the unbound fraction was performed using 100 μM t-butyl hydroperoxide for 30 min at 4°C. Washing and elution fractions were concentrated 10 times using acetone precipitation. Silver staining of gels was performed using the protocol of Heukeshoven and Dernick (1988).
Insulin and BAS1 Reduction Assays
Insulin reduction assays were performed according to a modified protocol from Holmgren (1979) as described by Rey et al. (1998) using E. coli thioredoxin (Sigma) as a control. Recombinant AtBAS1mC54S mutant protein was engineered by attaching a fluorescein derivative (FAM-SE, 5- and 6-isomers; Molecular Probes, Eugene, OR), via a disulfide bond, to Cys-175, as described by Poole et al. (2000a). Concentration of the modified protein, designated AtBAS1m-FAM, was measured using a modified Lowry procedure (Sigma). When bound to peroxiredoxins such as AtBAS1m, the fluorescein fluorescence is quenched. In the case of the AhpC-FAM complex, Poole et al. (2000a) estimated the percentage of quenched fluorescence at ∼70%.
Reduction of AtBAS1m-FAM results in fluorescein release, which can be monitored as an increase in fluorescence emission. Assays were performed in 1-mL reaction volume in 50 mM phosphate buffer, pH 7.0. Fluorescence emission was recorded using light excitation at 493 nm and a 510-nm filter on a LS50B spectrophotometer (Perkin-Elmer). Addition of CDSP32 started the reaction. Before use, the CDSP32 proteins were reduced overnight at 4°C in the presence of a 100-fold DTT excess in molarity, and excess DTT was removed by gel filtration chromatography.
Peroxidase Activity
AtBAS1m peroxidase activity in the presence of AtCDSP32m was measured in 500 μL of 50 mM phosphate buffer with 100 μM H2O2. AtCDSP32m was prereduced using DTT as described above. H2O2 concentration was measured using the FOX1 reagent (Wolff, 1994) (PeroXOquant quantitative peroxide assay; Pierce). Reaction aliquots (5 μL) were mixed with 250 μL of FOX1 reagent in microwell plate wells. After 15 min at room temperature, the absorbance at 595 nm was read using a microwell plate Dynatech MR5000 spectrophotometer (Saint-Cloud, France). A blank value without H2O2 was recorded, and a standard calibration curve was plotted using increasing H2O2 concentrations from 0 to 100 μM.
Plant Transformation Procedures
Potato CDSP32 (StCDSP32) cDNA from nucleotides 46 to 1049 was amplified by PCR using primers containing restriction site extensions and cloned as a BamHI-XbaI fragment in pBluescript KS+ (Stratagene). The cDNA was excised as a HindIII-SacI fragment and cloned in the sense orientation in the binary vector pKYLX71 (Schardl et al., 1987) containing a double 35S constitutive promoter from Cauliflower mosaic virus (Odell et al., 1985) and the ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit terminator. After introduction of the construct or of the empty vector in Agrobacterium tumefaciens C58, potato cv Désirée plants were transformed according to a modified protocol of Visser et al. (1989) using stem explants from the upper third part of 5-week-old plantlets grown in vitro. Roest and Bokelmann's medium (1976) was used during transformation and regeneration procedures, and kanamycin resistance was applied for selection of transformants 7 days after explant cocultivation with Agrobacterium. Transgenic and wild-type lines were propagated in vitro and transferred for growth in vivo in a phytotron (PPFD of 300 μmol·m−2·s−1, 12-h night, 23/19°C day/night).
Genomic DNA and RNA Preparation, and DNA and RNA Gel Blot Hybridizations
Potato genomic DNA was extracted from young leaves using the DNeasy Plant Mini Kit (Qiagen). After digestion (3 μg) with XbaI, restriction fragments were separated on 0.8% agarose gels and transferred onto positively charged nylon membranes (Roche Diagnostics, Meylan, France). Hybridization to a StCDSP32 cDNA probe labeled with digoxigenin (DIG)-dUTP was performed in DIG Easy Hyb mix (Roche Diagnostics) overnight at 42°C. Membranes were washed in 2 × SSC and 0.1% SDS twice for 5 min at room temperature and twice for 20 min at 55°C. Immunological revelation was performed using an anti-DIG–alkaline phosphatase conjugate and a chemiluminescent substrate according to Roche Diagnostics procedures.
For RNA gel blot analysis, total RNAs were prepared from young expanded potato leaves from 3-week-old plants using the RNeasy Plant Mini Kit (Qiagen) and separated on formaldehyde gels. Gels were stained with ethidium bromide to ensure that equal amounts of RNAs (6 μg) had been loaded. After blotting onto positively charged nylon membranes, hybridization to a DIG-labeled StCDSP32 cDNA was performed in DIG Easy Hyb mix (Roche Diagnostics) overnight at 50°C. Washing and immunological revelation were as described for DNA gel blot procedures.
Oxidative Treatments, Chlorophyll Content Determination, and Fluorescence Measurements
For incubation experiments, 1.4-cm-diameter discs were excised from young well-expanded leaflets of 3-week-old plants. Discs were incubated on 0.1% Tween 20 containing 1 μM methyl viologen (Sigma) or 2 mM t-butyl hydroperoxide (Sigma) in the dark for 16 h and then exposed to light (375 μmol·m−2·s−1) for 26 h under phytotron photoperiod conditions. Discs then were frozen in liquid nitrogen and assayed for chlorophyll content. After blending, the powder was resuspended in 80% acetone, and the chlorophyll content was calculated according to Lichtenthaler and Wellburn (1983).
Three-week-old plants were subjected to high light and low temperature (1100 μmol·m−2·s−1, 6/5°C day/night) in controlled growth chambers with 20% air renewal per hour (André and Du Cloux, 1993) for 25 h (10 h of light/12 h of dark/3 h of light).
Chlorophyll fluorescence emission from upper leaf disc surfaces was measured using a PAM-2000 modulated Walz fluorometer (Effeltrich, Germany; Havaux et al., 2000). After dark adaptation for 30 min, the initial level (Fo) of chlorophyll fluorescence was excited by a dim red light modulated at 600 Hz and determined after a 2-s illumination with far-red light. The maximal level of chlorophyll fluorescence (Fm) was induced by a 800-ms pulse of intense white light. The maximal quantum yield of photosystem II photochemistry was calculated as (Fm − Fo)/Fm = Fv/Fm, where Fv equals variable fluorescence.
Accession Numbers
The accession numbers for the sequences described and mentioned in this article are as follows: AtCDSP32 (EMBL, AJ318055); StCDSP32 cDNA (EMBL, Y09987); and the two overlapping genomic fragments from Arabidopsis chromosome I containing AtCDSP32 (GenBank, AC009978 and ATZ83321). The SWISS-PROT accession number for AtBAS1m is Q96291.
Acknowledgments
The authors thank Noëlle Becuwe for valuable assistance in cloning and sequencing experiments, Jacqueline Massimino and Véronique Cardettini for growing plant material, and the Groupe de Recherches Appliquées en Phytotechnologie team for technical assistance with experiments in controlled growth chambers. We are particularly grateful to Pierre Gans (Commissariat à l'Energie Atomique–Institute of Structural Biology, Grenoble, France) for helpful advice in circular dichroism experiments and for reading preliminary NMR data. We also are grateful to Christine Foyer (Institute of Arable Crops Research, Rothamsted, UK), Gilles Peltier, and Doan Luu for helpful and critical reading of the manuscript.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.001644.
References
- Altschul, S., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- André, M., and Du Cloux, H. (1993). Interaction of CO2 enrichment and water limitations on photosynthesis and water efficiency in wheat. Plant Physiol. Biochem. 31, 103–112. [Google Scholar]
- Asada, K. (1994). Mechanisms for scavenging reactive molecules generated in chloroplasts under light stress. In Photoinhibition of Photosynthesis, N.R. Baker and J.R. Bowyer, eds (Oxford, UK: Bios Scientific Publishers), pp. 129–142.
- Babbs, C.F., Pham, J.A., and Coolbaugh, R.C. (1989). Lethal hydroxyl radical production in paraquat-treated plants. Plant Physiol. 90, 1267–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier, M., and Dietz, K.-J. (1997). The plant 2-Cys peroxiredoxin BAS1 is a nuclear-encoded chloroplast protein: Its expressional regulation, phylogenetic origin, and implications for its specific physiological function in plants. Plant J. 12, 179–190. [DOI] [PubMed] [Google Scholar]
- Baier, M., and Dietz, K.-J. (1999. a). Protective function of chloroplast 2-cysteine peroxiredoxin in photosynthesis: Evidence from transgenic Arabidopsis. Plant Physiol. 119, 1407–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baier, M., and Dietz, K.-J. (1999. b). Alkyl hydroperoxide reductases: The way out of the oxidative breakdown of lipids in chloroplasts. Trends Plant Sci. 4, 166–168. [DOI] [PubMed] [Google Scholar]
- Baier, M., Noctor, G., Foyer, C., and Dietz, K.-J. (2000). Antisense suppression of 2-cysteine peroxiredoxin in Arabidopsis specifically enhances the activities and expression of enzymes associated with ascorbate metabolism but not glutathione metabolism. Plant Physiol. 124, 823–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer, Y., and Schürmann, P. (2001). Heterodimer formation between thioredoxin f and fructose 1,6-biphosphatase from spinach chloroplasts. FEBS Lett. 492, 58–61. [DOI] [PubMed] [Google Scholar]
- Broin, M., Cuiné, S., Peltier, G., and Rey, P. (2000). Involvement of CDSP32, a drought-induced thioredoxin, in the response to oxidative stress in potato plants. FEBS Lett. 467, 245–248. [DOI] [PubMed] [Google Scholar]
- Chae, H.Z., Chung, S.J., and Rhee, S.G. (1994). Thioredoxin-dependent peroxide reductase from yeast. J. Biol. Chem. 269, 27670–27678. [PubMed] [Google Scholar]
- Cho, M.-J., Wong, J.H., Marx, C., Jiang, W., Lemaux, P.G., and Buchanan, B.B. (1999). Overexpression of thioredoxin h leads to enhanced activity of starch debranching enzyme (pullulanase) in barley grain. Proc. Natl. Acad. Sci. USA 96, 14641–14646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clackson, T., Güssow, D., and Jones, P.T. (1991). Gene assembly by “splicing by overlap extension”. In PCR: A Practical Approach, M.J. McPherson, P. Quirke, and G.R. Taylor, eds (New York: Oxford University Press), pp. 187–214.
- Creissen, G., Firmin, J., Fryer, M., Kular, B., Leyland, N., Reynolds, H., Pastori, G., Wellburn, F., Baker, N., Wellburn, A., and Mullineaux, P. (1999). Elevated glutathione biosynthetic capacity in the chloroplasts of transgenic tobacco plants paradoxically causes increased oxidative stress. Plant Cell 11, 1277–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund, H., Gleason, F.K., and Holmgren, A. (1991). Structural and functional relations among thioredoxins of different species. Proteins Struct. Funct. Genet. 11, 13–28. [DOI] [PubMed] [Google Scholar]
- Ellis, H.R., and Poole, L.B. (1997). Roles for the two cysteine residues of AhpC in catalysis of peroxide reduction by alkyl hydroperoxide reductase from Salmonella typhimurium. Biochemistry 36, 13349–13356. [DOI] [PubMed] [Google Scholar]
- Eymery, F., and Rey, P. (1999). Immunocytolocalization of two chloroplastic drought-induced stress proteins in well-watered or wilted Solanum tuberosum L. plants. Plant Physiol. Biochem. 37, 305–312. [Google Scholar]
- Goyer, A., Decottignies, P., Lemaire, S., Ruelland, E., Issakidis-Bourguet, E., Jacquot, J.-P., and Miginiac-Maslow, M. (1999). The internal Cys-207 of sorghum leaf NADP-malate dehydrogenase can form mixed disulphides with thioredoxin. FEBS Lett. 444, 165–169. [DOI] [PubMed] [Google Scholar]
- Havaux, M., Bonfils, J.-P., Lütz, C., and Niyogi, K.K. (2000). Photodamage of the photosynthetic apparatus and its dependence on the leaf developmental stage in the npq1 Arabidopsis mutant deficient in the xanthophyll cycle enzyme violaxanthin de-epoxidase. Plant Physiol. 124, 273–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heukeshoven, J., and Dernick, R. (1988). Improved silver-staining procedure for fast staining in PhastSystem Development Unit. I. Staining of sodium dodecyl sulfate gels. Electrophoresis 9, 28–32. [DOI] [PubMed] [Google Scholar]
- Hideg, E., Kàlai, T., Hideg, K., and Vass, I. (2000). Do oxidative stress conditions impairing photosynthesis in the light manifest as photoinhibition? Philos. Trans. R. Soc. Lond. B 355, 1511–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren, A. (1979). Thioredoxin catalyses the reduction of insulin disulphides by dithiothreitol and dihydrolipoamide. J. Biol. Chem. 264, 9627–9632. [PubMed] [Google Scholar]
- Holmgren, A. (1985). Thioredoxin. Annu. Rev. Biochem. 54, 237–271. [DOI] [PubMed] [Google Scholar]
- Issakidis-Bourguet, E., Mouaheb, N., Meyer, Y., and Miginiac-Maslow, M. (2001). Heterologous complementation of yeast reveals a new putative function for chloroplast m-type thioredoxin. Plant J. 25, 127–135. [DOI] [PubMed] [Google Scholar]
- Jacquot, J.-P., Lancelin, J.-M., and Meyer, Y. (1997). Thioredoxins: Structure and function in plant cells. New Phytol. 136, 543–570. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler, H.K., and Wellburn, A.R. (1983). Determination of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 603, 591–592. [Google Scholar]
- Maxwell, K., and Johnson, G.N. (2000). Chlorophyll fluorescence: A practical guide. J. Exp. Bot. 51, 659–668. [DOI] [PubMed] [Google Scholar]
- Meyer, Y., Verdoucq, L., and Vignols, F. (1999). Plant thioredoxins and glutaredoxins: Identity and putative roles. Trends Plant Sci. 4, 388–394. [DOI] [PubMed] [Google Scholar]
- Mills, W.R., and Joy, K.W. (1980). A rapid method for isolation of purified physiologically active chloroplasts used to study the intracellular distribution of amino acids in pea leaves. Planta 148, 75–83. [DOI] [PubMed] [Google Scholar]
- Mishra, R.K., and Singhal, G.S. (1992). Function of photosynthetic apparatus of intact wheat leaves under high light and heat stress and its relationship with peroxidation of thylakoid lipids. Plant Physiol. 98, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi, K., Kondoh, A., Stumpp, M.T., and Hisabori, T. (2001). Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc. Natl. Acad. Sci. USA 98, 11224–11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouaheb, N., Thomas, D., Verdoucq, L., Montfort, P., and Meyer, Y. (1998). In vivo functional discrimination between plant thioredoxins by heterologous expression in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 95, 3312–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, E.G.D. (1991). Thioredoxin deficiency in yeast prolongs S phase and shortens G1 interval of the cell cycle. J. Biol. Chem. 266, 9194–9202. [PubMed] [Google Scholar]
- Odell, J.T., Nagy, F., and Chua, N.-H. (1985). Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313, 810–812. [DOI] [PubMed] [Google Scholar]
- Parker, J.E., Coleman, M.J., Szabo, V., Frost, L.N., Schmidt, R., van der Biezen, E.A., Moores, T., Dean, C., Daniels, M.J., and Jones, J.D.G. (1997). The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the Toll and interleukin 1 receptors with N and L6. Plant Cell 9, 879–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole, L.B., Godzik, A., Nayeem, A., and Schmitt, J.D. (2000. a). AhpF can be dissected into two functional units: Tandem repeats of two thioredoxin-like folds in the N-terminus mediate electron transfer from the thioredoxin reductase-like C-terminus to AhpC. Biochemistry 39, 6602–6615. [DOI] [PubMed] [Google Scholar]
- Poole, L.B., Reynolds, C.M., Wood, Z.A., Karplus, P.A., Ellis, H.R., and Calzi, M.L. (2000. b). AhpF and other NADH:peroxiredoxin oxidoreductases, homologues of low Mr thioredoxin reductase. Eur. J. Biochem. 267, 6126–6133. [DOI] [PubMed] [Google Scholar]
- Pruvot, G., Massimino, J., Peltier, G., and Rey, P. (1996). Effects of low temperature, high salinity and exogenous ABA on the synthesis of two chloroplastic drought-induced proteins in Solanum tuberosum. Physiol. Plant. 97, 123–131. [Google Scholar]
- Rey, P., Pruvot, G., Becuwe, N., Eymery, F., Rumeau, D., and Peltier, G. (1998). A novel thioredoxin-like protein located in the chloroplast is induced by water deficit in Solanum tuberosum L. plants. Plant J. 13, 97–107. [DOI] [PubMed] [Google Scholar]
- Ritz, D., Patel, H., Doan, B., Zheng, M., Aslund, F., Storz, G., and Beckwith, J. (2000). Thioredoxin 2 is involved in the oxidative stress response in Escherichia coli. J. Biol. Chem. 275, 2505–2512. [DOI] [PubMed] [Google Scholar]
- Roest, S., and Bokelmann, G.S. (1976). Vegetative propagation of Solanum tuberosum L. in vitro. Potato Res. 19, 173–178. [Google Scholar]
- Ruelland, E., and Miginiac-Maslow, M. (1999). Regulation of chloroplast enzyme activities by thioredoxins: Activation or relief from inhibition? Trends Plant Sci. 4, 136–141. [DOI] [PubMed] [Google Scholar]
- Saitoh, M., Nishitoh, H., Fujii, M., Takeda, K., Todiume, K., Sawada, Y., Kawabata, M., Miyazono, K., and Ichijo, H. (1998). Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. EMBO J. 17, 2596–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardl, C.L., Byrd, A.D., Benzion, G., Altschuler, M.A., Hildebrand, D.F., and Hunt, A.G. (1987). Design and construction of a versatile system for the expression of foreign genes in plants. Gene 61, 1–11. [DOI] [PubMed] [Google Scholar]
- Schenk, H., Klein, M., Erdbrugger, W., Droge, W., and Schluze-Osthoff, K. (1994). Distinct effects of thioredoxin and antioxidants on the activation of transcription factors NF-kappa B and AP-1. Proc. Natl. Acad. Sci. USA 91, 1672–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürmann, P., and Jacquot, J.-P. (2000). Plant thioredoxin systems revisited. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 371–400. [DOI] [PubMed] [Google Scholar]
- Smirnoff, N. (1993). The role of active oxygen in the response of plants to water deficit and desiccation. New Phytol. 125, 27–58. [DOI] [PubMed] [Google Scholar]
- Verdoucq, L., Vignols, F., Jacquot, J.-P., Chartier, Y., and Meyer, Y. (1999). In vivo characterisation of a thioredoxin h target protein defines a new peroxiredoxin family. J. Biol. Chem. 274, 19714–19722. [DOI] [PubMed] [Google Scholar]
- Visser, R.G.F., Jacoben, E., Hesseleng-Meinders, A., Schans, M.J., Witholt, B., and Feenstra, W.J. (1989). Transformation of homozygous diploid potato with an Agrobacterium tumefaciens binary vector system by adventitious shoot regeneration on leaf and stem segments. Plant Mol. Biol. 12, 329–337. [DOI] [PubMed] [Google Scholar]
- Wang, P.-F., Veine, D.M., Ahn, S.H., and Williams, C.H. (1996). A stable mixed disulphide between thioredoxin reductase and its substrate, thioredoxin: Preparation and characterisation. Biochemistry 35, 4812–4819. [DOI] [PubMed] [Google Scholar]
- Whatley, J.M. (1993). The endosymbiotic origin of chloroplasts. Int. Rev. Cytol. 144, 259–299. [Google Scholar]
- Wolff, S.P. (1994). Ferrous ion oxidation in the presence of ferric ion indicator xylenol orange for measurement of hydroperoxides. Methods Enzymol. 233, 182–189. [Google Scholar]
- Wynn, R., Cocco, M., and Richards, F.M. (1995). Mixed disulphide intermediates during the reduction of disulphides by Escherichia coli thioredoxin. Biochemistry 34, 11807–11813. [DOI] [PubMed] [Google Scholar]
- Yang, Y., and Wells, W.W. (1991). Catalytic mechanism of thioltransferase. J. Biol. Chem. 266, 12766–12771. [PubMed] [Google Scholar]