Bile is at the center of the traditional medicines of many cultures. For the ancient Greeks and Romans, each of the four macrocosmic elements that comprised the natural world, fire, earth, air, and water, had a specific microcosmic reflection in the four humors from which the body was constructed, yellow bile, black bile, blood, and phlegm. The view that imbalances in these components cause disease dominated Western medical thought for nearly 2000 years. Ayurvedic, or Indian traditional medicine is based on the similar belief that health is dependent on the balance of the three doshas, pitta (bile), kapha (wind), and vata (phlegm), each of which represents two of their five macrocosmic elements.
Bile has a much more humble place in current medical thinking. It is generally thought of as a detergent needed for the absorption of lipids and fat-soluble nutrients, as well as a potentially harmful agent responsible for adverse effects in cholestasis and other pathologic situations. This simplistic view was challenged by the recent discovery by three independent groups (1–3) that a member of the nuclear hormone receptor superfamily, farnesoid X receptor (FXR), functions as a specific receptor for a wide variety of bile acids. Since other members of this family include the receptors for steroids and thyroid hormone, this observation suggested that bile acids should be thought of as hormones.
Negative feedback regulation of bile acid synthesis
Such a direct regulatory function would be consistent with a well known negative feedback loop in which increased bile acid levels result in a compensatory decrease in the rate of bile acid synthesis. The specific role of FXR in this loop is thought to be the bile acid–dependent induction of expression of another nuclear hormone receptor, small heterodimer partner (SHP) (4, 5), which functions as a specific repressor of transcription (6). In this proposed nuclear receptor cascade, the increased SHP levels cause a decrease in expression of the cytochrome P450 enzyme cholesterol 7α-hydroxylase, Cyp7a1, the first and rate-limiting step in the complex pathway of bile acid biosynthesis. The importance of this regulatory pathway was supported by results with knockout mice lacking either the FXR (7) or SHP (6, 8) genes. Intriguingly, however, the results with the SHP knockouts suggested additional functional roles for bile acids. Specifically, the SHP-null animals showed no repression of Cyp7a1 expression in response to a synthetic FXR agonist, but high levels of bile acids themselves still inhibited Cyp7a1 and other bile acid biosynthetic enzymes. This result suggests that bile acids are able to signal via additional pathways outside of the FXR/SHP cascade.
Alternate signaling pathways
At least three such pathways have been identified, two of which are mediated by other nuclear hormone receptors. A limited number of toxic bile acids (e.g., lithocholic acid) have been identified as ligands for the xenobiotic receptor pregnane X receptor (PXR) (9, 10) and, quite surprisingly, the vitamin D receptor (11). Their activation by these hydrophobic bile acids induces expression of a detoxifying system thought to be based on induction of another P450 enzyme, Cyp3a, which can convert these compounds to less toxic forms. At least in the case of PXR, this induction also results in decreased expression of Cyp7a1. In addition, bile acids, particularly the taurine conjugate of cholic acid, have been reported to activate the stress kinase c-jun N-terminal kinase (JNK), which also represses Cyp7a1 expression (12).
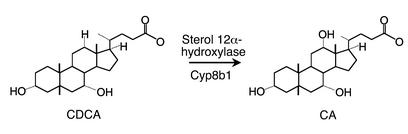
These results not only strongly support the proposed regulatory functions of bile acids, but also predict that distinct bile acids may have distinct biological activities. The basis for the production of the two primary bile acids, cholic acid (CA) and chenodeoxycholic acid (CDCA), is sterol 12α-hydroxylase (encoded by the Cyp8b1 gene), yet another P450 enzyme that specifically catalyzes the hydroxylation of the 12 position on the cholesterol derived bile acid backbone (Figure 1). It is evident that the absence of this enzyme would result in the failure to produce cholic acid and other secondary, 12α-hydroxylated bile acids, and would therefore significantly alter the composition of the bile acid pool. In the article by Li-Hawkins et al. in this issue of the JCI (13), the prediction that this could affect bile acid homeostasis was tested using a Cyp8b1-knockout mouse.
Figure 1.
Formation of cholic acid (CA) from chenodeoxycholic acid (CDCA) by the microsomal cytochrome P450 enzyme sterol 12α-hydroxylase, which is encoded by the Cyp8b1 gene. The enzyme can also hydryoxylate other 7α-hydroxylated intermediates in the bile acid pathways (see ref. 13).
Loss of cholic acid alters negative feedback regulation
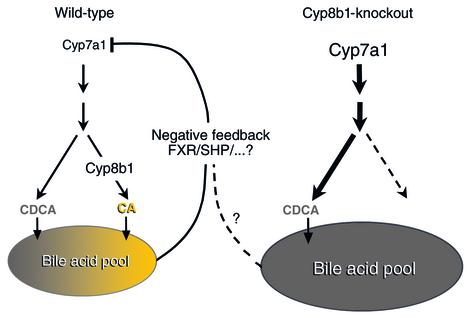
As expected, the Cyp8b1-null animals essentially lack cholic acid, which makes up more than half of the bile acid pool in wild-type animals. The size of this pool is modestly increased in the knockouts but, in contrast to the expectations from the negative feedback mechanism, Cyp7a1 expression is actually increased (Figure 2). This increase is attributed to decreased expression of the inhibitor, SHP. However, the expression of other hepatic genes known to be direct targets of FXR, such as the bile salt export protein (BSEP), is not altered. Thus, changing the composition of the bile acid pool results in rather complex alterations in bile acid metabolism that include a striking dysregulation of the negative feedback regulation. The authors conclude that cholic acid makes a particularly important contribution to this regulatory loop.
Figure 2.
Loss of negative feedback regulation in bile acid synthesis in Cyp8b1-knockout mice. While both wild-type (WT) and mutant mice produce chenodeoxycholic acid (CDCA), Cyp8b1 knockouts lack cholic acid (CA). Contrary to the expectation from the redundant negative feedback mechanisms mediated by the farnesoid X receptor (FXR) and small heterodimer partner (SHP), and also other factors in WT mice, Cyp7a1 expression in the knockout mice is actually increased. The mechanisms whereby the loss of CA results in the loss of negative feedback are unknown.
These results leave open the very interesting question of which of the potential bile acid–dependent pathways account for the observed effects. Although they acknowledge the potential role of the FXR-independent pathways, Li-Hawkins et al. speculate that various bile acids may have differential effects on FXR (13). This could be analogous to results with the estrogen receptor, which adopts different conformations when occupied by even closely related ligands (14). One problem with this suggestion is that in vitro approaches typically show cholate to be a relatively ineffective FXR activator, leading the authors to suggest that additional mechanisms, such as the presence of novel coactivators in the liver, may account for the observed effects.
Regardless of the mechanism, these results strongly support the idea that different bile acids have different regulatory effects. The chemical effects of the relative hydrophobicity of bile acids on processes such as the prevention of cholesterol gallstone formation and promotion of lipid absorption are well known. How the emerging hormonal or signaling effects of distinct bile acids are integrated with their chemical functions is a very open and interesting question.
It is ironic that these ideas seem new to us, but would have been self evident to Hippocrates, Galen, and their students for two thousand years. They would argue that Cyp8b1-null animals should have a serious humoral imbalance. It is difficult to predict the outcome of this imbalance, since the ancient literature is silent on whether cholic acid is yellow bile or black bile. However, the Merck Index correctly describes cholic acid as bitter tasting, a quality associated with yellow bile. Black bile, in contrast, is sour. One might therefore predict that a loss of yellow bile and a compensatory increase in black bile would make the Cyp8b1–/– mice melancholy (which of course is derived from Greek words meaning “black bile”). Alternatively, if all bile acids are yellow bile, the Cyp8b1 knockouts should be choleric. We await future studies on their mood.
Footnotes
See the related article beginning on page 1191.
Conflict of interest: No conflict of interest has been declared.
Nonstandard abbreviations used: farnesoid X receptor (FXR); small heterodimer partner (SHP); cholesterol 7α-hydroxylase (Cyp7a1); pregnane X receptor (PXR); c-jun N-terminal kinase (JNK); cholic acid (CA); chenodeoxycholic acid (CDCA); bile salt export protein (BSEP).
References
- 1.Parks DJ, et al. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999;284:1365–1368. doi: 10.1126/science.284.5418.1365. [DOI] [PubMed] [Google Scholar]
- 2.Makishima M, et al. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- 3.Wang H, Chen J, Hollister K, Sowers LC, Forman BM. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol Cell. 1999;3:543–553. doi: 10.1016/s1097-2765(00)80348-2. [DOI] [PubMed] [Google Scholar]
- 4.Goodwin B, et al. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell. 2000;6:517–526. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 5.Lu TT, et al. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. 2000;6:507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- 6.Lee YK, Moore DD. Dual mechanisms for repression of the monomeric orphan receptor liver receptor homologous protein-1 by the orphan small heterodimer partner. J Biol Chem. 2002;277:2463–2467. doi: 10.1074/jbc.M105161200. [DOI] [PubMed] [Google Scholar]
- 7.Sinal CJ, et al. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000;102:731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 8.Kerr TA, et al. Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev Cell. 2002;2:713–720. doi: 10.1016/s1534-5807(02)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Staudinger JL, et al. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci USA. 2001;98:3369–3374. doi: 10.1073/pnas.051551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie W, et al. An essential role for nuclear receptors SXR/PXR in detoxification of cholestatic bile acids. Proc Natl Acad Sci USA. 2001;98:3375–3380. doi: 10.1073/pnas.051014398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makishima M, et al. Vitamin D recep-tor as an intestinal bile acid sensor. Science. 2002;296:1313–1316. doi: 10.1126/science.1070477. [DOI] [PubMed] [Google Scholar]
- 12.Gupta S, Stravitz RT, Dent P, Hylemon PB. Down-regulation of cholesterol 7alpha-hydroxylase (CYP7A1) gene expression by bile acids in primary rat hepatocytes is mediated by the c-Jun N-terminal kinase pathway. J Biol Chem. 2001;276:15816–15822. doi: 10.1074/jbc.M010878200. [DOI] [PubMed] [Google Scholar]
- 13.Li-Hawkins J, et al. Cholic acid mediates negative feedback regulation of bile acid synthesis in mice. J Clin Invest. 2002;110:1191–1200. doi:10.1172/JCI200216309. doi: 10.1172/JCI16309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paige LA, et al. Estrogen receptor (ER) modulators each induce distinct conformational changes in ER alpha and ER beta. Proc Natl Acad Sci USA. 1999;96:3999–4004. doi: 10.1073/pnas.96.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]