Normal cardiac excitation and relaxation involves a delicate balance of complex dynamic interactions between ionic currents passing through a variety of membrane channels and the cellular environment. Genetic defects, polymorphisms, therapeutic intervention or structural abnormalities can disrupt this balance and underlie severe arrhythmogenic phenotypes that lead to sudden cardiac death. Inheritable gene defects give rise to phenotypic variation and an unpredictable manifestation of syndromes, ranging from silent gene carriers to profoundly symptomatic individuals, even within single families (1–7). As such, realizing the relationship between genetic mutations and clinical syndromes is becoming increasingly complex. In this issue of the JCI, Grant and colleagues (3) investigate the manifestations of phenotypically opposite and overlapping cardiac arrhythmogenic syndromes that surprisingly stem from the same mutation (1–4).
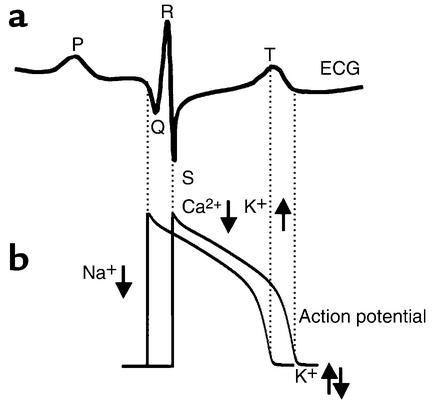
Cardiac excitation reflects membrane depolarization of cardiac myocytes, primarily due to the activation of fast voltage-dependent Na+ channels that underlie the action potential upstroke. Activation is followed by a long depolarized plateau phase that permits Ca2+-induced Ca2+ release from the sarcoplasmic reticulum, binding of Ca2+ to contractile proteins on the sarcomeres, and coordinated contraction. Repolarization follows due to the time- and voltage-dependent activation of repolarizing potassium currents. Relaxation of contraction is coupled to the electrical repolarization phase, which allows filling of the ventricles prior to the next excitation. Each of these electrical processes can be detected on the body surface electrocardiogram (ECG) as a signal average of the temporal and spatial gradients generated during each phase (8–11) (Figure 1a). Electrical excitation gradients in the atria (atrial depolarization) manifest on the ECG as P waves, while gradients of ventricular depolarization are seen as the QRS complex. Gradients in ventricular repolarization are reflected in the T wave (Figure 1).
Figure 1.
Electrical gradients in the myocardium can be detected on the body surface ECG. (a) An illustrative example of a single cardiac cycle detected as spatial and temporal electrical gradients on the ECG. The P wave is generated by the spread of excitation through the atria. The QRS complex represents ventricular activation and is followed by the T wave reflecting ventricular repolarization gradients. (b) Schematic representation of cellular electrical activity underlying the ECG (see text for details). Where downward arrows represent inward current and upward arrows represent outward current.
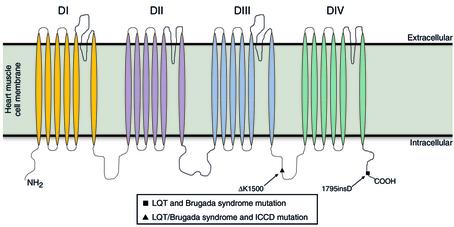
A recently described example of a multi-syndrome genetic defect in the SCN5A gene, encoding the cardiac Na+ channel (Figure 2), is the insertion of an aspartic acid, 1795insD, in the C-terminus of the cardiac Na+ channel that underlies both Brugada (BrS) and Long-QT (LQTs) cardiac arrhythmic syndromes (1, 2).
Figure 2.
The predicted transmembrane topology of domains I–IV of the cardiac Na+ channel α subunit encoded by SCN5A showing the location and nature of the mutations inducing LQTs, BrS, and isolated cardiac conduction disease.
Grant and colleagues investigate an even more complex mutation (3). The deletion of lysine, ΔK1500, in the III–IV linker of SCN5A (Figure 2) is associated with BrS, LQTs, and isolated cardiac conduction disease (ICCD). LQTs is typically associated with a gain of Na+ channel function that stems from mutation induced destabilization of channel inactivation, leading to a persistent inward Na+ current (INa) during the action potential (AP) plateau and prolonged repolarization (12, 13). Paradoxically, BrS and ICCD are linked to a loss of Na+ channel function and a resulting reduction in macroscopic current (3, 6, 7, 14, 15). How can multiple and seemingly contradictory arrhythmic syndromes arise from a mutation at a single locus?
The heterogeneous myocardial substrate
The solution seems to lie in the intrinsic heterogeneity of the underlying myocardial substrate with which mutant Na+ channels interact. The ventricular myocardium is comprised of at least three distinct cell types referred to as epicardial, midmyocardial (M), and endocardial cells, which exhibit distinct electrophysiological properties (8).
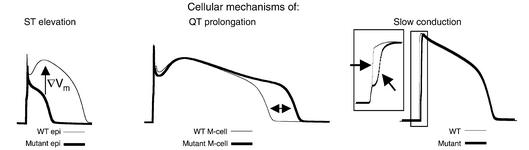
Epicardial cells display a characteristic spike and dome morphology (Figure 3, left panel, thin line) due to large transient outward K+ current (Ito), and short action potential duration (APD) resulting from a high density of the slowly activating component of the delayed rectifier K+ current (IKs). Mutations that act to reduce INa in the presence of these large repolarizing currents (Ito and IKs), result in premature plateau repolarization and APs with distinctive triangular morphology (Figure 3, left panel, thick line) (8–11, 16). Selective loss of the AP plateau in these cells, results in dispersion of plateau potentials across the ventricular wall. This gradient (∇) generates ST segment elevation on the ECG, which is a diagnostic indicator of BrS (9, 14). Clinically, ST segment elevation is observed in right precordial leads of BrS patients, consistent with the large Ito density in right ventricular epicardium (9, 11). In M cells Ito and IKs are smaller, and so unable to overwhelm the mutation induced reduction in INa (8, 9, 16). The non-inactivating component of INa, in the presence of smaller repolarizing currents, acts to prolong the plateau of the action potential (16) and may allow for the development of arrhythmogenic triggered activity, referred to as early afterdepolarizations (EADs) (Figure 3, middle panel). APD prolongation is reflected in a prolonged QT interval on the ECG, indicative of LQTs.
Figure 3.
Cellular electrical abnormalities and their relation to changes in the ECG. (Left) At fast pacing (heart) rates, mutation-induced changes in epicardial AP morphologies (thick line) cause dispersion of plateau potentials and a voltage gradient (∇Vm, arrow) (WT, thin line). This gradient will manifest on the ECG as ST segment elevation, indicative of BrS. (Middle) Mutations may prolong APD in M cells (thick line) compared to WT (thin line). The delay in repolarization (ΔAPD ∼ 60ms) is reflected as QT prolongation on the ECG, a hallmark of LQTs. (Right) A rightward shift in the Na+ channel activation curve is sufficient to reduce AP upstroke and slow cardiac impulse conduction (wide QRS segment) as observed in ICCD. WT, wild-type.
Brugada syndrome vs. isolated cardiac conduction disease
One of the more compelling aspects in attempting to elucidate the relationship between genotype and the clinical presentation of disease exists in the differentiation of BrS and ICCD (3, 5, 6, 7). Experimental evidence suggests that a reduction of INa is a common denominator of both syndromes (3, 5–7, 14, 15). Indeed, a number of mechanisms, including reduced rates of recovery from inactivation, faster inactivation subsequent to channel opening, and protein trafficking defects have been shown to underlie the loss of Na+ channel function in BrS. This explanation however, fails to reconcile the presence of ST segment elevation (observed in BrS) in the absence of slowed conduction (as seen in ICCD) and vice-versa.
A possible explanation for the phenotypic overlap (3) or exclusivity (5) may lie in the notable commonality among the relatively few characterized ICCD mutations; namely, a consistent rightward shift in the voltage dependence of the mutant Na+ channel activation curve (5–7) (see Figure 3b in Grant et al. in this issue). A depolarizing shift of the Na+ channel activation curve indicates that mutant channels require a more positive membrane potential to fully activate than wild-type (WT) channels (5). Most likely, this shift results from a reduction in the rate of channel activation or decreased channel sensitivity to the voltage required for activation. In either case, a rightward shift of the Na+ channel activation curve may not necessarily lead to decreased current, but rather a different voltage at which the maximum current amplitude occurs. For example, if an ICCD mutation results in mutant channels that obtain maximum current amplitude at 0 mV, as opposed to –10 mV for WT channels, mutant channels fully activate within the physiological range of the action potential upstroke. However, the mutant channels may require more time to reach the membrane potential at which the maximum current amplitude occurs. The lag in activation of ICCD mutant Na+ channels would result in a reduction of the action potential upstroke velocity (Figure 3, right panel; inset: magnification of AP upstroke), a primary determinant of conduction velocity (17, 18). A reduction in the velocity of electrical propagation will manifest on the ECG in a widening of the QRS interval.
The defect characterized by Grant and coauthors in this issue of the JCI is unique in that a single amino acid deletion in the cardiac Na+ channel III–IV linker gives rise to three distinct cellular phenotypes as well as three separate clinical syndromes (3). The ΔK1500 mutation leads to a significantly increased component of non-inactivating Na+ current, which results in prolongation of repolarization and the QT interval on the ECG. The mutation also results in faster inactivation, a property that is consistent with reduced Na+ current. In some epicardial cells, where the repolarizing potassium currents are large, the reduced Na+ current may be overwhelmed and AP plateaus heterogeneously suppressed, resulting in ST segment elevation, a hallmark of BrS. Finally, the ΔK1500 mutation results in a rightward shift of the channel activation curve, which may be responsible for the reduced conduction velocity seen clinically as the broadening of the QRS interval on the ECG of ICCD patients.
While the cellular characterization of the ΔK1500 mutation is consistent with the development of LQTs, BrS, and ICCD simultaneously, the clinical observations are less clear-cut. The four generations of mutation carriers evaluated by Grant et al. display a wide variety of clinical presentations and symptoms (3). The explanation for the variability in phenotypic manifestation of ΔK1500 may well lie in the underlying genetic substrate via mutant channel interactions with modifier genes and environmental factors.
Modifier genes: A role in the reconciliation of multiple phenotypes from a single genotype?
The broad diversity in phenotypic expression of gene defects among affected individuals with identical cardiac ion channel mutations is likely influenced by other genetic traits separate from the primary disease locus. Genotype-phenotype correlation studies show significant variability in phenotypes among affected individuals with identical causal mutations (6, 7, 19, 20). Clinical presentation is determined by complex interactions between causal genes, genetic background (modifier genes), and environmental factors. While individual modifier genes for Na+ channel–linked arrhythmias remain largely unknown, potential modifiers of genetically based arrhythmias include, but are not limited to, gender, febrile states, adrenergic stimulation, signaling molecules, channel-associated protein kinases (20–23), channel-associated protein phosphatases (20–22), and individual electrophysiological and morphological substrates.
Thus far, studies are limited to polymorphism association studies and electrophysiological characterization in expression systems, where channels are removed from the physiological interactive environment. Identification of modifier genes will complement the current studies that have identified and characterized causative genes, which may improve upon genetically based diagnosis, risk stratification, and implementation of preventive and therapeutic interventions in patients with Na+ channel linked arrhythmias.
Footnotes
See the related article beginning on page 1201.
Conflict of interest: No conflict of interest has been declared.
Nonstandard abbreviations used: electrocardiogram (ECG); Brugada syndrome (BrS); Long-QT syndrome (LQTs); isolated cardiac conduction disease (ICCD); inward Na+ current (INa); action potential (AP); midmyocardial (M); transient outward K+ current (Ito); action potential duration (APD); slowly activating component of the delayed rectifier current (IKs); early afterdepolarization (EAD); wild-type (WT).
References
- 1.Veldkamp MW, et al. Two distinct congenital arrhythmias evoked by a multidysfunctional Na(+) channel. Circ Res. 2000;86:E91–E97. doi: 10.1161/01.res.86.9.e91. [DOI] [PubMed] [Google Scholar]
- 2.Rivolta I, et al. Inherited brugada and LQT-3 syndrome mutations of a single residue of the cardiac sodium channel confer distinct channel and clinical phenotypes. J Biol Chem. 2001;276:30623–30630. doi: 10.1074/jbc.M104471200. [DOI] [PubMed] [Google Scholar]
- 3.Grant AO, et al. Long QT syndrome, Brugada syndrome, and conduction system disease are linked to a single sodium channel mutation. J Clin Invest. 2002;110:1201–1209. doi:10.1172/JCI200215570. doi: 10.1172/JCI15570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bezzina C, et al. A single Na(+) channel mutation causing both long-QT and Brugada syndromes. Circ Res. 1999;85:1206–1213. doi: 10.1161/01.res.85.12.1206. [DOI] [PubMed] [Google Scholar]
- 5.Tan HL, et al. A sodium-channel mutation causes isolated cardiac conduction disease. Nature (Lond) 2001;409:1043–1047. doi: 10.1038/35059090. [DOI] [PubMed] [Google Scholar]
- 6.Kyndt F, et al. Novel SCN5A mutation leading either to isolated cardiac conduction defect or Brugada syndrome in a large French family. Circulation. 2001;104:3081–3086. doi: 10.1161/hc5001.100834. [DOI] [PubMed] [Google Scholar]
- 7.Shirai N, et al. A mutant cardiac sodium channel with multiple biophysical defects associated with overlapping clinical features of Brugada syndrome and cardiac conduction disease. Cardiovasc Res. 2002;53:348–354. doi: 10.1016/s0008-6363(01)00494-1. [DOI] [PubMed] [Google Scholar]
- 8.Liu DW, Gintant GA, Antzelevitch C. Ionic bases for electrophysiological distinctions among epicardial, midmyocardial, and endocardial myocytes from the free wall of the canine left ventricle. Circ Res. 1993;72:671–687. doi: 10.1161/01.res.72.3.671. [DOI] [PubMed] [Google Scholar]
- 9.Yan GX, Antzelevitch C. Cellular basis for the brugada syndrome and other mechanisms of arrhythmogenesis associated with ST-segment elevation. Circulation. 1999;100:1660–1666. doi: 10.1161/01.cir.100.15.1660. [DOI] [PubMed] [Google Scholar]
- 10.Gima K, Rudy Y. Ionic current basis of electrocardiographic waveforms: a model study. Circ Res. 2002;90:889–896. doi: 10.1161/01.res.0000016960.61087.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dumaine R, et al. Ionic mechanisms responsible for the electrocardiographic phenotype of the brugada syndrome are temperature dependent. Circ Res. 1999;85:803–809. doi: 10.1161/01.res.85.9.803. [DOI] [PubMed] [Google Scholar]
- 12.Bennett PB, Yazawa K, Makita N, George AL. Molecular mechanism for an inherited cardiac arrhythmia. Nature (Lond) 1995;376:683–685. doi: 10.1038/376683a0. [DOI] [PubMed] [Google Scholar]
- 13.Clancy CE, Rudy Y. Linking a genetic defect to its cellular phenotype in a cardiac arrhythmia. Nature (Lond) 1999;400:566–569. doi: 10.1038/23034. [DOI] [PubMed] [Google Scholar]
- 14.Chen Q, et al. Genetic basis and molecular mechanism for idiopathic ventricular fibrillation. Nature (Lond) 1998;392:293–296. doi: 10.1038/32675. [DOI] [PubMed] [Google Scholar]
- 15.Wang DW, Makita N, Kitabatake A, Balser JR, George AL. Enhanced Na(+) channel intermediate inactivation in Brugada syndrome. Circ Res. 2000;87:E37–E43. doi: 10.1161/01.res.87.8.e37. [DOI] [PubMed] [Google Scholar]
- 16.Clancy CE, Rudy Y. Na(+) Channel mutation that causes both brugada and Long-QT syndrome phenotypes: a simulation study of mechanism. Circulation. 2002;105:1208–1213. doi: 10.1161/hc1002.105183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaw RM, Rudy Y. Ionic mechanisms of propagation in cardiac tissue. Roles of the sodium and L-type calcium currents during reduced excitability and decreased gap junction coupling. Circ Res. 1997;81:727–741. doi: 10.1161/01.res.81.5.727. [DOI] [PubMed] [Google Scholar]
- 18.Rohr S, Kucera JP, Kleber AG. Slow conduction in cardiac tissue, I: effects of a reduction of excitability versus a reduction of electrical coupling on microconduction. Circ Res. 1998;83:781–794. doi: 10.1161/01.res.83.8.781. [DOI] [PubMed] [Google Scholar]
- 19.Benhorin J, et al. Variable expression of long QT syndrome among gene carriers from families with five different HERG mutations. Ann Noninvasive Electrocardiol. 2002;7:40–46. doi: 10.1111/j.1542-474X.2001.tb00137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwartz PJ, et al. Long QT syndrome patients with mutations of the SCN5A and HERG genes have differential responses to NA+ channel blockade and to increases in heart rate: implications for gene-specific therapy. Circulation. 1995;92:3381–3386. doi: 10.1161/01.cir.92.12.3381. [DOI] [PubMed] [Google Scholar]
- 21.Marx SO, et al. Phosphorylation-dependent regulation of ryanodine receptors. A novel role for leucine/isoleucine zippers. J Cell Biol. 2001;153:699–708. doi: 10.1083/jcb.153.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marks AR, Marx SO, Reiken S. Regulation of ryanodine receptors via macromolecular complexes. A novel role for leucine/isoleucine zippers. Trends Cardiovasc Med. 2002;12:166–170. doi: 10.1016/s1050-1738(02)00156-1. [DOI] [PubMed] [Google Scholar]
- 23.Marx SO, et al. Requirement of a macromolecular signaling complex for beta adrenergic receptor modulation of the KCNQ1-KCNE1 potassium channel. Science. 2002;295:496–499. doi: 10.1126/science.1066843. [DOI] [PubMed] [Google Scholar]