Abstract
Rhabdomyosarcoma (RMS) in children occurs predominantly as two major histologically defined subtypes called embryonal RMS (RMS-E) and the prognostically less favorable alveolar RMS (RMS-A). Comparative genomic hybridization (CGH) was performed on 21 RMS and identified consistent gains affecting chromosomes 2 (8/10), 5 (5/10), 6 (3/10), 7 (7/10), 8 (9/10), 11 (6/10), and 12 (5/10) in RMS-E. Losses/deletions involved chromosomes 19 (2/10) and chromosomes 4, 9, 10, 17, 21 (1/10 each). High copy number amplification, involving the 2p24 region (5/11) and less frequently, the 12q13-21 (2/11), 9p22 (1/11), and 17q22-25 (1/11) regions, was detected in RMS-A. Gene amplification at band 2p24 was present in 6/12 alveolar tumors, and in each case, MYCN was amplified, together with the distally placed DDX1 gene. For these patients there was a shorter disease free interval and a higher mortality than patients with tumors without amplification. Detailed spectral karyotype analysis (SKY) was performed on two RMS cell lines (one of each subtype) and identified a surprisingly high level of structural change. Gene expression studies with the Atlas Human Cancer Array (588 genes) showed that 153 genes generated a signal of similar intensity in both cell lines, and 45 genes appeared to have subtype-specific expression. The chromosomal location of differentially expressed genes was compared to the pattern of genomic alteration in RMS as determined by CGH in this study and the literature.
Keywords: rhabdomyosarcoma, CGH, SKY, microarray
Introduction
Rhabdomyosarcoma (RMS) is a soft tissue sarcoma of skeletal muscle derivation with an incidence of 1 in 200,000 (1). This childhood tumor consists of several different subtypes recognized primarily by their characteristic histology. The alveolar (RMS-A) and embryonal (RMS-E) subtypes are the two most common forms, with the embryonal histology conferring a more favorable clinical outcome (2). Although the traditional prognostic indicators of stage, site, tumor size and histologic subtype remain the best predictors of clinical outcome, an improved characterization of the molecular genetics of RMS will help with subtype diagnosis as well as help identify the molecular pathways that could lead to variable prognosis (3–5).
Ninety percent of RMS-A have a structural rearrangement of the FKHR (also known as ALV) gene on chromosome 13 (5). In the majority of RMS-A cases, there is a translocation t(2;13)(q35;q14), which leads to the juxtaposition of the PAX3 gene on chromosome 2 with FKHR, and the production of a DNA-binding fusion oncoprotein derived from the 5′ end of the PAX3 and the 3′ end of FKHR (4). RMS-A that do not have this rearrangement usually have a variant translocation t(1;13)(p36;q14), in which rearrangement of FKHR occurs with PAX7 on chromosome 1 (6). In contrast to RMS-A, no primary molecular genetic aberration analogous to the FKHR rearrangements has yet been detected in RMS-E. A high frequency of loss of heterozygosity at 11p15 has been found in RMS-E, suggesting the location of an unidentified tumor suppressor gene (7). Cytogenetic analyses of RMS-E indicate a more general pattern of chromosomal gain leading to hyperdiploidy (8), but precise delineation of the genomic region(s) subject to gain in the genesis of this subtype have yet to be established.
Comparative genomic hybridization (CGH) has proved helpful in identifying regions of chromosomes associated with amplification, loss, and gain in specific tumors ((9); reviewed in Ref (10)). CGH is particularly useful for solid tumors where, it is often technically difficult to obtain sufficient G-banded metaphases of good quality for detailed analysis. One disadvantage of CGH is that it can only detect large blocks (5–10 Mb) of over- or underrepresented chromosomal DNA; balanced rearrangements such as inversions or translocations escape detection. This drawback is complemented by recent advances in the use of multicolor fluorescence in situ hybridization (FISH), which makes it possible to analyze all chromosomes simultaneously. Two different approaches have been developed: spectral karyotype analysis (SKY) (11) and combinatorial multifluor FISH (M-FISH) (12). Both techniques are based on the principle of the differential display of colored fluorescent chromosome-specific paints providing a complete analysis of the human chromosomal complement. Thus, extremely complex rearrangements in the karyotype of a tumor can be identified by the pattern of color distribution along aberrant chromosomes so that complex or cryptic rearrangements will lead to a transition from one color to another at the positions where the chromosome becomes abnormal. This technology is particularly suited to solid tumors such as RMS where the complexity of the karyotypes may often mask the presence of subtle chromosomal aberrations. In addition, application of SKY technology will help to characterize the type of chromosomal rearrangements leading to loss and gain of chromosome regions identified by CGH.
To date, no studies of the use of SKY or M-FISH in the analysis of RMS have been reported, and only one study of CGH analysis of 24 tumors (including four cell lines) has been performed (13). Two additional case studies of RMS have been reported by Bayani and colleagues (14) and Steilin-Gimbel and colleagues (15). CGH studies of RMS-E identified gain of part or all of chromosomes 2, 13, 12, 8, 7, 17, 18, and 19 and the loss of chromosomes 16, 10, 15, and 14 (13), which is consistent with the findings of hyperdiploidy by conventional cytogenetic analysis (8). CGH analysis of RMS-A indicates consistent evidence of copy number gains at 13q32, 17q, 1p36, 1q21, and 8q13-21. Gene amplification leads to copy number increases when double minute chromosomes (dmin) or homogeneously staining regions (hsr) form and usually have a greater copy number than gains resulting from aneuploidy caused by mitotic nondisjunction. Although amplification is not a consistent feature of RMS-E, the following regions have been shown to be amplified in RMS-A: 1p36 (PAX7); 1p31-32; 2p24 (MYCN); 12q13-15 (MDM2, GLI, CDK4); and 13q14 (FKHR). Of these, MYCN gene amplification has been detected most frequently in RMS-A but generally at lower copy numbers (<20) than seen in neuroblastoma (25–>100) (16,17). Recently, we have shown that another gene, DDX1, mapping to band 2p24, (18) is coamplified in 60% to 70% of the MYCN-amplified neuroblastomas (19–23). To date, there has been no systematic study of the incidence of coamplification of the MYCN and DDX1 genes in RMS.
The prognosis of alveolar RMS is poorer than that of classic embryonal RMS, mainly due to early tumor dissemination in alveolar RMS (1). In this study, we wish to extend CGH analysis of the overall incidence of cytogenetic aberration in RMS-E and RMS-A and to evaluate the use of SKY and microarray analysis on RMS cell lines derived from the two major subtypes. By characterizing the pattern of gene amplification and expression in RMS and by correlating the findings with potential prognostic markers such as coamplification of MYCN and DDX1, an improved and more comprehensive description of the profile of chromosomal change in RMS subtypes will become possible.
Material and Methods
Tumor Samples and Cell Lines
All RMS tumors included in this study were diagnosed by light microscopy and immunohistochemistry according to the Intergroup Rhabdomyosarcoma Study classification system (1) and are presented in Table 1. Sufficient DNA for Southern blot analysis only was obtained from one tumor (A12). Two established cell lines, RD, derived from a RMS-E tumor (24), and SJRH-30, derived from a RMS-A tumor (25), were obtained from ATCC at passage 37 and 39, respectively, and were used for SKY and microarray analysis.
Table 1.
CGH Changes in RMS-E and RMS-A.
| Patient No. | CGH Changes |
| E1 | +2; +3p24; +5; +8q24-qter; +11; +12; +20 |
| E2 | +2p22-pter; +3p; -4q; +7; +8q22-qter; -9; -10; -19 |
| E3 | +1p31; +8q24; +10q25 |
| E4† | +2p; +7; +8q; +11q14-qter; +12; +14q23-qter; +18q12-21; |
| E5 | +1; +2p; +3q24-qter; +5q23-qter; +7; +8; +9q22-qter; +11; +12q14-21*; +13; +19; +20 |
| E6 | +2; +5; +8; +13q; +14q21; -17p; +17q; -21q |
| E7† | +1q; +2; +3q24-qter; +5; +7; +8; +11p15; +12; +20 |
| E8† | +5q23-32; +6q; +8q; -19 |
| E9 | +2; +6; +7; +8; +11; +13q14-qter; +14q22-qter; +18q21-22; +19; +20 |
| E10 | +2q; +6; +7; +11q; +12; +14q24-qter |
| A1† | +1q31; +2q; +4p; +5p; +11p15; +11q22-qter; +12q13-21*; +13q22-qter; +14q31-32 |
| A2 | +1p22-31; +2; +12q13-21*; |
| A3 | +2p23-pter*; +5p; +9p21-23; +11p |
| A4 | +1; +3q26-qter; +6q21; +7; +8; -9; +11; +12; -13; +15q22-qter; +17q22-qter*; +21 |
| A5 | +2p23-pter*; +9p21-23*; +11p15 |
| A6 | +2p23-pter*; +11; +12; +20 |
| A7 | +2p22-24*; +18q11-21 |
| A8 | +2p23-24*; +3p22-pter; +6p22-pter; +12q13-21; +13q21-qter; +20 |
| A9 | +2; +4; +5; +8; +11p; +12q13-21; +13q; +17; +20 |
| A10 | +1q; +13q14 |
| A11 | +1p36; +14q32 |
Denotes amplification.
NOTE. All analyses carried out before treatment except those indicated by †.
Comparative Genomic Hybridization
Metaphase spreads from normal human lymphocytes were prepared by using standard protocols (26). The slides were aged for 2 to 3 days before denaturation at 70°C by 70% formamide in 2x standard saline citrate, followed by dehydration in an ethanol series. The slides were treated with proteinase K at a concentration of 0.1 µg/mL in 20 mmol/L Tris, pH 7.5, 2 mmol/L CaCl2, before hybridization. The CGH procedure was similar to published standard protocols (26,27). Twenty images were captured with a Nikon Labophot-2 microscope equipped with an automatic filter wheel and a 83,000 filter set (Chroma, Brattleboro VT), with single band pass exciter filters for UV/fluorescein isothiocyanate (FITC; 490 nm), DAPI (360 nm), and rhodamine (570 nm), and were analyzed by using the ISIS CGH software version 2.0 (MetaSystems, Heidelberg, Germany). With this software, the ratio of FITC to rhodamine signal is expressed as a green to red ratio and a deviation from a 1 to 1 ratio indicates gain or loss of chromosome material. The lower and upper limit for a gain and loss was established by performing control CGH experiments with IMR32, a well-characterized neuroblastoma cell line (28), and DNA derived from male and female normal tissue. Based on these findings, the cut-off values were set at 1.20 and 0.80, with a 95% confidence limit. Gene amplification was defined as gain ratios greater than 1.5.
SKY and Imaging
The cytogenetic preparations from the tumor cell lines were prepared as per standard protocols with colcemid and KCl hypotonic solutions. SKY was performed on five previously G-banded metaphases that were analyzed by conventional G-banding methods. Pretreatment of G-banded slides was performed as described previously (29). The commercially available SKY KIT from Applied Spectral Imaging (ASI, Carlsbad, CA) was used for SKY. The slide treatment, posthybridization detection, and washes were done as per standard protocols and the manufacturer's instruction (11,29). Spectral images were acquired and analyzed with a SD 200 spectral bioimaging system (ASI Ltd, MigdalHaemek, Israel) attached to a Zeiss microscope (Karl Zeiss, Canada) (Axioplan 2). The generation of a spectral image is achieved by acquiring ∼100 frames of the same image that differ from each other only in the optical path difference (OPD). The images were stored in a computer for further analysis by using SKYVIEW (ver 1.2) (ASI) software. To determine the position of chromosomal breakpoints in many of the rearrangements, the G-banded chromosomes were aligned with their spectrally analyzed RGB counterpart. For every chromosomal region, identity was determined by measuring the spectral emission. Pseudocolor classifications were made to aid in the delineation of specific structural aberrations.
cDNA Expression Microarray Blots
Total RNA was isolated from both cell lines with the RNeasy kit™ (Qiagen, Valencia CA). 32P-labeled cDNA probes were generated by using the CDS primer mix (Clontech, PaloAlto, CA) and hybridized (final probe concentration of ∼0.5−1 x 106 cpm/mL) to the Atlas Human Cancer Array (Clontech, Palo Alto, CA; Cat #7742-1) as per the manufacturer's instructions and previously published protocols (30,31). After three posthybridization washes in a solution of SSC and sodium dodecyl sulfate (SDS), the membranes were exposed to Kodak Biomax MS X-ray film at -70°C for different exposures. The complete list of genes present on the Atlas Human Cancer Array can be obtained from Clontech's web site: (http://www.clontech.com/archive/APR98UPD/Atlaslist.html). The autoradiograms were digitized with the AGFA T1200 duoscan scanner (Agfa Inc, Toronto, ON). The AtlasImage software (Clontech, Palo Alto, CA) was used to normalize TIFF images of each expression array with respect to levels of nine housekeeping genes (see Figure 5; GD 5–7, GE 12–14, and GF 19–21). Relative expression levels were obtained by comparing intensities for each gene in the array and assigning the following expression values in units x103 of signal intensity: ‘+’ (range, 14–20); ‘++’ (21–27); ‘+++’ (28–34); ‘++++’ (35 and above). Any signal intensity close to the background was considered as ‘-.’
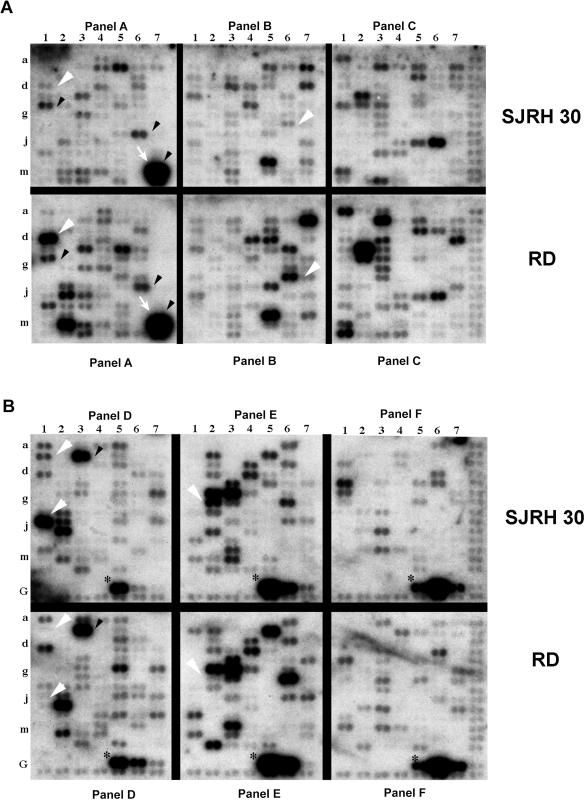
Figure 5.
The differential gene expression in RMS cell lines of RD and SJRH30 in the six different quadrants (A—Panel A–C and B—Panel D–F). Approximately 29% of the 588 genes present on the array generated readily detectable hybridization signals. Of these 77.3% was commonly expressed in the two tumor subtypes (black arrowheads), whereas 22.7% genes had dissimilar profiles (white arrowheads). Strong hybridization signals at A7m and A7n (white arrows) indicate a high expression of vimentin and desmin, respectively, which are overexpressed in rhabdomyosarcomas. Positive control housekeeping genes (*) present on the array are used by the AtlasImage software to normalize the intensity of the signals between the blots.
Molecular and FISH Analyses
The DNA from all tumors and controls were extracted by using the standard phenol/chloroform extraction method (32). Polyadenylated RNAs were extracted with the Quick Prep mRNA purification kit (Pharmacia-LKB Biochemical Co Baie d'Vrfe, Quebec, Canada) and were reverse transcribed with oligo dT (BRL). In cases where RNA was available, reverse transcription polymerase chain reaction (RT-PCR) analysis was performed to detect the presence of the PAX3-FKHR fusion transcript as described previously (4). Of the tumors tested, only the RMS-A tumors tested positive for the fusion transcript; all the RMS-E tumors tested were negative. Southern blot analysis was performed to determine the copy number of MYCN and DDX1 in these tumors by using standard procedures (32). Densitometric analysis was performed with ImageQuant software on a 300A computer densitometer (Molecular Dynamics, Sunnyville, CA). FISH interphase analysis was performed by using standard procedures (26). The DDX1 probe used was a 13.5-kb genomic clone in lambda 2001, which spans exon 4 to exon 10 of the gene (18).
Results
CGH Profiles of RMS-A and RMS-E Patient Tumors
A representative example of CGH profiles from one tumor is shown in Figure 1. The results are presented in Table 1, and summary CGH karyograms for both subtypes are illustrated in Figure 2. The overall trend for both subtypes was chromosomal gain, with this being more apparent in RMS-E than RMS-A. In contrast, chromosome loss only occurred in isolated cases, and there was no evidence of a consistent deletion or monosomy in either subtype. For RMS-E, there were consistent gains of whole or partial arms of several chromosomes, notably chromosomes 2 (8/10), 5 (5/10), 6 (3/10), 7 (7/10), 8 (9/10), 11 (6/10), and 12 (5/10). Losses/deletions were infrequent, involving chromosomes 19 (2/10) and chromosomes 4, 9, 10, 17, 21 (1/10 each). The chromosomes showing gains in RMS-A were 2 (8/11); 12 and 11 (6/11); 1 (5/11); 13 (4/11); 5 and 20 (3/11); 3, 4, 6, 8, 9, 14, and 17 (2/11); and 7, 15, 18, and 21 (1 case each). With the exception of one case of RMS-E, subchromosomal regions of amplification were confined to RMS-A as follows: 2p24 (5 RMS-A tumors); 9p21-23 (one RMS-A); 12q13-15 (two RMS-A), and 17q22-qter (one RMS-A). A single RMS-E tumor demonstrated amplification at 12q13-15. Detailed characterization of gene amplification at 2p24 in RMS-A is described later. Tables 2 and 3 compare the presence of chromosomal changes as determined by CGH and disease outcome. No overall trend can be determined in the RMS-E analysis, although it is noteworthy that the shortest disease remission was the only patient that had a tumor with high copy number amplification at 12q13-15. Pathologic review of this case indicated that the histology exhibited a typical morphology associated with RMS-E. RMS-A patients had a much shorter disease-free interval and a higher mortality than children with RMS-E did. Although the results suggest the existence of a substantial difference between the probability of CCR in the two groups, the sample size is quite small and the results are not statistically significant. The five patients with MYCN and DDX1 coamplification all had a poor response to treatment with limited or no remission. Two of the six RMS-A patients without amplification remain disease free.
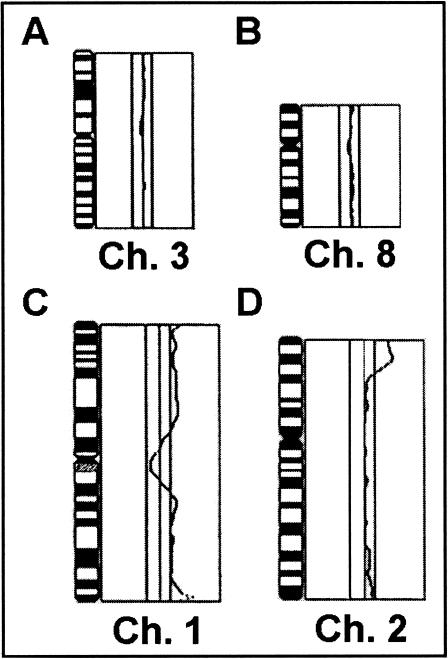
Figure 1.
Examples of CGH changes from a RMS tumor. The line in the middle of each profile indicates the base line ratio (1.0), and the lines on the left and right sides show the cut-off values of 0.80 (for loss) and 1.20 (gain), respectively. The profiles demonstrate a normal green-to-red ratio for chromosome 3 (A) and 8 (B), gain of chromosome 1 (C), and high level gain at 2p23-25 (D).
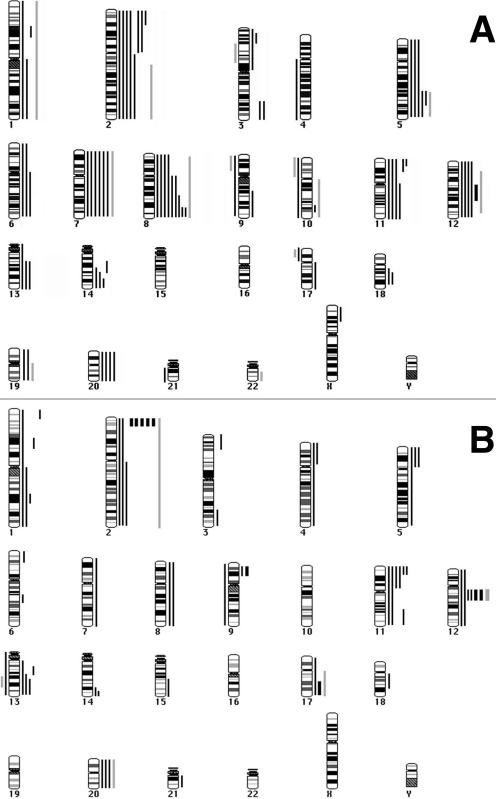
Figure 2.
Ideograms summarizing the results of CGH analysis of RMS. Gains are represented by vertical lines to the right of schematic chromosomes, whereas losses are represented by lines to the left. Solid lines indicate changes identified by using DNA derived from the 21 patient tumors, and the gray lines indicate changes identified by using DNA from the two RMS cell lines. Thicker lines to the right indicates the presence of a high copy amplification. A. CGH analysis of RMS-E. Copy number changes of 10 tumors and 1 cell line indicate a pattern of gains and gene amplification. In particular, gains of chromosomes 2, 7, 8, 11, and 12 are frequent. Only one tumor exhibited amplification (thicker solid line; chromosome 12). B. CGH Analysis of RMS-A. Copy number changes of 11 tumors and 1 cell line indicate a pattern of gains and gene amplification. High level gains/amplifications (∼>5 copy number) are illustrated by thicker lines. In contrast to RMS-E, there are fewer whole chromosome gains, and more tumors with localized gene amplification, involving both the MYCN region and, less frequently, the MDM2 region. Two other amplifications were detected, one on the short arm of chromosome 9 and one at the distal region of 17q.
Table 2.
Molecular Changes and Clinical Data for RMS-E.
| Year Diagnosed | Chromosomal Gains | Outcome | Disease/Duration of Remission | ||||
| (Patient #) | 2 | 7 | 8 | 11 | 12 | ||
| 1988 (E1) | +2 | +8q24-qter | +11 | +12 | CCR | Disease free | |
| 1989 (E2) | +2p22-pter | +7 | +8q22-qter | CCR | Disease free | ||
| 1987 (E3) | +8q24 | CCR | Disease free | ||||
| 1989 (E4) | +2p | +7 | +8q | +11q14-qter | +12 | D | 18 months |
| 1991 (E5) | +2p | +7 | +8 | +11 | +12q14-21* | D | 6 months |
| 1987 (E6) | +2 | +8 | D | 12 months | |||
| 1995 (E7) | +2 | +7 | +8 | +11p15 | +12 | CCR | Disease free |
| 1993 (E8) | +8q | D | 11 months | ||||
| 1992 (E9) | +2 | +7 | +8 | +11 | CCR | Disease free | |
| 1989 (E10) | +2q | +7 | +11q | +12 | CCR | Disease free | |
Denotes amplification.
Abbreviations: CCR, complete clinical remission; D, dead.
Table 3.
Molecular Cytogenetic Changes and Disease Outcome Data for RMS-A.
| Year Diagnosed | Amplification | Other Gains | Live/Dead | Disease/Duration of Remission | |
| (Patient #) | MYCN | DDX1 | |||
| 1989 (A1)ρ | No | No | +11; +12q13-21 | D | Progressive |
| 1991 (A2) | No | No | +12q13-21* | D | Progressive |
| 1989 (A3) | Yes; 13x | Yes | +11p | D | 7 |
| 1990 (A4)ρ | No | No | +11; +12; +17q22-qter* | CCR | Disease free |
| 1986 (A5) | Yes; 11x | Yes | +11p15 | D | 1 |
| 1982 (A6)ρ | Yes; 5x | Yes | +11; +12 | D | Progressive |
| 1994 (A7)ρ | Yes; 20x | Yes | D | 6 | |
| 1994 (A8)ρ | Yes; 6x | Yes | +12q13-21 | D | 4 |
| 1994 (A9)ρ | No | No | +11p; +12q13-21; +17 | CCR | Disease free |
| 1993 (A10) | No | No | D | Progressive | |
| 1980 (A11) | No | No | D | Progressive; 24 | |
Denotes amplification.
CCR, complete clinical remission; D, dead.
-t(2,13) positive.
SKY Analysis and Comparison Between SKY, CGH, and G-banding in RMS Cell Lines
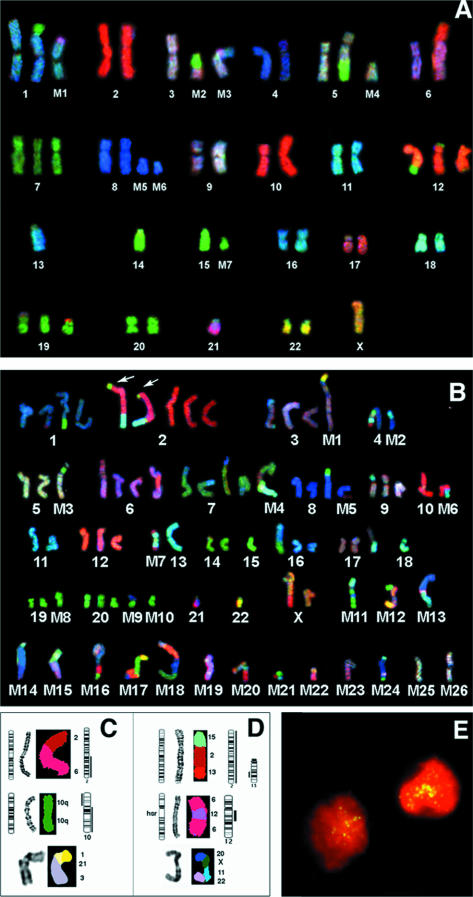
SKY analysis requires the availability of good quality metaphase chromosome preparations. Because cytogenetic preparations were not available for the series of tumors used for the previously mentioned CGH profile analysis, we used two established representative RMS cell lines, RD (RMS-E) and SJRH30 (RMS-A), to study the structural rearrangements in these two subtypes in detail. Furthermore, combining conventional G-banding analysis with SKY analysis on the same metaphase preparation provides more accurate positional information on highly abnormal karyotypes. A complete G-band analysis of the cell lines (Figure 3A and B) was performed, and RGB display images (Figure 4A and B) were used in combination with classification spectral results (not shown) to identify all chromosomal regions in both cell lines. In total, 17 structural aberrations were detected in RD and 42 in SJRH30. RD was observed to have undergone considerably fewer rearrangements overall than SJRH 30, with only seven markers being present. We identified several cryptic rearrangements (see Figure 3A [arrows], and Figure 4A) and marker chromosomes in RD that were not detected by G-banding (Table 4, panel A, boldface type]. The level of structural change in the near tetraploid SJRH30 karyotype identified by sequential G-banding (see Figure 3B) and SKY analysis (Figure 4B) was surprisingly high. Some marker chromosomes were present as duplicate aberrations, including the reciprocal translocation involving chromosomes 2 and 13, typical of RMS-A (Figure 4B [arrows] and D). Previously, this cell line has been shown to also have an amplification of GLI and other genes from 12q12-21 present as dmins (25). Our CGH analysis confirmed amplification in this region. SKY analysis did not identify dmins but detected an insertion of material from chromosome 12 in part of chromosome 6. G-banding analysis confirmed the presence of an hsr at this region of chromosome 6 (Figure 4D). The complexity of this karyotype is reflected in the ISCN descriptions of this tumor (Table 4, panel B).
Figure 3.
A. G-banding analysis of embryonal RMS cell line RD. The chromosomal origins were confirmed using SKY analysis (see Figure 4A). Examples of cryptic rearrangements, which were undetected by G band analysis but identified by SKY, are arrowed. B. G-banding analysis of alveolar RMS cell line SJRH30. The chromosomal origins were confirmed by using SKY analysis (see Figure 4B). Numerous marker (M1–M26) chromosomes and partially identified derivatives can be seen.
Figure 4.
SKY karyotypes for RD (A) and SJRH30 (B) are shown as RGB display, and individual rearrangements (panel C and D) are shown as classified SKY image display. A. SKY analysis of the embryonal RMS cell line RD. Identification is presented in Table 4 (Panel A). B. SKY analysis of the RMS-A cell line SJRH-30 Identification is presented in Table 4 (Panel B). C. Comparison between CGH and SKY analysis for RD. The schematic ideogram indicates the structural rearrangement based on a combination of SKY and G-banding. CGH profiles with loss on the left and gain on the right have been placed adjacent to each chromosome ideogram involved. For RD, the topmost rearrangement is an unbalanced 2;6 translocation, leading to 2q gain detected by CGH. The middle panel shows an isochromosome 10q with a gain of 10q and loss of 10p. The lower panel depicts a complex rearrangement detected by SKY observed in RD (M3 in Figure 3A and 4A). The chromosomes involved were 1, 21, and 3. D. Comparison between CGH and SKY for SJRH-30. The upper rearrangement is between regions of chromosomes 2, 13, and 15. This structural aberration has resulted in a partial loss and gain of the chromosomal segments involved as there is total gain of chromosome 2, and loss of part of the rearranged segment of distal 13q (also seen arrowed in panel B). The middle rearranged chromosome is an hsr-bearing chromosome 6, with amplified material mapping to the Gli and CDK4 region of chromosome 12 as identified by CGH. The lower panel displays a complex SKY chromosome marker (M12 on Figures 3B and 4B) with small segments of chromosomes involved (20, X, 11, 22). E. Interphase FISH analysis of a RMS-A tumor with DDX1 identifies gene amplification as dispersed ‘speckling’ of FISH signals within nuclei. This pattern of gene amplification is usually associated with the presence of dmin. The patient (A7 in Table 3) had 20 copies of MYCN and DDX1, which was confirmed by Southern blot analysis.
Table 4.
Summary of G-Banding and SKY Findings for Each RMS Cell Line Classified by ISCN Nomenclature.
| A. | |
| RD, G-Banding Karyotype: | |
| 50,X,-X,add(1p34),-3,der(5)t(5;14)(q22;q12),der(6)t(2;6)(q23;p22),+7,add(9q34),i(10q),+?12,add(12q24),-13,-14,-15,add(19p13), +del(19)(p13),-21 ,+mar1-7 | |
| RD, SKY Karyotype: | |
| 50,X,der(1)t(1;15)(p34;q22),+der(1)del(1)(q32)t(1;21)(p32;?q22),der(5)t(5;14)(q22;q12),+der(5),der(6)t(2;6)(q22;p22),+7,+der(8),+der(8),der(9)t(9;17)(q34;q?),i(10q),+?12,der(12)t(12;19)(p13;p13),der(12)t(12;20)(q24;q11),-13,-14,der(15), | |
| der(17)t(9;17)(?;p11),+der(19)t(12;19)(p11;?),-21, | |
| +mar2=3 and 19 | |
| +mar3=1, 21 and 3 | |
| B. | |
| SJRH30, near tetraploid karyotype G-Banding: | |
| 81,X,der(X),-Y,-Y,der(1),add(1)(q25),der(1)t(1;15)(q32;q13),der(2)t(2;13)(q35;q14)x2,+i(2)(p10),der(3),add(3)(p25),-4,-4,-4,der(4),-5.-5,add(6)(p15),del(6)(p15),hsr(6)(q15),add(7)(p22)x3,add(7)(q36),-8,add(8)(p21),-9,del(9)(p22),-10,-10,-10,add(10)(p11),-11,-11,-12,-13,-13,i(13)(q10),-14,-14,-15,-15,-15,-16,add(16)(q24),-17,add(17)(p11),-18,-18,-18,-19,-19,-20,-20,-21,-21,-21,-22,-22,-22,der(22),+mar1-26 | |
| SJRH30, Structural aberrations detected by SKY Analysis: | |
| 81,der(X)t(X;10)(q27;q?22),der(1)t(1;21)(q?;q?),der(1)t(1;15)(q32;q13),der(2)t(2;13;15)(q35;q14;q?)x2,i(2)(p10),der(3)t(3;11;16)(p25;?;?),der(4)t(2;13;3;19;4),der(6)t(4;6),hsr(6)(q15),der(7)t(7;8)(p22;?)x3,der(7)t(1;7)(?;q36),der(8)t(8;20)(p21;?),der(9)t(9;11)(p22;?),i(13q),der(16)t(14;16)(?;q24),der(17)t(11;17)(?;p11) | |
| mar1=22,1,3,1 and 3 | mar14=4 |
| mar2=4 and 11 | mar15=15 and 16 |
| mar3=5, 15 and 5 | mar16=9, 4, 20, 10 and 15 |
| mar4=7, 6 and 18 | mar17=22, 15, 12, 4 and 12 |
| mar5=17 and 8 | mar18=9, 12, 8 and 7 |
| mar6=10 and 11 | mar19=?21 and Y (isochromosome) |
| mar7=2, 13 and 2 (isochromosome) | mar20=X, 11, X, 11 and 10 |
| mar8=8 | mar21=2, 15 and 16 |
| mar9=1 and 20 | mar22=X and 21 |
| mar10=20 | mar23=4, 10, 1 and 10 |
| mar11=18, 3, 20, 8 and 20 | mar24=18, 4, 22 and 20 |
| mar12=20, X, 11 and 22 | mar25=3 |
| mar13=4 and 11 | mar26=3 |
cDNA Expression Array Analysis
The overall level of genomic change ascertained by molecular cytogenetic methods suggested that considerable alteration of gene expression might have taken place in RD and SJRH 30. To determine whether gene expression patterns were also markedly different in the two RMS cell lines, steady state gene expression levels were analyzed with a commercial cDNA expression array filter set (Atlas Human Cancer Array [Clontech #7742-1]) (30,31). The 588 bound cDNAs are arrayed within six quadrants on each membrane: panel A–F consisting of oncogenes and tumor suppressor genes, cell cycle and growth regulators, apoptosis genes, DNA damage and repair genes, and so on. Expression levels of the nine control housekeeping genes on both membranes exhibited hybridization signals of equal intensity. Nine negative control spots (Figure 5B: GD2–GD4, GE2–GE4, and GF2–GF4) did not show any hybridization signals on the autoradiogram, even after an exposure of 10 days. Overnight exposure of the filter revealed positive signals for 169 (28.74%) cDNAs in SJRH 30 and 198 (33.67%) in RD (Figure 5). Approximately 78% of the commonly expressed genes had a similar signal intensity (Figure 5 [black arrowheads], whereas 22% of the genes were differentially expressed (Figure 5 [white arrowheads]). As expected, high level expression of desmin and vimentin was observed (Figure 5A, [white arrows]), which is in accordance with the literature (33,34). Noteworthy differentially expressed genes (Table 5) included heparin-binding vascular endothelial growth factor (VEGF), DNA mismatch repair protein, retinoblastoma binding protein RBQ1, MYCC binding protein, and cyclins A and D. Some genes found to be overexpressed in the microarray analysis were also observed to be overrepresented by the CGH analysis. For example, oncogene c-MET, present on chromosome 7 (7q31), was both overexpressed (microarray) and overrepresented (CGH) in the cell line RD. The proto-oncogene MYCN, amplified in approximately 50% (see the next section) of the RMS-A tumors was not amplified (by CGH, Southern blot) or overexpressed (microarray) in SJRH 30.
Table 5.
Representative Autoradiograms Indicating Differential Expression of Genes on the Atlas Array.
| Gene Name (Chromosome Location) | GDB# | Expression | |
| RD | SJRH30 | ||
| Cell division protein kinase 4 (12) | M14505 | +++ | + |
| Cyclin A (4) | X51688 | +++ | - |
| Cyclin B1 (5) | M25753 | +++ | + |
| MAP kinase kinase; (MEK1) (15) | L05624 | - | ++++ |
| PCNA; cyclin (20) | M15796 | +++ | - |
| GRB-IR/GRB10 (7) | U69276 | - | + |
| ICH-2 protease precursor; caspase-4 (11) | U28014 | - | +++ |
| DNA mismatch repair protein MSH2 (2) | U04045 | +++ | + |
| MET (7) | J02958 | +++ | + |
| RBQ1 retinoblastoma binding protein (16) | X85133 | + | +++ |
| DNA topoisomerase II alpha (17) | J04088 | +++ | + |
| Byglycan (X) | J04599 | - | +++ |
| Collagen type III pro-alpha-1 (2) | X14420 | - | ++++ |
| Urokinase-type plasminogen activator precursor (UPA) (10) | M15476 | + | ++++ |
| Endothelial plasminogen activator inhibitor-1 precursor (PAI-1) (7) | X04429 | - | +++ |
| Heparin-binding vascular endothelial growth factor (VEGF) (6) | M32977 | ++ | +++ |
Gene Amplification in RMS
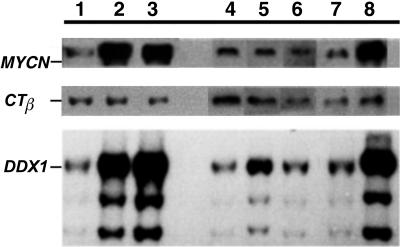
Amplification was a consistent feature of the RMS-A subtype with 6 of 12 of these tumors having gene amplification involving the 2p24 region. Southern blot analysis (Figure 6) confirmed that six RMS-A tumors were coamplified for MYCN and DDX1 with copy numbers in the range of 5 to 20x. The ratio of the MYCN to DDX1 was 1 to1, as determined by densitometric analysis (data not shown) and there were no examples of tumors having amplified MYCN but not DDX1 or vice versa (see Table 3). None of the seven RMS-E tested had amplification of this chromosomal region. Interphase FISH with methanol-acetic acid fixed tumor cells identified the presence of additional copies of MYCN and DDX1 in the RMS-A tumors and indicated that the additional copies resulted in paired signals dispersed through the interphase nucleus (see Figure 4E). This pattern of additional signal is typical of dmin in contrast to the clustering of signals as a discrete domain that is characteristic of amplification involving an hsr. In three tumors studied by interphase FISH, there was considerable range in MYCN and DDX1 gene copy number per nucleus. Sequential imaging of MYCN and DDX1 signals with two-color FISH (data not shown) indicated that each gene colocalized to the same discrete regions within nuclei suggesting that both MYCN and DDX1 exist as a single amplification unit on common dmins. Both tumor subtypes showed low copy number changes of chromosome 2. In RMS-E, four cases have additional copy of the whole chromosome, with two tumors having gains of 2p and one of 2q. There is no evidence of high copy gain (amplification) on chromosome 2 in RMS-E.
Figure 6.
Southern blot analysis was carried out on the alveolar lanes 1[(A1), 2(A3), 3(A7)] and the embryonal lanes [4(E3), 5(E4), 6(E5)] tumors by using the MYCN and DDX1 probe. The tumor and normal (thymus) DNA, together with DNA from a neuroblastoma cell line, IMR32 (coamplified with MYCN and DDX1), was digested with EcoR1 and separated on a 1% agarose gel in Tris borate EDTA buffer. After transfer to a nylon membrane, the filter was probed with MYCN, CTβ (a control probe), and DDX1. Coamplification of MYCN and DDX1 is seen in the alveolar tumors in lanes 2 (A3) and 3 (A5) and the control lane 8 (IMR32). Lane 7 is a normal control.
Discussion
RMS is a pediatric soft tissue sarcoma of the skeletal muscle where histologic classification of the tumor subtypes is sometimes equivocal (3). There are only limited numbers of classical cytogenetic studies of RMS, and most have centered on the early designation of the 2;13 chromosomal rearrangement (35–37). Molecular studies, such as Loss of Heterozygosity (LOH) analysis, have provided information about losses of DNA in particular genetic regions of interest (7), but they are too labor intensive to be used as a general screening method. The newer molecular cytogenetic methodologies and microarray expression analysis are convenient and rapid ways to screen for chromosomal and gene expression changes that may be of benefit in understanding the molecular basis of variable prognosis and lead to additional approaches for distinguishing between the subtypes.
An important differentiating feature between the two RMS subtypes is the presence of genomic amplifications. The RMS-A tumors appear to be much more susceptible to genomic amplification than the RMS-E subtype. The regions involved are the 2p24 region (MYCN and DDX1) and the 12q13 region (harboring MDM2, CDK4 and GLI). In isolated reports, amplification has also been observed by CGH on chromosomes 1, 2, 8, 12, and 13 (38). Reports published earlier by our group (17), Dias and colleagues (16), and more recently, Hachitanda and colleagues (39) have shown MYCN to be amplified in approximately 45% to 55% of the RMS-A patients. In this study, 6/12 (50%) tumors had coamplification of MYCN and DDX1. In most tumors this was revealed as a gain at band 2p24 by CGH, which was shown to involve amplification of MYCN when studied by interphase FISH and Southern blot analysis. Recently, DDX1 has been shown to be coamplified in retinoblastomas and neuroblastomas. DDX1 is a member of the DEAD box protein family of putative RNA helicases. DEAD box proteins are involved in translation initiation and RNA splicing and have been implicated in RNA stability, gene expression, and cell growth and differentiation (40–45). DDX1 by itself has recently been demonstrated to have oncogenic potential in both in vitro and in vivo assays (46), so its role as a coamplified gene, together with MYCN, may be an important contributing factor in more aggressive neoplastic disease. The screening of RMS tumors in this study showed DDX1 to be coamplified in all of the MYCN-amplified tumors. This finding appears to be somewhat different to observations made in neuroblastoma, in which only 60% to 70% of the MYCN-amplified tumors were coamplified with DDX1 (19–23). Significantly, the copy number observed in neuroblastoma MYCN-amplified tumors is considerably higher than that found in RMS-A: 50 to 100x compared with less than 20x. This finding indicates a possible role for coamplification of DDX1 and MYCN in the RMS-A subtype that is distinct to the established pattern of MYCN gene amplification in neuroblastoma and suggests that copy numbers less than 20x may be adequate to confer a selective advantage in RMS-A. The patient outcome data in this study indicated RMS-A patients had a shorter disease-free interval and a higher mortality than children with RMS-E, in keeping with previous epidemiological data (2). RMS-A patients with MYCN and DDX1 coamplification had a poor outcome as expected for tumors in this subtype but a larger sample size is required to demonstrate that the presence of coamplification as an independent prognostic indicator.
A chromosomal region that has aberrations in both subtypes is the 12q13 region. Amplification studies of known oncogenes mapping to this region in various sarcomas have identified diverse patterns of amplification, including MDM2, GLI, SAS, and GADD153 (47,48). Interestingly, the MDM2 gene amplification or overexpression is believed to provide an alternative route for inactivating the p53 pathway. Inactivation of the TP53 plays a key role in tumor progression, probably through a disturbed cell cycle control and an increased susceptibility to genomic instability. The absence of the p53 protein leads to genomic alterations through chromosome breakage and DNA aneuploidy. Studies have shown increased frequencies of amplification of the CAD and DHFR gene in cells with TP53 mutations (49,50). Some reports implicate preferential amplification of CDK4, a cell division cycle protein kinase also present at band 12q13-15, in sarcomas (including RMS). This gene has been shown to be coamplified with several genes, including MDM2, either as one large contiguous amplicon or as a second amplicon without an amplified intervening region (51–54). Further characterization of the amplicons and studies on these amplified genes are required to reveal the importance of disruption of the p53 and cell cycle pathways in RMS. Amplicons observed on chromosomal region 1q21, 8q13, and 13q32 by Weber-Hall and colleagues (13) were not observed in the tumor samples in this study, indicating that larger surveys are required to determine the full spectrum of amplification in RMS.
Consistent aneusomies or whole chromosome gains were observed by CGH in RMS-E and were in agreement with the earlier reported cytogenetic and CGH analysis (6,8,10,13,35,36,55,56). Gains were observed in most of the chromosomes but were prominent in chromosomes 2, 5, 7, 8, 11, and 12. Whole to partial gains of chromosome 8, predominantly in RMS-E (9/10), are consistent with conventional cytogenetic findings (8). Trisomy 8 is a common feature of a wide variety of malignancies (35) indicating a generalized proliferative advantage in cancer. Gain of chromosome 7 was frequent (7/10) and was associated specifically with RMS-E. This chromosomal gain may provide a useful subtype-specific aneusomic marker for future interphase FISH analysis. It is noteworthy that one of the genes that we found differentially expressed in our microarray analysis is the c-Met oncogene with overexpression in RMS-E compared to RMS-A. The c-MET oncogene (mapping to chr 7q31) has been associated with the invasive growth in RMS (57) and is also thought to be a downstream target of the PAX3-FKHR fusion protein (58). Microarray studies on RMS-A conducted by Khan and colleagues (59) revealed 37 of the 1238 genes tested were consistently expressed in RMS-A relative to a reference cell line. Other genes exhibiting differential gene expression in our microarray analysis include CDK4 (chr 12q13), MEK-1 (chr 15), and chromosome segregation homologue (chr. 20). As mentioned earlier, CDK4 is preferentially amplified and overexpressed in sarcomas, including RMS. Although it is difficult to draw specific conclusions regarding the expression levels observed by microarray analysis of in vitro material, it is clear that the ∼22% of genes on this panel that exhibit differential expression will provide starting points for future in vivo investigations of the critical biological pathways that determine the differences between RMS-E and RMS-A.
Newer FISH techniques such as CGH and SKY provide powerful tools for screening tumors to identify both the type of chromosomal aberrations present and the regions of the genome involved in rearrangement. When the techniques are applied to the same sample, the combined results provide a better overall picture of the genetic events present in a complex karyotype. The use of SKY in combination with G-banding identified many subtle translocations and complex genomic rearrangements, especially in the SJRH 30 RMS-A cell line. SKY analysis confirmed some of the changes reported in earlier cytogenetic studies conducted on the cell lines by Houghton and colleagues (24) and Robert and colleagues (25) but also identified with greater precision the origin and number of partner chromosomes present in all marker chromosomes. For example, add(1p34) was confirmed to be der(1)t(1;15)(p34;q22). Even in the most highly abnormal marker chromosomes present in SJRH 30, it was possible to recognize the chromosomal origins of each partner chromosome. In general, SKY analysis also demonstrated there was a greater frequency of structural aberration present in the RMS-A cell line in comparison to the RMS-E cell line in which whole chromosome loss and gain was noted. There is increasing knowledge about the role of genes that govern the acquisition of aneuploidy in neoplastic cells (60). It is conceivable that RMS-E subtype tumors have a greater tendency to undergo nondisjunctional mitotic segregation anomalies that lead to whole chromosome gains, whereas RMS-A appears to be subject to genomic alterations leading to chromosomal rearrangement and amplification. The presence of the distinct cytogenetic differences between RMS-E and RMS-A as determined by CGH, SKY and microarray expression aid in subtyping RMS and draw attention to previously unrecognized areas of the genome that need further study for identification of genes of potential importance in the progression and future treatment of RMS.
Acknowledgements
We thank Shawn Brennan, Sandra Johnson, Paula Marrano, Yim Kwan Ng, and Catherine Janish (ASI, Carlsbad, CA) for their excellent help with this project, and Anne Feltis for providing the clinical information for the RMS patients. We are also grateful to Dr. S. Minkin for his assistance with the statistics. This work was supported by the National Cancer Institute of Canada with funds from the Canadian Cancer Society.
Abbreviations
- RMS
rhabdomyosarcoma(s)
- RMS-A
alveolar rhabdomyosarcoma
- RMS-E
embryonal rhabdomyosarcoma
- CGH
comparative genomic hybridization
- SKY
spectral karyotyping
- FISH
fluorescence in situ hybridization
References
- 1.Newton WA, Jr, Gehan EA, Webber BL, Marsden HB, van Unnik AJ, Hamoudi AB, Tsokos MG, Shimada H, Harms D, Schmidt D. Classification of rhabdomyosarcomas and related sarcomas. Pathologic aspects and proposal for a new classification—an Intergroup Rhabdomyosarcoma Study. Cancer. 1995;76:1073–1085. doi: 10.1002/1097-0142(19950915)76:6<1073::aid-cncr2820760624>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 2.Diller L. Rhabdomyosarcoma and other soft tissue sarcomas of childhood. Curr Opin Oncol. 1992;4:689–695. doi: 10.1097/00001622-199208000-00014. [DOI] [PubMed] [Google Scholar]
- 3.Scrable H, Witte D, Shimada H, Seemayer T, Wang-Wuu S, Soukup S, Koufos A, Houghton P, Lampkin B, Cavenee W. Molecular Differential Pathology of Rhabdomyosarcoma. Genes Chromosomes Cancer. 1989;1:23–25. doi: 10.1002/gcc.2870010106. [DOI] [PubMed] [Google Scholar]
- 4.Galili N, Davis R, Fredericks W, Mukhopadhyay S, Rauscher F, III, Emanuel B, Rovera G, Barr F. Fusion of a fork head domain gene to PAX3 in the solid tumor alveolar rhabdomyosarcoma. Nat Genet. 1993;5:230–235. doi: 10.1038/ng1193-230. [DOI] [PubMed] [Google Scholar]
- 5.Barr F, Nauta L, Davis R, Schäfer B, Nycum L, Biegel J. In vivo amplification of the PAX3-FKHR and PAX7-FKHR fusion genes in alveolar rhabdomyosarcoma. Hum Mol Genet. 1996;5:15–21. doi: 10.1093/hmg/5.1.15. [DOI] [PubMed] [Google Scholar]
- 6.Davis R, D'Cruz C, Lovell M, Biegel J, Barr F. Fusion of PAX7 to FKHR by the variant t(1,13)(p36;q14) translocation in alveolar rhabdomyosarcoma. Cancer Res. 1994;54:2869–2872. [PubMed] [Google Scholar]
- 7.Scrable H, Cavenee W, Ghavimi F, Lovell M, Morgan K, Sapienza C. A model for embryonal rhabdomyosarcoma tumorigenesis that involves genome imprinting. Proc Natl Acad Sci USA. 1989;86:7480–7484. doi: 10.1073/pnas.86.19.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sandberg A, Bridge E. The cytogenetics of bone and soft tissue tumors. Austin, TX: RG Landes; 1994. [Google Scholar]
- 9.Forozan F, Karhu R, Kononen J, Kallioniemi A, Kallioniemi OP. Genome screening by comparative genomic hybridization. Trends Genet. 1997;13:405–409. doi: 10.1016/s0168-9525(97)01244-4. [DOI] [PubMed] [Google Scholar]
- 10.Knuutila S, Bjorkqvist A, Aautio K, Tarkkanen M, Wolf M, Monni O, Szymanska J, Larramendy M, Tapper J, Pere H, El-Rifai W, Hemmer S, Wasenius V, Vidgren V, Zhu Y. DNA copy number amplifications in human neoplasms: Review of comparative genomic hybridization. Am J Pathol. 1998;152:1107–1123. [PMC free article] [PubMed] [Google Scholar]
- 11.Schrock E, du Manoir S, Veldman T, Schoell B, Wienberg J, Ferguson-Smith MA, Ning Y, Ledbetter DH, Bar-Am I, Soenksen D, Garini Y, Ried T. Multicolor spectral karyotyping of human chromosomes [see comments] Science. 1996;273:494–497. doi: 10.1126/science.273.5274.494. [DOI] [PubMed] [Google Scholar]
- 12.Speicher MR, Gwyn Ballard S, Ward DC. Karyotyping human chromosomes by combinatorial multi-fluor FISH. Nat Genet. 1996;12:368–375. doi: 10.1038/ng0496-368. [DOI] [PubMed] [Google Scholar]
- 13.Weber-Hall S, Anderson J, McManus A, Abe S, Nojima T, Pinkerton R, Pritchard-Jones K, Shipley J. Gains, losses, and amplification of genomic material in rhabdomyosarcoma analyzed by comparative genomic hybridization. Cancer Res. 1996;56:3220–3224. [PubMed] [Google Scholar]
- 14.Bayani J, Thorner P, Zielenska M, Pandita A, Beatty B, Squire JA. Application of a simplified comparative genomic hybridization technique to screen for gene amplification in pediatric solid tumors. Pediatr Pathol Lab Med. 1995;15:831–844. doi: 10.3109/15513819509027020. [DOI] [PubMed] [Google Scholar]
- 15.Steilen-Gimbel H, Remberger K, Graf N, Steudel WI, Zang KD, Henn W. A novel site of DNA amplification on chromosome 1p32-33 in a rhabdomyosarcoma revealed by comparative genomic hybridization. Hum Genet. 1996;97:87–90. doi: 10.1007/BF00218839. [DOI] [PubMed] [Google Scholar]
- 16.Dias P, Kumar P, Marsden HB, Gattamaneni HR, Heighway J, Kumar S. N-myc gene is amplified in alveolar rhabdomyosarcomas (RMS) but not in embryonal RMS. Int J Cancer. 1990;45:593–596. doi: 10.1002/ijc.2910450403. [DOI] [PubMed] [Google Scholar]
- 17.Driman D, Thorner PS, Greenberg ML, MacNeil-Chilton S, Squire J. MYCN gene amplifications in rhabdomyosarcoma. Cancer. 1994;73:2231–2237. doi: 10.1002/1097-0142(19940415)73:8<2231::aid-cncr2820730832>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 18.Godbout R, Squire J. Amplification of a DEAD box protein gene in retinoblastoma cell lines. Proc Natl Acad Sci USA. 1993;90:7578–7582. doi: 10.1073/pnas.90.16.7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amler L, Schurmann J, Schwab M. The DDX1 gene maps within 400 kbp 5′ to MYCN and is frequently coamplified in human neuroblastoma. Genes Chromosomes Cancer. 1996;15:134–137. doi: 10.1002/(SICI)1098-2264(199602)15:2<134::AID-GCC9>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 20.George RE, Kenyon RM, McGuckin AG, Malcolm AJ, Pearson AD, Lunec J. Investigation of co-amplification of the candidate genes ornithine decarboxylase, ribonucleotide reductase, syndecan-1 and a DEAD box gene, DDX1, with N-myc in neuroblastoma. United Kingdom Children's Cancer Study Group. Oncogene. 1996;12:1583–1587. [PubMed] [Google Scholar]
- 21.Manohar C, Salwen H, Brodeur G, Cohn S. Co-amplification and concomitant high levels of expression of a DEAD box gene with N-MYC in human neuroblastoma. Genes Chromosomes Cancer. 1995:196–203. doi: 10.1002/gcc.2870140307. [DOI] [PubMed] [Google Scholar]
- 22.Noguchi T, Akiyama K, Yokoyama M, Kanda N, Matsunaga T, Nishi Y. Amplification of a DEAD box gene (DDX1) with the MYCN gene in Neuroblastomas as a result of cosegregation of sequences flanking the MYCN locus. Genes Chromosomes Cancer. 1996;15:129–133. doi: 10.1002/(SICI)1098-2264(199602)15:2<129::AID-GCC8>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 23.Squire J, Thorner P, Weitzman S, Maggi J, Dirks P, Doyle J, Hale M, Godbout R. Co-amplification of MYCN and a DEAD box gene (DDX1) in primary neuroblastoma. Oncogene. 1995;10:1417–1422. [PubMed] [Google Scholar]
- 24.Houghton JA, Houghton PJ, Brodeur GM, Green AA. Development of resistance to vincristine in a childhood rhabdomyosarcoma growing in immune-deprived mice. Int J Cancer. 1981;28:409–415. doi: 10.1002/ijc.2910280403. [DOI] [PubMed] [Google Scholar]
- 25.Roberts W, Douglass E, Peiper S, Houghton P, Look A. Amplification of the gli Gene in Childhood Sarcomas. Cancer Res. 1989;49:5407–5413. [PubMed] [Google Scholar]
- 26.Dracopoli NC. Current Protocols in Human Genetics. New York: John Wiley and Sons; 1999. [Google Scholar]
- 27.Kallioniemi O, Kallioniemi A, Sudar D, Rutovitz D, Gray J, Waldman F, Pinkel D. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 28.Tumilowicz JJ, Nichols WW, Cholon JJ, Greene AE. Definition of a continuous human cell line derived from neuroblastoma. Cancer Res. 1970;30:2110–2118. [PubMed] [Google Scholar]
- 29.Veldman T, Vignon C, Schrock E, Rowley JD, Ried T. Hidden chromosome abnormalities in haematological malignancies detected by multicolour spectral karyotyping. Nat Genet. 1997;15:406–410. doi: 10.1038/ng0497-406. [DOI] [PubMed] [Google Scholar]
- 30.DeRisi J, Penland L, Brown P, Bittner M, Meltzer P, Ray M, Chen Y, Su Y, Trent J. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 31.DeRisi J, Iyer V, Brown P. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 32.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Sturhl K. Current Protocols in Molecular Biology. New York: John Wiley and Sons; 1998. [Google Scholar]
- 33.Dodd S, Malone M, McCulloch W. Rhabdomyosarcoma in children: A histological and immunohistochemical study of 59 cases. J Pathol. 1989;158:13–18. doi: 10.1002/path.1711580105. [DOI] [PubMed] [Google Scholar]
- 34.Parham DM, Webber B, Holt H, Williams WK, Maurer H. Immunohistochemical study of childhood rhabdomyosarcomas and related neoplasms. Results of an Intergroup Rhabdomyosarcoma study project. Cancer. 1991;67:3072–3080. doi: 10.1002/1097-0142(19910615)67:12<3072::aid-cncr2820671223>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 35.Heim S, Mitelman F. Cancer Cytogenetics. New York: Alan R. Liss; 1994. [Google Scholar]
- 36.Trent J, Casper J, Meltzer P, Thompson F, Fogh J. Nonrandom chromosome alterations in rhabdomyosarcoma. Cancer Genet Cytogenet. 1985;16:189–197. doi: 10.1016/0165-4608(85)90045-7. [DOI] [PubMed] [Google Scholar]
- 37.Wang-Wuu S, Soukup S, Ballard E, Gotwals B, Lampkin B. Chromosomal analysis of sixteen human rhabdomyosarcoma. Cancer Res. 1988;48:983–987. [PubMed] [Google Scholar]
- 38.Weber-Hall S, McManus A, Anderson J, Nojima T, Abe S, Pritchard-Jones K, Shipley J. Novel formation and amplification of the PAX7-FKHR fusion gene in a case of alveolar rhabdomyosarcoma. Genes Chromosomes Cancer. 1996;17:7–13. doi: 10.1002/(SICI)1098-2264(199609)17:1<7::AID-GCC2>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 39.Hachitanda Y, Toyoshima S, Akazawa K, Tsuneyoshi M. N-myc gene amplification in rhabdomyosarcoma detected by fluorescence in situ hybridization: Its correlation with histologic features. Mod Pathol. 1998;11:1222–1227. [PubMed] [Google Scholar]
- 40.Schmid SR, Linder P. DEAD protein family of putative RNA helicases. Mol Microbiol. 1992;6:283–291. doi: 10.1111/j.1365-2958.1992.tb01470.x. [DOI] [PubMed] [Google Scholar]
- 41.Iost I, Dreyfus M. mRNA can be stabilized by DEAD-box proteins. Nature. 1994;372:193–196. doi: 10.1038/372193a0. [DOI] [PubMed] [Google Scholar]
- 42.Buelt MK, Glidden BJ, Storm DR. Regulation of p68 RNA helicase by calmodulin and protein kinase C. J Biol Chem. 1994;269:29367–29370. [PubMed] [Google Scholar]
- 43.Py B, Higgins CF, Krisch HM, Carpousis AJ. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996;381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- 44.Rozen F, Edery I, Meerovitch K, Dever TE, Merrick WC, Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol Cell Biol. 1990;10:1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Godbout R, Packer M, Bie W. Overexpression of a DEAD box protein (DDX1) in neuroblastoma and retinoblastoma cell lines. J Biol Chem. 1998;273:21161–21168. doi: 10.1074/jbc.273.33.21161. [DOI] [PubMed] [Google Scholar]
- 46.George R, Thomas H, McGuckin A, Angus B, Newell D, Pearson A, Lunec J. The DDX1 gene which is frequently co-amplified with MYCN in primary neuroblastoma is itself tumorigenic. Proc Am Assoc Cancer Res. 1998;39 Abstract # 3205. [Google Scholar]
- 47.Ladanyi M, Jhanwar S, Healey J, Huvos A. MDM2 amplification in Ewing's sarcoma. Lab Invest. 1994;70:147A. doi: 10.1002/path.1711750209. [DOI] [PubMed] [Google Scholar]
- 48.Oliner J, Kinzler K, Meltzer P, George D, Volgelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 49.Goker E, Waltham M, Kheradpour A, Trippett T, Mazumdar M, Elisseyeff Y, Schnieders B, Steinherz P, Tan C, Berman E. Amplification of the dihydrofolate reductase gene is a mechanism of acquired resistance to methotrexate in patients with acute lymphoblastic leukemia and is correlated with p53 gene mutations. Blood. 1995;86:677–684. [PubMed] [Google Scholar]
- 50.Hartwell L. Defects in a cell cycle checkpoint may be responsible for the genomic instability of cancer cells. Cell. 1992;71:543–546. doi: 10.1016/0092-8674(92)90586-2. [DOI] [PubMed] [Google Scholar]
- 51.Khatib ZA, Matsushime H, Valentine M, Shapiro DN, Sherr CJ, Look AT. Coamplification of the CDK4 gene with MDM2 and GLI in human sarcomas. Cancer Res. 1993;53:5535–5541. [PubMed] [Google Scholar]
- 52.Meddeb M, Valent A, Danglot G, Nguyen VC, Duverger A, Fouquet F, Terrier-Lacombe MJ, Oberlin O, Bernheim A. MDM2 amplification in a primary alveolar rhabdomyosarcoma displaying a t(2;13)(q35;q14) Cytogenet Cell Genet. 1996;73:325–330. doi: 10.1159/000134368. [DOI] [PubMed] [Google Scholar]
- 53.Thorner P, Squire J, Chilton-MacNeil S, Marrano P, Bayani J, Malkin D, Greenberg M, Lorenzana A, Zielenska M. Is the EWS/FLI-1 fusion transcript specific for Ewing sarcoma and peripheral primitive neuroectodermal tumor? A report of four cases showing this transcript in a wider range of tumor types. Am J Pathol. 1996;148:1125–1138. [PMC free article] [PubMed] [Google Scholar]
- 54.Waber PG, Chen J, Nisen PD. Infrequency of MDM2 gene amplification in pediatric solid tumors and lack of association with p53 mutations in adult squamous cell carcinomas. Cancer Res. 1993;53:6028–6030. [PubMed] [Google Scholar]
- 55.Wang-Peng J, Knutsen T, Theil K, Horowitz M, Triche T. Cytogenetic Studies in Subgroups of Rhabdomyosarcoma. Genes Chromosomes Cancer. 1992;5:299–310. doi: 10.1002/gcc.2870050405. [DOI] [PubMed] [Google Scholar]
- 56.Biegel JA, Nycum LM, Valentine V, Barr FG, Shapiro DN. Detection of the t(2;13)(q35;q14) and PAX3-FKHR fusion in alveolar rhabdomyosarcoma by fluorescence in situ hybridization. Genes Chromosomes Cancer. 1995;12:186–192. doi: 10.1002/gcc.2870120305. [DOI] [PubMed] [Google Scholar]
- 57.Ferracini R, Olivero M, Di Renzo MF, Mariano M, De Giovanni C, Nanni P, Basso G, Scotlandi K, Lollini PL, Comoglio PM. Retrogenic expression of the MET proto-oncogene correlates with the invasive phenotype of human rhabdomyosarcomas. Oncogene. 1996;12:1697–1705. [PubMed] [Google Scholar]
- 58.Ginsberg JP, Davis RJ, Bennicelli JL, Nauta LE, Barr FG. Up-regulation of MET but not neural cell adhesion molecule expression by the PAX3-FKHR fusion protein in alveolar rhabdomyosarcoma. Cancer Res. 1998;58:3542–3546. [PubMed] [Google Scholar]
- 59.Khan J, Simon R, Bittner M, Chen Y, Leighton SB, Pohida T, Smith PD, Jiang Y, Gooden GC, Trent JM, Meltzer PS. Gene expression profiling of alveolar rhabdomyosarcoma with DNA microarrays. Cancer Res. 1998;58:5009–5013. [PubMed] [Google Scholar]
- 60.Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B. Mutations of mitotic checkpoint genes in human cancers [see comments] Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]