Abstract
Malignant progression is accompanied by degradation of extracellular matrix proteins. Here we describe a novel confocal assay in which we can observe proteolysis by living human breast cancer cells (BT20 and BT549) through the use of quenched-fluorescent protein substrates. Degradation thus was imaged, by confocal optical sectioning, as an accumulation of fluorescent products. With the BT20 cells fluorescence was localized to pericellular focal areas that coincide with pits in the underlying matrix. In contrast, fluorescence was localized to intracellular vesicles in the BT549 cells, vesicles that also label for lysosomal markers. Neither intracellular nor pericellular fluorescence was observed in the BT549 cells in the presence of cytochalasin B, suggesting that degradation occurred intracellularly and was dependent or endocytic uptake of substrate. In the presence of a cathepsin B-selective cysteine protease inhibitor, intracellular fluorescence was decreased ∼90% and pericellular fluorescence decreased 67% to 96%, depending on the protein substrate. Matrix metallo protease inhibitors reduced pericellular fluorescence ∼50%, i.e., comparably to a serine and a broad spectrum cysteine protease inhibitor. Our results suggest that: 1) a proteolytic cascade participates in pericellular digestion of matrix proteins by living human breast cancer cells, and 2) the cysteine protease cathepsin B participates in both pericellular and intracellular digestion of matrix proteins by living human breast cancer cells.
Keywords: quenched fluorescent substrates, laser scanning confocal microscopy, cysteine proteases, extracellular matrix, endocytosis
Introduction
Proteases of the matrix metallo, serine, aspartic and cysteine classes have been implicated in the ability of tumor cells to invade through extracellular matrices (for reviews, see Refs. [1–5]). Evidence to support a role for proteases in tumor invasion is that proteases of all four classes are able to degrade extracellular matrix (ECM) proteins and intact basement membrane in vitro [3,6–9] and selective inhibitors of the four protease classes reduce degradation of ECM and tumor cell invasion [10,11]. In vivo, decreases in staining for ECM proteins (e.g., laminin, type IV collagen) are observed at the invasive edges of tumors [12–14], suggesting that the proteins are being degraded by the invading tumor cells.
Active proteases can be shown to be associated with tumors in vivo by using near-infrared probes and optical imaging [15,16]. These studies do not identify the tumor proteases that are cleaving ECM proteins in situ as they use small synthetic substrates designed for selectivity against individual proteases. Thus, we are not presently able to assess the ability of living tumors to digest ECM protein substrates that are of relevance in tumor invasion. Therefore, like others, we have used in vitro assays with fluorescently tagged ECM proteins to image proteolysis by living tumor cells [17,18]. These assays have limitations in that the cells and matrices are fixed for observation so that one images prior proteolysis by cells that are motile, and thus pericellular proteolysis may have occurred at a site no longer coincident with an absence of fluorescence. Furthermore, one images loss of fluorescence (which may pre-exist due to uneven tagging or coating of tagged proteins) rather than acquisition of fluorescence. In the present study, we have used confocal laser scanning microscopy. This technology has allowed us: 1) to analyze proteolysis in situ by living human breast cancer cells, and 2) to accurately localize the site of proteolysis by optically sectioning through both the cells and the matrix underneath them. By using this assay in combination with protease inhibitors, one can determine: 1) the ability of any given inhibitor to reduce or eliminate degradation, and 2) identify the protease or protease class responsible. A novel finding was that two breast cancer cell lines differ in their sites of digestion of protein substrates: in the BT20 line digestion occurred outside the cells and in the BT549 line inside the cells. Both intracellular and pericellular degradation were reduced by highly selective inhibitors of the cysteine protease cathepsin B.
Materials and Methods
Cell Culture
BT20 and BT549 human breast cancer cell lines were purchased from American Type Culture Collection and cultured in E-MEM and RPMI 1640 + insulin, respectively, with 10% fetal bovine serum as recommended by the American Type Culture Collection.
Assays for Proteolysis
Living cells Precooled glass coverslips were coated (1 to 1.5 mm in depth) with 25 µg/ml of the quenched fluorescent substrates DQ-BSA or DQ-collagen IV (Molecular Probes), 2% bovine skin gelatin (Type B, 225 Bloom) and 2% sucrose for 10 min at 4°C. Cells (50,000) were plated and observed over a period of 48 h for fluorescent degradation products by confocal microscopy on a Zeiss LSM 310 using a 40x water immersion objective. In studies with inhibitors, the cells were plated as above and grown in the presence of 100 nM of cystatin C (M. Abrahamson, University of Lund, Sweden), an endogenous inhibitor of cysteine proteases, 10 µM of the highly selective cathepsin B inhibitor CAO74 or its membrane-permeable derivative CA074Me (Peptides International), 10 µM of the broad spectrum cysteine protease inhibitor E-64 (Sigma) or its membrane-permeable derivative E-64d (Peptides International), 10µM of a broad spectrum MMP inhibitor (BB3103; a kind gift of British Biotech), 1 µM of the serine protease inhibitor aprotinin (Sigma), or 5 µg/ml of cytochalasin B (Sigma) over the entire period of observation. Cells were shown to be ≥99% viable under all of the assay conditions described. Fluorescence intensities and areas in the presence and absence of inhibitors were quantitated from confocal fluorescence images using Image Gauge V3.3 Software (Fuji Medical Systems).
Cathepsin B Purified human liver cathepsin B (2.5 µg; Athens Research and Technology) was diluted in 50 mM sodium acetate buffer, 1 mM EDTA, pH 5.2. Activation buffer of 5 mM EDTA and 10 mM DDT was added and samples incubated for 15 min at 37°C. For studies at pH 7.2, the pH was titrated with 0.2 M citrate phosphate buffer, pH 7.4. CAO74 (10 µM) was added for 30 min followed by addition of DQ-BSA or DQ-collagen IV (25 µg/ml). Samples were incubated overnight at 37°C and fluorescence recorded at an excitation wavelength of 485 nm and an emission wavelength of 538 nm using a Labsystems Fluoroskan II and Delta Soft 3 software.
Immunocytochemical Studies
We localized, according to our published procedures [19], intracellular and cell surface cathepsin B in saponin-permeabilized and unpermeabilized BT20 and BT549 cells, respectively, which were grown on coverslips coated with mouse laminin (Upstate Biotechnology). The primary antibodies used were rabbit anti-human liver cathepsin B IgG and mouse anti-human tubulin monoclonal (Developmental Studies Hybridoma Bank). Controls were run in the absence of primary antibodies. Coverslips were mounted upside-down with SlowFade anti-fade reagent (Molecular Probes) on glass slides and observed on a Zeiss LSM 310 confocal microscope. Forty optical sections were taken at 0.2-µm intervals from the top to the bottom of the cells and used to reconstruct the three-dimensional images as described previously [20].
Colocalization of DQ-substrate with Active Cathepsin B or a Lysosomal Marker
BT549 cells (50,000) were plated on DQ-BSA or DQ-collagen IV/gelatin-coated coverslips and grown for 48 h. Coverslips were placed on slides and 1) cathepsin B activity was assessed using 10 µM of a highly selective substrate for cathepsin B [Z-Arg-Arg]2 cresyl violet 2 HBr (Enzymes Systems Products) in RPMI 1640, or 2) lysosomes were imaged by the addition of 75 ng/ml of LysoTracker (Molecular Probes) to the living cells. Cells were pulsed with LysoTracker for 2 h and then chased for 1 h to insure that the LysoTracker had reached the lysosomal compartment. Cleavage of the cresyl violet substrate as well as localization of LysoTracker and the fluorescent degradation products of the DQ-substrate were imaged on a Zeiss LSM 310 confocal microscope using a 40x water immersion objective. Overlap of fluorescence (i.e., colocalization) was quantitated from confocal optical sections using Image Gauge V3.3 Software (Fuji Medical Systems).
Statistical Analysis
SuperANOVA version 1.11 (Abacus) was used to determine significant differences.
Results
Imaging Degradation of DQ-BSA by Living Cells
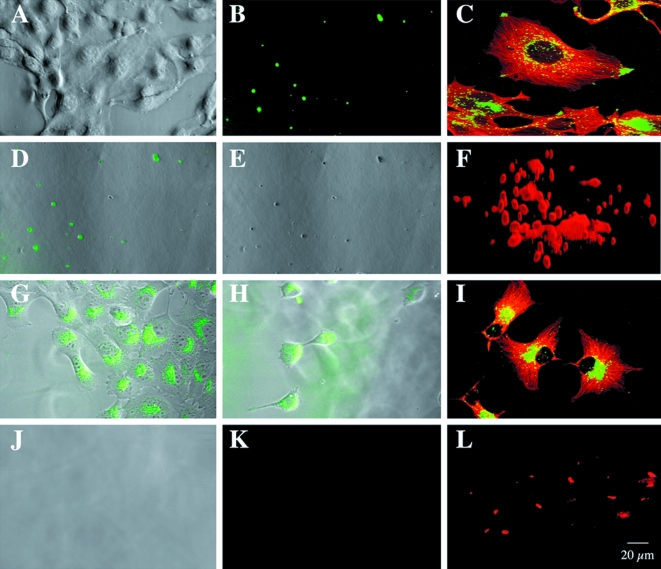
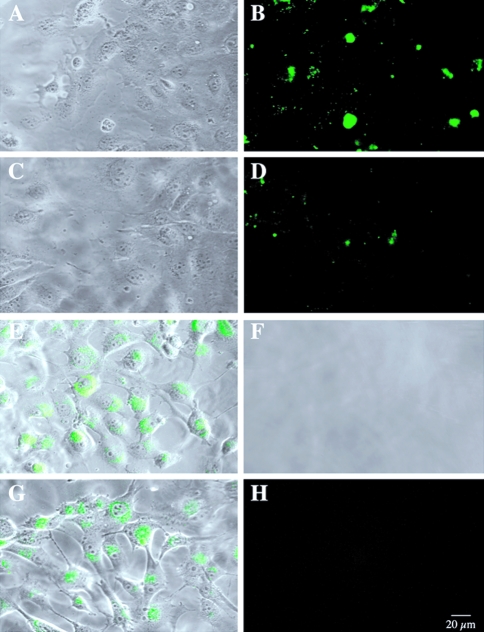
We plated BT20 and BT549 human breast cancer cells on an intramolecularly quenched fluorescent protein substrate, DQ-BSA, mixed with gelatin and coated onto glass coverslips. The cells and their underlying matrix were then analyzed by optical sectioning through both with a laser confocal microscope. Phase images of living BT20 cells revealed that these cells grew in clusters on the matrix with some cells extending long pseudopodia over the surfaces of others (Figure 1A). Degradation of the DQ-BSA by the living BT20 cells was visualized by confocal microscopy as small spots of green fluorescence in optical sections underneath the cells (Figure 1B). Fluorescent degradation products were not observed in optical sections taken at the level of the BT20 cells. In phase images of the gelatin matrix underneath the cells, finger-like pits or tunnels could be observed (Figure 1E). Superimposition of fluorescent optical sections and the phase image of the gelatin matrix revealed that the pits and sites of proteolysis coincided (Figure 1D), suggesting that proteolysis might occur on pseudopodia extending from the surface of the BT20 cells.
Figure 1.
Living BT20 and BT549 human breast cancer cells degraded DQ-BSA pericellularly and intracellularly, respectively. (A) Phase image of BT20 cells. (B) Optical section underneath the BT20 cells illustrating pericellular green fluorescent degradation products. E, phase image of matrix underneath the BT20 cells. (D) Coincidence of green fluorescent degradation products and pits in superimposition of optical section (B) and phase image (E). (G) Superimposition of optical and phase images through BT549 cells present near the top of the matrix, illustrating green fluorescent degradation products within the cells. (H) Superimposition of optical and phase images through BT549 cells that have moved into the matrix, also illustrating green fluorescent degradation products within the cells. (J) Phase image of matrix underneath BT549 cells illustrating absence of pits. (K) Optical section underneath BT549 cells illustrating absence of pericellular green fluorescent degradation products. Note that images are all of living cells and the matrices on which they are growing. Intracellular and surface localization of the cysteine protease cathepsin B differed between the BT20 and BT549 human breast cancer cells. Immunofluorescent staining for intracellular cathepsin B (green) and microtubules (red) in permeabilized BT20 (C) and BT549 (I) human breast cancers grown on laminin-coated coverslips. Immunofluorescent staining for cathepsin B (red) on the surface of unpermeabilized BT20 (F) and BT549 (L) cells. Forty optical sections at 0.2-µm intervals were used to reconstruct the surface staining in three dimensions.
The BT549 cells, like the BT20 cells, grew in clusters on the matrix, but long pseudopodia were not observed (Figure 1G). Pits in the underlying gelatin were not present (Figure 1J), nor were pericellular degradation products (i.e., green fluorescence; Figure 1K) observed in optical sections taken underneath the BT549 cells. Instead, fluorescent products were found to be present within cells in optical sections taken at the level of the BT549 cells (Figure 1G and H). This intracellular localization appeared to be vesicular and perinuclear, a localization similar to that of lysosomes. This was the case in BT549 cells that remained near the top of the matrix (Figure 1G) as well as those that had migrated through the matrix (Figure 1H). The proteolytic cleavage products of DQ-BSA within the BT549 cells may represent: 1) intracellular proteolysis of DQ-BSA that entered the cells by an endocytic mechanism, or 2) pericellular proteolysis of DQ-BSA followed by endocytosis of degraded DQ-BSA and its accumulation intracellularly.
Living Breast Cancer Cells Use Cathepsin B to Degrade DQ-BSA Pericellularly and Intracellularly
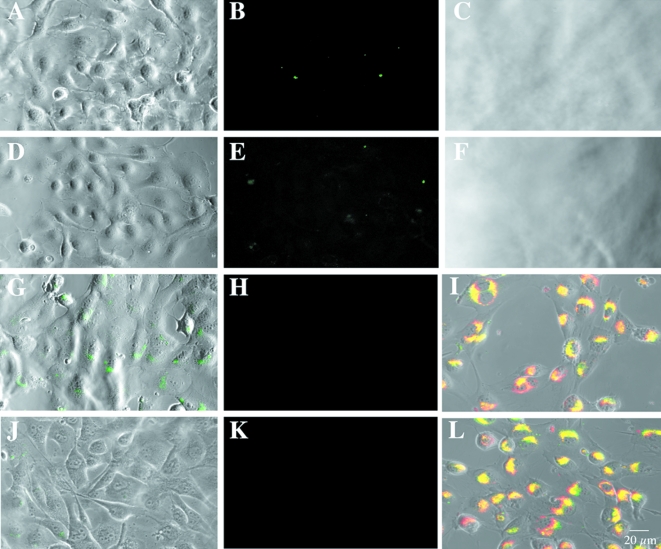
We have previously shown that the cysteine protease cathepsin B is localized on the surface of breast cancer cells as well as in the lysosomes of these cells [19]. As the localization of cleaved DQ-BSA in the BT549 cells appeared to be lysosomal, we immunolocalized cathepsin B in the BT20 and BT549 breast cancer cell lines. In the BT20 cells, cathepsin B was localized to vesicles heavily concentrated at the tips of cell processes (Figure 1C). The surface of BT20 cells also stained intensely for cathepsin B, particularly finger-like extensions (pseudopodia) from the cell surface (Figure 1F). In contrast, cathepsin B was localized predominantly to perinuclear vesicles in the BT549 cells (Figure 1I), with only a small amount of cathepsin B on the surface of these cells (Figure 1L). We had shown previously that surface staining for cathepsin B on U87 human glioma cells correlates with a functional role for this enzyme in invasion through Matrigel and into normal brain aggregates [10] and in degradation of the ECM protein laminin [17]. We therefore used protease inhibitors to determine whether cathepsin B on the surface of BT20 cells was responsible for the proteolysis of DQ-BSA by these cells and the accompanying formation of pits in the matrix underneath the cells. BT20 cells were incubated with two inhibitors: cystatin C (Figure 2A – C), an endogenous inhibitor of cysteine proteases [21], and CA074 (Figure 2D – F), a highly selective synthetic inhibitor of cathepsin B [22,23]. Both inhibited degradation of DQ-BSA as assessed by a reduction in the number of green fluorescent spots (Figure 2B and E compare with Figure 1B) and by an absence of pits in the underlying gelatin matrix (Figure 2C and F compare with Figure 1E). We scanned optical sections for fluorescent cleavage products in the absence and presence of inhibitors and showed that fluorescent degradation products were significantly reduced (≥90%) by either cystatin C or CA074 (Table 1: BT20). Such comparable inhibition by cystatin C and CA074 would be consistent with cathepsin B being the cysteine protease responsible for pericellular degradation of DQ-BSA by the BT20 cells.
Figure 2.
Inhibitors of cysteine proteases and cathepsin B reduced pericellular degradation of DQ-BSA by living BT20 cells (A–C and D–F, respectively) and intracellular degradation of DQ-BSA by living BT549 cells (G–H and J–K, respectively). Phase images of BT20 cells (A, D). Optical sections underneath BT20 cells (B, E). Phase images of the gelatin matrix underneath the BT20 cells (C, F). Optical sections through the BT549 cells superimposed on phase images of those cells (G, J). Optical sections underneath the BT549 cells (H, K). Pericellular degradation by the BT20 cells was inhibited by 100 nM cystatin C (B, C) or 10 µM CA074 (E, F) and intracellular degradation by the BT549 cells was inhibited by 100 nM cystatin C (G, H) or 10 µM CA074Me, a membrane-permeable form of CA074 (J, K). In the BT549 cells, fluorescent degradation products for DQ-BSA (I, green) or DQ-collagen IV (L, green) colocalized (yellow staining) with activity of the lysosomal cysteine protease cathepsin B as detected with a selective cresyl violet substrate (red fluorescence).
Table 1.
The Cysteine Protease Inhibitor Cystatin C and the Cathepsin B Inhibitors CA074 and CA074Me Reduce Pericellular Degradation of DQ-BSA by BT20 Cells and Intracellular Degradation of DQ-BSA by BT549 Cells.
| Fluorescence Intensity* | % Control | Area† | % Control | |
| BT20 | ||||
| Control | 315,568±372 | 100 | 3,572 | 100 |
| Cystatin C | 31,006±110 | 10 | 367 | 10 |
| CA074 | 11,581±031 | 4 | 106 | 3 |
| BT549 | ||||
| Control | 2,030,026±250 | 100 | 12,110 | 100 |
| Cystatin C | 782,062±1,091 | 38 | 4,064 | 34 |
| CA074Me | 157,431±235 | 8 | 852 | 7 |
Fluorescence intensity is in arbitrary units with each unit equal to the sum of the gray scale value for the pixels within an image; results are expressed as mean±SD. SD were calculated for all pixels in an image using Image Gauge V3.3 Software (Fuji).
Area is in Px2 units.
The intracellular vesicles in the BT549 cells that contain fluorescent degradation products resemble lysosomes. To determine whether degradation occurred intracellularly through lysosomal cysteine proteases, we incubated BT549 cells with cystatin C. We had previously shown that cystatin C can be phagocytosed and thus reach the lysosomal compartment [24]. Accumulation of degraded DQ-BSA within intracellular vesicles was reduced in the presence of cystatin C (Figure 2G compare with Figure 1G) fluorescent degradation products were decreased >60% (Table 1: BT549). Degradation products were not observed pericellularly (Figure 2H), nor were pits in the gelatin matrix (not illustrated). Thus, the accumulation of fluorescent products in intracellular vesicles appeared to be due to digestion within the BT549 cells. To determine whether cathepsin B was the cysteine protease responsible for degradation of DQ-BSA in the lysosomes, we incubated BT549 cells with a methylated, membrane-permeable derivative of CA074, i.e., CA074Me [23,25]. This proinhibitor is inactive extracellularly and is activated intracellularly by cytoplasmic esterases. In the presence of CA074Me, there was a reduction in the amount of DQ-BSA degradation products within the BT549 cells (Figure 2J) compared with the amount observed in the absence of inhibitor (Figure 1G) or in the presence of cystatin C (Figure 2G). Fluorescent degradation products were reduced by >90% (Table 1: BT549). Degraded DQ-BSA was not observed pericellularly (Figure 2K), nor were pits in the gelatin matrix (not illustrated). Thus, the accumulation of degradation products within the BT549 cells appeared to be due to intracellular degradation within the lysosomes by cathepsin B.
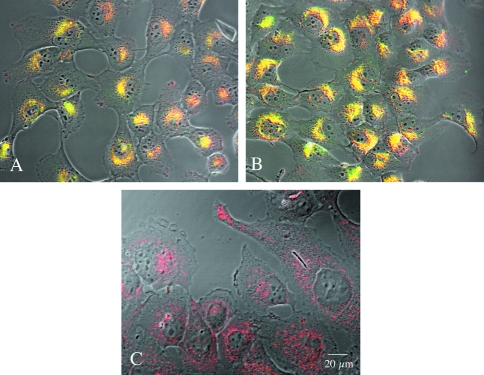
To confirm that the perinuclear vesicles staining for degraded DQ-BSA also contained active cathepsin B, we incubated the cells with DQ-BSA (or DQ-collagen IV, see below) and a cresyl violet substrate that fluoresces red on cleavage by cathepsin B [26]. Most of the perinuclear vesicles fluoresced yellow (Figure 2I), thus indicating a colocalization of active cathepsin B (red fluorescence) and degraded DQ-BSA (green fluorescence) in the BT549 cells. Some cells contained vesicles staining for cathepsin B activity that did not contain degraded DQ-BSA. As a further means to identify the intracellular vesicles containing degraded DQ-BSA, we incubated BT549 cells with two lysosomal markers: Lysotracker and Texas red-labeled dextran as well as with anti-cathepsin B antibodies. Degraded BSA colocalized with both lysosomal markers in the BT549 cells (Lysotracker, Figure 3A dextran, not illustrated). Fluorescence intensity and area, respectively, were 1,388,317±408 and 25,610 for Lysotracker and 1,362,547±642 and 26,112 for degraded BSA. Cathepsin B also colocalized with both lysosomal markers in the BT549 cells (Lysotracker, Figure 3B dextran, not illustrated). Some vesicles staining for cathepsin B but not for Lysotracker were also observed; these were most likely endosomes. Fluorescence intensity and area, respectively, were 2,699,861±1091 and 28,786 for Lysotracker and 3,191,686±1228 and 42,714 for cathepsin B. Thus, we have demonstrated that the lysosomes of BT549 cells contained both cathepsin B and fluorescent products resulting from degradation of DQ-BSA. In contrast, the lysosomes of BT20 cells could be identified with Lysotracker (Figure 3C), yet fluorescent products resulting from degradation of DQ-BSA were not present within these cells, an observation consistent with matrix degradation occurring pericellularly in BT20 cells.
Figure 3.
Lysotracker, a marker of lysosomes, colocalized with degraded DQ-BSA and with cathepsin B in the BT549 cells. (A) BT549 cells double-labeled for intracellular cathepsin B (green) and Lysotracker (red); vesicles staining yellow labeled for both cathepsin B and Lysotracker. (B) BT549 cells labeled for Lysotracker (red) and containing green fluorescent products due to degradation of DQ-BSA; vesicles staining yellow contain both degraded DQ-BSA and Lysotracker. No colocalization was observed for Lysotracker and degraded DQ-BSA in the BT20 cells. (C) BT20 cells labeled for Lysotracker (red) but did not contain green fluorescent products due to degradation of DQ-BSA, nor vesicles staining yellow due to presence of both degraded DQ-BSA and Lysotracker.
As further evidence that degradation of DQ-BSA took place intracellularly in BT549 cells, we incubated the cells with cytochalasin B to inhibit endocytosis of either intact DQ-BSA or the cleavage products of DQ-BSA [27]. In the presence of cytochalasin B, degraded DQ-BSA was not present intracellularly nor did it accumulate pericellularly (not illustrated), thus confirming that degradation of DQ-BSA occurred inside the BT549 cells. Taken together, our results suggest that cathepsin B degraded DQ-BSA in lysosomes within living BT549 cells.
Cathepsin B Can Degrade Protein Substrates In Vitro
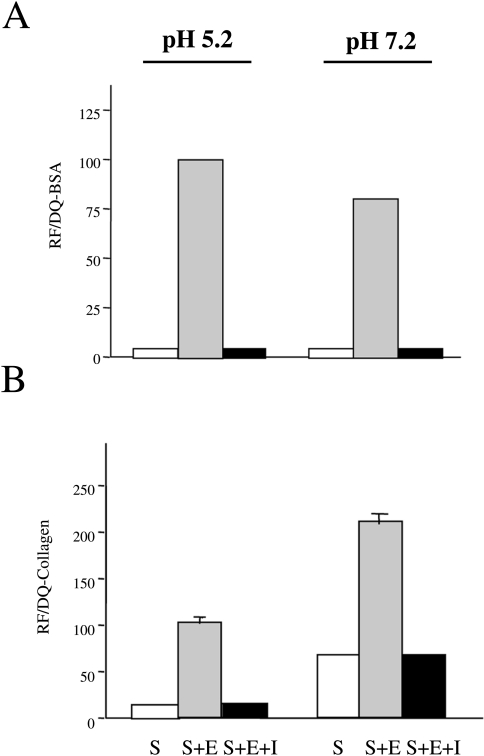
The present studies indicate that cathepsin B could degrade DQ-BSA intracellularly within lysosomes in BT549 cells as well as pericellularly at the surface of BT20 cells. The intracellular degradation of DQ-BSA presumably took place in a compartment at acidic pH and the pericellular degradation at neutral (or a less acidic) pH. To establish that DQ-BSA can be cleaved by cathepsin B at both acidic and neutral pHs, we incubated DQ-BSA with purified human liver cathepsin B at pH 5.2 and 7.2 using CA074 to prove that cathepsin B was responsible for the cleavage observed in situ. Cathepsin B degraded DQ-BSA at both pHs and CA074 abrogated this degradation (Figure 4A). Degradation of DQ-BSA by cathepsin B at pH 7.2 was ∼80% of that observed at pH 5.2.
Figure 4.
Purified human liver cathepsin B degraded DQ-BSA (A) and DQ-collagen IV (B) at both acidic and neutral pH. CA074, a highly selective inhibitor of cathepsin B, totally blocked degradation of the two DQ-substrates at both pHs. S=substrate: DQ-BSA (A) or DQ-collagen IV (B); S+E=substrate+enzyme (cathepsin B); S+E+I=substrate+enzyme+inhibitor (10 µM CAO74). Results represent three independent experiments performed in triplicate and are expressed as a percentage of 100% relative fluorescence (RF) for S+E at pH 5.2; mean±SD. In columns without standard deviations, they were too small to depict.
Bovine serum albumin, although it appears to be an excellent marker for the proteolytic activity of living cells, as shown above, would not be encountered by human breast cancer cells migrating or invading in vivo. Therefore, we determined whether a quenched fluorescent derivative of type IV collagen (DQ-collagen IV) might be a substrate for cathepsin B in situ (see below). We had previously shown that human liver and tumor cathepsin B can degrade purified type IV collagen in vitro at acidic and neutral pHs [6]. We reconfirmed these earlier findings by demonstrating that cathepsin B could digest DQ-collagen IV at both pH 5.2 and 7.2 (Figure 4B). CA074 abrogated degradation at both pHs. Interestingly, degradation of DQ-collagen IV by cathepsin B was ∼50% greater at neutral pH than at acidic pH, indicating that type IV collagen could well be a substrate for cell surface cathepsin B.
Imaging Degradation of DQ-Collagen IV by Living Cells
Proteolysis of DQ-collagen IV by living BT20 and BT549 cells paralleled their proteolysis of DQ-BSA (cf. Figure 1) in that BT20 cells (Figure 5B and D) and BT549 cells (Figure 5E and G) degraded DQ-collagen IV pericellularly and intracellularly, respectively. The ability of both cell lines to invade through the gelatin matrix appeared to be stimulated by DQ-collagen IV as two distinct layers of cells were visible in optical section confocal microscopy (BT20: Figure 5A and C BT549: Figure 5E and G). Green fluorescent spots, indicative of proteolysis of DQ-collagen IV, were present pericellularly underneath the BT20 cells (Figure 5B and D). Small spots similar to those under cells grown on DQ-BSA/gelatin were observed under the lower layer of BT20 cells (Figure 5D). In contrast, under the top layer of BT20 cells there were large pools of fluorescent degradation products (Figure 5B). Perhaps the initial movement of the BT20 cells involved an extension of pseudopodia digesting DQ-collagen IV at their surface. This might then have been followed by a more extensive degradation of DQ-collagen IV. Both the lower and upper layers of BT549 cells contained degraded DQ-collagen IV intracellularly within perinuclear vesicles (Figure 5E and G). Pericellular degradation was not observed (Figure 5F and H).
Figure 5.
Living BT20 (A–D) and BT549 (E–H) human breast cancer cells degraded DQ-collagen IV pericellularly and intracellularly, respectively. (A and C) Phase images of BT20 cells. (B and D) Optical sections underneath the BT20 cells illustrating pericellular green fluorescent degradation products. (E and G) Superimposition of optical and phase images through BT549 cells, illustrating green fluorescent products within the cells. (F) Phase image of the matrix underneath BT549 cells illustrating absence of pits. (H) Optical section underneath BT549 cells, illustrating absence of pericellular green fluorescent degradation products. Cells near the top of the matrix are illustrated in panels (A) and (E), whereas those that have moved into the matrix and are near the bottom of the matrix are illustrated in panels (C) and (G).
To demonstrate that the degraded DQ-collagen IV was localized in lysosomes and that the lysosomes contained active cathepsin B, we incubated the BT549 cells with DQ-collagen IV and LysoTracker, a fluorescent marker that accumulates in lysosomes [28] or a cresyl violet substrate to detect cathepsin B activity [26]. Most of the perinuclear vesicles fluoresced yellow, indicating that degraded DQ-collagen IV and active cathepsin B colocalized (Figure 2L) and that degraded DQ-collagen IV was present in vesicles containing LysoTracker (not illustrated).
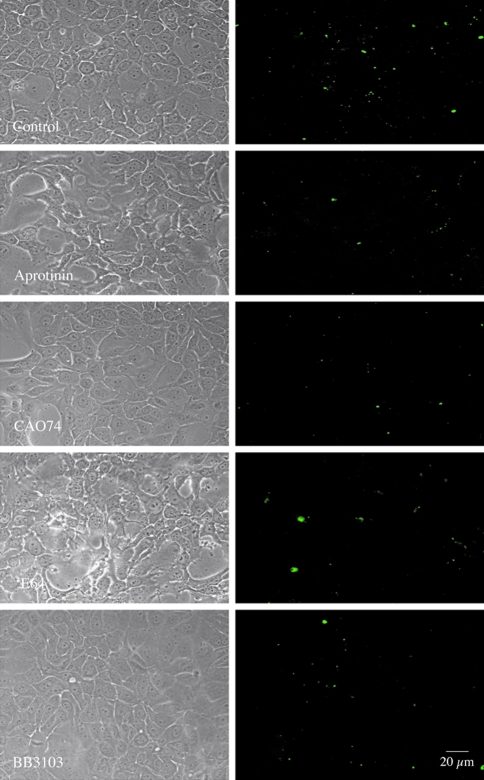
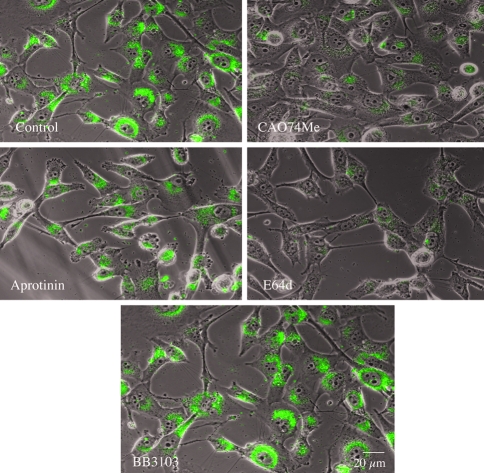
The pericellular degradation of DQ-collagen IV observed with living BT20 cells was inhibited comparably by the serine protease inhibitor aprotinin, a broad spectrum MMP inhibitor (BB3103) and a broad-spectrum cysteine protease inhibitor E-64 (Figure 6). In the presence of these three inhibitors, fluorescent degradation products were decreased 52% to 56% (Table 2: BT20). The highly selective cathepsin B inhibitor CA074 was more effective in reducing degradation (Figure 6), reducing degradation products by 67% (Table 2: BT20). The intracellular accumulation of degraded DQ-collagen IV in living BT549 cells was inhibited comparably by CA074Me and E-64d, less effectively by aprotinin and not by BB3103 (Figure 7). Fluorescence (i.e., DQ-collagen IV degradation) was reduced 87% to 89% by CA074Me and E64d, 28% by aprotinin and 0% by BB3103 (Table 2: BT549). These results suggest that: 1) lysosomal cathepsin B was the primary enzyme responsible for intracellular degradation of DQ-collagen IV by living BT549 cells, and 2) a proteolytic cascade may be responsible for pericellular degradation of DQ-collagen IV by living BT20 cells. Furthermore, cathepsin B was also an important component of the pericellular proteolytic cascade.
Figure 6.
Inhibitors of multiple protease classes reduced pericellular degradation of DQ-collagen IV by living BT20 human breast cancer cells. Left panels are phase images of living BT20 cells. Right panels are optical sections of the underlying matrix illustrating the green fluorescent products resulting from degradation of DQ-collagen IV. Aprotinin (1 µM), BB3103 (10 µM) and CA074 (10 µM) produced a comparable reduction in pericellular proteolysis. E-64 (10 µM) seemed to be slightly less effective.
Table 2.
Protease Inhibitors Reduce Degradation of DQ-Collagen IV by BT20 Cells and BT549 Cells.
| Fluorescence Intensity* | % Control | Area† | % Control | |
| BT20 | ||||
| Control | 162,462±1,154 | 100 | 2,665 | 100 |
| E-64 | 71,370±667 | 44 | 1,165 | 44 |
| CA074 | 53,132±575 | 33 | 911 | 34 |
| Aprotinin | 70,797±425 | 44 | 1,719 | 64 |
| BB3103 | 78,339±486 | 48 | 1,140 | 43 |
| BT549 | ||||
| Control | 1,513,900±818 | 100 | 26,953 | 100 |
| E-64d | 163,904±269 | 11 | 5,594 | 21 |
| CA074Me | 203,939±333 | 13 | 4,648 | 17 |
| Aprotinin | 1,095,071±619 | 72 | 23,399 | 87 |
| BB3103 | 1,535,287±973 | 101 | 22,644 | 84 |
Fluorescence intensity is in arbitrary units with each unit equal to the sum of the gray scale value for the pixels within an image; results are expressed as mean±SD. SD were calculated for all pixels in an image using Image Gauge V3.3 Software (Fuji).
Area is in Px2 units.
Figure 7.
Inhibitors of cysteine proteases and cathepsin B reduced intracellular degradation of DQ-collagen IV by living BT549 human breast cancer cells. Phase images of living BT549 cells are superimposed on optical sections through the cells. Green fluorescent products resulting from cleavage of DQ-collagen IV can be observed in the control cells and those treated with BB3103 (10 µM). A striking decrease in green fluorescent products can be observed in cells treated with E-64d (10 µM) or CA074Me (10 µM).
Discussion
The present studies demonstrate that living human breast cancer cells digest the protein substrates DQ-BSA and DQ-collagen IV pericellularly (BT20 cells) and intracellularly (BT549 cells). The ability to reduce intracellular degradation ∼90% with cysteine protease inhibitors, including a highly selective cathepsin B inhibitor, suggests that cathepsin B was the cysteine protease primarily responsible for intracellular degradation in the BT549 breast cancer cells. In contrast, the pericellular degradation observed with the BT20 cells appeared to involve a proteolytic cascade because comparable inhibition was observed with a serine protease inhibitor and broad spectrum inhibitors of MMPs and cysteine proteases and significantly more inhibition with a highly selective cathepsin B inhibitor. The ability of proteases to degrade the unlabeled proteins in this system, e.g., the gelatin in which the DQ substrates were embedded, would not be observed under the conditions described. The somewhat surprising ability of aprotinin to reduce intracellular degradation ∼30% may reflect a role for plasmin in release of the DQ-substrates from the gelatin and thus a reduction in their subsequent endocytosis. The confocal assay as described herein does not assess the contribution of proteases from other cell types associated with a tumor such as stromal and inflammatory cells. We, therefore, are presently analyzing the ability of mixtures of living tumor cells and stromal or inflammatory cells to degrade quenched fluorescent ECM proteins. These mixed populations of cells exhibit patterns of degradation that differ from either single-cell type alone and, thus, may provide an in vitro model for the study of tumor-associated proteolysis and the identification of proteases that may be responsible for degradation of specific ECM proteins by tumors in vivo.
A cursory reading of the cancer literature might suggest that MMPs are the only class of proteases needed for tumor invasion; however, serine proteases of the plasminogen cascade also have been strongly implicated in tumor invasion [3,9]. Nevertheless, the serine protease inhibitor aprotinin and the MMP inhibitor Batimastat are unable to prevent invasion of human esophageal and ovarian cancer cells in vivo [29], suggesting that other proteases such as cysteine proteases may participate in invasion by some cancers. Proteolytic cascades in which proteases activate one another appear to be involved in invasion. In ovarian cancers, for example, cathepsin B has been shown to initiate a proteolytic cascade by activating prourokinase plasminogen activator, a cascade resulting in degradation of ECM [30,31]. Cysteine protease inhibitors reduce both ECM degradation and invasion of the ovarian cancer cells through the ECM [32]. The present studies would be consistent with a proteolytic cascade of cathepsin B, serine proteases and MMPs participating in pericellular degradation of ECM by breast cancer cells.
The dogma is that proteolysis of ECM proteins occurs extracellularly. Receptors on the tumor cell surface for urokinase and the identification of integral membrane MMPs (MT-MMPs) have led to a hypothesis that ECM degradation occurs at the tumor cell membrane. In this regard, it is of interest that we have demonstrated that cathepsin B binds to the annexin II heterotetramer on the tumor cell surface [33], a complex that binds at separate sites plasminogen and tissue-type plasminogen activator [34], i.e., serine proteases of the plasminogen cascade. We have localized cathepsin B to the cell surface of a variety of human cancer cell lines, e.g., MCF-7 [19] and BT20 breast cancer cells, U87 glioma cells [10,17] and colon cancer cells (Sameni, Cavallo-Medved and Sloane, unpublished data), suggesting that cathepsin B may be part of a cell surface proteolytic cascade in these cancer cell lines as well as in ovarian cancer cells [30–32].
An unanticipated finding in the present study was that the BT549 cells degrade extracellular proteins intracellularly in lysosomes. Tumor cells had previously been hypothesized to take up protein substrates and degrade them intracellularly on the basis of static micrographs in which fragments of collagen are present in intracellular vesicles [35]. In addition, Montcourrier et al. [36,37] have hypothesized that breast cancer cells degrade ECM in large acidic vesicles in which the lysosomal aspartic protease cathepsin D and internalized ECM colocalize. Nonetheless, the concept of intracellular degradation contributing to tumor invasion has not been generally accepted. The results of the present study are not without precedence as in situ proteolysis of a Bodipy-labeled bovine serum albumin has been observed in living macrophages [38]. The present confocal assay imaged proteolysis by living cancer cells and thus unequivocally demonstrated that intracellular pathways and the cysteine protease cathepsin B contribute to degradation of ECM by cancer cells.
Modifications of this confocal assay could be used to screen frozen sections of animal or human tumors for proteolytic activity(ies) before treatment with protease inhibitors and thus provide a biologic endpoint for preclinical and clinical trials of protease inhibitors. With such an assay, an investigator/clinician might be able to choose protease inhibitors for therapy that could be shown to reduce proteolytic activity of the tumors ex vivo. A caveat is that such assays on frozen sections would only identify extracellular/pericellular proteolysis and not proteolysis dependent on endocytic uptake of protein substrates.
Acknowledgements
We thank Bruce Linebaugh for expert technical assistance and Linda Mayernik for assistance in preparation of figures.
Abbreviations
- DQ-BSA
DQ-bovine serum albumin
- ECM
extracellular matrix
- MMP
matrix metalloproteinase
Footnotes
This work was supported by National Institutes of Health (NIH) grant # 56586. The Microscopy and Imaging Resources Laboratory is supported, in part, by NIH Center grants P30ES06639 and P30ES22453.
References
- 1.Garcia M, Platet N, Liaudet E, Laurent V, Derocq D, Brouillet JP, Rochefort H. Biological and clinical significance of cathepsin D in breast cancer metastasis. Stem Cells. 1996;14:642–650. doi: 10.1002/stem.140642. [DOI] [PubMed] [Google Scholar]
- 2.Chambers AF, Matrisian LM. Changing views of the role of matrix metalloproteinases in metastasis. J Natl Cancer Inst. 1997;89:1260–1270. doi: 10.1093/jnci/89.17.1260. [DOI] [PubMed] [Google Scholar]
- 3.Johnsen M, Lund LR, Romer J, Almholt K, Dano K. Cancer invasion and tissue remodeling: common themes in proteolytic matrix degradation. Curr Opin Cell Biol. 1998;10:667–671. doi: 10.1016/s0955-0674(98)80044-6. [DOI] [PubMed] [Google Scholar]
- 4.Rooprai HK, McCormick D. Proteases and their inhibitors in human brain tumors. Anticancer Res. 1997;17:4151–4162. [PubMed] [Google Scholar]
- 5.Elliott E, Sloane BF. The cysteine protease cathepsin B in cancer. Perspect Drug Discovery Des. 1996;6:12–32. [Google Scholar]
- 6.Buck MR, Karustis DG, Day NA, Honn KV, Sloane BF. Degradation of extracellular matrix proteins by human cathepsin B from normal and tumor tissues. Biochem J. 1992;282:273–278. doi: 10.1042/bj2820273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guinec N, Dalet-Fumeron V, Pagano M. “In vitro” study of basement membrane degradation by the cysteine proteases, cathepsin B, B-like and L. Biol Chem Hoppe-Seyler. 1993;374:1135–1146. doi: 10.1515/bchm3.1993.374.7-12.1135. [DOI] [PubMed] [Google Scholar]
- 8.Montcourrier P, Mangeat PH, Valembois C, Salazar G, Sahuquet A, Duperray C, Rochefort H. Characterization of very acidic phagosomes in breast cancer cells and their association with invasion. J Cell Sci. 1994;107:2381–2391. doi: 10.1242/jcs.107.9.2381. [DOI] [PubMed] [Google Scholar]
- 9.Ossowski L, Clunie G, Masucci MT, Blasi F. In vivo paracrine interaction between urokinase and its receptor: effect on tumor invasion. J Cell Biol. 1991;115:1107–1112. doi: 10.1083/jcb.115.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demchik L, Sameni M, Nelson K, Rozhin J, Mikkelsen T, Sloane BF. Cathepsin B and glioma invasion. Int J Dev Neurosci. 1999;17:483–494. doi: 10.1016/s0736-5748(99)00011-8. [DOI] [PubMed] [Google Scholar]
- 11.DeClerck YA, Imren S, Montgomery AM, Muller BM, Reisfeld RA, Laug WE. Proteases and protease inhibitor in tumor progression. Adv Exp Med Biol. 1997;425:89–97. doi: 10.1007/978-1-4615-5391-5_9. [DOI] [PubMed] [Google Scholar]
- 12.Zeng ZS, Cohen AM, Guillem JG. Loss of basement membrane type IV collagen is associated with increased expression of metalloproteases 2 and 9 (MMP-2 and MMP-9) during human colorectal tumorigenesis. Carcinogenesis. 1999;20:749–755. doi: 10.1093/carcin/20.5.749. [DOI] [PubMed] [Google Scholar]
- 13.Hauptmann S, Zardi L, Siri A, Carnemolla B, Borsi L, Castellucci M, Klosterhalfen B, Hartung P, Weis J, Stocker G, Haubeck H-D, Kirkpatrick CJ. Extracellular matrix proteins in colorectal carcinomas. Expression of tenascin and fibronectin isoforms. Lab Invest. 1995;73:172–182. [PubMed] [Google Scholar]
- 14.Khan A, Krishna M, Baker SP, Malhothra R, Banner BF. Cathepsin B expression and its correlation with tumor-associated laminin and tumor progression in gastric cancer. Arch Pathol Lab Med. 1998;122:172–177. [PubMed] [Google Scholar]
- 15.Weissleder R, Tung CH, Mahmood U, Bogdanov A., Jr In vivo imaging of tumors with protease-activated near-infrared fluorescent probes. Nat Biotechnol. 1999;17:375–378. doi: 10.1038/7933. [DOI] [PubMed] [Google Scholar]
- 16.Mahmood U, Tung CH, Bogdanov A, Jr, Weissleder R. Near-infrared optical imaging of protease activity for tumor detection. Radiology. 1999;213:866–870. doi: 10.1148/radiology.213.3.r99dc14866. [DOI] [PubMed] [Google Scholar]
- 17.Sloane BF. Suicidal tumor proteases. Nat Biotechnol. 1996;14:826–827. doi: 10.1038/nbt0796-826b. [DOI] [PubMed] [Google Scholar]
- 18.Chen W-T, Chen JM, Parsons SJ, Parsons JT. Local degradation of fibronectin at sites of expression of the transforming gene product pp60src. Nature. 1985;316:156–158. doi: 10.1038/316156a0. [DOI] [PubMed] [Google Scholar]
- 19.Sameni M, Elliott E, Ziegler G, Fortgens PH, Dennison C, Sloane BF. Cathepsins B and D are localized at surface of human breast cancer cells. Pathol Oncol Res. 1995;1:43–53. doi: 10.1007/BF02893583. [DOI] [PubMed] [Google Scholar]
- 20.Moin K, Cao L, Day NA, Koblinski JE, Sloane BF. Tumor cell membrane cathepsin B. Biol Chem. 1998;379:1093–1099. doi: 10.1515/bchm.1998.379.8-9.1093. [DOI] [PubMed] [Google Scholar]
- 21.Abrahamson M, Mason RW, Hansson H, Buttle DJ, Grubb A, Ohlsson K. Human cystatin C. Role of the N-terminal segment in the inhibition of human cysteine proteinases and in its inactivation by leucocyte elastase. Biochem J. 1991;273:621–626. doi: 10.1042/bj2730621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murata M, Miiyashta S, Yokoo C, Tamai M, Hanada K, Hatayama K, Towatari T, Nikawa T, Katunuma N. Novel epoxysuccinyl peptides. Selective inhibitors of cathepsin B, in vitro. FEBS Lett. 1991;280:307–310. doi: 10.1016/0014-5793(91)80318-w. [DOI] [PubMed] [Google Scholar]
- 23.Bogyo M, Verhelst S, Bellingard-Dubouchaud V, Toba S, Greenbaum D. Selective targeting of lysosomal cysteine proteases with radiolabeled electrophilic substrate analogs. Chem Biol. 2000;7:27–38. doi: 10.1016/s1074-5521(00)00061-2. [DOI] [PubMed] [Google Scholar]
- 24.Ahram M, Sameni M, Qiu RG, Kim D, Sloane BF. Cathepsin B in Rac1-induced cell invasion. Mol Biol Cell. 1998;9:421a. [Google Scholar]
- 25.Buttle DJ, Murata M, Knight CG, Barrett AJ. CAO74 methy ester: a proinhibitor for intracellular cathepsin B. Arch Biochem Biophys. 1992;299:377–380. doi: 10.1016/0003-9861(92)90290-d. [DOI] [PubMed] [Google Scholar]
- 26.Van Noorden CJ, Jonges TG, Van Marie J, Bissell ER, Griffini P, Jans M, Snel J, Smith RE. Heterogeneous suppression of experimentally induced colon cancer metastasis in rat liver lobes by inhibition of extracellular cathepsin B. Clin Exp Metastasis. 1998;16:159–167. doi: 10.1023/a:1006524321335. [DOI] [PubMed] [Google Scholar]
- 27.Everts V, Korper W, Niehof A, Jansen I, Beertsen W. Type VI collagen is phagocytosed by fibroblasts and digested in the lysosomal apparatus: involvement of collagenase, serine proteinases and lysosomal enzymes. Matrix Biol. 1994;14:665–676. doi: 10.1016/s0945-053x(05)80030-7. [DOI] [PubMed] [Google Scholar]
- 28.Via LE, Fratti RA, McFalone M, Pagan-Ramos E, Deretic D, Deretic V. Effects of cytokines on mycobacterial phagosome maturation. J Cell Sci. 1998;111:897–905. doi: 10.1242/jcs.111.7.897. [DOI] [PubMed] [Google Scholar]
- 29.Della Porta P, Soeltl R, Krell HW, Collins K, O'Donoghue M, Schmitt M, Kruger A. Combined treatment with serine protease inhibitor aprotinin and matrix metalloproteinase inhibitor Batimastat (BB-94) does not prevent invasion of human esophageal and ovarian carcinoma cells in vivo. Anticancer Res. 1999;19:3809–3816. [PubMed] [Google Scholar]
- 30.Kobayashi H, Schmitt M, Goretzki L, Chucholowski N, Calvete J, Kramer M, Gunzler WA, Janicke F, Graeff H. Cathepsin B efficiently activates the soluble and the tumor cell receptor-bound form of the proenzyme urokinase-type plasminogen activator (Pro-uPA) J Biol Chem. 1991;266:5147–5152. [PubMed] [Google Scholar]
- 31.Kobayashi H, Moniwa N, Sugimura M, Shinohara H, Ohi H, Terao T. Effects of membrane-associated cathepsin B on the activation of receptor-bound prourokinase and subsequent invasion of reconstituted basement membranes. Biochim Biophys Acta. 1993;1178:55–62. doi: 10.1016/0167-4889(93)90109-3. [DOI] [PubMed] [Google Scholar]
- 32.Kobayashi H, Ohi H, Sugimura M, Shinohara H, Fujii T, Terao T. Inhibition of in vitro ovarian cancer cell invasion by modulation of urokinase-type plasminogen activator and cathepsin B. Cancer Res. 1992;52:3610–3614. [PubMed] [Google Scholar]
- 33.Mai J, Finley RL, Jr, Waisman DM, Sloane BF. Human procathepsin B interacts with annexin II tetramer on the surface of tumor cells. J Biol Chem. 2000;275:12806–12812. doi: 10.1074/jbc.275.17.12806. [DOI] [PubMed] [Google Scholar]
- 34.Kassam G, Choi KS, Ghuman J, Kang HM, Fitzpatrick SL, Zackson T, Zackson S, Toba M, Shinomiya A, Waisman DM. The role of annexin II tetramer in the activation of plasminogen. J Biol Chem. 1998;273:4790–4799. doi: 10.1074/jbc.273.8.4790. [DOI] [PubMed] [Google Scholar]
- 35.Levine AM, Reddick R, Triche T. Intracellular collagen fibrils in human sarcomas. Lab Invest. 1978;39:531–540. [PubMed] [Google Scholar]
- 36.Montcourrier P, Mangeat PH, Salazar G, Morisset M, Sahuquet A, Rochefort H. Cathepsin D in breast cancer cells can digest extracellular matrix in large acidic vesicles. Cancer Res. 1990;50:6045–6054. [PubMed] [Google Scholar]
- 37.Montcourrier P, Mangeat PH, Valembois C, Sahuquet A, Duperray C, Rochefort H. Characterization of very acidic phagosomes in breast cancer and their association with invasion. J Cell Sci. 1994;107:2381–1391. doi: 10.1242/jcs.107.9.2381. [DOI] [PubMed] [Google Scholar]
- 38.Reis RCM, Sorgine MHF, Coelho-Sampaio T. A novel methodology for the investigation of intracellular proteolytic processing in intact cells. Eur J Cell Biol. 1998;75:192–197. doi: 10.1016/S0171-9335(98)80061-7. [DOI] [PubMed] [Google Scholar]